Abstract
The avian nucleus laminaris (NL) is a brainstem nucleus necessary for binaural processing, analogous in structure and function to the mammalian medial superior olive. In chickens (Gallus gallus), NL is a well-studied model system for activity-dependent neural plasticity. Its neurons have bipolar extension of dendrites, which receive segregated inputs from two ears and display rapid and compartment-specific reorganization in response to unilateral changes in auditory input. More recently, FMRP (fragile X mental retardation protein), an RNA-binding protein that regulates local protein translation, has been shown to be enriched in NL dendrites, suggesting its potential role in the structural dynamics of these dendrites. To explore the molecular role of FMRP in this nucleus, we performed proteomic analysis of NL, using micro laser capture and liquid chromatography tandem mass spectrometry. We identified 657 proteins, greatly represented in pathways involved in mitochondria, translation and metabolism, consistent with high levels of activity of NL neurons. Of these, 94 are potential FMRP targets, by comparative analysis with previously proposed FMRP targets in mammals. These proteins are enriched in pathways involved in cellular growth, cellular trafficking and transmembrane transport. Immunocytochemistry verified the dendritic localization of several proteins in NL. Furthermore, we confirmed the direct interaction of FMRP with one candidate, RhoC, by in vitro RNA binding assays. In summary, we provide a database of highly expressed proteins in NL and in particular a list of potential FMRP targets, with the goal of facilitating molecular characterization of FMRP signaling in future studies.
Keywords: autism spectrum disorders, gene ontology, RNA binding protein, dendritic plasticity, cytoskeletal proteins, RhoC, RRID: AB_94856, RRID: AB_776174, RRID: AB_297884, RRID: AB_357520, RRID: AB_309663, RRID: AB_2277755, RRID: AB_2155806, RRID: AB_1859928, RRID: AB_10615780, RRID: AB_2620155
Introduction
Local translation of mRNAs occurs in dendrites in response to changes in neuronal activity (Martin and Zukin, 2006; Steward and Falk, 1985; Steward et al., 2014). Fragile X mental retardation protein (FMRP) is an RNA binding protein, which plays an important role in proper dendrite development by mechanisms of local translation (Santoro et al., 2012). Loss of FMRP leads to fragile X syndrome (FXS), a neurodevelopmental disorder with sensory, learning and social difficulties (Penagarikano et al., 2007). We have yet to understand the exact mechanism underlying these difficulties, but evidence suggests that abnormal dendritic plasticity is involved. Postmortem analyses of the brains of FXS patients as well as knock-out animal models reveal abnormalities of dendrites (Galvez et al., 2003; Galvez and Greenough, 2005; Hinton et al., 1991; Irwin et al., 2000; Zarnescu et al., 2005).
We have previously shown that FMRP is enriched in the dendrites of chick nucleus laminaris (NL) as well as in its equivalent structure, the medial superior olive (MSO), in mammals, including humans (Wang et al., 2014). NL and MSO are important for detecting temporal coincidence of afferent input between the two ears, which is believed to be essential for encoding the location of sound source along the azimuth (reviewed in Ashida and Carr, 2011; Grothe, 2000; Joris and Yin, 2007; Kuba, 2007). As a fundamental substrate for computing interaural time differences (ITDs), NL and MSO neurons form bipolar dendrites that are innervated differentially by segregated afferent inputs from the two ears. The evolutionary conservation in FMRP expression in NL and MSO dendrites suggests its importance in auditory temporal processing. Consistently, FXS patients exhibit problems with communication, hypersensitivity to auditory stimuli and inability to habituate to repeated auditory stimuli (Rotschafer and Razak, 2014). FMRP knock-out mice also display hypersensitivity to auditory stimuli (Chen and Toth, 2001).
NL/MSO dendrites display compartment-specific plasticity in structure and in protein levels, consistent with the functional role of FMRP in regulating dendritic and synaptic modifications via spatially controlling translation of its mRNA targets. Dendrites of NL and MSO neurons are highly dynamic. For example, unilateral deafening causes retraction of the dendrites receiving input from the affected side but not the other dendrites of the same neurons (Benes et al., 1977; Deitch and Rubel, 1984). Changes in dendrites, including the shortening of terminal dendritic branches, occur rapidly, within hours, and importantly, these changes are reversible in response to changes in afferent activity (Sorensen and Rubel, 2006, 2011; Wang and Rubel, 2012). Accompanying the structural changes, altered expression levels of a number of proteins in these dendrites also occur within a similar timescale (Wang and Rubel, 2008). These rapid and highly localized changes suggest that very rapid mechanisms are taking place perhaps by local protein translation or degradation.
Thus, NL/MSO provides a suitable model for studying neuroplasticity in dendrites, particularly for studying the molecular and cellular events regulated by FMRP signaling, with functional and clinical relevance to understanding the auditory and communication related behavioral deficits in FXS. Several hundred potential mRNA targets of FMRP have been predicted from tissue samples homogenized from whole mouse brains (Brown et al., 2001; Darnell et al., 2011; Ascano et al., 2012). In the current study, we take advantage of the simple anatomy of the chicken NL, which allows for the specific tissue collection from one neuronal cell type and its incoming afferent axons (Fig. 1). We first identified the proteins expressed in NL by mass spectrometry using a shotgun proteomic approach and then performed comparative analyses to identify potential FMRP targets in NL. In addition, we verified the dendritic localization of a number of the identified candidates using immunocytochemistry and confocal imaging. Finally, we confirmed the direct interaction of one such candidate with FMRP using in vitro RNA electrophoretic mobility shift assay (REMSA). Our results provide a list of promising FMRP targets in NL dendrites that may be important to structural dynamics of NL dendrites.
Figure 1.
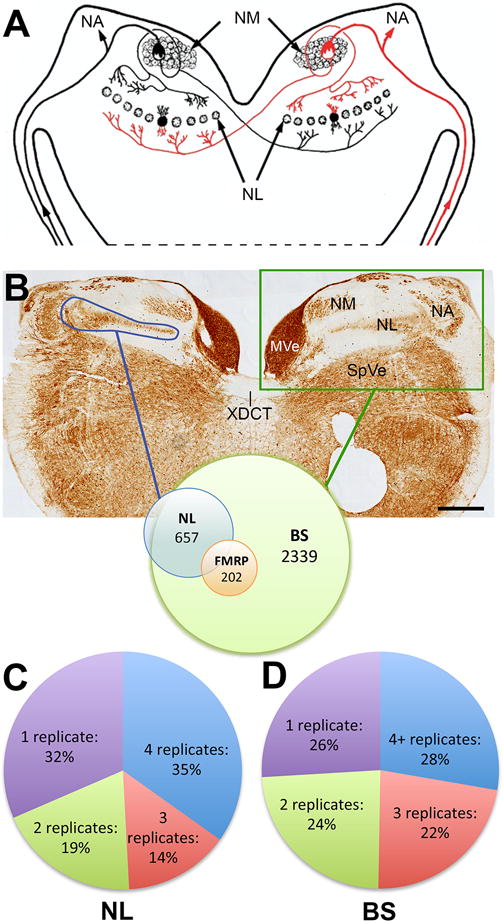
Proteins identified from nucleus laminaris (NL) and caudal brainstem (BS). A, Schematic of a coronal section of the brainstem is shown. Signals from the cochlea are transmitted via the eighth cranial nerve to NM. NM neurons then project to the dorsal aspect of ipsilateral NL and decussate to target the ventral aspect of contralateral NL. Axons and neurons innervated by the right cochlea are indicated in red color. B, A low-magnification image taken from the caudal brainstem at the level of NL in the coronal plane. The section is immunostained for microtubule-associated protein 2 (MAP2) to illustrate the location of the cell groups. Proteomic analysis was performed on unstained tissue block containing auditory brainstem (green box) or just NL collected by laser microdissection of cryosections (blue lines). 657 proteins identified from NL are a subset of 2339 proteins identified from BS. 202 proteins are putative FMRP targets. C–D, Pie charts demonstrate the distribution of identified proteins from NL (C) and BS (D) that were identified in one, two, three or four biological samples. Abbreviations: NM, nucleus magnocellularis; NL, nucleus laminaris; NA, nucleus angularis; MVe, medial vestibular nucleus; SpVe, spinal vestibular nucleus; XDCT, dorsal crossed cochlear track. Scale bars = 500 μm in A.
Materials and Methods
This study was performed on White Leghorn chick hatchlings (Gallus gallus) of less than ten-day age. All procedures were approved by the Florida State University and University of Washington Institutional Animal Care and Use Committees, and carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Tissue collection for mass spectrometry
Two types of tissue samples were collected from the brainstem of newly hatched chickens (postnatal day 0–4; n=18 total). The first sample type was collected specifically from the NL cell group (Fig. 1B; blue lines). After decapitation, the brains were quickly removed from the skull. The brainstem was blocked and immersed in optimal culture temperature (OCT) compound (Thermo Scientific, Inc., Waltham, MA), snap frozen in liquid nitrogen and stored at −80°C. Coronal sections (20 μm thick) containing NL were collected from the brainstem block using a cryostat and mounted on 0.9 μm Polyesther (POL)-membrane slides (Leica Microsystems, Buffalo Grove, IL). Under a laser micro-dissection microscope (LMD-6000; Leica Microsystems), the NL was outlined at 20× magnification, containing cell bodies and dendritic layers of NL with minimal inclusion of surrounding tissues. The outlined tissue was dissected with a laser (laser line width of 2 μm; laser power at 128, cutting speed at 1 μm per section) and dropped into a sterile 0.6 ml microtube. Dissected NL pieces from both sides of the brainstem from 2 animals were combined in one microtube and considered as one biological sample labeled as “NL”. In total, we collected 4 NL biological samples from 8 animals. These samples were stored at −80°C until protein extraction. The number of animals needed was based on how much protein was needed to identify ~500–2000 proteins.
The second type of tissue sample was collected from the dorsal brainstem at the caudorostral level of NL (Fig. 1B; green lines). After the brain was removed from the skull, a three-millimeter (3 mm) thick block was manually collected from the brainstem and placed flat in a petri dish. The dorsal brainstem at the level of the crossed dorsal cochlear tract was separated from the ventral brainstem. This sample contains three auditory nuclei (NL, nucleus magnocellularis, and nucleus angularis) as well as several vestibular cell groups (medial, lateral, spinal and ventral vestibular nuclei). Similarly, tissue samples from 2 animals were combined as one biological sample labeled as “BS” (brainstem). In total, we collected 5 BS biological samples from 10 animals. These samples were snap frozen in OCT in liquid nitrogen and stored at −80°C until protein extraction.
Protein preparation for mass spectrometry
For BS samples, tissues were thawed on ice in T-buffer (20mM Tris, 150mM NaCl, 1mM EDTA, 1mM EGTA), sonicated in 2×10 sec pulses in 500–1000 μl volume, centrifuged at 2300 × g for 10 minutes to remove cellular debris. Supernatant was diluted to 1 mg/ml and then ultracentrifuged at 100,000 × g for one hour. To improve the identification of low abundance membrane proteins, the membrane pellet and cytosolic supernatant were collected separately and processed independently as one biological sample each. Both were solubilized at 60°C in sodium dodecyl sulfate (SDS) at a final concentration of 0.1%, incubated in 5 mM dithiothreitol (DTT) for 30 minutes at 60°C followed by 15 mM Iodoacetamide (IAA), an alkylating agent, for 30 minutes at the room temperature in the dark, and then digested with 2 μg trypsin (Sigma, proteomic grade) overnight at 37°C to hydrolyze specifically at the carboxyl side of arginine and lysine residues. Samples were hydrolyzed with 100 mM HCl for 5 minutes at 37°C. Detergent was then removed with Oasis MCX cleanup kit (Waters Corporation; Milford, MA).
For NL samples which were attached to POL membrane pieces, samples were incubated with 0.1% rapigest/Tris-buffer for 5 minutes at 95°C. As described above, tissues were treated with DTT and IAA. Proteins were digested with trypsin and then the detergent was hydrolyzed with HCl. Samples were centrifuged twice at 16,000 × g for 5 minutes each to remove the POL membrane debris. The solution was dried to a volume of 20 μl with a speedvac before mass spectrometry.
