Summary
The hexameric AAA+ ATPases Rvb1 and Rvb2 (Rvbs) are essential for diverse processes ranging from metabolic signaling to chromatin remodeling but their functions are unknown. While originally thought to act as helicases, recent proposals suggest that Rvbs act as protein assembly chaperones. However, experimental evidence for chaperone-like behavior is lacking. Here, we identify a potent protein activator of the Rvbs, a domain (Ino80INS) in the Ino80 ATPase subunit, of the INO80 chromatin-remodeling complex. Ino80INS stimulates Rvbs’ ATPase activity by 16-fold while concomitantly promoting their dodecamerization. Using mass spectrometry, cryo-EM, and integrative modeling, we find that Ino80INS binds asymmetrically along the dodecamerization interface, resulting in a conformationally flexible dodecamer that collapses into hexamers upon ATP addition. Our results demonstrate the chaperone-like potential of Rvb1/Rvb2 and suggest a model where binding of multiple clients like Ino80 stimulates ATP-driven cycling between hexamers and dodecamers, providing iterative opportunities for correct subunit assembly.
eTOC blurb
Rvb1 and Rvb2 (Rvbs) are AAA+ ATPases that have been hypothesized to assemble multi-subunit protein complexes, though this activity has not been experimentally demonstrated. Zhou et al. find that a domain within the chromatin remodeling Ino80 ATPase is a potent activator of Rvb1/Rvb2, creating a metastable dodecamer of Rvb/1Rvb2 that dissociates upon ATP addition
Introduction
AAA+ ATPases are a highly conserved family of molecular motors that use ATP binding and hydrolysis to drive a diverse set of structural rearrangements in their substrates (Singleton et al., 2007; White and Lauring, 2007). Rvb1 and Rvb2 (Rvbs) are two AAA+ ATPases from S. cerevisiae that are essential for many cellular processes including transcription, cellular differentiation, cell signaling, mitosis, snoRNP assembly, and DNA damage repair (Matias et al., 2015, Nano and Houry et al., 2013). Despite their extensive involvement in biology, the role of the Rvbs in these processes is unknown. Even the basic enzymatic function of these motors is poorly defined. While the Rvbs have ATPase activity (Gribun et al., 2008), there are competing models for how this activity contributes to the function of the Rvbs. Based on structural similarity of the Rvbs to the bacterial helicase RuvAB, it was initially hypothesized that the Rvbs are DNA helicases. While a few groups have been able to detect DNA unwinding activity (Gribun et al., 2008; Kanemaki et al., 1999; Makino et al., 1999), other groups have reported a lack of such an activity (Ikura et al., 2000; Matias et al., 2006; Qiu et al., 1998). Recently, it has been proposed that the Rvbs act as protein assembly chaperones, based on the observation that the Rvbs co-purify with several multi-subunit protein complexes including the R2TP complex, telomerase, snoRNP complexes and chromatin remodeling complexes (Nano and Houry, 2013). Yet, no chaperone-like properties of the Rvbs have been experimentally demonstrated.
To investigate the possibility that the Rvbs act as assembly chaperones, we chose the yeast INO80 complex as a model system due to the wealth of biochemical and structural information available on this remodeling complex. We found that a small insertion within the Ino80 chromatin remodeling ATPase subunit of INO80 (Ino80INS) stimulates the ATPase activity of Rvb1/Rvb2 by 16-fold. To study how Ino80INS regulates Rvb structure and activity we used a combination of crosslinking-MS, native-MS, cryo-EM and integrative modeling approaches. Our results suggest a model in which (i) the activated dodecameric Rvb1/Rvb2/Ino80INS is poised to bring together several protein clients in close proximity and (ii) nucleotide binding and/or hydrolysis collapses the activated intermediate, helping assemble multi-subunit complexes containing a stably associated and less active Rvb1/Rvb2 hexamer.
Results
The insertion domain of the Ino80 ATPase (Ino80INS) is a potent activator of Rvb1/Rvb2
An early study showed that the ATP hydrolysis activity of Rvb2 is required for formation of active INO80 complexes in yeast (Jónsson et al., 2004). Recently, it was shown that a small insertion within the Ino80 ATPase subunit (Ino80INS, Figure 1A, top) is required for association with the Rvbs and other subunits of the complex that are critical for activity (Chen et al., 2013). These results suggested that Ino80INS is used by the Rvbs as a scaffold for assembling an active INO80 complex. To test this possibility, we asked if Ino80INS on its own could directly interact with the Rvb1/Rvb2 complex and whether or not this interaction influences the ATPase activity of the Rvbs.
Figure 1. The insertion domain of the Ino80 ATPase (Ino80INS) is a direct interactor and activator of Rvb1/Rvb2.
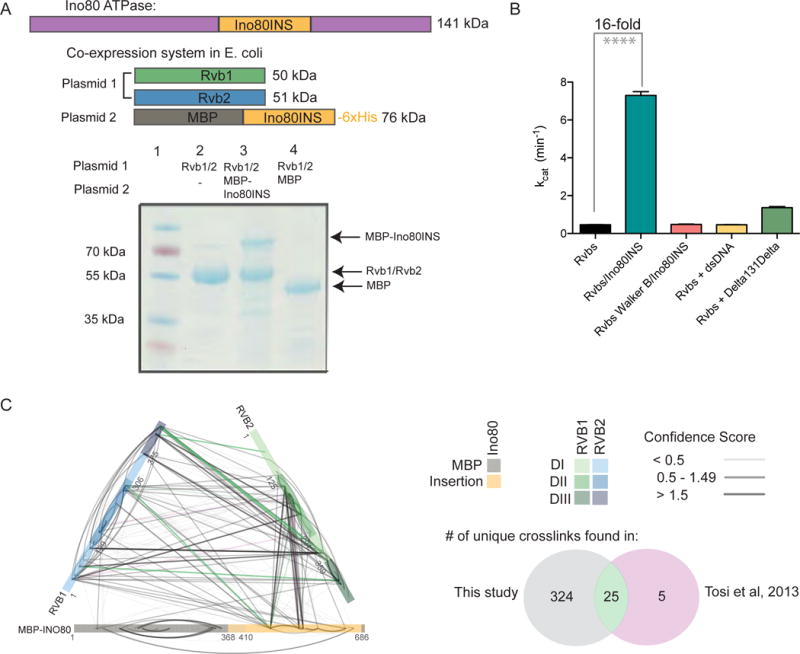
(A) Top, Sequence architecture of the Ino80 remodeling ATPase from yeast, with Ino80INS in yellow. Below, co-expression system for Rvb1/Rvb2/Ino80INS in E. coli. Plasmid 1 contains Rvb1 and Rvb2. Plasmid 2 contains the Ino80 insertion (Ino80INS, residues 1022–1294) expressed as an MBP-fusion and with a C-terminal histidine tag. Bottom, SDS-PAGE gel showing results of pull down with co-expressed MBP-Ino80INS and Rvb1/Rvb2 (Lane 3).
(B) Comparison of kcat values for ATP hydrolysis by different Rvb1/Rvb2 and Rvb1/Rvb2/Ino80INS complexes. Stimulation of 5 M Rvbs with either 25 M double stranded DNA or MBP-Δ131Δ are shown in yellow and green, respectively. Bars represent the mean ± SEM of 3 replicates. **** = p < 0.0001.
(C) XL-MS data for the Rvb1/Rvb2/Ino80INS complex. Rvb1 (green) and Rvb2 (blue) are shown with different shades representing DI, DII, and DIII. Crosslinks found only in our dataset, in only the native INO80 complex (Tosi et al., 2013), or both datasets are shown in gray, pink, and green respectively (see venn diagram). The confidence of the MS identification (SVM score) is represented by the thickness and transparency of the lines.
See also Figure S1 and Table S1.
