Abstract
Purpose
Little is known about the molecular signatures associated with specific metastatic sites in breast cancer. Using comprehensive multi-omic molecular profiling, we assessed whether alterations or activation of the PI3K–AKT–mTOR pathway is associated with specific sites of breast cancer metastasis.
Experimental Design
Next-generation sequencing–based whole-exome sequencing was coupled with reverse-phase protein microarray (RPPA) functional signaling network analysis to explore the PI3K–AKT–mTOR axis in 32 pretreated breast cancer metastases. RPPA-based signaling data were further validated in an independent cohort of 154 metastatic lesions from breast cancer and 101 unmatched primary breast tumors. The proportion of cases with PI3K–AKT–mTOR genomic alterations or signaling network activation were compared between hepatic and nonhepatic lesions.
Results
PIK3CA mutation and activation of AKT (S473) and p70S6K (T389) were detected more frequently among liver metastases than nonhepatic lesions (P < 0.01, P = 0.056, and P = 0.053, respectively). However, PIK3CA mutations alone were insufficient in predicting protein activation (P = 0.32 and P = 0.19 for activated AKT and p70S6K, respectively). RPPA analysis of an independent cohort of 154 tumors confirmed the relationship between pathway activation and hepatic metastasis [AKT (S473), mTOR (S2448), and 4EBP1 (S65); P < 0.01, P = 0.02, and P = 0.01, respectively]. Similar results were also seen between liver metastases and primary breast tumors [AKT (S473) P < 0.01, mTOR (S2448) P < 0.01, 4EBP1 (S65) P = 0.01]. This signature was lost when primary tumors were compared with all metastatic sites combined.
Conclusions
Breast cancer patients with liver metastasis may represent a molecularly homogenized cohort with increased incidence of PIK3CA mutations and activation of the PI3K–AKT–mT OR signaling network.
Introduction
Despite the introduction of more effective detection methods and therapeutic regimens, breast cancer still represents the second leading cause of female-specific cancer-related mortality, accounting for approximately 40,000 deaths every year (1). Breast cancer is a heterogeneous malignant phenotype originating in mammary tissues and is intrinsically subdivided into luminal hormone receptor–positive tumors, Her2-neu–positive tumors, and basal-like tumors (2). The development of distant metastases is the strongest negative prognostic factor associated with breast cancer mortality (1). Although breast cancer metastases are detected in approximately 5%–10% newly diagnosed cases, 25%–50% of patients affected by early-stage lesions or by locally advanced tumors may develop metachronous metastatic disease (3, 4).
Data from the Cancer Genome Atlas (TCGA) indicate that PI3K–AKT–mTOR signaling pathway mutations (PIK3CA, PTEN, PIK3R1, AKT1) commonly occur in breast cancer (5). PIK3CA is the second most commonly mutated gene in hormone receptor–positive breast cancer (5, 6). The inhibitor everolimus, a compound that specifically targets mTOR, a downstream member of the PI3K axis, is selected as an early therapeutic option for metastatic breast cancer patients presenting with well-defined molecular characteristics like ER expression (7–9). The therapeutic efficacy of a number of AKT, PI3K, and mTOR inhibitors is under extremely active clinical investigation for the treatment of breast cancer and many other cancers.
The PI3K–AKT–mTOR signaling pathway is known for playing a major role in cancer progression as it represents a central node for regulating a number of cellular functions including cell growth and survival, motility, apoptosis, cell cycle, and various metabolic functions (10, 11). Signaling network analyses have indicated that this pathway is highly activated in liver metastases from colorectal cancer compared with matched primary tumors (12, 13). Deregulation of the PI3K–AKT–mTOR signaling axis has also emerged as a pivotal molecular mechanism in primary hepato- and cholangiocarcinomas, even though genomic alterations of the members of the PI3K pathway are relatively infrequent in this group of tumors (14–17). Although a large number of genomic and proteomic studies have demonstrated that at the molecular level metastatic lesions significantly differ from matched primary tumors (12, 18, 19), the impact of the microenvironment at different organ sites has been only partially explored. On the basis of these previous observations, we postulated that alterations of the PI3K–AKT-mTOR signaling network may be especially important for cancer cells to survive in the liver microenvironment and that this pathway may represent an organ-specific targetable signature for liver metastases in breast cancer.
Using a “multi-omic” approach, including NGS-based whole exome sequencing coupled with reverse-phase protein microarray (RPPA)-based functional phosphoprotein signaling network activation analysis, we assessed the frequency and role of the PI3K–AKT–mTOR signaling alterations in breast cancer hepatic metastases. Functional RPPA-based phosphoprotein signaling data were then used to validate the initial findings in an independent cohort of primary and metastatic breast cancers.
Materials and Methods
Patient populations
Breast cancer patients were prospectively enrolled in the Side Out 2 Trial, a clinical study wherein NGS/protein/phosphoprotein “multi-omic” profiling was used to identify therapeutic targets for patients with metastatic disease refractory to at least one line of standard therapy (20). The study protocol was approved by the local Institutional Review Board (IRB). Patients entered the trial voluntarily and provided informed consent before sample collection and treatment administration. At the time of enrollment, metastatic lesions were sampled under CT/US guidance or via surgical excision. For patients presenting with multiple metastatic lesions, only one lesion was biopsied. Samples were snap frozen within 20 minutes of collection and shipped to the laboratories overnight in dry ice. Matched whole blood samples were collected along with the metastatic lesions for germ line subtraction of somatic sequencing data.
Formalin-fixed, paraffin-embedded (FFPE) tissues consisting of 154 breast cancer metastases and 101 primary breast tumors prospectively collected were used for validation. Tissues included in Study Set 2 were collected both under IRB-approved protocols (J.A. O’Shaughnessy, M. Cristofanilli, and B. Leyland-Jones) and used for patient treatment decisions using a commercially available CAP/CLIA–approved test, TheraLink (TM; Theranostics Health, Inc.; ref. 21). This platform measures the activation level of 24 drug targets and downstream substrates. For the purpose of this work, only data concerning the AKT–mTOR pathway are reported.
All tissue samples were obtained by core needle biopsy and processed immediately into FFPE or snap frozen within 20 minutes of biopsy. Such operating principals are within the time limits previously shown to minimize the effects of cold ischemia on phosphorylation levels (22, 23).
Whole-exome sequencing and RNA-seq analysis
DNA/RNA extractions
Fresh tumor tissue biopsy specimens were evaluated histologically to assess tumor cellularity when available (Supplementary Table S1). DNA and total RNA was extracted from tumor biopsy specimens using Qiagen Allprep DNA/RNA/miRNA Universal Kit. Genomic germline DNA was extracted from patient blood samples using QIAamp DNA blood Mini Kit. Quality of DNA and RNA was assessed using Agilent 2200 TapeStation Instrument as per manufacturer protocol.
Library preparation and sequencing
Tumor and normal germline exome libraries were prepared from 200 ng of genomic DNA using Kapa Library Preparation Kit and Agilent SureSelectXT Human All Exon V5+ UTRs capture probes. Unstranded mRNA libraries were prepared with 1000 ng of tumor RNA (eRIN > 8) using Illumina TruSeq RNA Library Prep Kit V2. All sequencing libraries were quantified and quality assessed with the Qubit Fluorometer and Agilent 2200 TapeStation Instrument, respectively. Exome and RNA libraries were paired-end sequenced on the Illumina HiSeq 2500 platform using Rapid Mode with a 109 × 7 × 109 read length.
