SUMMARY
Extracellular cues that regulate cellular shape, motility, and navigation are generally classified as growth-promoting (i.e., growth factors/chemoattractants and attractive guidance cues) or growth-preventing (i.e., repellents and inhibitors). Yet, these designations are often based on complex assays and undefined signaling pathways, and thus may misrepresent direct roles of specific cues. Herein, we find that a recognized growth-promoting signaling pathway amplifies the F-actin disassembly and repulsive effects of a growth-preventing pathway. Focusing on Semaphorin/Plexin repulsion, we identified an interaction between the F-actin-disassembly enzyme Mical and the Abl tyrosine kinase. Biochemical assays revealed Abl phosphorylates Mical to directly amplify Mical Redox-mediated F-actin disassembly. Genetic assays revealed Abl allows growth factors and Semaphorin/Plexin repellents to combinatorially increase Mical-mediated F-actin disassembly, cellular remodeling, and repulsive axon guidance. Similar roles for Mical in growth factor/Abl-related cancer cell behaviors further revealed contexts in which characterized positive effectors of growth/guidance stimulate such negative cellular effects as F-actin disassembly/repulsion.
eTOC Blurb
Semaphorin/plexin signaling repels cellular growth and promotes actin disassembly. Surprisingly, Yoon et al. find that these effects are amplified by growth-promoting factors acting via Abl tyrosine kinase. These results reveal a role for chemoattractant cues in promoting the effects of chemorepellents, and suggest complex interactions among growth-suppressing and -promoting pathways.
INTRODUCTION
Preventing cellular growth, motility, and navigation – in cancer, for example – or stimulating it in the diseased brain is crucial for curing many devastating pathologies. These cellular behaviors are specified in both physiological and pathological contexts when signals from outside of cells impinge through signal transduction pathways on the proteins that directly regulate the assembly, organization, and stability of the actin filament (F-actin) cytoskeleton (Pollard and Cooper, 2009; Bray, 2001). The search for the extracellular signals underlying these behaviors has now yielded a vast array of molecules that are generally grouped into two classes based on their positive or negative effects on cell shape and movement (Kolodkin and Tessier-Lavigne, 2011; Berzat and Hall, 2010; Swaney, et al., 2010). Extracellular signals such as growth factors, some of which are also chemoattractants (referred to as growth factors/chemoattractants below), and attractive guidance cues, for example, are linked together based on their positive effects on cell shape and motility – effects that are believed to be the result of their ability to promote F-actin formation and stability (Gomez and Letourneau, 2014; Vitriol and Zheng, 2012; Kolodkin and Tessier-Lavigne, 2011; Berzat and Hall, 2010; Swaney, et al., 2010). Cues have also been identified and classified as inhibitors/repellents based on their negative influences on cellular form and mobility – effects that are believed to result from their ability to disassemble the F-actin cytoskeleton (Gomez and Letourneau, 2014; Vitriol and Zheng, 2012; Hung and Terman, 2011; Kolodkin and Tessier-Lavigne, 2011; Mayor and Carmona-Fontaine, 2010). Such classifications have triggered a plethora of specific therapeutic strategies such as using growth factors/attractants to stimulate cellular movement (e.g., using nerve growth factors/neurotrophins to encourage axon regeneration) and repellents to limit the movement of cells (e.g., using repulsive cues to limit cancer cell metastasis). Yet, these extracellular signals and their positive or negative effects have often been defined solely on the basis of complex in vitro and in vivo cellular assays where outcomes may be indirect and obscure the direct roles of specific cues. Likewise, there remains a lack of defined signaling pathways from the cell surface to the actin cytoskeleton for many of these extracellular signals, further confounding our understanding of their specific physiological and pathological functions. Such gaps in our knowledge are therefore likely to mask critical concepts and biomedically-relevant distinctions among these cues, their actions, and their use in clinical settings.
To aid in the understanding of how extracellular signals affect the actin cytoskeleton and elicit cellular behaviors, we have been employing simple high-resolution model systems and one of the largest families of guidance cues, the Semaphorins (Semas) (Taylor Alto and Terman, 2017). Semas, with over twenty members conserved from invertebrates to humans, play critical functional roles in numerous tissues and have been best characterized as repellents – destabilizing the F-actin cytoskeleton to negatively regulate the shape, movement, and navigation of cells and their membranous extensions (Hung and Terman, 2011; Kolodkin and Tessier-Lavigne, 2011). Recently, our approaches uncovered a direct pathway from the cell surface repulsive Semaphorin receptor Plexin (Plex) to the actin cytoskeleton – identifying an oxidation-reduction (redox) enzyme, Mical, that directly associates with both Plex and actin filaments, and induces F-actin disassembly via the posttranslational oxidation of actin (Hung, et al., 2011; Hung, et al., 2010; Terman, et al., 2002). The MICALs are now becoming widely recognized as using this F-actin disassembly Redox activity to alter the behaviors of multiple cell types (reviewed in (Manta and Gladyshev, 2017; Wilson, et al., 2016)) but little is known if other signal transduction pathways may intersect with Mical to direct actin cytoskeletal disassembly.
We now find that the Abl non-receptor protein tyrosine kinase and oncoprotein signaling pathway activates Mical to direct multiple cellular effects – including extending and shaping cellular processes, guiding axons, and orchestrating cancer cell invasion, colony formation, and survival. Our results go on to reveal that Abl directly associates and phosphorylates a conserved tyrosine residue (Y500) on Mical, stimulating Mical’s enzymatic activity to disassemble F-actin and increase cellular repulsion. Moreover, we find that these Mical and Abl effects are amplified by the combinatorial action of both Growth Factors/Chemoattractants and Sema/Plexin repellents. Our findings, therefore, uncover contexts in which a recognized “growth-promoting” signaling pathway functions to magnify the F-actin disassembly/repulsive effects of a “growth-preventing” pathway.
RESULTS
The Abl Non-receptor Protein Tyrosine Kinase is a Mical-interacting Protein
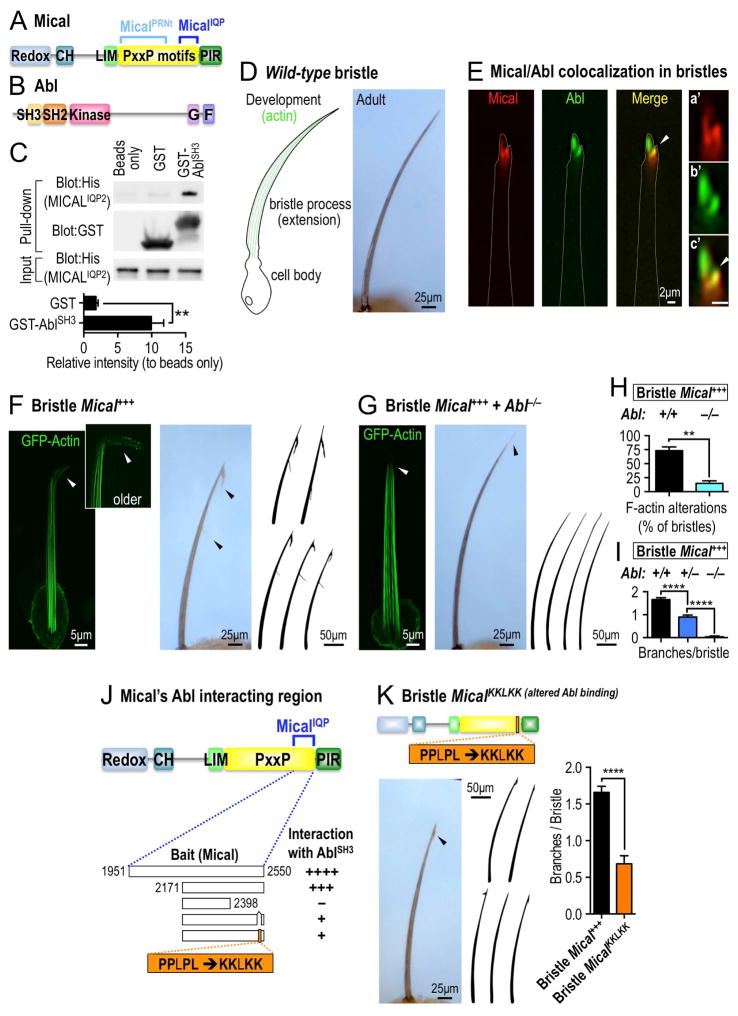
MICALs are large multidomain proteins that contain an enzymatically active monooxygenase (redox) domain, a calponin homology (CH) domain, a LIM domain, a long proline (P) - rich region, and an Ezrin-Radixin-Moesin (ERM) α-like domain (Figure 1A). The MICALs use their redox domain to exert their effects in multiple cell types (Manta and Gladyshev, 2017; Wilson, et al., 2016), but less is known of how this redox activity is regulated. Interestingly, MICALs have conserved proline (P)-rich PxxP motifs within their proline-rich region (Figures 1A and S1A), which are protein interaction modules known to serve as ligands for SH3 domains and regulate the biochemical function of different proteins. Employing different portions of the Mical proline–rich region, we conducted a yeast two-hybrid screen and identified the non-receptor protein tyrosine kinase Abl as a Mical interactor (Figures 1B, S1B–C). Further analysis revealed that the Abl SH3 domain specifically interacted with a particular portion of the Mical proline-rich region (Figures 1A, 1B, S1B–C) and that this Mical–Abl SH3 domain interaction was specific since the SH3 domains of other proteins that were tested did not interact in the yeast two-hybrid system (Figures S1C–D). Purifying proteins containing the interacting regions of Mical and Abl and performing GST pull down assays (Figure 1C) also supported our yeast two-hybrid results – and together with related yeast two-hybrid, co-localization, co-immunoprecipitation, and enzyme assays (as shown below) indicate that Mical and Abl associate together.
Figure 1. The Abl non-receptor tyrosine kinase interacts with and modulates Mical-mediated actin disassembly and cellular remodeling.
(A) Mical protein organization. Mical has a long proline (P) -rich region (yellow) containing well-conserved PxxP motifs (where x is any amino acid). Portions of Mical’s PxxP motif region (MicalPRNt, amino acids 861-1539; MicalIQP, amino acids 1951-2550) were baits in yeast two-hybrid screens. Redox, monooxygenase enzymatic domain; CH, calponin homology domain; LIM, LIM domain; PIR, plexin interacting region. (B) Drosophila Abl protein organization. G, G-actin binding domain; F, F-actin binding domain. (C) GST pull-down assays. Purified GST-AblSH3 protein, but not beads or GST protein alone, significantly interacts with purified His-MicalIQP2 protein (a partial portion of the MicalIQP region, amino acids 2127-2550). Note that equal amounts of purified GST tagged proteins (GST blot) and purified His-MicalIQP2 protein (His blot) were used. The interaction intensity was normalized to that of beads only. n=3; mean±s.e.m.; t-test; **P≤0.01. (D) The Drosophila bristle model. During wild-type bristle development (left), F-actin (green) pushes-out a branchless extension from the bristle cell body – and the morphology of this bristle process/extension is preserved in the adult (right). (E) Mical (red) and Abl (green) prominently colocalize (yellow, arrowhead) within the tips of bristles at sites of F-actin reorganization and bristle branch formation (see Figure 1F). See also higher magnification images of the bristle tips in a′-c′. Abl showed a wider distribution than Mical in the bristle but like Mical, was enriched in the bristle tip (as seen in this single plane confocal image). Scale bar=2μm. (F–I) Abl is required for Mical-mediated F-actin disassembly and cellular remodeling. (F) Drosophila bristles remodel and become branched as the result of F-actin disassembly and reorganization (arrowheads) when Mical is overexpressed (Mical+++) specifically within them. (G) Decreasing Abl levels (Abl homozygous mutant [Abl−/−] background) strongly suppresses Mical-mediated F-actin disassembly and bristle branching to generate wild-type looking bristles (right, compare with wild-type in Figure 1D). Abl−/−=Abl2/Abl4. (H–I) Quantification of F-actin alterations and cellular remodeling following genetic manipulations of Mical and Abl. n≥29 bristles/genotype; mean ± s.e.m.; t-test; **P≤0.01; ****P≤0.0001. Abl+/−=Abl4/+. Similar interactions were also seen with Abl1/+, Abl2/+, and Abl4/+. (J–K) Alteration to Mical’s Abl interacting region suppresses Mical-mediated F-actin alterations/cellular remodeling. (J) Representation of protein constructs containing different portions of the Mical proline-rich region and their ability to interact with Abl’s SH3 domain (AblSH3), as assessed using a yeast two-hybrid assay in which the numbers of + equals the strength of the interaction. –, no interaction. The MicalIQP construct (blue bracket) was the bait construct used for the initial yeast two-hybrid screen described in Figure S1B–D. The region to the right of Mical residue 2398 is necessary for the interaction with AblSH3 and this region contains a stretch of PxxP/Y/L motifs predicted to interact with Abl’s SH3 domain (Hou, et al., 2006). Substituting the P/L residues in PPLPL with K residues decreases the interaction with AblSH3. (K) Expressing Mical with an alteration to its Abl interacting PxxP/Y/L motif (MicalKKLKK) decreases Mical-mediated F-actin alterations/cellular remodeling (arrowhead, drawings, and graph). n≥19 bristles/genotype; mean±s.e.m.; t-test; ****P≤0.0001.
Abl Regulates Mical-mediated F-actin Disassembly and Cellular Remodeling
Abl family kinases are distinguished from other non-receptor tyrosine kinases by the presence of SH2, SH3, and actin binding domains (Figure 1B), and they play critical but still incompletely understood roles in multiple cellular events (Khatri, et al., 2016; Wang, 2014). The MICALs also play essential cellular roles through their ability to control actin organization in multiple cell types (Manta and Gladyshev, 2017; Wilson, et al., 2016), including within developing Drosophila bristle cells (Hung, et al., 2010), which have long provided a simple high-resolution single cell model for characterizing actin dependent events in vivo (Figure 1D; (Hung and Terman, 2011)). We therefore employed the bristle cell to determine if Abl is involved in Mical-mediated actin alterations.