We co-expressed untagged Rvb1 and Rvb2 along with a his-tagged, MBP-Ino80INS fusion in Escherichia coli. These three proteins form a stable complex that purified as a single peak over a size exclusion column (Figure 1A). Efforts to purify the complex without an MBP tag on Ino80INS, or MBP-Ino80INS alone were unsuccessful, suggesting Ino80INS is inherently unstable, but can be stabilized via interactions with Rvb1 and Rvb2. Next, we tested whether the activity of the Rvbs is affected by Ino80INS and found that Rvb1/Rvb2/Ino80INS has a 16-fold greater kcat for ATP hydrolysis than Rvbs alone. In contrast, an excess of DNA did not stimulate the Rvbs’ ATPase activity, consistent with previously published results with human Rvb1/Rvb2 (Puri et al., 2007). Interestingly, the partially unfolded region of staphylococcal nuclease △131△, routinely used as a model substrate for the chaperone Hsp90 (Street et al., 2011), modestly stimulated the ATPase activity of the Rvbs (2–3 fold, Figure 1B). This result further suggests that protein, not DNA, is the biologically relevant substrate for yeast Rvb1/Rvb2.
We next used crosslinking mass spectrometry (XL-MS) (Leitner et al., 2016) to investigate residue-level interactions between the Rvbs and Ino80INS. From two separate XL-MS experiments with the Rvb1/Rvb2/Ino80INS (Figure 1C) and the Rvb1/Rvb2 alone (Figure S1A), we identified 519 unique cross-linked residue pairs (“crosslinks”) at a false discovery rate of 1.7% (Figure S1B, Table S1). Strikingly, a majority of the high confidence crosslinks containing Ino80INS mapped to the DII regions of Rvb1 and Rvb2 (Figure 1C). The DII domains are of particular interest due to their role in regulating the activity and oligomeric state of the Rvbs (Gorynia et al., 2011; Niewiarowski et al., 2010). Very few residues from the Rvbs crosslinked to MBP (Figure 1C), confirming that the complex is predominantly stabilized by direct interactions between the DII regions of Rvb1/Rvb2 and Ino80INS. Also, our data recapitulate over 80% of the interactions found in a cross-linking study of the complete yeast INO80 complex (Tosi et al., 2013), suggesting that the Rvb1/Rvb2/Ino80INS subcomplex maintains the same interactions as seen within the native INO80 complex.
The Ino80 insertion promotes dodecamerization of Rvb1/Rvb2
Rvbs are predominantly hexameric, but a small population of naturally occurring dodecamers has also been reported (Jeganathan et al., 2015). Our observation that Ino80INS binds predominantly to the DII regions of the Rvbs raised the possibility that Ino80INS may activate the Rvbs’ ATPase activity by altering the oligomeric state of Rvb1/Rvb2. To investigate this possibility, we used size exclusion chromatography-multi angle light scattering (SEC-MALS) to determine the molecular weight of Rvb1/Rvb2/Ino80INS. We found that the complex has a molecular weight that is most consistent with a dodecamer of Rvb1/Rvb2 bound to at least two copies of the Ino80 insertion (Figure 2A). In contrast, Rvb1/Rvb2 alone gave a molecular weight most consistent with hexamers.
Figure 2. The Ino80 insertion promotes dodecamerization of Rvb1/Rvb2.
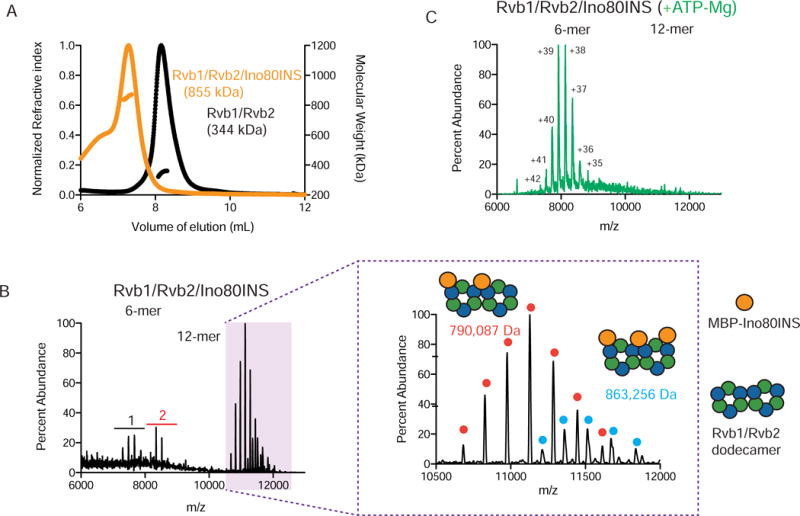
(A) SEC-MALS of Rvb1/Rvb2/Ino80INS complexes. Normalized refractive index (left y-axis) and molecular weight of the major peak (right y-axis) are plotted against the volume of elution. For each experiment, either 10 M of Rvb1/Rvb2/Ino80INS (orange) or 30 M Rvb1/Rvb2 (black) was injected. The molecular weights shown in corresponding colors are an average of the calculated molecular weight values across the main peak.
(B) Left, raw spectrum of Rvb1/Rvb2/Ino80 complexes using native-MS. Charge envelopes for the hexameric species are shown in black (1) and red (2). Right, an expanded view of the charge envelopes representing the two dodecameric species. Cartoons representing the MBP-Ino80INS and Rvb1/Rvb2 illustrate the predicted stoichiometries represented by the two charge envelopes.
(C) Raw spectrum of 1 M Rvb1/Rvb2/Ino80INS complexes with 20 M ATP-Mg added, by native-MS.
See also Figure S2 and Table S2.
To further analyze the distribution of species in the Rvb1/Rvb2/Ino80INS sample, we turned to native mass spectrometry (native-MS). Using this technique (Rose et al., 2012), we found two distinct charge envelopes with masses consistent with either two or three copies of Ino80INS bound to a single Rvb1/Rvb2 dodecamer (Figure 2B, Table S2, Figure S2A–B). Along with the SEC-MALS results, these data suggest that more than one Ino80INS can bind to a single dodecamer of Rvb1/Rvb2. In addition to the dodecamer, we also observed a small population of Rvb1/Rvb2 hexamer (Species 1 in Figures 2B, S2B, Table S2) and Rvb1/Rvb2 hexamer bound to a single MBP-Ino80INS (Species 2 in Figures 2B, S2B, Table S2). Intriguingly, addition of ATP-Mg2+ to the Rvb1/Rvb2/Ino80INS complex caused the dodecameric species to convert to a hexameric species (Figure 2C). We also observed dissociation into hexamers by negative-stain EM, as barrel shaped Rvb1/Rvb2 dodecamers fall apart to ring-shaped hexamers upon addition of ATP-Mg2+ (Figure S2C). These results suggest that destabilization of the Rvb1/Rvb2/Ino80INS dodecamer is coupled to the nucleotide state of the Rvbs.
Nucleotide state drives the switch between hexameric and dodecameric forms of Rvb1/Rvb2
Our observation that the Rvb1/Rvb2/Ino80INS complex collapses in an ATP-dependent manner raised the possibility that the transition between dodecamer and hexamer is a core activity of the Rvbs, and may be related to the Rvbs’ ability to function as a chaperone. To better elucidate how the oligomeric state of the Rvbs alone is regulated by ATP state, we used native-MS to analyze the relative populations of oligomers of Rvb1/Rvb2 in different nucleotide states. The high mass resolution of this method also allowed us to more accurately determine subunit stoichiometry and the nucleotide binding state of each oligomer. Untagged yeast Rvb1/Rvb2 ionize as hexamers with a mass that is most consistent with a 3:3 stoichiometry of Rvb1: Rvb2 (Figure 3A, Figure S3A–B, Table S3), in contrast to a 4:2 stoichiometry reported for human Rvbs (Niewiarowski et al., 2010). Intact denatured mass spectrometry on the same sample confirmed the 3:3 stoichiometry (Figure S3C).
Figure 3. Nucleotide state drives the switch between hexameric and dodecameric forms of the Rvb1/Rvb2 complex.