Mass spectrometry (MS) and protein identification
Purified peptide samples were loaded onto 30 cm by 75 μm width silica columns packed with 4 μm Jupitor 90Å beads and 2 mm length Kasil trap with 3 μm Jupitor 90Å beads. Flow pressure was kept ~1500 psi at 0.250 μl/min flow. The LTQ-FT Ultra (ThermoFisher Scientific) mass spectrometer was used. Spectra were acquired using a cycle of one high-resolution MS scan (400–1400 m/z) followed by five data-dependent MS/MS scans at low resolution, repeated continuously throughout the analysis. Spectra were matched to peptide sequences using SEQUEST (Eng et al., 1994) (non-tryptic ends). Peptide-spectrum match (PSM) and peptide identifications were obtained from Percolator (v2.01) (Käll et al., 2007). Peptides with Percolator with q-value <0.01 were given as input to ID Picker (Zhang et al., 2007) for protein identification. We used a decoy database using scrambled Gallus gallus genome sequence (build 12/17/11). We required at least 2 peptides per protein, each with a q-value (false discovery rate) of <0.01 (estimated 1% rate of false discoveries among the accepted peptides). At least four biological replicates with three technical replicates each were performed. We required each peptide to present in every technical replicate (n=3) and at least 2 peptides per protein for identification. Because of the multiple technical replicates, we pooled the proteins identified in each of the biological samples. Fig. 1C–D demonstrates the percentages of proteins identified in multiple biological samples. Protein identification comparisons among different experiments were made using MSDaPl (Sharma et al., 2012).
Gene ontology analyses
Several software programs were used to perform gene ontology analyses of the identified proteins. The first is the DAVID Bioinformatics Resources 6.8 (National Institute of Allergy and Infectious Diseases, National Institute of Health; https://david.ncifcrf.gov/). We used this resource for protein functional annotation and gene ID conversion. Ensembl Bio-mart software (http://www.ensembl.org/biomart/martview/) was also used for gene ID conversion. For identifying transmembrane proteins, we used the TMHMM program version 2.0 located at: http://www.cbs.dtu.dk/services/TMHMM/. Finally, we used the Ingenuity pathway analysis at http://www.ingenuity.com/ as an alternative approach to DAVID for identifying enriched pathways.
Western blot
Protein samples were harvested from flash frozen chicken brainstem tissue. Samples were homogenized in EDTA buffer (62.5 mM Tris-HCl pH 6.8, 2% SDS, 10% Glycerol, 5% β-ME, 10 mM EDTA) using the Ultra-Turrax® T10 homogenizer (IKA® Works, Inc., Wilmington, NC). 50 μg of protein lysate in SDS buffer (2% SDS, 50 mM Tris pH 7.6, 5% glycerol, and 0.025% bromophenol blue) was incubated at 70°C for 10 minutes, resolved in NuPAGE 4–12% Bis-Tris Gels (Life Technologies, Carlsbad, CA), and then transferred onto PDVF membranes (GE Healthcare, Chicago, IL). After blocking in 5% milk in PBS with 0.05% Tween (PBS-T) for 30 minutes at room temperature, membranes were probed against the primary antibodies (Tab. 1) overnight at 4°C in 1% milk in PBS-T. Specific secondary HRP-conjugated antibodies were used at 1:2500 dilution (Santa Cruz, Biotechonology®, Inc., Dallas, TX) and blots were developed with SuperSignal™ West Pico Chemiluminescent Substrate (Thermo Scientific, Inc., Waltham, MA) and exposed to X-ray film.
Table 1.
Primary antibodies used for immunocytochemistry and other staining. Abbreviations: ICC: immunocytochemistry; WB: Western blot.
| Immunogen | Manufacturer, catalog number, Host, monoclonal or polyclonal; RRID, | Concentration | |
|---|---|---|---|
| Anti-MAP2 | Bovine brain MAP2 (aa 997–1332) | Millipore (Billerica, MA), MAB3418; Mouse monoclonal; RRID:AB_94856 |
1:1000 (ICC); 1:1000 (WB) |
| Anti-MAP2 | Synthetic peptide rat MAP2 (aa 1-100) | Abcam (Cambridge, MA), ab32454; Rabbit polyclonal; RRID:AB_776174 |
1:1000 (ICC) |
| Anti-MAP1B | Full length rat brain MAP1B | Abcam, Ab11266; Mouse monoclonal IgG1 clone AA6; RRID:AB_297884 |
1:10,000 (ICC); 1:1000 (WB) |
| Anti-TuJ-1 | Rat brain tubulin beta-3 | R&D Systems (Minneapolis, MN), MAB1195; Mouse monoclonal IgG2a; RRID:AB_357520 |
1:10,000 (ICC); 1:1000 (WB) |
| Anti-eEF1a | Crude calmodulin-binding proteins from Trypanosoma brucei chromatography | Millipore, 05-235; Mouse monoclonal IgG1k clone CBP-KK1; RRID:AB_309663 |
1:1000 (ICC); 1:1000 (WB) |
| Anti-p-eEF2 | Synthetic phosphopeptide surrounding Thr56 of human eEF2, GETRFtDTRK | Cell Signaling Technology (Danvers, MA), No. 2331; Rabbit polyclonal; RRID:AB_2277755 |
1:1000 (ICC) |
| Anti-NSF-1 | Recombinant human full length NSF. | Abcam, ab16681; Mouse monoclonal IgG; RRID:AB_2155806 |
1:1000 (ICC); 1:1000 (WB) |
| Anti-RhoC | Synthetic peptide human RhoC (aa 100-C-term) | Abcam, ab64659; Rabbit polyclonal; RRID:AB_1859928 |
1:1000 (ICC); 1:1000 (WB) |
| Anti-SERCA2 | Purified canine cardiac sarcoplasmic reticulum. | Millipore, MAB2636; Mouse monoclonal IgG1k clone IID8; RRID:AB_10615780 |
1:1000 (ICC); 1:1000 (WB) |
Immunostaining
Immunocytochemistry was used to verify the expression of a number of MS-identified proteins in NL neurons and examine their subcellular localization. Chickens (P0–P4) were transcardially perfused with 0.9% saline followed by 4% paraformaldehyde in 0.1 M phosphate buffer (PB). The brains were removed from the skull, post fixed overnight in the same fixative, and transferred to 30% sucrose in 0.1 M PB. Brains were then sectioned in the coronal plane at 20–30 μm on a freezing sliding microtome or a cryostat and collected in PBS. Sections containing NL were immunostained using the primary antibodies listed in Table 1. The staining procedure was described previously (Wang et al., 2009). Briefly, blocking was performed by 4% normal goat serum in PBS overnight at 4°C or with a blocking solution (PerkinElmer FP1020; Waltham, MA) at room temperature for 30 minutes. Sections were incubated with primary antibody solutions diluted in PBS with Triton X-100 (at a concentration of 0.3% or 0.5%) overnight at 4°C followed by AlexaFluor secondary antibodies (1:200) for 1–2 hours at room temperature. Secondary antibodies were purchased from either Molecular Probes (Eugene, OR) or Jackson ImmunoResearch (West Grove, PA). Sections were coverslipped with Fluoromount-G (SouthernBiotech, Birmingham, AL).
Antibody characterization
Table 1 lists the primary antibodies used in the current study, including the immunogen, host species, clone type, manufacturer’s information, as well as dilution used for each antibody.
We used the microtubule-associated protein 2 (MAP2) as a dendritic marker. MAP2 expression and immunohistochemistry using a mouse monoclonal antibody (Millipore, MAB3418) in NL dendrites has been well characterized (Wang and Rubel, 2008; Wang et al., 2014). In this study, we also used a second rabbit polyclonal antibody for MAP2 (Abcam, ab32454) to facilitate double staining with other antibodies raised in the mouse. Double staining with both the rabbit and mouse anti-MAP2 antibodies demonstrated identical staining pattern in NL (see Fig. 5). In this study, we verified the specificity of the mouse anti-MAP2 antibody with Western blot in chicken brain tissue (Fig. 2).
Figure 5.
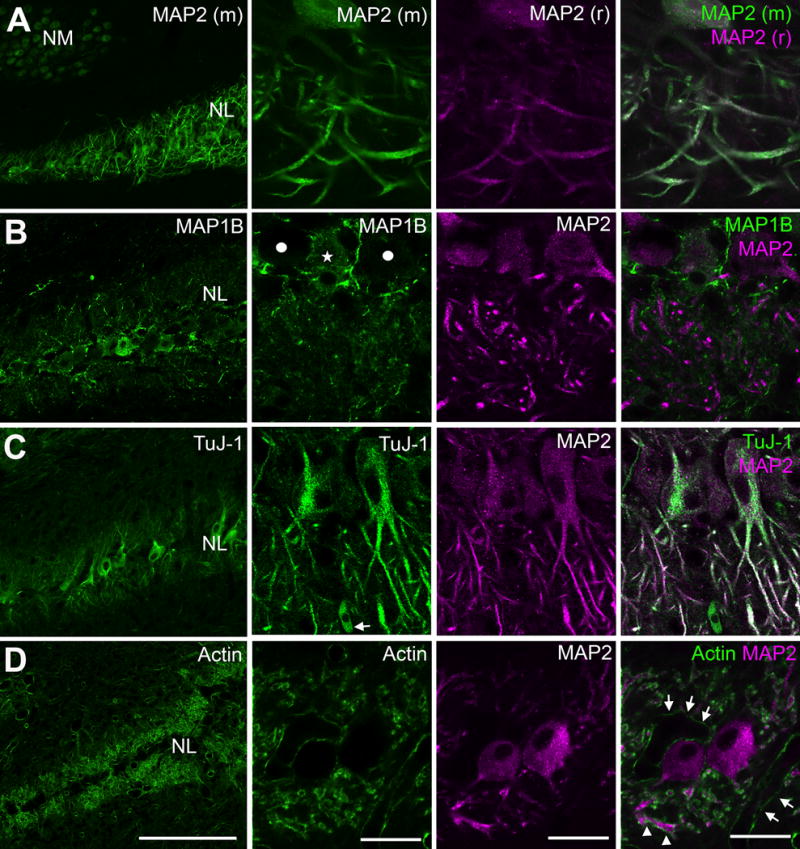
Subcellular distribution of cytoskeletal elements and their associated proteins in NL examined by immunocytochemistry. A: Two antibodies for MAP2 raised in rabbit (r) or mouse (m) display identical dendritic labeling in NL. These two antibodies are subsequently used as dendritic markers for examining the localization of other protein candidates. B: Double labeling of MAP1B and MAP2. Strong MAP1B immunoreactivity in NL neuropil regions does not overlap with MAP2-labeled dendrites. Detectable MAP1B immunoreactivity is found in some (white star) but not other NL cell bodies (solid white circles). C: Double labeling of TuJ-1 and MAP2. TuJ-1 immunoreactivity is strong in the cell bodies and the primary portions of dendrites, and relatively weaker in the more distal dendritic branches. D: Phalloidin stain visualizing the distribution of F-actin surrounding MAP2 labeled dendritic branches (arrowheads). Arrows point to phalloidin stain along blood vesicles. Abbreviations: NM, nucleus magnocellularis; NL, nucleus laminaris; TuJ-1, neuron-specific class III beta-tubulin; MAP2, microtubule-associated protein 2; MAP1B, microtubule-associated protein 1B. Scale bar = 100 μm (left column) and 20 μm (all other columns).
Figure 2.