Sequencing data analysis
Raw sequencing reads were converted to .fastq files using bcl2fastq 1.8.4. A pipeline consisting of published software tools was used to analyze the .fastq data to generate a genetic profile to determine the presence of somatic single nucleotide variations (SNV), insertion/deletion events (INDELS), gene fusions, and differential expression for each tumor. Briefly, exome data were aligned to the human reference genome hs37d5 using Burrows-Wheeler Alignment (BWA v0.7.8) Tool (24). After alignment, the base quality scores were recalibrated and joint INDEL realignment was performed on the BAM files using GATK v3.1-1. Germline SNPs and INDELS were identified using GATK HaplotypeCaller in the constitutional samples (25). Final BAM files were then used to identify germline and somatic events. SNVs and INDELS were detected using three algorithms including, Strelka 1.0.13, MuTect 1.1.4, and Seurat (26–28). Filtering was performed on exome data to give greatest confidence to somatic coding variants that were called by two of the three mutation callers. Somatic copy number detection was based on a log2 comparison of normalized physical coverage (or clonal coverage) across tumor and normal whole exome sequencing data, where physical coverage was calculated by considering the entire region a paired-end fragment span. Normal and tumor physical coverage was then normalized, smoothed, and alltered for highly repetitive regions prior to calculating the log2 comparison. Loss of heterozygosity (LOH) was also deduced by calculating alternate allele frequencies for SNPs. Briefly, B-allele frequencies (BAF) are allele fraction of nonreference reads in the tumor at heterozygote common polymorphic SNPs (minor allele frequency >5%) from 1,000 genomes in the patient’s germline (29). Only heterozygous SNPs were plotted and determined from that patient’s germline calls. The calculation alt/(ref + alt) where alt is B was used. This should be 50/50 unless LOH or allele imbalance has occurred at that site. The BAF were then plotted against map position to identify regions of LOH. Copy number analysis was also performed using the circular binary segmentation algorithm DNA copy within the BioConductor package (30).
RNA-seq data were aligned to GRCh37.74 using STAR 2.3.1 Aligner. Gene counts were done with HTSeq and FPKM values were generated using Cuffiinks 2.2.1. Differential gene expression of RNA-seq data was generated against a pooled normal breast RNA reference using Cuffdiff from the Cuffiinks 2.2.1 suite and the DESeq2 package.
Somatic SNVs, germline SNPs, genes related to copy number events, and differentially expressed genes were annotated using SnpEff 3.5 h (31).
Functional protein signaling network analysis of AKT–mTOR pathway by RPPA
Isolation of pure tumor cells via laser capture microdissection
Frozen samples were immediately processed into 8 μm cryosections and mounted on uncharged glass slides upon arrival. Mounted samples were stored at −80°C for at least 12–18 hours before further processing. Adequate sample preservation (e.g., absence of freeze–thaw artifacts) and presence of sufficient amount of tumor cells was verified by a certified pathologist (L.A. Liotta) on one or more hematoxylin and eosinstained slides.
To preserve protein phosphorylation during LCM: (i) cells were first fixed in 70% ethanol, (ii) protease inhibitors were added to the 70% ethanol, dH2O, hematoxylin, and Scott’s tap water, (iii) cells were isolated using an infrared laser and each stained slide was microdissected for less than 30 minutes. This protocol has previously been described as sufficient to preserve the phosphoproteome and neither the microdissection nor the tissue fixation process significantly impacts the data generated (32, 33).
Pure subpopulations of cancer cells were isolated from the surrounding microenvironment via direct visualization using an Arcturus PixCell II instrument or an Arcturus XT automated system. Microdissected cells were lysed in a 1:1 mixture of T-PER (Tissue Protein Extraction Reagent; Pierce) and 2× Tris-glycine SDS sample buffer (Invitrogen) containing 5% 2-mercaptoethanol as previously described, and boiled for 8 minutes before printing arrays (34).
A similar protocol was followed for the isolation of the tumor cells from the FFPE samples included in the validation set. Differences between the two protocols include: (i) deparaffinization of the FFPE slides in xylene twice for 5 minutes followed by rehydration in 100%, 90%, and 70% ethanol, respectively; (ii) microdissected cells lysates were boiled for 60 minutes before printing arrays.
Reverse-phase protein microarray preparation and immunostaining
Using a 2470 Aushon arrayer outfitted with 185-μm pins, experimental samples were printed onto nitrocellulose covered slides (GRACE Bio-Labs; Sartorius Stedim Biotech) along with standard curves and internal controls. Samples, reference standards and internal controls were printed in three or four technical replicates on each slide. To verify that each sample was in the linear dynamic range of the assay, a BSA serial dilution curve was added to the array to estimate the protein concentration of each sample.
Immunostaining was performed on an automated DAKO system using a commercially available Catalyzed Signal Amplification (CSA) Kit (Dako). Samples included in the training set were probed using primary antibodies targeting AKT (S473) (Cell Signaling Technology, catalog no. 9871; dilution 1:100), and the mTOR downstream substrate p70S6K (T389) (Cell Signaling Technology, catalog no. 9205; dilution 1:100). The validation set was probed with AKT (S473) (Cell Signaling Technology, catalog no. 9871; dilution 1:100), mTOR (S2448) (Cell Signaling Technology, catalog no. 2971; dilution 1:100), and two downstream substrates: 4EBP1 (S65) (Cell Signaling Technology, catalog no. 9451; dilution 1:100), and S6 Ribosomal Protein (S6RP) (S235/236; Cell Signaling Technology, catalog no. 4858; dilution 1:100). Each antibody was rigorously validated on a panel of cell lines and human samples using conventional Western blotting technique and tested on the arrays to assure the linear dynamic range of the analytes was captured (35).
Signal detection was achieved using a biotinylated goat antirabbit secondary antibody (1:7,500 and 1:5,000 for the discovery and validation set, respectively; Vector Laboratories) coupled with a biotinyl-tyramide–based amplification system (34). The IRDye 680RD Streptavidin (LI-COR Biosciences; dilution 1:50 in PBS supplemented with 1% BSA) or the Cy5 Strepatavidin (KPL; dilution 1:100 in PBS supplemented with 1% BSA) fluorescent detection systems were used for the discovery and validation set, respectively. Finally, to quantify the amount of protein in each sample, selected slides were stained with Sypro Ruby Protein Blot Stain (Molecular Probes) following manufacturer’s recommendation (34).
A laser scanner was used to acquire antibody and Sypro Ruby stained slides (TECAN or Genepix 4200 AL, Molecular Devices). Images were analyzed using the commercially available microarray software MicroVigene Version 5.1.0.0 (Vigenetech) or Genepix Pro 6.1 (Molecular Devices). Final intensity values were generated after: (i) subtraction of background and unspecific binding generated by the secondary antibody; (ii) normalization to the corresponding amount of protein derived from the Sypro Ruby–stained slides; and (iii) average of the technical replicates. Experimental sample values were interpolated from the reference standards using standard linear interpolation techniques. Each patient value was compared to the reference population and scored on a categorical scale (above or below the cut-off point).
