Abstract
Purpose
To characterize the clinical and molecular genetic characteristics of a large, multigenerational Chinese family showing different phenotypes.
Methods
A pedigree consisted of 56 individuals in 5 generations was recruited. Comprehensive ophthalmic examinations were performed in 16 family members affected. Mutation screening of CYP4V2 was performed by Sanger sequencing. Next-generation sequencing (NGS) was performed to capture and sequence all exons of 47 known retinal dystrophy-associated genes in two affected family members who had no mutations in CYP4V2. The detected variants in NGS were validated by Sanger sequencing in the family members.
Results
Two compound heterozygous CYP4V2 mutations (c.802-8_810del17insGC and c.992A>C) were detected in the proband who presented typical clinical features of BCD. One missense mutation (c.1482C>T, p.T494M) in the PRPF3 gene was detected in 9 out of 22 affected family members who manifested classical clinical features of RP.
Conclusions
Our results showed that two compound heterozygous CYP4V2 mutations caused BCD, and one missense mutation in PRPF3 was responsible for adRP in this large family. This study suggests that accurate phenotypic diagnosis, molecular diagnosis, and genetic counseling are necessary for patients with hereditary retinal degeneration in some large mutigenerational family.
1. Introduction
Retinitis pigmentosa (RP) (MIM 268000) is the most common form of hereditary retinal degeneration (HRD), with a worldwide prevalence of 1 in 4000 [1]. The disease can be inherited in an autosomal recessive (AR), autosomal dominant (AD), or X-linked manner [2]. Autosomal dominant RP (adRP) is the most common form of RP and typically begins with night blindness in the early teens, followed by progressive loss in the peripheral visual field, subsequent loss of vision, and eventually legal blindness. To date, mutations in 22 genes have been associated with adRP (RetNet: http://www.sph.uth.tmc.edu/retnet/sum-dis.htm, last updated November 16, 2016), of which five genes have been reported in Chinese adRP patients [3–7].
Bietti's crystalline corneoretinal dystrophy (BCD) (MIM 210370) is an autosomal recessive retinal dystrophy that is characterized by numerous tiny glistening yellow-white crystals that are scattered at the posterior pole of the retina, progressive atrophy of the retinal pigment epithelium (RPE), and choroidal sclerosis. Patients with BCD are usually present in the 2nd or 3rd decade of life and progress to legal blindness by the 5th or 6th decade [8]. Mutations in the CYP4V2 gene (MIM 608614) are associated with BCD [9]. BCD is relatively common in the East Asian populations, especially in Chinese and Japanese populations [9–18].
BCD and RP are considered as two different types of retinal dystrophies with distinct clinical courses and features during its early stage. However, the fundus features at the later stage of BCD are occasionally similar to a severe form of RP. The interaction or coexistence of the two clinical phenotypes thus requires further elucidation. In the present study, we distinguish the inheritance patterns, clinical phenotype, and molecular genetic characteristics of the patients in a large, multigeneration Chinese family with RP and BCD.
2. Methods
2.1. Pedigree
A pedigree consisted of 56 individuals in 5 generations was recruited. The Ethics Review Board of the Southwest Hospital (Chongqing, China) approved the research protocol (number 2012-11), which adhered to the tenets of the Declaration of Helsinki, and informed consent was obtained from all participants.
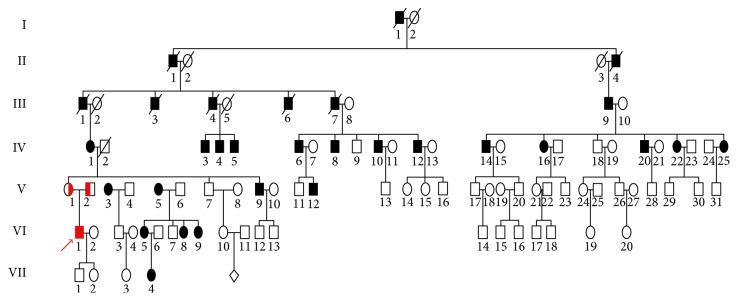
The proband (Figure 1, VI:1) was initially presented to our medical institution for genetic counseling based on the observation that most of the family members developed night blindness and visual loss and even complete blindness, resulting in an inability to work. This five-generation family from Southwest of China was assessed in terms of RP and BCD. Thirty-nine participants were ascertained at the Southwest Eye Hospital, Southwest Hospital, Chongqing, China (Figure 1). No consanguineous marriage in the family was declared.