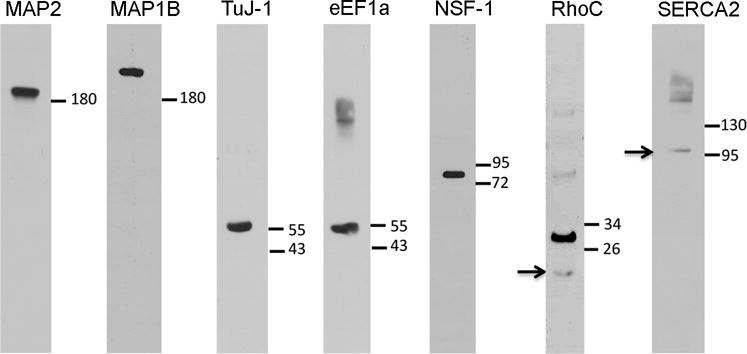
Western blotting for antibody characterization in the chicken brain. Fifty micrograms (μg) of protein lysate from brain tissue was loaded to each lane. Molecular weight standards (right to each lane) were used to determine relative sizes of labeled protein bands. Arrows point to the bands of the protein of interest with predicted molecular weight. See Table 1 and the Methods for more information on these antibodies.
Mouse monoclonal anti-MAP1B (clone AA6) was produced using rat brain MAP. This antibody recognizes a conserved nonphosphorylated and nonphosphorylatable epitope on MAP1B (DiTella et al., 1996; Paglini et al., 1998). It reacts with all isoforms of MAP1B, in both Western blot and immunofluorescence applications (Franzen et al., 2001; Impens et al., 2008; Eriksson et al., 2010). Based on the datasheet provided by the manufacturer, this antibody does not react with tubulin or other microtubule associated proteins. In chicken brain tissue, we verified the specificity of the antibody recognizing the chicken MAP1B protein by western blot (Fig. 2).
The clone TuJ-1 of the anti-beta-III tubulin monoclonal antibody was raised against rat microtubules and purified from hybridoma culture supernatant. This antibody recognizes neuron-specific beta-III tubulin specifically, as verified by both Western blot and immunocytochemistry in human, mouse and rat brains (see the datasheet provided by the manufacturer). In this study, we further verified the specificity of the antibody in chicken brain tissue, demonstrating a single Western blot band at approximately 55 Kilodaltons (KDa) (Fig. 2), the predicted molecular weight for chicken beta-III tubulin.
The clone CBP-KK1 of the anti-eEF1a monoclonal antibody was produced against crude calmodulin-binding proteins from Trypanosoma brucei (T brucei) isolated using calmodulin-affinity chromatography. This antibody binds to eEF1 alpha by lambda library from T brucei (Kaur and Ruben, 1994). In this study, we confirmed that the antibody recognizes the chicken eEF1a at ~50 kDa protein (Fig. 2), corresponding to the predicted molecular weight.
The anti-phosphorylated-eEF2 (anti-p-eEF2) antibody was raised against the phosphor peptide surrounding Thr56 of human eEF2. We have previously characterized this antibody by Western blot in chicken brain samples (McBride et al., 2013). This anti-p-eEF2 antibody recognizes chicken p-eEF2 at ~95 kDa.
Monoclonal antibody to NSF protein was raised against recombinant human NSF and previously characterized by Western blotting (Boström et al., 2007). In this study, we identified a single band at approximately 80 KDa on Western blot for the chicken brain tissue (Fig. 2), corresponding to the predicted molecular weight.
Anti-RhoC rabbit polyclonal antibody was raised against the synthetic peptide corresponding to the C-terminal end (aa100-C-term) of human RhoC. The manufacturer tested the antibody in human and mouse cells by Western blot. In this study, we verified that this antibody recognizes a 21 KDa band on Western blotting (Fig. 2), corresponding to the predicted chicken RhoC protein. In addition, there is a second, strong unexpected band of higher molecular weight at ~30 KDa.
The IgG1k clone IID8 of the anti-SERCA2 monoclonal antibody was developed against canine cardiac sarcoplasmic reticulum and recognizes human SERCA2 (Chami et al., 2001). In this study, we confirmed the specificity of the antibody in the chicken brain by Western blot, showing a band of approximately 105 KDa (Fig. 2), corresponding to the predicted molecular weight of the chicken SERCA2.
Phalloidin staining
Phalloidin is a well characterized chemical used for staining filamentous actin (F-actin). Following immunocytochemistry for a MAP2 antibody, the sections were incubated in Alexa Fluor 647 Phalloidin (Life Technology; Eugene, OR) diluted in 1:100 in PBS for 30 minutes in the dark at the room temperature. Staining was stopped by washing the sections in PBS.
Imaging
Images were captured either with a Zeiss M2 microscope for bright-field and epi-fluorescent images, or with the Zeiss LSM 880 confocal microscope. Epi-fluorescent images taken with the M2 microscope were treated with the Zeiss Apotome, an optical sectioning approach using structured illumination for reducing out-of-focus information in epi-fluorescent images (Neil et al., 1997; Neil et al., 2000). Photomontages were applied in the Zeiss Zen blue software. Image brightness, gamma, and contrast adjustments were performed in Adobe Photoshop (Adobe Systems, Mountain View, CA). All adjustments were applied equally to all images of the same set of staining from the same animal unless stated otherwise.
Recombinant chicken HIS tagged FMRP expression and purification
Chicken FMRP isoform 2, sequence-optimized for expression in E. coli, was cloned into the NdeI/XhoI restriction sites in the expression vector pET-21a (+) (Merck Millipore Corp., Billerica, MA) containing a histidine (His) tag. BL21 E. coli cells were transformed via electroporation and glycerol stocks were stored at −80°C. Cells were plated on 2YT/Amp plates and incubated for 16 hours at 37°C. 1 L of 2YT/Amp culture was inoculated from fresh confluent plate and grown to OD595 between 0.6–0.7 at 37°C with vigorous shaking. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to final concentration of 1 mM and cells were grown at 30°C with vigorous shaking for 3 hours. Cells were pelleted by centrifugation at 4000 rpm for 15 minutes at 4°C. Cell pellets were resuspended in Lysis buffer (20mM HEPES pH 7.5, 200 mM NaCl, 10 mM β-mercaptoethanol, 25 mM Imidazole) plus protein inhibitors: 0.5 mM PMSF, 2μg/mL of Aprotinin, 0.5 μg/mL of Leupeptin and 1 μg/mL of Pepstatin, frozen in dry ice and stored at −80°C freezer. Cells were thawed at 37°C and kept on ice for 15 minutes. Protease inhibitors were added and Bugbuster® plus Lysonase (Merck Millipore) were added and incubated at 4°C for 20 minutes in a nutator. Triton X-100 was added to final concentration of 1% and incubated for 10 minutes at 4°C in the nutator. Cell lysate was centrifuged at 15000g for 30 minutes at 4°C. Supernatant was added to 1 mL of Ni-NTA resin (Qiagen, Inc., Hilden, Germany) pre-washed in lysis buffer and batch binding was performed at 4°C for 2 hours. Resin was washed with 40 volumes of Lysis buffer plus 0.5% NP40 and resuspended in 40 vol of Lysis buffer containing 40mM of Imidazole. The ressuspended beads were added to a column and settled by flow gravity. His-FMRP protein was eluted with 4 × 1 mL of Lysis buffer plus 100 mM Imidazole. Eluates were pooled and dialyzed against dialysis buffer (20 mM HEPES pH 7.5, 1 mM EDTA, 2 mM DTT, 100 mM NaCl, 0.05% NP40 and 20% Gycerol). Protein concentration was measured using standard techniques, aliquoted and stored at 4°C.
RNA gel electrophoretic mobility shift assay
To test for direct interaction between FMRP and RhoC, we looked at the RNA sequence for RhoC and identify several of putative FMRP binding domains based on previous work by Anderson et al. (2016). We then chose a region that contained at least 6 putative binding sites and designed an RNA probe for RhoC containing these putative binding sites. The 5′ biotin-labeled and unlabeled RhoC RNAs were chemically synthesized (GenScript, Picataway NJ) with the following sequence: 5′ GAACUACAUCGCCGACAUUGAGGUGGAUGGGAAGCAGGUGGAGCUGGCG-3′. RNA binding was carried out using the LightShift® Chemiluminescent RNA EMSA kit (Thermo Fisher Scientific, Waltham, MA). Briefly, 5mM of biotin-labeled RhoC RNA was incubated with 125 ng of purified HIS-FMRP in the absence or presence of increasing concentrations of unlabeled RhoC RNA as well as with increasing concentrations of a non-specific unlabeled RNA probe, for 30 minutes at room temperature. The reactions were electrophoresed in 6% polyacrylamide gel in 0.5X Tris Borate EDTA (TBE) and transferred onto a nylon membrane. Blot was UV cross-linked at 120mJ/cm2 using a CL-1000 UV Cross linker (UVP LLC, Upland, CA). Blot was blocked, probed with Streptavidin-Horseradish Peroxidase conjugate, and exposed to X-ray film.
Results
Identification of proteins from dorsal brainstem (BS) and nucleus laminaris (NL)
Using mass spectrometry, we identified 657 proteins from 4 biological samples of NL (blue, NL laser capture) (Fig. 1B). Approximately two thirds of these proteins (450; 68%) are identified in more than one biological replicates and more than one third of total proteins (229; 35%) are identified in all 4 biological replicates (Fig. 1C). The total spectral count for and the number of biological replicates in which each protein was identified are listed in supplemental Table S1.
Although laser capture provides specificity of tissue collection from NL, we needed to verify that the additional steps in tissue processing do not cause a significant increase in false positives. A comparison with traditional approach of snap frozen tissue served as a straightforward and powerful strategy to address this issue. We identified 2339 proteins from 5 biological samples of BS (green, dorsal brainstem en bloc; snap frozen tissue, Fig. 1B). Among these proteins, 1732 (74%) proteins are identified in more than one biological replicates and 648 (28%) proteins are identified in at least 4 out of 5 biological replicates (Fig. 1D), demonstrating comparable reproducibility as NL samples. The total spectral count for and the number of biological replicates in which each protein was identified, are listed in supplemental Table S2. The majority (96%; 632/657) of NL proteins were also identified in BS. This near complete overlap is consistent with NL proteins being a subset of BS proteins and further validates the reproducibility of the protein identification strategy.
On the other hand, a number of proteins that are known to be expressed in NL neurons were not identified in NL samples but included in BS proteins, such as FMRP itself, indicating that the 657 protein list of NL is only a fraction of the entire proteomics of this nucleus. Using the THMM software, we found 13% of NL and 20% of BS proteins with predicted transmembrane features. As a comparison, human genome predictions have estimated ~20% are transmembrane proteins, suggesting that our proteomic analysis of NL may be biased towards soluble proteins. In support of this suggestion, NL neurons express a number of voltage-gated potassium and sodium channels as evidenced by previous immunocytochemical studies (Lu et al., 2004b; Kuba et al., 2005; 2014), which were not identified by the mass spectrometry in this study.
NL is known to be highly metabolic. NL neurons generate high rates of action potentials, both in conditions of quiet and of acoustic stimulation (Born et al., 1991). Consistently, NL neurons contain a high density of mitochondria throughout their dendrites (Deitch and Rubel, 1989). Functionally, NL dendrites display high levels of energy consumption indicated by a high level of cytochrome oxidase and glucose uptake as measured by 2-deoxyglucose method (Dezso et al., 1993; Heil and Scheich, 1986; Lippe et al., 1980). Using DAVID software (Huang da et al., 2008), we obtained corresponding gene IDs for the proteins identified from NL and BS. We looked for enriched pathways represented by the gene list compared to what would be expected from the Gallus gallus genome. Enrichment score of >1.3 is considered meaningful (Huang da et al., 2008). Among the top ten enriched pathways in the BS and NL samples, mitochondrial, metabolic and translation pathways were on the top in both sets of samples (Table 2), indicating that the protein identification of NL in this study reflects this prominent feature of high activity in NL.
Table 2.
