Figure 1D of the above article was published with an insufficient resolution resulting in the loss of some important details. This had an impact on the understanding of the content. The enlarged figure is reproduced in all detail below.
Figure 1.
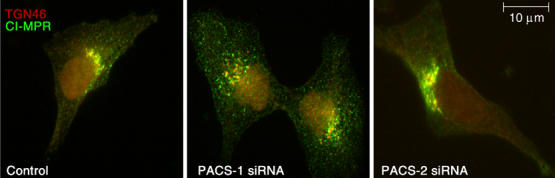
Identification and characterization of PACS-2, a sorting protein found on the ER and mitochondria. (A) Top: Schematic and Kyte–Doolittle hydrophobicity plot of the human PACS-1 and PACS-2 proteins. FBR, cargo/adaptor-binding region; ARR, atrophin-1-related region. Radiation hybrid and genome database analyses mapped the PACS-1 gene to chromosome 11q13.1 (Genbank AY320283) and the PACS-2 gene to chromosome 14q32.33 (Genbank AY320284). Bottom: Northern blot analysis of the tissue distribution for PACS-1 and PACS-2 transcripts. (B) Confocal immunofluorescence of endogenous PACS-1 and PACS-2 in A7 cells. PACS-1/PACS-2 were visualized with Alexa488 (green) and markers were visualized with Alexa546 or mitotracker (red). Scale bar, 10 μm. (C) A7 cells transfected with PACS-1 or PACS-2 siRNAs were analyzed by Western blot 48 h post-transfection. (D) A7 cells were transfected or not with PACS-1 or PACS-2 siRNAs. After 48 h, cells were processed for immunofluorescence microscopy using anti-CI-MPR (green) and anti-TGN46 (red). (E) A7 cells were transfected with control (scrambled), PACS-1, or PACS-2 siRNAs for 48 h and processed for confocal immunofluorescence localization of mitochondria (mitotracker, red) and ER (PDI, green). (F) A7 cells were transfected with the corresponding siRNAs and assayed for cell death by Annexin V/propidium iodide staining and FACS analysis. Treatment of cells with the proapoptotic CtBP siRNA served as a positive control (Zhang et al, 2003). Error for all graphs=s.d.