Bristle process elongation is actin dependent such that bristles are formed when a bristle cell orchestrates actin assembly to extend an unbranched, slightly curved, cellular protrusion (Figure 1D). Mical localizes prominently to growing bristles tips (Figure 1E; (Hung, et al., 2010)), and Abl, likewise, localized to these extending tips, strongly co-localizing with Mical (Figure 1E). Turning to genetic analyses, we also found that Abl homozygous mutants displayed the straight, thick, blunt, and/or bent bristles that characterize Mical mutants and their altered F-actin organization (Figures S1E–G). Transheterozygous Abl and Mical (double heterozygotes (Abl+/− and Mical+/−)) mutants, likewise, displayed increased cell morphological defects when compared to either Mical or Abl heterozygotes alone (Figure S1H), providing additional genetic evidence that Abl and Mical function in the same signaling pathway in these cells. To further examine the cellular effects of this Mical and Abl genetic interaction, we increased Mical levels specifically in bristles, which generates F-actin disassembly and reorganization that produces a branched bristle (Figures 1F, 1H, and 1I; (Hung, et al., 2010)). Notably, decreasing Abl levels (Abl heterozygous mutants (Abl+/−)) suppressed this Mical-mediated F-actin disassembly/cellular remodeling, decreasing the length and number of bristle branches (Figures 1I and S1I). Loss of Abl (Abl homozygous mutants, Abl−/−) further suppressed Mical-mediated F-actin disassembly and remodeling, dramatically restoring bristles to a near-normal unbranched appearance (Figures 1F–I and S1J and compare to wild-type in Figure 1D). Narrowing down the specific PxxP-SH3 domain interacting region between Mical and Abl (Figure 1J) and generating transgenic flies with mutations altering these Mical-Abl interactions (which we called MicalKKLKK; Figure 1J–K), also significantly decreased Mical’s ability to induce F-actin/cellular remodeling (Figures 1K and S2A–B). Collectively, therefore, our results reveal that Abl is required in vivo for Mical-mediated F-actin disassembly and cellular remodeling, and support a hypothesis that an interaction with Abl is critical for Mical’s ability to orchestrate actin alterations and cellular remodeling.
Abl’s Kinase Activity Directly Enhances Mical-mediated F-actin Disassembly
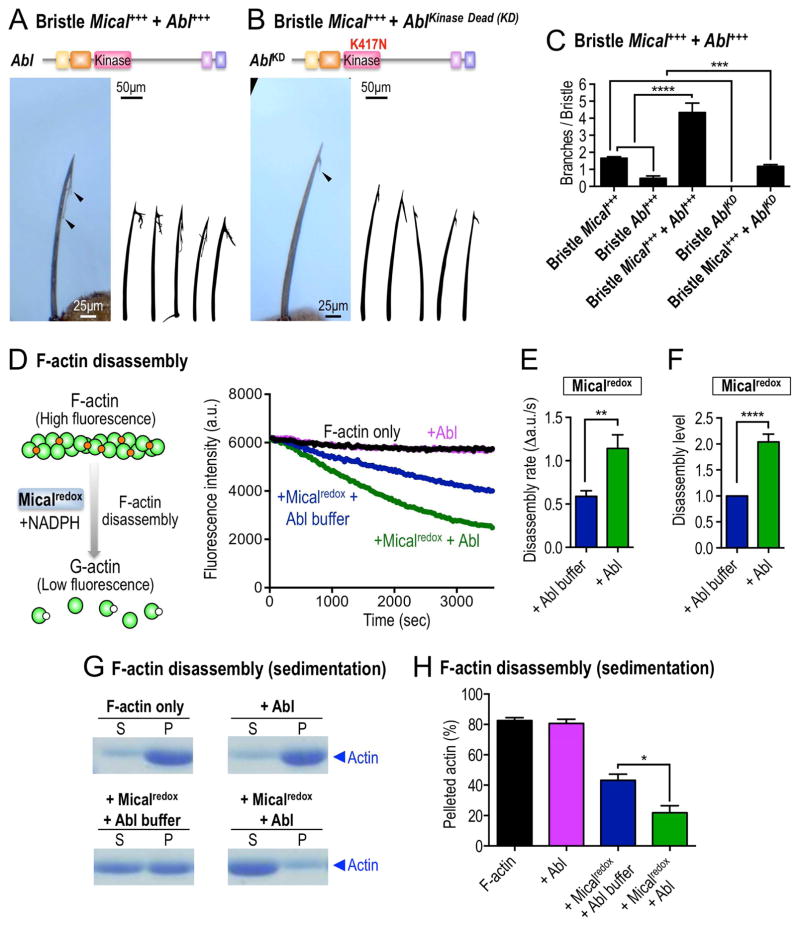
A number of known Abl-SH3-binding proteins are also substrates for the Abl kinase (Colicelli, 2010), so in light of the physical and functional interaction we observed between Abl and Mical, we investigated if Abl uses its kinase activity to regulate Mical’s effects on actin/cellular remodeling. We first examined if Abl’s kinase domain is required for Mical-mediated F-actin alterations in vivo. Employing the bristle system, we found that increasing Abl levels specifically in bristles dramatically enhanced Mical-mediated actin alterations/cellular remodeling – generating bristles with more and longer branches (Figures 2A and 2C and compare to Figure 1F). In contrast, bristle-specific expression of a kinase dead version of Abl (AblKD), in which Abl’s catalytically critical lysine 417 was mutated to asparagine (K417N), not only did not enhance Mical-dependent actin alterations/cellular remodeling, it suppressed Mical’s effects (Figures 2B–C and compare to Figure 1F). Furthermore, raising Abl levels alone, but not AblKD alone, in bristles was sufficient to generate branching (Figures 2C, S2C–D), and these cellular effects were suppressed by decreasing Mical levels (Figure S2E). Decreasing Mical levels in an AblKD background, in contrast, generated bent bristle defects that resembled both Mical and Abl homozygous mutants (Figure S2F). These results therefore support the necessity and sufficiency for Abl and its kinase function in Mical-mediated actin alterations and cellular remodeling.
Figure 2. Abl augments Mical-mediated actin disassembly.
(A–C) Abl modulates Mical-mediated actin alterations and cellular remodeling in vivo in a kinase-dependent manner. (A, C) Increasing the bristle levels of Abl enhances Mical-induced F-actin/cellular remodeling and generates bristles with longer and more complex branches (e.g., arrowhead, compare with Figure 1F). (B–C) Bristle specific expression of a kinase inactive mutant of Abl (AblKD) suppresses Mical-induced F-actin alterations/cellular remodeling (e.g., arrowhead; compare with Figure 1F). n≥33 bristles/genotype; mean±s.e.m; t-test; ****P≤0.0001; ***P≤0.001. (D–F) Abl uses its kinase activity to directly promote Mical-mediated F-actin disassembly. Pyrene-labeled actin depolymerization assays using purified proteins, in which Mical’s ability to disassemble F-actin could be followed in real-time, reveal that F-actin disassembles (decreasing fluorescence) in the presence of Micalredox (blue, preincubated with Abl kinase buffer). Pre-incubation of Micalredox with the Abl kinase increases the rate and extent of Mical-mediated actin depolymerization (green, preincubated with Abl kinase). Abl kinase alone does not affect actin depolymerization (pink). Mical’s co-enzyme NADPH was present in all conditions. n=7; mean±s.e.m.; t-test; **P≤0.01; ****P≤0.0001. [Mical]=50nM, [NADPH]=100μM. (G–H) Actin sedimentation assays, using the same experimental strategy described in Figures 2D–F, also reveal that Abl enhances Micalredox-mediated F-actin disassembly (in which the actin present in the S fraction is from disassembled filaments). S, supernatant (G-actin); P, pellet (F-actin); n=3; mean±s.e.m.; t-test; *P≤0.05. [Mical]=50nM, [NADPH]=100μM.
To more directly examine the Abl kinase effect on Mical, we turned to in vitro actin biochemical assays, where we had previously identified that purified Mical protein in the presence of its NADPH co-enzyme directly induces F-actin disassembly (Figure 2D (left); (Hung, et al., 2010)). Strikingly, using purified proteins corresponding to the F-actin regulatory redox domain of Mical and the kinase domain of Abl, we found that the active Abl kinase (in the presence of ATP) significantly increased the rate and extent of Mical-mediated F-actin disassembly (Figures 2D–F). Further analyses revealed that purified active Abl kinase alone had no effects on F-actin disassembly (Figure 2D), but its effects on Mical-mediated F-actin disassembly were concentration dependent (Figure S2G). We saw similar results using purified proteins and F-actin sedimentation assays (Figures 2G–H), further demonstrating that the Abl kinase directly increases Mical-mediated F-actin disassembly.
Abl Phosphorylates Mical’s Actin-Regulatory Redox Domain to Amplify Mical’s Catalytic F-actin Disassembly Activity
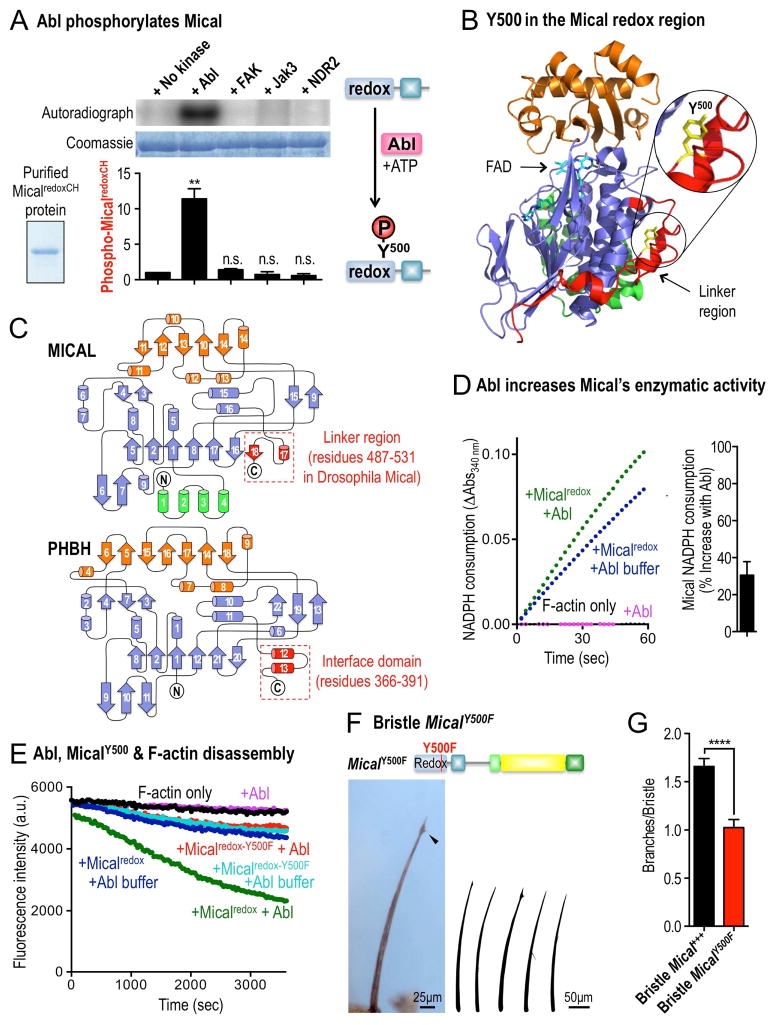
To better understand this Abl kinase-mediated enhancement of Mical F-actin disassembly, we turned to in vitro kinase assays to determine if the Abl kinase directly phosphorylates Mical. Using purified proteins, we found that Mical is specifically and selectively phosphorylated by Abl (Figures 3A and S3A–C). Furthermore, mass spectrometry revealed that a site of this phosphorylation occurs on a phylogenetically-conserved tyrosine (Y500) residue within the linker region of the actin-regulatory redox domain of Mical (Figures 3B, S3D–E, and S4A). While nothing is known of the importance of this residue in MICAL family redox enzymes, residues within the related linker/interface region of the Redox domain of p-hydroxybenzoate hydroxylase (PHBH) (Schreuder, et al., 1989) – the enzyme most closely related to Mical (Siebold, et al., 2005; Terman, et al., 2002) – play a critical role in regulating the enzymatic activity of PHBH (Figure 3C; (Moran, et al., 1997; van der Bolt, et al., 1997; Lah, et al., 1994; Palfey, et al., 1994; Eschrich, et al., 1993; Entsch, et al., 1991)). Therefore, we wondered if phosphorylation of Y500 might regulate Mical’s catalytic activity. Since Mical is a redox enzyme that utilizes the co-enzyme NADPH to oxidize its F-actin substrate (Hung, et al., 2011), we monitored Mical’s catalytic activity by following its NADPH consumption. Notably, Mical’s F-actin dependent enzyme activity increased 30% in the presence of the Abl kinase (Figures 3D and S4B), indicating a biochemical means by which Abl phosphorylation could affect Mical-mediated F-actin alterations. To further test this hypothesis, we purified Mical protein with a mutation of the Y500 residue to the chemically related phenylalanine (F) residue, and examined the effect of this Micalredox-Y500F protein on F-actin disassembly. Notably, we found that although purified Micalredox-Y500F protein behaved similarly to purified unmodified Micalredox protein (Figures 3E, S4C–D), it was no longer affected by Abl (Figures 3E and S4D). Moreover, expression of a full-length Mical with a single substitution of this residue (MicalY500F) also suppressed Mical-mediated effects on F-actin alterations/cellular remodeling in vivo (Figures 3F–G, S4E). Thus, Abl phosphorylates Mical’s Y500 residue and this phosphorylation enhances Mical’s F-actin disassembly activity.
Figure 3. Abl phosphorylates Mical’s Redox enzymatic domain to potentiate Mical-mediated actin disassembly.
