Abstract
Background
An understanding of the activated protein signaling architecture in non–small-cell lung cancer (NSCLC) is of critical importance to the development of new therapeutic approaches and identification of predictive and prognostic biomarkers for patient stratification.
Methods
We used reverse-phase protein microarrays to map the activated protein signaling networks of 47 NSCLC tumors, 28 of which were node negative, which were subjected to tumor cellular enrichment using laser capture microdissection. The phosphorylation/cleavage levels of 111 key signaling proteins and total levels of 17 proteins were measured for broadscale signaling analysis.
Results
Pathway activation mapping of NSCLC revealed distinct subgroups composed of epidermal growth factor receptor (ERBB1), v-erb-b2 erythroblastic leukemia viral oncogene homolog 2 (ERBB2), v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (ERBB3), v-erb-a erythroblastic leukemia viral oncogene homolog 4 (ERBB4), v-akt murine thymoma viral oncogene homolog 1- mammalian target of rapamycin (AKT-mTOR), protein kinase, AMP-activated, alpha 2 catalytic subunit (AMPK), and autophagy-related signaling, along with transforming growth factor-beta-signaling protein 1 (SMAD), insulin-line growth factor receptor (IGFR), rearranged during transfection proto-oncogene (RET), and activated CDC42-associated kinase (ACK) activation. Investigation of epidermal growth factor receptor (EGFR)-driven signaling identified a unique cohort of tumors with low EGFR protein expression yet high relative levels of phosphorylated EGFR and high EGFR total protein with low relative levels of phosphorylation. Last, mapping analysis of patients with NSCLC with N0 disease revealed a pilot pathway activation signature composed of linked epidermal growth factor receptor family (HER)-AMPK-AKT-mTOR signaling network along with focal adhesion kinase- LIM domain kinase-1 (FAK-LIMK) and janus kinase (JAK)-signal transducers and activators of transcription (STAT) pathways that correlated with short-term survival and aggressive disease.
Conclusions
Functional protein pathway activation mapping of NSCLC reveals distinct activation subgroups that are underpinned by important therapeutic targets and that patients with early-stage node negative disease and poor prognosis may be identified by activation of defined, biochemically linked protein signaling events. Such findings, if confirmed in larger study sets, could help select and stratify patients for personalized targeted therapies.
Keywords: Cell signaling, Protein pathway activation mapping, Reverse-phase protein microarrays, Non–small-cell lung cancer
Lung cancer is the leading cause of cancer-related mortality in the United States and worldwide.1 In 2004, lung cancer caused 20% of all cancer-related deaths in Europe and 29% in the United States.2,3 Lung tumors are routinely classified in two major histological subtypes: small-cell lung carcinoma (SCLC) and non–small-cell lung carcinoma (NSCLC), and NSCLC accounts for approximately 85% of all cases of lung cancer. The majority of NSCLC is further divided into squamous-cell carcinoma, adenocarcinoma, and large-cell carcinoma.4 Adenocarcinoma has become the most prevalent subtype of NSCLC in recent decades.5,6 Although early-stage lung cancer has a higher 5-year survival, the prognosis of stage I lung cancer is highly variable. Postoperative recurrence of stage I NSCLC leads to an early mortality rate of approximately 40%,7 and current clinical pathology techniques cannot distinguish stage I patients into long-term (survivors) and short-term survival (fatality) groups.
Numerous studies have used genomic-based approaches to better characterize the molecular underpinnings of NSCLC and develop new taxonomical means to describe the disease.8–11 Although there have been some recent attempts to use novel discovery-based proteomic approaches for NSCLC cell line studies12–14 and limited protein signaling analysis of clinical material by us and others,15,16 there is yet to be a systematic broadscale analysis of the functional protein signaling architecture of NSCLC clinical samples and of aggressive early-stage disease. A deeper understanding of the ongoing functional protein signaling events within the tumor is of critical importance because protein-expression levels cannot mostly be predicted by gene transcript expression,17–19 and protein signaling events mediated principally by phosphorylation-driven posttranslational modifications are modulated by ongoing kinase activities that are at the nexus of molecularly targeted inhibitors that now comprise a large portion of the current oncology drug pipeline.
A specific kinase-driven pathway that has well-known significance in lung cancer is the epidermal growth factor receptor (EGFR) family signaling network. Overexpression of EGFR has been observed in 40% to 80% of the NSCLC,20–22 which is often associated with aggressive clinical behaviors, such as advanced stage, increased metastatic rate, higher tumor proliferation rate, and poor prognosis.23–25 EGFR overexpression in NSCLC provided a rationale to develop EGFR tyrosine kinase inhibitors (TKIs) that block either receptor extracellular domains or the intracellular kinase activity, and a number of these TKIs have been cleared by the United States Food and Drug Administration for treatment of advanced or metastatic NSCLC.26–30
The observation that certain subgroups of patients, particularly female patients, nonsmokers, East Asians, or patients with lung adenocarcinoma, have a higher response rate and clinical benefit with certain targeted therapies, motivated researchers to elucidate the molecular mechanism responsible for this increased response.31–34 Recent findings have revealed a positive relationship between the presence of activating mutations in the EGFR tyrosine kinase domain and clinical response.31–34 These somatic mutations cause constitutive activation of the EGFR tyrosine kinase by destabilizing its autoinhibited conformation, which is normally maintained in the absence of ligand stimulation.35 However, most of those patients with mutational portraits that predict best response to EGFR TKIs usually relapse and become resistant to further treatment with these inhibitors.36–41 Thus, although most of the clinical research on EGFR has been focused on receptor overexpression and gene mutation profiling/status, only recently have we begun to understand the relationships between EGFR phosphorylation patterns in lung cancer tissue, relationships between EGFR mutations and EGFR phosphorylation levels at defined sites, and an understanding of which EGFR phosphorylation sites are activated in individual patient tumors.15,42
In the current study, we analyzed the activated (phosphorylated) protein signaling architecture in laser capture microdissected (LCM) NSCLC epithelial cells from individual biopsy specimens using reverse phase protein microarray (RPMA) to interrogate over a hundred key signaling proteins in patients with node-negative and node-positive disease. The goals of this study were to use a functional signal pathway activation mapping approach to develop a pilot NSCLC signaling taxonomy knowledge base, a deeper understanding of EGFR signaling architecture, and to determine whether there were protein signaling network activation events that could be found in early-stage N0 disease that correlated with an aggressive phenotype.
MATERIALS AND METHODS
NSCLC Tissue Study Set
The study population consisted of 47 fully informed patients (36 men and 11 women; mean age 66 years; range, 43–83), who underwent surgery for lung NSCLC at Clinica Chirurgica 2, Padova, Italy, between 1993 and 2005. Twenty-seven of the tumors were adenocarcinoma and 20 were squamous carcinoma. According to TNM (tumor, node, metastasis) staging system, 24 (51%) were classified T1, 20 (42.5%) T2, and 3 (6.5%) T3; 28 (59.6%) were N0, 14 (29.8%) were N1, 4 (8.5%) were N2, and 1 (2.1%) Nx. The histological grade was assessed according to World Health Organization criteria: 7 tumors (14.9%) were well differentiated, 25 (53.2%) moderately differentiated, 11 (23.4%) poorly differentiated, and in 4 tumors (8.5%), histological grade was not available (Table 1). All specimens were snap-frozen in liquid nitrogen within 5 minutes of surgical removal to preserve molecular information and stored at −80°C for the entire duration. None of the patients underwent preoperative chemotherapy or radiotherapy, and all tumor specimens were obtained before any therapy from treatment-naive patients.
TABLE 1.