Figure 1.
Pedigree plot. The proband is indicated by an arrow. One affected patient with red solid box showed clinical findings compatible to the diagnosis of Bietti's crystalline corneoretinal dystrophy (VI:1), and the other affected members with black solid box presented clinical features of retinitis pigmentosa. Males and females are represented by squares and circles, respectively. Filled symbols: affected members; open symbols: unaffected members.
The proband presented clinical features that were compatible with a diagnosis of BCD (VI:1), and the family members were subsequently evaluated. Twenty-two living individuals in the family had the clinical features of RP and presented similar symptoms of night blindness and progressive reduction in their field of vision. The RP phenotype followed an autosomal dominant pattern of inheritance in this pedigree (Figure 1).
Thirteen affected individuals (III:9, IV:1, IV:3, IV:5, IV:6, IV:10, IV:12, IV:14, IV:20, V:3, V:9, V:12, and VI:1) and twenty-six unaffected family members (Table 1) underwent examination, including best-corrected visual acuity testing with the Snellen vision chart, fundoscopy, slit-lamp biomicroscopy, spectral domain optical coherence tomography (SD-OCT, Spectralis OCT, Version 6.0; Heidelberg Engineering, Germany), full-field electroretinogram (FERG), and multifocal electroretinogram (mfERG). For the ages 6 months, 1, 2, 3, and 4 years old, the visual acuity was assessed using Teller acuity cards and then converted into Snellen vision chart.
Table 1.
Demographic and clinical features and genotypes of PRPF3 and CYP4V2 mutations in a family with adRP and arBCD.
| Number | Gender | Age | First symptom | Phenotype | BCVA | Eye complications | Systemic diseases | PRPF3 mutation | CYP4V2 mutations | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OD | OS | c.1481C>G | c.802-8_810del17insGC | c.992A>C | |||||||
| III:8 | Female | 76 | None | Normal | 20/250 | 20/250 | Cataract | ND | ND | ND | |
| III:9 | Male | 80 | Night blindness | RP | LP | LP | Cataract | MT | Wild | Wild | |
| III:10 | Female | 80 | None | Normal | 20/50 | 20/50 | Cataract | ND | ND | ND | |
| IV:1 | Female | 82 | Night blindness | RP | LP | LP | Cataract | MT | Wild | Wild | |
| IV:3 | Male | 57 | Night blindness | RP | LP | LP | Cataract | ND | Wild | Wild | |
| IV:5 | Male | 57 | Night blindness | RP | LP | LP | Cataract | ND | ND | ND | |
| IV:6 | Male | 57 | Night blindness | RP | LP | LP | Cataract | MT | Wild | Wild | |
| IV:7 | Female | 54 | None | Normal | 20/30 | 20/30 | None | Wild | Wild | Wild | |
| IV:10 | Male | 39 | Night blindness | RP | 20/200 | 20/200 | None | MT | Wild | Wild | |
| IV:12 | Male | 35 | Night blindness | RP | 20/400 | LP | None | ND | Wild | Wild | |
| IV:14 | Male | 55 | Night blindness | RP | 20/100 | 20/200 | Cataract | MT | Wild | Wild | |
| IV:15 | Female | 50 | None | Normal | 20/35 | 20/35 | None | ND | ND | ND | |
| IV:16 | Female | 51 | None | Normal | 20/35 | 20/20 | None | ND | ND | ND | |
| IV:18 | Male | 49 | None | Normal | 20/20 | 20/20 | None | ND | ND | ND | |
| IV:20 | Male | 45 | Night blindness | RP | 20/100 | 20/200 | None | MT | Wild | Wild | |
| V:1 | Female | 57 | None | Normal | 20/20 | 20/25 | None | Breast cancer | ND | MT | Wild |
| V:2 | Male | 60 | None | Normal | 20/30 | 20/30 | Cataract | Fatty liver disease | ND | Wild | MT |
| V:3 | Female | 55 | Night blindness | RP | 20/10 | 20/200 | None | Breast cancer | MT | Wild | Wild |
| V:7 | Male | 45 | None | Normal | 20/30 | 20/30 | Myopia | Wild | Wild | Wild | |
| V:8 | Female | 46 | None | Normal | 20/18 | 20/18 | None | Wild | Wild | Wild | |
| V:9 | Male | 43 | Night blindness | RP | 20/200 | LP | Myopia | MT | Wild | Wild | |