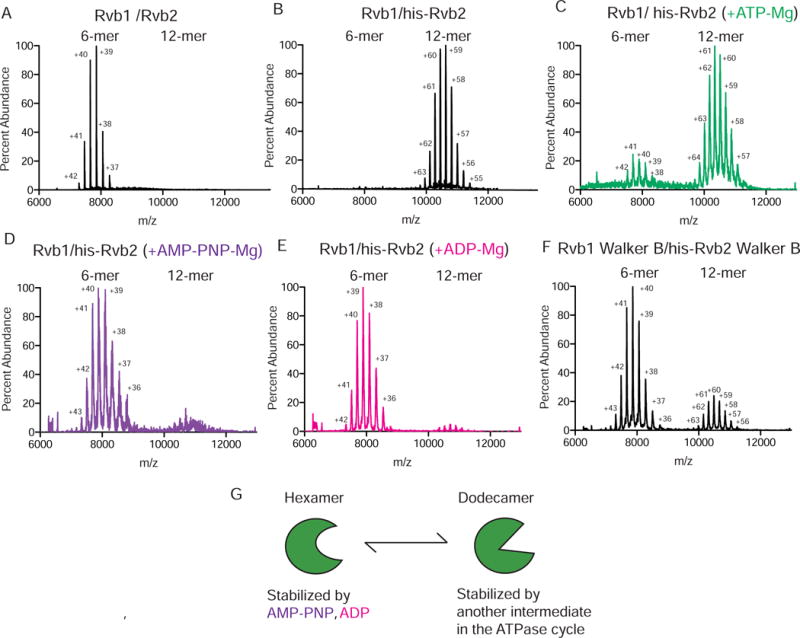
(A) Raw spectrum of 1 M of untagged Rvb1/Rvb2 by native-MS.
(B) Raw spectrum of 1 M Rvb1/his-Rvb2.
(C) Raw spectrum of 1 M Rvb1/Rvb2-his tag after addition of 20 M ATP-Mg.
(D) Raw spectrum of 1 M Rvb1/Rvb2-his tag after addition of 20 M AMP-PNP-Mg.
(E) Raw spectrum of 1 M Rvb1/Rvb2-his tag after addition of 20 M ADP-Mg, using native-MS.
(F) Raw spectrum of 1 M his-tagged Walker B mutants of Rvb1 (D311N) and Rvb2 (D296N).
(G) Model for how hexameric and dodecameric states of Rvbs are regulated by nucleotide state.
See also Figure S3, Table S3, and Table S4.
We next addressed how the Rvb1/Rvb2 dodecamer might be affected by nucleotide state. This experiment could not be done with untagged Rvb1/Rvb2, as the complex is predominantly a hexamer at concentrations compatible with native-MS (1 M) with and without nucleotide (Figs. 3A and S3D). Previous work with yeast Rvb1/Rvb2 showed that an N-terminal his-tag promotes dodecamerization, providing a means to study the dodecameric state by native-MS (Cheung et al., 2010; Jeganathan et al., 2015). As expected, his-tagged Rvbs ionized as a dodecamer (Figure 3B). When an excess of ATP-Mg2+ was added, a small proportion (14%) of the dodecamer dissociated into hexamers (Figure 3C, Table S4), consistent with our observation for the Rvb1/Rvb2/Ino80INS complex (Figure 2C).
To test which stage(s) of the ATPase cycle affects oligomerization, we used non-hydrolyzable ATP analogs AMP-PNP and ATP-γ-S to mimic the ATP bound state and ADP to mimic the post-hydrolysis state. Surprisingly, AMP-PNP, ATP-γ-S and ADP all substantially increased the proportion of the hexameric state (Figures 3D–E, Figure S3E). Importantly, this increase was greater than what was observed with ATP (Figures 3C vs. 3D–E, Table S4). The quantitative difference in the effects we observed with AMP-PNP vs. ATP does not appear to be due to differences in affinity of the nucleotide for Rvb1/Rvb2, as similar amounts of nucleotide loading was observed for both ATP-Mg2+ and AMP-PNP-Mg2+ (Table S4). To further investigate the role of nucleotide binding vs. hydrolysis, we mutated the Walker B motifs of Rvb1 and his-Rvb2, which diminished ATP hydrolysis by >20-fold (Figure S3F). Compared to wild-type Rvb1/his-Rvb2, the population of hexamers in the Walker B Rvb1/his-Rvb2 mutants increased dramatically, from 0% to 84%, and addition of ATP gave similar results (Figure 3F and S3I, Table S4). These results suggest a model in which nucleotide binding promotes a conformation of the Rvbs that stabilizes the hexameric state, while an intermediate in the ATP hydrolysis reaction stabilizes a conformation that promotes formation of a metastable dodecamer (Figure 3G).
For these studies, it is important to note that the his-tag is not present on the Rvbs in vivo, so the biological significance of the his-tagged dodecameric species is unclear. However, since the basic observation we derive from the his-tagged construct, namely disassembly into hexamers in the presence of ATP, is recapitulated in the context of untagged Rvbs bound to a native protein activator (Ino80INS) we suggest that the behavior of the his-tagged complex captures some of the core features of a native Rvb complex.
ATPase activity of the Rvb1/Rvb2 complex is enhanced by dodecamerization
We previously observed that the stimulation of Rvbs by Ino80INS is concurrent with dodecamerization of Rvb1/Rvb2. We next asked whether this correlation exists in a context outside of Ino80INS. Similar to the N-terminal his-tag, replacing the DII domain with a flexible linker also increases dodecamerization of Rvbs, but there is conflicting evidence for how these mutants affect the Rvbs’ ATPase activity in the yeast system (Gorynia et al., 2011; Jeganathan et al., 2015; Niewiarowski et al., 2010). When we deleted the DII domains in Rvb1 and Rvb2, we found that this Rvb1ΔDII/Rvb2ΔDII complex has a molecular weight (341 kDa) that is in between what is expected for a dodecamer (474 kDa) and hexamer (237 kDa) by SEC-MALS (Figure 4A and Table S5). We reasoned that the intermediate molecular weight could reflect either a rapidly exchanging mixture of hexamer and dodecamer or a homogeneous population of an oligomeric state in between a dodecamer and hexamer (Table S5). Increasing the concentration of Rvb1ΔDII/Rvb2ΔDII from 10 M to 30 M resulted in a 437 kDa complex that is approximately the expected molecular weight of a dodecamer (Figure S4A, Table S5). Native-MS with 1 M Rvb1ΔDII/Rvb2ΔDII indicated predominantly hexamers with a small (3%) population of dodecamer (Figure S4C), consistent with a concentration-dependent oligomerization of Rvb1ΔDII/Rvb2ΔDII. In contrast, wild-type Rvb1/Rvb2 remained hexameric at 80 M (Figure S4B).
Figure 4. The ATPase activity of the Rvb1/Rvb2 complex is enhanced by dodecamerization.
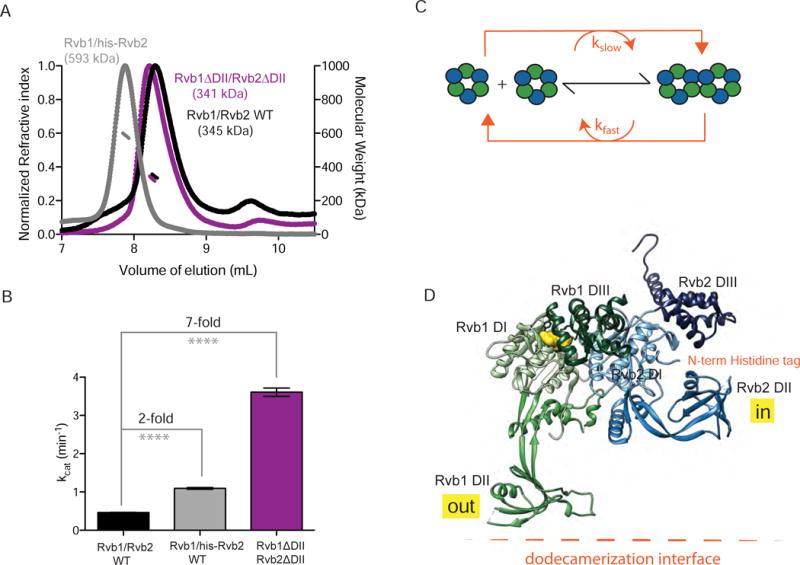
(A) SEC-MALS of different Rvb1/Rvb2 complexes. Normalized refractive index (left y-axis) and molecular weight of the major peak (right y-axis) are plotted against the volume of elution. 10 μM of either Rvb1/Rvb2 (black), Rvb1/Rvb2-his (gray), or Rvb1ΔDII, Rvb2ΔDII (purple) was injected. The molecular weights shown in corresponding colors are an average of the calculated molecular weight values across the main peak.