Calibration of the RPPA assay and cut-off point determination
To directly compare samples individually processed, standard curves were added to each microarray slide. In brief, standard curves were created using two commercially available cell lines containing different amounts of the analytes of interest. Because most of the analytes involved in the AKT–mTOR pathway are serine/threonine kinases, Jurkat cells stimulated with Calyculin A, a specific phospho-serine/-threonine phosphatase inhibitor, were selected as the cell line with high expression of the analytes of interest (36). Untreated HeLa cells, on the other hand, were chosen because of the low/absent signal of the phosphoprotein measured. Standard curves were created by adding a decreasing amount of Jurkat cells treated with Calyculin A to a progressively increasing amount of untreated HeLa cells. Protein content across the two cell lines was matched before standard curves were constructed so that each point of the calibrator had equal protein concentration while the amount of the analytes of interest decreased progressively.
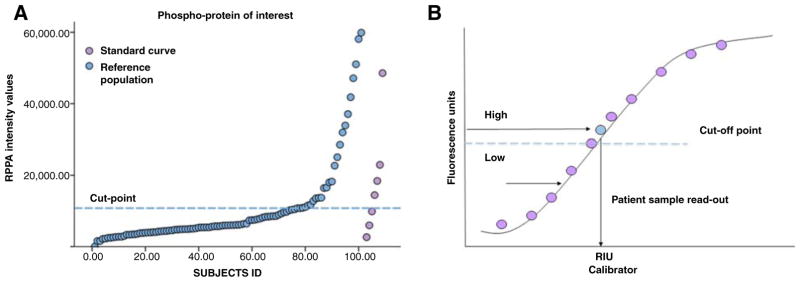
To test whether these standard curves covered the linear dynamic range of the analyte of interest, reference standards were tested on the RPPA platform along with large populations of microdissected human cancers. Reference populations, including samples that were similar to the one described in this article, were used to establish cut-off points that allow identification of patients with high activation of the analytes of interest (Fig. 1A). Using conventional linear interpolation models, sample read outs were transformed from absolute RPPA intensity values to relative units of the standard curve (Fig. 1B). Experimental samples matching the activation level of the top 25% to 40% of the reference population were classified as highly activated. For AKT (S473), the only protein measured by both institutions, the threshold above which AKT activation was classified as positive was set independently by the two laboratories for study set one (GMU) and study set two (Theranostics Health, Inc.) at the same level of the standard curve. Both laboratories shared the same internal standard curve for AKT (S473), which was printed on all RPPA slides. Cut-off points were set before the initiation of the trial and tested weekly along with internal controls for QC/QA. RPPA inter- and intra-assay reproducibility has been previously described (37, 38).
Figure 1.
Standardization of the RPPA platform for clinical use. Standard curves are printed along with experimental samples to explore the dynamic range of the phosphoproteins of interest and set cut-off points for the identification of patients with high activation of any given protein (A). Once standard curves are added to the arrays, the intensity value of each sample is interpolated from the linear dynamic range of the standard curve and reported in terms of relative units of the standard curve (B).
Data analysis
Molecular markers were classified as dichotomous variables. Genomic markers were classified as mutant/amplified versus wild-type. Signaling data were classified as activated (above the cut-off point) or nonactivated (below the cut-off point). Fisher exact test was performed to compare the molecular profile of: (i) liver metastases versus other metastatic sites; (ii) liver metastases versus primary tumors; and (iii) all metastatic sites versus primary tumors. Comparisons with P ≤ 0.05 were considered significant. Data were displayed using mosaic plots generated in JMP version 5.1 (SAS institute Inc.) and adapted for publication in Photoshop version 11.0.
Results
PIK3CA mutations are more prevalent in liver metastases compared to other metastatic sites
The role of the AKT–mTOR signaling pathway in promoting metastasis to different host organs was first evaluated across 32 metastatic breast cancer patients enrolled in the Side Out 2 trial. Metastatic lesions were classified as hepatic (n = 11) and others (n = 21), the latter including cutaneous (n = 10), lung (n = 4), lymph nodes (n = 4), and intra-abdominal lesions (n = 3). Of the 32 patients enrolled, 31 had sufficient tumor content for ER, PR, and Her2 determination. Patients were classified as ER+/Her2−(n = 19; 61.3%), ER/PR/Her2−(n = 8; 25.8%), and ER/PR−/Her2+ (n = 4; 12.9%; Table 1). Subtype distribution did not differ between liver metastases and lesions developed at the other organ sites (P = 0.14).
Table 1.
Molecular characteristics of the patients included in the discovery set
| Case ID | Metastatic site | ER | PR | Her2 |
|---|---|---|---|---|
| N02-03-003 | Liver | Pos | Neg | Not amplified |
| N02-03-011 | Liver | Neg | Neg | Not amplified |
| N02-03-012 | Liver | Pos | Pos | Not amplified |
| N02-03-010 | Liver | Pos | Pos | Not amplified |
| N02-03-017 | Liver | Pos | Neg | Not amplified |
| N02-03-018 | Liver | Pos | Neg | Not amplified |
| N02-03-020 | Liver | Pos | Neg | Not amplified |
| N02-02-023a | Liver | Neg | Neg | Not amplified |
| N02-02-036 | Liver | Pos | Neg | Not amplified |
| N02-03-037 | Liver | Pos | Pos | Not amplified |
| N02-03-043 | Liver | Pos | Neg | Not amplified |
| N02-03-001 | Lung | Neg | Neg | Not amplified |
| N02-01-005 | Lung | Pos | Neg | Not amplified |
| N02-04-014 | Lung | Pos | Neg | Not amplified |
| N02-03-039 | Lung | Neg | Neg | Amplified |
| N02-02-006 | Lymph node | Pos | Neg | Not amplified |
| N02-02-023* | Lymph node | Neg | Neg | Not amplified |
| N02-01-025 | Lymph node | Pos | Neg | Not amplified |
| N02-04-022 | Lymph node | NA | NA | NA |
| N02-02-021 | Omentum | Pos | Neg | Not amplified |
| N02-02-027 | Omentum | Pos | Neg | Not amplified |
| N02-04-009 | Intra-abdominal mass | Pos | Pos | Not amplified |
| N02-01-004 | Chest wall/skin | Neg | Neg | Amplified |
| N02-04-007b | Chest wall/skin | Pos | Neg | Not amplified |
| N02-04-008 | Chest wall/skin | Pos | Pos | Not amplified |
| N02-02-019 | Chest wall/skin | Neg | Neg | Amplified |
| N02-01-026 | Chest wall/skin | Neg | Neg | Amplified |
| N02-02-028 | Chest wall/skin | Pos | Neg | Not amplified |
| N02-02-029 | Chest wall/skin | Neg | Neg | Not amplifiedc |
| N02-03-032 | Chest wall/skin | Neg | Neg | Not amplified |
| N02-03-038 | Chest wall/skin | Neg | Neg | Not amplified |
| N02-02-041 | Chest wall/skin | Neg | Neg | Not amplified |
NOTE: The site of metastasis along with ER, PR, and Her2 status are reported of each lesion.
