Abstract
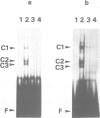
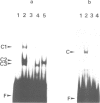
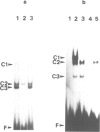
A systematic search for upstream controlling elements necessary for efficient expression of the yeast fatty acid synthase genes FAS1 and FAS2 revealed identical activation sites, UASFAS, in front of both FAS genes. The individual element confers, in a heterologous yeast test system, an approximately 40-fold stimulation of basal gene expression. The UASFAS motifs identified have the consensus sequence TYTTCACATGY and function in either orientation. The same sequence motif is found in the upstream regions of all so far characterized yeast genes encoding enzymes of phospholipid biosynthesis. In gel retardation assays, a protein factor, Fbf1 (FAS binding factor), was identified which interacted with UASFAS. The UASFAS motif proved to be an inositol/choline responsive element (ICRE) conferring strict repression by exogenous inositol and choline on a heterologous reporter gene. Its core sequence perfectly matches the CANNTG motif typical of basic helix-loop-helix DNA-binding proteins. In contrast to the individual UASFAS element, the intact yeast FAS promoters are not significantly influenced by inositol and choline, and thus allow nearly constitutive fatty acid synthase production. Available evidence suggests that additional cis- and trans-acting elements, other than UASFAS and Fbf1, are involved in this constitutive FAS gene expression.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alber T., Kawasaki G. Nucleotide sequence of the triose phosphate isomerase gene of Saccharomyces cerevisiae. J Mol Appl Genet. 1982;1(5):419–434. [PubMed] [Google Scholar]
- Anderson M. S., Yarger J. G., Burck C. L., Poulter C. D. Farnesyl diphosphate synthetase. Molecular cloning, sequence, and expression of an essential gene from Saccharomyces cerevisiae. J Biol Chem. 1989 Nov 15;264(32):19176–19184. [PubMed] [Google Scholar]
- Arcangioli B., Lescure B. Identification of proteins involved in the regulation of yeast iso- 1-cytochrome C expression by oxygen. EMBO J. 1985 Oct;4(10):2627–2633. doi: 10.1002/j.1460-2075.1985.tb03980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R. E., Fitzgerald-Hayes M., O'Brien T. C. Purification of the yeast centromere binding protein CP1 and a mutational analysis of its binding site. J Biol Chem. 1989 Jun 25;264(18):10843–10850. [PubMed] [Google Scholar]
- Beacham I. R., Schweitzer B. W., Warrick H. M., Carbon J. The nucleotide sequence of the yeast ARG4 gene. Gene. 1984 Sep;29(3):271–279. doi: 10.1016/0378-1119(84)90056-8. [DOI] [PubMed] [Google Scholar]
- Beckmann H., Kadesch T. Identification of a yeast protein with properties similar to those of the immunoglobulin heavy-chain enhancer-binding protein NF-muE3. Mol Cell Biol. 1989 Oct;9(10):4535–4540. doi: 10.1128/mcb.9.10.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann H., Su L. K., Kadesch T. TFE3: a helix-loop-helix protein that activates transcription through the immunoglobulin enhancer muE3 motif. Genes Dev. 1990 Feb;4(2):167–179. doi: 10.1101/gad.4.2.167. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990 Nov 23;250(4984):1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- Brindle P. K., Holland J. P., Willett C. E., Innis M. A., Holland M. J. Multiple factors bind the upstream activation sites of the yeast enolase genes ENO1 and ENO2: ABFI protein, like repressor activator protein RAP1, binds cis-acting sequences which modulate repression or activation of transcription. Mol Cell Biol. 1990 Sep;10(9):4872–4885. doi: 10.1128/mcb.10.9.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M., Davis R. W. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990 May 4;61(3):437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- Carman G. M., Henry S. A. Phospholipid biosynthesis in yeast. Annu Rev Biochem. 1989;58:635–669. doi: 10.1146/annurev.bi.58.070189.003223. [DOI] [PubMed] [Google Scholar]