Patient Characteristics of Tissue Study Set
| Primary Lung Cancer Samples | |
|---|---|
|
| |
| No of Patients | Total |
| Female | 11 |
| Male | 36 |
| Histological subtype | |
| Adenocarcinoma | 27 |
| Squamous | 20 |
| T stage | |
| T1 | 24 |
| T2 | 20 |
| T3 | 3 |
| N stage | |
| N0 | 28 |
| N1 | 14 |
| N2 | 4 |
| NX | 1 |
| Tumor grade | |
| GX | 4 |
| G1 | 7 |
| G2 | 25 |
| G3 | 11 |
| Months of survival | |
| <31 | 23 |
| >31 | 24 |
| Status | |
| Died | 38 |
| Alive | 9 |
| Age mean at diagnosis (yr) | 66 (48–85) |
Laser Capture Microdissection and RPMA
Highly enriched (>95%) tumor epithelium cell populations were obtained using LCM as described previously.43,44 Approximately 20,000 cells, taken from several tissue sections to ensure tumor coverage and control for cellular heterogeniety, were procured for each patient sample. Pathway activation mapping was performed by reverse-phase protein microrray (RPMA) as previously described.15,45–47 In brief, LCM procured tumor epithelia were subjected to lysis with 2.5% solution of 2-mercaptoethanol (Sigma, St. Louis, MO) in Tissue Protein Extraction Reagent (t-PER™ Pierce)/2× sodium dodecyl sulfate Tris-Glycine 2× sodium dodecyl sulfate buffer (Invitrogen, Carlsbad, CA). The lysates were printed on glass-backed nitrocellulose array slides (Schott, Louisville, KY) using an Aushon 2470 arrayer (Aushon BioSystems, Burlington, MA) equipped with 185 μm pins. Each primary NSCLC lysate was printed in triplicate. Arrays were blocked (I-Block, Applied BioSystems, Foster City, CA) for 1 hour and subsequently probed with 128 phosphorylated, cleaved or total protein antibodies. Detection was performed using a fluorescence-based tyramide signal amplification strategy using Streptavidin-conjugated IRDye680 (LI-COR Biosciences, Lincoln, NE) as detection reagent. All antibodies were validated for single band specificity and for ligand induction (for phospho-specific antibodies) by Western blotting before use on the arrays, as described previously.47 Each array was scanned using a Vidar scanner (Vidar Systems Corporation, Herndon, VA). After scanning, spot intensity was analyzed, data were normalized to total protein, and a standardized, single data value was generated for each sample on the array by MicroVigene software V2.999 (VigeneTech, North Billerica, MA). A full list of all 128 signaling proteins (including total protein/phosphoprotein and cleaved proteins) is shown in Supplementary Table 1 (Supplemental Digital Content 1, http://links.lww.com/JTO/A346). Protein signaling analytes were chosen for analysis based on their previously described involvement in key aspects of tumorigenesis: growth, survival, autophagy, apoptosis, differentiation, adhesion, motility, and inflammation.
EGFR Mutational Analysis
Mutation status of EGFR kinase domain (exons 18–21) was assessed for each tissue sample. DNA was directly extracted from 10-mm frozen sections by using Maxwell 16 Tissue DNA Purification Kit (Promega Corporation, Madison, WI) according to the manufacturer’s instruction. Amplification of exons 18–21 and fragments sequencing were accomplished using primer sequences previously described.29,32 Polymerase chain reaction (PCR) was executed with Phusion High-Fidelity DNA Polymerases (Finnzymes, Espoo, Finland) with an annealing temperature of 58°C. Amplified fragments were purified using MultiScreen PCRm96 Plate (EMD Millipore, Billerica, MA) according to the manufacturer’s instruction. Purified PCR products were sequenced by using Big Dye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems) to determine the presence or absence of EGFR mutations. Cycle sequencing reactions were performed in 96-well format at 25 cycles of 96°C for 10 seconds, 50°C for 5 seconds, 60°C for 4 minutes. Sequencing reactions were precipitated using Montage SEQ96 Sequencing Reaction Cleanup kit (EMD Millipore, Billerica, MA) heated 3 minutes at 96°C and analyzed with ABI PRISM 3100 Genetic Analyzer with the Sequencing Analysis software (Applied Biosystems).
Statistical Analysis
The continuous variable RPMA data generated were subjected to both unsupervised and supervised statistical analysis. Statistical analyses were performed on final RPMA intensity values obtained using SAS version 9 software (supervised analysis) or JMP v5.0 (SAS Institute, Cary, NC) (for unsupervised clustering analysis). For unsupervised two-way hierarchical clustering, we used the Ward method in JMP. Subgroup cuttings were chosen based on distance plots (shown in bottom right of the heatmaps shown) whereby a rapid rise in the centroid distance defined the cutting point. For supervised analysis, initially, the distribution of variables was checked. If the distribution of variables for the analyzed groups (e.g., primary versus metastasis or metastatic versus nonmetastatic) was normal, a two-sample t test was performed. If the variances of two groups were equal, two-sample t test with a pooled variance procedure was used to compare the means of intensity between two groups. Otherwise, two-sample t test without a pooled variance procedure was adopted. For non-normally distributed variables, the Wilcoxon rank sum test was used. All significance levels were set at a p value less than 0.05. Hazard ratios of Cox proportional hazard model were calculated using R version 2.9.2 software (The R Foundation for Statistical Computing). Hazard ratio h of one variable v1 to another variable v2 means the rate of progression to death of v1 is h times that of v2. p Value of χ2 test tests the null hypothesis that the coefficient of a variable is equal to zero. If the p value for the test is less than 0.05, the null hypothesis is rejected, and this variable, such as pathway signature, sex, age, grade, or stage, is significantly related to survival time. Kaplan–Meier (log-rank) survival estimates were used for univariate survival analysis.
RESULTS
Protein Pathway Activation Mapping of NSCLC
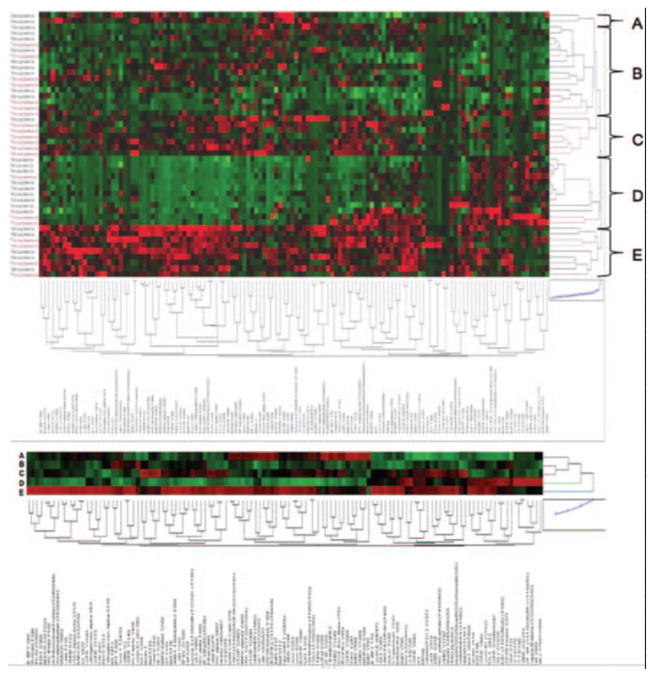
Unsupervised hierarchical clustering analysis (Fig. 1) revealed the presence of five different pathway activation-based subgroups of patients with lung cancer. Group A was composed of only two adenocarcinoma cases and was characterized by the activation of growth-factor–driven signaling, namely activation of ERBB2 and ERBB3, platelet-derived growth factor receptor, vascular endothelial growth factor receptor (VEGFR), and a large number of known linked downstream cytoplasmic signaling networks such as AMPK, phosphoinositide 3-kinase- mammalian target of rapamycin (PI3K)-mTOR, JAK-STAT, and extracellular signal-regulated kinase (ERK) signaling modules. Group B was underpinned by AKT network activation and increased relative Cyclin D1 expression. Group C contained tumors that shared ERBB2-3–based signaling, increased ERBB4 along with activation of SMAD signaling, and Notch homolog 1 (NOTCH) signaling and increased relative expression of autophagy proteins such as Beclin. Group D was dominated by high relative levels of proapoptosis proteins such as nearly all cleaved Caspases measured3,7,9 and cleaved PARP along with increased expression and/or activation of EGFR receptor family, namely EGFR, ERBB3, and ERBB4. Other important tryrosine kinase molecules were activated in group D such as IGFR and RET and ACK. Finally, group E showed a systemic activation of nearly all signaling networks measured, except for ERBB2- and ERBB3-driven signaling, which was comparably lower. These pathway activation mapping results indicate that NSCLC seems to segregate into distinct pathway-driven molecular phenotypes each with a unique molecular signature and shed further light on the molecular heterogeneity of the disease. However, because some of these subgroups (e.g., subgroup A) are composed of a small number of patients, further validation of these pathway modules is absolutely necessary to determine significance and verification of these cohorts.
