Abstract
Eukaryotic proteins that terminate in a CaaX motif undergo three processing events: isoprenylation, C-terminal proteolytic cleavage, and carboxyl methylation. In Saccharomyces cerevisiae, the latter step is mediated by Ste14p, an integral endoplasmic reticulum membrane protein. Ste14p is the founding member of the isoprenylcysteine carboxyl methyltransferase (ICMT) family, whose members share significant sequence homology. Because the physiological substrates of Ste14p, such as Ras and the yeast a-factor precursor, are isoprenylated and reside on the cytosolic side of membranes, the Ste14p residues involved in enzymatic activity are predicted to be cytosolically disposed. In this study, we have investigated the topology of Ste14p by analyzing the protease protection of epitope-tagged versions of Ste14p and the glycosylation status of Ste14p-Suc2p fusions. Our data lead to a topology model in which Ste14p contains six membrane spans, two of which form a helical hairpin. According to this model most of the Ste14p hydrophilic regions are located in the cytosol. We have also generated ste14 mutants by random and site-directed mutagenesis to identify residues of Ste14p that are important for activity. Notably, four of the five loss-of-function mutations arising from random mutagenesis alter residues that are highly conserved among the ICMT family. Finally, we have identified a novel tripartite consensus motif in the C-terminal region of Ste14p. This region is similar among all ICMT family members, two phospholipid methyltransferases, several ergosterol biosynthetic enzymes, and a group of bacterial open reading frames of unknown function. Site-directed and random mutations demonstrate that residues in this region play a critical role in the function of Ste14p.
INTRODUCTION
Proteins that terminate in a CaaX motif (C is a cysteine, A is an aliphatic amino acid, and X is one of several amino acids) undergo a series of posttranslational modifications that include isoprenylation, C-terminal cleavage of the last three amino acids, and carboxyl methylation. In Saccharomyces cerevisiae, Ste14p mediates the last step of the CaaX modification pathway. Expression of an enzymatically active recombinant Ste14p fusion protein in Escherichia coli along with the demonstration that a Δste14 strain lacks carboxyl methyltransferase activity provided evidence that Ste14p is an isoprenylcysteine carboxyl methyltransferase (Hrycyna and Clarke, 1990; Hrycyna et al., 1991). The analysis of Ste14p activity in vitro with the use of N-acetyl-S-farnesyl-l-cysteine (AFC) as a substrate indicated that a prenylated cysteine with a free carboxyl group is the minimal substrate for Ste14p (Hrycyna et al., 1991). Known physiological substrates for Ste14p in vivo include the mating pheromone a-factor, Ras1p, and Ras2p (Hrycyna and Clarke, 1990; Hrycyna et al., 1991).
Ste14p was the first prenylcysteine carboxyl methyltransferase to be cloned and sequenced (Blair, 1979; Wilson, 1985; Ashby et al., 1993; Sapperstein et al., 1994). Recently, the CaaX methyltransferases from Schizosaccharomyces pombe, Xenopus laevis, and humans (mam4p, Xmam4p, and pcCMT, respectively) have been cloned. These proteins share significant amino acid similarity with Ste14p, are similar to Ste14p in their hydropathy profiles, and have been shown to function as CaaX methyltransferases in vitro (Imai et al., 1997; Dai et al., 1998). These methyltransferases have also been shown to transcomplement each other in vivo (Imai et al., 1997; Dai et al., 1998; Romano et al., 1998). A mouse cell line was recently generated that is homozygous for the Icmt knockout mutation and lacks the corresponding methyltransferase activity. Localization of K-Ras was shown to be abnormal in these cells, indicating the importance of the methyltransferase in Ras protein targeting in mammalian cells (Bergo et al., 2000). Finally, database searches have identified a rat open reading frame (accession number AF0755595.1), and two Caenorhabditis elegans open reading frames (accession numbers U88175 and U80450) that have significant amino acid homology to Ste14p and similar hydropathy profiles. These proteins form a novel family of protein methyltransferases, designated the isoprenylcysteine carboxyl methyltransferase (ICMT) family. Interestingly, Ste14p and its homologues, along with a short list of additional methyltransferases, do not contain any of the conserved S-adenosyl-l-[methyl-3H]methionine (AdoMet) binding motifs common to most other DNA and protein methyltransferases (Kagan and Clarke, 1994; Imai et al., 1997; Dai et al., 1998). The identification of functional domains for members of the ICMT family is important, particularly because the requirement of ICMT in Ras modification and localization suggests that this enzyme could represent an excellent target for chemotherapeutic agents. A drug capable of blocking ICMT activity would be expected to render hyperactive oncogenic Ras inactive. An understanding of which regions of ICMT are required for function would considerably facilitate development of such drugs.
In this study we have analyzed the topology of Ste14p. Our results lead to a model in which Ste14p contains six membrane spans and has a majority of its hydrophilic regions located in the cytosol. As a first step in identifying residues important for Ste14p activity, we have also mutagenized Ste14p. Five of six loss-of-function mutations obtained by random mutagenesis lie in residues that are conserved between Ste14p and its ICMT homologues. In addition, by database searches and sequence comparisons we have identified a C-terminal region of Ste14p that is conserved between the ICMT family of protein methyltransferases, two phospholipid methyltransferases, three sterol reductases, and a series of bacterial open reading frames of unknown function. This region defines a consensus sequence that we have designated the RHPxY-hyd-EE motif. Site-directed and random mutations indicate that residues in this region play a critical role in Ste14p function.
MATERIALS AND METHODS
Yeast Strains, Media, and Growth Conditions
The S. cerevisiae strains used in this study are listed in Table 1. Complete (YEPD), synthetic (SD), and synthetic dropout (SC-Ura, SC-Trp) media were prepared as described previously (Michaelis and Herskowitz, 1988), except that dropout media lacked cysteine. All experiments were performed at 30°C. Yeast transformations were performed by the lithium acetate method (Ito et al., 1983).
Table 1.
Strains used in this study
| Strains | Genotypea | Reference or source |
|---|---|---|
| DGY505 | MATα suc2-Δ9 trp1 his3 ade2 ura3 leu2 | Gift of Eitan Bibi (Weizmann Institute of Science, Israel) |
| SM1068 | MATα lys1 | Michaelis and Herskowitz (1988) |
| SM1086 | MATα sst2 | Michaelis and Herskowitz (1988) |
| SM1188 | MATa trp1 leu2 ura3 his4 can1 ste14-3∷TRP1 | Sapperstein et al. (1994) |
| SM2515 | [p2μ LEU2 STE14∷HA (Q3)] | Transformant of SM1188 with pSM757 |
| SM2705 | MATα suc2-Δ9 trp1 his3 ade2 ura3 leu2 [p2μ TRP1 PPGK] | Transformant of DGY505 with pRS424-PGK (Schonberger et al. (1996) |
| SM2894 | MATα suc2-Δ9 trp1 his3 ade2 ura3 leu2 [p2μ TRP1 PPGK SUC2] | Transformant of DGY505 with pRS/SUC2 Geller et al. (1996) |
| SM2926 | [pCEN URA3] | Romano et al. (1998) |
| SM3185 | [pCEN URA3 STE14] | Romano et al. (1998) |
| SM3191 | [pCEN URA3 STE14∷HA (V75)] | Transformant of SM1188 with pSM1252 |
| SM3194 | [pCEN URA3 STE14∷HA (K110)] | Transformant of SM1188 with pSM1254 |
| SM3410 | [pCEN URA3 ste14-E213Q] | Transformant of SM1188 with pSM1241 |
| SM3412 | [pCEN URA3 ste14-E214D] | Transformant of SM1188 with pSM1243 |
| SM3413 | [pCEN URA3 ste14-E213D] | Transformant of SM1188 with pSM1244 |
| SM3428 | [pCEN URA3 STE14∷HA (K86)] | Transformant of SM1188 with pSM1256 |
| SM3429 | [pCEN URA3 STE14∷HA (G144)] | Transformant of SM1188 with pSM1258 |
| SM3874 | [pCEN URA3 STE14∷myc (6x) (Q3)] | Transformant of SM1188 with pSM1504 |
| SM3876 | [pCEN URA3 STE14∷myc (6x) (I239)] | Transformant of SM1188 with pSM1506 |
| SM3798 | [p2μ LEU2 STE14∷myc (6x) (Q3)] | Transformant of SM1188 with pSM1472 |
| SM3800 | [p2μ LEU2 STE14∷myc (6x) (I239)] | Transformant of SM1188 with pSM1474 |
| SM4278 | [pCEN URA3 ste14-G31E] | Transformant of SM1188 with pSM1637 |
| SM4279 | [pCEN URA3 ste14-L217S] | Transformant of SM1188 with pSM1638 |
| SM4306 | MATα suc2-Δ9 trp1 his3 ade2 ura3 leu2 [p2μ TRP1 PPGK STE14 (1-25)∷SUC2] | Transformant of DGY505 with pSM1657 |
| SM4307 | MATα suc2-Δ9 trp1 his3 ade2 ura3 leu2 [p2μ TRP1 PPGK STE14 (1-57)∷SUC2] | Transformant of DGY505 with pSM1660 |
| SM4308 | MATα suc2-Δ9 trp1 his3 ade2 ura3 leu2 [p2μ TRP1 PPGK STE14 (1-87)∷SUC2] | Transformant of DGY505 with pSM1658 |
| SM4309 | MATα suc2-Δ9 trp1 his3 ade2 ura3 leu2 [p2μ TRP1 PPGK STE14 (1-239)∷SUC2] | Transformant of DGY505 with pSM1656 |
| SM4321 | [p2μ LEU2 ste14∷HA (Q3)-S148F] | Transformant of SM1188 with pSM1668 |
| SM4322 | [p2μ LEU2 ste14∷HA (Q3)-P173L] | Transformant of SM1188 with pSM1669 |
| SM4324 | [p2μ LEU2 ste14∷HA (Q3)-G132R] | Transformant of SM1188 with pSM1671 |
| SM4326 | [p2μ LEU2 ste14∷HA (Q3)-L81F] | Transformant of SM1188 with pSM1673 |
| SM4361 | MATα suc2-Δ9 trp1 his3 ade2 ura3 leu2 [p2μ TRP1 PPGK STE14 (1-113)∷SUC2] | Transformant of DGY505 with pSM1675 |
| SM4363 | MATα suc2-Δ9 trp1 his3 ade2 ura3 leu2 [p2μ TRP1 PPGK STE14 (1-156)∷SUC2] | Transformant of DGY505 with pSM1677 |
| SM4470 | MATα suc2-Δ9 trp1 his3 ade2 ura3 leu2 [p2μ TRP1 PPGK STE14 (1-239) N191L, P192L∷SUC2] | Transformant of DGY505 with pSM1726 |
All strains used in this study are isogenic to SM1188 unless otherwise indicated.