- Dean-Johnson M., Henry S. A. Biosynthesis of inositol in yeast. Primary structure of myo-inositol-1-phosphate synthase (EC 5.5.1.4) and functional analysis of its structural gene, the INO1 locus. J Biol Chem. 1989 Jan 15;264(2):1274–1283. [PubMed] [Google Scholar]
- Diffley J. F., Stillman B. Purification of a yeast protein that binds to origins of DNA replication and a transcriptional silencer. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2120–2124. doi: 10.1073/pnas.85.7.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Henry S. A. myo-Inositol-1-phosphate synthase. Characteristics of the enzyme and identification of its structural gene in yeast. J Biol Chem. 1981 Jul 10;256(13):7077–7085. [PubMed] [Google Scholar]
- Dorsman J. C., Doorenbosch M. M., Maurer C. T., de Winde J. H., Mager W. H., Planta R. J., Grivell L. A. An ARS/silencer binding factor also activates two ribosomal protein genes in yeast. Nucleic Acids Res. 1989 Jul 11;17(13):4917–4923. doi: 10.1093/nar/17.13.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsman J. C., van Heeswijk W. C., Grivell L. A. Identification of two factors which bind to the upstream sequences of a number of nuclear genes coding for mitochondrial proteins and to genetic elements important for cell division in yeast. Nucleic Acids Res. 1988 Aug 11;16(15):7287–7301. doi: 10.1093/nar/16.15.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsman J. C., van Heeswijk W. C., Grivell L. A. Yeast general transcription factor GFI: sequence requirements for binding to DNA and evolutionary conservation. Nucleic Acids Res. 1990 May 11;18(9):2769–2776. doi: 10.1093/nar/18.9.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor P. D., Sawadogo M., Roeder R. G. The adenovirus major late transcription factor USF is a member of the helix-loop-helix group of regulatory proteins and binds to DNA as a dimer. Genes Dev. 1990 Oct;4(10):1730–1740. doi: 10.1101/gad.4.10.1730. [DOI] [PubMed] [Google Scholar]
- Guarente L., Ptashne M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Hahn S., Buratowski S., Sharp P. A., Guarente L. Isolation of the gene encoding the yeast TATA binding protein TFIID: a gene identical to the SPT15 suppressor of Ty element insertions. Cell. 1989 Sep 22;58(6):1173–1181. doi: 10.1016/0092-8674(89)90515-1. [DOI] [PubMed] [Google Scholar]
- Halfter H., Müller U., Winnacker E. L., Gallwitz D. Isolation and DNA-binding characteristics of a protein involved in transcription activation of two divergently transcribed, essential yeast genes. EMBO J. 1989 Oct;8(10):3029–3037. doi: 10.1002/j.1460-2075.1989.tb08453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi N., Oshima Y. Specific cis-acting sequence for PHO8 expression interacts with PHO4 protein, a positive regulatory factor, in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Feb;11(2):785–794. doi: 10.1128/mcb.11.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. Transcriptional and translational regulation of gene expression in the general control of amino-acid biosynthesis in Saccharomyces cerevisiae. Prog Nucleic Acid Res Mol Biol. 1990;38:195–240. doi: 10.1016/s0079-6603(08)60712-6. [DOI] [PubMed] [Google Scholar]
- Hjelmstad R. H., Bell R. M. The sn-1,2-diacylglycerol cholinephosphotransferase of Saccharomyces cerevisiae. Nucleotide sequence, transcriptional mapping, and gene product analysis of the CPT1 gene. J Biol Chem. 1990 Jan 25;265(3):1755–1764. [PubMed] [Google Scholar]