Enriched pathways of NL and BS proteins revealed by DAVID analysis. Proteins identified from each tissue set were compared to the genome of Gallus gallus to determine if there is enrichment for proteins from certain pathways, using DAVID program. Score of >1.3 is significant. The top 5 enriched pathways suggest a highly metabolic mileu.
| Nucleus Laminaris (NL) | Dorsal Brainstem (BS) | ||||
|---|---|---|---|---|---|
|
| |||||
| Cluster | Enrichment Score | Cluster | Enrichment Score | ||
| 1 | Mitochondrial | 16.28 | 1 | Mitochondrial | 12.50 |
| 2 | Ribosomal/Translation | 13.98 | 2 | Metabolic | 12.24 |
| 3 | Protein folding | 9.94 | 3 | Ribosomal/Translation | 9.48 |
| 4 | Cytoskeletal | 9.18 | 4 | Respiration | 7.49 |
| 5 | Metabolic | 9.08 | 5 | Oxidative | 7.29 |
| 6 | Protein transport | 8.79 | 6 | Protein folding | 6.39 |
| 7 | Cellular respiration | 8.21 | 7 | Nucleotide binding | 5.95 |
| 8 | Nucleotide binding | 7.37 | 8 | Cytoskeletal | 5.89 |
| 9 | Neuronal projection | 7.12 | 9 | Nucleotide binding | 5.09 |
| 10 | RNA recognition | 6.69 | 10 | Oxidoreductase | 4.97 |
Comparative analyses identified 94 putative FMRP targets in NL
We were interested in determining which of the proteins identified were most likely to be translated locally in dendrites. In particular, we were interested in those proteins that may be products of mRNAs regulated by FMRP. We obtained a combined list of FMRP targets previously identified in mouse brains (Brown et al., 2001; Darnell et al., 2011). To allow comparative analyses between different species (mouse vs. chicken) and between RNA and amino acid sequence, we first created databases for each list with corresponding RNA sequence, protein sequence, gene ID and gene name using a combination of DAVID and Ensembl Bio-mart software (Fig. 3). We performed comparative analyses first at the gene level by finding orthologous matches based on the aforementioned curated database of Gene IDs, and then by BLAST analysis to manually identify additional orthologs by sequence homology. In total, we found a unique list of 94 proteins that are products of putative FMRP targets (Table. 3). Consistent with a recent study that described the characteristics of top FMRP binding transcripts as having >18 RNA WGGA or ACUK sequences (Ascano et al., 2012), all 94 putative FMRP candidates have these sequences and the majority of them (88 out of 94 candidates; 94%) have >18 copies of such sequences.
Figure 3.
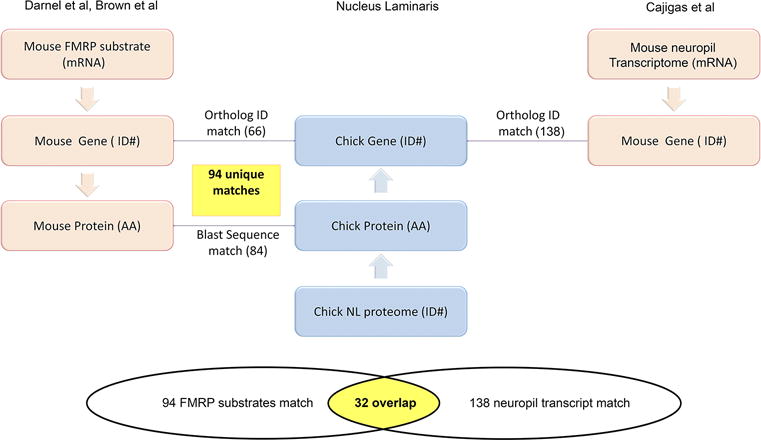
Schematic chart flow showing the approach used for identifying 94 unique proteins as candidate targets of FMRP in NL. We first combined the predicted mRNA targets of FMRP in the mouse brain from two studies (Brown et al., 2001; Darnell et al., 2011). We then created two databases with corresponding gene (ID#) and protein sequences (AA) using NCBI, ENSEMBL, BIOMART, and DAVID programs. Similarly, we generated two databases for protein sequence (AA) and corresponding gene (ID#) from the MS-identified proteins from the chicken NL. Finally, in order to find known equivalent FMRP from mice in our chicken NL proteome data, we employed the following strategy: find annotated orthologs (top line) or identify protein sequence matches (bottom line) by BLASTP. In total, 66 ortholog ID and 84 BLAST sequence matches were found, which together generate 94 unique matches. We also performed a comparison with a list of transcripts found in hippocampal neuropil (Cajigas et al., 2012). Of the 657 NL proteins, 138 targets match this list. Among the 138 matches, 32 candidates overlap with the 94 FMRP target candidates, presenting a selective list of proteins with a high likelihood of local translation in NL dendrites.
Table 3.
A list of 94 FMRP candidate substrates. Entrez gene identification number, official gene symbol and description are provided. Also provided are functional categorization (F), subcellular localization (S), the total number of spectral counts (SC) matching the protein and the number of biological replicates (BR) in which protein was identified (n=4).
| GeneID | Symbol (alias) | Description | F | S | SC | BR | NCBI Reference |
|---|---|---|---|---|---|---|---|
| 395373 | ACLY | ATP citrate lyase | m | c | 29 | 3 | NP_001025711 |
| 374009 | ACO2 | aconitase 2, mitochondrial | m | m | 246 | 4 | NP_989519 |
| 396526 | ACTB | actin, beta | c | c | 286 | 3 | NP_990849 |
| 423298 | ACTC1 | actin, alpha, cardiac muscle 1 | c | c | 213 | 3 | NP_001072949 |
| 415296 | ACTG1 | Actin, gamma 1, cytoplasmic type 5 | c | c | 418 | 4 | NP_001007825 |
| 422882 | ADD1 | adducin 1 (alpha) | c | c | 17 | 2 | NP_001073198 |
| 395492 | ALDOC | aldolase C, fructose-bisphosphate | m | c | 229 | 4 | NP_001193425 |
| 420761 | AMPH | amphiphysin | v | v | 27 | 4 | NP_001004398 |
| 423102 | AP2A2 | adaptor-related protein complex 2, alpha 2 subunit | v | p | 20 | 2 | NP_001012914 |
| 417525 | AP2B1 | adaptor-related protein complex 2, beta 1 subunit | v | p | 21 | 3 | XP_415772 |
| 420398 | ARF1 | ADP-ribosylation factor 1 | v | v | 25 | 2 | NP_001006352 |
| 769725 | ARF4 | ADP-ribosylation factor 4 | v | c | 30 | 3 | XP_001232784 |
| 396530 | ATP1A1 | ATPase, Na+/K+ transporting, alpha 1 polypeptide | t | p | 294 | 4 | NP_990852 |
| 396468 | ATP1A2 | ATPase, Na+/K+ transporting, alpha 2 polypeptide | t | p | 607 | 4 | NP_990807 |
| 396467 | ATP1A3 | ATPase, Na+/K+ transporting, alpha 3 polypeptide | t | v | 773 | 4 | NP_990806 |
| 396529 | ATP1B1 | ATPase, Na+/K+ transporting, beta 1 polypeptide | t | p | 149 | 4 | NP_990851 |
| 396446 | ATP2A2 (SERCA2) | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2; sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | t | v | 8 | 2 | XP_003642224 |
| 415958 | ATP2B2 | ATPase, Ca++ transporting, plasma membrane 2 | t | p | 43 | 4 | XP_001231642 |
| 374159 | ATP5A1 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle | t | p | 370 | 4 | NP_989617 |
| 431564 | ATP5A1W | ATP synthase subunit alpha | t | m | 349 | 4 | XP_429118 |
| 426673 | ATP5B | ATP synthase subunit beta, mitochondrial | t | m | 366 | 4 | NP_001026562 |
| 395497 | ATP6V1B2 (VATB) | ATPase, H+ transporting, V-type proton ATPase subunit B | t | v | 56 | 4 | XP_424534 |
| 425976 | BCAN | brevican | g | e | 15 | 2 | XP_423655 |
| 395855 | CALM | calmodulin | s | c | 125 | 4 | NP_990336 |
| 417837 | CAND1 | cullin-associated and neddylation-dissociated 1 | p | c | 6 | 1 | XP_416078 |
| 396248 | CKB | creatine kinase, brain | m | m | 628 | 4 | NP_990641 |
| 395272 | CLTC (CHC) | clathrin heavy chain 1 | v | p | 204 | 4 | NP_001073586 |
| 416765 | CLTCL1 | clathrin, heavy chain-like 1 | v | p | 33 | 1 | XP_415060 |
| 395921 | CNP | 2′,3′-cyclic nucleotide 3′ phosphodiesterase | g | p | 639 | 4 | NP_990381 |
| 427286 | CPLX1 | complexin 1 | v | v | 20 | 2 | XP_424869 |
| 395156 | CRMP1 (CRMP1B) | collapsin response mediator protein 1 | g | c | 66 | 4 | NP_989826 |
| 417217 | DNM1 | dynamin 1 | v | p | 14 | 1 | XP_415501 |
| 395155 | DPYSL2 (CRMP2A) | dihydropyrimidinase-like 2, collapsin response mediator protein-2A | g | c | 169 | 4 | NP_989825 |
| 423461 | DYNC1H1 | dynein, cytoplasmic 1, heavy chain 1 | v | c | 57 | 3 | XP_421371 |
| 373963 | EEF1A1 | eukaryotic translation elongation factor 1 alpha 1 | tx | c | 143 | 4 | NP_989488 |
| 419244 | EEF1A2 | eukaryotic translation elongation factor 1 alpha 2 | tx | c | 98 | 4 | NP_001027570 |
| 396325 | EEF2 | eukaryotic translation elongation factor 2 | tx | c | 80 | 4 | NP_990699 |
| 419117 | EPB41L1 | erythrocyte membrane protein band 4.1-like 1 | c | p | 10 | 2 | XP_417304 |
| 396061 | FASN | fatty acid synthase, thioesterase | m | c | 12 | 2 | NP_990486 |
| 416485 | FSCN1 | fascin | c | c | 172 | 4 | NP_001171603 |
| 396489 | GLUL | glutamine synthetase | m | m | 247 | 4 | NP_990824 |
| 419402 | GNB1 | guanine nucleotide binding protein (G protein), beta polypeptide 1 | s | c | 86 | 4 | NP_001012853 |
| 424974 | GNB4 | guanine nucleotide binding protein (G protein), beta polypeptide 4 | s | c | 47 | 3 | XP_003641822 |
| 373889 | HK1 | hexokinase 1 | m | m | 89 | 4 | NP_989432 |
| 423463 | HSP90AA1 | heat shock protein HSP 90-alpha | p | c | 333 | 4 | NP_001103255 |
| 396188 | HSP90AB1 (HSPCB) | heat shock cognate protein HSP 90-beta | p | c | 23 | 1 | NP_996842 |
| 408046 | KIF5C | kinesin family member 5C | r | c | 9 | 1 | XP_422155 |
| 100857214 | LOC100857214 | tubulin alpha-4A chain-like | c | c | 47 | 1 | XP_003641691 |
| 100857247 | LOC100857247 | tubulin alpha-5 chain-like | c | c | 254 | 4 | XP_003641692 |
| 100857345 | LOC100857345 | V-type proton ATPase subunit d 1-like | t | v/p | 3 | 1 | XP_414041 |
| 100857582 | LOC100857582 | ubiquitin-like modifier-activating enzyme 1-like | p | c | 2 | 1 | XP_003643588 |
| 100858165 | LOC100858165 | V-type proton ATPase subunit d 1-like | t | v/p | 3 | 1 | XP_003643353 |
| 425049 | LOC425049 | tubulin alpha-3 chain-like | c | c | 147 | 3 | XP_422851 |
| 768337 | LOC768337 | tubulin beta-2 chain-like | c | c | 187 | 2 | XP_001231210 |
| 415588 | MAP1A | microtubule-associated protein 1A | c | c | 54 | 4 | XP_003641886 |
| 396174 | MAP1B | microtubule-associated protein 1B | c | c | 5 | 1 | XP_001231729 |
| 396097 | MAP4 | microtubule-associated protein 4 | c | c | 7 | 1 | XP_418480 |