Plasmid Constructions
In general, plasmids in this study were generated by homologous recombination, as described previously (Ma et al., 1987; Oldenburg et al., 1997). Briefly, this method involves the following steps: A linearized or gapped target plasmid containing a selectable marker (i.e., URA3) is cotransformed into yeast, together with a polymerase chain reaction (PCR) product or a restriction fragment bounded by at least 40 base pairs with homology to sites on the target plasmid. Homologous recombination between these pieces of DNA results in the recircularization of the linearized target plasmid to yield Ura+ yeast transformants. A high percentage of these transformants contain the desired construct, as ascertained by colony PCR. DNA is isolated from yeast, transformed into E. coli, reisolated, and analyzed by restriction analysis and DNA sequencing. For further phenotypic analysis, a plasmid containing the verified construct is retransformed into fresh yeast cells. Passage of the DNA through E. coli, followed by retransformation is important in this procedure, because many of the initial yeast Ura+ transformants contain multiple and heterogeneous plasmids. In certain cases, as specified, plasmids used in this study were constructed with the use of conventional cloning procedures.
STE14 was epitope-tagged at either its N or C terminus after residues Q3 and I239, respectively, with six copies of the myc epitope. First, a BglII site was created in STE14 with the use of site-directed mutagenesis (Kunkel et al., 1987) to create pSM1343 (2μ LEU2 STE14 BglII Q3) and pSM1344 (2μ LEU2 STE14 BglII I239). Next, a BamHI fragment containing the triply iterated myc epitope from pSM937 (Berkower, 1995) was subcloned into the BglII sites of pSM1343 and pSM1344 to yield pSM1472 [2μ LEU2 STE14::myc (Q3)] and pSM1474 [2μ LEU2 STE14::myc (I239)], respectively. All epitope tag insertions were confirmed by DNA sequencing and each plasmid was found to contain six, rather than three, copies of the myc epitope. CEN versions of these plasmids [pSM1504 (CEN URA3 STE14::myc (Q3)] and pSM1506 [CEN URA3 STE14::myc (I239)] were constructed in vivo by homologous recombination with the use of a PvuI fragment from pSM1472 or pSM1474 that was cotransformed with PvuII-gapped pSM1237 (CEN URA3 STE14) (Romano et al., 1998) into SM1188. Plasmids from Ura+ transformants were isolated, transformed into E. coli, reisolated, and subjected to restriction enzyme digestion to verify the construction.
For protease protection experiments, Ste14p was tagged with three copies of the hemagglutinin (HA) epitope at one of four locations (after residue V75, K86, K110, and G144). For each construct, a BglII site was first inserted into STE14 with the use of site-directed mutagenesis (Kunkel et al., 1987). Next, the triply iterated HA tag from pSM492 (Berkower et al., 1994) was subcloned into the BglII sites with the use of standard subcloning procedures, to create pSM1252 (CEN URA3 STE14::HA V75), pSM1254 (CEN URA3 STE14::HA K110), pSM1256 (CEN URA3 STE14::HA K86), and pSM1258 (CEN URA3 STE14::HA G144), respectively.
The ste14 mutants E213Q, E214D, and E213D were generated by site-directed mutagenesis (Kunkel et al., 1987) of pSM187 (CEN URA3 STE14) (Sapperstein et al., 1994). The mutagenized region of ste14 was transferred to a fresh plasmid, pSM1237 (CEN URA3 STE14) (Romano et al., 1998), to generate pSM1241 (CEN URA3 ste14- E213Q), pSM1243 (CEN URA3 ste14-E214D), and pSM1244 (CEN URA3 ste14-E213D), with the use of standard subcloning procedures. DNA sequence analysis of the STE14 coding sequence was carried out to verify the presence of the mutations.
Construction of Ste14p-Suc2p Fusions
To generate in-frame STE14::SUC2 fusions, we replaced STE6 in pRS424-STE6-SUC2 (Geller et al., 1996) with regions of STE14, with the use of homologous recombination. We replaced STE6 with STE14 codons 1–25, 1–57, 1–87, 1–113, 1–156, and 1–1239, which were amplified by PCR and cotransformed with SalI, EcoRI linearized pRS424-STE6-SUC2, to generate pSM1657 [2μ TRP1 PPGK STE14(1-25)::SUC2], pSM1660 [2μ TRP1 PPGK STE14(1-57)::SUC2], pSM1658 [2μ TRP1 PPGK STE14(1-87)::SUC2], pSM1675 [2μ TRP1 PPGK STE14(1-113)::SUC2], pSM1677 [2μ TRP1 PPGK STE14(1-156)::SUC2], and pSM1656 [2μ TRP1 PPGK STE14(1-239)::SUC2], respectively.
Mutagenesis
To generate random ste14 mutants, we mutagenized a STE14 plasmid, pSM757 [2μ LEU2 STE14::HA (Q3)] with hydroxylamine (7%) according to published procedures (Rose et al., 1990). The mutagenized plasmid DNA was transformed into bacteria and ∼17,600 transformants were isolated. Plasmid DNA was prepared from the bacterial transformants and transformed into the yeast strain SM1188 (Δste14-3) (Sapperstein et al., 1994). Yeast transformants were screened for mating defects with the use of the plate-mating assay described below. We screened ∼2600 transformants and obtained eleven down-maters, of which seven represented novel ste14 alleles and four were duplicates. Four of the seven novel alleles were missense mutations in STE14 and are listed in Table 2. Three of the seven were nonsense mutations (Q43Z, W180Z, W181Z) and were not studied further.
Table 2.
ste14 mutants isolated in this study
| Strain name | Mutation | Method | Quantitative mating (% of WT) | Methyltransferase activity (pmol/mg/min) | Protein levela |
|---|---|---|---|---|---|
| SM3185 | Ste14p | NA | 100 | 45.8 (±8.3) | + |
| SM2515 | Ste14p-HA | NA | 100 | 37.3 (±3.3) | + |
| SM2926 | Δste14 | NA | <0.001 | 2.7 (±0.9) | − |
| SM4278 | G31E | EMS | <0.001b | 3.0 (±1.3) | + |
| SM4326 | L81F | Hydroxlamine | 7.4c (±1.7) | 1.6 (±0.05) | + |
| SM4324 | G132R | Hydroxlamine | <0.001c | ND | ± |
| SM4321 | S148F | Hydroxlamine | 18.8c (±1.7) | 2.3 (±0.3) | + |
| SM4322 | P173L | Hydroxlamine | <0.001c | ND | ± |
| SM3413 | E213D | Site-directed | 0.45b (±0.25) | 2.9 (±1.3) | + |
| SM3410 | E213Q | Site-directed | <0.001b | 2.9 (±1.2) | + |
| SM3412 | E214D | Site-directed | 88.4b (±28.5) | 5.2 (±1.3) | + |
| SM4279 | L217S | EMS | 51.7b (±9.4) | 3.8 (±0.6) | + |
Protein levels were compared to the WT parent strain using immunoblots probed with anti-Ste14p antisera.
Mating percentage was calibrated to the isogenic parent strain SM3185, which contains a plasmid expressing untagged Ste14p.
Mating percentage was calibrated to the isogenic parent strain SM2515, which contains a plasmid expressing Ste14p-HA.
ND, not determined; NA, not applicable; WT, wild-type.
Five chromosomal mutations presumed to reside in STE14 because they were complemented by a STE14 plasmid had been previously isolated in a screen for new sterile mutants generated by ethyl methanesulfonate mutagenesis (Fujimura-Kamada et al., 1997). To transfer the ste14 mutation from the chromosome onto a plasmid, we transformed the original mutants (ste14-11, ste14-12, ste14-13, ste14-14, and ste14-15) with either ClaI and AflII gapped pSM1237 (CEN URA3 STE14), PflmI and EcoRI gapped pSM1237, or Nsi1 and NdeI gapped pSM1316 (CEN LEU2 STE14) (Romano et al., 1998). Transformants were screened by mating and plasmids were prepared from the down-maters. One of the mutants did not yield down-mater plasmids, presumably because the mutation lay outside of the gapped region in a noncoding region of the gene. The remaining four plasmids were transformed into SM1188 (Δste14-3), a mating defect was confirmed, and the ste14 mutation in each plasmid was determined by DNA sequence analysis of the STE14 coding region. Three new ste14 missense mutations were identified, and one mutation was a duplicate. One of the three missense mutations, M1I, alters the ATG initiation codon of STE14, presumably interfering with Ste14p protein production, and was not studied further. The other two missense mutations G31E and L217S are listed in Table 2 and discussed in the text.
Plasmids pSM1637 (CEN URA3 ste14-G31E) and pSM1638 (CEN URA3 ste14-L217S) were constructed with the use of homologous recombination to transfer a ste14 mutation from the chromosome to the plasmid (Orr-Weaver et al., 1981). Strains containing the uncharacterized ste14 alleles described above were transformed with ClaI- and AflII gapped pSM1237 (CEN URA3 STE14) (Romano et al., 1998) and Ura+ transformants were selected. The resulting plasmids, pSM1637 and pSM1638, were shown by DNA sequence analysis to contain the ste14 mutant alleles G31E (ste14-11) and L217S (ste14-13), respectively.
Mating and a-Factor Halo Assays
Qualitative plate mating tests were performed as described previously (Michaelis and Herskowitz, 1988). Briefly, patches or colonies of MATa cells grown on selective media were replica-plated onto a SD plate spread with a lawn of the MATα tester strain SM1068 and 0.3 ml of YEPD. Growth of prototrophic diploids is indicative of mating.