- Hjelmstad R. H., Bell R. M. sn-1,2-diacylglycerol choline- and ethanolaminephosphotransferases in Saccharomyces cerevisiae. Nucleotide sequence of the EPT1 gene and comparison of the CPT1 and EPT1 gene products. J Biol Chem. 1991 Mar 15;266(8):5094–5103. [PubMed] [Google Scholar]
- Hosaka K., Kodaki T., Yamashita S. Cloning and characterization of the yeast CKI gene encoding choline kinase and its expression in Escherichia coli. J Biol Chem. 1989 Feb 5;264(4):2053–2059. [PubMed] [Google Scholar]
- Hosaka K., Murakami T., Kodaki T., Nikawa J., Yamashita S. Repression of choline kinase by inositol and choline in Saccharomyces cerevisiae. J Bacteriol. 1990 Apr;172(4):2005–2012. doi: 10.1128/jb.172.4.2005-2012.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshizaki D. K., Hill J. E., Henry S. A. The Saccharomyces cerevisiae INO4 gene encodes a small, highly basic protein required for derepression of phospholipid biosynthetic enzymes. J Biol Chem. 1990 Mar 15;265(8):4736–4745. [PubMed] [Google Scholar]
- Inokuchi K., Nakayama A., Hishinuma F. Identification of sequence elements that confer cell-type-specific control of MF alpha 1 expression in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Sep;7(9):3185–3193. doi: 10.1128/mcb.7.9.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987 Dec;51(4):458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyono K., Miura K., Kushima Y., Hikiji T., Fukushima M., Shibuya I., Ohta A. Primary structure and product characterization of the Saccharomyces cerevisiae CHO1 gene that encodes phosphatidylserine synthase. J Biochem. 1987 Nov;102(5):1089–1100. doi: 10.1093/oxfordjournals.jbchem.a122147. [DOI] [PubMed] [Google Scholar]
- Kodaki T., Hosaka K., Nikawa J., Yamashita S. Identification of the upstream activation sequences responsible for the expression and regulation of the PEM1 and PEM2 genes encoding the enzymes of the phosphatidylethanolamine methylation pathway in Saccharomyces cerevisiae. J Biochem. 1991 Feb;109(2):276–287. [PubMed] [Google Scholar]
- Kodaki T., Yamashita S. Yeast phosphatidylethanolamine methylation pathway. Cloning and characterization of two distinct methyltransferase genes. J Biol Chem. 1987 Nov 15;262(32):15428–15435. [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Köttig H., Rottner G., Beck K. F., Schweizer M., Schweizer E. The pentafunctional FAS1 genes of Saccharomyces cerevisiae and Yarrowia lipolytica are co-linear and considerably longer than previously estimated. Mol Gen Genet. 1991 Apr;226(1-2):310–314. doi: 10.1007/BF00273618. [DOI] [PubMed] [Google Scholar]
- Lopes J. M., Hirsch J. P., Chorgo P. A., Schulze K. L., Henry S. A. Analysis of sequences in the INO1 promoter that are involved in its regulation by phospholipid precursors. Nucleic Acids Res. 1991 Apr 11;19(7):1687–1693. doi: 10.1093/nar/19.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J., Jiang W., Funk M., Rathjen J., Barnes C. A., Hinz T., Hegemann J. H., Philippsen P. CPF1, a yeast protein which functions in centromeres and promoters. EMBO J. 1990 Dec;9(12):4017–4026. doi: 10.1002/j.1460-2075.1990.tb07623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncollin V., Stalder R., Verdier J. M., Sentenac A., Egly J. M. A yeast homolog of the human UEF stimulates transcription from the adenovirus 2 major late promoter in yeast and in mammalian cell-free systems. Nucleic Acids Res. 1990 Aug 25;18(16):4817–4823. doi: 10.1093/nar/18.16.4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers A. M., Tzagoloff A., Kinney D. M., Lusty C. J. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene. 1986;45(3):299–310. doi: 10.1016/0378-1119(86)90028-4. [DOI] [PubMed] [Google Scholar]