A second biopsy was collected from the same patient after recurrence.
Metastatic lesion from a male breast tumor.
Data retrieved from whole exome sequencing analysis.
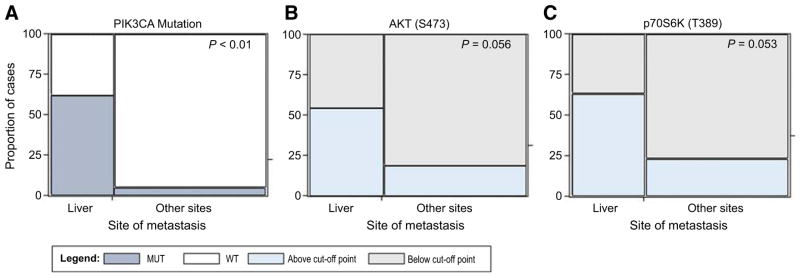
PIK3CA mutations along with genomic alterations of other members of the AKT–mTOR pathway were first evaluated (Supplementary Fig. S1). Whole exome sequencing and RNA-Seq data were available for 8 of the 11 liver metastases and 19 of the 21 nonhepatic lesions. PIK3CA somatic mutation frequency was significantly greater among liver metastases compared to the other metastatic sites (P = 0.01) with PIK3CA mutation being detected in 5/8 liver metastases (62.5%) and 1/19 non-hepatic lesions (5.3%; Fig. 2A and Supplementary Table S2). Although four of the PIK3CA mutant liver metastases had mutations involving the helical and/or kinase domain, the PIK3CA mutation detected in the omental lesion was of unknown significance (Table 2). Therefore, the frequency of known activating mutations in PIK3CA among liver metastases was 50%. Among patients with liver metastases and PIK3CA mutation, one patient presented with a concomitant RPS6KB1 and RPTOR amplification and a second patient with RPS6KB1 amplification only. Amplifications of RPS6KB1 and/or RPTOR were not detected in nonhepatic metastases.
Figure 2.
Mosaic plots representing PIK3CA mutation incidence and activation of AKT (S473) and p70S6K (T389) for the 30 patients enrolled in the Side Out 2 trial. Proportion of positive patients is plotted along with the site of metastasis. P values were calculated using a Fisher exact test.
Table 2.
Molecular characteristics of the six patients harboring a PIK3CA mutation
| Case ID | Metastatic site | AKT (S473) | P70S6K (T389) | PIK3CA mutation | Mutation description |
|---|---|---|---|---|---|
| N02-03-003 | Liver | Low | High | E542K | Helical domaina |
| N02-03-011 | Liver | High | High | C420R | Kinase domain |
| N02-03-012 | Liver | High | High | H1047L | Kinase domain |
| N02-03-020 | Liver | High | High | H1047Y, E545K | Kinase, helical domainb |
| N02-03-037 | Liver | Low | Low | N345K | Noncanonical |
| N02-02-027 | Omentum | Low | Low | R1023Q | Unknown role |
NOTE: Site of metastasis along with the type of mutation and activation of AKT (S473) and downstream substrate p70S6K (T389) are reported for each patient.
Concomitant RPTOR and RPS6KB1 amplification.
Concomitant RPS6KB1 amplification.
Among PIK3CA wild-type liver metastases, PTEN gene deletion was detected in one patient. Within the PIK3CA wild-type non-hepatic metastases, two patients had an AKT2 amplification and another two patients presented with a PTEN deletion. In these cases, the two PTEN deletions were a 56-base deletion and a deletion affecting the splice site, although both the wild-type and variant PTEN were expressed.
AKT–mTOR protein signaling network is highly activated in liver metastases from breast cancer compared with other metastatic sites
Because the proportion of patients with PIK3CA mutations was significantly higher in the liver metastases compared to the other metastatic sites, functional signaling analysis of the AKT–mTOR axis was further explored across different host organs. Phosphorylation of the PI3K downstream substrate AKT (S473) and of the well-known mTOR target p70S6K (T389) were used to directly and quantitatively measure the activation level of the AKT–mTOR signaling network. We observed a trend, only marginally significant, of increased activation of AKT (S473) (P = 0.06) across metastatic sites. AKT (S473) was activated in 6 of the 11 liver metastases (54.5%) and 4 of the 21 nonhepatic lesions (19.0%), respectively (Fig. 2B and Supplementary Table S2). Moreover, the frequency of activation of p70S6K (T389) was significantly higher (P = 0.05) in the liver metastases (63.6%) compared to the other organ sites (23.8%; Fig. 2C and Supplementary Table S2).
Finally, we explored whether PIK3CA mutation status alone was sufficient in predicting AKT and p70S6K activation. As expected, all three patients with a PIK3CA mutation involving the kinase domain had high activation of AKT (S473) and p70S6K (T389) (Table 2). One patient with PIK3CA mutation of the helical domain and concomitant RPTOR and RPS6KB1 amplification presented with high activation of p70S6K (T389), but not of AKT (S473), suggesting that the activation of the AKT downstream substrate p70S6K (T389) in this specific patient may have been directly modulated by the amplification of RPTOR or RPS6KB1. The two patients with noncanonical derangement or mutation with unknown significance presented with low activation of both AKT (S473) and p70S6K (T389). Nonetheless, overall concordance between PIK3CA mutation and AKT (S473) and p70S6K (T389) activation did not reach statistical significance (P = 0.32 and 0.19, respectively) indicating that PIK3CA status alone is not sufficient for identifying patients with activated AKT–mTOR signaling network (Supplementary Fig. S2). A recent study exploring the concordance between PIK3CA mutation and AKT–mTOR protein signaling activation in large breast cancer study sets have found little to no correlation (39), which provide evidence that these results are likely generalizable to larger study sets. When associations between AKT–mTOR protein activation and genomic alterations were evaluated by combining all genomic alterations affecting PIK3CA, AKT, and PTEN (e.g., any of the 3 genomic alteration being positive), concordance between genomic alteration and protein activation was P = 0.01 and 0.25 for AKT (S473) and p70S6K (T389), respectively. Activation of AKT (S473) and p70S6K (T389) based on individual alterations of AKT and PTEN was not performed due to low frequency of these genomic events.
Independent validation confirms high activation of AKT–mTOR signaling axis in liver metastases from breast cancer
To validate further, the increased prevalence of AKT–mTOR signaling pathway activation in liver metastases, a second independent study set was used. The validation set included 154 metastatic lesions of which 58 were hepatic and 96 originated at other organ sites. The extra-hepatic lesions of the validation set included: 42 lymph nodes, 21 skin/chest wall, 22 brain, and 11 lung metastases. Because the distribution of the different breast cancer subtypes was not statistically different between liver metastases and other metastatic sites in the training set, ER/PR, and Her2 determination was not evaluated in the validation set.