Quantitative mating tests were performed as described previously (Rose et al., 1990). Briefly, 0.2 OD600 units of MATa cells were mixed with 0.25 OD600 units of MATα cells and concentrated together on a filter (Millipore, Bedford, MA), which was incubated at 30°C on a YEPD plate for 4 to 6 h. The cells were released from the filter by vortexing and plated onto either selective media, to determine the number of diploids formed, or on nonselective media, to determine the total number of cells subjected to mating. The percentage of mating of the mutant MATa strains was calculated as the ratio of diploids to the total number of cells, compared with the value for the isogenic wild-type MATa strain, which was set to 100%. The assay was performed in triplicate.
For a-factor halo assays, 1 ml of a saturated overnight culture of cells, grown in synthetic complete drop-out medium, was pelleted, washed twice with water, and the pellet was resuspended in 25 μl of water. Two microliters of each cell pellet was spotted onto a YPD plate that had been spread with a lawn of SM1086 cells, which are super-sensitive to a-factor. Plates were incubated at 30°C overnight. A clear zone, or halo, surrounding the spot of MATa cells reflects the presence of mature secreted a-factor, which inhibits the growth of the underlying lawn.
Yeast Cell Membrane Preparations
Logarithmically growing cells (120 OD600 units) were harvested by centrifugation at 1300 × g for 5 min at 4°C in a JA-14 rotor (Beckman, Fullerton, CA), washed with 10 mM NaN3, and resuspended in 10 mM NaN3, 250 mM β-mercaptoethanol to a final concentration of 1 OD600/10 μl. After a 10-min incubation on ice, an equal volume of oxalyticase buffer was added (2.8 M sorbitol, 100 mM potassium phosphate, pH 7.5, 10 mM NaN3) and Oxalyticase (Enzogenetics, Corvallis, OR) was added to a final concentration of 0.5 μg/OD600 units of cells. Samples were incubated at 30°C for 60 min. All subsequent steps occurred on ice. Spheroplasts were collected by centrifugation for 10 min at 500 × g at 4°C in a JA-20 rotor (Beckman) through a cushion of 2 M sorbitol. Spheroplasts were resuspended in lysis buffer (0.3 M mannitol, 0.1 M KCl, 50 mM Tris, pH 7.5, 1 mM EGTA) and homogenized with a Dounce homogenizer (20 strokes, tight). The homogenates were cleared of intact cells and debris twice by centrifugation for 5 min (500 × g). To collect a membrane fraction, homogenates were centrifuged at 200,000 × g for 30 min in a TLA100.2 rotor (Beckman) at 4°C. Membranes were resuspended in lysis buffer and protein concentrations were determined with the use of the Bio-Rad protein assay reagent (Bio-Rad, Richmond, CA).
In Vitro Methyltransferase Assay
Methyltransferase assays were carried out as described previously (Philips and Pillinger, 1995; Volker et al., 1995). Briefly, 0.5 μg of membrane proteins was incubated with 30 μM AdoMet and 100 μM AFC in 100 mM Tris, pH 7.4, 2 mM EDTA and for 20 min at 30°C. Each reaction was processed according to Philips and Pillinger (1995) to determine the amount of methylated AFC. S-Adenosyl-l-[methyl-3H]methionine was purchased from PerkinElmer Life Science Products (Boston, MA) and N-acetyl-S-farnesyl-l-cysteine was purchased from BIOMOL Research Laboratories (Plymouth Meeting, PA) and prepared as a 10–50 mM stock solution in dimethyl sulfoxide. The final concentration of dimethyl sulfoxide in the reactions was 1%.
Antibodies
The mouse anti-myc monoclonal antibody (mAb) (9E10) was obtained from the monoclonal antibody facility at the Johns Hopkins University School of Medicine, Baltimore, MD. The mouse anti-HA (12CA5) mAb was purchased from Babco (Richmond, CA). The rabbit polyclonal anti-Kar2p and anti-Suc2p antibodies were gifts from Dr. Jeffrey Brodsky (University of Pittsburgh, Pittsburgh, PA) and Dr. Susan Ferro-Novick (Yale University, New Haven, CT), respectively. The horseradish peroxidase-conjugated secondary antibody (sheep anti-mouse Ig and donkey anti-rabbit Ig) used for immunoblotting was purchased from Amersham Pharmacia Biotech (Arlington Heights, IL). The Cy3-conjugated goat anti-mouse and the fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit secondary antibodies used for immunofluorescence were purchased from Jackson ImmunoResearch (West Grove, PA).
Protease Protection Assay
Yeast membrane extracts were prepared as described above. Protease protection assays were performed as described previously (Abeliovich et al., 1998), with the use of proteinase K (Roche Molecular Biochemicals, Indianapolis, IN). Membrane proteins (80 μg) were incubated with combinations of 0.3 mg/ml proteinase K and/or 0.4% Triton X-100 for 5 min on ice. Reactions were terminated by the addition of phenylmethylsulfonyl fluoride (PMSF) to a final concentration of 1 mM and proteins were precipitated with 10% trichloroacetic acid. Samples were resolved by 12.5% SDS-PAGE and analyzed by immunoblotting for Ste14p-myc, Ste14p-HA, or Kar2p.
Preparation of Cell Extracts and Immunoblotting Analysis
Cell extracts were prepared for immunoblots as described previously except that 5 OD600 units of cells were grown logarithmically in synthetic drop-out media (Fujimura-Kamada et al., 1997). Immunoblots were prepared and probed as described previously (Romano et al., 1998) The primary antibodies used were anti-myc, anti-HA, or anti-Kar2p (all diluted 1:10,000) or anti-Suc2p (diluted 1:500).
Immunofluorescence
Cells were prepared for immunofluorescence as described previously (Berkower et al., 1994; Romano et al., 1998), except that the primary antibodies were anti-myc antibody (diluted 1:1000) or anti-Kar2p antibody (diluted 1:5000). The secondary antibodies were Cy3-conjugated anti-mouse (diluted 1:2000) or FITC-conjugated anti-rabbit (diluted 1:500). The slides were viewed at 100× magnification with the use of a Zeiss Axiovert microscope equipped with fluorescence optics. Images were captured with the use of a Photometrics PXL CCD camera (Photometrics, Tucson, AZ) and IP Lab Spectrum Software.
RESULTS
Three Models of Ste14p Topology
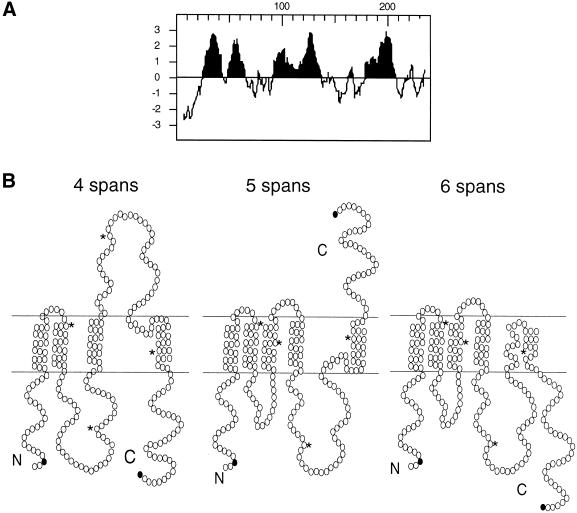
Based on the hydropathy plot shown in Figure 1A, three possible models for the topology of Ste14p can be predicted. Each model depicts a different number of transmembrane segments: four, five, or six membrane spans (Figure 1B). In the four-membrane-span model, only the four highest hydrophobic peaks (>2 U) were chosen as membrane spans (Figure 1B, left). In the five-membrane-span model, the third hydrophobic peak, which had a hydrophobicity value >1.5 U but <2 U, was included as a membrane span (Figure 1B, center). In the six-membrane-span model, the last hydrophobic segment was predicted to span the membrane twice (Figure 1B, right). In both the four- and six (i.e., even)-span models, the N and C termini of Ste14p are on the same side of the membrane. In the five-span model, however, they reside on opposite sides of the membrane. In the experiments described below, the four-, five-, and six-span models for Ste14p topology were evaluated, with the use of protease protection assays and gene fusion approaches.
Figure 1.
Predicted membrane spans of Ste14p. (A) Hydropathy plot of Ste14p was generated according to the algorithm of Kyte and Doolittle (1982) with a window of 11 amino acids. Hydrophobic regions are shown in black. (B) Three topology models for Ste14p are shown, with circles indicating individual amino acids. The N and C termini of Ste14p are marked. The black circles indicate the amino acid immediately preceding the insertion site of the myc epitope tags in Ste14p-myc Q3 and I239. The asterisks mark the locations of amino acids 50, 100, 150, and 200 for orientation.
N- and C-terminally Myc-tagged Ste14p Constructs Are Functional and Endoplasmic Reticulum (ER)-localized
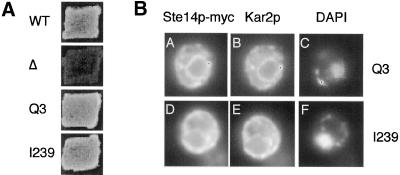
To facilitate the analysis of Ste14p topology, we tagged Ste14p at its N and C termini (at amino acids Q3 and I239, respectively; Figure 1B, black circles) with six copies of the myc epitope. A 37-kDa protein species was detected by anti-myc antibodies in immunoblots of extracts from strains expressing Ste14p-myc (Q3) and (I239) (our unpublished results). To determine whether the insertion of the myc tag affected function, we performed qualitative and quantitative mating assays. Ste14p methyltransferase activity is necessary for the production and secretion of the a-factor mating pheromone. Therefore, the mating assay provides an indirect but sensitive measure of Ste14p activity. As shown by qualitative patch mating in Figure 2A, a Δste14 strain is unable to mate. In contrast, both the Ste14p-myc (Q3) and Ste14p-myc (I239) constructs permit mating at a level indistinguishable from wild type. We also tested these strains by a quantitative filter mating assay and again found that mating was essentially wild-type [95.7 ± 5.4 and 105.3 ± 4.7% of wild-type mating for (Q3) and (I239), respectively]. These results indicate that the insertion of the myc epitope did not significantly impair Ste14p activity.
Figure 2.