FIGURE 1.
Protein pathway activation map of NSCLC by unsupervised hierarchical clustering. A, Clustering analysis of 128 signaling proteins (x axis) whose activation/expression levels determined by RPMA are shown for all 47 patient tumor samples obtained by LCM (y axis). The five major pathway-driven subgroups A–E are highlighted. Squamous tumors are shown in red and adenocarcinoma tumors in black (y axis). In the map, red is highest relative level of expression/activation, black is mean values, and green is lowest relative level of expression/activation. B, Clustering analysis as in Panel A except average values for each protein/phosphoprotein within each of the five subgroups were used. NSCLC, non–small-cell lung cancer; LCM, laser capture microdissection.
To fully elucidate the nature of the molecular heterogeneity, we explored the extent of differences in the protein signaling architecture between the two major histological types of NSCLC, adenocarcinoma and squamous cell lung cancers. Mean comparison analysis (Table 2) between squamous and adenocarcinoma groups revealed 26 proteins differently expressed between the two types (p < 0.05). Interestingly, a large majority of these proteins were more highly activated in squamous compared with the adenocarcinoma cases. Only two proteins, total ERBB3/HER3 and protein kinase C (PKC) alpha/beta II T638/641, had higher intensity levels in adenocarcinomas compared with squamous carcinomas. The analysis revealed a systemic EGFR-AKT pathway activation in squamous cell carcinomas compared with adenocarcinomas, with increased phosphorylation of EGFR, ERBB3, and a large number of AKT substrates (BCL2-associated agonist of cell death [BAD], forkhead homolog in rhabdomyosarcoma [FOXO1], forkhead homolog in rhabdomyosarcoma/forkhead box O3 [FOXO1/O3], proline-rich Akt-substrate 1 [PRAS40], eukaryotic translation initiation factor 4E binding protein 1 [4E-BP1], cyclin-dependent kinase inhibitor 1B [p27], and glycogen synthase kinase 3 [GSK3]).
TABLE 2.
Statistically Significant Analytes Squamous versus Adenocarcinoma NCLC
| Proteins/Phosphoproteins | p. Value | Expression/Activation Trend |
|---|---|---|
| PKCalpha/betaII (T638/641) | 0.0296 | S▼ |
| ERBB3/HER3 | 0.0002 | S▼ |
| FOXO1 (T24)/FOXO3a (T32) | 0.0003 | S▲ |
| LKB1 S334 | 0.0023 | S▲ |
| 4E-BP1 T70 | 0.0042 | S▲ |
| PRAS40 T246 | 0.0052 | S▲ |
| p27 T187 | 0.0058 | S▲ |
| Akt S473 | 0.0062 | S▲ |
| PKC zeta/lamda (T410/403) | 0.0091 | S▲ |
| ACK1 Y284 | 0.0093 | S▲ |
| Src Family Y416 | 0.0112 | S▲ |
| MST1 (T183)/MST2 (T180) | 0.0131 | S▲ |
| SMAD1(S/S)/SMAD5(S/S)/SMAD8(S/S) | 0.0174 | S▲ |
| Total EGFR | 0.0174 | S▲ |
| AURORA A (T288)/B (T232)/C (T198) | 0.0174 | S▲ |
| GSK3 alpha-beta S21/9 | 0.0235 | S▲ |
| BAD S136 | 0.0002 | S▲ |
| PKC theta T538 | 0.0412 | S▲ |
| FOXO1 S256 | 0.0491 | S▲ |
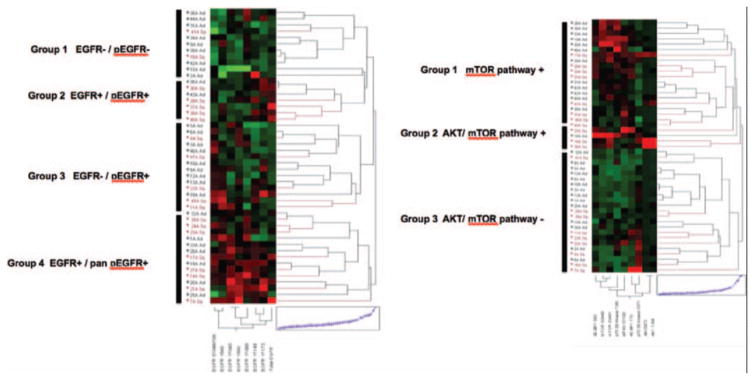
On the basis of these results, we next explored more specifically the signaling aspects of the EGFR and AKT-mTOR networks within squamous and adenocarcinomas, leveraging the multiplexed nature of the reverse phase protein microarray (RPMA) format. Unsupervised hierarchical clustering analysis (Fig. 2A) of total EGFR and seven independent phosphorylation sites of the receptor (Y1173, Y1148, Y1068, Y992, Y845, Y1045, S1046/1047) revealed that the EGFR signaling architecture clustered into four major groups. These groupings were characterized by overt lack of EGFR protein and concomitant lack of EGFR activation at any site (group 1), or high expression of EGFR protein along with EGFR activation/phosphorylation (group 2), low or absent total EGFR yet relatively high levels of EGFR activation/phosphorylation at one or more of the seven sites measured (group 3), and high levels of EGFR protein and high levels of phosphorylation of EGFR at nearly all of the seven phosphorylation sites measured (group 4). This analysis revealed the molecular heterogeneity of EGFR signaling in NSCLC, and that there is a subset of patients with NSCLC that harbor low EGFR yet relatively high levels of receptor activation (group 3). However, the distribution of adenocarcinoma (black label, Fig. 2A) and squamous NSCLC (red label, Fig. 2A) was fairly even among the signaling clusters. In keeping with the statistical analysis that revealed increased activation of EGFR and AKT-mTOR in squamous cell carcinomas, of the four subgroups identified based on EGFR signaling (Fig. 2 A), the one subgroup (subgroup 1) that is characterized by lack of EGFR expression and EGFR signaling/phosphorylation is composed of a majority of adenocarcinomas (9 of 11, 82%). Moreover, the signaling subgroups were underpinned neither by a statistically significant association with nodal status nor by EGFR mutational status (data not shown).
FIGURE 2.