| 373953 | MAPK1 | mitogen-activated protein kinase 1 | s | c | 6 | 1 | NP_989481 |
| 396217 | MBP | myelin basic protein | s | p | 726 | 4 | NP_990611 |
| 396465 | MYH10 | myosin, heavy chain 10, non-muscle | c | c | 8 | 1 | NP_990805 |
| 428253 | NCAM1 | neural cell adhesion molecule 1 | g | p | 20 | 2 | NP_001122300 |
| 768618 | NDRG4 | NDRG family member 4 | s | c | 9 | 2 | XP_001231665 |
| 419972 | NSF | N-ethylmaleimide-sensitive factor, vesicle-fusing ATPase | v | v | 23 | 3 | NP_001019627 |
| 426429 | OGDH | 2-oxoglutarate dehydrogenase, mitochondrial | m | m | 58 | 4 | NP_001026553 |
| 396214 | PLP1 | myelin proteolipid protein | g | p | 231 | 4 | NP_990608 |
| 395602 | PSAP | prosaposin | m | v | 2 | 1 | NP_990142 |
| 374204 | QKI | QKI, KH domain containing, RNA binding; protein quaking | s | p | 13 | 2 | NP_989641 |
| 395869 | RHOC | ras homolog gene family, member C | s | c | 29 | 3 | NP_001025020 |
| 378791 | RTN1 | reticulon 1 | g | v | 13 | 3 | NP_001001466 |
| 378790 | RTN4 (NOGO) | reticulon 4 | g | v | 74 | 4 | XP_003640941 |
| 416778 | SEPT5 | septin 5 | g | c | 36 | 4 | NP_001025825 |
| 422954 | SFXN5 | sideroflexin 5 | t | m | 9 | 2 | XP_420891 |
| 422971 | SLC17A6 (VGLUT2) | solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6; vesicular glutamate transporter 2 | t | p | 8 | 1 | NP_001161855 |
| 423156 | SLC1A2 | solute carrier family 1 (glial high affinity glutamate transporter), member 2; excitatory amino acid transporter 2 | t | p | 80 | 4 | NP_001012917 |
| 422649 | SLC4A4 | solute carrier family 4, sodium bicarbonate cotransporter, member 4 | t | p | 27 | 4 | XP_420603 |
| 396444 | SNAP25 | synaptosomal-associated protein, 25kDa | v | v | 6 | 1 | NP_990789 |
| 428635 | SNAP91 | clathrin coat assembly protein AP180, synaptosomal-associated protein 91 | v | p | 13 | 2 | NP_001012969 |
| 374234 | SPTAN1 (SPECA) | spectrin alpha chain, non-erythrocytic 1 | c | c | 135 | 4 | NP_001036003 |
| 421216 | SPTBN1 | spectrin beta chain, non-erythrocytic 1 | c | c | 137 | 4 | NP_001186354 |
| 404293 | STXBP1 | syntaxin binding protein 1, Unc18-1 | v | v | 67 | 4 | NP_996859 |
| 418015 | SYNGR1 | synaptogyrin 1 | v | v | 11 | 3 | NP_001239207 |
| 418498 | SYNJ1 | synaptojanin 1 | v | v | 7 | 1 | XP_416706 |
| 420800 | TPPP | tubulin polymerization promoting protein | c | c | 50 | 4 | XP_418894 |
| 421169 | TUBA3E | tubulin, alpha 3e | c | c | 44 | 1 | XP_419249 |
| 396254 | TUBB | tubulin beta-7 chain | c | c | 393 | 4 | NP_990646 |
| 420883 | TUBB2B (TUBB2) | tubulin beta-2 chain | c | c | 331 | 4 | NP_001004400 |
| 417255 | TUBB2C (TUBB4) | tubulin, beta 2C; tubulin beta-3 chain | c | c | 354 | 4 | NP_001074329 |
| 431043 | TUBB3 | tubulin, beta 3 class III; tubulin beta-4 chain | c | c | 303 | 4 | NP_001026769 |
| 421037 | TUBB6 | tubulin, beta 6 class V; tubulin beta-5 chain | c | c | 118 | 2 | NP_001026183 |
| 416013 | UQCRC1 | ubiquinol-cytochrome c reductase core protein I | m | m | 120 | 4 | XP_414356 |
| 418290 | USP5 | ubiquitin specific peptidase 5 (isopeptidase T) | p | c | 4 | 2 | XP_003640490 |
| 418273 | VAMP1 | vesicle-associated membrane protein 1 (synaptobrevin 1) | v | v | 21 | 3 | NP_001034575 |
| 419368 | VAMP3 | vesicle-associated membrane protein 3 (cellubrevin) | v | v | 24 | 3 | NP_001034578 |
| 427820 | YWHAG | 14-3-3 protein gamma, tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | s | c | 164 | 4 | NP_001026648 |
F – Functional categorization: c (cytoskeleton), g (cell growth), m (metabolism), p (protein modification), r (RNA transport), s (signaling), tx (translation), v (vesicle transport), t (ion/amine transport)
S – Subcellular categorization: c (cytosol), e (extracellular), m (mitochondrion), p (plasma membrane), v (vesicle)
We performed DAVID analysis on the identified 94 proteins as putative FMRP targets (Table 4). In contrast to the previous analysis on all identified NL proteins, which revealed enrichment of pathways involved in metabolism, FMRP putative targets were enriched in pathways involved in cellular growth and transport (ribonucleotide binding, GTPases, ATPases, microtubules and neuronal projection) and transmembrane transport.
Table 4.
Enriched pathways of the 94 putative FMRP targets revealed by DAVID analysis.
| FMRP substrate from NL | ||
|---|---|---|
|
| ||
| Cluster | Enrichment Score | |
| 1 | Ribonucleotide Binding | 6.66 |
| 2 | GTPase/microtubule | 3.47 |
| 3 | ATPase/transmembrane transport | 3.46 |
| 4 | Neuronal projection | 2.89 |
| 5 | Methylation | 2.67 |
Gene ontology analysis of these 94 select proteins suggests a role in ribonucleotide binding, transmembrane transport and cellular growth, pathways needed for structural changes.
Further categorizing by subcellular localization (Fig. 4B), we found that the majority of these 94 proteins (72%) were localized to the cytosol (49%) and vesicles (19%), although a sizeable portion (21%) were localized to the plasma membrane. Categorization by function (Fig. 4A) reveals that the largest group (34% of proteins) was involved in cytoskeleton and cell growth, consistent with potential involvement of FMRP in regulating structural changes. The next most highly represented groups were those involved in intracellular trafficking (20%), useful in delivery of proteins needed to effect immediate changes, and those involved in transport across membranes (17%) including glutamate transporters.
Figure 4.
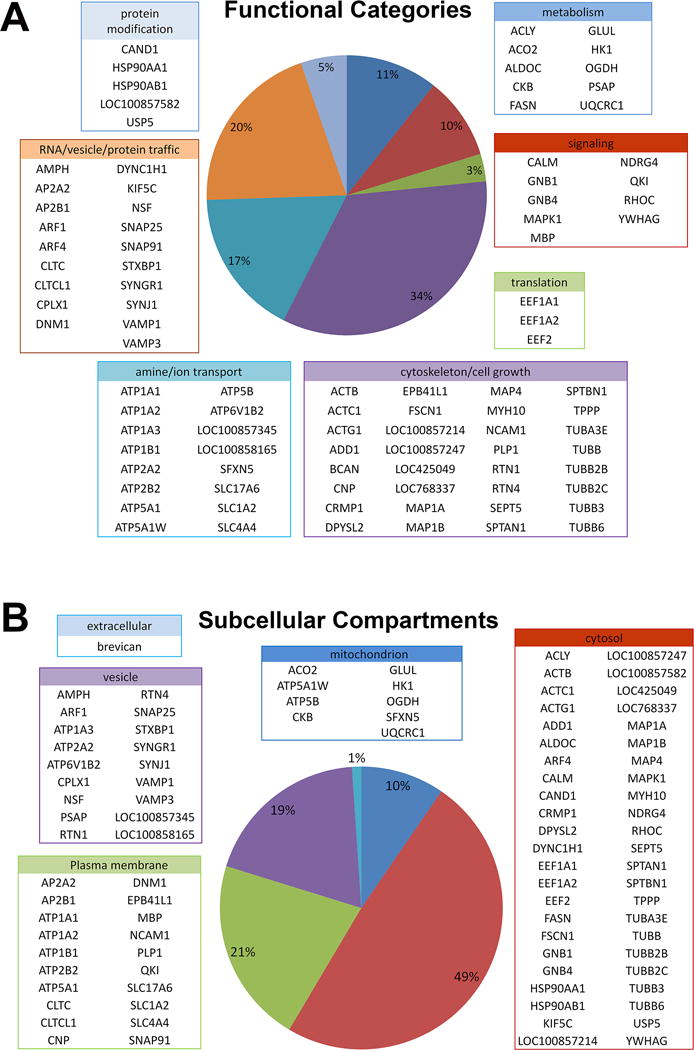
Characterization of 94 FMRP candidate targets. Using DAVID program, the proteins were subdivided by function (A) and subcellular compartments (B). The names of individual proteins are listed in specific category boxes with corresponding colors. A majority of the proteins are involved in cell growth 34%, intracellular trafficking 20% and transmembrane transport 17%. About half (49%) of the proteins are cytosolic proteins with the second half associated with plasma membrane, mitochondria, and vesicles. One protein was predicted to have an extracellular localization.
Comparative analyses identified 32 putative FMRP targets that maybe locally translated
Another group had previously published a list of transcripts found in hippocampal neuropil to determine locally translated proteins (Cajigas et al., 2012). We performed a comparison with this list and found that of the 657 NL proteins, 138 match this list (Fig. 3). Of the 94 FMRP target candidates, 32 also match this list, which is a unique and selective list of proteins with a high likelihood of local translation in NL dendrites (Table 5).
Table 5.
Thirty-two (32) of the 94 FMRP targets likely to be translated locally.
| Cell Growth/Cytoskeleton | |
|
| |
| CRMP1 | collapsin response mediator protein 1 |
| MAP1B | microtubule-associated protein 1B |
| ACTG1 | Actin, gamma 1 |
| SPTBN1 | spectrin, beta, non-erythrocytic 1 |
| TUBB | tubulin, beta class I |
| TUBB2C | tubulin, beta 2C |
| TUBB2B | tubulin, beta 2B class IIb |
| TUBB3 | tubulin, beta 3 class III |
|
| |
| Transport of Ions/Amines (Integral membrane proteins) | |
|
| |
| ATP6V1B2 (VATB) | ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 |
| ATP2A2 (SERCA2) | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| ATP1B1 | ATPase, Na+/K+ transporting, beta 1 polypeptide |
| ATP1A1 | ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| LOC100857345 | V-type proton ATPase subunit d 1-like |
| LOC100858165 | V-type proton ATPase subunit d 1-like |
|
| |
| Metabolism | |
|
| |
| HK1 | hexokinase 1 |
| ACO2 | aconitase 2, mitochondrial |
|
| |
| Protein Modification | |
|
| |
| USP5 | ubiquitin specific peptidase 5 (isopeptidase T) |
|
| |
| Trafficking of RNA, proteins or vesicles | |
|
| |
| KIF5C | kinesin family member 5C |
| DYNC1H1 | dynein, cytoplasmic 1, heavy chain 1 |
| DNM1 | dynamin 1 |
| ARF1 | ADP-ribosylation factor 1 |
| AMPH | amphiphysin |
| SNAP25 | synaptosomal-associated protein, 25kDa |
| SYNJ1 | synaptojanin 1 |
| NSF | N-ethylmaleimide-sensitive factor |
| SNAP91 | synaptosomal-associated protein, 91kDa homolog (mouse) |
|
| |
| Signaling | |
|
| |
| CALM | calmodulin 2 (phosphorylase kinase, delta) |
| MBP | myelin basic protein |
| NDRG4 | N-myc downstream regulated gene family member 4 |
|
| |
| Translation | |
|
| |
| EEF2 | eukaryotic translation elongation factor 2 |
| EEF1A1 | eukaryotic translation elongation factor 1 alpha 1 |
| EEF1A2 | eukaryotic translation elongation factor 1 alpha 2 |
The NL samples contain not only the somata and dendrites of NL neurons but also incoming axons of NM neurons and glia. The tissue samples used to generate the list of locally translated proteins in hippocampus (Cajigas et al., 2012) also contain dendrites, axons and glial cells. As expected, a number of proteins in the 32 protein list are known to be specific to glial cells including the myelin basic protein (MBP) and N-myc downstream regulated gene (NDRG) family member 4 or proteins primarily localized in presynaptic compartments such as SNAP25. Other proteins such as MAP1B are expressed in both post and presynaptic ends. To further narrow down the list of promising FMRP targets in NL dendrites, we next examined the subcellular localization of a number of selected proteins included in the 94-protein list, in particular those included in the short 32-protein list.