Ste14p-myc Q3 and I239 are functional and ER-localized. (A) Mating phenotype of Ste14p-myc Q3 and I239. Patches of the indicated MATa strains were replica-plated onto a lawn of the MATα mating tester strain SM1068 on an SD plate containing 0.3 ml of YPD. Plates were incubated at 30°C for 2 d. Strains tested were SM2926 (Δste14-3) (Δ), SM3185 (CEN STE14) (WT), SM3874 (CEN STE14::myc Q3) (Q3), and SM3876 (CEN STE14::myc I239) (I239). (B) Indirect immunofluorescence of strains expressing Ste14p-myc Q3 (top) and I239 (bottom). A single cell is shown for each construct. Fixed and permeabilized cells were stained with a 1:1000 dilution of anti-myc antiserum followed by secondary decoration with Cy3-conjugated goat anti-mouse antiserum (A and D), a 1:5000 dilution of anti-Kar2p antiserum followed by secondary decoration with FITC-conjugated goat anti-rabbit antiserum (B and E), and with DAPI (4′,6-diamino-2-phenylindole) (C and F). Strains visualized were SM3798 (2μ STE14::myc Q3) and SM3800 (2μ STE14::myc I239).
In a previous study we found that the insertion of a 3x HA epitope into Ste14p at positions Q3 and I239 resulted in the mislocalization of a significant subpopulation of the protein from the ER to the Golgi (Romano et al., 1998). To determine whether the 6x myc epitope influenced the localization of Ste14p, we performed indirect immunofluorescence with the use of an anti-myc mAb. In cells expressing Ste14p-myc (Q3) and (I239) we observed a ring-like staining pattern that mainly surrounded the nucleus (Figure 2B, A and D) as marked by 4′,6-diamidino-2-phenylindole fluorescence (Figure 2B, C and F). Both Ste14p-myc (Q3) and (I239) colocalized with the ER marker protein Kar2p (Figure 2B, compare A and D with B and E). Therefore, unlike the insertion of the 3x HA epitope, the insertion of the 6x myc epitope at positions Q3 and I239 of Ste14p did not result in the mislocalization of the protein.
Orientation of the N and C Termini of Ste14p
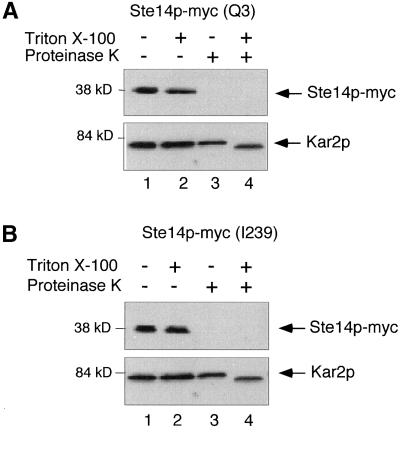
To determine whether the N and C termini of Ste14p are present on the cytosolic face of the ER membrane or in the ER lumen, we used a protease protection assay. After gentle lysis of yeast spheroplasts and collection of the membrane fraction, the sensitivity of ER proteins to proteinase K could be monitored in the absence and presence of detergent. In this system, a lumenal protein such as Kar2p, or a lumenally disposed epitope tag on a membrane protein, should be protected from protease in the absence of detergent. In contrast, an epitope present on the cytosolic face of the ER membrane should be sensitive to protease in the absence of detergent.
The data indicate that for Ste14p-myc, both the N- and C-terminal epitope tags were sensitive to protease in the absence of detergent, whereas the ER lumenal protein Kar2p was only sensitive to protease in the presence of detergent (Figure 3, A and B, lanes 3 and 4; compare top [Ste14p-myc] and bottom [Kar2p] panels). In the case of Kar2p, proteinase K clipped the protein to a smaller size, rather than resulting in its disappearance (Figure 3, A and B, compare lanes 3 and 4; bottom panels). The loss of the Ste14p-myc species was apparently due solely to the digestion of the myc epitope, as opposed to the digestion of an interior site of Ste14p, because a protein species that migrated at the position of untagged Ste14p was detected with anti-Ste14p antibodies in extracts of Ste14p-myc (Q3) treated with protease (our unpublished results). From these data we conclude that both the N and C termini of Ste14p are located in the cytosol instead of in the ER lumen. These results eliminate the five-span-topology model (Figure 1B, center) and strongly suggest that Ste14p contains an even number of membrane spans, in accordance with the four-span- or six-span-topology models (Figure 1B, left and right).
Figure 3.
Protease protection of Ste14p-myc Q3 and I239. Yeast membranes were treated with proteinase K in the presence and absence of 0.4% Triton X-100. After 5 min on ice, the reactions were terminated with 1 mM PMSF and 10% trichloroacetic acid. Samples were resolved by 12.5% SDS-PAGE and transferred to nitrocellulose. Ste14p-myc and Kar2p were detected with anti-myc and anti-Kar2p antisera, respectively. (A) SM3874 (CEN STE14::myc Q3). (B) SM3876 (CEN STE14::myc I239).
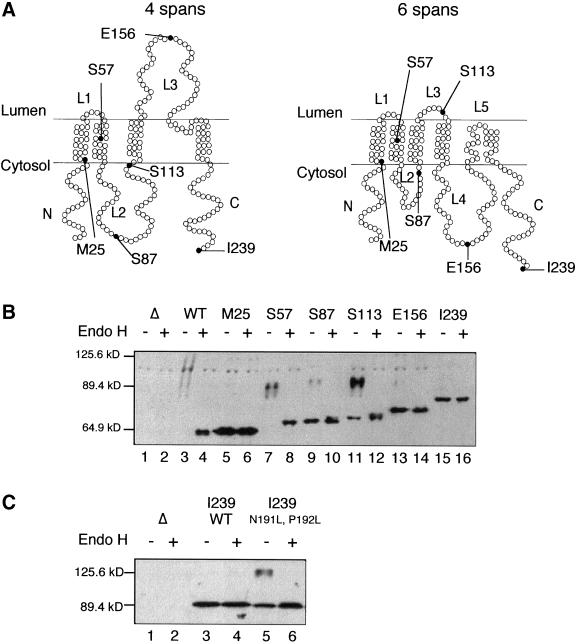
Analysis of Ste14p-Suc2p Fusion Protein Topology
Suc2p (invertase) has been well-used as a “topology sensor” for multispanning membrane proteins (i.e., Sec62p and the N-terminal half of Ste6p) (Deshaies and Schekman, 1990; Geller et al., 1996). Its utility as a topology reporter is based on the capacity of Suc2p to become highly glycosylated when translocated to the ER lumen, on its own or as part of a fusion protein, a modification that is detectable as a mobility shift by SDS-PAGE. To analyze the topology of Ste14p in more detail, we created a series of Ste14p-Suc2p fusions. This fusion series contains increasingly longer portions of Ste14p at the N terminus, fused to mature Suc2p, lacking its native signal sequence, at the C terminus. The sites of the Suc2p fusions to Ste14p are indicated in the context of the four- and six-span models (Figure 4A). We found that a strain expressing full-length Ste14p fused to Suc2p [Ste14p-Suc2p (I239)] could mate as well as strains expressing wild-type Ste14p (our unpublished results), suggesting that the presence of Suc2p per se does not interfere with Ste14p folding.
Figure 4.
Diagram and detection of Ste14p-Suc2p fusions. (A) Topology models of Ste14p (4 span and 6 span), with the sites of Suc2p fusions denoted by a black circle. (B) Immunoblot of Ste14p-Suc2p fusions. Crude yeast cell extracts were incubated at 37°C in the presence or absence of 1000 U of endoglycosidase H for 12 h. The cell extracts (0.4 OD600 cell equivalents) were resolved by 8% SDS-PAGE and transferred to nitrocellulose. Ste14p-Suc2p fusions were detected with anti-Suc2p antibodies. Lanes 1 and 2, SM2705 (Δsuc2); lanes 3 and 4, SM2894 (2 μ PPGKSUC2); lanes 5 and 6, SM4306 [2 μ PPGKSTE14(1-25)::SUC2]; lanes 7 and 8, SM4307 [2 μ PPGKSTE14(1-57)::SUC2]; lanes 9 and 10, SM4308 [2 μ PPGKSTE14(1-87)::SUC2]; lanes 11 and 12, SM4361 [2 μ PPGKSTE14(1-113)::SUC2]; lanes 13 and 14, SM4363 [2 μ PPGKSTE14(1-156)::SUC2]; and lanes 15 and 16, SM4309 [2 μ PPGKSTE14(1-239)::SUC2]. (C) Introduction of the mutations N191L, P192L into Ste14p-Suc2p (239) flips the orientation of Suc2p from cytosolic to partially lumenal. Cell extracts were prepared, treated, and analyzed as in B. Lanes 1 and 2, SM2705 (Δsuc2); lanes 3 and 4, SM4309 [2 μ PPGKSTE14(1-239)::SUC2]; and lanes 5 and 6, SM4470 [2 μ PPGKSTE14(1-239)::SUC2, N191L, P192L].
To determine the orientation of Suc2p in relation to the ER membrane, we examined the gel migration pattern and endoglycosidase H (Endo H) sensitivity of the fusion proteins. Only fusions with their Suc2 portions present in the lumen would be substrates for glycosylation (Deshaies and Schekman, 1990; Traxler et al., 1993; Geller et al., 1996). Cellular extracts were treated in the presence or absence of Endo H, which cleaves high-mannose structures, and the Ste14p-Suc2p fusions were detected in immunoblots with the use of anti-Suc2p antibodies (Figure 4B).
The control lanes (Figure 4B, lanes 1–4) contain extracts from strains expressing wild-type SUC2 or deleted for this gene (Δsuc2). A faint series of heterogeneous high-molecular-weight species (migrating between the 89.4- and 125.6-kDa markers) is detectable in the wild-type SUC2 extract, but absent in the Δsuc2 extract (Figure 4B, compare lane 3 with lane 1). On treatment with endoglycosidase H, the higher molecular weight species in lane 3 collapse into a single band, comigrating with the 64.9-kDa marker (Figure 4B, lane 4). The heterogeneity, slow mobility, and Endo H sensitivity of the species in lane 3 are indicative of glycosylated Suc2p. With the use of these same criteria, we analyzed our complete Ste14p-Suc2p fusion series. Below, we first consider the four fusions that provided straightforward results concerning Ste14p topology. Next, two fusions that yielded ambiguous results are considered.
