Abstract
Telomerase is an RNP that synthesizes the 3′ ends of linear chromosomes and is an important regulator of telomere length. It contains a single long non-coding telomerase RNA (TER), telomerase reverse transcriptase (TERT), and other proteins that vary among organisms. Recent progress in structural biology of telomerase includes reports of the first cryo-electron microscopy structure of telomerase, from Tetrahymena, new crystal structures of TERT domains, telomerase RNA structures and models, and identification in Tetrahymena telomerase holoenzyme of human homologues of telomere-associated proteins that have provided a more unified view of telomerase interaction at telomeres as well as insights into the role of telomerase RNA in activity and assembly.
Graphical Abstract

Introduction
Telomerase is a ribonucleic acid-protein complex (RNP) composed of a single long non-coding (lnc) RNA, called telomerase RNA (here abbreviated TER), and associated proteins, including telomerase reverse transcriptase (TERT). Telomerase functions to extend the 3′ ends of linear chromosomes by synthesizing multiple copies of the species-specific DNA telomere repeat sequence (G-strand), utilizing a complementary template contained in the TER [1]. Telomerase may also play a role in regulating synthesis of the complementary strand (C-strand) [2]. TERT and TER assembled in vitro can catalyze some level of telomere repeat synthesis, but in vivo telomerase activity requires association with proteins involved in biogenesis, recruitment to telomeres, and telomere G- or C-strand DNA binding and synthesis [3]. Telomerase activity is mostly undetectable in somatic cells, moderate in stem and germ line cells, and high in most cancer cells [4]. Telomerase insufficiency due to mutations in TER, telomerase proteins, or promoter elements manifests as genetic diseases such as dyskeratosis congenita (DC) and idiopathic pulmonary fibrosis, while its aberrant upregulation is a prerequisite for the immortal phenotype of most cancer cells [5–7].
Telomerase has been a challenging target for structural studies for a variety of reasons, including its low cellular abundance, sequence and structural variability of TER among species, divergent protein composition except for TERT, and difficulties in protein expression, purification, and RNP assembly in vitro. Starting around 2003, structural studies focused on domains of TER, particularly the essential pseudoknot, TERT, and TERT-TER domains complexes [8,9]. In 2013, negative stain electron microscopy (EM) structures at about 25 Å resolution of human telomerase [10] and the ciliated protozoan Tetrahymena were reported [11]. Two TERTs were located in a human telomerase dimer, while for the monomeric Tetrahymena telomerase, the architecture of the complete holoenzyme was defined. Somewhat more rapid structural progress had been made with telomere binding proteins, especially from yeasts [12]. During the past couple of years, a surge of progress has been made on structural biology of telomerase, including reports of the first cryo-electron microscopy (cryo-EM) structure of telomerase, from Tetrahymena [13], crystal structures of TERT and TERT-interacting protein domains [14–17], telomerase RNA structures and models [18,19], and identification in Tetrahymena telomerase holoenzyme of human homologues of telomere-associated proteins [13,20] that have provided a more unified view of telomerase interaction at telomeres [21]. Here we review these and other studies published since 2015 on structural investigations of primarily Tetrahymena and vertebrate telomerase and its interaction at telomeres. We also refer the interested reader to more comprehensive recent reviews on telomerase mechanism [3], proteins and RNA structure and function [8,22,23], and single-molecule FRET (smFRET) studies [24], including studies of plant, fungal, and yeast telomerase not discussed here [25–28] due to space limitations and limited structural information.
The first cryo-EM structure of telomerase, from Tetrahymena
The publication of ~9 Å resolution cryo-EM structures of Tetrahymena telomerase holoenzyme and pseudoatomic model of TERT, TER, and most of the other proteins marked a major breakthrough in our understanding of telomerase structure and interaction at telomeres [13]. The study employed an integrative structural biology approach, combining information from negative stain and cryo-EM, X-ray crystallography, NMR spectroscopy, and mass spectrometry. Tetrahymena telomerase holoenzyme is constitutively assembled and comprises 9 proteins and TER (Figure 1a,b). Only TERT and the La-related group 7 protein p65 contact TER, defining the RNP core required for biogenesis and catalysis. The structure provided the first view of the path of TER on TERT (discussed below). The accessory proteins p50 and a complex of p75-p45-p19, previously thought to have no homologues in other species, were found to have structural and functional homologies to human telomere associated proteins TPP1 and CTC1-STN1-TEN1 (CST), respectively (Figure 1d,e). Teb1, a paralog of the large subunit of Replication Protein A (RPA70), was found to form a ternary complex with two previously unknown proteins, Teb2 and Teb3, whose identities were confirmed by mass spectrometry. The p50-Teb1 complex promotes telomerase repeat addition processivity (RAP) in a manner homologous to the activation function of human TPP1-POT1 (Figure 1c,e) [21]. The TERT TEN domain, which had been implicated in TER binding, single-stranded DNA (ssDNA) handling, and RNA-DNA duplex stabilization was revealed to have numerous interactions – with p50, Teb1, the TERT insertion-in-fingers domain (IFD), and potentially TER and the template-DNA duplex (Figure 1a).
Figure 1.
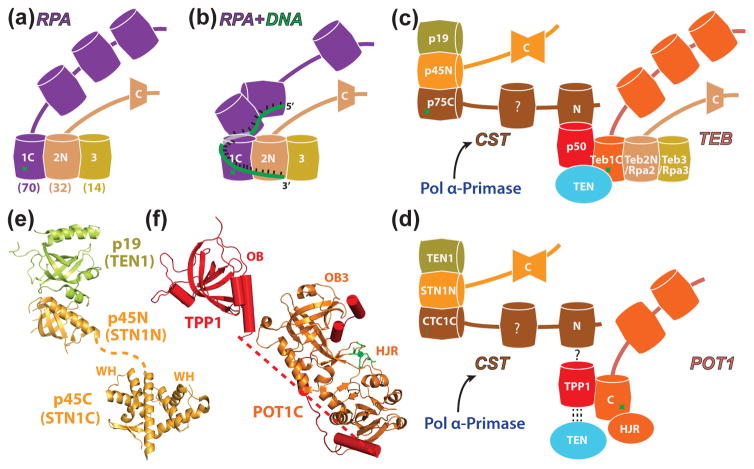
Tetrahymena and human telomerase holoenzymes. (a) Front (left) and back (right) views of Tetrahymena telomerase cryo-EM map at ~9Å resolution (RNP core, blue; CST, tan; TEB, straw; and p50, red) and pseudoatomic models of the RNP core and TEB and CST trimerization domains of 3 OB-folds [13]. Tetrahymena telomerase protein domains for which structures have been determined are rendered as ribbons and with bases, respectively; homology models are rendered with ribbons and cylinders. TER single-stranded regions are shown as ribbons except the template which includes bases; folded domains are shown with bases. (b, d) Schematics illustrating (b) Tetrahymena and (d) human TER secondary structure and domains, TER interacting proteins, and proteins that interact with the RNP core. Proteins or complexes with generally homologous functions between organisms are colored the same, except H/ACA scaRNP proteins which are distinguished from each other. (c, e) Schematics of (c) Tetrahymena and (e) human interactions at telomere 3′ ends. Dashed green line indicates extension of the 3′ end by telomerase. In (e), lines with arrowheads indicate transient interactions and lines with bars indicate inhibitory interactions. Panels b-e are modified from [8].