NSCLC EGFR signaling analysis. A, Clustering analysis of total EGFR and seven EGFR phosphorylation sites signaling proteins (in order from left to right: S1046/1047, Y845, Y1045, Y992, Y1068, Y1148, Y1173,) (x axis) whose activation/expression levels determined by RPMA are shown for all 47 patient tumor samples obtained by LCM (y axis). The four major pathway-driven subgroups1–4 are highlighted. Squamous tumors are shown in red and adenocarcinoma tumors in black (y axis). In the map, red is highest relative level of expression/activation, black is mean values, and green is lowest relative level of expression/activation. B, Clustering analysis of AKT-mTOR pathway network, showing three major cohorts (groups 1–3). Pathway activation analytes on x axis from left to right: 4EBP1 (S65), mTOR (S2448), mTOR (S2481), p70 S6 (S371), eIF4G (S1108), 4EBP1 (T70), p70 S6 (T389), AKT (S473), AKT (T308). NSCLC, non–small-cell lung cancer; LCM, laser capture microdissection; 4E-BP1, eukaryotic translation initiation factor 4E binding protein 1; mTOR, mammalian target of rapamycin; AKT, v-akt murine thymoma viral oncogene homolog 1.
Pathway activation mapping of the AKT-mTOR signaling axis, whereby activation of AKT, mTOR, 4EBP1, p70S6K, and eIF4G was measured, revealed three overarching subgroups of activation (Fig. 2B). The results showed both AKT and mTOR networks having high relative activation (group 1), mainly mTOR pathway activation (group 2), or neither pathway significantly activated (group 3). In keeping with the statistical results that revealed increased relative levels of AKT-mTOR pathway signaling proteins in squamous cell NSCLC compared with adenocarcinomas, the AKT-mTOR pathway activation subgrouping analysis (Fig. 2 B) revealed that a majority (14 of 23, 61%) of the AKT-mTOR pathway negative subgroup (subgroup 3) were adenocarcinomas, and 75% (3 of 4) of the highly activated AKT-mTOR samples in subgroup 2 were squamous cell NSCLC tumors. These results support those seen for EGFR network activation, namely that NSCLC is composed of distinct, yet heterogeneous, subgroups underpinned by pathway activation differences. Generally, AKT activation was seen concomitant with mTOR pathway activation, which is known to lie downstream of AKT. No statistically significant correlation with EGFR mutation status was seen with AKT-mTOR activation/phosphorylation subgroups (data not shown).
Protein Pathway Activation Mapping of Node-Negative NSCLC
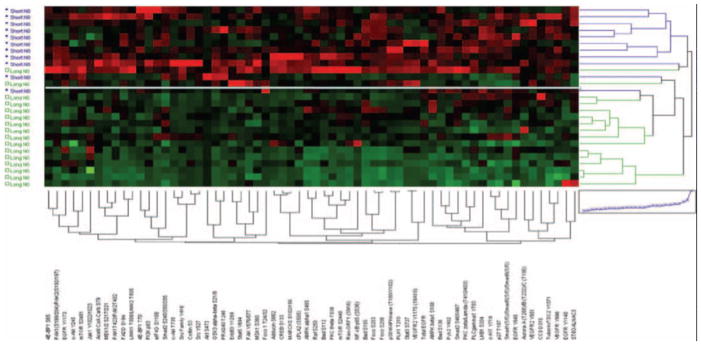
Although the signaling architecture of pathways that underpin important current molecular targets for NSCLC such as EGFR, AKT, and mTOR provide evidence of distinct heterogeneity that has implications for therapeutic stratification and response to these therapies, of critical importance in the management of lung cancer is the identification of patients with node-negative disease, who have aggressive tumors. Our study set of tumors contained 28 N0 tumors of both adenocarcinoma (n = 18) and squamous (n = 9) carcinoma lineage, and this set provided a unique opportunity to determine whether any protein signaling network(s) correlated with overall survival (OS). As shown in Table 3, using a median OS value of 31 months as a cutpoint for short- versus long-term survival for 27 N0 patients from whom the OS data were obtained, RPMA analysis revealed 65 signaling analytes that were significantly more highly activated/phosphorylated in the short-term OS N0 cases (n = 11; range, 2–31 months, median 9 months) versus long-term OS (n = 16; range, 31–120 months, median 90 months). Unsupervised clustering analysis of the data reveals, as expected, near complete segregation of the short-term OS from long-term OS node-negative group (Fig. 3) with the notable high systemic activation/phosphorylation of many of the signaling proteins seen.
TABLE 3.
Statistically Significant Analytes for N0 NSCLC with Short-Term Survival
| Proteins | No p Value |
|---|---|
| 4E-BP1 S65 | 0.0394 |
| 4E-BP1 T70 | 0.0462 |
| Acetil CoA Carb S79 | 0.0256 |
| Adducin S662 | 0.0049 |
| Akt S473 | 0.0849 |
| AMPK alpha1 S485 | 0.0041 |
| AMPK beta1 S108 | 0.0286 |
| Aurora A (T288)/B (T232)/C (T198) | 0.0107 |
| Bad S112 | 0.0057 |
| Bad S136 | 0.0035 |
| Bad S155 | 0.0228 |
| B-Raf S445 | 0.0273 |
| c-Abl T735 | 0.0049 |
| c-Abl Y245 | 0.0343 |
| CC9 D330 | 0.0122 |
| Chk1 S345 | 0.0273 |
| c-KIT Y719 | 0.0128 |
| cPLA2 (S505) | 0.031 |
| CREB S133 | 0.0162 |
| EGFR Y1148 | 0.0497 |
| EGFR Y1173 | 0.0097 |
| eIF4E S209 | 0.0452 |
| eIF4G S1108 | 0.0179 |
| ErbB3 Y1197 | 0.0273 |
| ErbB3 Y1289 | 0.035 |
| FADD S194 | 0.0057 |
| FAK Y576/577 | 0.0185 |
| FoxO1/3a T24/32 | 0.0029 |
| FoxO3A S253 | 0.0047 |
| FoxO1 S256 | 0.0003 |
| GSK3 alpha-beta S21/9 | 0.0041 |
| Jak1 Y1022/1023 | 0.0538 |
| LIMK1 T508/LIMK2 T505 | 0.0161 |
| LKB1 S334 | 0.0241 |
| MARCKS S152/156 | 0.0011 |
| MEK1/2 S217/221 | 0.014 |
| MSK1 S360 | 0.0043 |
| mTOR S2448 | 0.0106 |
| p27 T187 | 0.0101 |
| p38 MAPKinase (T180/Y182) | 0.0078 |
| PAK1(S199/204)/PAK2(S192/197) | 0.0426 |
| PAK1T423/PAK2T402 | 0.0122 |
| PKC theta T538 | 0.0011 |
| PKC zeta/Lamda (T410/403) | 0.0049 |
| PLCgamma1 Y783 | 0.0114 |
| PLK1 T210 | 0.0248 |
| PRAS40 T246 | 0.0001 |
| Pyk2 Y402 | 0.0014 |
| Raf S259 | 0.0139 |
| Ras-GRF1 (S916) | 0.031 |
| Smad1(S/S)/Smad5(S/S)/Smad8(S/S) | 0.0168 |
| Smad2 S245/250/255 | 0.0037 |
| Smad2 S465/467 | 0.0443 |
| Src Family Y416 | 0.0014 |
| Src Y527 | 0.0162 |
| ST6GALNAC5 | 0.0885 |
| Stat3 S727 | 0.0055 |
| Stat5 Y694 | 0.0185 |
| Tuberin/TSC2 Y1571 | 0.0088 |
| VEGFR Y996 | 0.0016 |
| VEGFR2 Y1175 (19A10) | 0.0018 |
| VEGFR2 Y951 | 0.0001 |
FIGURE 3.