| V:11 | Male | 24 | None | Normal | 20/18 | 20/18 | None | Wild | Wild | Wild | |
| V:12 | Male | 22 | Night blindness | RP | 20/100 | 20/100 | Myopia | MT | Wild | Wild | |
| V:13 | Male | 13 | None | Normal | 20/30 | 20/20 | None | ND | Wild | Wild | |
| V:15 | Female | 8 | None | Normal | 20/20 | 20/20 | None | ND | Wild | Wild | |
| V:17 | Male | 37 | None | Normal | 20/25 | 20/25 | None | ND | Wild | Wild | |
| V:18 | Female | 35 | None | Normal | 20/18 | 20/18 | High myopia | ND | ND | ND | |
| V:19 | Female | 35 | None | Normal | 20/25 | 20/20 | Myopia | ND | ND | ND | |
| V:20 | Male | 33 | None | Normal | 20/18 | 20/18 | None | Wild | Wild | Wild | |
| V:31 | Male | 15 | None | Normal | 20/18 | 20/18 | Myopia | ND | ND | ND | |
| VI:1 | Male | 33 | Decreased vision | BCD | 20/25 | 20/30 | Myopia, uveitis | Wild | MT | MT | |
| VI:2 | Female | 29 | None | Normal | 20/40 | 20/50 | High myopia | ND | ND | ND | |
| VI:10 | Female | 22 | None | Normal | 20/18 | 20/18 | Myopia | ND | Wild | Wild | |
| VI:12 | Male | 15 | None | Normal | 20/20 | 20/20 | None | ND | Wild | Wild | |
| VI:14 | Male | 13 | None | Normal | 20/20 | 20/20 | None | ND | Wild | Wild | |
| VI:15 | Male | 10 | None | Normal | 20/20 | 20/20 | None | ND | Wild | Wild | |
| VI:16 | Male | 9 | None | Normal | 20/20 | 20/20 | None | ND | Wild | Wild | |
| VII:1 | Male | 5 | None | Normal | 20/30 | 20/30 | None | ND | MT | Wild | |
| VII:2 | Female | 1 | None | Normal | 20/30 | 20/200 | Hyperopia | ND | Wild | Wild | |
MT: mutation; ND: not detected; LP: light perception; NLP: nonlight perception.
2.2. Mutation Screening
Genomic DNA was extracted from peripheral blood samples of 39 family members (Table 1) using a QIAamp DNA Blood Midi Kit (Qiagen, Hilden, Germany) following the manufacturer's standard procedure. All coding exons and intron-exon boundaries of the CYP4V2 gene were amplified by polymerase chain reaction (PCR) using primers described by Li et al. [9]. The PCR products were subsequently purified with a TIANgen Mini Purification Kit (Tiangen Biotech Co. Ltd., Shanghai, China) and sequenced by Sanger sequencing with an ABI BigDye Terminator Cycle Sequencing Kit v3.1 (Applied Biosystems (ABI), Foster City, CA). CYP4V2 sequencing was performed in eight patients (III:9, IV:1, IV:6, IV:12, V:1, V:12, VI:1, and VI:2), and the detected mutation was further screened in 12 affected family members and 11 unaffected members.
Next-generation sequencing (NGS) was then applied to two affected family members with RP (III:9 and IV:1), who did not have CYP4V2 mutations, then to identify disease-causing variants in 47 RP-related genes including the PRPF31, CRB1, PRPF8, CA4, TULP1, PRPF3, ABCA4, RPE65, EYS, CERKL, NRL, FAM161A, FSCN2, TOPORS, SNRNP200, SEMA4A, PRCD, NR2E3, MERTK, USH2A, PDE6B, PROM1, KLHL7, PDE6A, RGR, CNGB1, IDH3B, SAG, GUCA1B, CNGA1, BEST1, TTC8, C2orf71, ARL6, IMPG2, PDE6G, ZNF513, DHDDS, PRPF6, CLRN1, MAK, CDHR1, FLVCR1, RLBP1, SPATA7, AIPL1, and LRAT genes. The detected variants in NGS were validated by Sanger sequencing and screened in other 6 affected and 7 unaffected individuals in the family.
3. Results
3.1. Clinical Features
The demographic and clinical features of the living affected members and mutation carriers are summarized in Table 1. The age of enrollment ranged from 1 to 82 years. The visual acuity ranged from 20/30 to nonlight perception (NLP). Seven family members had refractive errors, including myopia (ranging −0.75 to −8 diopters) and astigmatism, and twelve members presented with cataract. All affected individuals except for the proband had congenital night blindness, and seven affected members already presented legal blindness. The proband's mother (V:1) and aunt (V:3) had breast cancer, and his father (V:2) had fatty liver disease.