(A) Purified MicalredoxCH protein (inset, Coomassie staining) is phosphorylated by purified Abl tyrosine kinase as determined by incorporation of 32P (left) – and mass analyses identified that the Y500 residue within Mical’s redox enzymatic portion is phosphorylated by Abl (right, see also Figures S3D–E). Coomassie staining shows the total amount of MicalredoxCH protein included. Other kinases (FAK, Jak3, and NDR2) show no appreciable phosphorylation of MicalredoxCH. n=3/kinase; mean±s.e.m.; t-test; **P≤0.01; n.s.=not significant. (B) The Y500 residue of Mical (yellow) that is phosphorylated by Abl resides in the linker region (red) of Mical’s redox enzymatic portion. Protein Data Bank (PDB) ID is 2BRY. (C) Mical’s Y500 residue within the linker region of its redox domain. Both the redox domain of mouse MICAL-1 (upper) and the Redox region of PHBH (lower) are shown. Mical’s linker region is comparable to the interface domain of PHBH, whose topology most closely resembles Mical’s redox domain. The interface domain is necessary for the activation and stability of PHBH’s enzyme function, while nothing is known of the role of the linker region in MICAL family enzymes. The linker region in mouse MICAL-1 corresponds to amino acid residues 445-489. (D) Mical’s enzymatic activity (as determined by conversion of NADPH to NADP+, which is measured by a change in absorbance at 340 nm [NADPH Consumption]) is markedly accelerated by pre-incubation with Abl kinase. [Mical]=50nM, [Abl kinase]=200U, [NADPH]=100μM, n=4; mean±s.e.m. (E) Pyrene-labeled actin depolymerization assays as in Figure 2D. Substitution of Mical’s Y500 residue to F500 (phenylalanine) abolishes Abl’s ability to enhance Micalredox-mediated F-actin disassembly (compare green and red). Note also that in the absence of Abl, the F-actin disassembly activity of Micalredox-Y500F is similar to Micalredox (compare blue and light blue), indicating that the Micalredox-Y500F protein is equally active and has been added at a similar activity level as wild-type Micalredox protein. (F–G) Expressing full-length Mical with a single substitution mutation of the Abl Y500 phosphorylation site (MicalY500F) decreases Mical-mediated F-actin alterations/cellular remodeling (arrowhead, drawings, and graph). n≥41 bristles/genotype; mean±s.e.m.; t-test; ****P≤0.0001.
Abl Amplifies Mical-mediated Cellular Remodeling in Response to Semas/Plexins and Growth Factors
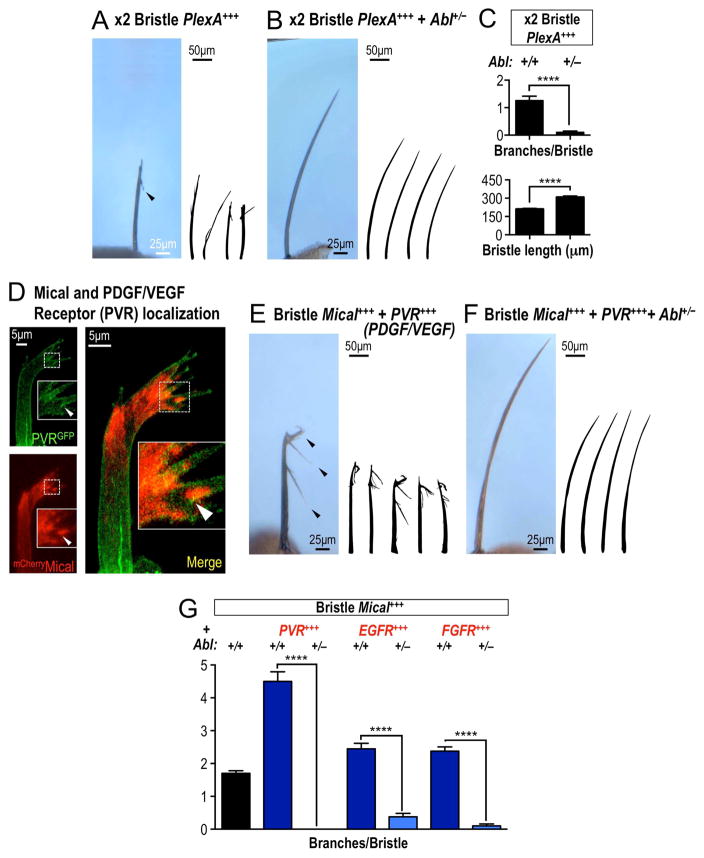
Abl family kinases are best known as functioning with a host of different growth factors/chemoattractants including PDGF, VEGF, HGF, NGF, and EGF (reviewed in (Khatri, et al., 2016; Wang, 2014)), which activate Abl to phosphorylate its targets. Interestingly, Mical has also been implicated in working with different growth factors including HGF, VEGF, EGF, and NGF (Deng, et al., 2016; Hou, et al., 2015; Lundquist, et al., 2014; Ashida, et al., 2006), but the role of these growth factors in regulating Mical-mediated F-actin disassembly is unknown. In a reciprocal way, Mical is best known to carry out the F-actin destabilizing effects of the cellular repellent Sema and its Plexin receptor (Hung, et al., 2010; Terman, et al., 2002) – and Abl, likewise, has been implicated in Sema/Plex signaling but its role in this process is unclear (Procaccia, et al., 2014; O’Connor, et al., 2008; Shimizu, et al., 2008; Moresco, et al., 2005; Toyofuku, et al., 2004). Therefore, we sought to better define these interactions, and did so by first using the bristle process, since our previous results revealed that increasing the Plexin A receptor (PlexA) specifically in bristles generates F-actin disassembly/cellular remodeling in a Mical-dependent manner (Figures 4A and 4C; (Hung, et al., 2010)). Notably, we found that these cellular changes were also dependent on Abl, such that decreasing Abl levels (Abl+/−) suppressed PlexA-mediated F-actin alterations/cellular remodeling, decreasing the number of branches and restoring bristles to a more normal length (Figures 4B–C). We also found that increasing Abl levels enhanced PlexA-induced bristle cellular changes (Figures S5A–C), revealing that Abl is both necessary and sufficient to modulate the actin disassembly/cellular remodeling abilities of Sema/Plex/Mical signaling. We therefore next asked if growth factor signaling could also regulate Abl/Mical-mediated F-actin disassembly and cellular remodeling. Since loss-of-function mutations in growth factor receptors such as PDGF/VEGF (PVR), EGF, and FGF lead to defects in bristle morphology and development (e.g., (Mummery-Widmer, et al., 2009; Culi, et al., 2001; Abdelilah-Seyfried, et al., 2000; Price, et al., 1997)) – we again employed the simple single cell bristle as an in vivo “F-actin disassembly/turning/branching assay” to determine the effects of increasing growth factor signaling on Mical-mediated F-actin alterations. Our results revealed that Mical and growth factor receptors are enriched in close proximity (including areas of overlap) in bristles (Figure 4D), and increasing the levels of signaling-competent growth factor receptors including PVR, EGF, or FGF in bristles significantly enhanced Mical-mediated F-actin alterations/cellular remodeling (Figures 4E and 4G, S5D). Dramatically, decreasing Abl levels (Abl+/−) suppressed these growth factor receptor-dependent effects on Mical-mediated F-actin/cellular alterations (Figures 4F–G). Together, these results indicate that Abl works to carryout the effects of both Semas/Plexins and growth factors on Mical-mediated F-actin disassembly and cellular remodeling.
Figure 4. Abl amplifies Mical-mediated cellular remodeling in response to Sema/Plexin repellents and growth factors.
(A–C) Abl is required for Sema/Plex-mediated actin alterations/cellular remodeling. (A, C) Bristles become branched (arrowhead) and shorter when high levels (x2) copies of PlexA are expressed specifically within them. (B–C) Decreasing Abl levels (Abl+/−) strongly suppresses these Sema/Plex-mediated effects. (C, upper) n≥39 bristles/genotype; mean±s.e.m.; t-test; ****P≤0.0001. (C, lower) n≥9 bristles/genotype; mean±s.e.m.; t-test; ****P≤0.0001. Abl+/−=Abl4/+. (D) The transmembrane receptor PVR (green) localizes along the plasma membrane of extending bristles and is enriched in close approximation and overlap (yellow areas, e.g., arrowhead in higher magnification image in inset) with sites of prominent Mical cytosolic localization (red). (E–G) Growth factor receptors enhance Mical-mediated F-actin alterations/cellular remodeling, and these effects are suppressed by decreasing Abl levels. (E, G) Bristles become heavily branched (e.g., arrowheads) when growth factor receptors and Mical are co-expressed in bristles. (F–G) Decreasing Abl levels (Abl+/−) strongly suppresses these growth factor receptor/Mical-mediated effects. Bristle-specific expression of these growth factor receptors alone has no effects on bristle branching under these conditions (not shown). n≥40 bristles/genotype; mean±s.e.m.; t-test; ****P≤0.0001. Abl+/−=Abl4/+.
Growth Factor/Abl Signaling Amplifies Sema/Plexin/Mical Signaling in Repulsive Axon Guidance
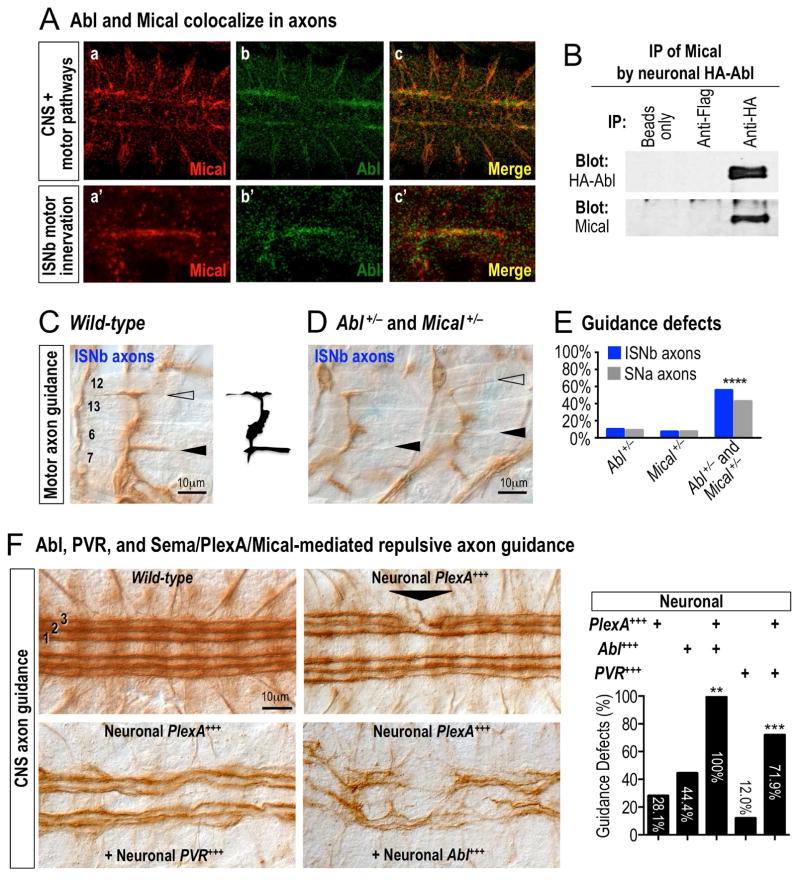
Since both Mical and Abl are broadly expressed (Khatri, et al., 2016; Wilson, et al., 2016; Thompson and Van Vactor, 2006; Moresco and Koleske, 2003), including within similar patterns in the developing nervous system (e.g., (Terman, et al., 2002; Bennett and Hoffmann, 1992)), we also employed the Drosophila nervous system as a model to further examine the convergence of Mical, Abl, Sema/Plex, and growth factor signaling. Similar to what we had observed in the bristle model, both Mical and Abl colocalized within developing neurons and their axons (Figure 5A) – and they could also be coimmunoprecipitated from them (Figure 5B). We next turned to functional assays in neurons, since Mical repulsive signaling is required for Drosophila motor axons to selectively defasciculate and innervate their muscle targets (Figure S5E and compare to wild-type in Figure 5C; (Terman, et al., 2002)). Abl−/− mutants, likewise, exhibited these same types of motor axon guidance defects (Figure S5F; (O’Donnell and Bashaw, 2013; Wills, et al., 1999)) – defects that were not only enhanced by decreasing Mical levels (Figure S5G), but were also observed in Mical and Abl transheterozygous mutants (Figures 5D–E). Thus, these loss-of-function genetic assays indicate that Mical and Abl function in the same signaling pathway to mediate axon guidance.
Figure 5. Abl amplifies Mical-mediated repulsive axon guidance in response to Sema/Plexin repellents and growth factors.
(A) Mical and Abl exhibit overlapping expression and co-localization in the developing Drosophila embryonic nervous system. Mical (Mical antibody, red) and Abl (genomic GFP-Abl, green) co-localize (yellow) in both CNS and motor axon pathways. (B) Mical and Abl associate within neurons in vivo. Drosophila embryos expressing HA-Abl specifically in neurons were subjected to immunoprecipitation (IP), and antibodies against HA-Abl (top) IP Mical (bottom), while controls (beads only or FLAG antibodies) do not. (C–E) Abl and Mical work together to guide axons in vivo. (C) As depicted in the image and the drawing, wild-type intersegmental nerve b (ISNb) motor axons defasciculate from the main nerve and characteristically innervate muscles 6 and 7 (filled arrowhead) and muscles 12 and 13 (open arrowhead). (D, E) In comparison to either heterozygote alone (which are similar to wild-type), note the absence/abnormal (arrowheads) innervation of muscles 6/7 and 12/13 in embryos heterozygous for mutations in both Abl (Abl1) and Mical (MicalDf(3R)swp2) (Abl+/− and Mical+/−) (D) and quantified in (E) for ISNb motor axons and another motor axon pathway (segmental nerve a (SNa)). n≥100; X2 test; ****p≤0.0001. (F) Abl, PVR growth factor receptor, and Sema/Plex/Mical work together in repulsive axon guidance. Axons within the Drosophila CNS exhibit a characteristic organizational pattern of three (1, 2, 3) longitudinal fasciculated bundles of Fasciclin II (1D4)-positive axons (wild-type). Increasing PlexA levels in neurons (Neuronal PlexA+++) alters the fasciculation and pathfinding of these longitudinal axons (arrowhead). Increasing the levels of PVR or Abl in neurons significantly enhances these Sema/Plex/Mical-dependent repulsive guidance effects. All genotypes are heterozygous (ELAV-GAL4/+, UAS:HAPlexA/+, and/or transgenes/+). UAS:Abl only was similar to that previously described by others (e.g., (Hsouna, et al., 2003; Bashaw, et al., 2000)). Graph=percentage (%) of animals with defects in the 1st or 2nd longitudinals. n=16–75 animals/genotype; X 2 test; **p≤0.01; ***p≤0.001. Scale bar applies to all images.