Clustering analysis of protein signaling architecture in node-negative NSCLC. Proteins from Table 4 were used in unsupervised hierarchical clustering analysis of 27 tumors from patients with N0 NSCLC. Patients with short-term survival (median OS = 9 months) are shown in blue (y axis) and long-term survival (median OS = 90 months) in green. Phosphoproteins/proteins are shown on the x axis. In the map, red is highest relative level of expression/activation, black is intermediate values, and green is lowest relative level of expression/activation. White line is shown that segregates the two clusters formed. NSCLC, non–small-cell lung cancer.
On the basis of this observation, an optimal cutpoint of 2.8 relative units based on a protein pathway activation signature was calculated using a combination of normalized and scaled relative intensity values of the 65 analytes shown in Table 3. The optimal cutpoint was determined by receiver operating characteristic curve analysis, which gave a sensitivity of 91% (10 of 11) and specificity of 88% (14 of 16) for distinguishing short-term (median 9 months) from long-term (median 90 months) survival in the 27 N0 patient population from which the OS was known. The score was determined by first normalizing the relative intensity values of each of the 65 analytes that were increased in patients with N0 and short-term survival. Within each analyte, the intensity value of every sample was divided by the highest patient’s value within the entire cohort of 65 patients. Normalized intensity values of each analyte were then summed for every patient. Thus, the final pathway signature score did not weigh any significant endpoint as more important than another. Cox proportional hazard model analysis of the data (Table 4) revealed that only the pathway signature score (p = 0.0001) and histology for adenocarcinoma (p = 0.01) had statistical significance for OS compared with other clinical variables measured, including nodal status, sex, grade, site, and stage. Because only four of the 27 N0 patients harbored an EGFR mutation, statistical correlation with OS was not determined.
TABLE 4.
Cox Proportional Analysis
| Characteristic | Cases | Deaths | Median Survival (mo) | p-Value Log-Rank Test |
|---|---|---|---|---|
| Sex | ||||
| Male | 20 | 15 | 42 | |
| Female | 7 | 5 | 42 | 0.99 |
| Age (yr) | ||||
| <70 | 16 | 12 | 42 | |
| ≥70 | 11 | 8 | 33 | 0.56 |
| Pathway activation signature | ||||
| 0 | 11 | 11 | 9 | |
| 1 | 16 | 9 | 90 | 0.0001 |
| T | ||||
| 1 | 18 | 13 | 46 | |
| 2–3 | 9 | 7 | 9 | 0.27 |
| Grade | ||||
| 1 | 5 | 4 | 42 | |
| 2 | 15 | 10 | 54 | |
| 3 | 7 | 6 | 15 | 0.18 |
| Histology | ||||
| Squamo | 9 | 8 | 15 | |
| Adeno | 18 | 12 | 63 | 0.01 |
Characterization of the underpinning repertoire of the statistically significant activated proteins that comprised the aggressive signature revealed a large number of receptor tyrosine kinases (RTK), EGFR family members, with multiple independent phosphorylation sites on a number of the receptors that were measured (EGFR (Y1173), (Y1148), and (Y845); ERBB3 (Y1289) and (Y1197); VEGFR (Y996), (Y951), and (Y1175); c-KIT Y7194). Concomitantly, many other proteins known to be downstream in RTK/growth factor–driven cellular signaling pathways were coordinately activated. In particular, the hierarchical clustering analysis revealed the combined expression of many proteins that belonged to the AKT signaling pathway, including AKT (S473), GSK3αβ (S21/9), PRAS40 (T246), multiple forkhead family members FOXO1/3A (T24/32), FOXO3A S253, FOXO1 S256, along with BAD (S155, S136, and S112) and p27 (T187), TSC2 (Y1571) as well as collateral mTOR networks (mTOR (S2448), 4EBP1 (S65 and T70)). Other linked networks seen coordinately activated in aggressive N0 tumors at multiple nodes were the AMPK signaling pathway (AMPK α (S485), AMPK β (S108), ACC (S79), LKB1 (S334)), SMAD signaling, pathways regulating motility and adhesion (e.g., FAK (Y576/577), LIMK 1/2 (T508/T505)), and the JAK-STAT pathway (JAK1 (Y1022/1023), STAT3 (S727), and STAT5 (Y694)).
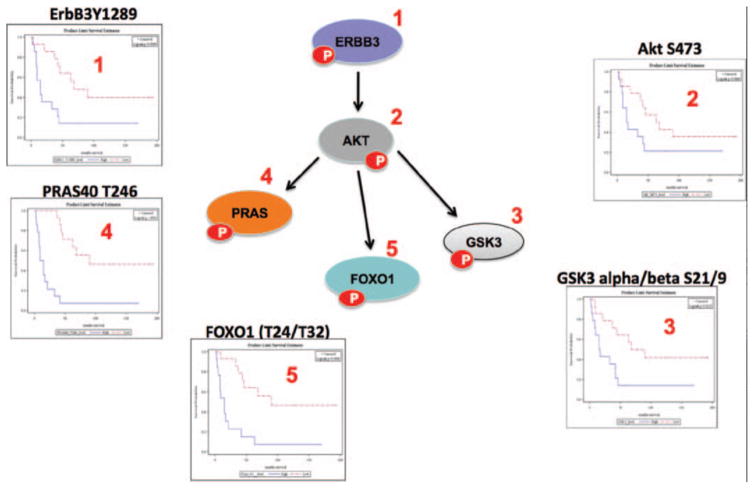
The coordinate nature of the activated kinase-substrate linkage within the signaling architecture of aggressive N0 tumors is visually revealed in Fig. 4, where a selection of the independent statistically significant nodes within the RTK-AKT pathway was mapped to a signaling diagram (reproduced courtesy of Cell Signaling, Inc., www.cellsignal.com). For each of the five interconected signaling pathway protein nodes selected, independent Kaplan–Meier plots are shown for N0 patients whose tumors have relative phosphorylation/activation levels above (HIGH, in blue) or below (LOW, in red) the median level of the selected analyte across the study set population. The data reveal both the nature of the discrimination achieved by the specific individual protein activation and the interlinked nature of the data, which indicates a coordinate activation of the pathways in patients with N0 disease and an aggressive intrinsic phenotype.
FIGURE 4.
Activated c-erbB3-AKT signaling network in node-negative NSCLC correlates with poor overall survival. A focused analysis of five biochemically linked signaling proteins (ERBB3, PRAS40, FOXO1, GSK3, and AKT) found within the overarching protein activation signature shown in Figure 3 is shown using a signaling cartoon of AKT signaling network (reproduced courtesy of Cell Signaling Technology, Inc., www.cellsignal.com) along with the individual statistically significant KM plots (ERBB3: p = 0.0089; PRAS40: p < 0.0001; FOXO1: p = 0.0006; GSK3: p = 0.0125; and AKT: p = 0.089) of each phosphoprotein shown numbered in the map. For the KM plots, red shows low levels of activation of the cognate protein and blue shows high relative phosphorylation levels (median intensity value of each protein was used as the cutpoint for statistical analysis). ERBB3,; PRAS40,; FOXO1,; GSK3,; AKT,; NSCLC, non–small-cell lung cancer; KM, Kaplan–Meier.