3.2. Mutations in the CYP4V2 Gene
Two previously reported CYP4V2 mutations (c.802-8_810del17insGC and c.992A>C (p.H331P)) were detected in this family. The proband (VI:1) was compound heterozygous for both mutations. The c.802-8_810del17insGC mutation was maternally derived (V:1), whereas the c.992A>C mutation was paternally inherited (V:2). Two other family members (V:9 and VII:1) were heterozygous for the c.802-8_810del17insGC mutation.
3.3. Mutations in the PRPF3 Gene
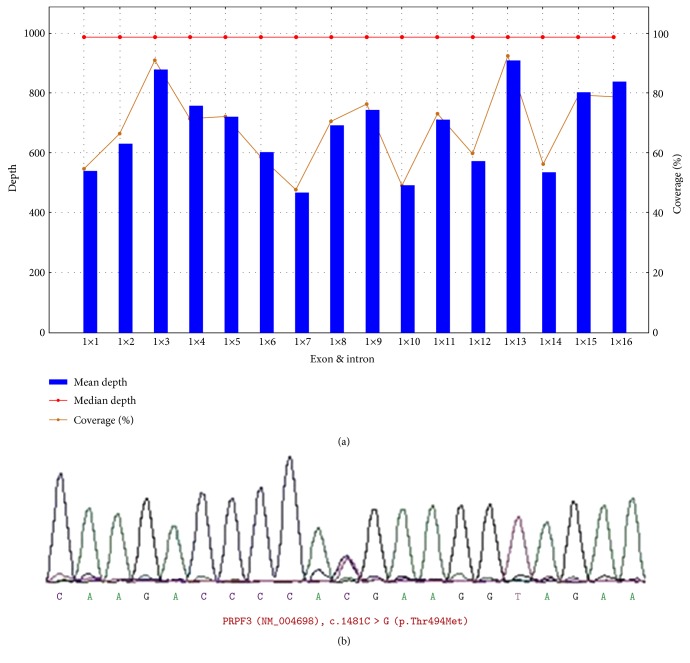
Targeted NGS of two affected members (IV:1 and III:9) revealed one common missense mutation in the PRPF3 gene (c.1481C>T) (Figure 2), which was then screened by Sanger sequencing in 8 affected (III:9, IV:1, IV:3, IV:6, IV:14, V:9, V:12, and VI:1) and seven unaffected family members (IV:9, V:1,V:7, V:11, V:16, V:20, and VI:13) for cosegregation analysis.
Figure 2.
The depth and coverage of next-generation sequencing of the PRPF3 gene and the chromatogram obtained by Sanger sequencing (patient III:9). (a) The rectangle shows the averaged sequencing depth and coverage of the family for all 16 exons of the PRPF3 gene as screened by next-generation sequencing. (b) Sanger sequencing detected a heterozygous mutation (c.1481C>T, p.Thr494Met) in PRPF3.
3.4. Clinical and Molecular Manifestations of Affected Family Members
Two types of clinical and molecular manifestations were observed in this family: (i) a BCD phenotype that was related to the compound heterozygous CYP4V2 mutations and (ii) a RP phenotype that was associated with the PRPF3 mutation and followed an autosomal dominant pattern of inheritance.