TER is a rapidly evolving lncRNA with a size range of ~150 nt to >10,000 nt in some yeasts (~159 nt in Tetrahymena, 451 nt in human, 1157 nt in Saccharomyces cerevisiae [22,29]. Almost universally, there are two domains (template/pseudoknot (t/PK) and stem-terminus element (STE)) that independently bind TERT and are required for catalysis, and a biogenesis domain that binds species-specific proteins involved in TER stabilization and assembly (Figure 1) [22,30]. By analogy to the constitutively assembled Tetrahymena telomerase holoenzyme, the human telomerase RNP core is minimally TER, TERT, and the H/ACA scaRNP proteins (Figure 1d). Several telomere-associated proteins also interact transiently with the telomerase RNP core to promote G-strand and/or C-strand synthesis, specifically TPP1-POT1 and CTC1-STN1-TEN1 [3,8,12].
New structures of TERT domains
TERT contains the palm and fingers (RT) and thumb (CTE) domains common to reverse transcriptases and a telomerase specific TEN (telomerase essential N-terminal domain) and TRBD (telomerase RNA binding domain) (Figure 2). The TEN and TRBD domains are separated by a long linker of low sequence complexity [31]. The first structure of a TERT, from the flour beetle (Tribolium casteneum) that atypically lacks a TEN domain, revealed that TRBD-RT-CTE form a ring (Figure 2a) [32]. Here we illustrate the structure in the classic right hand view for polymerases with palm, fingers, and thumb. The pseudoatomic model of TERT built into the cryo-EM map from crystal structures of Tetrahymena TEN and partial TRBD and the flour beetle RT and CTE domains revealed that the TEN domain stacks over the thumb (CTE) with a single-stranded region of TER 3′ to the template between the two domains (Figures 1a,b and 2b).
Figure 2.
Structure of TERT and interaction with TER. (a) Structure of flour beetle TERT with bound inhibitor BIBR1532 (PDB ID: 5CQG). The model is shown in the classical polymerase hand view (pale green) with RT palm (purple), fingers (orange), IFD (gold), CTE thumb (light blue), and TRBD (blue). Active site on RT is space-filled red. (b) Hand view of structure of Tetrahymena TERT with bound TER. Note that this view is rotated 180° at the Y-axis from the cryo-EM model back view in Figure 1a. (c) Tetrahymena TRBD-TBE complex (PDB ID: 5C9H), TBE is red, T motif is cyan, CP is gold, and CP2 is purple. (d) Human CTE (thumb) (PDB ID: 5UGW). (e) Model of human TERT bound to human TER t/PK based on the model of the human t/PK and the positions of homologous domains of Tetrahymena TR in the cryo-EM map. The dash line indicates the proposed location of P6.1. Subdomains of TERT and TR are colored as in Figure 1, except human TER J2a/b is colored as green.
Several new TERT domain crystal structures were recently reported, the beetle TERT in complex with the inhibitor BIBR1532 [33] (Figure 2a), a Tetrahymena TRBD-TER complex (Figure 2c) [14], and the thumb (CTE) domain of human TERT (Figure 2d) [15]. There is also a preliminary NMR study of a TEN domain from the thermostable yeast Ogataea polymorpha that indicates it forms a folded domain [34]. TERT TRBD specifically binds two domains of TER, the template boundary element (TBE) and the STE (loop 4 in Tetrahymena and CR4/5 in vertebrates) (Figure 2b), and examples of each of these interactions have now been solved. The Tetrahymena TRBD-TBE structure encompasses the complete TRBD (aa 217-516 vs 254-519 in the earlier structure [35]), which includes the CP2 motif important for TER binding [14]. This CP2 motif was shown to be sequence and structurally homologous to the TFLY motif identified in the vertebrate TRBD [36]. A vertebrate (Medaka) STE-TRBD complex showed complex interactions at the CR4/5 three-way junction, but the loop of P6.1, which is proposed to insert between the TRBD and CTE is not visible in the crystal structure [37]. In the beetle TERT-inhibitor structure, BIBR1532 binds on the thumb domain at a novel FVYL motif, and it is proposed that in human telomerase this interaction would disrupt binding of the P6.1 loop. The human CTE crystal structure is the first of any domain of human TERT [15]. This domain, comprising 6 α-helices, is highly similar to the beetle TERT (Figure 2d). The structure shows that some mutations associated with a plastic anemia and idiopathic pulmonary fibrosis are on a region of the CTE predicted to interact with the template-DNA duplex.
TERTs have an IFD that is a short helix in beetle TERT, but in Tetrahymena is much larger and could not be homology modeled in the cryo-EM map [13]. Nevertheless, the density attributed to the IFD contacts both the TEN domain and p50. Recent studies show that the IFD affects telomerase processivity and recruitment to telomeres by TPP1, suggesting that the IFD interacts with TPP1 [38,39]. Finally, we note that crystal structures of the RT domain of two group II intron-encoded reverse transcriptases and a cryo-EM structure of a group II intron and its bound reverse transcriptase have been reported [40,41], and possible ancestral relationships to TERT are discussed. An IFD is one of the common elements of TERT and group II intron reverse transcriptases.
TER structure and assembly with TERT
Telomerase assembly and activity requires extensive TER interactions with TERT in addition to the template. The cryo-EM structure of Tetrahymena telomerase revealed that the template/pseudoknot domain, which forms a circle closed by stem 1, encircles the TERT ring with the template on one side and the pseudoknot on the other. The STE (loop 4) inserts between the TRBD and CTE on the opposite side of the TERT ring from the pseudoknot, positioned by the interactions of p65 with stem-loop 4 (Figures 1a,b and 2b).