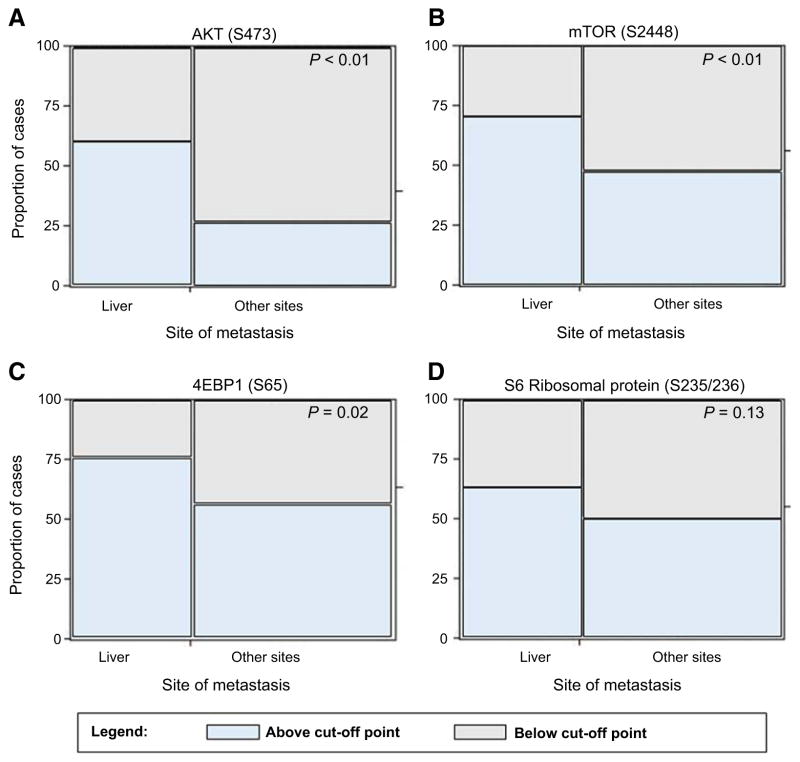
In keeping with the results in the discovery set, AKT (S473) and mTOR (S2448) activation was significantly higher (P < 0.01) in liver metastases (60.3% and 70.7%, respectively) compared to the other metastatic sites (27.1% and 47.9%, respectively) (Fig. 3A and B and Supplementary Table S2). Activation of the mTOR downstream effector 4EBP1 (S65) was also significantly higher (P = 0.02) in the hepatic lesion (75.9%) compared to the other metastatic sites (56.2%; Fig. 3C and Supplementary Table S2). On the contrary, activation of S6RP (S235/236), an indirect read out of mTOR activity, was not significant across the different sites of metastases (Fig. 3D and Supplementary Table S2). Taken together, these data confirm that liver metastases have increased activation of the AKT–mTOR signaling pathway compared to metastases that develop at different organ sites.
Figure 3.
Mosaic plots representing activation of AKT (S473) and downstream substrates mTOR (S2448), S6RP (S235/236), and 4EBP1 (S65) across 154 metastatic breast cancer. Proportion of positive patients is plotted along with the site of metastasis. P values were calculated using a Fisher exact test.
The relationship between ER/Her2 expression and AKT activation, using both binary (+/−) and continuous H-score data, found no statistical relationship between the receptor expression and AKT (S473) in metastatic lesions (Supplementary Tables S3 and S4). These data indicate that, while AKT activation may correlate with ER status in the primary tumor, it does not appear to be related to underpinning ER status in secondary metastatic lesions.
AKT–mTOR signaling network is highly activated in liver metastases from breast cancer compared to primary breast tumors
To validate further the AKT–mTOR liver-specific signature, the activation levels of AKT (S473), mTOR (S2448), 4EBP1 (S65), and S6RP (S235/236) were compared between hepatic lesions and unmatched primary breast tumors (n = 101). The proportion of patients with high activation of AKT (S473), mTOR (S2448), and 4EBP1 (S65) was significantly higher in liver metastases compared to primary breast tumors (P < 0.01, P < 0.01, and P = 0.01, respectively). In particular, activation of AKT (S473), mTOR (S2448), and 4EBP1 (S65) was seen in 60.3%, 70.7%, and 75.9% liver metastases compared to 29.7%, 46.5%, and 55.4% primary tumors respectively (Supplementary Table S2). When the analysis was repeated combining the liver metastasis with all other metastatic sites, none of the biomarkers reached statistical significance indicating that the activation of the AKT–mTOR axis is specific to the liver metastases only and not to breast cancer secondary lesions in general [AKT (S473) P = 0.11, mTOR (S2448) P = 0.13, 4EBP1 (S65) P = 0.19, and S6RP (S235/236) P = 0.44]. Finally, when primary tumors were compared to the other metastatic sites individually (lung, brain, lymph nodes, and chest wall/skin), none of the biomarkers reached statistical significance (P > 0.05).
Discussion
This is one of the first studies in which a combined genomic/phosphoproteomic or “multi-omic” approach was used to explore the molecular landscape of metastatic lesions originating from primary breast cancer. The data presented here suggest that the activation of the AKT–mTOR pathway may represent an organ-specific drug target signature for liver metastases in breast cancer. Specifically, whole exome sequencing detected a significantly higher incidence of activating PIK3CA mutations in hepatic lesions compared to metastases developed at other organ sites. Alongside, functional proteomics/phosphoproteomic data collected from two independent study sets confirmed the increased functional activation of the AKT–mTOR signaling axis in liver metastases compared to primary tumors and other metastatic sites.
Because a universal definition of phosphorylation/pathway activation does not exist, the cut-off points used in this study were set to capture the activation level of AKT and direct downstream kinase substrates like mTOR, p70S6K, 4EBP1, and S6RP. Reference population including hundreds of primary and metastatic breast cancers were analyzed internally along with standard curves and controls for comparisons across study sets. This approach allowed us to explore not only the activation state of individual proteins, but to undertake a pathway-oriented type of approach. As a consequence, the concomitant increase in terms of AKT activation along with increased phosphorylation of its downstream substrates suggest that the AKT–mTOR axis is more globally activated in breast cancer cells within the hepatic parenchyma compared to metastatic lesions growing within different organ sites as well as primary tumors.
Because the AKT–mTOR pathway is a central player in sustaining cell survival, its role in tumor progression, including metastasis, and its value as a therapeutic target have all been amply scrutinized over the years, in many types of cancer (40–42). Previous studies have found that patient-matched metastatic lesions have increased activation of the AKT-mTOR pathway, as measured by phosphorylation of p70S6K, mTOR, and 4EBP1, compared to the primary cancer in women who received adjuvant endocrine therapy (43). Primary gastric tumors harboring a PIK3CA mutation have shown increased tendency for hematogenic dissemination and high tropism for the hepatic parenchyma (44). Similarly, Li and colleagues have suggested that primary KRAS mutant colorectal cancers harboring a PIK3CA mutation may have a higher incidence of liver metastases suggesting that specific characteristics of the primary tumor related to the AKT–mTOR axis may favor the development of secondary lesions in the liver (45). High activation levels of the AKT–mTOR pathway have also been described in primary hepatic lesions (14, 15, 46). Whether the hepatic microenvironment directly activates this signaling network or favors the growth of lesions that already have high activation of this signaling architecture needs to be further explored using matched primary and hepatic lesions collected from the same patient. Previous studies in colorectal cancer indicated that some protein pathway activation signatures of the primary tumor are retained and amplified in the metastatic microenvironment (e.g., “seed” signatures). Meanwhile, other signaling activation present in nearly every metastatic lesion appear to be unique to the metastatic microenvironment (12, 13, 47).