Fusions at M25, S57, E156, and I239 yielded clear-cut results (Figure 4A). For the fusion of Suc2p to residues M25 or I239 in the N- and C-terminal regions of Ste14p, a single protein species that was insensitive to endoglycosidase H was detected, indicative of the absence of glycosylation (Figure 4B, lanes 5, 6, 15, and 16). These data corroborate our findings with the proteolysis of myc-tagged Ste14p (discussed in the section above and shown in Figure 3), and thus provide independent evidence that the N and C termini of Ste14p are located in the cytosol. For the fusion of Suc2p to residue S57 of Ste14p, a series of heterogeneous high-molecular-weight bands of ∼90 kDa was detected (Figure 4B, lane 7) that collapses to a single species of ∼70 kDa upon Endo H treatment (Figure 4B, lane 8), indicative of glycosylation. Thus, the Suc2p portion of this fusion protein must be lumenal. Because the M25 and S57 fusion proteins are cytosolically and lumenally disposed, respectively, a membrane span must be present between residues M25 and S57, as predicted in both the four- and six-span-topology models (Figure 4A). For the fusion of Suc2p to Ste14p at E156, a single, Endo H-insensitive band is detected (Figure 4B, lanes 13 and 14), suggesting that this fusion protein was unglycosylated and hence facing the cytosol. These data support the six-membrane-span model of Ste14p and not the four-membrane-span model (Figure 4A).
Two of the fusions (S87 and S113) yielded mixed results with regard to Ste14p topology, because these fusions existed as a mixture of glycosylated and unglycosylated species (Figure 4B, lanes 9–12). For the S87 fusion, two populations of fusion proteins were detected (Figure 4B, lanes 9 and 10). The major population comprised a single species of ∼71 kDa that was insensitive to Endo H treatment, suggesting that residue S87 resides in the cytosol (in agreement with both the 4- and 6-span models). However, a minor population of the S87 fusion existed as a 90-kDa species that was sensitive to Endo H treatment (Figure 4B, lanes 9 and 10). Similarly, two populations were detected for the Ste14-Suc2p (S113) fusion (Figure 4B, lanes 11 and 12). For this fusion the majority of molecules existed as a series of high-molecular-weight species (88–94 kDa) that were sensitive to Endo H treatment, suggesting that S113 is located in the lumen, although some molecules migrated as single, Endo H-sensitive 72-kDa species.
Taken together, the results from the Ste14p-Suc2p fusions favor the six-membrane-span-topology model, rather than the four-membrane-span model. However, because of the ambiguity in the results from the S87 and S113 fusions, we turned to a different approach to examine the topology of the middle region of Ste14p.
Protease Protection versus Susceptibility of Internal Epitope Tags in Ste14p
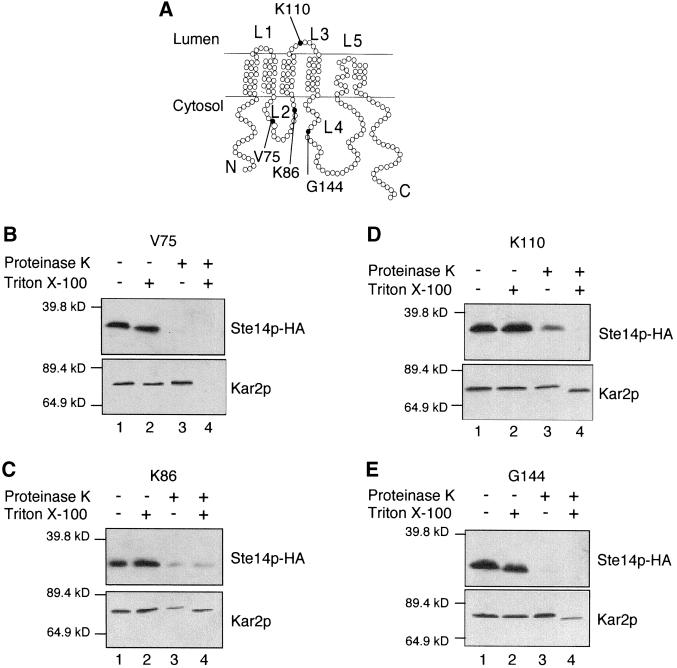
To further investigate the topology of the middle region of Ste14p (specifically, loops L2, L3, and L4 in Figure 5A), we chose to use protease protection assays. We inserted the triple HA epitope tag into several internal sites in Ste14p, adjacent to residues V75, K86, K110, and G144, as indicated in Figure 5A. To determine whether insertion of the HA epitope affected Ste14p function, we tested the ability of Ste14p-HA to produce active a-factor with the use of a plate mating assay. Strains expressing Ste14p-HA (K86) and (K110) mated as well as wild type, whereas strains expressing Ste14p-HA (V75) and (G144) were unable to mate (our unpublished results).
Figure 5.
Protease Protection of Ste14p-HA. (A) Topology model of Ste14p (6 span), with the site of the HA insertions denoted by a black circle. (B–E) Yeast membranes were treated with proteinase K in the presence and absence of 0.4% Triton X-100. After 5 min on ice, the reactions were terminated with 1 mM PMSF and 10% trichloroacetic acid. Samples were resolved by 12.5% SDS-PAGE and transferred to nitrocellulose. Ste14p-HA and Kar2p were detected with anti-HA and anti-Kar2p antisera, respectively. (B) SM3191 (CEN STE14::HA V75) (C) SM3428 (CEN STE14::HA K86) (D) SM4316 (2 μ STE14::HA K110) (E) SM3429 (CEN STE14::HA G144).
With insertions at V75 and K86 in loop L2, the HA epitope tag was sensitive to proteinase K digestion in the presence and absence of detergent (Figure 5, B and C, compare lanes 1–4). These results suggest that loop L2 of Ste14p is located on the cytosolic side of the membrane, as depicted in Figure 5A.
Unlike the unambiguous pattern described above, the protease protection results for the epitope insertion at K110 (loop L3) were not clear-cut. A fraction of the Ste14p-HA (K110) molecules was accessible to proteinase K in the absence of detergent, whereas a second fraction was inaccessible (Figure 5D, lane 3, top). The possibility that the membranes were compromised is unlikely, because the ER lumenal protein Kar2p was inaccessible to proteinase K in the absence of detergent (Figure 5D, lane 3, bottom). This “mixed topology” result for Ste14p-HA (K110) in the protease protection assay is strikingly analogous to the mixed topology we observed in the previous section for the Ste14p-Suc2 (S113) fusion protein by the glycosylation assay. In both cases, although loop L3 appeared to be lumenally oriented in some of the molecules, a fraction of molecules exhibited the opposite orientation, rendering definitive conclusions about topology difficult for loop L3.
The HA epitope tag inserted at G144 in loop L4 was susceptible to digestion by proteinase K in the presence and absence of detergent, whereas the lumenal ER marker Kar2p was only susceptible to proteinase K in the presence of detergent (Figure 5E, compare lanes 3 and 4). This result indicated that loop L4 is located in the cytosol in Ste14p-HA (G144), agreeing with the results obtained with the use of the Ste14p-Suc2p (E156) fusion (Figure 4B, lanes 13 and 14), and thereby also supporting the six-membrane-span model.
Evidence For a Hairpin Turn Formed by Residues N191 and P192 Between Transmembrane Spans 5 and 6 in Ste14p
The results from the Ste14p-Suc2p fusion and protease protection experiments described above clearly revealed that loop L4 (6-membrane span model) and the C terminus of Ste14p are both located on the cytosolic side of the membrane (Figure 4B, lanes 13 and 14; Figure 5E for loop L4; Figure 3B, lanes 3 and 4; Figure 4B, lanes 15 and 16 for the C terminus). For these regions to be on the same side of the membrane, the hydrophobic stretch of 31 amino acids between them (residues L176-F206) must span the membrane twice, as indicated in Figure 5A. However, because 20 amino acids is often considered as the minimal length for a true transmembrane span, and this segment of Ste14p contains only 31 amino acids, we considered alternative possibilities. The most attractive of these is that these 31 amino acids form a helix-turn-helix, also called a “helical hairpin,” within the membrane. Indeed, a recent study has shown that 31 residues is the minimum length required to form such a helical hairpin, provided a strong turn-inducing amino acid is present (Monne et al., 1999b). Alternatively, the entire L176-F206 segment of Ste14p could simply reside in the cytosol, or associate with the membrane peripherally. However this latter possibility is highly unlikely, based on the high degree of hydrophobicity of this region of Ste14p (>2 in the hydrophobicity plot shown in Figure 1A).
Von Heijne and coworkers have generated a “turn propensity scale” by measuring the tendency for each of the 20 amino acids to generate a tight turn in a membrane helix and thus induce the formation of a helical hairpin (Monne et al., 1999a,b). The two residues with the greatest propensity for generating a turn in a membrane span are asparagine and proline. Interestingly, an asparagine-proline pair (N191 and P192) is located approximately midway through the stretch of 31 hydrophobic amino acids between loop L4 and the C terminus of Ste14p (Figures 5A and 7). It thus seemed reasonable to hypothesize that residues N191 and P192 generate a turn in this stretch of 31 amino acids, thereby creating a hairpin loop in the membrane.
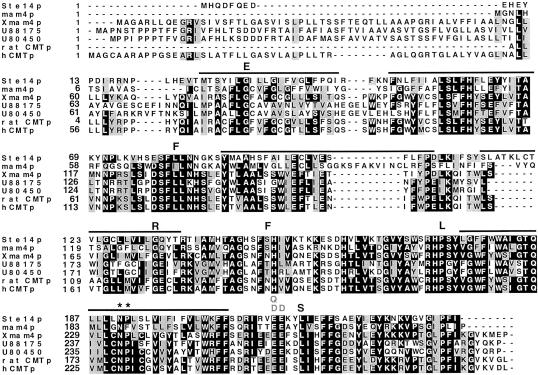
Figure 7.
Alignment of the ICMT family depicting the location of the ste14 mutants identified by random and site-directed mutagenesis. Alignments of S. cerevisiae Ste14p, S. pombe mam4p, X. laevis Xmam4p, 2 C. elegans open reading frames (accession numbers U88175 and U80450), a rat open reading frame (accession number AF0755595.1), and human pcCMTp are shown. Protein sequences were aligned with the use of Clustal W software (Thompson et al., 1994). The default parameters of the program were used (gap extension penalty of 0.05, gap opening penalty of 10.0, protein weight matrix - Blosum series). Black boxes denote amino acid identity and gray boxes denote amino acid similarity as determined with the Boxshade server. The bars above the sequence denote the putative membrane spans of Ste14p. The black letters above the sequence denote the random mutations described in Table 2. The site-directed mutations described in Table 2 are depicted in gray. The asterisks indicate the positions of residues N191 and P192.