DISCUSSION
A broadscale analysis of the functional protein signaling architecture of NCSLC, quantitatively measuring 128 key signaling proteins concomitantly, revealed the presence of distinct molecular subgroups underpinned by distinct interlinked signaling pathways and at the same time displaying unique patient-specific signaling heterogeneity. Such pathways are revealed because we are measuring enzymatically driven protein phosphorylation as direct read-out of the preceding kinase-phosphatase activity. This activity is based on the aggregate of pathway use in asynchratic tissue cells whose combined snapshots in time provide a window into the ongoing signaling activity. The results showed that the overall signaling profiles of squamous and adenocarcinoma tumors seemed to co-mingle, although the two were composed of distinct underpinning signaling motifs. A deeper investigation into EGFR signaling cascades identified unique molecular subgroups of an EGFR-low/pEGFR-high cohort and an EGFR-high/pEGFR-low cohort of NSCLC. Last, pathway activation mapping analysis identified biochemically interlinked RTK-driven protein pathways that could distinguish N0 patients with short-term OS (median 9 months) from long-term (median 61 months) survival. Analysis of the AKT-mTOR pathway activation network also revealed distinct molecular subgroups of activation with both concomitant linked activation of AKT and mTOR signaling and independent activation of either module.
The study used a protein microarray–driven platform, the RPMA, which has been extensively used by us in other analyses of human malignancies,45–47 to perform the most comprehensive mapping of the protein signaling architecture of human NSCLC clinical specimens to date. The study set used a collection of frozen specimens that were carefully chosen based on the control of preanalytical variables such as sample collection, handling, storage, and time-to-freezing after surgical removal. Furthermore, based on past evidence that upfront cellular enrichment is required for accurate protein measurement/activation determination of cellular tissue compartments,46,48 we used LCM to significantly enrich for tumor epithelium (>95% purity based on pre- and post-LCM microscopic visualization) as the cellular input for all analysis.
The combination of these efforts has produced an important preliminary data set that will require much further efforts to verify the signaling subtypes identified, and the pathway activation found in tumors from patients with node-negative disease who have poor OS. Despite the powerful methodology used combined with the LCM enrichment techniques, the study set used comprised too small a number (47 cases) to generate final conclusive data about what pathway activation phenotypes underpin NSCLC and aggressive protein pathway activation signatures. However, on the basis of methodological workflow used, we expect that this pilot data can be confidently used as a launch-point for further hypothesis-driven analysis and verification of the results herein. Moreover, although the signaling analysis was extensive, the full repertoire of the human kinome was certainly not surveyed and many specific signaling proteins were not queried, and a number of important signaling pathways untested. We fully expect that even further coverage of the signal transduction machinery will yield new discoveries. However, our rationale for this study was to select key signaling proteins known to be involved in tumorigenesis and metastasis, regulating growth and energy metaboism, survival, apoptosis, differentiation, motility, and inflammation and which were key surrogates and direct targets for the many kinase inhibitors that populate current phase I–III clinical trial pipeline. Furthermore, although signaling can be regulated by a number of post-translational modification–driven events (e.g., glycosylation and acetylation), we chose to study these proteins at the functional level by measuring protein phosphorylation (the principal regulator of signal transduction and the key analyte endpoint for the recording of ongoing cellular kinase activity) so that we could generate a direct knowledge snapshot of the ongoing signaling cascades within the tumor cells. Moreover, we postulate that we are able to identify signaling derangements even in unmeasured individual signaling proteins based on the fact that we are measuring many of the key principal “hub” locations within the signaling architecture which integrate signaling activation from a multitude of inputs.
Past and recent work involving the analysis of NSCLC using both genomic and proteomic approaches has found evidence for molecular heterogeniety and distinct cohorts of patients underpinned by specific gene expression and protein expression differences.8–15 Recent signaling analysis of NSCLC clinical samples using a much smaller number of phosphoprotein endpoints than what was used in this study found that tumor signaling portraits could be accurately distinguished from matched normal tissue and that energy-sensing signaling networks correlated with recurrence.16
Our analysis indicated that the signaling architecture of NSCLC makes it highly amenable to targeted therapy-based inhibition and consideration of new combinations of therapeutics. We identified distinct cohorts of patients whose tumor portraits contained relatively high levels of activation of RTKs such as platelet-derived growth factor receptor, EGFR concomitant with ERBB2 and ERBB3 activation, along with RET and ACK, and downstream pathway activation through cytoplasmic signaling such as AKT-mTOR, RAS-ERK, and JAK-STAT activation. Although it is not known at this time whether the activation levels are driven by activating mutations or exogenous receptor-ligand signaling cascades and tumor-stroma interactions, therapeutic targeting, and modulation of the activation could test the causal role the high levels of pathway activation have in each patient tumor.
The identification of cohorts of patients with NSCLC with relatively high and low levels of total receptor proteins such as EGFR, yet disconcordant phosphorylation levels of the same protein, could be clinically important if validated in further study sets. Although identification of patients who respond to EGFR inhibitors based on mutational analysis and alternate pathway activation has been a poster child for pharmacogenomics,31–41 recent reports indicate that mutational status may be accurately predicted by EGFR phosphorylation patterns, especially Y1068 and Y104515,49 and Y1173,42 and thus, protein phosphorylation profiling of EGFR may provide a new companion diagnostic assay method for selection and stratification for therapy.50 Development of quantitative approaches to measure EGFR phosphorylation, such as by RPMA, can be used to identify optimal cutpoints for molecular correlates, and a more comprehensive quantitative survey of the many EGFR phosphorylation sites (we measured seven different sites) along with downstream signaling analysis through AKT and ERK could yield better response prediction to EGFR inhibitors. The fact that AKT and mTOR pathway activation were found in patient subgroups in which both modules were activated simultaneously or alone indicates the potential to stratify patients for combination therapeutics that target PI3K-AKT and downstream mTOR together, or PI3K and mTOR inhibitors alone.
Identification of patients with early-stage NSCLC, who have aggressive tumors that are predestined to a more rapid disease progression would be of critical importance, especially those patients with N0 disease who are mostly left untreated after surgery. These patients could be stratified to more aggressive paths of treatment and monitoring. Thus, pathway activation mapping provides data that could extend utility beyond prognostic signatures of recurrence and progression as has been done by mRNA and miRNA profiling51–55 and provides data that could rationally specify the best therapies that would work to mitigate the rapid course of the disease in N0 patients with the most aggressive disease course. Our preliminary finding points to a systemic activation of interlinked pathway networks in patients with aggressive N0 disease composed of a number of important drug targets. Members of the AMPK-LKB1 pathway, vascular epidermal growth factor receptor (VEGFR), EGFR/ERBB3, PTK2B protein tyrosine kinase 2 beta- focal adhesion kinase (PYK2-FAK), aurora kinase A (AURORA) and polo-like kinase 1 (PLK1), and JAK-STAT, concomitant with many members of the AKT-mTOR downstream signaling modules, which integrate these upstream activating events, were all found systemically activated in these aggressive tumors. Each of these is under intense investigation for the development of targeted therapy inhibitors, so this signature, if validated in larger independent study sets, could form the basis not only for prognostic determination but also for clinicians so that they have an armamentarium of molecularly targeted inhibitors to use for these patients in prospective clinical trials. This signature was based on a single pilot study and would require much further verification in independent study sets to understand any potential clinical significance. However, some of the molecules we identified in the aggressive signature have also been recently reported as being implicated in aggressive early-stage NSCLC and patients with NSCLC with worse overall outcome, which, although not minimizing the existing need for independent validation, does provide some level of confidence to the overall fidelity of our initial findings. Analysis of 134 patients with resected stage IA–IIB NSCLC revealed that total levels of mTOR protein as measured by immunohistochemistry (IHC) were significantly higher in those patients with node-negative or stage IA disease who had poor outcome.56 Proteomic analysis of recurrent node-negative tumors identified activation of AMPK and adhesion molecules.13 Analysis by IHC of FoxM1 total protein levels from squamous cell NSCLC revealed a statistical correlation with outcome and an aggressive clinical course.57 Recently, it was reported that patients with NSCLC whose tumors had high relative levels of AKT phosphorylation and loss of PTEN expression showed significantly worse 5-year survival rates,58 and that increased levels of phosphorylated eIF4E are associated with survival through AKT pathway activation in NSCLC.59 Interestingly, we found that activation/phosphorylation of multiple SMAD family members (SMAD1/5/8 and SMAD2) was increased in patients with N0 tumors and poor survival. Given the dichotomous nature of transforming growth factor–driven signaling,60 our preliminary evidence that activation of SMAD signaling occurs in patients with early-stage, node-negative lung cancer with short survival warrants further investigation.