Previous studies have indicated that ER+ tumors may be enriched for PIK3CA mutation, although this enrichment may not be directly associated with activation of AKT (48, 49). Although no association was found between ER/Her2 expression and AKT activation in this cohort of samples, subanalysis where ER/Her2 expression, AKT activation and site of metastases are concomitantly evaluated, was not conducted due to the relatively low number of patients in each comparison group. The hepatic signature here described needs to be further validated in a larger set of samples where patients are classified based on their molecular subtype. Nonetheless, to our knowledge this is one of the first studies for which molecular information was collected directly from the metastatic lesions, a type of lesion for which extensive tissue banks are not available. This approach allowed us to directly explore whether different host organs in which the metastatic lesions develop may favor the adaption of specific molecular alterations. By exploring the role of different organs this approach may shed new light on the biological mechanism underlying of metastatic progression and may lead to the development of organ specific therapeutic interventions.
Because of the relatively low sample size and mutation frequency for some of the gene members of the PI3K–AKT–mTOR axis (e.g., PTEN alterations, AKT2 amplification), this analysis did not allow us to statistically explore the impact at the signaling level of some key genomic alterations. Based on the rare appearance of these alterations, even when the pathway was functionally activated, our data suggest these alterations by themselves were not significant drivers of AKT–mTOR functional pathway activation.
When we analyzed the concordance of AKT–mTOR protein pathway activation with a PIK3CA–AKT gene “signature” wherein a genomic alteration in any one of the members constituted a positive event, we found a trend towards concordance with AKT activation. However, even in this instance there were 4 patients whose tumors had AKT–mTOR functional signaling activation in the absence of any PIK3CA–AKT genomic alteration. Previous analysis exploring PIK3CA and PTEN genomic alterations along with AKT–mTOR protein activation/phosphorylation found little concordance between measurements (15, 39, 48, 50). When these alterations were evaluated in the context of response to endocrine therapy, only the phosphorylation levels of the AKT–mTOR signaling, but not the underpinning genomic alterations in those proteins, were predictive of endocrine response (51).
Given the centrality of the AKT–mTOR signaling “hub” in the cellular biochemistry, activation of the pathway in the absence of any underpinning genomic alteration of PI3K–AKT–mTOR genes should not be surprising. Cross-talk between AKT-mTOR signaling and many other pathways, secretion of soluble ligands that activate upstream drivers of AKT-mTOR signaling (such as receptor tyrosine kinases like Her2, Her3, ALK, EGFR, IGFR, MET), or upstream pathway activation based on genomic alterations of those components, and establishment of feedback loops are all known mechanisms that can shape the signaling architecture of any tumor even in the absence of pathway-specific genomic alterations (40, 52, 53). Finally, the interconnection between pathways can lead to the activation of mTOR downstream substrates in an mTOR-independent manner. For example, the integration of different signaling cascades may explain the lack of significance of activation of the S6RP in liver metastases, even though activation of the upstream proteins p70S6K and 4EPB1 was highly significant in this group of lesions (54).
The combination of high throughput genomic and functional proteomic/phosphoproteomic data generated in this study allowed us to comprehensively explore of the role of the AKT–mTOR signaling pathway in the development of metastatic lesions at different organ sites and to identify a signature of a hepatic metastasis based on AKT–mTOR signaling activation. The introduction of molecularly rationalized treatments for metastatic disease based on molecular genotyping of the primary tumor and/or the metastatic lesion has created new opportunities for identifying targetable alterations underlying tumor growth and progression. Nonetheless, the identification of the true molecular drivers in any given patient’s tumor can be extremely challenging because genomic analysis often fails to identify actionable alterations due to low prevalence rates, or uncovers multiple genomic alterations within the same tumor making it impossible to know which, if any, of the alterations found is causally significant as a targetable event. However, the addition of functional, phosphoprotein-based, drug target activation mapping could provide key missing information to the identification and selection of a molecularly rationalized therapy regimen (55). In this context, the development of quantitative, standardized, CAP/CLIA-accredited platforms suitable for measuring the activation level of drug targets and downstream substrates, including RPPA-based analysis of specific phosphoprotein levels, are becoming invaluable for capturing the molecular landscape of malignant lesions (21, 56, 57).
By using our multi-omic approach, this study successfully measured functional protein activation, measured as phosphorylation, of the AKT–mTOR signaling network along with broad-scale NGS profiling of a large cohort of metastatic breast cancers. Because alterations of this signaling pathway appear to be predominant in the hepatic lesions, these data indicate that the AKT–mTOR pathway may represent an organ-specific drug target signature for liver metastases in breast cancer. The predictive or therapeutic value of these findings should be prospectively evaluated. These findings reinforce the growing evidence that not only are the metastatic lesions molecularly different from the primary tumor, but that molecular analysis of the specific affected organ site of the metastatic lesion may also be essential in the accurate selection of targeted therapy for any given patient and a necessary component of the precision medicine ecosystem.
Supplementary Material
Translational Relevance.
Customization of treatment based on molecular profiling of a patient’s primary tumor has yielded promising results and has opened a new paradigm for treating patients with advanced disease. Nonetheless, the identification of the specific molecular drivers of any given patient’s tumor based solely on genomic alterations can be challenging without functional information. The addition of protein-based drug target activation mapping to genomic profiling of tumors may provide the functional synergy needed to more fully uncover the molecular landscape of cancer and optimal therapeutic selection. The identification of organ-specific targetable signatures based on multi-omic analysis, such as the activation of the PI3K–AKT–mTOR signaling network within liver metastases, may help both design more effective treatment for patients with metastatic breast cancer and identify patients that can be molecularly matched to a given therapy.
Acknowledgments
The authors thank the patients who participated in the Side Out 2 Trial and their families, and the Side Out Foundation who sponsored and provided the funding for the Side Out 2 Trial. Side Out Foundation and TD2 were responsible for the study design. TD2 coordinated the collection, monitoring and management of the trial. This trial is registered with ClinicalTrials.gov, number NCT01919749.
Grant Support
This work was supported by the College of Science, George Mason University, and by the Side Out Foundation who sponsored and provided the funding for the Side Out 2 Trial.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
M. Pierobon holds ownership interest (including patents) in Theranostics Health and is a consultant/advisory board member for Perthera. M. Cristofanilli reports receiving speakers bureau honoraria from Agendia and is a consultant/advisory board member for Dompe and Vortex. B. Leyland-Jones reports receiving speakers bureau honoraria from Genentech. L.A. Liotta reports receiving royalties on LCM or RPPA patents owned by NIH or George Mason University. J.A. O’Shaughnessy is a consultant/advisory board member for AbbVie, AstraZeneca, Celgene, Eli Lilly and Company, Novartis, and Pfizer. E.F. Petricoin is an employee of Perthera, holds ownership interest (including patents) in Avant Diagnostics, Ceres Nanosciences, and Perthera, and is a consultant/advisory board member for Ceres Nanosciences and Perthera. No potential conflicts of interest were disclosed by the other authors.