3.4.1. Type 1 (Proband VI:1)
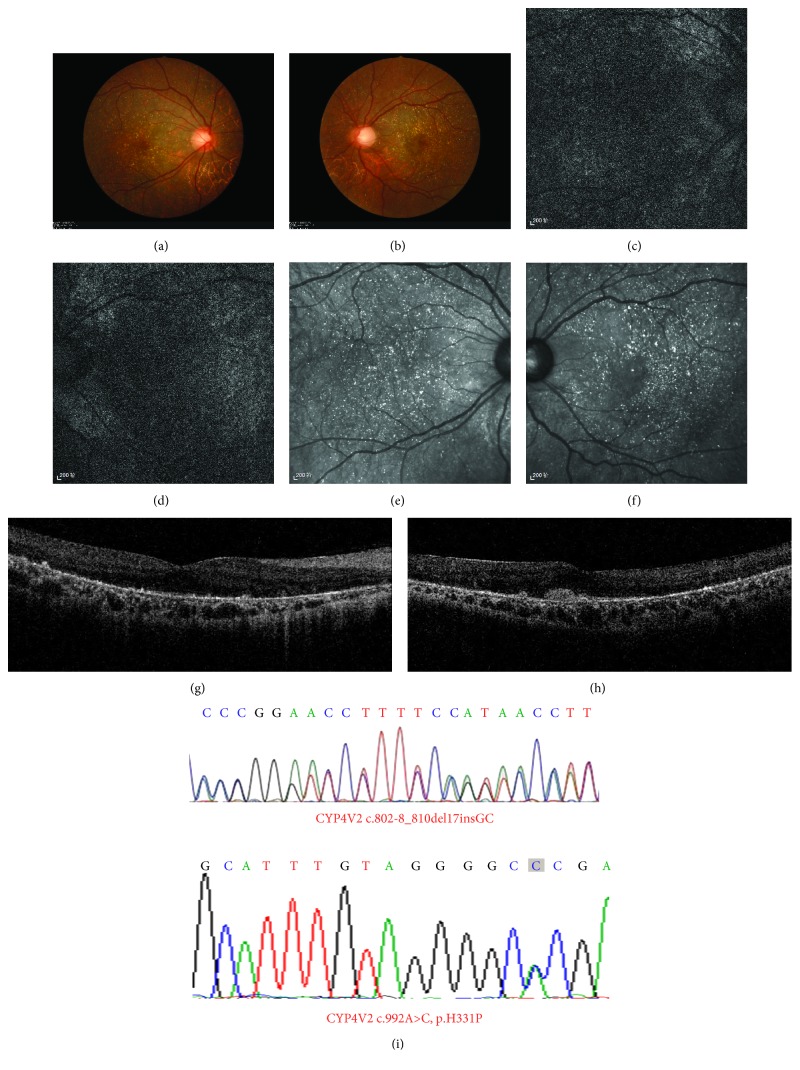
The proband was a 33-year-old man referred to us for genetic counseling based on a significant decrease in visual acuity starting at the age of 17 years. The patient developed night blindness in his early 30s. He had high myopia (−7.00 D) in both eyes, and best-corrected Snellen visual acuity was 20/30 in his both eyes. There was a history of chronic uveitis in his left eye since age 28. He was diagnosed with BCD based on clinical findings that included numerous tiny glistening yellow-white crystals scattered at the posterior pole of the retina, RPE atrophy (Figure 3), and decreased responses in FERGs and mfERGs.
Figure 3.
Fundal images and chromatograms of the proband with a clinical diagnosis with Bietti's crystalline corneoretinal dystrophy and harboring compound heterozygous mutations in the CYP4V2 gene (patient VI:1). Fundal photographs (a, b), autofluorescence images (c, d), and near-infrared images (e, f) of both eyes are shown on the left, and chromatograms of two mutations are demonstrated on the right (g, h).
Two previously reported disease-causing mutations in CYP4V2 (c.802-8_810del17insGC in exon 7 and c.992A>C (p.H331P) in exon 8) were identified in the proband [19]. The compound heterozygosity was confirmed by screening his unaffected parents; his mother (V:1) carried the c.802-8_810del17insGC variant, and his father (V:2) harbored the c.992A>C mutation. The proband's unaffected son (VII:1) had the c.802-8_810del17insGC mutation, whereas no pathogenic CYP4V2 mutations were detected in the apparently normal daughter (VII:2). Notably, no PRPF3 mutations were detected in the proband.
3.4.2. Type 2
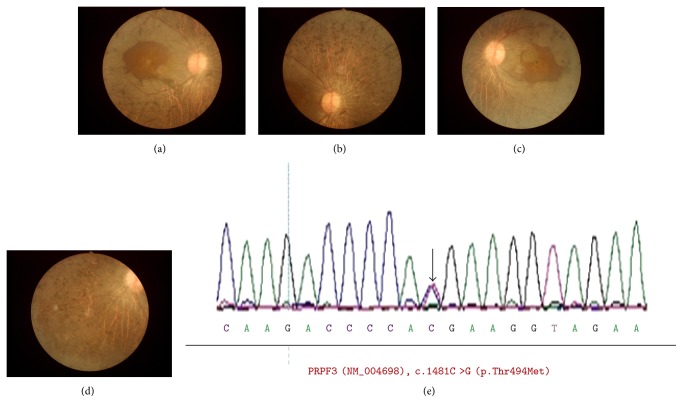
In addition to the proband, other family members affected with adRP presented with night blindness since birth. Best-corrected visual acuity was from 200/400 to NLP. Fundus examination showed severe features of RP, with a mass of bone-spicule pigmentation depositions, more severe RPE atrophy involving the macular and choroidal sclerosis extending to the midperipheral retina, whereas partial attenuation of the retinal blood vessels, slight waxy pallor of the optic disc, was presented (Figure 4). FERG demonstrated undetectable responses both in scotopic and photopic conditions and extinguished mfERG.