We therefore turned to gain-of-function axon guidance assays that are dependent on the repulsive effects of Sema/Plex/Mical signaling to further define these interactions. Increasing Mical levels specifically in neurons generates increased motor axon-axon defasciculation/repulsion (Hung, et al., 2010; Terman, et al., 2002) – and we found that decreasing Abl levels suppressed these Mical-mediated repulsive effects (Figure S5H). In a similar way, increasing PlexA levels specifically in neurons generates increased central nervous system (CNS) axon-axon defasciculation/repulsion (Figure 5F; (Hung, et al., 2013; Yang and Terman, 2012)) – and we found that raising the levels of Abl or growth factor receptors enhanced these Sema/PlexA/Mical-dependent repulsive effects (Figure 5F). Our neuronal results therefore support our work in the bristle model and with purified proteins, and indicate that Mical and Abl functionally interact, converging with both Semas/Plexins and growth factors to direct repulsive axon guidance.
MICAL functions with Abl in tumor growth and progression – sensitizing cancer cells to treatment with the Abl kinase inhibitor Gleevec
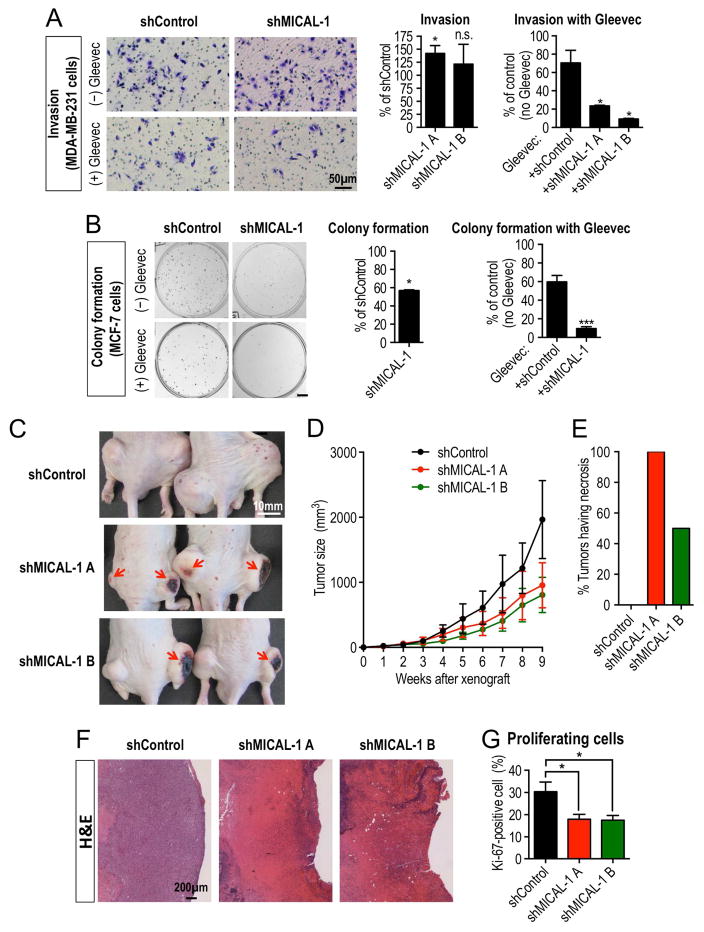
In addition to Abl and Mical’s critical role in multiple physiological processes, Abl kinases contribute pathologically to multiple different cancers including leukemia, where Gleevec (imatinib mesylate, STI571), an Abl kinase inhibitor, is a well-known first-line therapeutic for Abl-related cancers (Greuber, et al., 2013). Interestingly, Mical also has ties to several of these Abl-related cancers (Mariotti, et al., 2016; Loria, et al., 2015; Ashida, et al., 2006), so we examined if MICAL might be linked to Abl’s carcinogenic effect. We tested this hypothesis using breast cancer cell lines as a model, because active Abl kinases are required for the invasion, growth, proliferation, and survival of these cancer cells (Ganguly and Plattner, 2012) – and genetic alterations including gene amplifications and mutations in MICALs have also been found in breast cancer cells/patient-derived xenografts (Figure S6A; (Eirew, et al., 2015; Rodenhiser, et al., 2008; Lin, et al., 2007; Sjoblom, et al., 2006)). First, using a highly invasive breast cancer cell line (MDA-MB-231 cells) (Ganguly and Plattner, 2012), we found that one of the human MICALs, MICAL-1, was expressed within these cancer cells (Figure S6B), and also co-immunoprecipitated with Abl in a growth factor serum-stimulation and PDGF-dependent manner (Figures S6C–D). Likewise, knockdown of MICAL-1 generated actin aggregation and morphological defects within these cancer cells (Figures S6E–F) – and made them dramatically more sensitive to the Abl kinase inhibitor Gleevec, including promoting an ~60% reduction in invasion when compared to Gleevec treatment alone (Figures 6A and S6G). We also saw similar effects following MICAL-1 knockdown in another breast cancer cell line – non-invasive, colony-forming MCF-7 cells (Greuber, et al., 2013) – including altered morphology (Figures S6H–I) and a dramatic increase in sensitivity to Gleevec treatment (Figure 6B).
Figure 6. Mical and Abl interactions regulate cancer cell biology.
(A–B) Decreasing MICAL-1 levels makes cancer cells more susceptible to the Abl kinase inhibitor Gleevec. (A) The Abl cancer drug Gleevec is more efficacious on inhibiting MDA-MB-231 cancer cell invasion when MICAL-1 levels are also decreased. MDA-MB-231 breast cancer cells infected with a lentiviral vector expressing an shRNA against non-silencing or MICAL-1 (see also Figure S6B) were pre-treated with either vehicle (DMSO) or Gleevec (10μM) in serum-free medium for 18 hrs. Cells were then plated in the upper well of matrigel-coated transwell (see Figure S6G), and after 48 hrs of incubation, the invaded cells were stained and counted. (A, left graph): n=3 independent experiments; t-test (compared with shControl). (A, right graph): n=4 random field view from 2 independent experiments; t-test (compared with no Gleevec); mean±s.e.m; *P≤0.05; n.s. not significant. (B) Gleevec inhibits MCF-7 cancer cell colony formation more efficiently when MICAL-1 is decreased. To measure the ability of a single cell to grow into a colony (colony formation assay), MCF-7 cells, which are highly differentiated, noninvasive breast cancer cells, were cultured after infection with a lentiviral vector expressing an shRNA against non-silencing or MICAL-1 (see also Figure S6H). Either vehicle (DMSO) or Gleevec (6μM) was added 24 hrs after plating. After 11 days of incubation, the colonies were stained and counted. Colonies were defined as clusters of >50 cells. (B, middle graph): n=3 independent experiments; t-test (compared with no shControl). (B, right graph): n=3 independent experiments; t-test (compared with no Gleevec); mean±s.e.m; *P≤0.05; ***P≤0.001. Scale bar=5mm. (C–G) Similar to treatment with the Abl kinase inhibitor Gleevec, MICAL-1 knockdown decreases xenograft tumor size – increasing tumor necrosis and decreasing cancer cell proliferation in vivo. (C–E) MICAL-1 knockdown decreases MDA-MB-231 cancer cell tumor size and increases tumor necrosis. (C) Following injection of MDA-MB-231 cancer cells stably expressing shRNA against non-silencing (control) or MICAL-1 into the flanks of nude mice, MICAL-1 knockdown tumors appear smaller in size (imaged at nine weeks following injection) compared to control knockdown tumors and have distinct dark areas of necrosis (arrows). (D–E) Isolated MICAL-1 knockdown tumors dissected nine weeks following injection also show a decreased size compared to controls (D, n=4/genotype; mean±s.e.m) and have severe surface necrosis (E, n=4/genotype). (F–G) MICAL-1 knockdown tumors are characterized by cell death and decreased cellular proliferation. (F) Hematoxylin and eosin (H&E) histological staining from control or MICAL-1 knockdown tumors. Control tumors are enriched with viable cancer cells with intact nuclei (hematoxylin, purple). In contrast, MICAL-1 knockdown tumors have large regions, which are enriched with cell debris/ghost cells (eosin, pink) that are consistent with cell death. (G) MICAL-1 knockdown tumors have a significant decrease in the percentage of proliferating cells (Ki-67 positive cells) compared to control tumors (see also Figure S7B). n=6/genotype; mean±s.e.m.; t-test (compared with shControl); *P≤0.05.
We therefore turned to robust in vivo assays, where injection of MDA-MB-231 breast cancer cells into immunodeficient nude mice produce tumors that are significantly reduced in size by treatment with the Abl kinase inhibitor Gleevec (e.g., (Blanchard, et al., 2014; Gil-Henn, et al., 2013; Schito, et al., 2012; Hiraga and Nakamura, 2009)). Likewise, injections of two independent MICAL-1 stable knockdown MDA-MB-231 breast cancer cell lines into immunodeficient nude mice revealed tumors that were significantly smaller than those formed from control cells (Figures 6C–D and S7A). Further histological examination revealed that MICAL-1 knockdown tumors had distinctive dark regions that were indicative of severe tumor necrosis (Figures 6C and 6E) – including a loss of nuclear staining, prominent cell death, and a decrease in the percentage of proliferating breast cancer cells (Figures 6F–G and S7B). Taken together, our observations indicate that MICAL proteins also function with Abl in tumor growth and progression, and decreasing MICAL levels sensitizes cancer cells to treatment with the Abl kinase inhibitor Gleevec.
DISCUSSION
Cellular behaviors are controlled by extracellular cues that have been historically classified into two independently acting and antagonistic groups: growth-promoting/attractants and growth-preventing/repellents. Our biochemical, genetic, and cellular assays herein now raise important concepts related to the role of these two groups of cues and how cells interpret and respond to them. Namely, we have identified a convergence of Mical and Abl signaling – with effects that serve to amplify Mical-mediated F-actin disassembly and repulsion in response to Sema/Plexin repellents and growth factors. Paradoxically, our results thus reveal contexts in which growth factor/chemoattractant signaling promotes repulsion/inhibition, thereby joining together these two disparate classes of cues for a common effect and reshaping our understanding of the role that chemoattractants and chemorepellents play in orchestrating cellular behaviors.
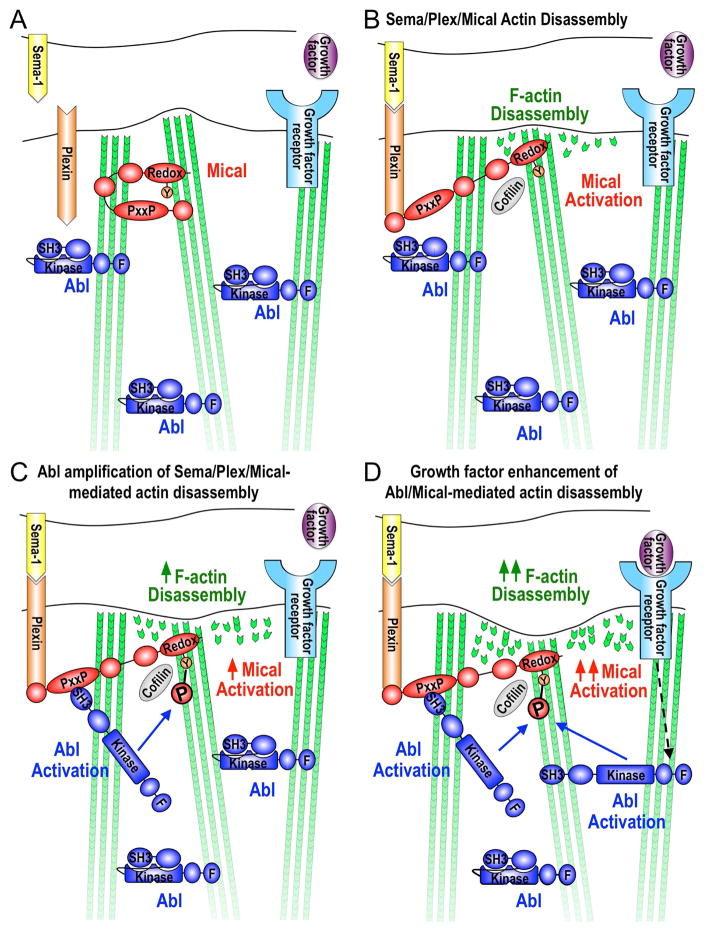
Abl is known to be activated by a diverse array of cell surface receptors, cytosolic proteins, and intrinsically generated stimuli such as DNA damage and oxidative stress (Khatri, et al., 2016; Wang, 2014). The specificity in Abl’s action is thought to be accomplished through its restricted activation, such that while being broadly expressed, Abl’s SH3 domain binds an internal PxxP motif and this intramolecular SH3/PxxP interaction locks Abl in an autoinhibitory state (Figure 7A; (Wang, 2014; Nagar, et al., 2003)). Activation of Abl occurs when a PxxP-containing protein binds to Abl’s SH3 domain, disrupting Abl’s intramolecular PxxP/SH3 interaction and thereby allowing Abl to specifically phosphorylate different proteins and regulate multiple different biological processes in a localized manner (Wang, 2014; Nagar, et al., 2003). Our results are consistent with such a model and support that Abl is positioned such that when Mical is activated by Sema-Plex signaling, Mical uses its PxxP motif to bind to Abl’s SH3 domain, and thereby (in a similar manner to other proteins binding to Abl’s SH3 domain; (Wang, 2014; Nagar, et al., 2003)) we propose activating Abl to phosphorylate Mical and enhance Mical’s F-actin disassembly/repulsive activity (Figures 7B–C). We also find that in the presence of other activators of Abl signaling, such as specific growth factor receptors, Mical-mediated F-actin disassembly/repulsion can be further amplified and this is dependent on the presence of Abl (Figure 7D). Our results therefore support a hypothesis that under conditions in which Sema/Plex activation is coupled with growth factor signaling, additional Abl becomes activated, allowing for the further amplification of Mical-mediated F-actin disassembly/cellular repulsion (Figure 7D).