(B) kcat for ATP hydrolysis activity for different Rvb1/Rvb2 complexes. Experiments were performed using 10 mM Rvbs and saturating ATP. Bars represent the mean ± SEM of 3 replicates. **** = p < 0.0001.
(C) Model for how Rvb1/Rvb2 oligomeric state is coupled to ATPase cycle. Rvb1/Rvb2 complexes exist at equilibrium between hexamer and dodecamer (black arrows). In the presence of nucleotide (orange arrows), Rvb1/Rvb2 complexes are constantly interchanging between the two oligomeric states, creating a steady state distribution of the hexamer and dodecamer, where hexamers hydrolyze ATP slower (kslow) than dodecamers (kfast).
(D) Top, domain architecture of Rvb1 and Rvb2 from S. cerevisiae. Domains of Rvb1 are colored as different shades of green and domains of Rvb2 are colored as different shades of blue. Bottom, model of Rvb1 (bound to ATP) and Rvb2 (apo) adapted from the crystal structure of Rvb1/Rvb2 complex from Chaetomium theramophilum (Lakomek et al, 2015, PDB: 4WVY). Domains I and III contain the catalytic residues while domain II folds as a separate module either in the “in” or “out” state. The location of the N-terminal histidine tag, which does not appear in the density, is shown in orange, and ATP is in yellow.
See also Figure S4 and Table S5.
We next compared the maximal ATPase activity (kcat) of the different Rvb1/Rvb2 mutants. We found that adding a his-tag to Rvb2 increased the kcat for ATP hydrolysis only 2-fold, while deleting the DII domains of Rvb1 and Rvb2 increased the specific activity by 7-fold (Figure 4B). Taken together with the data in Figure 2, these data suggest that the ATPase cycle of Rvb1/Rvb2 drives a constant inter-conversion between hexamers and dodecamers, resulting in a defined steady state distribution of the two oligomeric states with different specific activities (Figure 4C). The regulation of Rvbs’ activity by Ino80INS serves as a natural extension of this model, as binding of Ino80INS to the DII domains of the Rvbs promotes an activated, metastable dodecamer that easily dissociates into hexamers upon addition of ATP.
Cryo-EM reconstruction of the Rvb1/Rvb2/Ino80INS complex suggests high degree of conformational flexibility
We were next curious about the structural basis of how Ino80INS engages the DII domains of Rvb1/Rvb2 to create a metastable dodecamer. Previous studies of Rvb1/Rvb2 have reported a high degree of conformational flexibility within the DII regions (Ewens et al., 2016; Gorynia et al., 2011; Jeganathan et al., 2015; López-Perrote et al., 2012; Petukhov et al., 2012), which appear to form the dodecameric interface. A crystal structure of the Rvb1/Rvb2 complex from the thermophillic fungus Chaetomium thermophilum (Lakomek et al., 2015) shows one possible arrangement of the Rvb1 and Rvb2 DII domains: the Rvb2 DII domains fold back toward the hexameric ring (“in” state), and the Rvb1 DII domains extend toward a second hexamer (“out” state) (Figure 4D). We therefore imagined two extreme models for how Ino80INS binding may alter the structure of the Rvbs: (i) Ino80INS “locks” the conformationally flexible dodecameric interface into one predominant conformation, or (ii) Ino80INS binds in a way that maintains the natural conformational flexibility of the DII domains.
To test these hypotheses, we determined the cryo-EM map of the Rvb1/Rvb2/Ino80INS complex at ~12.0 resolution (Figure 5B, S5B). Both raw images and 2D class averages show a distinct barrel shape created by the two hexameric rings flanking the DII domains of the Rvbs (Figure 5A, S5A). We also observed both bent and straight conformations of the Rvb1/Rvb2/Ino80INS dodecamer in our 2D classes (Figure 5A). These features are similar to those reported in a recently published cryo-EM structure of Rvb1/Rvb2 dodecamers without an activator bound (Ewens et al., 2016; Silva-Martin et al., 2016), suggesting that Ino80INS binding does not substantially diminish the natural dynamics of the Rvbs.
Figure 5. Cryo-EM of the Rvb1/Rvb2/Ino80INS complex reveals conformational heterogeneity.
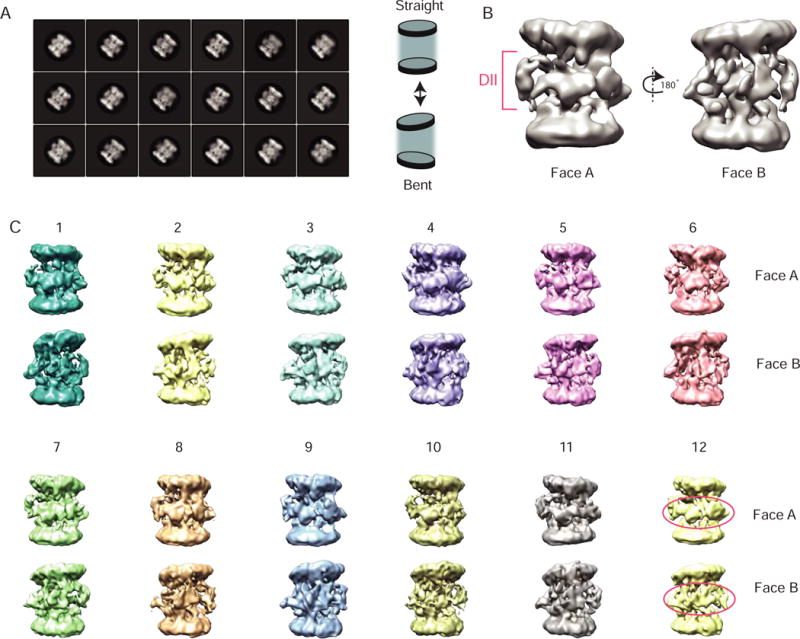
(A) Left: Side views of reference-free 2D class averages of Rvb1/Rvb2/Ino80INS particles. Right: Scheme displaying “bent” and straight” features of the Rvb1/Rvb2/Ino80INS dodecamer.
(B) Rvb1/Rvb2/Ino80INS consensus model generated by refinement of all 366,000 particles used for 2D classification in (A).
(C) Twelve 3D classes representing all of the particles used for the consensus structure in (B). The top and bottom rows of each class represent two opposite faces of the dodecamer. Each class contains approximately 30,000 particles.
See also Figure S5.
To further investigate the extent of the conformational dynamics in the Rvb1/Rvb2/Ino80INS complex, all particles used for 2D classification were also used for 3D refinement to generate a consensus cryo-EM map (Figure 5B), with no imposed symmetry. Interestingly, the bent feature observed in the 2D classes is retained in the consensus map, suggesting that it is a common feature of most of the particles. To find higher-resolution subpopulations of the dataset, we further subclassified particles to generate twelve different 3D classes (Figure 5C). Each class contained a similar number of particles, and had resolutions that were similar to one another and to that for the consensus map. Despite many rounds of further subclassification, we were unable to achieve higher resolution, implying a high degree of conformational heterogeneity in the system. This conformational heterogeneity is most prominent in the densities surrounding the DII domains, as well as in the degree of tilt observed between the two hexameric rings (Figure 5C). In addition to the conformational heterogeneity, a second general feature of the Rvb1/Rvb2/Ino80INS system is its asymmetry at the dodecameric interface. In both the consensus map and each of the twelve subclasses, one face of the dodecamer consistently shows bulkier density than the opposite face of the dodecamer (Face A vs. Face B, Figures 5B–C).
Ino80INS bind asymmetrically on the Rvb1/Rvb2 dodecamer
To obtain more insight into the conformational flexibility and asymmetry of the Rvb1/Rvb2/Ino80INS complex, we mapped all unique Rvb-Rvb crosslinks observed in the Rvb1/Rvb2/Ino80INS complex onto a comparative structural model of the yeast Rvb1/Rvb2 dodecamer. We created this model based on sequence identity between the S. cerevisiae Rvb1 and Rvb2 subunits and the S. thermophilus structure of the Rvb1/Rvb2 dodecamer (Lakomek et al., 2015). Strikingly, this mapping revealed that only 58% of the unique 226 Rvb-Rvb crosslinks are satisfied (Figure S7A). It is unlikely that the low percentage of satisfied crosslinks is due to inaccurate spectral assignments given the low false discovery rate (FDR) of our crosslinking experiment (less than 2%, Figure S3B). It is also unlikely that our comparative model of the Rvb1/Rvb2 dodecamer is insufficiently accurate to explain the XL violations, given the high sequence similarity between the S. cerevisiae and S. thermophilus Rvbs (73% for Rvb1 and 72% for Rvb2) and the high structural conservation in this family (Iyer et al., 2004). Thus, the high proportion of crosslinks violated by the model is likely due to the additional conformations that the Rvb1/Rvb2/Ino80INS complex can adopt beyond the one exhibited in the comparative model of the Rvb1/Rvb2 dodecamer.