To test this hypothesis, we made use of the full-length Ste14p-Suc2p (I239) fusion construct, in which we had shown that Suc2p is cytosolically disposed (Figure 4). We reasoned that if a hairpin turn were formed by residues N191 and P192, then mutational alteration of these residues to the nonturn-inducing residue leucine (N191L P192L) should “flip” Suc2p to a lumenal disposition where it could undergo glycosylation. Strikingly, this prediction was borne out to a significant extent (Figure 4C). For the wild-type Ste14p-Suc2p (I239) fusion protein, a single 90-kDa protein species that is insensitive to Endo H was detected (Figure 4C, lanes 3 and 4), as was observed earlier (Figure 4B, lanes 15 and 16). However, for the mutant fusion protein, in addition to a population of 90-kDa Endo H-insensitive molecules, a new heterogeneous high-molecular-weight (∼125-kDa) species was detected that is sensitive to Endo H and represents a portion of the fusion protein population (Figure 4C, lanes 5 and 6). Thus, for the mutant fusion Ste14p-Suc2p (N191L P192L), a subpopulation of the Suc2p moiety was glycosylated and located on the lumenal side of the membrane, whereas a second subpopulation of Suc2 remained unglycosylated (i.e., cytosolic). Thus, the introduction of the leucine mutations was at least partially, if not fully, able to flip the orientation of Suc2p relative to the membrane. Overall, these results provide strong support for the notion that residues N and P promote the formation of a hairpin turn in Ste14p, essentially permitting the L176-F206 region to function topologically as a pair of membrane spans (spans 5 and 6) but without an intervening hydrophilic loop.
Mutagenesis of STE14 Defines Residues That Are Critical for Function
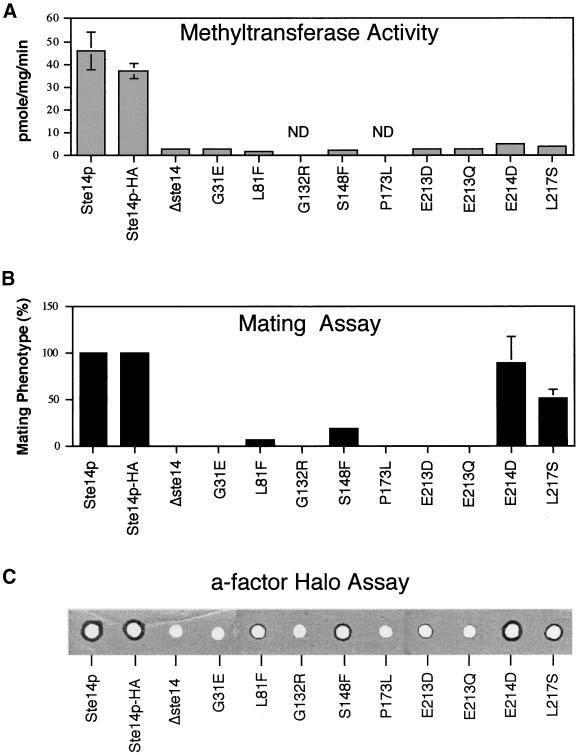
To identify residues critical for the activity of Ste14p, we generated 9 ste14 mutants with the use of a combination of several methods (Table 2). First, we carried out random mutagenesis of a plasmid containing STE14 with the use of hydroxylamine. Transformation of the mutagenized DNA into a Δste14 strain, followed by a screen for transformants with a mating-defective phenotype yielded four unique ste14 missense mutations (L81F, G132R, S148F, and P173L; Table 2) and three nonsense mutations that were not characterized further (see MATERIALS AND METHODS). Second, we analyzed several chromosomal ste14 mutant alleles obtained in a previous random ethyl methanesulfonate mutagenesis screen for MATa-specific mating-defective mutants (Fujimura-Kamada et al., 1997). The mutations were transferred by recombination from the chromosome onto a plasmid and subjected to DNA sequence analysis, yielding two unique mutations (G31E and L217S; Table 2). Finally, three additional mutations were generated by site-directed mutagenesis. These mutations (E213Q, E213D, and E214D; Table 2) altered two highly conserved glutamate residues found in the consensus motif described in the next section. Together, these strategies yielded a set of nine mutants (6 obtained by random mutagenesis and 3 by site-directed mutagenesis). We analyzed these mutants by an in vitro methyltransferase assay, an in vivo quantitative mating assay, and an in vivo a-factor halo assay. In addition, the level of expression of each mutant protein was evaluated by immunoblotting. A summary of the data is presented in Table 2 and Figure 6.
Figure 6.
Comparison of the methyltransferase activity, mating efficiency, and a-factor halo production in strains expressing wild-type and mutant forms of Ste14p. The data for the methyltransferase activity (A) and mating efficiency (B) of strains bearing wild-type and mutant Ste14p plasmids are shown graphically and is derived from Table 2. The a-factor halo assay (C) was performed as described in MATERIALS AND METHODS. The strains used for these assays are listed in Table 2.
Several general features of the mutants are notable. First, and most significantly, five of the six mutants identified by random mutagenesis (G31E, L81F, G132R, P173L, L217S) lie in residues conserved among all members of the ICMT family (Figure 7), indicating that conserved residues in this family are indeed critical for function. Second, all nine mutations have a significant impact on Ste14p enzymatic activity, as judged by the in vitro methyltransferase assay (Table 2, column 5; and Figure 6A). In all cases, the methyltransferase activity is essentially indistinguishable from the “background” level of a Δste14 mutant (with the possible exception of E214D and L217S, which are very slightly elevated compared with the other mutants). It should be noted that the relatively high background of the Δste14 strain in the in vitro methyltransferase assay precludes the assessment of the true extent of the mutant defect. Third, four mutants (G31E, G132R, P173L, and E213Q) are completely defective in function, based on their complete loss of activity in the highly sensitive in vivo quantitative mating assay (<0.001% mating compared with a wild-type strain, Table 2, column 4; Figure 6B). The analogous result is evident in the equally sensitive, but qualitative a-factor halo assay, in which these four mutants fail to generate any hint of a halo (Figure 6C). Four of the remaining mutants exhibit low (L81F, S148F, E213D) or intermediate (L217S) residual activity in the mating and halo assays (Table 2, column 4; Figure 6, B and C). Mutant E214D, containing a conservative acidic substitution, has a high level of residual function, as indicated by its near normal mating and halo levels (despite low methyltransferase activity). Based on the highly sensitive mating and halo tests, the residues G31, G132, P173, and E213, which when mutated are completely defective in activity, are the best candidates for defining structurally and/or catalytically critical residues of Ste14p.
Consensus Motif Identified Through Sequence Comparisons and Database Searches
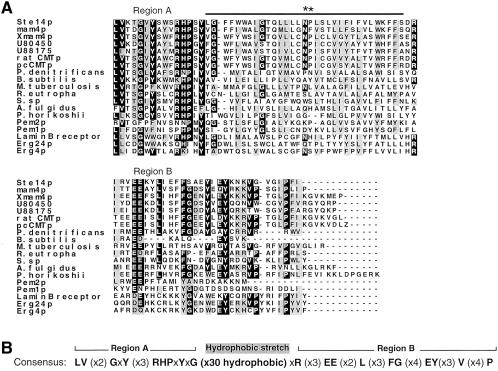
Sequence comparisons often point out regions of potential functional importance. Because the ICMT family contains numerous regions of extended homology that exist throughout the length of the proteins, pinpointing a specific region of functional importance is difficult. To uncover possible sequence motifs involved in Ste14p catalysis, we carried out two types of sequence comparisons. The C-terminal portion of Ste14p (residues 161–239) emerged as a region of particular interest, because a number of proteins shared homology to Ste14p in this region. First, we compared the sequence of Ste14p to other methyltransferases. Specifically, we focused on those methyltransferases pointed out by Kagan and Clarke, that like Ste14p, lack a tripartite consensus sequence present in a large group of protein and nucleic acid methyltransferases; this sequence is believed to be involved in AdoMet binding. Of seven proteins examined by pairwise alignment with Ste14p, we discovered that two of these shared some sequence similarity to the C-terminal region of Ste14p (residues 161–239). Interestingly, these are also methyltransferases, namely, the yeast phosphatidylethanolamine methyltransferases, Pem1p (residues 259–337) and Pem2p (residues 137–206) (Figure 8A).
Figure 8.
The C-terminal region of Ste14p and its ICMT homologues is similar to several bacterial open reading frames, two phosphatidylethanolamine methyltransferases, and human and yeast reductases involved in ergosterol biosynthesis. (A) Alignment of the C-terminal region of S. cerevisiae Ste14p and its homologues (S. pombe mam4p, X. laevis Xmam4p, 2 C. elegans open reading frames [accession numbers U88175 and U80450], a rat open reading frame [accession number AF0755595.1], and human pcCMTp), open reading frames from Pseudomonas denitrificans (M62866), Bacillus subtilis (L77246), Mycobacterium tuberculosis (Z81451), Ralstonia eutropha (X98451), Synechocystis sp. (D90917), Archaeoglobus fulgidus (AAB91272), and Pyrococcus horikoshii (BAA30325), the S. cerevisiae lipid methyltransferases Pem1p and Pem2p, the sterol biosynthesis enzymes from S. cerevisiae Erg4p and Erg24p, and the human lamin B receptor, which is an Erg24p homologue. Protein sequences were aligned with the use of Clustal W software (Thompson et al., 1994). Black boxes denote amino acid identity and gray boxes denote amino acid similarity as determined with the use of Boxshade. The bar above the sequence denotes the putative membrane span of Ste14p. (B) Consensus sequence is based on the alignment shown in A. Only residues that are identical in ≥11 of the proteins in the alignment were scored as consensus residues; other residues are shown as an x. The hydrophobic region was determined by a hydropathy plot.