The length of the pseudoknot stems, especially stem 2, varies greatly among organisms [22]. The structures of a minimal human telomerase pseudoknot, containing the stem and loop nucleotides that define the fold, revealed a conserved triple-helix around the stem-stem junction [42]. These interactions, including several major groove U-A-U triples, have been shown to be important for activity in human and yeast telomerase [42,43]. The solution NMR structure of the Tetrahymena pseudoknot, which has a much shorter stem 2 than yeasts and vertebrates, was more recently reported [13,19]. The free Tetrahymena pseudoknot is much less stable than those of yeasts and vertebrates, and includes only two major groove triples: a U-A-U and a novel C-G-A. The location of the pseudoknot on the opposite side of the TERT ring from the template apparently precludes any direct interaction of the pseudoknot in catalysis. Instead, the pseudoknot may act like a watch-band ratchet clasp to tighten the t/PK around TERT during a late step in assembly.
A model of the t/PK domain of human TER in complex with TERT was previously calculated by combining solution NMR structures of 3 sub-domains of the full-length pseudoknot [44,45], i.e. the minimal pseudoknot, a central helical region containing a 5 nt bulge-loop, and a terminal helical region containing several mismatches and nucleotide bulges. A similar approach was recently used to model the t/PK of medaka fish (Orzyis lapites), which has the smallest vertebrate TER [46]. This structural study of medaka pseudoknot domains showed that the tertiary stem-loop interactions are identical to human TER except it lacks a single nt bulge near the stem-stem junction, the predicted 6 nt bulge-loop is actually a 5 nt loop with the same structure as in hTER, and the adjacent predicted single-strand region forms a small hairpin that stacks on the adjacent helix [18]. Based on the model structures of the medaka and human t/PK and the locations of TER elements in Tetrahymena, models of medaka and human TERT in complex with the t/PK were presented (Figure 2e). Interestingly, the bulge region, which creates a 90° bend between helices, appears to allow the long helical region of the hTER PK to fold around TERT, whereas in Tetrahymena this region is single-stranded. A similar model for human t/PK bound to TERT was independently determined based on Rosetta modeling using restraints from smFRET data and the solution NMR structures of human TER domains and modeled human TERT [47].
The Tetrahymena STE (loop 4) appears to be positioned near the TRBD by bending of stem-loop 4 by p65 C-terminal xRRM. In contrast, the higher affinity binding of the human STE (CR4/5 P5-P6-P6.1 three-helix junction) apparently does not require additional proteins, as a minimized hTER with the H/ACA protein binding region deleted assembles an RNP in vivo that maintains stable telomere length homeostasis [48].
smFRET studies and SHAPE chemical probing indicated that the Tetrahymena pseudoknot does not fold in the free TER [49,50]. Solution NMR studies of the entire t/PK as well as smaller RNA constructs provided additional details of the t/PK secondary structure and determined that the entire pseudoknot sequence is sequestered in an alternative helical structure that includes the template nucleotides (Figure 3a) [19]. This free TER structure has both of the high affinity TERT TRBD binding sites exposed (TBE and STE/loop 4); the authors propose that TERT assembly with TER could be initiated at the TBE. A similar alternative structure for free human TER was also proposed based on sequence analysis [19], and this structure is consistent with smFRETdata [51]. SHAPE analysis supports pseudoknot folding for S. cerevisiae and human TER t/PK [52]. A recent study using in vivo chemical probing (DMS) also apparently showed that the pseudoknot does form in the protein-free human TER, but the template is sequestered and becomes more exposed on TERT binding [53]. However, the chemical probing data may be equally consistent with the alternative model for free human TER discussed above.
Figure 3.
Models for steps in Tetrahymena telomere DNA synthesis and assembly of TER with TERT. (a) Schematic illustration of secondary structure of Tetrahymena TER bound to p65 (left) and to p65 and TERT (right). In the p65 bound TER, the pseudoknot is sequestered in a helix that prevents it from folding, but the TBE and loop 4 remain accessible for subsequent binding to TERT. (b) Schematic illustrating DNA exit pathway and interaction with Teb1, based on [13]. (c) Schematics illustrating template movement through the active site at beginning and end of synthesis of a telomere repeat. The template boundary element interacts with TERT to physically restrain the RNA so that residues beyond the template are not copied (right). A proposed sstDNA retention surface (SRS) within the CTE is colored as dark blue [56]. (d) Schematic illustrating the DNA hairpin proposed to form after translocation as part of the telomere repeat synthesis cycle [55]. This model assumes a large movement of the CTE (thumb) domain during translocation, indicated by the arrowheads connected by dotted line. The “hands” in the panels indicate the orientation of TERT in the panels relative to the views in figure 2.
Mechanism and dynamics of telomere repeat synthesis
In most species, the G-strand telomere repeat is 4–8 nt long and is copied off the RNA template that contains about 1.5 sequence-complementary repeats. The 3′-end of the telomere DNA initially pairs with the 5′ end of the template leaving one full repeat unpaired and a complete telomere repeat is then synthesized, followed by strand-separation and pairing of the newly synthesized 3′ end with the 5′ end of the template (Figure 3c). Processive synthesis of multiple telomere repeats also requires that the template translocate in the active site to the start site of synthesis. Elegant biochemical experiments have been conducted to try to define the steps that allow proper template alignment, copying, strand separation, translocation, and the path of the exiting DNA [3]. Mutational studies identified a template boundary element (TBE), i.e. nucleotides important for preventing copying beyond the end of the template, that in Tetrahymena is the bottom base pairs of stem 2 and single-strand nucleotides on either side [54]. Both the cryo-EM model (Figures 1a and 2b) and the crystal structure of the TRBD-TBE complex (Figure 2c) showed that the single-strands flanking stem 2 wrap around either side of the TRBD, and suggested a model where the TBE interactions act as an anchor to prevent nts beyond the template from being pulled into the active site (Figure 3c).
The structural basis of the strand separation-translocation steps still remains to be determined. An intriguing model based in part on the large conformational dynamics of translation DNA polymerases was proposed whereby the DNA forms a hairpin at the end of a telomere-repeat cycle, the template translocates, the hairpin unfolds and reforms base pairs with the 3′ end of the template (Figure 3d) [55]. This model requires a large movement of the thumb (CTE) relative to the palm (RT). The smFRET data of human TERT-TER complexes (discussed above) with primer bound (stalled) and actively synthesizing DNA showed three different positions of the pseudoknot fold relative to the template [47]. The authors propose that the pseudoknot moves during the telomere repeat synthesis cycle (pseudoknot tracking model); however, since they assumed a rigid structure for TERT in their Rosetta modeling they also suggest that the PK fold motion stabilizes the alternative conformation of TERT CTE proposed by Wang [18]. Most recently, a comprehensive mutational screen of TERT identified a ssDNA retention surface (SRS) within the CTE (Figure 3c), which is proposed to maintain placement of the 3′ end of the nascent DNA telomere repeat in the active site during template translocation, thereby contributing to repeat addition processivity [56]. A detailed pathway for telomere repeat synthesis is proposed that is compatible with but does not require DNA hairpin formation.