Figure 4.
Fundal photographs and chromatogram of a patient with severe phenotype of retinitis pigmentosa and harboring PRPF3 mutations (patient IV:12). Fundal photographs of the right eye (a, b) and the left eye (c, d), and chromatogram of the PRPF3 mutation (c.1481C>T, p.T494M) is shown (e).
One PRPF3 mutation, c.1481C>T (p.T494M), was detected in 13 family members, including 11 males and 2 females. No novel mutation and previously reported mutations were detected in the other 45 genes in the panel. The identified mutation (c.1481C>T) cosegregated with the RP phenotype in 11 affected family members tested and was not observed in 9 unaffected family members (Figure 1). This mutation was observed across four generations. Taken together, the c.1481C>T mutation was considered to be the main cause of adRP in this family.
4. Discussion
PRPF3 (MIM 607301) is a precursor mRNA-processing factor gene that was first identified for adRP in 2002 [20]. In the present study, a pathogenic mutation (c.1481C>T, p.T494M) in the PRPF3 gene was identified in 11 individuals presenting an adRP phenotype in a five-generation Chinese family. The molecular genetic features of a Chinese pedigree with a PRPF3 mutation have been previously reported. The c.1481C>T mutation is considered to be one of the most common mutations in PRPF3 [20–25]. Previous reports have shown that patients harboring the c.1481C>T mutation develop early-onset night blindness, visual field loss, and visual acuity loss between the ages of 30 and 40, as well as loss of ERG responses after the age of 30. Compared to those in previously reported Japanese, Spanish, Korean, Swiss, and North American families, members of this Chinese family with the c.1481C>T mutation presented a more severe disease phenotype, which included congenital blindness, severe visual acuity loss, extended RPE atrophy, and completely extinguished ERG responses.
Mutations in the CYP4V2 gene (MIM 608614) are the only known causative factor for BCD to date. The CYP4V2 gene consists of 11 exons and encodes a 525 amino acid protein belonging to the CYP450 family. CYP4V2 is widely expressed in tissues, including the retina, RPE, lymphocytes, heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas, which has been thought to play a crucial role in fatty acid and corticosteroid metabolism. In the present study, two compound heterozygous mutations in CYP4V2 (c.802-8_810del17insGC and c.992A>C) were identified in the proband who presented typical BCD. In our previous study, CYP4V2 mutation screening among 92 Chinese patients with BCD showed that c.802-8_810del17insGC and c.992A>C are common pathogenic mutations in Chinese with BCD [26]. The parents of the proband are not a consanguineous marriage couple. So we speculate that these heterozygous mutations in Chinese population may be universal. This phenomenon may be related to the common ancestor based on the huge population of China. The heterozygous state of the same gene carried by parents is consistent with the autosomal recessive inheritance pattern. This will be important for prenatal testing for family planning, early finding carrier status, and determining risk of inheritance in Chinese.
Coexistence of variants in two or three genes associated with retinal degeneration has rarely been reported in a family [3]. In the present study, we identified the coexistence of two distinct phenotypes in one family, namely, BCD and RP, which were caused by the pathogenic variants in the CYP4V2 and PRPF3 genes, respectively. The mode of inheritance of the two diseases was maintained in this family, in which BCD demonstrated an autosomal recessive trait and RP showed an autosomal dominant trait.
Two types of clinical and molecular manifestations identified in this study include (i) a BCD phenotype related to CYP4V2 mutations and (ii) an RP phenotype related to PRPF3 variants. Clinical features for (i) BCD and (ii) RP of this family were similar to those in previous reports. The proband affected with BCD in this family had a later onset for night blindness and relatively slow progression, with a predominantly affected choroid at the posterior pole. On the other hand, family members with RP caused by the PRPF3 mutation showed a more severe phenotype. Our study provides a better understanding of the genotype-phenotype correlation in a family with two independent pathogenic gene mutations and may be used in clinics for the differential diagnosis of retinal degenerations.