Immunocytochemistry confirms the expression of selected protein candidates in NL and their dendritic localization
We chose to focus on cytosolic proteins whose translation does not require Golgi, as no Golgi apparatus has been detected in NL dendrites at the electron microscope level (Deitch and Rubel, 1984). Since the 94 putative FMRP targets are enriched in pathways that facilitate structural changes, we first examined the distribution of cytoskeletal elements and their associated proteins. The NL dendrites were visualized by a neuronal somatodendritic marker, the microtubule associated protein 2 (MAP2). We used two antibodies for MAP2, raised in different hosts (rabbit and mouse), which show complete overlap of strong dendritic staining as well as weaker somatic staining (Fig. 5A). The distribution of another microtubule associated protein, MAP1B, is illustrated in Fig. 5B. Interestingly, the intensity of somatic MAP1B immunoreactivity varies among NL neurons. In the neuropil regions, prominent MAP1B immunoreactivity was largely located outside of MAP2-stained NL dendrites. The distribution of the beta tubulin class III (TUBB3) was examined using TUJ1 immunoreactivity, a specific marker for this type of tubulin. TUJ1 staining was strong in both cell bodies and the primary dendrites. Diffuse staining also overlapped with MAP2-stained dendrites further from the cell bodies (Fig. 5C). Fluorescent conjugated phalloidin was used to visualize F-actin. As expected, F-actin stain displayed a peri-membrane pattern, surrounding MAP2-stained cell bodies and dendritic structure (Fig. 5D).
We next examined the three eukaryotic elongation factor proteins (eEF) that were included in the 32-protein list, eEF1a1, eEF1a2 and eEF2. Using an antibody that specifically recognizes the phosphorylated form of eEF2 (p-eEF2), we identified the distribution of p-eEF2 in NL cell bodies as well as MAP2-stained dendrites at varied levels (Fig. 6A). In contrast, we only observed a somatic distribution of eEF1a in NL using an antibody recognizing both eEF1a1 and eEF1a2 (Fig. 6B).
Figure 6.
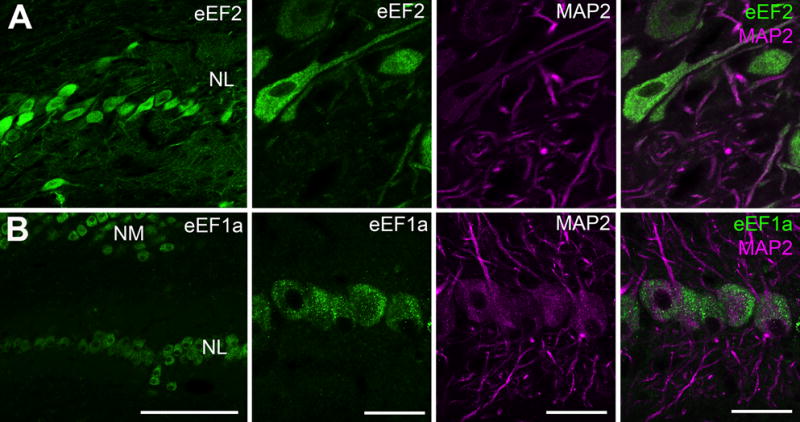
Subcellular distribution of elongation factors 1a and 2 in NL examined by immunocytochemistry. A: Double labeling of phosphorylated eEF2 (p-eEF2) and MAP2, showing the distribution of eEF2 in both cell bodies and dendrites of NL neurons. Dendritic level of p-eEF2 varies between branches. B: Double labeling of eEF1a and MAP2. Strong immunoreactivity for eEF1a was observed in NL cell bodies, while no detectable staining was found in NL neuropil regions containing dendrites. Abbreviations: NM, nucleus magnocellularis; NL, nucleus laminaris; MAP2, microtubule-associated protein 2; eEF1a, eukaryotic elongation factor 1a; eEF2, eukaryotic elongation factor 2. Scale bar = 100 μm (left column) and 20 μm (all other columns).
Finally, we examined three proteins involved in signaling regulation. As an important mechanism for calcium regulation, SERCA2 immunoreactivity was detected in some MAP2-labeled NL dendrites in addition to expected somatic staining (Fig. 7A). N-ethylmaleimide-sensitive factor (NSF) is a vesicle fusion protein. As expected (Serwin, 2012), NSF was strongly localized along perinuclear and cytoplasmic regions (Fig. 7B). In addition, significant NSF immunoreactivity was detected in the NL neuropil regions. Ras homolog gene family member C (RhoC) is a small G-protein that can promote remodeling of actin cytoskeleton. Immunocytochemistry revealed a particularly high level of RhoC protein in NL dendrites (Fig. 7C).
Figure 7.
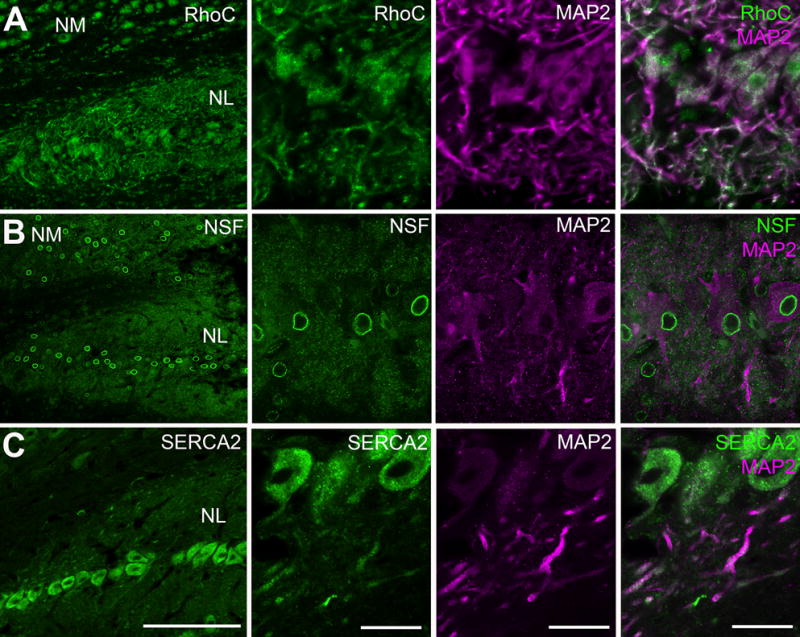
Subcellular distribution of NSF, SERCA2 and RhoC in NL examined by immunocytochemistry. A: Double labeling of NSF and MAP2, showing localization of NSF in NL dendrites in addition to intense perinuclear staining. B: Double labeling of SERCA2 and MAP2. In addition to the cell bodies, SERCA2 is also localized in NL dendrites. C: Double labeling of RhoC and MAP2, showing an intense localization of RhoC overlapping with MAP2-labeled NL dendrites. Abbreviations: NM, nucleus magnocellularis; NL, nucleus laminaris; MAP2, microtubule-associated protein 2; SERCA2, sarco/endoplasmic reticulum Ca2+ ATPase. Scale bar = 100 μm (left column) and 20 μm (all other columns).
RNA electrophoretic-mobility shift assay reveals FMRP interaction with RhoC RNA
RhoC is of particular interest as a potential FMRP target due to its function in signal transduction and cytoskeleton regulation, as well as its intense localization in NL dendrites. In order to validate the interaction of RhoC with FMRP, we performed RNA binding assays with recombinant chicken FMRP. Fig. 8 shows specific interaction between FMRP and RhoC RNA. When biotin-labeled RhoC RNA probe was incubated with purified FMRP, we detected a shift on the probe indicating their interaction in vitro (lanes 1 and 2). The specificity of this interaction was demonstrated when unlabeled RhoC probe competed for FRMP binding in a dose dependent manner (lanes 3–6), which was not observed if RhoC probe was replaced with a non-specific RNA probe (lanes 7–10).
Figure 8.
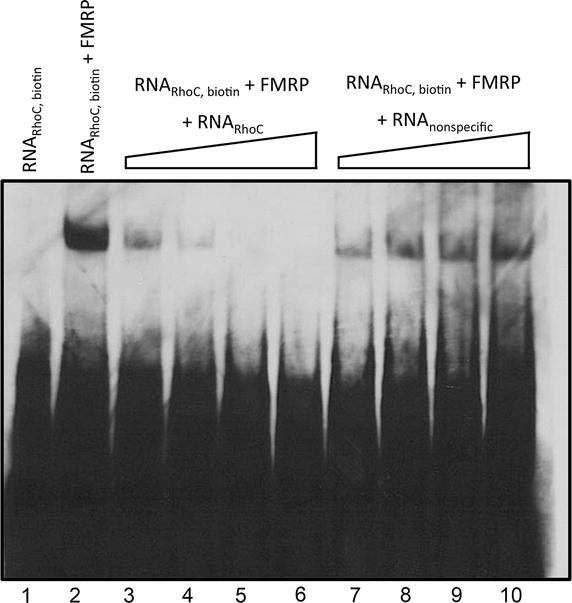
FMRP binds specifically to RhoC RNA. RNA electrophoretic mobility shift assay was performed to verify direct interaction between RhoC RNA and recombinant FMRP as described in the Material and Methods. Lane 1, biotin labeled RhoC RNA alone. Lane 2, biotin labeled RhoC RNA plus FMRP. Lanes 3–6, biotin labeled RhoC RNA plus FMRP with increasing concentrations of unlabeled RhoC RNA. Lanes 7–10 biotin labeled RhoC RNA plus FMRP with increasing concentrations of non-specific competitor RNA. Both unlabeled RNA competitors are at 10X, 20X, 40X, 100X fold excess of the biotin labeled RhoC RNA.
Discussion
NL is a critical structure for localizing the source of sound, necessary processing for acoustic scene analysis. Given its unique bipolar dendritic structure and highly dynamic structural properties of these dendrites (Sorensen and Rubel, 2006), NL is also a particularly useful model for studying the neuroplasticity of dendrites in response to afferent stimulation. In view of the growing acceptance for a critical role of local protein translation in dendritic neuroplasticity (Steward et al., 2014), the proteome of NL dendrites would be useful for determining local proteins that are being differentially translated during periods of dendritic remodeling. In this study, we present an initial proteomic analysis of NL with identification of 657 proteins, of which 94 are putative targets of FMRP, an mRNA-binding protein that regulates local protein translation in neural dendrites.
Methodological consideration
Two limitations need to be taken into consideration before applying the data generated in this study into future studies. First, the 657 proteins identified for NL is a conservative list of all proteins expressed by NL neurons and these proteins may be biased towards soluble proteins. This bias may be attributed to several factors. One is the lack of sufficient tissue, given the small size of the nucleus, to perform subcellular fractionation to enrich for membrane proteins, which are usually low abundance proteins. It is also possible that laser capture of dehydrated tissues may hinder the extraction of lipophilic molecules. Lastly, the hydrophobicity of membrane proteins renders them less accessible for trypsin digestion given that their hydrophobic regions are not accessible. On the other hand, the list of NL proteins we provide is a consistent list of proteins sampled reproducibly in multiple replicates. Due to the stochastic sampling of tandem mass spectrometry data using data dependent acquisition, the repeatability of peptides identified between technical replicates has been reported to range between 35 and 60% (Tabb et al., 2010). This variability is increased with greater complexity of the protein sample. We collected proteins from a single brainstem nucleus and we required each peptide to present in every technical replicate (n=3) and at least 2 peptides per protein for identification. With these strict requirements, we have 96% overlap of NL samples with BS list. This suggests a conservative list of proteins but likely biased towards more abundant proteins.