The telomeric ssDNA exits from the template, which lays along the RT on the inner edge of the TERT ring, toward the TEN domain and over Teb1C on the side facing away from TEN (Figure 3b). The EM model suggests that the template-DNA duplex buts up against the TEN domain [13]. smFRET and earlier biochemical studies of Tetrahymena and human TERT-TER complexes, respectively, support a role for the TEN domain in stabilizing the short template-DNA hybrids that initiate each round of telomere repeat synthesis [57,58], thereby enhancing RAP. Details and physical confirmation of the models discussed above await high resolution structures of telomerase with bound DNA to provide snap-shots of the telomere repeat synthesis cycle.
RPA-related proteins bridge the telomerase RNP core and telomere ssDNA ends
RPA is heterotrimeric complex that binds ssDNA sequence non-specifically and plays essential roles in DNA replication and repair, e.g. recruitment of DNA polymerase α-primase (Pol α) and DNA repair proteins [59]. Its 3 subunits (Rpa1-Rpa2-Rpa3; in humans, RPA70-RPA32-RPA14) associate via three OB-folds (Figure 4a). The large subunit, RPA70, contains four OB- folds, N, A, B, C. OB-C includes a Zn+2-ribbon motif, and ssDNA binds the A, B, and C domains while N is involved in protein recruitment (Figure 4b). The Tetrahymena telomerase cryo-EM map and crystal structures of protein domains revealed that the constitutively assembled Tetrahymena telomerase contains two RPA-like complexes: TEB and p74-p45-p19 [13]. Each of these complexes is tethered to p50, which is tethered to the RNP core by interactions with the TERT TEN domain (Figure 4c). RPA-related proteins have now been universally found associated with telomeres and telomerase in ciliates, yeasts, plants, and vertebrates [12].
Figure 4.
RPA, TEB, CST and their interactions. (a) Schematic of the RPA heterotrimer of Rpa1-Rpa2-Rpa3 (Rpa70, 32, and 14 in humans). Shared subunits Teb2/Rpa2 and Teb3/Rpa3 found in Tetrahymena are indicated by common colors with panel c. (b) Schematic of the structure of a RPA-ssDNA complex, based on [81]. (c) Schematic of the two RPA-like complexes, CST (p75-p45-p19) and TEB (Teb1-Teb2-Teb3), in Tetrahymena telomerase and their interactions with p50 and TEN domain; (d) Schematic of the two RPA-like complexes/proteins that transiently associate with human telomerase, CST (CTC1-Stn1-Ten1) and Pot1 (paralog of Rpa1), their interaction with TPP1 (putative for CTC1), and the interaction of TPP1 with the TEN domain. The human telomerase RNP core is recruited to telomere by TPP1-POT. CST interaction with TPP1-POT1 inhibits telomerase activity; (e,f) Crystal structures of (e) p45N-p19 complex (PDB ID: 5DOI) and p45C domain (PDB ID: 5DFN) and (f) POT1C-TPP1 complex which is modeled based on crystal structures of POT1C-TPP1 PBM complex (PDB ID: 5UN7), TPP1 (PDB ID: 2I46), and TEBPα–β–ssDNA complex (PDB ID: 1OTC) [65]. In (a-d), barrels indicate OB-fold domains, single trapezoid is WH domain, double trapezoid is tandem WH-WH domains, green star is a zinc-binding motif. Homologous domains for Tetrahymena and human telomerase/telomere proteins are colored the same (c-d); for RPA (a, b) the two smaller subunits are identical with those of TEB.
TEB contains the same domain structure as RPA, including a C-terminal winged-helix (WH) on Teb2 and a Zn+2-ribbon in Teb1C (Figure 4c). In Tetrahymena, the two smaller subunits of RPA had not been previously identified, and expression levels of Teb2 and Teb3, which are much higher than other telomerase proteins, suggested the possibility that these might be shared subunits with Tetrahymena RPA. Remarkably, this is the case, despite the sequence divergence for Teb1 vs Rpa1 [60]. RPA binds ssDNA sequence non-specifically and Rpa2 enhances the binding, while TEB is specific for single-stranded telomere repeat DNA (sstDNA) and Teb2 does not enhance DNA binding. TEB binds the sstDNA as it exits the template (Figure 3b), facilitates RAP, and recruits Tetrahymena telomerase to telomeres in a p50-dependent fashion [11,61,62]. Like Teb1, Pot1, which is homologous to the large subunit of RPA (RPA70 or Rpa1), binds sstDNA (at the 3′ ends of telomeres).
Telomere end replication not only requires G-strand synthesis but also synthesis of the complementary C-strand. The C-strand is filled in after synthesis of the G-strand by Pol α, which is recruited by Cdc13/Ctc1 and Stn1 (Figure 4d) [63]. CST in humans has been proposed to negatively regulate telomerase activity [2]. The cryo-EM map and crystal structures of Tetrahymena p19 and p45 revealed that Tetrahymena p75-p45-p19 as a ciliate CST complex (Figure 4e) [13,20]. The Tetrahymena CST complex interacts with the catalytic core as a subcomplex hinged on p50 via p75 (Figure 1a) [13]. Biochemical evidence supports a role for p75-p45-p19 in recruiting Pol α [20].
Recruitment of telomerase to telomeres
In vertebrates and plants, the 3′ sstDNA binds POT1, which forms a complex with TPP1 that bridges to the other shelterin proteins via TIN2 [64]. The ciliate Sterkiella nova (formerly Oxytricha nova) telomere end-binding proteins TEBPα and TEBPβ [65] were subsequently identified as POT1 and TPP1 homologues, respectively, based on the crystal structures [66,67]. Two crystal structures of human POT1 C-terminal domain in complex with TPP1 POT1-binding motif (PBM) were recently reported [16,17]. POT1C has an unusual architecture consisting of an OB-fold (OB3) that includes a Zn+2-ribbon motif and a Holliday junction resolvase (HJR)-like domain, formed from an insertion in the C-terminal end of the primary sequence, that pack tightly against one another. In the complex, TPP1 PBM (residues 255-337), which forms 4 helices connected by structured linkers, interacts extensively with both domains (Figure 4f) [17]. Based on the crystal structure of the Oxytricha nova TEBPα–β–ssDNA complex and SAXS data on the TPP1-POT1 complex, the overall architecture of the POT1–TPP1 complex has also been modeled (Figure 4f) [16]. Comparison of this model structure with the relative positions of p50, whose structure has not been determined, and Teb1 in the cryo-EM map of Tetrahymena telomerase reveals that they are oriented similarly relative to each other.