- Nikawa J., Hosaka K., Tsukagoshi Y., Yamashita S. Primary structure of the yeast choline transport gene and regulation of its expression. J Biol Chem. 1990 Sep 15;265(26):15996–16003. [PubMed] [Google Scholar]
- Nikawa J., Kodaki T., Yamashita S. Primary structure and disruption of the phosphatidylinositol synthase gene of Saccharomyces cerevisiae. J Biol Chem. 1987 Apr 5;262(10):4876–4881. [PubMed] [Google Scholar]
- Nikawa J., Tsukagoshi Y., Yamashita S. Isolation and characterization of two distinct myo-inositol transporter genes of Saccharomyces cerevisiae. J Biol Chem. 1991 Jun 15;266(17):11184–11191. [PubMed] [Google Scholar]
- Ogawa N., Oshima Y. Functional domains of a positive regulatory protein, PHO4, for transcriptional control of the phosphatase regulon in Saccharomyces cerevisiae. Mol Cell Biol. 1990 May;10(5):2224–2236. doi: 10.1128/mcb.10.5.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oulmouden A., Karst F. Isolation of the ERG12 gene of Saccharomyces cerevisiae encoding mevalonate kinase. Gene. 1990 Apr 16;88(2):253–257. doi: 10.1016/0378-1119(90)90039-t. [DOI] [PubMed] [Google Scholar]
- Schneider J. C., Guarente L. The untranslated leader of nuclear COX4 gene of Saccharomyces cerevisiae contains an intron. Nucleic Acids Res. 1987 Apr 24;15(8):3515–3529. doi: 10.1093/nar/15.8.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer M., Lebert C., Höltke J., Roberts L. M., Schweizer E. Molecular cloning of the yeast fatty acid synthetase genes, FAS1 and FAS2: illustrating the structure of the FAS1 cluster gene by transcript mapping and transformation studies. Mol Gen Genet. 1984;194(3):457–465. doi: 10.1007/BF00425558. [DOI] [PubMed] [Google Scholar]
- Schweizer M., Roberts L. M., Höltke H. J., Takabayashi K., Höllerer E., Hoffmann B., Müller G., Köttig H., Schweizer E. The pentafunctional FAS1 gene of yeast: its nucleotide sequence and order of the catalytic domains. Mol Gen Genet. 1986 Jun;203(3):479–486. doi: 10.1007/BF00422073. [DOI] [PubMed] [Google Scholar]
- Shore D., Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987 Dec 4;51(5):721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- Shore D., Stillman D. J., Brand A. H., Nasmyth K. A. Identification of silencer binding proteins from yeast: possible roles in SIR control and DNA replication. EMBO J. 1987 Feb;6(2):461–467. doi: 10.1002/j.1460-2075.1987.tb04776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Nix J., Schweizer M., Jäger D., Schweizer E. Mapping of the trifunctional fatty acid synthetase gene FAS2 on chromosome XVI of Saccharomyces cerevisiae. Yeast. 1990 Sep-Oct;6(5):411–415. doi: 10.1002/yea.320060506. [DOI] [PubMed] [Google Scholar]
- Singh N., Wakil S. J., Stoops J. K. Yeast fatty acid synthase: structure to function relationship. Biochemistry. 1985 Nov 5;24(23):6598–6602. doi: 10.1021/bi00344a044. [DOI] [PubMed] [Google Scholar]
- Stillman D. J., Bankier A. T., Seddon A., Groenhout E. G., Nasmyth K. A. Characterization of a transcription factor involved in mother cell specific transcription of the yeast HO gene. EMBO J. 1988 Feb;7(2):485–494. doi: 10.1002/j.1460-2075.1988.tb02836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichert U., Mechler B., Müller H., Wolf D. H. Lysosomal (vacuolar) proteinases of yeast are essential catalysts for protein degradation, differentiation, and cell survival. J Biol Chem. 1989 Sep 25;264(27):16037–16045. [PubMed] [Google Scholar]
- Tsay Y. H., Robinson G. W. Cloning and characterization of ERG8, an essential gene of Saccharomyces cerevisiae that encodes phosphomevalonate kinase. Mol Cell Biol. 1991 Feb;11(2):620–631. doi: 10.1128/mcb.11.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]