In summary, this is the first report on PRPF3-associated adRP and CYP4V2-associated arBCD in a large multigenerational Chinese family. The inheritance pattern of each gene mutation is independent. Our study provides an insight into the clinical effects of two independent gene mutations in a large family to facilitate accurate diagnosis and disease counseling.
Acknowledgments
The authors are grateful to Chen Sun, Mingfang Zhang, Gang Wang, and Min Wang for their great support. This work was supported by the National Basic Research Program of China (973 Program, 2013CB967002 and 2013CB967003) and the National Natural Science Foundation of China (81200710).
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Boughman J. A., Conneally P. M., Nance W. E. Population genetic studies of retinitis pigmentosa. American Journal of Human Genetics. 1980;32(2):223–235. [PMC free article] [PubMed] [Google Scholar]
- 2.Ayuso C., Millan J. M. Retinitis pigmentosa and allied conditions today: a paradigm of translational research. Genome Medicine. 2010;2(5):p. 34. doi: 10.1186/gm155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim K. P., Yip S. P., Cheung S. C., Leung K. W., Lam S. T., To CH Novel PRPF31 and PRPH2 mutations and co-occurrence of PRPF31 and RHO mutations in Chinese patients with retinitis pigmentosa. Archives of Ophthalmology. 2009;127(6):784–790. doi: 10.1001/archophthalmol.2009.112. [DOI] [PubMed] [Google Scholar]
- 4.Alvarez B. V. V. E., Yang Z., Koh A. H., et al. Identification and characterization of a novel mutation in the carbonic anhydrase IV gene that causes retinitis pigmentosa. Investigative Ophthalmology & Visual Science. 2007;48(8):3459–3468. doi: 10.1167/iovs.06-1515. [DOI] [PubMed] [Google Scholar]
- 5.Wang L., Ribaudo M., Zhao K., et al. Novel deletion in the pre-mRNA splicing gene PRPF31 causes autosomal dominant retinitis pigmentosa in a large Chinese family. American Journal of Medical Genetics Part A. 2003;121A(3):235–239. doi: 10.1002/ajmg.a.20224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo H., Qin Y., Meng Q., Zhang H., Jin H., Chen Y. Linkage analysis and mutation screening of the rhodopsin gene in a Chinese Bai family with autosomal dominant retinitis pigmentosa. Journal of Human Genetics. 2010;55(9):571–576. doi: 10.1038/jhg.2010.68. [DOI] [PubMed] [Google Scholar]
- 7.Zhao C., Bellur D. L., Lu S., et al. Autosomal-dominant retinitis pigmentosa caused by a mutation in SNRNP200, a gene required for unwinding of U4/U6 snRNAs. American Journal of Human Genetics. 2009;85(5):617–627. doi: 10.1016/j.ajhg.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaiser-Kupfer M. I., Chan C. C., Markello T. C., et al. Clinical biochemical and pathologic correlations in Bietti’s crystalline dystrophy. American Journal of Ophthalmology. 1994;118(5):569–582. doi: 10.1016/S0002-9394(14)76572-9. [DOI] [PubMed] [Google Scholar]
- 9.Li A., Jiao X., Munier F. L., et al. Bietti crystalline corneoretinal dystrophy is caused by mutations in the novel gene CYP4V2. American Journal of Human Genetics. 2004;74(5):817–826. doi: 10.1086/383228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gekka T., Hayashi T., Takeuchi T., Goto-Omoto S., Kitahara K. CYP4V2 mutations in two Japanese patients with Bietti’s crystalline dystrophy. Ophthalmic Research. 2005;37(5):262–269. doi: 10.1159/000087214. [DOI] [PubMed] [Google Scholar]
- 11.Lee K. Y., Koh A. H., Aung T., et al. Characterization of Bietti crystalline dystrophy patients with CYP4V2 mutations. Investigative Ophthalmology & Visual Science. 2005;46(10):3812–3816. doi: 10.1167/iovs.05-0378. [DOI] [PubMed] [Google Scholar]