To better characterize this conformational heterogeneity, we applied an integrative structure determination approach based on all information available for the Rvb1/Rvb2/Ino80INS complex including the XL-MS dataset (Figures 3C, Table S4), the consensus cryo-EM map (Figure 5B), the comparative model of the Rvb1/Rvb2 dodecamer, the crystal structure of MBP (Kim, 2013), and the predicted secondary structure of Ino80INS (Figure S6, Table S6). The resulting integrative models are expected to be more accurate and precise than models based on a subset of data (Alber et al., 2007; Robinson et al., 2015; Russel et al., 2012; Shi et al., 2014). We imposed a stoichiometry of two copies of Ino80INS bound to one Rvb1/Rvb2 dodecamer based on the major species found by native-MS (Figure 2C). The ensemble of models, consistent with input information, was computed by satisfying spatial restraints implied by the data, allowing us to quantify the structural heterogeneity and asymmetry observed in the cryo-EM data as well as to predict the binding site of Ino80INS.
The ensemble contains three clusters of models, each of which represent possible conformations of the Rvb1/Rvb2/Ino80INS dodecamer. For Cluster 1, the precision (average pairwise RMSD of the models in the cluster) is 25 Å, and the precisions of the Rvb1/Rvb2 dodecamer and Ino80INS in particular are 15.8 and 39.3 Å, respectively. These values represent the average fluctuations of the individual residues or beads in 3-D space across the ensemble of solutions. The precisions of the other two clusters are comparable (Table S6). Each cluster of models satisfies the XLs approximately within the expected tolerance (>90%) (Table S6, Figure 6A), and 98% of all crosslinks are satisfied by at least one structure in the three clusters. The remaining 2% unsatisfied cross-links are likely explained by the 2% false positive rate of cross-linking, sample heterogeneity, insufficient conformational sampling, and coarse-grained representation of the modeled components. Each cluster matches the EM density map, as demonstrated by the overlaps of the cluster localization density maps with the consensus cryo-EM density map when all maps are contoured to match their volumes (Figure 6B, Table S6). A localization density map for a set of models is defined as the probability of observing a model component at any point in space (Robinson et al., 2015). Because each cluster satisfies the cross-links and EM map comparably well, and each cluster contains a comparable number of solutions, it is likely that the localization density maps represent the underlying conformational heterogeneity in the sample rather than a lack of information used for modeling.
Figure 6. Ino80INS binds to Rvb1/Rvb2 dodecamer asymmetrically.
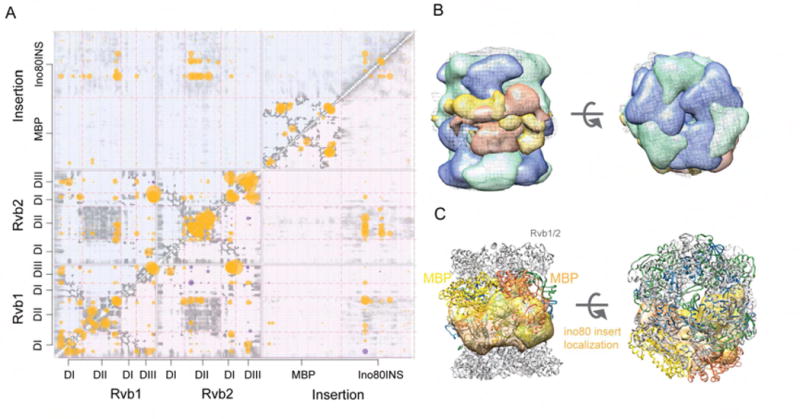
(A) Characterization of Clusters 1 (top half, shaded light blue) and 2 (bottom half, shaded light red). The gray bins indicate pairs of proximal beads representing the model, with the intensity of grey proportional to the fraction of models in the cluster whose distance is closer than the cutoff of 12 Å. The yellow circles correspond to the cross-links satisfied by at least one model in a cluster, with the size of the circle proportional to the spectral count. The purple circles indicate cross-links that are not satisfied by any model in the cluster.
(B) Side and top views of the localization density maps for Rvb1 (green), Rvb2 (blue), and MBP-Ino80INS (orange and yellow) calculated for Cluster 1 (thresholded at 0.15). The grey mesh shows the consensus EM density map.
(C) Top and side views of a representative model from Cluster 1. The distributions of the two copies of Ino80INS are shown by their localization densities (orange and yellow, respectively).
Also see Figure S6, Figure S7, Table S6.
In all clusters, Ino80INS binds to the central region of the barrel, contacting the DII domains (Figure 6A and S7C). Furthermore, the two Ino80INS domains bind asymmetrically on the Rvb1/Rvb2 dodecamer, contacting the same side of the barrel. Examination of the MBP crosslinks shows that some crosslinks are necessarily inter-molecular (Figure S7B), suggesting the two MBP domains are close to each other. Due to the proximity of the Ino80 and MBP domains in the linear sequence, this observation further supports a model of asymmetric binding of the two INO80INS to the Rvbs dodecamer.
Discussion
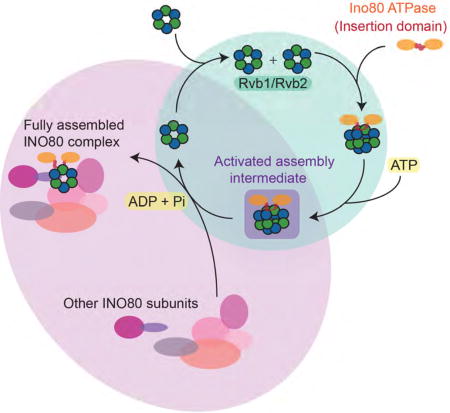
In addition to the INO80 complex, Rvb1 and Rvb2 have been found to associate with a number of multi-subunit complexes, including snoRNP complexes (McKeegan et al., 2009), PIKK signaling complexes (Izumi et al., 2010) and telomerase (Venteicher et al., 2008), leading to the hypothesis that Rvbs may act as chaperones for assembling and/or remodeling these complexes (Nano and Houry, 2013). Based on our data and prior work, we propose the following model for how the Rvbs may help assemble multi-subunit complexes such as INO80 (Figure 7). In this model, nucleotide binding is coupled to a conformation of the Rvb that is autoinhibitory for ATP hydrolysis and incompatible with dodecamerization, while ATP hydrolysis (either the transition state or the ADP-Pi product state) is coupled to a conformation of the Rvb that promotes dodecamerization (Figure 3G). Binding of clients such as Ino80INS then drives the Rvbs into the more active dodecameric state, yet one that is metastable and is more likely to dissociate into hexamers (Figure 2). Thus, a window of opportunity is created for other client proteins to bind to adjacent DII domains of the Rvb1/Rvb2 complex. We speculate that the Rvb1/Rvb2 dodecamer is able to “sense” co-localization of multiple client proteins by coupling the association of these proteins to nucleotide-driven dissociation of the dodecamer into hexamers. This model is consistent with our observation that two copies of Ino80INS can bind asymmetrically to one side of the Rvb1/Rvb2 dodecamer (Figure 6), which we speculate reflects the ability of the Rvb1/Rvb2 dodecamer to bind to multiple different client proteins in vivo.
Figure 7. Model for activation of Rvb1/Rvb2 by client proteins.