Figure 7. Model of Abl amplification of Mical-driven F-actin disassembly.
(A) Intramolecular interaction between Mical’s N-terminal and C-termini keeps Mical inactive (Manta and Gladyshev, 2017; Wilson, et al., 2016). Abl is also kept inactive through intramolecular interactions between Abl’s SH3 domain and an internal PxxP motif (Wang, 2014; Nagar, et al., 2003). (B) Activation of Sema-Plex signaling induces Mical’s activation, which directly disassembles F-actin through oxidation of the Met44/Met47 residues at the pointed end of actin subunits (Hung, et al., 2013; Hung, et al., 2011; Hung, et al., 2010; Terman, et al., 2002). Mical-oxidized actin also recruits cofilin to help in F-actin disassembly (Grintsevich, et al., 2016). (C) Herein, our results propose a model that Mical uses its proline-rich region to associate with Abl’s SH3 domain. We also propose that (in a similar manner to other proteins binding to Abl’s SH3 domain; (Wang, 2014; Nagar, et al., 2003)) this intermolecular interaction activates Abl to phosphorylate the Y500 residue within Mical’s actin regulatory redox domain and enhance Mical oxidation-mediated F-actin disassembly. (D) Our results also support that growth factor receptor signaling enhances Mical-mediated actin disassembly in an Abl-dependent manner. These observations are consistent with a model that under conditions when growth factor signaling is coupled with Sema/Plex activation, additional Abl becomes activated, allowing for the phosphorylation of additional amounts of Mical protein and further amplification of Mical-mediated F-actin disassembly. These results therefore also support a model that Mical activation through Sema/Plex signaling can be amplified when it takes place in combination with other extracellular signals such as growth factors. It is also interesting to note that Abl is inhibited through its interactions with actin filaments (Wang, 2014). These previous observations provide an intriguing addition to the model presented herein: that the Mical-induced dismantling (loss) of F-actin may also serve to activate additional Abl, which would allow for further phosphorylation/amplification of Mical-mediated F-actin disassembly. This Mical-induced disassembly of F-actin may also serve to activate Abl to phosphorylate additional F-actin regulatory proteins such as cofilin, cortactin, Ena/VASP, and WAVE (Khatri, et al., 2016; Courtemanche, et al., 2015; Wang, 2014). In addition, it should also be noted that while exuberant growth in response to the application of a particular extracellular cue in culture has historically led to the classification of a cue as a growth-promoting factor (i.e., growth factor/chemoattractant or attractive guidance cue), our results indicate that such a classification may be misleading or not tell the full story for that particular cue. For example, what looks like exuberant growth in culture could be the result of an excessive dismantling of F-actin and the sending out of filopodia/neurites in new directions. Thus, such an extracellular cue might not be a growth promoter (F-actin assembler) but an F-actin disassembler that limits growth in a particular direction. Another example would be using a culture assay to find that a given extracellular cue could amplify the effects of a given growth-promoting cue. Such a cue might then be classified as a growth promoter. However, given a different context, this same extracellular cue might amplify the effects of a growth-preventing cue. Thus, classification of such a cue as a growth-promoter or a growth-preventer would be inaccurate (because it can do both depending on the context and what other cues are present). Our results using simple in vivo, in vitro, and biochemical systems with purified proteins support contexts in which growth factors/chemoattractant signaling can enhance F-actin disassembly. Thus, this raises concerns about indiscriminately applying growth factors/chemoattractants in order to stimulate axon growth (for example), because the results could amplify F-actin disassembly (and enhanced branching in a given area, for example) and not lead to the intended effect of promoting F-actin assembly-driven long-term growth/extension of axons.
Our observations therefore raise questions into the simplistic groupings of attractive and repulsive cues by discovering contexts in which growth factor/chemoattractant signaling cascades work in an opposite manner to how they are thought to act and promote repellent-linked effects such as F-actin disassembly. In particular, in order to better understand the signal transduction pathways that form, elongate, shape, and orient/guide the membranous extensions of cells, we have sought to couple sensitive biochemical assays using purified proteins with clear single cell in vivo genetic and cellular models. Herein, our biochemical and in vivo observations support a role for Abl in promoting Mical-mediated F-actin disassembly. Likewise, our in vivo data support a role for Mical and Abl in Sema/Plex repulsive signaling. Yet, our in vivo cellular and genetic data also support a role for growth factors/chemoattractants – which are factors that are typically considered positive effectors of cytoskeletal assembly/growth/movement (i.e., enhancers of F-actin assembly/motility) – in amplifying the F-actin disassembly effects of Mical. Thus, our results are consistent with the idea that cues thought to function primary as positive effectors of movement (growth factors/attractants) can at least in some contexts serve to enhance the F-actin disassembly activity of repellents. Likewise, our observations may add to our understanding of why specific extracellular guidance cues have been characterized as having dual-functionality – being described as both attractants and repellents (Kolodkin and Tessier-Lavigne, 2011). Historically, this dual-functionality has been thought to be due to the activation of different receptors but our results indicate that such effects may also be related to the context in which they are functioning. Indeed, proteins such as Abl have long been considered to have different biological effects in different cells and in a context-dependent manner (Khatri, et al., 2016; Wang, 2014). In conclusion, therefore, understanding these context-dependent differences are not only critical for deciphering developmental events and physiological processes, but also for selecting appropriate therapeutic combinations that promote positive effects on cell movement versus inadvertently promoting cell repulsion/inhibition (e.g., for spinal cord injury/neurodegenerative disease treatment) – or vice versa (e.g., for cancer treatment) (Figure 7D).
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jonathan R. Terman (jonathan.terman@utsouthwestern.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Mice were housed and treated in accordance with The University of Texas Southwestern Institutional Animal Care and Use Committee. Likewise, in vivo tumor xenograft experiments were done on 5- to 6-week old female athymic nude mice – and all procedures and experiments were approved by The University of Texas Southwestern Institutional Animal Care and Use Committee and conducted according to the institutional guidelines using standard approaches as previously employed (Mender, et al., 2015) with both sexes of littermate or age-matched mice.
Fruit Flies
Flies were maintained and genetic crosses were performed at 25’C following standard procedures.
Yeast
Yeast growth, approaches, and protocols were conducted at 30’C using standard techniques.
Cell Lines
Cells were cultured in basal X media (the mixture of 4 parts of high glucose DMEM and 1 part of HyClone™ medium 199 (GE Healthcare Life Sciences, Logan, UT) supplemented with 10% HyClone™ Cosmic Calf™ serum (GE Healthcare Life Sciences, Logan, UT) and were maintained in a humidified incubator at 37°C with 5% CO 2. Culture work was done using standard approaches for working with MCF-7, MDA-MB-231, and 293FT cells, which are female in origin, and all cell lines were negative for mycoplasma.
METHOD DETAILS
Yeast Two-Hybrid Screening and Interaction Assays
Yeast two-hybrid protocols were conducted using standard techniques (Golemis, et al., 2011; Terman, et al., 2002). Portions of the PxxP motif region of Drosophila Mical including amino acids 861-1539 (MicalPRNt), amino acids 1951-2550 (MicalIQP), amino acids 2171-2550, amino acids 2171-2398, or amino acids 2171-2524 and 2533-2550 were inserted into the yeast bait vector (pEG202). The QuickChange® Multi Site-Directed Mutagenesis Kit (Stratagene) was used to introduce point mutations and generate MicalKKLKK (amino acids P2529 to K2529, P2530 to K2530, P2532 to K2532, and L2533 to K2533) constructs. All resulting constructs were sequenced on both strands. cDNAs of Abl (LD03455), shot (SD19937), Trio (LD19830), Vav (LD25754), and Calcium channel β subunit (RH74212) were used to PCR-amplify portions corresponding to their SH3 domains and inserted into the prey vector (pJG4-5). The levels of bait and prey interactions were determined using standard approaches (Yang and Terman, 2012; Golemis, et al., 2011; Terman, et al., 2002) by observing color development and growth rate from day 3 to day 5 after plating on both Glucose and Galactose/Raffinose-containing media with 0.05mg/ml of X-gal.
GST pull-down assays
In vitro interaction between GST-AblSH3 and His-MicalIQP2 (amino acids 2171-2550) was examined using conventional GST pull-down assays (Yang and Terman, 2012). GST and His-tagged proteins were purified using standard approaches (e.g., (Yang and Terman, 2012)). 380μM of GST recombinant protein (GST only or GST-AblSH3) was incubated with 24μg (230μM) of His-MicalIQP2 recombinant protein in GST lysis buffer (20mM Tris-HCl, pH8.0, 1mM EDTA, pH8.8, 200mM NaCl, and 0.25% NP-40). Then, GST proteins and interacting proteins were pulled down by adding immobilized glutathione beads (Thermo Scientific) for 3hrs at 4°C on a tube rotator. Boun d proteins were detected by Western blot analysis using a His antibody (1:3,000; Roche) or a GST antibody (1:1,000; Genscript). 2% of each reaction was also analyzed with a His antibody (1:3,000; Roche) to determine that equal amounts of His-MicalIQP2 were used. Band intensity was measured using GeneTools (Syngene) and normalized to a beads-only negative control.
Co-immunoprecipitation
Co-immunoprecipitation assays were performed using standard approaches (Yang and Terman, 2012; Terman, et al., 2002). To look for co-immunoprecipitation between Abl and Mical in neurons, lysates from embryos expressing HA-Abl in neurons (ELAV-GAL4, UAS:HA-Abl embryos) were prepared in RIPA buffer (50mM Tris-HCl pH8.0, 150mM NaCl, 1% NP-40, 0.5% deoxycholic acid, 0.1% SDS) supplemented with complete protease inhibitor cocktail tablets (Roche) and PhosSTOP tablets (Roche). Co-immunoprecipitation assays were performed by adding mouse HA monoclonal antibodies (5μg; 12CA5, Roche) into the embryonic lysates. Beads only (no antibody) or mouse Flag monoclonal antibodies (5μg; M2, Sigma) were used as negative controls for immunoprecipitation. Western analysis was performed using HA antibodies (1:3,000; 3F10, Roche) and the Mical-CT antibodies (1:2,000; (Terman, et al., 2002)).
For co-immunoprecipitation assays from MDA-MB-231 breast cancer cells, cells were washed with PBS and replaced with serum-free medium overnight for serum starvation. To stimulate the cells with serum or growth factor (PDGF), the serum-free medium was replaced with 10% serum medium or medium with PDGF (5ng/ml; abcam) and the cells were incubated at 37°C for the indicated time. Serum-stimulated cells or PDGF stimulated cells were washed with PBS and lysed in IP buffer (50mM Tris-HCl, pH7.4, 120mM NaCl, 0.5% NP-40, 1mM EDTA) supplemented with complete protease inhibitor cocktail tablets (Roche) and PhosSTOP tablets (Roche). Co-immunoprecipitation assays were performed by adding rabbit Abl polyclonal antibodies (5μg; K-12, Santa Cruz Biotechnology) into the cell lysates. Normal rabbit serum (5μg; Santa Cruz Biotechnology) was used as a negative control for immunoprecipitation, as was immunoprecipitating as described above with an antibody to Abl but using lysates from MICAL-1 stable knockdown cells. Western analysis was performed using MICAL-1 antibodies (1:1,000; Proteintech) and Abl antibodies (1:1,000; BP Pharmingen).
Genetics, Molecular Biology, and Transgenic Lines
Mical pUAST flies, GFP-Mical pUAST flies, mCherry-Mical pUAST flies, Mical deficiency flies (MicalDf(3R)Swp2), and HA-PlexA flies were generated as described (Hung, et al., 2013; Hung, et al., 2010; Terman, et al., 2002). To generate MicalKKLKK pUAST flies, a DNA fragment (nucleotides 6894-7978) was removed from the Mical “short” isoform cDNA using SapI. The removed region of cDNA was replaced with a DNA fragment derived from the pEG202 MicalKKLKK construct that was used for the yeast two-hybrid interacting assays, which corresponds to the same region (nucleotides 6894-7978) including point mutations. The resultant cDNA was then moved into the pUAST vector to generate the transgenic flies. To generate MicalY500F pUAST flies, the A1499 nucleotide base was changed to T in Mical in the pOT2A vector using the QuickChange® Multi Site-Directed Mutagenesis Kit (Stratagene) following the manufacturer’s instructions. A DNA fragment (nucleotides 476-4540) including A1499 was removed from Mical in the pUAST vector using AsiSI and SpeI and was replaced with the corresponding DNA fragment derived from MicalY500F pOT2A. HA-Abl pUAST flies were generated by inserting a 3x HA tag at the N-terminus of a full-length Drosophila Abl cDNA in the pUAST vector. Multiple fly lines of UAS:MicalKKLKK, UAS:MicalY500F and UAS:HA-Abl were generated and transgenic fly lines of the same genotypes showed similar bristle defects when expressed with the bristle-specific B11-GAL4 driver. Drosophila embryo injections were done by BestGene, Inc. All other Mical and Abl stocks had been used previously in multiple other studies. All other stocks were obtained from the Bloomington Stock Center except Mical point mutation stocks (kind gifts from Hermann Aberle), B11-Gal4 (a kind gift from John Merriam), endogenous-Abl::GFP (a kind gift from Mark Peifer), DAbl pUAST and DAblK417N (DAblKD) pUAST fly lines (kind gifts from Frances Fogerty), PVR-GFP and PVRDN (kind gifts from Pernille Rørth), and GFP-Actin pUAST (Drosophila Genetic Resource Center, Japan).