- 12.Lin J., Nishiguchi K. M., Nakamura M., Dryja T. P., Berson E. L., Miyake Y. Recessive mutations in the CYP4V2 gene in East Asian and Middle Eastern patients with Bietti crystalline corneoretinal dystrophy. Journal of Medical Genetics. 2005;42(6, article e38) doi: 10.1136/jmg.2004.029066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shan M., Dong B., Zhao X., et al. Novel mutations in the CYP4V2 gene associated with Bietti crystalline corneoretinal dystrophy. Molecular Vision. 2005;11:738–743. [PubMed] [Google Scholar]
- 14.Wada Y., Itabashi T., Sato H., Kawamura M., Tada A., Tamai M. Screening for mutations in CYP4V2 gene in Japanese patients with Bietti’s crystalline corneoretinal dystrophy. American Journal of Ophthalmology. 2005;139(5):894–899. doi: 10.1016/j.ajo.2004.11.065. [DOI] [PubMed] [Google Scholar]
- 15.Jin Z. B., Ito S., Saito Y., Inoue Y., Yanagi Y., Nao-i N. Clinical and molecular findings in three Japanese patients with crystalline retinopathy. Japanese Journal of Ophthalmology. 2006;50(5):426–431. doi: 10.1007/s10384-006-0350-0. [DOI] [PubMed] [Google Scholar]
- 16.Lai T. Y., Ng T. K., Tam P. O., et al. Genotype phenotype analysis of Bietti’s crystalline dystrophy in patients with CYP4V2 mutations. Investigative Ophthalmology & Visual Science. 2007;48(11):5212–5220. doi: 10.1167/iovs.07-0660. [DOI] [PubMed] [Google Scholar]
- 17.Zenteno J. C., Ayala-Ramirez R., Graue-Wiechers F. Novel CYP4V2 gene mutation in a Mexican patient with Bietti’s crystalline corneoretinal dystrophy. Current Eye Research. 2008;33(4):313–318. doi: 10.1080/02713680801983217. [DOI] [PubMed] [Google Scholar]
- 18.Liu D. N., Liu Y., Meng X. H., Yin Z. Q. The characterization of functional disturbances in Chinese patients with Bietti’s crystalline dystrophy at different fundus stages. Graefe’s Archive for Clinical and Experimental Ophthalmology. 2012;250(2):191–200. doi: 10.1007/s00417-011-1809-3. [DOI] [PubMed] [Google Scholar]
- 19.Giuffre G. Progression of Bietti’s crystalline dystrophy. Journal Francais d’Ophtalmologie. 1991;14(4):249–254. [PubMed] [Google Scholar]
- 20.Chakarova C. F., Hims M. M., Bolz H., et al. Mutations in HPRP3, a third member of pre-mRNA splicing factor genes, implicated in autosomal dominant retinitis pigmentosa. Human Molecular Genetics. 2002;11(1):87–92. doi: 10.1093/hmg/11.1.87. [DOI] [PubMed] [Google Scholar]
- 21.Vaclavik V., Gaillard M. C., Tiab L., Schorderet D. F., Munier F. L. Variable phenotypic expressivity in a Swiss family with autosomal dominant retinitis pigmentosa due to a T494M mutation in the PRPF3 gene. Molecular Vision. 2010;16:467–475. [PMC free article] [PubMed] [Google Scholar]
- 22.Wada Y., Itabashi T., Sato H., Tamai M. Clinical features of a Japanese family with autosomal dominant retinitis pigmentosa associated with a Thr494Met mutation in the HPRP3 gene. Graefe’s Archive for Clinical and Experimental. 2004;242(11):956–961. doi: 10.1007/s00417-004-0923-x. [DOI] [PubMed] [Google Scholar]
- 23.Kim C., Kim K. J., Bok J., et al. Microarray-based mutation detection and phenotypic characterization in Korean patients with retinitis pigmentosa. Molecular Vision. 2012;18:2398–2410. [PMC free article] [PubMed] [Google Scholar]
- 24.Martínez-Gimeno M., Gamundi M. J., Hernan I., et al. Mutations in the pre-mRNA splicing-factor genes PRPF3, PRPF8, and PRPF31 in Spanish families with autosomal dominant retinitis pigmentosa. Investigative Ophthalmology & Visual Science. 2003;44(5):2171–2177. doi: 10.1167/iovs.02-0871. [DOI] [PubMed] [Google Scholar]
- 25.Blanco-Kelly F., García-Hoyos M., Cortón M., et al. Genotyping microarray: mutation screening in Spanish families with autosomal dominant retinitis pigmentosa. Molecular Vision. 2012;18:1478–1483. [PMC free article] [PubMed] [Google Scholar]
- 26.Meng X. H., Guo H., Xu H. W., et al. Identification of novel CYP4V2 gene mutations in 92 Chinese families with Bietti’s crystalline corneoretinal dystrophy. Molecular Vision. 2014;20:1806–1814. [PMC free article] [PubMed] [Google Scholar]