TPP1-POT1 switches from telomerase inhibitory to telomerase activating on interaction with telomerase core RNP [66,68–71]. A so-called TEL patch surface on the conserved OB-fold in TPP1 (and S. cerevisiae Est3) has been shown to interact directly with TERT TEN domain for recruitment to telomeres [72] and to stabilize the association between telomerase and sstDNA [73]. A crystal structure of TPP1 K170Δ, a mutation associated with DC, showed that the deletion restructures a loop in the TEL patch [74]. There is apparently an equivalent but constitutive interaction to that of human TPP1 and TEN between Tetrahymena p50 and the TEN domain [13]. In Tetrahymena p50-TEB and Tpt1-Pot1a [75] may have evolved to separate the telomerase activator and inhibitor functions, respectively, found together in TPP1-POT1.
Telomerase is a functional monomer with diverse biogenesis proteins among species
Many polymerases function as dimers, and the stoichiometry of TERT and TER in telomerase has been extensively investigated. While functional and structural studies have established Tetrahymena telomerase as a monomer [11,76], questions have remained about the dimerization state of yeast and human telomerase. Using fluorescence in situ hybridization experiments of two differentially tagged S. cerevisiae TERs (TLC1) Bajon et al. were able to show that there is only one molecule of TER in yeast telomerase [77]. Because of its low cellular abundance (estimated at ~240 RNPs/cell [78]), most functional and structural studies of human telomerase have utilized a telomerase RNP generated by transient transfection and overexpression of TERT and TER in human cells. The negative stain EM structure of human telomerase showed two areas of density that could be fit with TERT and evidence for two bound DNA primers consistent with biochemical data supporting two molecules of TERT and TER [10]. A recent study employing single-molecule imaging of fluorescently labeled TERT in TERT-TER complexes assembled both in vitro and in human cells showed remarkably heterogeneous complexes with TERT composition varying with purification method [31]. TERT self-association was attributed to a region of low sequence complexity in the long linker between the TEN and TRBD domains, and could be suppressed by removing or modifying this sequence without affecting activity. Thus, it appears that a monomeric architecture for telomerase is evolutionarily conserved among species [31].
Recent studies indicate that there are previously unidentified proteins components in yeast telomerase that are involved in biogenesis. These include the discovery in S. cerevisiae of Pop1/Pop6/Pop7 proteins that are common to RNase P/MRP and identification of a homologous binding domain in TER [79,80]. These and other studies that have established the minimal active human telomerase RNP in vivo [31] should facilitate future investigations of human and yeast telomerase structure.
Highlights.
First cryo-EM structure of telomerase, from Tetrahymena, at ~9 Å resolution.
New TERT and TER domain structures and interactions.
RPA-like proteins bridge the telomerase RNP core and telomere DNA 3′ ends.
Insights into mechanism and dynamics of telomere repeat synthesis.
Acknowledgments
Research on telomerase in the Feigon lab is supported by NIH grant GM048123 and NSF grant MCB 1517629 to J.F.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Blackburn EH, Collins K. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol. 2011;3:a003558. doi: 10.1101/cshperspect.a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen LY, Redon S, Lingner J. The human CST complex is a terminator of telomerase activity. Nature. 2012;488:540–544. doi: 10.1038/nature11269. [DOI] [PubMed] [Google Scholar]
- 3.Wu RA, Upton HE, Vogan JM, Collins K. Telomerase Mechanism of Telomere Synthesis. Annu Rev Biochem. 2017;86:4.1–4.22. doi: 10.1146/annurev-biochem-061516-045019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armanios M, Blackburn EH. The telomere syndromes. Nat Rev Genet. 2012;13:693–704. doi: 10.1038/nrg3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shay JW. Role of Telomeres and Telomerase in Aging and Cancer. Cancer Discov. 2016;6:584–593. doi: 10.1158/2159-8290.CD-16-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarek G, Marzec P, Margalef P, Boulton SJ. Molecular basis of telomere dysfunction in human genetic diseases. Nat Struct Mol Biol. 2015;22:867–874. doi: 10.1038/nsmb.3093. [DOI] [PubMed] [Google Scholar]
- 7.Townsley DM, Dumitriu B, Young NS. Bone marrow failure and the telomeropathies. Blood. 2014;124:2775–2783. doi: 10.1182/blood-2014-05-526285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan H, Wang Y, Feigon J. Progress in Human and Tetrahymena Telomerase Structure. Annu Rev Biophys. 2017;46:199–225. doi: 10.1146/annurev-biophys-062215-011140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Theimer CA, Feigon J. Structure and function of telomerase RNA. Curr Opin Struct Biol. 2006;16:307–318. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Sauerwald A, Sandin S, Cristofari G, Scheres SH, Lingner J, Rhodes D. Structure of active dimeric human telomerase. Nat Struct Mol Biol. 2013;20:454–460. doi: 10.1038/nsmb.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang J, Miracco EJ, Hong K, Eckert B, Chan H, Cash DD, Min B, Zhou ZH, Collins K, Feigon J. The architecture of Tetrahymena telomerase holoenzyme. Nature. 2013;496:187–192. doi: 10.1038/nature12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis KA, Wuttke DS. Telomerase and telomere-associated proteins: structural insights into mechanism and evolution. Structure. 2012;20:28–39. doi: 10.1016/j.str.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **13.Jiang J, Chan H, Cash DD, Miracco EJ, Ogorzalek Loo RR, Upton HE, Cascio D, O’Brien Johnson R, Collins K, Loo JA, et al. Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science. 2015;350:aab4070. doi: 10.1126/science.aab4070. First cryoEM structure of telomerase, at ~9 Å resolution, provides pseudoatomic model of the RNP core (TERT-TER-p65) and reveals that it contains two RPA-like complexes, TEB (Teb1-Teb2-Teb3) and CST (p75-p45-p19), tethered to p50, a TPP1 homologue. Reports the crystal structures of p19 and p45C and NMR structure of telomerase RNA pseudoknot. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **14.Jansson LI, Akiyama BM, Ooms A, Lu C, Rubin SM, Stone MD. Structural basis of template-boundary definition in Tetrahymena telomerase. Nat Struct Mol Biol. 2015;22:883–888. doi: 10.1038/nsmb.3101. Reports the crystal structure of the Tetrahymena TERT telomerase RNA binding domain to the TER template boundary element, revealing atomic details of TER-TRBD structure and interactions. Presents a model for template boundary definition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **15.Hoffman H, Rice C, Skordalakes E. Structural Analysis Reveals the Deleterious Effects of Telomerase Mutations in Bone Marrow Failure Syndromes. J Biol Chem. 2017;292:4593–4601. doi: 10.1074/jbc.M116.771204. Reports the first structure of a domain from human TERT, the CTE (thumb) domain. Proposes an FVYL pocket as important for TER binding or telomerase assembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **16.Chen C, Gu P, Wu J, Chen X, Niu S, Sun H, Wu L, Li N, Peng J, Shi S, et al. Structural insights into POT1-TPP1 interaction and POT1 C-terminal mutations in human cancer. Nat Commun. 2017;8:14929. doi: 10.1038/ncomms14929. Reports a crystal structure of a complex of human POT1 C-terminal domain and TPP1 peptide comprising the POT1 binding motif (PBM, 266–320). Reveals that POT1 C-terminal domain comprises an OB-fold with an unexpected Holliday junction resolvase-like domain, both of which interact with the PBM. XL-MS and SAXS analysis provides a model of the POT1C with TPP1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **17.Rice C, Shastrula PK, Kossenkov AV, Hills R, Baird DM, Showe LC, Doukov T, Janicki S, Skordalakes E. Structural and functional analysis of the human POT1-TPP1 telomeric complex. Nat Commun. 2017;8:14928. doi: 10.1038/ncomms14928. Reports a crystal structure of a complex human POT1 C-terminal domain and TPP1 peptide comprising the POT1 binding domain (PBD, 255–337). Reveals that POT1 C-terminal domain comprises an OB-fold and Holliday junction resolvase-like domain, both of which interact with the PBM. The longer PBD in this study has an additional helix not present in ref 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *18.Wang Y, Yesselman JD, Zhang Q, Kang M, Feigon J. Structural conservation in the template/pseudoknot domain of vertebrate telomerase RNA from teleost fish to human. Proc Natl Acad Sci USA. 2016;113:E5125–5134. doi: 10.1073/pnas.1607411113. Reports the structure of medaka fish TER domain P2ab and an NMR-based model of the full-length pseudoknot, highlighting the evolutionary structural conservation among vertebrate TER. Presents a model of human TERT-template/pseudoknot based the cryoEM model of Tetrahymena telomerase RNP core and model of human template/pseudoknot. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *19.Cash DD, Feigon J. Structure and folding of the Tetrahymena telomerase RNA pseudoknot. Nucleic Acids Res. 2017;45:482–495. doi: 10.1093/nar/gkw1153. Reports details of the solution NMR structure of Tetrahymena TER pseudoknot. Determined the secondary structure of the free full-length TER by NMR, which shows that the bases from the pseudoknot sequence are sequestered in an alternate helix that includes the template, and an assembly pathway for the RNP core is proposed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **20.Wan B, Tang T, Upton H, Shuai J, Zhou Y, Li S, Chen J, Brunzelle JS, Zeng Z, Collins K, et al. The Tetrahymena telomerase p75-p45-p19 subcomplex is a unique CST complex. Nat Struct Mol Biol. 2015;22:1023–1026. doi: 10.1038/nsmb.3126. Reports the crystal structures of Tetrahymena telomerase p19, p45C, and p19-p45N complex. Structures and biochemical data support the proposal that p75-p45-p19 is Tetrahymena CST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feigon J, Chan H, Jiang J. Integrative structural biology of Tetrahymena telomerase - insights into catalytic mechanism and interaction at telomeres. FEBS J. 2016;283:2044–2050. doi: 10.1111/febs.13691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Podlevsky JD, Chen JJ. Evolutionary perspectives of telomerase RNA structure and function. RNA Biology. 2016;13:720–732. doi: 10.1080/15476286.2016.1205768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price CM, Boltz KA, Chaiken MF, Stewart JA, Beilstein MA, Shippen DE. Evolution of CST function in telomere maintenance. Cell Cycle. 2010;9:3157–3165. doi: 10.4161/cc.9.16.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parks JW, Stone MD. Single-Molecule Studies of Telomeres and Telomerase. Annu Rev Biophys. 2017;46:357–377. doi: 10.1146/annurev-biophys-062215-011256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armstrong CA, Tomita K. Fundamental mechanisms of telomerase action in yeasts and mammals: understanding telomeres and telomerase in cancer cells. Open Biol. 2017:7. doi: 10.1098/rsob.160338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vasianovich Y, Wellinger RJ. Life and Death of Yeast Telomerase RNA. J Mol Biol. 2017 doi: 10.1016/j.jmb.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 27.McKnight TD, Riha K, Shippen DE. Telomeres, telomerase, and stability of the plant genome. Plant Mol Biol. 2002;48:331–337. doi: 10.1023/a:1014091032750. [DOI] [PubMed] [Google Scholar]
- 28.Nelson AD, Shippen DE. Evolution of TERT-interacting lncRNAs: expanding the regulatory landscape of telomerase. Front Genet. 2015;6:277. doi: 10.3389/fgene.2015.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Podlevsky JD, Bley CJ, Omana RV, Qi X, Chen JJ. The telomerase database. Nucleic Acids Res. 2008;36:D339–343. doi: 10.1093/nar/gkm700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Podlevsky JD, Li Y, Chen JJ. The functional requirement of two structural domains within telomerase RNA emerged early in eukaryotes. Nucleic Acids Res. 2016;44:9891–9901. doi: 10.1093/nar/gkw605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *31.Wu RA, Dagdas YS, Yilmaz ST, Yildiz A, Collins K. Single-molecule imaging of telomerase reverse transcriptase in human telomerase holoenzyme and minimal RNP complexes. Elife. 2015:4. doi: 10.7554/eLife.08363. Reports that human telomerase TERT-TER complex is active as a monomer. Shows that TERT and TER can assemble as monomers or dimers, depending on conditions and mediated in part by a long linker of low-complexity sequence between the TEN and TRBD domain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 33.Bryan C, Rice C, Hoffman H, Harkisheimer M, Sweeney M, Skordalakes E. Structural Basis of Telomerase Inhibition by the Highly Specific BIBR1532. Structure. 2015;23:1934–1942. doi: 10.1016/j.str.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polshakov VI, Petrova OA, Parfenova YY, Efimov SV, Klochkov VV, Zvereva MI, Dontsova OA. NMR assignments of the N-terminal domain of Ogataea polymorpha telomerase reverse transcriptase. Biomol NMR Assign. 2016;10:183–187. doi: 10.1007/s12104-015-9663-6. [DOI] [PubMed] [Google Scholar]