Bristle Morphology and F-actin Analysis
Adult fly bristle phenotypic analyses were done as described previously (Hung, et al., 2013; Hung, et al., 2010) by crossing flies at 25°C and subjecting only recently emerged adult offspring to collection, genotyping, and qualitative and quantitative analysis under a dissecting microscope (Leica Stereo Zoom S8 APO). Specifically, bristles were scored for defects in morphology including branching, bending, and alterations to their tips as described previously (Hung, et al., 2010). We considered any deviation from the main bristle shaft as a branch. For example, a bent bristle would not be considered to have a branch because it is simply bent without any deviation. However, a “cross” shaped bristle would be considered to have a branch. The number of branches on each mutant bristle was counted and the results were presented as the mean number of branches per bristle (± the standard error of the mean (SEM)). All large bristles (macrochaetae) were examined for defects in Mical−/− and Abl−/− mutant flies. For all genetic interaction experiments, adult flies were examined for alterations to their posterior scutellar bristles, and thereby allowing precise comparison between single bristle cells from animal to animal. The levels of enhancement/suppression of Mical-mediated bristle effects were also measured in a category from −2 to 2 (e.g., Figure S5D and similar to (Hung, et al., 2013), where 0 is similar to bristle morphological alterations with Mical only, −2 is strong and −1 is mild suppression of Mical-mediated bristle morphological alterations (such as, less numbers and shorter bristle branches), and +1 is mild and +2 is strong enhancement of Mical-mediated bristle morphological alterations (such as increases in the number, length, and complexity of bristle branches). Adult bristle imaging and drawings were done with the aid of a Zeiss Discovery M2 Bio stereomicroscope, a Zeiss Axiocam HR camera, a motorized focus and zoom, three-dimensional reconstruction software (Zeiss Axiovision software and Extended Focus Software [a kind gift from Bernard Lee]), and Microsoft Office Powerpoint, as described (Hung, et al., 2010). Bristle length was determined by drawing a line on each bristle image and measuring the length of the line with the aid of Image J. Mutant analyses and genetic interactions were done with multiple different alleles.
Analysis of pupae were done similar to that described previously (Hung, et al., 2013; Hung, et al., 2010). Pupae were genotyped with the aid of Tb balancers on the second and third chromosomes and placed on double-sided tape (3M) within Petri-dishes containing wet Kimwipes to keep the pupae moist. The pupae were incubated in a 25°C incubator until they reached the desired developmental stage. For mCherry-Mical and GFP-Abl co-localization analyses and mCherry-Mical and GFP-PVR localization analyses in bristles, pupae were dissected and placed in concave well slides in Vectashield mounting medium (Vector Labs) and imaged using a Zeiss LSM 510 confocal microscope. For F-actin analysis, age-matched GFP-actin expressing pupae whose scutellar bristles had reached the length of 50–80μm were transferred to concave well slides with Vectashield mounting medium (Vector Labs) and imaged using a Zeiss LSM 510 confocal microscope. Any deviation of F-actin organization within the main shaft was considered an F-actin alteration.
Drosophila Embryonic Axon Guidance Assays
Drosophila embryos were collected, processed, staged, immunostained and dissected as previously described (Yang and Terman, 2012; Terman, et al., 2002). Embryos were collected with PBS containing 0.1% Triton (PBT) and dechorionated in 50% bleach for 5mins. Embryos were then fixed in 4% paraformaldehyde containing 50% heptane for 12mins. Following devitellization with hard shaking in methanol, embryos were then washed in PBT and incubated with the appropriate antibodies. For colocalization experiments, endogenous-Abl::GFP embryos were incubated with the Mical-CT antibody (1:2,000; (Terman, et al., 2002)). Immunostained embryos were dissected in the mounting media (50% PBS and 50% Vectashield (Vector Labs)) and analyzed for endogenous-Abl::GFP and Mical localization. Embryo imaging was done with the aid of a Zeiss LSM 510 confocal microscope. Standard approaches were used to assess axon guidance defects, where embryos were incubated with N-CAM/Fasciclin II antibodies (1:4; 1D4 supernatant, Developmental Studies Hybridoma Bank). Immunostained embryos were then incubated and dissected in 70% glycerol and examined for motor axon guidance defects using detailed criteria as previously reported (Yang and Terman, 2012; Hung, et al., 2010; Terman, et al., 2002) using a Zeiss Axioimager microscope equipped with DIC optics, a Zeiss Axiocam HR camera, and Zeiss Axiovision software. Semaphorin/Plexin/Mical repulsive CNS guidance defects were as described previously (e.g., (Grintsevich, et al., 2016; Hung, et al., 2013; Yang and Terman, 2012)), where embryos were examined for defects in CNS axonal pathfinding including discontinuous or missing first or second CNS longitudinal connectives and/or axons crossing the midline. Midline guidance defects with UAS:Abl only were also similar to that described previously (e.g., (Hsouna, et al., 2003; Bashaw, et al., 2000)). As with our analyses in bristles, mutant analyses and genetic interactions were done with multiple different alleles.
Protein Purification
The Micalredox (redox domain, amino acids 44-531) and MicalredoxCH proteins were purified in a similar manner to that previously described (Wu, et al., 2016; Hung, et al., 2010). To generate Micalredox protein with a point mutation on the tyrosine 500 (Y500) residue, the QuickChange® Multi Site-Directed Mutagenesis Kit (Stratagene) was used following the manufacturer’s instructions. Micalredox in pET28a was used as a PCR template of site-directed mutagenesis and the forward primer 5′-ggacatcagtgcctttaccgtggatccgg-3′ and the reverse primer 5′-ccggatccacggtaaaggcactgatgtcc-3′ were used. The generated cDNA construct was sequenced to confirm that the tyrosine 500 residue was mutated to phenylalanine and the rest of the sequence was not mutated. Micalredox-Y500F protein was purified using the same methodology as wild type Micalredox. In brief, the Micalredox-Y500F expression vector was transformed into ArcticExpress competent cells (Agilent Technologies) and grown in TB (Terrific broth) and 0.2mM IPTG (isopropyl-β-D-thiogalactoside) was used to induce protein expression. Cells were then processed in lysis buffer (50mM Tris-HCl, pH8.0, 500mM NaCl, 20mM imidazole, and 10mM β-mercaptoethanol) and cleared by centrifugation. Cleared cell lysates were loaded on to a HisTrap FF column (GE Healthcare) and bound proteins were eluted. The eluted protein from the HisTrap FF column was concentrated and diluted in S-A buffer (20mM MES, pH6.5, 150mM NaCl and 2mM DTT) and then loaded onto a Mono S 5/50 GL column (GE Healthcare). The purified Micalredox-Y500F protein was placed in storage buffer (20mM Tris-HCl, pH8.0, 150mM NaCl, 5% glycerol, and 2mM DTT) and kept at −80°C until use.
In Vitro Kinase Assays
Phosphorylation of Mical protein was determined by standard in vitro kinase assays using [γ-32P] ATP (PerkinElmer) as a phosphate donor following the manufacturer’s recommendation for Abl tyrosine kinase and as described (e.g., (Yang and Terman, 2012; Xiong, et al., 2009)). We found similar results with two different independently generated and purified Abl kinases (containing the kinase domain [amino acids 254-516] of Abl), including one each from New England Biolabs (Abl amino acids 237-643) and Amsbio (Abl amino acids 248-531). Western analysis was also performed using standard approaches to determine the phosphorylation of the Mical protein using a phosphotyrosine antibody (1:2,000; 4G10, Millipore) in combination with a His antibody (1:3,000; His-2, Roche) or a GST antibody (1:1,000; polyclonal GST-tag antibody, GenScript). In particular, purified MicalredoxCH protein, Micalredox protein, or GST protein was incubated with Abl tyrosine kinase (NEB) in a tyrosine kinase reaction buffer (50mM Tris-HCl pH7.5, 10mM MgCl2, 2mM DTT, 0.1mM EDTA, 0.01% Brij 35) supplemented with 0.4mM ATP (or [γ-32P] ATP) at 30°C for 1hr. Other kinases were obtaine d as indicated: FAK (Sigma-Aldrich), Jak3 (Sigma-Aldrich), and NDR2 (Origene) and diluted in each kinase reaction buffer. The reactions were stopped by adding 2X Laemmli sample buffer and boiling. The results of the kinase reactions were resolved by SDS-PAGE. In the case of the in vitro kinase assays using [γ-32P] ATP, the gels were stained with Coomassie Brilliant Blue solution and dried gels were exposed to a PhosphorImager screen and the level of incorporated γ-32P was measured using a Storm PhosphorImager scanner (Molecular Dynamics). In cases where the samples were immunostained, SDS-PAGE gels were transferred to membrane using standard approaches and subjected to Western blotting with the antibodies described above. The concentrations of Mical and Abl we used in these assays were based off of previous studies (e.g., (Grintsevich, et al., 2016; Hung, et al., 2010; Xiong, et al., 2009)) and ranged from 50nM – 3.5μM for Mical and 8.6nM – 560nM for Abl. We had similar observations over that range of protein concentrations.
Mass Spectrometry
Five different LC/MS/MS analyses of different kinase reactions were performed. Three LC/MS/MS analyses were performed in combination with David Trudgian at The University of Texas Southwestern Proteomics Core and two analyses were performed in combination with MS Bioworks, LLC (Ann Arbor, Michigan). Similar results and phosphorylation of Y500 and Y612 residues were obtained from both facilities. Purified MicalredoxCH protein was incubated at 30°C for 1hr with the Abl tyrosine kinase (NEB) as described above for the in vitro kinase assays. The results of the kinase reaction were resolved by SDS-PAGE and the gels were stained with Coomassie Brilliant Blue solution. The stained gel bands were washed with 25mM ammonium bicarbonate followed by acetonitrile. Then, the gel bands were reduced with 10mM dithiothreitol at 60°C followed by alkyla tion with 50mM iodoacetamide at RT and digested with sequencing grade trypsin (Promega) at 37°C for 4hrs or chymotrypsin/elastase (Promega) at 37°C for overnight. The digested samples were quenched with formic acid and the supernatant was analyzed by nano LC/MS/MS with a Waters NanoAcquity HPLC system interfaced to a ThermoFisher Q Exactive. The peptides were loaded on a trapping column and eluted over a 75μm analytical column at 350nL/min. Both columns were packed with Jupiter Proteo resin (Phenomenex). The injection volume was 30μL. The mass spectrometer was operated in data dependent mode, with the Orbitrap operating at 60,000 FWHM and 17,500 FWHM for MS and MS/MS respectively. The fifteen most abundant ions were selected for MS/MS. The data were searched using a local copy of Mascot. Mascot DAT files were parsed into the Scaffold software for validation, filtering and to create a non-redundant list per sample. Data were filtered using at 1% protein and peptide FDR and requiring at least two unique peptides per protein.
Actin Depolymerization and Sedimentation Assays
Actin depolymerization assays were performed using standard approaches (Grintsevich, et al., 2016; Hung, et al., 2010). Pyrene-labeled rabbit skeletal muscle actin (Cytoskeleton, Inc.) was resuspended in 1x G-actin buffer (5mM Tris-HCl, pH 8.0, 0.2mM CaCl2, 0.2mM ATP, and 1mM DTT) and then polymerized by adding 10x actin polymerization buffer (50mM Tris-HCl, pH7.5, 500mM KCl, 20mM MgCl2, 10mM EGTA, 2mM ATP, and 5mM DTT) and incubating for 1hr at 25°C to generate a final concentration of 23.25μM actin. Depolymerization of F-actin was initiated by a five-fold dilution with 1x G-actin buffer to a final concentration of 4.65μM actin at 25°C.
To examine the ability of Abl to affect Mical’s actin depolymerizing activity, Abl kinase only, Micalredox protein + Abl kinase, Micalredox protein + Abl kinase buffer, Micalredox-Y500F protein + Abl kinase, or Micalredox-Y500F protein + Abl kinase buffer was incubated in a tyrosine kinase reaction buffer (50mM Tris-HCl pH7.5, 10mM MgCl2, 2mM DTT, 0.1mM EDTA, 0.01% Brij 35) supplemented with 0.4mM ATP at 30°C for 1hr as described above. After the kinase r eaction, each of the samples was then added to the final F-actin concentration as described above and fluorescence intensity was monitored at 407nm with excitation at 365nm using a fluorescence spectrophotometer (Spectra max M2; Molecular Devices). Multiple independent experiments were performed for each condition. The concentrations of Mical and Abl we used in these assays were based off of previous studies (e.g., (Grintsevich, et al., 2016; Hung, et al., 2010; Xiong, et al., 2009)) and were 50nM for the different Mical proteins and ranged from 8.6nM – 1.1 μM for Abl.
To determine the composition of G-actin and F-actin after 1hr of actin depolymerization using the approach described above (pyrene-labeled actin fluorescence monitoring), actin cosedimentation assays were performed as described previously (Hung, et al., 2013; Hung, et al., 2011; Hung, et al., 2010). Each reaction was then transferred into polycarbonate centrifuge tubes and centrifuged at 156,565g for 20mins at room temperature. Supernatants containing G-actin were carefully transferred into new tubes and mixed with sample buffer. The pellet containing F-actin was resuspended in pure deionized water with pipetting and incubating on ice for 10mins. The resuspended pellet was transferred into new tubes and mixed with sample buffer. The supernatants and resuspended pellets were run on an SDS-PAGE gel and then the gel was stained with Coomassie Brilliant Blue gel staining solution to visualize actin.