To identify additional sequence motifs of interest, we performed a database search with the use of the BLAST algorithm, and querying with Ste14p. In addition to the known ICMT family members, this analysis revealed 10 gene products with significant sequence similarity to the C-terminal region of Ste14p (Figure 8A). Seven of these are open reading frames of unknown function from diverse bacteria. The other three are ergosterol biosynthetic enzymes: the yeast reductases Erg4p and Erg24p, and an Erg24p homologue from humans (also designated the lamin B receptor). The latter protein was originally identified as a lamin B binding protein, but has since been shown to have an Erg24p activity (Silve et al., 1998). It should be noted that our BLAST search revealed additional proteins with short stretches of homology to other regions of Ste14p, but these were not analyzed further.
An alignment of the C-terminal region of Ste14p with members of the ICMT family, as well as the more distantly related proteins discussed above that share homology in this region, is shown in Figure 8A. For each of these proteins the hydropathy profile in this region is similar to that of Ste14p (our unpublished results). We derived a consensus sequence for this region that we designate the RHPxY-hyd-EE motif, named according to the most highly conserved residues in the core of this region (Figure 8B). The consensus sequence is tripartite in nature, with two regions of homology (region A and region B) flanking a hydrophobic stretch of 30 amino acids (Figure 8B). Notably, site-directed mutagenesis of the highly conserved glutamates in region B of the consensus sequence (mutations E213D,Q and E214D) reveals that these are critical functional residues for Ste14p, as discussed in the section above and shown in Table 2 and Figure 6. In addition, random mutagenesis identified two other conserved residues within this consensus motif as being critical for function. These are P173 in region A and L217 in region B (Table 2 and Figure 6). The conservation of the RHPxY-hyd-EE motif and the finding that several of its residues are critical for activity, implicate this region of Ste14p as a key region for further functional analysis.
DISCUSSION
The ICMT family of proteins, as exemplified by yeast Ste14p, comprises a novel group of ER membrane-localized carboxyl methyltransferases whose substrates are prenylated CaaX proteins. The ICMTs contain multiple predicted membrane spans (Imai et al., 1997; Dai et al., 1998), and lack the tripartite consensus sequence shared by most other nucleic acid and protein methyltransferases (Kagan and Clarke, 1994; Imai et al., 1997; Dai et al., 1998). Carboxyl methylation is critical for the proper localization of Ras proteins (Hrycyna et al., 1991; Choy et al., 1999; Bergo et al., 2000), activated forms of which are implicated in numerous types of cancer. Hence, ICMT enzymatic activity represents an attractive target for chemotherapeutic intervention. Structure-function insights obtained from studying the yeast ICMT Ste14p are expected to apply to its human homolog pcCMT, and thereby facilitate the development of pharmacological agents that block methyltransferase activity. In this study we sought to establish fundamental structural and functional properties of Ste14p. In particular, we focused on elucidating the topology of Ste14p and determining residues critical for its function. Ultimately, because of its small size, well-characterized enzymatic activity, and highly hydrophobic nature, Ste14p may represent an ideal “test case” for developing methodologies to crystallize multispanning membrane proteins, a critically neglected area of current biology.
Based on the topology studies carried out here, we propose that Ste14p contains six membrane spans, with the majority of its hydrophilic portions present in the cytosol and only two short loops exposed in the ER lumen. In this study we have identified a number of key residues throughout the length of Ste14p, which, when mutated, result in a loss of methyltransferase activity. Our topology model for Ste14p and the position of mutationally altered residues are shown in Figure 9. In addition to defining important residues of Ste14p, our studies demonstrate the utility of two highly sensitive in vivo assays (mating and a-factor halo tests) for examining very low levels of Ste14p methyltransferase activity. These points are discussed in more detail below.
Figure 9.
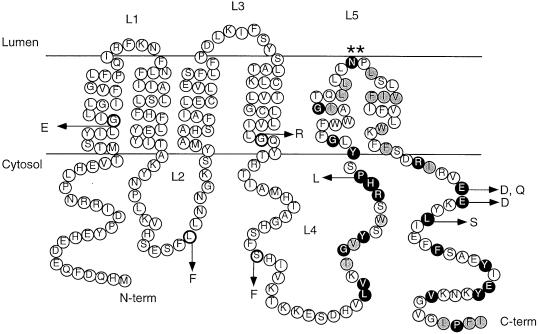
Topology model of Ste14p indicating the locations of the ste14 mutants described in this study. The six-membrane-span model for Ste14p, supported by the results of this study, is shown. In this model, the N and C termini of Ste14p extend into the cytosol. The mutations described in this study are indicated. The conserved residues found in the RHPxY-hyd-EE consensus region described in Figure 8 are indicated; identical residues are shaded black and similar residues are shaded gray.
Topology of Ste14p
At the outset of our studies, we considered three topology models for Ste14p (4, 5, or 6 membrane spans; Figure 1) based on hydropathy analysis. However, early on we could eliminate the five-span model. This was because protease protection assays (Figure 3) and Endo H treatment of Ste14p-Suc2p fusions (Figure 4) demonstrated that both the N and C termini of Ste14p were cytosolic, as had also been previously suggested from experiments with ubiquitin fusions (Wittke et al., 1999). Thus, Ste14p must contain an even number of membrane spans.
A major difference in the two remaining models (4 or 6 span) is the orientation of amino acids R136 through G177 (loop L3 in the 4-span model, which is lumenal, and loop L4 in the 6-span model, which is cytosolic) (Figure 4A). The gel mobility and lack of Endo H sensitivity of the Ste14p-Suc2p (E156) fusion (Figure 4, lanes 13 and 14) and the protease sensitivity of the epitope-tagged construct Ste14p-HA (G144) (Figure 5, A and E) indicate that this region of Ste14p is located in the cytosol. Only the six-membrane-span model is consistent with the cytosolic localization of these residues, and we thus designate this region loop L4 (Figures 4, 5, and 8). Our remaining results are also consistent with the six-membrane-span model: The protease protection results of Ste14p-HA V75 and K86 (Figure 5, B and C) and the gel mobility and endoglycosidase H insensitivity of the Ste14p-Suc2p S87 fusion (Figure 4B, lanes 9 and 10) indicate that residues V75, K86, and S87 are cytosolic, which is consistent with the placement of loop L2 in the six-span model.
The only region of Ste14p that yielded ambiguous results was the short ER lumenal loop L3 (Figure 5A). Our analysis of Ste14p-Suc2p fusions and protease protection of Ste14p-HA constructs in this region both yielded “mixed topology” results: The Ste14p-Suc2p S113 fusion (Figure 4B, lanes 11 and 12) showed a mostly, but not exclusively, lumenal pattern. Likewise, protease protection was only partial for Ste14p-HA (K110) (Figure 5D, lane 3), suggesting that K110 is partly, but not completely lumenal. This type of mixed topology is not uncommon to encounter in hydropathy analysis (Zhang and Ling, 1991; Geller et al., 1996). Such behavior may result because the proper topology of Ste14p is easily perturbed in this particular region. Alternatively, such behavior may reflect a functionally significant property, such as mobility of this region of Ste14p within the membrane. Despite the minor ambiguity, overall our data strongly favor the six-span model for Ste14p topology. Ultimately, proof for this model will require some type of structural analysis of Ste14p.
Topology of Other Members of the ICMT Family
Hydropathy analysis suggests that the overall topology of Ste14p is preserved among all members of the ICMT family, although the ICMTs of higher organisms contain two additional putative membrane spans at their N terminus, compared with Ste14p and another fungal family member, mam4p (our unpublished data). It is unlikely that these two N-terminal membrane spans contribute to catalytic function, given their absence in Ste14p. In contrast to their differing N-terminal topologies, ICMT family members share similar C-terminal hydropathy profiles.
An interesting aspect of Ste14p relates to the nature of the hydrophobic stretch of 31 amino acids that represent transmembrane spans 5 and 6. The length of this region is shorter than the standard length of two transmembrane alpha helices (generally >40 residues). The short length of this region as well as the presence of two adjacent residues (N191, P192) with a high propensity to induce turns in a transmembrane helix led us to hypothesize that these residues (L176-F206) probably form a helical hairpin structure. If this were the case, then mutating N191 and P192 to leucine residues would be predicted to flip a C-terminally fused Suc2p moiety from the cytosol to the lumen. Indeed, upon introduction of the double mutation N191L, P192L into the Ste14p-Suc2p (239) fusion, this is precisely what we observed for at least a portion of the fusion molecules. Thus, it is probable that N191 and P192 are essential for inducing a turn in the C-terminal hydrophobic segment of Ste14p. Notably, residue N191 is strictly conserved among all of the ICMT family members (Figures 7 and 8). In all but one case, the adjacent proline is also conserved. Therefore, it is likely that this hairpin loop is a significant feature of the ICMT family of methyltransferases.
We identified a consensus sequence (RHPxY-hyd-EE) in the C terminus of Ste14p that is characteristic of a group of proteins that include ICMTs, two yeast phospholipid methyltransferases (Pem1p and Pem2p), ergosterol biosynthesis enzymes, and a series of bacterial open reading frames of unknown function (Figure 8). This consensus sequence consists of a stretch of hydrophobic amino acids (hyd) that forms the predicted helical hairpin region described in the section above, flanked by two regions of amino acid conservation, regions A and B. Nearly all of the proteins aligned in Figure 8 contain within their hydrophobic stretch a residue with a strong propensity for helix turn production (P and N are best then R, D, H, Q, K; Monne et al., 1999a) at roughly the same position as N191 and P192 of Ste14p (Figure 8). Thus, it is possible that this stretch of hydrophobic amino acids forms a hairpin turn in most or all of the proteins that share this consensus sequence. Interestingly, a helical hairpin turn places the conserved regions A and B on the same side of the membrane, where they may form a single functional domain (Figures 8 and 9).
Residues Critical for Ste14p Function Identified by Mutational Analysis
To identify residues critical for the activity of Ste14p, we generated ste14 mutants with the use of random mutagenesis in conjunction with a screen for loss of mating (Table 2). Sequence analysis revealed that five of the six missense mutations obtained by random mutagenesis are found in residues that are conserved among the ICMT family of methyltransferases (G31E, L81F, G132R, P173L, and L217S; Figure 7). Two of these (G31E and G132R) introduce charged residues into transmembrane spans, which may disrupt the topology or structure of Ste14p. It is noteworthy that two random mutations (P173L and L217S) are located in regions A and B of the RHPxY-hyd-EE consensus sequence, respectively (Figure 9). We also used site-directed mutagenesis to probe the function of a pair of highly conserved glutamates in region B of consensus region (E213D, E213Q, and E214D). All of these mutations disrupted Ste14p function (Table 2, column 5; and Figure 6A).