Second, the comparative analyses were performed to identify the most promising protein candidates that may be involved in potential FMRP regulation of NL dendrites. These comparisons did not take into consideration of potential interspecies variation of FMRP signals and function (Kwan et al., 2012). As a result, our 94 and 32 protein lists likely contain both false positive and false negative candidates. In addition, we did not attempt to unambiguously validate any FMRP target in this study. Immunocytochemical and in vitro RNA binding experiments aimed to further narrow down the list of protein candidates. Dendritic localization and the ability of binding FMRP in vitro are necessary, although not sufficient, as a FMRP target in neuronal dendrites. Functional manipulations combined with co-immunoprecipitation analyses are required for validating each FMRP target in future studies.
Proteins of interest
Many identified proteins in the 94-list are cytoskeletal elements (Actin and Tubulin) and their associated proteins (MAPs). Immunocytochemistry confirmed the dendritic localization of F-actin, beta 3 class III tubulins, and MAP2, suggesting that FMRP may regulate both actin and microtubule function in NL dendrites. Interestingly, MAP1B does not display significant dendritic localization, although it is detected in a subpopulation of NL cell bodies. Genes encoding MAP1B and MAP2 are both predicted to have strong associations with FMRP (ranking 5 and 62 among 842 FMRP-associated genes, Darnell et al., 2011). Extensive evidence has confirmed the colocalization of FMRP with MAP1B mRNA in both dendrites and axons (Antar et al., 2005; Antar et al., 2006). MAP1B protein level is elevated in hippocampus of Fmr1 knockout mice although the subcellular localization of the elevated MAP1B is unknown (Lu et al., 2004a). In particular, it has been shown that FMRP regulates MAP1B translation and controls microtubule stability in vertebrate neurons in vivo (Lu et al., 2004a; Zalfa et al., 2003). Lack of significant amount of MAP1B protein in NL dendrites may indicate the FMRP-MAP1B mRNA is a less prominent pathway in regulating microtubule in NL dendrites, as compared to potential FMRP regulation of MAP2 signaling. We did observe MAP1B positive fibers in the NL neuropil layer, presumably on incoming NM axons. Direct targeting of FMRP to MAP1B has been observed in axons and to regulate axon elongation (Wang et al., 2015). These observations further support the possibility that FMRP signaling is cell type specific.
In our analysis, we identified three elongation factors as FMRP targets, two isoforms of eEF1a and eEF2. Furthermore, we visualized dendritic localization of eEF2 but not of eEF1a in NL neurons, suggesting that FMRP may affect dendritic protein synthesis by modulating a subset of translational regulatory machinery. Although a direct link between FMRP and eEF1a and eEF2 mRNAs has not been demonstrated, both of these two elongation factors have been associated with potential roles in cellular growth/proliferation and signal transduction (Lin et al., 2010; Morrissey et al., 2015) as well as synaptic regulation (Becker et al., 2013; Mateyak and Kinzy, 2010; Rosenblum et al., 1993). In particular, eEF2 has recently been considered as a biochemical sensor that couples neuronal transmission to spine plasticity, a major proposed function of FMRP signaling (Verpelli et al., 2010).
This study also identified a number of dendritic localizing proteins that are predicted to be FMRP targets (Brown et al., 2001; Darnell et al., 2011), but their interaction with FMRP has yet to be validated. SERCA2 is an intracellular calcium ATPase pump located in the sarcoplasmic reticulum. Although a possible link between SERCA2 and FMRP has not been reported, recent studies demonstrated that FMRP regulates depolarization-induced calcium signal in critical periods of drosophila brain development (Doll and Broadie, 2016). Interestingly, another calcium regulator, the plasmid membrane calcium ATPases type 2 (PMCA2) is also a predicted FMRP target with highest binding affinity with FMRP (ranking 10 among 842; (Darnell et al., 2011)). Importantly, PMCA2 is intensely localized in NL dendrites and its protein levels rapidly change in response to afferent deprivation in shortening NL dendrites (Wang et al., 2009).
Another newly identified protein, as a potential FMRP target in the dendritic region of NL is NSF. NSF is a SNARE protein, reported to be in postsynaptic SNARES involved in synapses and synaptic transmission. NSF binding to AMPA receptor GluA2 intracellular domain has been shown to regulate the plasma membrane insertion of GluA2 (Araki et al., 2010). Recent reports suggest an indirect role of NSF in synaptic plasticity by way of regulating glutamate receptor plasma membrane expression (Huganir and Nicoll, 2013).
RhoC is a GTPase localized near cell membranes and is thought to share overlapping roles with RhoA, which is better characterized and shown to be important in neuronal migration through effects on actin and microtubule cytoskeleton (Stankiewicz and Linseman, 2014). There is also evidence that inactivation of RhoA results in increased dendrite arbor growth rate (Govek et al., 2005). In addition, in vitro studies of neonatal neurons indicate that reduction of RhoA and RhoC leads to the impaired dendritic growth (Calvet et al., 1998), which suggests that RhoC may be required for normal dendritic development. The intense localization of RhoC in NL dendrites may imply an important role of RhoC in NL dendritic dynamics, likely through effects on actin polymerization. It is possible that FMRP locally regulates the translation of targets such as RhoC, within NL dendrites, to control dendritic structural changes in response to neuronal activity. In support of this notion, we verified the FMRP interaction in vitro with a partial RhoC RNA containing several predicted FMRP binding sites (Anderson et al., 2016). This result indicates that the comparative analysis performed in this study indeed generates promising FMRP target candidates and has great potential for identifying novel FMRP signaling pathways. It would be interesting, as future studies, to explore the interaction of the identified putative FMRP targets with FMRP in NL dendrites in vivo in response to auditory stimulation or unilateral auditory deafferentation.
Conclusion
In summary, the data generated in this study provides a first proteomic analysis of NL. Comparison analyses generated a list of 94 proteins as potential FMRP targets. As initiated in the current study, subcellular localization of individual protein and its mRNA as well as their interaction with FMRP need to be confirmed and characterized using multiple approaches, including immunocytochemistry, in situ hybridization, immunoprecipitation, as well as loss-of-function studies.
Data Accessibility
The Full lists of MS-identified proteins will be deposited onto the Biolucida Cloud server maintained by MBF Biosciences upon the acceptance of the manuscript.
Supplementary Material
Acknowledgments
This study was supported by NIDCD Grants DC000018, DC013074, DC03829; NIGMS Grants P41 GM103533; NIGMS R01 GM121818, and the Genentech Advanced Neurodegenerative Disease Research grant.
Table of Abbreviations used in the figures
- eEF1a
eukaryotic elongation factor 1a
- eEF2
eukaryotic elongation factor 2
- MAP1B
microtubule-associated protein 1B
- MAP2
microtubule-associated protein 2
- MVe
medial vestibular nucleus
- NM
nucleus magnocellularis
- NL
nucleus laminaris
- NA
nucleus angularis
- SpVe
spinal vestibular nucleus
- SERCA
sarco/endoplasmic reticulum Ca2+-ATPase
- TuJ-1
neuron-specific class III beta-tubulin
- XDCT
dorsal crossed cochlear track
Footnotes
Associate Editor: Deanna Benson
References
- Anderson BR, Chopra P, Suhl JA, Warren ST, Bassel GJ. Identification of consensus binding sites clarifies FMRP binding determinants. Nucleic Acids Res. 2016;44:6649–6659. doi: 10.1093/nar/gkw593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antar LN, Dictenberg JB, Plociniak M, Afroz R, Bassel GJ. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 2005;4:350–359. doi: 10.1111/j.1601-183X.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- Antar LN, Li C, Zhang H, Carroll RC, Bassel GJ. Local functions for FMRP in axon growth cone motility and activity-dependent regulation of filopodia and spine synapses. Mol Cell Neurosci. 2006;32:37–48. doi: 10.1016/j.mcn.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Araki Y, Lin DT, Hunganir RL. Plasma membrane insertion of the AMPA receptor GluA2 subunit is regulated by NSF binding and Q/R editing of the ion pore. Proc Natl Acad Sci U S A. 2010;107:11080–11085. doi: 10.1073/pnas.1006584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida G, Carr CE. Sound localization: Jeffress and beyond. Curr Opin Neurobiol. 2011;21:745–751. doi: 10.1016/j.conb.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascano MJ, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Tuschl T. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M, Kuhse J, Kirsch J. Effects of two elongation factor 1A isoforms on the formation of gephyrin clusters at inhibitory synapses in hippocampal neurons. Histochem Cell Biol. 2013;140:603–609. doi: 10.1007/s00418-013-1122-9. [DOI] [PubMed] [Google Scholar]
- Benes FM, Parks TN, Rubel EW. Rapid dendritic atrophy following deafferentation: an EM morphometric analysis. Brain Res. 1977;122:1–13. doi: 10.1016/0006-8993(77)90658-8. [DOI] [PubMed] [Google Scholar]
- Born DE, Durham D, Rubel EW. Afferent influences on brainstem auditory nuclei of the chick: nucleus magnocellularis neuronal activity following cochlea removal. Brain Res. 1991;557:37–47. doi: 10.1016/0006-8993(91)90113-a. [DOI] [PubMed] [Google Scholar]
- Boström P, Anderson L, Rutberg M, Perman J, Lidberg U, Johansson BR, Olofsson SO. SNARE proteins mediate fusion between cytosolic lipid droplets and are implicated in insulin sensitivity. Nat Cell Biol. 2007;9:1286–1293. doi: 10.1038/ncb1648. [DOI] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Warren ST. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Cajigas IJ, Tushev T, Will TJ, tom Dieck S, Fuerst N, Schuman EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;453:453–466. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvet S, Doherty P, Prochiantz A. Identification of a signaling pathway activated specifically in the somatodendritic compartment by a heparan sulfate that regulates dendrite growth. J Neurosci. 1998;18:9751–9765. doi: 10.1523/JNEUROSCI.18-23-09751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chami M, Gozuacik D, Lagorce D, Brini M, Falson P, Peaucellier G, Paterlini-Bréchot P. SERCA1 truncated proteins unable to pump calcium reduce the endoplasmic reticulum calcium concentration and induce apoptosis. J Cell Biol. 2001;153:1301–1314. doi: 10.1083/jcb.153.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Toth M. Fragile X mice develop sensory hyperreactivity to auditory stimuli. Neuroscience. 2001;103:1043–1050. doi: 10.1016/s0306-4522(01)00036-7. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Darnell RB. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitch JS, Rubel EW. Afferent influences on brain stem auditory nuclei of the chicken: time course and specificity of dendritic atrophy following deafferentation. J Comp Neurol. 1984;229:66–79. doi: 10.1002/cne.902290106. [DOI] [PubMed] [Google Scholar]
- Deitch JS, Rubel EW. Changes in neuronal cell bodies in N. laminaris during deafferentation-induced dendritic atrophy. J Comp Neurol. 1989;281:259–268. doi: 10.1002/cne.902810208. [DOI] [PubMed] [Google Scholar]