NADPH consumption
NADPH (the reduced form of the coenzyme), which is Mical’s coenzyme, absorbs light at 340nm, while the oxidized form (NADP+) does not (Hung, et al., 2011). Therefore, this difference between the oxidized and reduced forms of the coenzyme makes it straightforward to measure the conversion of one to another in enzyme assays. To do this, Abl kinase only, Micalredox protein + Abl kinase, or Micalredox protein + Abl kinase buffer was incubated in a tyrosine kinase reaction buffer (50mM Tris-HCl pH7.5, 10mM MgCl2, 2mM DTT, 0.1mM EDTA, 0.01% Brij 35) supplemented with 0.4mM ATP at 30°C for 1hr as described above. The samples from the kinase reaction were then added to NADPH after basal NADPH consumption had been measured in the presence of F-actin [18.4μM] and NADPH [200μM] for 3mins. Then, the absorbance was monitored every 2secs at 340nm using a fluorescence spectrophotometer (Spectra max M2; Molecular Devices) for 3mins.
Human MICAL-1 Stable Knockdown Cell Line Generation
Lentiviruses were collected from the culture medium of 293FT cells, which were transfected with pGIPZ-shRNA against MICAL-1 (Open Biosystems), pMD2.G, and psPAX2 plasmids using Effectene Transfection Reagent (Qiagen). To establish a human MICAL-1 knockdown cell line, two different breast cancer cell lines (MDA-MB-231 and MCF-7) were infected with lentiviruses expressing an shRNA against human MICAL-1 in the presence of 2μg/ml Polybrene (Sigma). Viral infected cells were selected using puromycin. To check the efficiency of knockdown, Western analysis was performed using cell lysates and human MICAL-1 polyclonal antibodies (1:1,000; Protein Tech).
Cytochemistry of Cultured Cells
Cell were processed, plated, cultured, and analyzed using standard approaches (Kim, et al., 2013). Specifically, MDA-MB-231 cells and MCF-7 cells were plated on cover slips coated with 1% gelatin. Then, cells were fixed using 3.7% paraformaldehyde for 10mins, permeabilized with 0.5% Triton X-100 in PBS on ice for 5mins and blocked with blocking solution (10% goat serum and 3% BSA in PBS containing 0.1% Triton X-100) for 30mins. Cells were then incubated with rhodamine phalloidin (1:40; Invitrogen) for an hour. Mounting solution (ProLong® Gold antifade reagent with DAPI, Life Technologies™) was then added to the cells and slides were coverslipped. Images were captured using a fluorescence microscope, Axiovert 200M (Carl Zeiss, Gottingen, Germany) and clarified using the deconvolution plug-in by ImageJ (NIH).
Matrigel Invasion Assay
Matrigel invasion assays were performed using standard approaches (Ly, et al., 2012) with minor modifications. Specifically, MDA-MB-231 cells were washed with PBS and replaced with serum-free medium with or without 10μM of Gleevec (Selleckchem) in dimethyl sulfoxide (DMSO) overnight for serum starvation. 24-well matrigel-coated transwell filters (BD Biosciences) were thawed and rehydrated according to the manufacturer’s instructions. Cells were collected, resuspended in 1000μl of serum-free medium, and 300μl were added to the top chamber in triplicates. The bottom chamber was filled with 600μl of 10% serum medium as a chemoattractant. After 48hrs incubation at 37°C, non-invaded cells were scraped off with a cotton swab and wells were washed with PBS. Invaded cells were fixed and stained with 6% glutaraldehyde and 0.5% crystal violet solution.
Colony Formation Assay/Clonogenic Survival Assay
Clonogenic survival assays were performed using standard approaches (Kim, et al., 2012). Trypsinized MCF-7 cells were counted and 100 cells of each genotype were seeded for colony formation assays in 6-well plates in triplicates. 24hrs after plating, vehicle (DMSO) or 6μM of Gleevec (Selleckchem) were added to cultures. After incubation for 11 days, colonies were stained with a mixture of 6.0% glutaraldehyde and 0.5% crystal violet and counted. Colonies were defined as clusters of >50 cells.
Mouse Tumor Xenograft Model
In vivo tumor xenograft experiments were done on 5- to 6-week old female athymic nude mice – and all procedures and experiments were approved by The University of Texas Southwestern Institutional Animal Care and Use Committee and conducted according to the institutional guidelines using standard approaches as previously employed (Mender, et al., 2015). A total of 2 × 106 MDA-MB-231 cells stably expressing control shRNA or MICAL-1 shRNA were harvested and resuspended in 100μl of PBS and subcutaneously injected into the flanks of 5- to 6-week old athymic nude mice. The tumor sizes were measured weekly for nine weeks. Nine weeks after injection, the mice were sacrificed and the tumors were dissected out. The tumors were fixed with 4% paraformaldehyde and embedded in paraffin. Sections from the tumors were stained with hematoxylin and eosin (H & E) or immunostained with Ki67 antibodies (1:200; Cell Signaling). The stained sections were examined and imaged with the aid of a Zeiss Axioskop 2 Plus upright microscope.
QUANTIFICATION AND STATISTICAL ANALYSIS
For each representative image, gel, graph, immunoblot, or in vivo experiment, the experiments were repeated at least two separate independent times with similar results. No statistical method was used to predetermine the sample size, which was based on what is published in the field. Differences between experimental and control animal conditions were large, with little variability and so the sample size was larger than needed to ensure adequate power to detect an effect. Animal studies were based on pre-established criteria to compare against age-matched animals. Animal experiments were not randomized: animals of the correct genotype were determined and those collected of that genotype were included as data. Blinding was done in initial genetic interaction assays (heterozygous genetic interactions in Bristle Mical assays) by two independent researchers. Other genetic experiments, in which the genotype needed to be determined on the basis of different Drosophila genetic/chromosome markers, blinding was not employed. Data are presented as means ± s.e.m. Statistical comparisons were performed with Student’s t-test or X2 test with the aid of GraphPad PRISM 7 and the statistical details including n can be found in the figure legends. A P value of >0.05 is not considered statistically significant. * indicates P≤0.05, ** indicates P≤0.01, *** indicates P≤0.001 and **** indicates P≤0.0001.
Supplementary Material
Table S1. List of oligonucleotides. Related to STAR Methods.
Highlights.
Abl binds Mical Redox enzymes to direct cellular remodeling and axon guidance
Abl phosphorylates Mical to enhance Mical’s catalytic F-actin disassembly activity
Abl amplifies Mical action in response to growth factors and Semaphorin repellents
Decreasing Mical levels sensitizes cancer cells to the Abl cancer drug Gleevec
Acknowledgments
We thank M. Cobb, M. Henkemeyer, and H. Krämer for comments, R.-J. Hung, T. Yang, U. Yazdani, other Terman lab members, and B.-C. Jeong for assistance, and H. Aberle, F. Fogerty, J. Merriam, M. Peifer, P. Rørth, C. Spaeth, Bloomington Stock Center, Drosophila Genetic Resource Center, and FlyBase for reagents. Supported by NIH (MH085923) and Welch Foundation (I-1749) grants to J.R.T.
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, J.Y. and J.R.T.; Methodology, J.Y., S.B.K and J.R.T.; Formal Analysis: J.Y. and S.B.K.; Investigation: J.Y., S.B.K., G.A. and J.R.T.; Resources: J.Y., S.B.K., G.A., J.W.S. and J.R.T.; Writing – Original Draft: J.Y.; Writing – Review & Editing: S.B.K., G.A., J.W.S. and J.R.T.; Visualization: J.Y., S.B.K. and J.R.T.; Supervision: J.W.S. and J.R.T.; Funding Acquisition: J.R.T.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelilah-Seyfried S, Chan YM, Zeng C, Justice NJ, Younger-Shepherd S, Sharp LE, Barbel S, Meadows SA, Jan LY, Jan YN. A gain-of-function screen for genes that affect the development of the Drosophila adult external sensory organ. Genetics. 2000;155:733–52. doi: 10.1093/genetics/155.2.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida S, Furihata M, Katagiri T, Tamura K, Anazawa Y, Yoshioka H, Miki T, Fujioka T, Shuin T, Nakamura Y, et al. Expression of novel molecules, MICAL2-PV (MICAL2 prostate cancer variants), increases with high Gleason score and prostate cancer progression. Clin Cancer Res. 2006;12:2767–73. doi: 10.1158/1078-0432.CCR-05-1995. [DOI] [PubMed] [Google Scholar]
- Bashaw GJ, Kidd T, Murray D, Pawson T, Goodman CS. Repulsive axon guidance: Abelson and Enabled play opposing roles downstream of the roundabout receptor. Cell. 2000;101:703–15. doi: 10.1016/s0092-8674(00)80883-1. [DOI] [PubMed] [Google Scholar]
- Bennett RL, Hoffmann FM. Increased levels of the Drosophila Abelson tyrosine kinase in nerves and muscles: subcellular localization and mutant phenotypes imply a role in cell-cell interactions. Development. 1992;116:953–66. doi: 10.1242/dev.116.4.953. [DOI] [PubMed] [Google Scholar]
- Berzat A, Hall A. Cellular responses to extracellular guidance cues. Embo Journal. 2010;29:2734–45. doi: 10.1038/emboj.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuchle D, Schwarz H, Langegger M, Koch I, Aberle H. Drosophila MICAL regulates myofilament organization and synaptic structure. Mech Dev. 2007;124:390–406. doi: 10.1016/j.mod.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Blanchard Z, Mullins N, Ellipeddi P, Lage JM, McKinney S, El-Etriby R, Zhang X, Isokpehi R, Hernandez B, Elshamy WM. Geminin overexpression promotes imatinib sensitive breast cancer: a novel treatment approach for aggressive breast cancers, including a subset of triple negative. PLoS One. 2014;9:e95663. doi: 10.1371/journal.pone.0095663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray D. Cell Movements: From Molecules to Motility Garland Science. 2001. [Google Scholar]
- Colicelli J. ABL tyrosine kinases: evolution of function, regulation, and specificity. Sci Signal. 2010;3:re6. doi: 10.1126/scisignal.3139re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtemanche N, Gifford SM, Simpson MA, Pollard TD, Koleske AJ. Abl2/Abl-related Gene Stabilizes Actin Filaments, Stimulates Actin Branching by Actin-related Protein 2/3 Complex, and Promotes Actin Filament Severing by Cofilin. J Biol Chem. 2015;290:4038–46. doi: 10.1074/jbc.M114.608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culi J, Martin-Blanco E, Modolell J. The EGF receptor and N signalling pathways act antagonistically in Drosophila mesothorax bristle patterning. Development. 2001;128:299–308. doi: 10.1242/dev.128.2.299. [DOI] [PubMed] [Google Scholar]
- Deng W, Wang Y, Gu L, Duan B, Cui J, Zhang Y, Chen Y, Sun S, Dong J, Du J. MICAL1 controls cell invasive phenotype via regulating oxidative stress in breast cancer cells. BMC Cancer. 2016;16:489. doi: 10.1186/s12885-016-2553-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirew P, Steif A, Khattra J, Ha G, Yap D, Farahani H, Gelmon K, Chia S, Mar C, Wan A, et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature. 2015;518:422–6. doi: 10.1038/nature13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entsch B, Palfey BA, Ballou DP, Massey V. Catalytic function of tyrosine residues in para-hydroxybenzoate hydroxylase as determined by the study of site-directed mutants. J Biol Chem. 1991;266:17341–9. [PubMed] [Google Scholar]
- Eschrich K, van der Bolt FJ, de Kok A, van Berkel WJ. Role of Tyr201 and Tyr385 in substrate activation by p-hydroxybenzoate hydroxylase from Pseudomonas fluorescens. Eur J Biochem. 1993;216:137–46. doi: 10.1111/j.1432-1033.1993.tb18125.x. [DOI] [PubMed] [Google Scholar]
- Fogerty FJ, Juang JL, Petersen J, Clark MJ, Hoffmann FM, Mosher DF. Dominant effects of the bcr-abl oncogene on Drosophila morphogenesis. Oncogene. 1999;18:219–32. doi: 10.1038/sj.onc.1202239. [DOI] [PubMed] [Google Scholar]
- Fox DT, Peifer M. Abelson kinase (Abl) and RhoGEF2 regulate actin organization during cell constriction in Drosophila. Development. 2007;134:567–78. doi: 10.1242/dev.02748. [DOI] [PubMed] [Google Scholar]
- Ganguly SS, Plattner R. Activation of abl family kinases in solid tumors. Genes Cancer. 2012;3:414–25. doi: 10.1177/1947601912458586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Henn H, Patsialou A, Wang Y, Warren MS, Condeelis JS, Koleske AJ. Arg/Abl2 promotes invasion and attenuates proliferation of breast cancer in vivo. Oncogene. 2013;32:2622–30. doi: 10.1038/onc.2012.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis EA, Serebriiskii I, Finley RL, Jr, Kolonin MG, Gyuris J, Brent R. Interaction trap/two-hybrid system to identify interacting proteins. Curr Protoc Cell Biol. 2011;Chapter 17(Unit 17):3. doi: 10.1002/0471143030.cb1703s08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TM, Letourneau PC. Actin dynamics in growth cone motility and navigation. J Neurochem. 2014;129:221–34. doi: 10.1111/jnc.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greuber EK, Smith-Pearson P, Wang J, Pendergast AM. Role of ABL family kinases in cancer: from leukaemia to solid tumours. Nat Rev Cancer. 2013;13:559–71. doi: 10.1038/nrc3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grintsevich EE, Yesilyurt HG, Rich SK, Hung RJ, Terman JR, Reisler E. F-actin dismantling through a redox-driven synergy between Mical and cofilin. Nat Cell Biol. 2016;18:876–85. doi: 10.1038/ncb3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga T, Nakamura H. Imatinib mesylate suppresses bone metastases of breast cancer by inhibiting osteoclasts through the blockade of c-Fms signals. Int J Cancer. 2009;124:215–22. doi: 10.1002/ijc.23903. [DOI] [PubMed] [Google Scholar]