In the course of our analysis, we found that mating and a-factor halo assays were very valuable for assessing the severity of ste14 mutations. Mutations that result in no activity versus a low level of residual methyltransferase activity cannot be distinguished by the in vitro methyltransferase assay due to a high level of background counts, even in strains bearing a ste14 deletion (Table 2, column 5; and Figure 6A). This background may be due to methyltransferase activities other than Ste14p present in the crude extracts used for the assay. These unrelated methyltransferases can effectively mask any low-level Ste14p-dependent methylation events. The biological assays (mating and halo) are not associated with any background and can therefore reliably detect exceedingly low levels of residual Ste14p function (compare the results from the methyltransferase assay [Figure 6A] to those from the mating [Figure 6B] and halo assays [Figure 6C]). Thus, we can use these sensitive in vivo assays with confidence to distinguish between weak and strong ste14 alleles (i.e., E214D vs. E214Q, respectively). We found that Ste14p mutants altered in residues G31, G132, P173, or E213 are completely defective in function (Figure 6, B and C), suggesting that these are excellent candidates residues for defining sites within Ste14p essential for carrying out catalysis or maintaining the proper folding of Ste14p. These results suggest that a large-scale mutagenesis of Ste14p, screening by mating and halo assays, could generate an excellent functional map of Ste14p. In addition, just as the yeast mating assay has been used to screen for prenyltransferase inhibitors, an analogous drug screen to search for methyltransferase inhibitors could potentially be developed.
Ste14p As a Test Case for Structural Analysis of Multispanning Membrane Proteins
The topological and mutational analysis presented here provides the groundwork for further structural and functional studies of the Ste14p methyltransferase. Such studies will require the purification of active Ste14p, the identification of its catalytic sites, and the determination of the AdoMet and isoprenylcysteine binding sites. It will be important to determine whether catalysis occurs on the cytosolic face of the membrane or within the plane of the membrane. The latter possibility is likely, based on the high number of membrane spans found in Ste14p and the membrane association of the prenylated substrates of Ste14p. In addition, further structural studies of Ste14p will be important in determining the structure of the AdoMet and isoprenylcysteine binding sites, and in verifying the orientation of loop L3 of Ste14p. Ste14p has a number of properties that make it very valuable as a candidate protein for structural determination, which include the fact that its topology is experimentally established and that several residues critical for function have already been identified.
ACKNOWLEDGMENTS
S.M. was supported by a grant from the National Institutes of Health (GM-41223). We are grateful to W. Piluek for performing the a-factor halo assay shown in Figure 7 and to B. Baum for calling our attention to the similarity between Ste14p and Pem1p and Pem2p.
REFERENCES
- Abeliovich H, Grote E, Novick P, Ferro-Novick S. Tlg2p, a yeast syntaxin homolog that resides on the Golgi and endocytic structures. J Biol Chem. 1998;273:11719–11727. doi: 10.1074/jbc.273.19.11719. [DOI] [PubMed] [Google Scholar]
- Ashby MN, Errada PR, Boyartchuk VL, Rine J. Isolation and DNA sequence of the STE14gene encoding farnesyl cysteine: carboxyl methyltransferase. Yeast. 1993;9:907–913. doi: 10.1002/yea.320090810. [DOI] [PubMed] [Google Scholar]
- Bergo MO, Leung GK, Ambroziak P, Otto JC, Casey PJ, Young SG. Targeted inactivation of the isoprenylcysteine carboxyl methyltransferase gene causes mislocalization of K-Ras in mammalian cells. J Biol Chem. 2000;275:17605–17610. doi: 10.1074/jbc.C000079200. [DOI] [PubMed] [Google Scholar]
- Berkower C. Analysis of STE6, a Saccharomyces cerevisiae ATP Binding Cassette (ABC) Protein. Ph.D. Thesis. Baltimore, MD: Johns Hopkins University; 1995. [Google Scholar]
- Berkower C, Loayza D, Michaelis S. Metabolic instability and constitutive endocytosis of STE6, the a-factor transporter of Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:1185–1198. doi: 10.1091/mbc.5.11.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair LC. Genetic Analysis of Mating Type Switching in Yeast. Ph.D. Thesis. Eugene, OR: University of Oregon; 1979. [Google Scholar]
- Choy E, Chiu VK, Silletti J, Feoktistov M, Morimoto T, Michaelson D, Ivanov IE, Philips MR. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell. 1999;98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- Dai Q, Choy E, Chiu V, Romano J, Slivka SR, Michaelis S, Philips MR. Human prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. J Biol Chem. 1998;273:15030–15034. doi: 10.1074/jbc.273.24.15030. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Schekman R. Structural and function dissection of Sec62p, a membrane-bound component of the yeast endoplasmic reticulum protein import machinery. Mol Cell Biol. 1990;10:6024–6035. doi: 10.1128/mcb.10.11.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura-Kamada K, Nouvet FJ, Michaelis S. A novel membrane-associated metalloprotease, Ste24p, is required for the first step of NH2-terminal processing of the yeast a-factor precursor. J Cell Biol. 1997;136:271–285. doi: 10.1083/jcb.136.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller D, Taglicht D, Edgar R, Tam A, Pines O, Michaelis S, Bibi E. Comparative topology studies in Saccharomyces cerevisiae and in Escherichia coliof the N-terminal half of the yeast ABC protein, Ste6. J Biol Chem. 1996;271:13746–13753. doi: 10.1074/jbc.271.23.13746. [DOI] [PubMed] [Google Scholar]
- Hrycyna CA, Clarke S. Farnesyl cysteine C-terminal methyltransferase activity is dependent upon the STE14 gene product in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:5071–5076. doi: 10.1128/mcb.10.10.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrycyna CA, Sapperstein SK, Clarke S, Michaelis S. The Saccharomyces cerevisiae STE14 gene encodes a methyltransferase that mediates C-terminal methylation of a-factor and RAS proteins. EMBO J. 1991;10:1699–1709. doi: 10.1002/j.1460-2075.1991.tb07694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Davey J, Kawagishi-Kobayashi M, Yamamoto M. Genes encoding farnesyl cysteine carboxyl methyltransferase in Schizosaccharomyces pombe and Xenopus laevis. Mol Cell Biol. 1997;17:1543–1551. doi: 10.1128/mcb.17.3.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan RM, Clarke S. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch Biochem Biophys. 1994;310:417–427. doi: 10.1006/abbi.1994.1187. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Ma H, Kunes S, Schatz PJ, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- Michaelis S, Herskowitz I. The a-factor pheromone of Saccharomyces cerevisiaeis essential for mating. Mol Cell Biol. 1988;8:1309–1318. doi: 10.1128/mcb.8.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monne M, Hermansson M, von Heijne G. A turn propensity scale for transmembrane helices. J Mol Biol. 1999a;288:141–145. doi: 10.1006/jmbi.1999.2657. [DOI] [PubMed] [Google Scholar]
- Monne M, Nilsson I, Elofsson A, von Heijne G. Turns in transmembrane helices: determination of the minimal length of a “helical hairpin” and derivation of a fine-grained turn propensity scale. J Mol Biol. 1999b;293:807–814. doi: 10.1006/jmbi.1999.3183. [DOI] [PubMed] [Google Scholar]
- Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivoin yeast. Nucleic Acids Res. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver TL, Szostak JW, Rothstein RJ. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci USA. 1981;78:6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips MR, Pillinger MH. Prenylcysteine-directed carboxyl methyltransferase activity in human neutrophil membranes. Methods Enzymol. 1995;256:49–63. doi: 10.1016/0076-6879(95)56009-2. [DOI] [PubMed] [Google Scholar]
- Romano JD, Schmidt WK, Michaelis S. The Saccharomyces cerevisiaeprenylcysteine carboxyl methltransferase Ste14p is in the endoplasmic reticulum membrane. Mol Biol Cell. 1998;9:2231–2247. doi: 10.1091/mbc.9.8.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1990. [Google Scholar]
- Sapperstein S, Berkower C, Michaelis S. Nucleotide sequence of the yeast STE14 gene, which encodes farnesylcysteine carboxyl methyltransferase, and demonstration of its essential role in a-factor export. Mol Cell Biol. 1994;14:1438–1449. doi: 10.1128/mcb.14.2.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonberger O, Knox C, Bibi E, Pines O. Split invertase polypeptides form functional complexes in the yeast periplasm in vivo. Proc Natl Acad Sci USA. 1996;93:9612–9617. doi: 10.1073/pnas.93.18.9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silve S, Dupuy PH, Ferrara P, Loison G. Human lamin B receptor exhibits sterol C14-reductase activity in Saccharomyces cerevisiae. Biochim Biophys Acta. 1998;1392:233–244. doi: 10.1016/s0005-2760(98)00041-1. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighing, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler B, Boyd D, Beckwith J. The topological analysis of integral cytoplasmic membrane proteins. J Membr Biol. 1993;132:1–11. doi: 10.1007/BF00233047. [DOI] [PubMed] [Google Scholar]
- Volker C, Pillinger MH, Philips MR, Stock JB. Prenylcysteine analogs to study function of carboxylmethylation in signal transduction. Methods Enzymol. 1995;250:216–225. doi: 10.1016/0076-6879(95)50074-x. [DOI] [PubMed] [Google Scholar]
- Wilson KW. Identification and Regulation of Cell-Type-Specific Genes Required for Mating in Saccharomyces cerevisiae. Ph.D. Thesis. San Francisco, CA: University of California; 1985. [Google Scholar]
- Wittke S, Lewke N, Muller S, Johnsson N. Probing the molecular environment of membrane proteins in vivo. Mol Biol Cell. 1999;10:2519–2530. doi: 10.1091/mbc.10.8.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J-T, Ling V. Study of membrane orientation and glycosylated extracellular loops of mouse P-glycoprotein by in vitrotranslation. J Biol Chem. 1991;266:18224–18232. [PubMed] [Google Scholar]