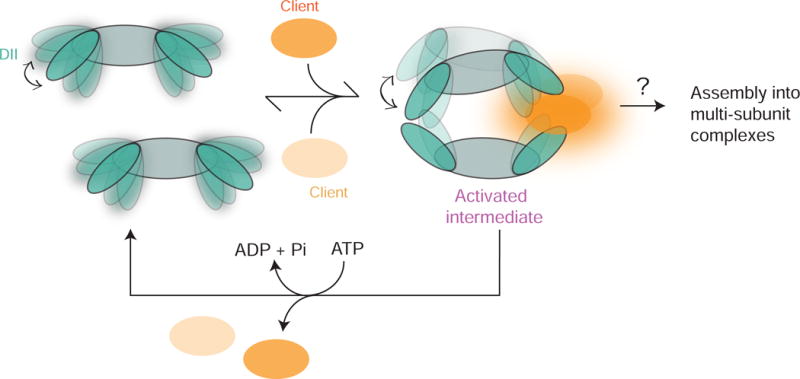
Binding of client proteins to DII domains of Rvb1/Rvb2 promotes the dodecameric state by stabilizing the flexible DII regions that comprise the dodecamerization interface. Two clients bind asymmetrically to the dodecamer, creating an activated intermediate that is highly competent for ATP hydrolysis. Addition of ATP causes the dodecamer to fall apart, allowing the two client proteins to be assembled into a multi-subunit complex. Meanwhile, Rvb1/Rvb2 hexamers are recycled and poised to repeat the assembly reaction.
A major feature of our model is that binding of a client protein such as Ino80INS promotes formation of a metastable dodecamer that is prone to collapsing into hexamers upon nucleotide binding. Our cryo-EM and integrative modeling data provide structural insight into how such a metastable dodecamer is created, as the dodecameric interface is highly flexible and dynamic (Figures 5 and 6). The ability of ATP to further destabilize this interface is consistent with recently published cryo-EM structures of the Rvb1/Rvb2 dodecamers alone, which suggest that addition of nucleotide causes a significant reconfiguration of the DII domains, resulting in a shift in the relative orientations of the two hexameric rings (Ewens et al., 2016). We therefore speculate that creation of a metastable dodecamer though reconfiguration of the DII domains is important for both the ATP-driven cycling between hexamers and dodecamers and the chaperone-like activity of the Rvbs.
Based our model, we would most simply expect that a hexameric Rvb1/Rvb2 bound to Ino80INS would be created during the ATPase cycle of the Rvbs. While we do observe a small amount of this species upon addition of ATP, we mainly observe Rvb1/Rvb2 hexamers without Ino80INS bound by native-MS (Figure 2C). We speculate that the different steps in ATP hydrolysis cause the Rvbs to cycle between strong and weak binding for their client proteins, analogous to other chaperones such as Hsp90 (Street et al., 2011; Verba et al., 2016). In the context of full-length Ino80, additional interactions may keep it bound to the Rvbs during the ATPase cycle, whereas in the context of the Ino80 insert, these additional binding interactions could be missing, causing it to dissociate.
Our model also addresses the potentially separate functions of the hexameric and dodecameric forms of the Rvbs. While both of these oligomeric states been observed in native Rvb1/Rvb2 systems (Ewens et al., 2016; Jeganathan et al., 2015), mechanistic roles for either oligomeric state have not yet been proposed. We speculate that the dodecameric Rvb1/Rvb2/Ino80INS complex is representative of an intermediate that is on pathway toward assembling a multi-subunit INO80 complex. Completion of assembly would then result in the stable binding of the less active, hexameric state of Rvb1/Rvb2 to the rest of the INO80 complex. In this model, the transition from dodecamer to hexamer is governed by both the presence of bound client proteins as well as the ATPase cycle, ultimately resulting in an equilibrium among dodecameric assembly intermediates and fully assembled complexes containing a hexamer of Rvb1/Rvb2. Importantly, the predominant stoichiometry of the components within the Rvb1/Rvb2/Ino80INS complex (6:6:2) is consistent with recently published EM data of the full INO80 complex, where a large number of the INO80 complexes contain a single Ino80 ATPase bound to a hexamer of Rvb1/Rvb2 (Watanabe et al., 2015).
We favor the model described above to explain our results because previous work has suggested that the Rvbs are required for assembly of active INO80 complexes (Chen et al., 2013; Jónsson et al., 2004). However, we propose two alternative models that are also compatible with our results. In the first alternative model, the ATPase stimulation caused by Ino80INS serves to limit the number of Ino80 proteins bound, as increasing the number of bound Ino80 proteins would further destabilize the already metastable dodecamer. In this model, the Rvbs would primarily play a role in regulating subunit stoichiometry. In the second alternative model, the ATPase stimulation caused by Ino80INS may reflect the Rvbs’ potential role for remodeling protein complexes, as Rvb1 and Rvb2 have been hypothesized to be involved in disaggregation of amyloid fibrils (Zaarur et al., 2015) and chromosome decondensation (Magalska et al., 2014). Importantly, our observation that protein, not nucleic acid, is a robust activator of Rvb1/Rvb2 helps exclude models involving a helicase-like role of the Rvbs in vivo.
While we believe we have identified the first client of Rvb1/Rvb2, further studies are required to fully demonstrate the chaperone-like potential of the yeast Rvbs. In the future, an ability to reconstitute all the steps in the reaction catalyzed by the Rvbs would be invaluable for obtaining insights into the mechanism of the process. We propose that the yeast INO80 complex can serve as a powerful model system for future mechanistic studies of the Rvbs for the following reasons: (i) the organization and stoichiometry of the INO80 subunits within the native complex, down to residue-level interactions, has been fully characterized (Tosi et al., 2013) and (ii) the proper assembly of INO80 is required for in vitro nucleosome remodeling activity (Chen et al., 2013; Jónsson et al., 2001). Thus, the INO80 complex provides an ideal opportunity for a complete in vitro reconstitution of the steps required in assembling an active multi-subunit complex. These experiments, while technically challenging, would provide unprecedented mechanistic insight into this class of essential yet poorly understood AAA+ ATPases.
Experimental Procedures
Also see supplemental experimental procedures for additional information.
Protein expression and purification
Rvb1 and Rvb2 were co-expressed in Rosetta (DE3) E. coli on a pet28a plasmid (a kind gift from the Hopfner lab). Rvb2 had a TEV-cleavable N-terminal His8-tag, which was cleaved unless otherwise noted. Rvb1/Rvb2 was purified using cobalt affinity resin followed by size exclusion chromatography.
SEC-MALS
Molecular weight was determined using SEC (Wyatt 050S5 column) with an Ettan LC (GE Healthcare) and in-line DAWN HELEOS MALS and Optilab rEX differential refractive index detectors (Wyatt Technology Corporation). SEC was performed in 80 mM PIPES (pH 6.9), 150 mM KCl, 5 mM MgCl2. Data were analyzed by the ASTRA VI software package (Wyatt).
XL-MS
Preparations of Rvb1/Rvb2 and Rvb1/Rvb2/Ino80INS were crosslinked with BS3, digested with trypsin, fractionated by size exclusion chromatography and analyzed by LC-MS using a QExactive Plus mass spectrometer. Spectra were searched for crosslinked peptides using Protein Prospector and further classified using a support vector machine (SVM) model (Robinson et al., 2015; Trnka et al., 2014).
Native-MS
Native-MS was performed on the Rvb1/Rvb2 protein complexes using the Exactive Plus EMR instrument (Thermo Scientific, San Jose, CA). Samples were buffer-exchanged into 150 mM ammonium acetate, pH 7.5 and then sprayed from Au/Pd-coated borosilicate emitters into mass spectrometer using static infusion.
Negative stain EM
Rvb1/Rvb2/Ino80INS complexes were prepared as described above and dialyzed into 25 mM Hepes pH 7.5, 150 mM KCl. 2.5 L of 5–10 M of Rvbs were adsorbed onto 30-second glow discharged copper grids coated with carbon. Samples were blotted and stained with 3 L 0.75% uranyl formate solution for five applications with blotting in between each application. Images were collected on a Technai T12 microscope (FEI Company) equipped with a LaB6 filament and operated at 120 kV. Images were recorded with an Ultrascan 4096 × 4096 pixel CCD camera (Gatan) at a nominal magnification of 52,000x.