- Hou ST, Nilchi L, Li X, Gangaraju S, Jiang SX, Aylsworth A, Monette R, Slinn J. Semaphorin3A elevates vascular permeability and contributes to cerebral ischemia-induced brain damage. Sci Rep. 2015;5:7890. doi: 10.1038/srep07890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou T, Chen K, McLaughlin WA, Lu B, Wang W. Computational analysis and prediction of the binding motif and protein interacting partners of the Abl SH3 domain. PLoS Comput Biol. 2006;2:e1. doi: 10.1371/journal.pcbi.0020001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsouna A, Kim YS, VanBerkum MF. Abelson tyrosine kinase is required to transduce midline repulsive cues. J Neurobiol. 2003;57:15–30. doi: 10.1002/neu.10232. [DOI] [PubMed] [Google Scholar]
- Hung RJ, Pak CW, Terman JR. Direct redox regulation of F-actin assembly and disassembly by Mical. Science. 2011;334:1710–3. doi: 10.1126/science.1211956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung RJ, Spaeth CS, Yesilyurt HG, Terman JR. SelR reverses Mical-mediated oxidation of actin to regulate F-actin dynamics. Nat Cell Biol. 2013;15:1445–54. doi: 10.1038/ncb2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung RJ, Terman JR. Extracellular inhibitors, repellents, and semaphorin/plexin/MICAL-mediated actin filament disassembly. Cytoskeleton (Hoboken) 2011;68:415–33. doi: 10.1002/cm.20527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung RJ, Yazdani U, Yoon J, Wu H, Yang T, Gupta N, Huang Z, van Berkel WJ, Terman JR. Mical links semaphorins to F-actin disassembly. Nature. 2010;463:823–7. doi: 10.1038/nature08724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaki M, Vishnu S, Cliffe A, Rorth P. Effective guidance of collective migration based on differences in cell states. Proc Natl Acad Sci U S A. 2012;109:2027–32. doi: 10.1073/pnas.1115260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri A, Wang J, Pendergast AM. Multifunctional Abl kinases in health and disease. J Cell Sci. 2016;129:9–16. doi: 10.1242/jcs.175521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SB, Ly P, Kaisani A, Zhang L, Wright WE, Shay JW. Mitigation of radiation-induced damage by targeting EGFR in noncancerous human epithelial cells. Radiat Res. 2013;180:259–67. doi: 10.1667/RR3371.1. [DOI] [PubMed] [Google Scholar]
- Kim SB, Pandita RK, Eskiocak U, Ly P, Kaisani A, Kumar R, Cornelius C, Wright WE, Pandita TK, Shay JW. Targeting of Nrf2 induces DNA damage signaling and protects colonic epithelial cells from ionizing radiation. Proc Natl Acad Sci U S A. 2012;109:E2949–55. doi: 10.1073/pnas.1207718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin AL, Tessier-Lavigne M. Mechanisms and molecules of neuronal wiring: a primer. Cold Spring Harb Perspect Biol. 2011;3:a001727. doi: 10.1101/cshperspect.a001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lah MS, Palfey BA, Schreuder HA, Ludwig ML. Crystal structures of mutant Pseudomonas aeruginosa p-hydroxybenzoate hydroxylases: the Tyr201Phe, Tyr385Phe, and Asn300Asp variants. Biochemistry. 1994;33:1555–64. doi: 10.1021/bi00172a036. [DOI] [PubMed] [Google Scholar]
- Lin J, Gan CM, Zhang X, Jones S, Sjoblom T, Wood LD, Parsons DW, Papadopoulos N, Kinzler KW, Vogelstein B, et al. A multidimensional analysis of genes mutated in breast and colorectal cancers. Genome Res. 2007;17:1304–18. doi: 10.1101/gr.6431107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loria R, Bon G, Perotti V, Gallo E, Bersani I, Baldassari P, Porru M, Leonetti C, Di Carlo S, Visca P, et al. Sema6A and Mical1 control cell growth and survival of BRAFV600E human melanoma cells. Oncotarget. 2015;6:2779–2793. doi: 10.18632/oncotarget.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist MR, Storaska AJ, Liu TC, Larsen SD, Evans T, Neubig RR, Jaffrey SR. Redox modification of nuclear actin by MICAL-2 regulates SRF signaling. Cell. 2014;156:563–76. doi: 10.1016/j.cell.2013.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly P, Eskiocak U, Parker CR, Harris KJ, Wright WE, Shay JW. RNAi screening of the human colorectal cancer genome identifies multifunctional tumor suppressors regulating epithelial cell invasion. Cell Res. 2012;22:1605–8. doi: 10.1038/cr.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manta B, Gladyshev VN. Regulated methionine oxidation by monooxygenases. Free Radic Biol Med. 2017 doi: 10.1016/j.freeradbiomed.2017.02.010. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti S, Barravecchia I, Vindigni C, Pucci A, Balsamo M, Libro R, Senchenko V, Dmitriev A, Jacchetti E, Cecchini M, et al. MICAL2 is a novel human cancer gene controlling mesenchymal to epithelial transition involved in cancer growth and invasion. Oncotarget. 2016;7:1808–1825. doi: 10.18632/oncotarget.6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor R, Carmona-Fontaine C. Keeping in touch with contact inhibition of locomotion. Trends Cell Biol. 2010;20:319–28. doi: 10.1016/j.tcb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mender I, Gryaznov S, Dikmen ZG, Wright WE, Shay JW. Induction of telomere dysfunction mediated by the telomerase substrate precursor 6-thio-2′-deoxyguanosine. Cancer Discov. 2015;5:82–95. doi: 10.1158/2159-8290.CD-14-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran GR, Entsch B, Palfey BA, Ballou DP. Electrostatic effects on substrate activation in para-hydroxybenzoate hydroxylase: studies of the mutant lysine 297 methionine. Biochemistry. 1997;36:7548–56. doi: 10.1021/bi9706327. [DOI] [PubMed] [Google Scholar]
- Moresco EM, Donaldson S, Williamson A, Koleske AJ. Integrin-mediated dendrite branch maintenance requires Abelson (Abl) family kinases. J Neurosci. 2005;25:6105–18. doi: 10.1523/JNEUROSCI.1432-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moresco EM, Koleske AJ. Regulation of neuronal morphogenesis and synaptic function by Abl family kinases. Curr Opin Neurobiol. 2003;13:535–44. doi: 10.1016/j.conb.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Mummery-Widmer JL, Yamazaki M, Stoeger T, Novatchkova M, Bhalerao S, Chen D, Dietzl G, Dickson BJ, Knoblich JA. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458:987–92. doi: 10.1038/nature07936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagar B, Hantschel O, Young MA, Scheffzek K, Veach D, Bornmann W, Clarkson B, Superti-Furga G, Kuriyan J. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112:859–71. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- O’Connor BP, Eun SY, Ye Z, Zozulya AL, Lich JD, Moore CB, Iocca HA, Roney KE, Holl EK, Wu QP, et al. Semaphorin 6D regulates the late phase of CD4+ T cell primary immune responses. Proc Natl Acad Sci U S A. 2008;105:13015–20. doi: 10.1073/pnas.0803386105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell MP, Bashaw GJ. Distinct functional domains of the Abelson tyrosine kinase control axon guidance responses to Netrin and Slit to regulate the assembly of neural circuits. Development. 2013;140:2724–33. doi: 10.1242/dev.093831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palfey BA, Entsch B, Ballou DP, Massey V. Changes in the catalytic properties of p-hydroxybenzoate hydroxylase caused by the mutation Asn300Asp. Biochemistry. 1994;33:1545–54. doi: 10.1021/bi00172a035. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–12. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JV, Savenye ED, Lum D, Breitkreutz A. Dominant enhancers of Egfr in Drosophila melanogaster: genetic links between the Notch and Egfr signaling pathways. Genetics. 1997;147:1139–53. doi: 10.1093/genetics/147.3.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procaccia V, Nakayama H, Shimizu A, Klagsbrun M. Gleevec/imatinib, an ABL2 kinase inhibitor, protects tumor and endothelial cells from semaphorin-induced cytoskeleton collapse and loss of cell motility. Biochem Biophys Res Commun. 2014;448:134–8. doi: 10.1016/j.bbrc.2014.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenhiser DI, Andrews J, Kennette W, Sadikovic B, Mendlowitz A, Tuck AB, Chambers AF. Epigenetic mapping and functional analysis in a breast cancer metastasis model using whole-genome promoter tiling microarrays. Breast Cancer Res. 2008;10:R62. doi: 10.1186/bcr2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schito L, Rey S, Tafani M, Zhang H, Wong CC, Russo A, Russo MA, Semenza GL. Hypoxia-inducible factor 1-dependent expression of platelet-derived growth factor B promotes lymphatic metastasis of hypoxic breast cancer cells. Proc Natl Acad Sci U S A. 2012;109:E2707–16. doi: 10.1073/pnas.1214019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreuder HA, Prick PA, Wierenga RK, Vriend G, Wilson KS, Hol WG, Drenth J. Crystal structure of the p-hydroxybenzoate hydroxylase-substrate complex refined at 1.9 A resolution. Analysis of the enzyme-substrate and enzyme-product complexes. J Mol Biol. 1989;208:679–96. doi: 10.1016/0022-2836(89)90158-7. [DOI] [PubMed] [Google Scholar]
- Shimizu A, Mammoto A, Italiano JE, Jr, Pravda E, Dudley AC, Ingber DE, Klagsbrun M. ABL2/ARG tyrosine kinase mediates SEMA3F-induced RhoA inactivation and cytoskeleton collapse in human glioma cells. J Biol Chem. 2008;283:27230–8. doi: 10.1074/jbc.M804520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebold C, Berrow N, Walter TS, Harlos K, Owens RJ, Stuart DI, Terman JR, Kolodkin AL, Pasterkamp RJ, Jones EY. High-resolution structure of the catalytic region of MICAL (molecule interacting with CasL), a multidomain flavoenzyme-signaling molecule. Proc Natl Acad Sci U S A. 2005;102:16836–41. doi: 10.1073/pnas.0504997102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Song JK, Kannan R, Merdes G, Singh J, Mlodzik M, Giniger E. Disabled is a bona fide component of the Abl signaling network. Development. 2010;137:3719–27. doi: 10.1242/dev.050948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney KF, Huang CH, Devreotes PN. Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu Rev Biophys. 2010;39:265–89. doi: 10.1146/annurev.biophys.093008.131228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor Alto L, Terman JR. Semaphorins and their Signaling Mechanisms. Methods Mol Biol. 2017;1493:1–25. doi: 10.1007/978-1-4939-6448-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terman JR, Mao T, Pasterkamp RJ, Yu HH, Kolodkin AL. MICALs, a family of conserved flavoprotein oxidoreductases, function in plexin-mediated axonal repulsion. Cell. 2002;109:887–900. doi: 10.1016/s0092-8674(02)00794-8. [DOI] [PubMed] [Google Scholar]
- Thompson CL, Van Vactor D. Abl family protein tyrosine kinases and the formation of neural connectivity. In: Koleske A, editor. In Abl family kinases in development and disease. Georgetown, Texas: Landes Bioscience; 2006. pp. 105–122. [Google Scholar]
- Toyofuku T, Zhang H, Kumanogoh A, Takegahara N, Yabuki M, Harada K, Hori M, Kikutani H. Guidance of myocardial patterning in cardiac development by Sema6D reverse signalling. Nat Cell Biol. 2004;6:1204–11. doi: 10.1038/ncb1193. [DOI] [PubMed] [Google Scholar]
- van der Bolt FJ, van den Heuvel RH, Vervoort J, van Berkel WJ. 19F NMR study on the regiospecificity of hydroxylation of tetrafluoro-4-hydroxybenzoate by wild-type and Y385F p-hydroxybenzoate hydroxylase: evidence for a consecutive oxygenolytic dehalogenation mechanism. Biochemistry. 1997;36:14192–201. doi: 10.1021/bi971213c. [DOI] [PubMed] [Google Scholar]
- Vitriol EA, Zheng JQ. Growth cone travel in space and time: the cellular ensemble of cytoskeleton, adhesion, and membrane. Neuron. 2012;73:1068–81. doi: 10.1016/j.neuron.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY. The capable ABL: what is its biological function? Mol Cell Biol. 2014;34:1188–97. doi: 10.1128/MCB.01454-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills Z, Marr L, Zinn K, Goodman CS, Van Vactor D. Profilin and the Abl tyrosine kinase are required for motor axon outgrowth in the Drosophila embryo. Neuron. 1999;22:291–9. doi: 10.1016/s0896-6273(00)81090-9. [DOI] [PubMed] [Google Scholar]
- Wilson C, Terman JR, Gonzalez-Billault C, Ahmed G. Actin filaments – a target for redox regulation. Cytoskeleton (Hoboken) 2016;73:577–595. doi: 10.1002/cm.21315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Hung RJ, Terman JR. A simple and efficient method for generating high-quality recombinant Mical enzyme for in vitro assays. Protein Expr Purif. 2016;127:116–24. doi: 10.1016/j.pep.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, Dabbouseh NM, Rebay I. Interactions with the Abelson tyrosine kinase reveal compartmentalization of eyes absent function between nucleus and cytoplasm. Dev Cell. 2009;16:271–9. doi: 10.1016/j.devcel.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Terman JR. 14-3-3epsilon couples protein kinase A to semaphorin signaling and silences plexin RasGAP-mediated axonal repulsion. Neuron. 2012;74:108–21. doi: 10.1016/j.neuron.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of oligonucleotides. Related to STAR Methods.