Authors’ Contributions
Conception and design: M. Pierobon, S.P. Anthony, L. Vocila, L.A. Liotta, J.A. O’Shaughnessy, J.D. Carpten, E.F. Petricoin
Development of methodology: M. Pierobon, G. Gambara, L.A. Liotta, J.D. Carpten, E.F. Petricoin
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): C. Ramos, S. Wong, S.P. Anthony, N.J. Robert, D.W. Northfelt, M. Jahanzeb, L. Vocila, J. Wulfkuhle, R.I. Gallagher, D.W. Craig, M. Cristofanilli, L.A. Liotta, J.A. O’Shaughnessy, J.D. Carpten
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): M. Pierobon, C. Ramos, J. Aldrich, S. Byron, S.P. Anthony, D.W. Northfelt, M. Jahanzeb, J. Wulfkuhle, R.I. Gallagher, N. Hoke, T. Dong, B. Leyland-Jones, L.A. Liotta, J.A. O’Shaughnessy, J.D. Carpten, E.F. Petricoin
Writing, review, and/or revision of the manuscript: M. Pierobon, S. Byron, S.P. Anthony, N.J. Robert, D.W. Northfelt, M. Jahanzeb, L. Vocila, J. Wulfkuhle, G. Gambara, D.W. Craig, M. Cristofanilli, B. Leyland-Jones, L.A. Liotta, J.A. O’Shaughnessy, J.D. Carpten, E.F. Petricoin
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): S. Wong, K.A. Hodge, S. Byron, L. Vocila, L.A. Liotta, J.D. Carpten, E.F. Petricoin
Study supervision: C. Ramos, M. Jahanzeb, L. Vocila, L.A. Liotta, J.D. Carpten, E.F. Petricoin
Other (foundation support): B. Dunetz
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Rivenbark AG, O’Connor SM, Coleman WB. Molecular and cellular heterogeneity in breast cancer: challenges for personalized medicine. Am J Pathol. 2013;183:1113–24. doi: 10.1016/j.ajpath.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawood S, Broglio K, Ensor J, Hortobagyi GN, Giordano SH. Survival differences among women with de novo stage IV and relapsed breast cancer. Ann Oncol. 2010;21:2169–74. doi: 10.1093/annonc/mdq220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng YC, Ueno NT. Improvement of survival and prospect of cure in patients with metastatic breast cancer. Breast Cancer. 2012;19:191–9. doi: 10.1007/s12282-011-0276-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–9. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baselga J, Campone M, Piccart M, Burris HA, Rugo HS, Sahmoud T, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366:520–9. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paplomata E, O’Regan R. The PI3K/AKT/mTOR pathway in breast cancer: targets, trials and biomarkers. Ther Adv Med Oncol. 2014;6:154–66. doi: 10.1177/1758834014530023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Generali D, Venturini S, Rognoni C, Ciani O, Pusztai L, Loi S, et al. A network meta-analysis of everolimus plus exemestane versus chemotherapy in the first- and second-line treatment of estrogen receptor-positive metastatic breast cancer. Breast Cancer Res Treat. 2015;152:95–117. doi: 10.1007/s10549-015-3453-9. [DOI] [PubMed] [Google Scholar]
- 10.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 11.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–44. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silvestri A, Calvert V, Belluco C, Lipsky M, De Maria R, Deng J, et al. Protein pathway activation mapping of colorectal metastatic progression reveals metastasis-specific network alterations. Clin Exp Metastasis. 2013;30:309–16. doi: 10.1007/s10585-012-9538-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scartozzi M, Giampieri R, Maccaroni E, Mandolesi A, Biagetti S, Alfonsi S, et al. Phosphorylated AKT and MAPK expression in primary tumours and in corresponding metastases and clinical outcome in colorectal cancer patients receiving irinotecan-cetuximab. J Transl Med. 2012;10:71. doi: 10.1186/1479-5876-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matter MS, Decaens T, Andersen JB, Thorgeirsson SS. Targeting the mTOR pathway in hepatocellular carcinoma: current state and future trends. J Hepatol. 2014;60:855–65. doi: 10.1016/j.jhep.2013.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villanueva A, Chiang DY, Newell P, Peix J, Thung S, Alsinet C, et al. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 2008;135:1972–83. doi: 10.1053/j.gastro.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sahin F, Kannangai R, Adegbola O, Wang J, Su G, Torbenson M. mTOR and P70 S6 kinase expression in primary liver neoplasms. Clin Cancer Res. 2004;10:8421–5. doi: 10.1158/1078-0432.CCR-04-0941. [DOI] [PubMed] [Google Scholar]
- 17.Ewald F, Grabinski N, Grottke A, Windhorst S, Nörz D, Carstensen L, et al. Combined targeting of AKT and mTOR using MK-2206 and RAD001 is synergistic in the treatment of cholangiocarcinoma. Int J Cancer. 2013;133:2065–76. doi: 10.1002/ijc.28214. [DOI] [PubMed] [Google Scholar]
- 18.de Gramont A, Watson S, Ellis LM, Rodón J, Tabernero J, Hamilton SR. Pragmatic issues in biomarker evaluation for targeted therapies in cancer. Nat Rev Clin Oncol. 2015;12:197–212. doi: 10.1038/nrclinonc.2014.202. [DOI] [PubMed] [Google Scholar]
- 19.Loupakis F, Pollina L, Stasi I, Ruzzo A, Scartozzi M, Santini D, et al. PTEN expression and KRAS mutations on primary tumors and metastases in the prediction of benefit from cetuximab plus irinotecan for patients with metastatic colorectal cancer. J Clin Oncol. 2009;27:2622–9. doi: 10.1200/JCO.2008.20.2796. [DOI] [PubMed] [Google Scholar]
- 20.Side Out Foundation. Clinical-Trials.gov. Bethesda, MD: National Library of Medicine (US); A pilot study utilizing proteomic and genomic profiling for patients with metastatic breast cancer (SO2) [ https://clinicaltrials.gov/ct2/show/NCT01919749?term=Side+out&rank=2] [cited 2016 Jul 20]. Available from: URL of the record NLM Identifier: NCT01919749. [Google Scholar]
- 21.Arnedos M, Vicier C, Loi S, Lefebvre C, Michiels S, Bonnefoi H, et al. Precision medicine for metastatic breast cancer—limitations and solutions. Nat Rev Clin Oncol. 2015;12:693–704. doi: 10.1038/nrclinonc.2015.123. [DOI] [PubMed] [Google Scholar]
- 22.Espina V, Edmiston KH, Heiby M, Pierobon M, Sciro M, Merritt B, et al. A portrait of tissue phosphoprotein stability in the clinical tissue procurement process. Mol Cell Proteomics. 2008;7:1998–2018. doi: 10.1074/mcp.M700596-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wulfkuhle JD, Berg D, Wolff C, Langer R, Tran K, Illi J, et al. Molecular analysis of HER2 signaling in human breast cancer by functional protein pathway activation mapping. Clin Cancer Res. 2012;18:6426–35. doi: 10.1158/1078-0432.CCR-12-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–7. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 27.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christoforides A, Carpten JD, Weiss GJ, Demeure MJ, Von Hoff DD, Craig DW. Identification of somatic mutations in cancer through Bayesian-based analysis of sequenced genome pairs. BMC Genomics. 2013;14:302. doi: 10.1186/1471-2164-14-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.1000 Genomes Project Consortium. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–72. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 31.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms. SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Espina V, Wulfkuhle JD, Calvert VS, VanMeter A, Zhou W, Coukos G, et al. Laser-capture microdissection. Nat Protoc. 2006;1:586–603. doi: 10.1038/nprot.2006.85. [DOI] [PubMed] [Google Scholar]