Although the causal significance of the pathway activation modules identified in this initial survey of the pathway activation architecture of NSCLC remains unknown, we hope that this initial survey will serve as a beginning for mapping signaling events in clinical specimens that could be used to rationally decide on targeted therapy choices and even prioritize combinations of therapeutics.61,62 Because our analysis did not include chemoresponse or targeted therapy prediction, our results are limited to molecular characterization of underpinning signaling network architecture. Because the architecture provided by the RPMA is focused around druggable targets in purified NSCLC tumor epithelium, it is our hope that this inaugural data set will form the basis by which predictive marker analysis/discovery could occur within the context of chemo/targeted therapeutic response. Moreover, our ongoing validation work in larger independent tumor sets where both chemo/targeted therapies were used and outcomes known, will provide the basis for selection and ranking those baseline signaling networks that will be chosen for further analysis in both animal models for functional workup and prospective clinical assay development for personalized medicine-based opportunities.
Supplementary Material
Acknowledgments
The authors appreciate the generous support of Dr. Vikas Chandhoke and the College of Life Sciences at George Mason University. This work was partly supported by the Italian Istituto Superiore di Sanità within the framework Italy/USA cooperation agreement between the U.S. Department of Health and Human Services, George Mason University, and the Italian Ministry of Public Health.
Footnotes
Disclosure: J.W., L.L. and E.P. are inventors on U.S. government and/or University assigned patents and patent applications that cover aspects of the technologies discussed. As inventors, they are entitled to receive royalties as provided by U.S. law and George Mason University policy. J.W., L.L., and E.P. are consultants and shareholders of Theranostics Health, Inc. The authors declare no conflicts of interest.
References
- 1.Parkin DM. Global cancer statistics in the year 2000. Lancet Oncol. 2001;2:533–543. doi: 10.1016/S1470-2045(01)00486-7. [DOI] [PubMed] [Google Scholar]
- 2.Boyle P, Ferlay J. Cancer incidence and mortality in Europe 2004. Ann Oncol. 2005;6:481–488. doi: 10.1093/annonc/mdi098. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Murray T, Ward E, et al. Cancer statistics, 2005. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 4.Brambilla E, Travis WD, Colby TV, Corrin B, Shimosato Y. The new World Health Organization classification of lung tumours. Eur Respir J. 2001;18:1059–1068. doi: 10.1183/09031936.01.00275301. [DOI] [PubMed] [Google Scholar]
- 5.Travis WD, Travis LB, Devesa SS. Lung cancer. Cancer. 1995;75(1 Suppl):191–202. doi: 10.1002/1097-0142(19950101)75:1+<191::aid-cncr2820751307>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 6.Franceschi S, Bidoli E. The epidemiology of lung cancer. Ann Oncol. 1999;10(Suppl 5):S3–S6. doi: 10.1093/annonc/10.suppl_5.s3. [DOI] [PubMed] [Google Scholar]
- 7.Goldstraw P, Crowley J, Chansky K, et al. International Association for the Study of Lung Cancer International Staging Committee; Participating Institutions. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007;2:706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- 8.Kurie JM, Shin HJ, Lee JS, et al. Increased epidermal growth factor receptor expression in metaplastic bronchial epithelium. Clin Cancer Res. 1996;2:1787–1793. [PubMed] [Google Scholar]
- 9.Hayes DN, Monti S, Parmigiani G, et al. Gene expression profiling reveals reproducible human lung adenocarcinoma subtypes in multiple independent patient cohorts. J Clin Oncol. 2006;24:5079–5090. doi: 10.1200/JCO.2005.05.1748. [DOI] [PubMed] [Google Scholar]
- 10.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryant CM, Albertus DL, Kim S, et al. Clinically relevant characterization of lung adenocarcinoma subtypes based on cellular pathways: an international validation study. PLoS ONE. 2010;5:e11712. doi: 10.1371/journal.pone.0011712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Machida K, Eschrich S, Li J, et al. Characterizing tyrosine phosphorylation signaling in lung cancer using SH2 profiling. PLoS ONE. 2010;5:e13470. doi: 10.1371/journal.pone.0013470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu G, Xiao CL, Lu CH, et al. Phosphoproteome profile of human lung cancer cell line A549. Mol Biosyst. 2011;7:472–479. doi: 10.1039/c0mb00055h. [DOI] [PubMed] [Google Scholar]
- 15.Wang YT, Tsai CF, Hong TC, et al. An informatics-assisted label-free quantitation strategy that depicts phosphoproteomic profiles in lung cancer cell invasion. J Proteome Res. 2010;9:5582–5597. doi: 10.1021/pr100394u. [DOI] [PubMed] [Google Scholar]
- 16.VanMeter AJ, Rodriguez AS, Bowman ED, et al. Laser capture microdissection and protein microarray analysis of human non-small cell lung cancer: differential epidermal growth factor receptor (EGPR) phosphorylation events associated with mutated EGFR compared with wild type. Mol Cell Proteomics. 2008;7:1902–1924. doi: 10.1074/mcp.M800204-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nanjundan M, Byers LA, Carey MS, et al. Proteomic profiling identifies pathways dysregulated in non-small cell lung cancer and an inverse association of AMPK and adhesion pathways with recurrence. J Thorac Oncol. 2010;5:1894–1904. doi: 10.1097/JTO.0b013e3181f2a266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson L, Seilhamer J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis. 1997;18:533–537. doi: 10.1002/elps.1150180333. [DOI] [PubMed] [Google Scholar]
- 19.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shankavaram UT, Reinhold WC, Nishizuka S, et al. Transcript and protein expression profiles of the NCI-60 cancer cell panel: an integromic microarray study. Mol Cancer Ther. 2007;6:820–832. doi: 10.1158/1535-7163.MCT-06-0650. [DOI] [PubMed] [Google Scholar]
- 21.Piyathilake CJ, Frost AR, Manne U, et al. Differential expression of growth factors in squamous cell carcinoma and precancerous lesions of the lung. Clin Cancer Res. 2002;8:734–744. [PubMed] [Google Scholar]
- 22.Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol. 1995;19:183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- 23.Scagliotti GV, Selvaggi G, Novello S, Hirsch FR. The biology of epidermal growth factor receptor in lung cancer. Clin Cancer Res. 2004;10(12 Pt 2):4227s–4232s. doi: 10.1158/1078-0432.CCR-040007. [DOI] [PubMed] [Google Scholar]
- 24.Ohsaki Y, Tanno S, Fujita Y, et al. Epidermal growth factor receptor expression correlates with poor prognosis in non-small cell lung cancer patients with p53 overexpression. Oncol Rep. 2000;7:603–607. doi: 10.3892/or.7.3.603. [DOI] [PubMed] [Google Scholar]
- 25.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37(Suppl 4):S9–15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 26.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 27.Mendelsohn J, Baselga J. The EGF receptor family as targets for cancer therapy. Oncogene. 2000;19:6550–6565. doi: 10.1038/sj.onc.1204082. [DOI] [PubMed] [Google Scholar]