ATPases
ATP hydrolysis rates were measured using an NADH enzyme coupled reaction assay (Lindsley, 2001). The final reaction included 10 U/mL of pyruvate kinase (Sigma) and 10 U/mL of lactate dehydrogenase (LDH, Sigma), 2 mM phosphoenylpyruvate (PEP, Sigma), between 2–10 M Rvb1/2 complex, 0.18 mM NADH, 4 mM ATP, 4.5 mM MgCl2, 150 mM KCl, and 25 mM Hepes pH 7.5. ATP concentration was empirically determined to be saturating for all constructs tested. All components other than Rvb1/2 were assembled and incubated at 30°C in a 384 well plate (clear bottom, Corning) for 10 minutes. Enzyme was mixed in, and absorbance at 340 nm was monitored using a SpectraMax M5e plate reader. After background subtraction, the data were fit to a linear regression, with the slope being the rate of hydrolysis (M/min).
Cryo-electron microscopy image acquisition
Rvb1/Rvb2/Ino80INS complexes were prepared as described for negative stain. For cryo-EM grid preparation, 2.5 L samples of 10 M Rvb monomer were applied to Quantifoil holey carbon grids (1.2/1.3 spacing) that had been glow discharged for 30 seconds. Using a Mark I Vitrobot (FEI Company), grids were blotted 5.5 or 6 s. with −3 mm offset at 100% humidity and plunge frozen into liquid ethane cooled by liquid nitrogen.
Images were collected on a Titan Krios microscope (FEI Company) operated at 300 kV under low-dose conditions at Janelia Research Campus. All images were recorded with a K2 Summit direct electron detector camera (Gatan) using super resolution counting mode, with a pixel size of 1.31 Å/pixel after 2×2 binning for motion correction and subsequent processing. The camera dose rate was set to ~1.4 e-/pixel/frame. Dose-fractionated stacks were collected over an exposure time of 8 s. with a sub-frame exposure of 200 ms. for a total of 40 subframes, resulting in a total dose of ~32.6 e-/Å2 for the entire stack and 0.8 e-/Å for each subframe.
Cryo-electron microscopy image processing
All dose-fractionated image stacks were motion corrected using MotionCor2 and the sum of all subframes was used for subsequent image processing (Zheng et al., 2016). Estimation of the Contrast Transfer Function and defocus was done using Gctf, a GPU-accelerated CTF estimation program (Zhang, 2016). A set of ~1400 particles were manually picked and extracted from corrected micrographs using RELION 1.4. Manually picked particles were subjected to 2D classification in RELION 1.4 and good classes were selected as a template for reference-based automatic particle picking. Multiple rounds of 2D classification yielded a particle stack of ~366,000 particles that was used to generate a consensus model with the RELION 3D auto-refine function, using C1 symmetry. The structure of the T. acidophilum 20S proteasome (EMDB 5623) filtered to 50 Å was used an initial model. All subsequent 3D classification was performed using either the consensus model, a model generated by prior 3D classification, or the C. thermophilum Rvb1/Rvb2 crystal structure (PDB 4WVY), all filtered to at least 40 Å, as an initial model. Classes of interest were subjected to further 3D classification with C1 symmetry and refined using gold-standard procedures within RELION 1.4 (Scheres, 2012).
Integrative modeling of the Rvb1/Rvb2/Ino80INS complex
We computed a structural model of the yeast Rvb1/Rvb2/Ino80INS complex using an integrative approach based on data from X-ray crystallography, cross-linking mass spectrometry and cryo-EM. The modeling protocol (Figure S6) was scripted using the Python Modeling Interface (PMI), a library for modeling macromolecular complexes based on our open-source Integrative Modeling Platform (IMP) package, version 2.5 (http://integrativemodeling.org) (Russel et al., 2012). Complete details of the integrative modeling procedure are in the Supplemental. Files containing the input data, scripts, and output structures are available online (https://github.com/integrativemodeling/Rvbs).
Supplementary Material
Highlights.
The insertion domain of the Ino80 ATPase (Ino80INS) activates yeast Rvb1/Rvb2.
Ino80INS activates Rvb1/Rvb2 by promoting dodecamerization of Rvb1/Rvb2.
Two Ino80INS bind asymmetrically along the dodecameric interface of the Rvbs.
Addition of ATP to Rvb1/Rvb2/Ino80INS collapses the dodecamer into hexamers.
Acknowledgments
We thank the Hopfner lab (Ludwig-Maximilians University in Munich) for the expression plasmid for the Rvbs. We thank L.R.P, S.M.C, P.A.D, L.H., S.J., A.G.L. and D.C. for critical reading of the manuscript, and members of the Narlikar lab for helpful discussions. We thank Daniel and Andrew for technical support. This work was funded by an NIH grant (RO1GM073767) to G.J.N., an NIH Ruth L. Kirschstein National Research Service Award (5F31CA180651-02) to C.Y.Z., an NSF graduate fellowship (1144247) to C.I.S., NIH grants (8P41GM103481 and 1S10D016229) and the UCSF Enabling Technology Advisory Committee to A.L.B, NIH grants (R01GM083960 and P41GM083960) to A.S. and NIH grants (1S10OD020054 and R01GM082893) to Y.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
C.Y.Z. conceptualized the study with guidance from G.J.N. C.Y.Z. cloned and purified the proteins, performed ATPase assays and SEC-MALS experiments. J.B.J. performed the native-MS experiments. M.J.T. performed the XL-MS experiments, created homology models of Rvb1/Rvb2, and analyzed the data. C.I.S. performed the EM experiments and analyzed the data with help from E.P. I.E. performed the integrative modeling. All authors contributed to preparation of the manuscript. Y.C., A.S., and A.L.B. provided expertise and support for the project.
Accession Numbers
The EMDataBank accession code for the cryo-EM map of the Rvb1/Rvb2/Ino80INS complex is EMDB-8696.
References
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Rout MP, Sali A. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- Chen L, Conaway RC, Conaway JW. Multiple modes of regulation of the human Ino80 SNF2 ATPase by subunits of the INO80 chromatin-remodeling complex. Proc Natl Acad Sci USA. 2013;110:20497–20502. doi: 10.1073/pnas.1317092110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung KLY, Huen J, Kakihara Y, Houry WA, Ortega J. Alternative oligomeric states of the yeast Rvb1/Rvb2 complex induced by histidine tags. J Mol Biol. 2010;404:478–492. doi: 10.1016/j.jmb.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens CA, Su M, Zhao L, Nano N, Houry WA, Southworth DR. Architecture and Nucleotide-Dependent Conformational Changes of the Rvb1-Rvb2 AAA+ Complex Revealed by Cryoelectron Microscopy. Structure. 2016;24:657–666. doi: 10.1016/j.str.2016.03.018. [DOI] [PubMed] [Google Scholar]
- Gorynia S, Bandeiras TM, Pinho FG, McVey CE, Vonrhein C, Round A, Svergun DI, Donner P, Matias PM, Carrondo MA. Structural and functional insights into a dodecameric molecular machine - the RuvBL1/RuvBL2 complex. J Struct Biol. 2011;176:279–291. doi: 10.1016/j.jsb.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Gribun A, Cheung KLY, Huen J, Ortega J, Houry WA. Yeast Rvb1 and Rvb2 are ATP-dependent DNA helicases that form a heterohexameric complex. J Mol Biol. 2008;376:1320–1333. doi: 10.1016/j.jmb.2007.12.049. [DOI] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1038/35036020. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Izumi N, Yamashita A, Iwamatsu A, Kurata R, Nakamura H, Saari B, Hirano H, Anderson P, Ohno S. AAA+ proteins RUVBL1 and RUVBL2 coordinate PIKK activity and function in nonsense-mediated mRNA decay. Sci Signal. 2010;3:ra27–ra27. doi: 10.1126/scisignal.2000468. [DOI] [PubMed] [Google Scholar]