- 33.Baldelli E, Haura EB, Crinò L, Cress DW, Ludovini V, Schabath MB, et al. Impact of upfront cellular enrichment by laser capture microdissection on protein and phosphoprotein drug target signaling activation measurements in human lung cancer: Implications for personalized medicine. Proteomics Clin Appl. 2015;9:928–37. doi: 10.1002/prca.201400056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pin E, Federici G, Petricoin EF. Preparation and use of reverse protein microarrays. Curr Protoc Protein Sci. 2014;75(Unit 27.7) doi: 10.1002/0471140864.ps2707s75. [DOI] [PubMed] [Google Scholar]
- 35.Signore M, Reeder KA. Antibody validation by Western blotting. Methods Mol Biol. 2012;823:139–55. doi: 10.1007/978-1-60327-216-2_10. [DOI] [PubMed] [Google Scholar]
- 36.Harhaj EW, Sun SC. The serine/threonine phosphatase inhibitor calyculin A induces rapid degradation of IkappaBbeta.Requirement of both the N-and C-terminal sequences. J Biol Chem. 1997;272:5409–12. doi: 10.1074/jbc.272.9.5409. [DOI] [PubMed] [Google Scholar]
- 37.Rapkiewicz A, Espina V, Zujewski JA, Lebowitz PF, Filie A, Wulfkuhle J, et al. The needle in the haystack: application of breast fine-needle aspirate samples to quantitative protein microarray technology. Cancer. 2007;111:173–84. doi: 10.1002/cncr.22686. [DOI] [PubMed] [Google Scholar]
- 38.Pierobon M, Silvestri A, Spira A, Reeder A, Pin E, Banks S, et al. Pilot phase I/II personalized therapy trial for metastatic colorectal cancer: evaluating the feasibility of protein pathway activation mapping for stratifying patients to therapy with imatinib and panitumumab. J Proteome Res. 2014;13:2846–55. doi: 10.1021/pr401267m. [DOI] [PubMed] [Google Scholar]
- 39.Beelen K, Opdam M, Severson TM, Koornstra RH, Vincent AD, Wesseling J, et al. PIK3CA mutations, phosphatase and tensin homolog, human epidermal growth factor receptor 2, and insulin-like growth factor 1 receptor and adjuvant tamoxifen resistance in postmenopausal breast cancer patients. Breast Cancer Res. 2014;16:R13. doi: 10.1186/bcr3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov. 2014;13:140–56. doi: 10.1038/nrd4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RV, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–1000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 42.Cai J, Xu L, Tang H, Yang Q, Yi X, Fang Y, et al. The role of the PTEN/PI3K/Akt pathway on prognosis in epithelial ovarian cancer: a meta-analysis. Oncologist. 2014;19:528–35. doi: 10.1634/theoncologist.2013-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beelen K, Hoefnagel LD, Opdam M, Wesseling J, Sanders J, Vincent AD, et al. PI3K/AKT/mTOR pathway activation in primary and corresponding metastatic breast tumors after adjuvant endocrine therapy. Int J Cancer. 2014;135:1257–63. doi: 10.1002/ijc.28769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fang WL, Huang KH, Lan YT, Lin CH, Chang SC, Chen MH, et al. Mutations in PI3K/AKT pathway genes and amplifications of PIK3CA are associated with patterns of recurrence in gastric cancers. Oncotarget. 2016;7:6201–20. doi: 10.18632/oncotarget.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li HT, Lu YY, An YX, Wang X, Zhao QC. KRAS, BRAF and PIK3CA mutations in human colorectal cancer: relationship with metastatic colorectal cancer. Oncol Rep. 2011;25:1691–7. doi: 10.3892/or.2011.1217. [DOI] [PubMed] [Google Scholar]
- 46.Nakanishi K, Sakamoto M, Yamasaki S, Todo S, Hirohashi S. Akt phosphorylation is a risk factor for early disease recurrence and poor prognosis in hepatocellular carcinoma. Cancer. 2005;103:307–12. doi: 10.1002/cncr.20774. [DOI] [PubMed] [Google Scholar]
- 47.Pierobon M, Calvert V, Belluco C, Garaci E, Deng J, Lise M, et al. Multiplexed cell signaling analysis of metastatic and nonmetastatic colorectal cancer reveals COX2-EGFR signaling activation as a potential prognostic pathway biomarker. Clin Colorectal Cancer. 2009;8:110–7. doi: 10.3816/CCC.2009.n.018. [DOI] [PubMed] [Google Scholar]
- 48.Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–91. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Creighton CJ, Fu X, Hennessy BT, Casa AJ, Zhang Y, Gonzalez-Angulo AM, et al. Proteomic and transcriptomic profiling reveals a link between the PI3K pathway and lower estrogen-receptor (ER) levels and activity in ER+ breast cancer. Breast Cancer Res. 2010;12:R40. doi: 10.1186/bcr2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zuurbier L, Petricoin EF, Vuerhard MJ, Calvert V, Kooi C, Buijs-Gladdines JG, et al. The significance of PTEN and AKT aberrations in pediatric T-cell acute lymphoblastic leukemia. Haematologica. 2012;97:1405–13. doi: 10.3324/haematol.2011.059030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beelen K, Opdam M, Severson TM, Koornstra RH, Vincent AD, Wesseling J, et al. Phosphorylated p-70S6K predicts tamoxifen resistance in postmenopausal breast cancer patients randomized between adjuvant tamoxifen versus no systemic treatment. Breast Cancer Res. 2014;16:R6. doi: 10.1186/bcr3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 2011;36:320–8. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–8. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roux PP, Shahbazian D, Vu H, Holz MK, Cohen MS, Taunton J, et al. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J Biol Chem. 2007;282:14056–64. doi: 10.1074/jbc.M700906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Conrads TP, Petricoin EF. The Obama Administration’s Cancer Moonshot: a call for proteomics. Clin Cancer Res. 2016;22:4556–8. doi: 10.1158/1078-0432.CCR-16-0688. [DOI] [PubMed] [Google Scholar]
- 56.Pierobon M, Gandhi L, Cristofanilli M, Tripathy D, Petricoin EF. Protein pathway activation mapping for multi-omic based precision medicine. AJHO. 2016;12:20–24. [Google Scholar]
- 57.Jameson GS, Petricoin EF, Sachdev J, Liotta LA, Loesch DM, Anthony SP, et al. A pilot study utilizing multi-omic molecular profiling to find potential targets and select individualized treatments for patients with previously treated metastatic breast cancer. Breast Cancer Res Treat. 2014;147:579–88. doi: 10.1007/s10549-014-3117-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.