- Dezso A, Schwarz DW, Schwarz IE. A survey of auditory brainstem nuclei in the chicken (Gallus domesticus) with cytochrome oxidase histochemistry. J Otoralyngol. 1993;22:385–390. [PubMed] [Google Scholar]
- DiTella MC, Feiguin F, Carri N, Kosik KS, Cáceres A. MAP-1B/TAU functional redundancy during laminin-enhanced axonal growth. J Cell Sci. 1996;109:467–477. doi: 10.1242/jcs.109.2.467. [DOI] [PubMed] [Google Scholar]
- Doll CA, Broadie K. Neuron class-specific requirements for Fragile X Mental Retardation Protein in critical period development of calcium signaling in learning and memory circuitry. Neurobiol Dis. 2016;89:76–87. doi: 10.1016/j.nbd.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Eriksson M, Samuelsson H, Björklund S, Tortosa E, Avila J, Samuelsson EB, Sundström E. MAP1B binds to the NMDA receptor subunit NR3A and affects NR3A protein concentrations. Neurosci Lett. 2010;471(1):33–37. doi: 10.1016/j.neulet.2010.03.039. [DOI] [PubMed] [Google Scholar]
- Franzen R, Tanner SL, Dashiell SM, Rottkamp CA, Hammer JA, Quarles RH. Microtubule-associated protein 1B: a neuronal binding partner for myelin-associated glycoprotein. J Cell Biol. 2001;155:893–898. doi: 10.1083/jcb.200108137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez R, Gopal AR, Greenough WT. Somatosensory cortical barrel dendritic abnormalities in a mouse model of the fragile X mental retardation syndrome. Brain Res. 2003;971:83–89. doi: 10.1016/s0006-8993(03)02363-1. [DOI] [PubMed] [Google Scholar]
- Galvez R, Greenough WT. Sequence of abnormal dendritic spine development in primary somatosensory cortex of a mouse model of the fragile X mental retardation syndrome. Am J Med Genet A. 2005;135:155–160. doi: 10.1002/ajmg.a.30709. [DOI] [PubMed] [Google Scholar]
- Govek EE, Newey SE, Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- Grothe B. The evolution of temporal processing in the medial superior olive, an auditory brainstem structure. Prog Neurobiol. 2000;61:581–610. doi: 10.1016/s0301-0082(99)00068-4. [DOI] [PubMed] [Google Scholar]
- Heil P, Scheich H. Effects of unilateral and bilateral cochlea removal on 2-deoxyglucose patterns in the chick auditory system. J Comp Neurol. 1986;252:279–301. doi: 10.1002/cne.902520302. [DOI] [PubMed] [Google Scholar]
- Hinton VJ, Brown WT, Wisniewski K, Rudelli RD. Analysis of neocortex in three males with the fragile X syndrome. Am J Med Genet. 1991;41:289–294. doi: 10.1002/ajmg.1320410306. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID gene ID conversion tool. Bioinformation. 2008;2:428–430. doi: 10.6026/97320630002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunganir RL, Nicoll RA. AMPARs and synaptic plasticity: the last 25 years. Neuron. 2013;80:704–717. doi: 10.1016/j.neuron.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impens F, Van Damme P, Demol H, Van Damme J, Vandekerckhove J, Gevaert K. Mechanistic insight into taxol-induced cell death. Oncogene. 2008;27:4580–4591. doi: 10.1038/onc.2008.96. [DOI] [PubMed] [Google Scholar]
- Irwin SA, Galvez R, Greenough WT. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb Cortex. 2000;10:1038–1044. doi: 10.1093/cercor/10.10.1038. [DOI] [PubMed] [Google Scholar]
- Joris P, Yin TC. A matter of time: internal delays in binaural processing. Trends Neurosci. 2007;30:70–78. doi: 10.1016/j.tins.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- Kaur KJ, Ruben L. Protein translation elongation factor-1 alpha from Trypanosoma brucei binds calmodulin. J Biol Chem. 1994;269:23045–23050. [PubMed] [Google Scholar]
- Kuba H, Yamada R, Fukui I, Ohmori H. Tonotopic specialization of auditory coincidence detection in nucleus laminaris of the chick. J Neurosci. 2005;25(8):1924–1934. doi: 10.1523/JNEUROSCI.4428-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba H. Cellular and molecular mechanisms of avian auditory coincidence detection. Neursci Res. 2007;59:370–376. doi: 10.1016/j.neures.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Kuba H, Adachi R, Ohmori H. Activity-dependent and activity-independent development of the axon initial segment. J Neurosci. 2014;34(9):3443–4353. doi: 10.1523/JNEUROSCI.4357-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Lam MM, Johnson MB, Dube U, Shim S, Rašin MR, Sestan N. Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell. 2012;149(4):899–911. doi: 10.1016/j.cell.2012.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KW, Yakymovych I, Jia M, Yakymovych M, Souchelnytskyi S. Phosphorylation of eEF1A1 at Ser300 by TbetaR-I results in inhibition of mRNA translation. Curr Biol. 2010;20:1615–1625. doi: 10.1016/j.cub.2010.08.017. [DOI] [PubMed] [Google Scholar]
- Lippe WR, Steward O, Rubel EW. The effect of unilateral basilar papilla removal upon nuclei laminaris and magnocellularis of the chick examined with [3H]2-deoxy-D-glucose autoradiography. Brain Res. 1980;196:43–58. doi: 10.1016/0006-8993(80)90715-5. [DOI] [PubMed] [Google Scholar]
- Lu R, Wang H, Liang Z, Ku L, O’Donnell WT, Li W, Feng Y. The fragile X protein controls microtubule-associated protein 1B translation and microtubule stability in brain neuron development. Proc Natl Acad Sci U S A. 2004a;101:15201–15206. doi: 10.1073/pnas.0404995101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Monsiavis P, Tempel BL, Rubel EW. Activity-dependent regulation of the potassium channel subunits Kv1.1 and Kv3.1. J Comp Neurol. 2004b;470:93–106. doi: 10.1002/cne.11037. [DOI] [PubMed] [Google Scholar]
- Martin KC, Zukin RS. RNA trafficking and local protein synthesis in dendrites: an overview. J Neurosci. 2006;26:7131–7134. doi: 10.1523/JNEUROSCI.1801-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateyak MK, Kinzy TG. eEF1A: thinking outside the ribosome. J Biol Chem. 2010;285:21209–21213. doi: 10.1074/jbc.R110.113795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride EG, Rubel EW, Wang Y. Afferent regulation of chicken auditory brainstem neurons: rapid changes in phosphorylation of elongation factor 2. J Comp Neurol. 2013;521:1165–1183. doi: 10.1002/cne.23227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey C, Schwefel D, Ennis-Adeniran V, Taylor IA, Crow YJ, Webb M. The eukaryotic elongation factor eEF1A1 interacts with SAMHD1. Biochem J. 2015;466:69–76. doi: 10.1042/BJ20140203. [DOI] [PubMed] [Google Scholar]
- Neil MA, Juskaitis R, Wilson T. Method of obtaining optical sectioning by using structured light in a conventional microscope. Opt Letter. 1997;22:1905–1907. doi: 10.1364/ol.22.001905. [DOI] [PubMed] [Google Scholar]
- Neil MA, Squire A, Juskaitis R, Bastiaens PI, Wilson T. Wide-field optically sectioning fluorescence microscopy with laser illumination. J Microsc. 2000;197:1–4. doi: 10.1046/j.1365-2818.2000.00656.x. [DOI] [PubMed] [Google Scholar]
- Paglini G, Pigino G, Kunda P, Morfini G, Maccioni R, Quiroga S, Cáceres A. Evidence for the participation of the neuron-specific CDK5 activator P35 during laminin-enhanced axonal growth. J Neurosci. 1998;18(23):9858–9869. doi: 10.1523/JNEUROSCI.18-23-09858.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penagarikano O, Mulle JG, Warren ST. The pathophysiology of fragile × syndrome. Annu Rev Genomics Hum Genet. 2007;8:109–129. doi: 10.1146/annurev.genom.8.080706.092249. [DOI] [PubMed] [Google Scholar]
- Rosenblum K, Meiri N, Dudai Y. Taste memory: the role of protein synthesis in gustatory cortex. Behav Neural Biol. 1993;59:49–56. doi: 10.1016/0163-1047(93)91145-d. [DOI] [PubMed] [Google Scholar]
- Rotschafer SE, Razak KA. Auditory processing in fragile × syndrome. Front Cell Neurosci. 2014;8:19. doi: 10.3389/fncel.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro MR, Bray SM, Warren ST. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu Rev Pathol. 2012;7:219–245. doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- Serwin M. Cell and System Biology. University of Toronto; 2012. Nuclear Localization of N-Ethylamaleimide Sensitive Factor (Master’s thesis) [Google Scholar]
- Sharma V, Eng JK, MacCoss MJ, Riffle M. A mass spectrometry proteomics data management platform. Mol Cell Proteomics. 2012;11:824–831. doi: 10.1074/mcp.O111.015149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen SA, Rubel EW. The level and integrity of synaptic input regulates dendrite structure. J Neurosci. 2006;26:1539–1550. doi: 10.1523/JNEUROSCI.3807-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen SA, Rubel EW. Relative input strength rapidly regulates dendritic structure of chick auditory brainstem neurons. J Comp Neurol. 2011;519:2838–2851. doi: 10.1002/cne.22656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz TR, Linseman DA. Rho family GTPases: key players in neuronal development, neuronal survival, and neurodegeneration. Front Cell Neurosci. 2014;8:314. doi: 10.3389/fncel.2014.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Falk PM. Polyribosomes under developing spine synapses: growth specializations of dendrites at sites of synaptogenesis. J Neurosci Res. 1985;13:75–88. doi: 10.1002/jnr.490130106. [DOI] [PubMed] [Google Scholar]
- Steward O, Farris S, Pirbhoy PS, Darnell J, Driesche SJ. Localization and local translation of Arc/Arg3.1 mRNA at synapses: some observations and paradoxes. Front Mol Neurosci. 2014;7:101. doi: 10.3389/fnmol.2014.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, Vega-Montoto L, Rudnick PA, Variyath AM, Ham AJ, Bunk DM, Spiegelman C. Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J Proteome Res. 2010;9(2):761–776. doi: 10.1021/pr9006365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verpelli C, Piccoli G, Zibetti C, Zanchi A, Gardoni F, Huang K, Sala C. Synaptic activity controls dendritic spine morphology by modulating eEF2-dependent BDNF synthesis. J Neurosci. 2010;30:5830–5842. doi: 10.1523/JNEUROSCI.0119-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Rubel EW. Rapid regulation of microtubule-associated protein 2 in dendrites of nucleus laminaris of the chick following deprivation of afferent activity. Neuroscience. 2008;154:381–389. doi: 10.1016/j.neuroscience.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cunningham DE, Tempel BL, Rubel EW. Compartment-specific regulation of plasma membrane calcium ATPase type 2 in the chick auditory brainstem. J Comp Neurol. 2009;514:624–640. doi: 10.1002/cne.22045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Rubel EW. In vivo reversible regulation of dendritic patterning by afferent input in bipolar auditory neurons. J Neurosci. 2012;32:11495–11504. doi: 10.1523/JNEUROSCI.1737-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sakano H, Beebe K, Brown MR, de Laat R, Bothwell M, Rubel EW. Intense and specialized dendritic localization of the fragile X mental retardation protein in binaural brainstem neurons: a comparative study in the alligator, chicken, gerbil, and human. J Comp Neurol. 2014;522:2107–2128. doi: 10.1002/cne.23520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Pan L, Wei M, Wang Q, Liu WW, Wang N, Bao L. FMRP-Mediated Axonal Delivery of miR-181d Regulates Axon Elongation by Locally Targeting Map1b and Calm1. Cell Rep. 2015;13:2794–2807. doi: 10.1016/j.celrep.2015.11.057. [DOI] [PubMed] [Google Scholar]
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- Zarnescu DC, Shan G, Warren ST, Jin P. Come FLY with us: toward understanding fragile X syndrome. Genes Brain Behav. 2005;4:385–392. doi: 10.1111/j.1601-183X.2005.00136.x. [DOI] [PubMed] [Google Scholar]
- Zhang B, Chambers MC, Tabb DL. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J Proteome Res. 2007;6:3549–3557. doi: 10.1021/pr070230d. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Full lists of MS-identified proteins will be deposited onto the Biolucida Cloud server maintained by MBF Biosciences upon the acceptance of the manuscript.