- 28.Engelman JA, Cantley LC. The role of the ErbB family members in non-small cell lung cancers sensitive to epidermal growth factor receptor kinase inhibitors. Clin Cancer Res. 2006;12(14 Pt 2):4372s–4376s. doi: 10.1158/1078-0432.CCR-06-0795. [DOI] [PubMed] [Google Scholar]
- 29.Rosell R, Viteri S, Molina MA, Benlloch S, Taron M. Epidermal growth factor receptor tyrosine kinase inhibitors as first-line treatment in advanced nonsmall-cell lung cancer. Curr Opin Oncol. 2010;22:112–120. doi: 10.1097/CCO.0b013e32833500d2. [DOI] [PubMed] [Google Scholar]
- 30.Mendelsohn J. The epidermal growth factor receptor as a target for cancer therapy. Endocr Relat Cancer. 2001;8:3–9. doi: 10.1677/erc.0.0080003. [DOI] [PubMed] [Google Scholar]
- 31.Cohen MH, Williams GA, Sridhara R, Chen G, Pazdur R. FDA drug approval summary: gefitinib (ZD1839) (Iressa) tablets. Oncologist. 2003;8:303–306. doi: 10.1634/theoncologist.8-4-303. [DOI] [PubMed] [Google Scholar]
- 32.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 33.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 34.Mitsudomi T, Kosaka T, Endoh H, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23:2513–2520. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- 35.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yun CH, Boggon TJ, Li Y, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carey KD, Garton AJ, Romero MS, et al. Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res. 2006;66:8163–8171. doi: 10.1158/0008-5472.CAN-06-0453. [DOI] [PubMed] [Google Scholar]
- 38.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riely GJ, Pao W, Pham D, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12(3 Pt 1):839–844. doi: 10.1158/1078-0432.CCR-05-1846. [DOI] [PubMed] [Google Scholar]
- 40.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 41.Linardou H, Dahabreh IJ, Kanaloupiti D, et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008;9:962–972. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- 42.Dahabreh IJ, Linardou H, Siannis F, Kosmidis P, Bafaloukos D, Murray S. Somatic EGFR mutation and gene copy gain as predictive biomarkers for response to tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2010;16:291–303. doi: 10.1158/1078-0432.CCR-09-1660. [DOI] [PubMed] [Google Scholar]
- 43.Zhang G, Fang B, Liu RZ, et al. Mass spectrometry mapping of epidermal growth factor receptor phosphorylation related to oncogenic mutations and tyrosine kinase inhibitor sensitivity. J Proteome Res. 2011;10:305–319. doi: 10.1021/pr1006203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emmert-Buck MR, Bonner RF, Smith PD, et al. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 45.Espina V, Wulfkuhle JD, Calvert VS, et al. Laser-capture microdissection. Nat Protoc. 2006;1:586–603. doi: 10.1038/nprot.2006.85. [DOI] [PubMed] [Google Scholar]
- 46.Paweletz CP, Charboneau L, Bichsel VE, et al. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene. 2001;20:1981–1989. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- 47.Wulfkuhle JD, Speer R, Pierobon M, et al. Multiplexed cell signaling analysis of human breast cancer applications for personalized therapy. J Proteome Res. 2008;7:1508–1517. doi: 10.1021/pr7008127. [DOI] [PubMed] [Google Scholar]
- 48.Pierobon M, Calvert V, Belluco C, et al. Multiplexed cell signaling analysis of metastatic and nonmetastatic colorectal cancer reveals COX2-EGFR signaling activation as a potential prognostic pathway biomarker. Clin Colorectal Cancer. 2009;8:110–117. doi: 10.3816/CCC.2009.n.018. [DOI] [PubMed] [Google Scholar]
- 49.Silvestri A, Colombatti A, Calvert VS, et al. Protein pathway biomarker analysis of human cancer reveals requirement for upfront cellular-enrichment processing. Lab Invest. 2010;90:787–796. doi: 10.1038/labinvest.2010.47. [DOI] [PubMed] [Google Scholar]
- 50.McMillen E, Ye F, Li G, Wu Y, Yin G, Liu W. Epidermal growth factor receptor (EGFR) mutation and p-EGFR expression in resected non-small cell lung cancer. Exp Lung Res. 2010;36:531–537. doi: 10.3109/01902148.2010.482176. [DOI] [PubMed] [Google Scholar]
- 51.Havaleshko DM, Smith SC, Cho H, et al. Comparison of global versus epidermal growth factor receptor pathway profiling for prediction of lapatinib sensitivity in bladder cancer. Neoplasia. 2009;11:1185–1193. doi: 10.1593/neo.09898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patnaik SK, Kannisto E, Knudsen S, Yendamuri S. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res. 2010;70:36–45. doi: 10.1158/0008-5472.CAN-09-3153. [DOI] [PubMed] [Google Scholar]
- 53.Kadara H, Behrens C, Yuan P, et al. A five-gene and corresponding protein signature for stage-I lung adenocarcinoma prognosis. Clin Cancer Res. 2011;17:1490–1501. doi: 10.1158/1078-0432.CCR-10-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gordon GJ, Richards WG, Sugarbaker DJ, Jaklitsch MT, Bueno R. A prognostic test for adenocarcinoma of the lung from gene expression profiling data. Cancer Epidemiol Biomarkers Prev. 2003;12:905–910. [PubMed] [Google Scholar]
- 55.Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 56.Larsen JE, Pavey SJ, Passmore LH, Bowman RV, Hayward NK, Fong KM. Gene expression signature predicts recurrence in lung adenocarcinoma. Clin Cancer Res. 2007;13:2946–2954. doi: 10.1158/1078-0432.CCR-06-2525. [DOI] [PubMed] [Google Scholar]
- 57.Dhillon T, Mauri FA, Bellezza G, et al. Overexpression of the mammalian target of rapamycin: a novel biomarker for poor survival in resected early stage non-small cell lung cancer. J Thorac Oncol. 2010;5:314–319. doi: 10.1097/JTO.0b013e3181ce6604. [DOI] [PubMed] [Google Scholar]
- 58.Yang DK, Son CH, Lee SK, Choi PJ, Lee KE, Roh MS. Forkhead box M1 expression in pulmonary squamous cell carcinoma: correlation with clinicopathologic features and its prognostic significance. Hum Pathol. 2009;40:464–470. doi: 10.1016/j.humpath.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 59.Tang JM, He QY, Guo RX, Chang XJ. Phosphorylated Akt overexpression and loss of PTEN expression in non-small cell lung cancer confers poor prognosis. Lung Cancer. 2006;51:181–191. doi: 10.1016/j.lungcan.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 60.Yoshizawa A, Fukuoka J, Shimizu S, et al. Overexpression of phospho-eIF4E is associated with survival through AKT pathway in non-small cell lung cancer. Clin Cancer Res. 2010;16:240–248. doi: 10.1158/1078-0432.CCR-09-0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jeon HS, Jen J. TGF-beta signaling and the role of inhibitory Smads in non-small cell lung cancer. J Thorac Oncol. 2010;5:417–419. doi: 10.1097/JTO.0b013e3181ce3afd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Janku F, Stewart DJ, Kurzrock R. Targeted therapy in non-small-cell lung cancer–is it becoming a reality? Nat Rev Clin Oncol. 2010;7:401–414. doi: 10.1038/nrclinonc.2010.64. [DOI] [PubMed] [Google Scholar]
- 63.Maione P, Gridelli C, Troiani T, Ciardiello F. Combining targeted therapies and drugs with multiple targets in the treatment of NSCLC. Oncologist. 2006;11:274–284. doi: 10.1634/theoncologist.11-3-274. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.