- 35.Rouda S, Skordalakes E. Structure of the RNA-binding domain of telomerase: implications for RNA recognition and binding. Structure. 2007;15:1403–1412. doi: 10.1016/j.str.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 36.Harkisheimer M, Mason M, Shuvaeva E, Skordalakes E. A motif in the vertebrate telomerase N-terminal linker of TERT contributes to RNA binding and telomerase activity and processivity. Structure. 2013;21:1870–1878. doi: 10.1016/j.str.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang J, Brown AF, Wu J, Xue J, Bley CJ, Rand DP, Wu L, Zhang R, Chen JJ, Lei M. Structural basis for protein-RNA recognition in telomerase. Nat Struct Mol Biol. 2014;21:507–512. doi: 10.1038/nsmb.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chu TW, MacNeil DE, Autexier C. Multiple Mechanisms Contribute to the Cell Growth Defects Imparted by Human Telomerase Insertion in Fingers Domain Mutations Associated with Premature Aging Diseases. J Biol Chem. 2016;291:8374–8386. doi: 10.1074/jbc.M116.714782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *39.Chu TW, D’Souza Y, Autexier C. The Insertion in Fingers Domain in Human Telomerase Can Mediate Enzyme Processivity and Telomerase Recruitment to Telomeres in a TPP1-Dependent Manner. Mol Cell Biol. 2016;36:210–222. doi: 10.1128/MCB.00746-15. Provides biochemical evidence that human TERT IFD is involved in the regulation of telomerase activity and processivity through an interaction with TPP1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao C, Pyle AM. Crystal structures of a group II intron maturase reveal a missing link in spliceosome evolution. Nat Struct Mol Biol. 2016;23:558–565. doi: 10.1038/nsmb.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qu G, Kaushal PS, Wang J, Shigematsu H, Piazza CL, Agrawal RK, Belfort M, Wang HW. Structure of a group II intron in complex with its reverse transcriptase. Nat Struct Mol Biol. 2016;23:549–557. doi: 10.1038/nsmb.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 43.Cash DD, Cohen-Zontag O, Kim NK, Shefer K, Brown Y, Ulyanov NB, Tzfati Y, Feigon J. Pyrimidine motif triple helix in the Kluyveromyces lactis telomerase RNA pseudoknot is essential for function in vivo. Proc Natl Acad Sci USA. 2013;110:10970–10975. doi: 10.1073/pnas.1309590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Q, Kim NK, Feigon J. Architecture of human telomerase RNA. Proc Natl Acad Sci USA. 2011;108:20325–20332. doi: 10.1073/pnas.1100279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Q, Kim NK, Peterson RD, Wang Z, Feigon J. Structurally conserved five nucleotide bulge determines the overall topology of the core domain of human telomerase RNA. Proc Natl Acad Sci USA. 2010;107:18761–18768. doi: 10.1073/pnas.1013269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie M, Mosig A, Qi X, Li Y, Stadler PF, Chen JJ. Structure and function of the smallest vertebrate telomerase RNA from teleost fish. J Biol Chem. 2008;283:2049–2059. doi: 10.1074/jbc.M708032200. [DOI] [PubMed] [Google Scholar]
- *47.Parks JW, Kappel K, Das R, Stone MD. Single-molecule FRET-Rosetta reveals RNA structural rearrangements during human telomerase catalysis. RNA. 2017;23:175–188. doi: 10.1261/rna.058743.116. Reports structural models of Tetrahymena telomerase TERT ring and TER template/pseudoknot based on single-molecule FRET and Rosetta modeling. Suggests that the pseudoknot is located on the CTE and moves relative to the TERT ring during different steps in telomere repeat synthesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **48.Vogan JM, Zhang X, Youmans DT, Regalado SG, Johnson JZ, Hockemeyer D, Collins K. Minimized human telomerase maintains telomeres and resolves endogenous roles of H/ACA proteins, TCAB1, and Cajal bodies. Elife. 2016:5. doi: 10.7554/eLife.18221. Reports a minimized human TER without the H/ACA scaRNP motif but with a modified 3′ end to confer stability in vivo can form an active enzyme with TERT in vitro and assembles in vivo as a biologically active enzyme that maintains telomere length, indicating that interaction with the Cajal body chaperone TCAB1 or the Cajal body scaffold protein Coilin is not required for RNP assembly or telomere length maintenance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mihalusova M, Wu JY, Zhuang X. Functional importance of telomerase pseudoknot revealed by single-molecule analysis. Proc Natl Acad Sci U S A. 2011;108:20339–20344. doi: 10.1073/pnas.1017686108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cole DI, Legassie JD, Bonifacio LN, Sekaran VG, Ding F, Dokholyan NV, Jarstfer MB. New models of Tetrahymena telomerase RNA from experimentally derived constraints and modeling. J Am Chem Soc. 2012;134:20070–20080. doi: 10.1021/ja305636u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hengesbach M, Kim NK, Feigon J, Stone MD. Single-molecule FRET reveals the folding dynamics of the human telomerase RNA pseudoknot domain. Angew Chem Int Ed. 2012;51:5876–5879. doi: 10.1002/anie.201200526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niederer RO, Zappulla DC. Refined secondary-structure models of the core of yeast and human telomerase RNAs directed by SHAPE. RNA. 2015;21:254–261. doi: 10.1261/rna.048959.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zemora G, Handl S, Waldsich C. Human telomerase reverse transcriptase binds to a pre-organized hTR in vivo exposing its template. Nucleic Acids Res. 2016;44:413–425. doi: 10.1093/nar/gkv1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lai CK, Miller MC, Collins K. Template boundary definition in Tetrahymena telomerase. Genes Dev. 2002;16:415–420. doi: 10.1101/gad.962602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *55.Yang W, Lee YS. A DNA-hairpin model for repeat-addition processivity in telomere synthesis. Nat Struct Mol Biol. 2015;22:844–847. doi: 10.1038/nsmb.3098. The authors present a model for the translocation step of telomere repeat synthesis that involves formation of a hairpin by the telomeric DNA, based on primer loopout in translesion DNA polymerase ν, unusual DNA hairpin structures, and large motions of the TERT thumb domain (CTE) relative to the palm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **56.Wu RA, Tam J, Collins K. DNA-binding determinants and cellular thresholds for human telomerase repeat addition processivity. EMBO J. 2017 doi: 10.15252/embj.201796887. Using extensive mutagenesis and nuclease protection assays propose a detailed model for cycle of telomere repeat synthesis and repeat addition that helps explain decades of biochemical data. Found a ssDNA retention surface (SRS) that binds and retains ssDNA in the active site. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *57.Akiyama BM, Parks JW, Stone MD. The telomerase essential N-terminal domain promotes DNA synthesis by stabilizing short RNA-DNA hybrids. Nucleic Acids Res. 2015;43:5537–5549. doi: 10.1093/nar/gkv406. Single-molecules telomerase binding assays that monitor DNA dynamics within telomerase-primer complexes by FRET provide evidence that the TEN domain stabilizes the initial short RNA-DNA hybrids. An L14A mutation that strongly decreases repeat addition processivity is shown to decrease the the ability to form a short RNA-DNA hybrids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu RA, Collins K. Human telomerase specialization for repeat synthesis by unique handling of primer-template duplex. EMBO J. 2014;33:921–935. doi: 10.1002/embj.201387205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sugitani N, Chazin WJ. Characteristics and concepts of dynamic hub proteins in DNA processing machinery from studies of RPA. Prog Biophys Mol Biol. 2015;117:206–211. doi: 10.1016/j.pbiomolbio.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Min B, Collins K. An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol Cell. 2009;36:609–619. doi: 10.1016/j.molcel.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Upton HE, Hong K, Collins K. Direct single-stranded DNA binding by Teb1 mediates the recruitment of Tetrahymena thermophila telomerase to telomeres. Mol Cell Biol. 2014;34:4200–4212. doi: 10.1128/MCB.01030-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **62.Upton HE, Chan H, Feigon J, Collins K. Shared Subunits of Tetrahymena Telomerase Holoenzyme and Replication Protein A Have Different Functions in Different Cellular Complexes. J Biol Chem. 2017;292:217–228. doi: 10.1074/jbc.M116.763664. This paper shows that Teb2 and Teb3, originally identified as components of Tetrahymena telomerase RPA-related TEB complex (Teb1-Teb2-Teb3) are shared subunits with Tetrahymena RPA. Specificity and binding to single-stranded telomeric DNA is conferred by Teb1, while in RPA, Rpa1 binds to single-stranded DNA sequence non-specifically and Teb2 contributes to binding affinity for longer substrates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lue NF, Chan J, Wright WE, Hurwitz J. The CDC13-STN1-TEN1 complex stimulates Pol alpha activity by promoting RNA priming and primase-to-polymerase switch. Nat Commun. 2014;5:5762. doi: 10.1038/ncomms6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frescas D, de Lange T. TRF2-tethered TIN2 can mediate telomere protection by TPP1/POT1. Mol Cell Biol. 2014;34:1349–1362. doi: 10.1128/MCB.01052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 66.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 67.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 68.Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O’Connor MS, Songyang Z. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 69.Sexton AN, Youmans DT, Collins K. Specificity requirements for human telomere protein interaction with telomerase holoenzyme. J Biol Chem. 2012;287:34455–34464. doi: 10.1074/jbc.M112.394767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhong FL, Batista LF, Freund A, Pech MF, Venteicher AS, Artandi SE. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell. 2012;150:481–494. doi: 10.1016/j.cell.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, Cech TR. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature. 2012;492:285–289. doi: 10.1038/nature11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmidt JC, Dalby AB, Cech TR. Identification of human TERT elements necessary for telomerase recruitment to telomeres. Elife. 2014:3. doi: 10.7554/eLife.03563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *73.Dalby AB, Hofr C, Cech TR. Contributions of the TEL-patch amino acid cluster on TPP1 to telomeric DNA synthesis by human telomerase. J Mol Biol. 2015;427:1291–1303. doi: 10.1016/j.jmb.2015.01.008. Using telomerase competition assay shows that TEL-patch amino acid cluster of TPP1 stabilizes telomerase on telomeric DNA throughout the catalytic cycle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *74.Bisht K, Smith EM, Tesmer VM, Nandakumar J. Structural and functional consequences of a disease mutation in the telomere protein TPP1. Proc Natl Acad Sci U S A. 2016;113:13021–13026. doi: 10.1073/pnas.1605685113. Reports the crystal strucutre of TPP1 K170Δ, a mutation associated with dyskeratosis congenita. The mutation restructures a loop that interacts with TERT and is shown to cause telomere shortening in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Linger BR, Morin GB, Price CM. The Pot1a-associated proteins Tpt1 and Pat1 coordinate telomere protection and length regulation in Tetrahymena. Mol Biol Cell. 2011;22:4161–4170. doi: 10.1091/mbc.E11-06-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bryan TM, Goodrich KJ, Cech TR. Tetrahymena telomerase is active as a monomer. Mol Biol Cell. 2003;14:4794–4804. doi: 10.1091/mbc.E03-07-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *77.Bajon E, Laterreur N, Wellinger RJ. A Single Templating RNA in Yeast Telomerase. Cell Rep. 2015;12:441–448. doi: 10.1016/j.celrep.2015.06.045. This report shows that only a single copy of telomerase RNA per yeat S. cerevisiae telomerase RNP is detected by multi-color fluorescence in situ hybridization. [DOI] [PubMed] [Google Scholar]
- 78.Xi L, Cech TR. Inventory of telomerase components in human cells reveals multiple subpopulations of hTR and hTERT. Nucleic Acids Res. 2014;42:8565–8577. doi: 10.1093/nar/gku560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **79.Lemieux B, Laterreur N, Perederina A, Noel JF, Dubois ML, Krasilnikov AS, Wellinger RJ. Active Yeast Telomerase Shares Subunits with Ribonucleoproteins RNase P and RNase MRP. Cell. 2016;165:1171–1181. doi: 10.1016/j.cell.2016.04.018. Discovered that the RNaseP/MRP protein components Pop1, Pop6, and Pop7 are shared subunits with yeast S. cerevisiae telomerase and identified the P3-like binding domain in the telomerase RNA. This complex is essential for telomerase activity at telomeres in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *80.Lin KW, McDonald KR, Guise AJ, Chan A, Cristea IM, Zakian VA. Proteomics of yeast telomerase identified Cdc48-Npl4-Ufd1 and Ufd4 as regulators of Est1 and telomere length. Nat Commun. 2015;6:8290. doi: 10.1038/ncomms9290. Used mass spectrometry to identify a complete set of yeast S. cerevisiae proteins associated with telomerase, and evaluated a few that were deemed to be significant. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fan J, Pavletich NP. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 2012;26:2337–2347. doi: 10.1101/gad.194787.112. [DOI] [PMC free article] [PubMed] [Google Scholar]