- Jeganathan A, Leong V, Zhao L, Huen J, Nano N, Houry WA, Ortega J. Yeast Rvb1 and Rvb2 Proteins Oligomerize As a Conformationally Variable Dodecamer with Low Frequency. J Mol Biol. 2015 doi: 10.1016/j.jmb.2015.01.010. [DOI] [PubMed] [Google Scholar]
- Jónsson ZO, Dhar SK, Narlikar GJ, Auty R, Wagle N, Pellman D, Pratt RE, Kingston R, Dutta A. Rvb1p and Rvb2p are essential components of a chromatin remodeling complex that regulates transcription of over 5% of yeast genes. J Biol Chem. 2001;276:16279–16288. doi: 10.1074/jbc.M011523200. [DOI] [PubMed] [Google Scholar]
- Jónsson ZO, Jha S, Wohlschlegel JA, Dutta A. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol Cell. 2004;16:465–477. doi: 10.1016/j.molcel.2004.09.033. [DOI] [PubMed] [Google Scholar]
- Kanemaki M, Kurokawa Y, Matsu-ura T, Makino Y, Masani A, Okazaki K, Morishita T, Tamura TA. TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J Biol Chem. 1999;274:22437–22444. doi: 10.1074/jbc.274.32.22437. [DOI] [PubMed] [Google Scholar]
- Kim M. Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues. Prion. 2013;7:221–228. doi: 10.4161/pri.23807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakomek K, Stoehr G, Tosi A, Schmailzl M, Hopfner KP. Structural Basis for Dodecameric Assembly States and Conformational Plasticity of the Full-Length AAA+ ATPases Rvb1·Rvb2. Structure. 2015 doi: 10.1016/j.str.2014.12.015. [DOI] [PubMed] [Google Scholar]
- Leitner A, Faini M, Stengel F, Aebersold R. Crosslinking and Mass Spectrometry: An Integrated Technology to Understand the Structure and Function of Molecular Machines. Trends Biochem Sci. 2016;41:20–32. doi: 10.1016/j.tibs.2015.10.008. [DOI] [PubMed] [Google Scholar]
- López-Perrote A, Muñoz-Hernández H, Gil D, Llorca O. Conformational transitions regulate the exposure of a DNA-binding domain in the RuvBL1-RuvBL2 complex. Nucleic Acids Res. 2012;40:11086–11099. doi: 10.1093/nar/gks871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalska A, Schellhaus AK, Moreno-Andrés D, Zanini F, Schooley A, Sachdev R, Schwarz H, Madlung J, Antonin W. RuvB-like ATPases function in chromatin decondensation at the end of mitosis. Dev Cell. 2014;31:305–318. doi: 10.1016/j.devcel.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Makino Y, Kanemaki M, Kurokawa Y, Koji T, Tamura TA. A rat RuvB-like protein, TIP49a, is a germ cell-enriched novel DNA helicase. J Biol Chem. 1999;274:15329–15335. doi: 10.1074/jbc.274.22.15329. [DOI] [PubMed] [Google Scholar]
- Matias PM, Gorynia S, Donner P, Carrondo MA. Crystal structure of the human AAA+ protein RuvBL1. J Biol Chem. 2006;281:38918–38929. doi: 10.1074/jbc.M605625200. [DOI] [PubMed] [Google Scholar]
- McKeegan KS, Debieux CM, Watkins NJ. Evidence that the AAA+ proteins TIP48 and TIP49 bridge interactions between 15.5K and the related NOP56 and NOP58 proteins during box C/D snoRNP biogenesis. Mol Cell Biol. 2009;29:4971–4981. doi: 10.1128/MCB.00752-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nano N, Houry WA. Chaperone-like activity of the AAA+ proteins Rvb1 and Rvb2 in the assembly of various complexes. Philos Trans R Soc Lond, B, Biol Sci. 2013;368:20110399. doi: 10.1098/rstb.2011.0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiarowski A, Bradley AS, Gor J, McKay AR, Perkins SJ, Tsaneva IR. Oligomeric assembly and interactions within the human RuvB-like RuvBL1 and RuvBL2 complexes. Biochem J. 2010;429:113–125. doi: 10.1042/BJ20100489. [DOI] [PubMed] [Google Scholar]
- Petukhov M, Dagkessamanskaja A, Bommer M, Barrett T, Tsaneva I, Yakimov A, Queval R, Shvetsov A, Khodorkovskiy M, Käs E, Grigoriev M. Large-scale conformational flexibility determines the properties of AAA+ TIP49 ATPases. Structure. 2012;20:1321–1331. doi: 10.1016/j.str.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Puri T, Wendler P, Sigala B, Saibil H, Tsaneva IR. Dodecameric structure and ATPase activity of the human TIP48/TIP49 complex. J Mol Biol. 2007;366:179–192. doi: 10.1016/j.jmb.2006.11.030. [DOI] [PubMed] [Google Scholar]
- Qiu XB, Lin YL, Thome KC, Pian P, Schlegel BP, Weremowicz S, Parvin JD, Dutta A. An eukaryotic RuvB-like protein (RUVBL1) essential for growth. J Biol Chem. 1998;273:27786–27793. doi: 10.1074/jbc.273.43.27786. [DOI] [PubMed] [Google Scholar]
- Robinson PJ, Trnka MJ, Pellarin R, Greenberg CH, Bushnell DA, Davis R, Burlingame AL, Sali A, Kornberg RD. Molecular architecture of the yeast Mediator complex. Elife. 2015;4:44. doi: 10.7554/eLife.08719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose RJ, Damoc E, Denisov E, Makarov A, Heck AJR. High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nat Methods. 2012;9:1084–1086. doi: 10.1038/nmeth.2208. [DOI] [PubMed] [Google Scholar]
- Russel D, Lasker K, Webb Ben, Velázquez-Muriel J, Tjioe E, Schneidman-Duhovny D, Peterson B, Sali A. Putting the Pieces Together: Integrative Modeling Platform Software for Structure Determination of Macromolecular Assemblies. PLOS Biol. 2012;10:e1001244. doi: 10.1371/journal.pbio.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Fernandez-Martinez J, Tjioe E, Pellarin R, Kim SJ, Williams R, Schneidman-Duhovny D, Sali A, Rout MP, Chait BT. Structural characterization by cross-linking reveals the detailed architecture of a coatomer-related heptameric module from the nuclear pore complex. Mol Cell Proteomics. 2014;13:2927–2943. doi: 10.1074/mcp.M114.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Martin N, Daudén MI, Glatt S, Hoffmann NA, Kastritis P, Bork P, Beck M, Müller CW. The Combination of X-Ray Crystallography and Cryo-Electron Microscopy Provides Insight into the Overall Architecture of the Dodecameric Rvb1/Rvb2 Complex. PLoS ONE. 2016;11:e0146457. doi: 10.1371/journal.pone.0146457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- Street TO, Lavery LA, Agard DA. Substrate binding drives large-scale conformational changes in the Hsp90 molecular chaperone. Mol Cell. 2011;42:96–105. doi: 10.1016/j.molcel.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi A, Haas C, Herzog F, Gilmozzi A, Berninghausen O, Ungewickell C, Gerhold CB, Lakomek K, Aebersold R, Beckmann R, Hopfner K-P. Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell. 2013;154:1207–1219. doi: 10.1016/j.cell.2013.08.016. [DOI] [PubMed] [Google Scholar]
- Trnka MJ, Baker PR, Robinson PJJ, Burlingame AL, Chalkley RJ. Matching cross-linked peptide spectra: only as good as the worse identification. Mol Cell Proteomics. 2014;13:420–434. doi: 10.1074/mcp.M113.034009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verba KA, Wang RY-R, Arakawa A, Liu Y, Shirouzu M, Yokoyama S, Agard DA. Atomic structure of Hsp90-Cdc37-Cdk4 reveals that Hsp90 traps and stabilizes an unfolded kinase. Science. 2016;352:1542–1547. doi: 10.1126/science.aaf5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Tan D, Lakshminarasimhan M, Washburn MP, Erica Hong EJ, Walz T, Peterson CL. Structural analyses of the chromatin remodelling enzymes INO80-C and SWR-C. Nat Commun. 2015;6:7108. doi: 10.1038/ncomms8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SR, Lauring B. AAA+ ATPases: achieving diversity of function with conserved machinery. Traffic. 2007;8:1657–1667. doi: 10.1111/j.1600-0854.2007.00642.x. [DOI] [PubMed] [Google Scholar]
- Zaarur N, Xu X, Lestienne P, Meriin AB, McComb M, Costello CE, Newnam GP, Ganti R, Romanova NV, Shanmugasundaram M, Silva ST, Bandeiras TM, Matias PM, Lobachev KS, Lednev IK, Chernoff YO, Sherman MY. RuvbL1 and RuvbL2 enhance aggresome formation and disaggregate amyloid fibrils. EMBO J. 2015;34:2363–2382. doi: 10.15252/embj.201591245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Meth. 2016;14:331–332. doi: 10.1101/061960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


