Abstract
The cause of adaptive protein evolution includes internal (for example, co-evolution of ligand-receptor pairs) and external (for example, adaptation to different ecological niches) mechanisms. Host defense peptides (HDPs) are a class of vertebrate-specific cationic antimicrobial peptides evolving under positive selection. Besides their antibiotic activity, HDPs also exert an effect on multiple host immune cells, thus providing an ideal model to study selective agents driving their evolution. On the basis of a combination of computational and experimental approaches, we studied the evolution of LL-37-type HDPs in mammals, the mature peptide of cathelicidin CAP18 (herein termed CAP18-MP) and investigated the driving force behind the evolution. Using codon-substitution maximum likelihood models, we analyzed CAP18-MPs in 40 species belonging to nine mammalian Orders and identified four positively selected sites (PSSs) that all are located on two terminal unordered regions of CAP18-MPs. Grafting the two positively selected regions of human or whale CAP18-MP to the α-helical scaffold of a rabbit homolog (substituting its corresponding parts) led to no alterations in antibacterial activity, spectrum and action mode. Likewise, further deletion of the two terminal regions did not alter its functional features. Evolutionary conservation analysis of mammalian FPR2, a receptor known to interact with the C-terminal positively selected region of LL-37, revealed high evolutionary variability in its ligand-binding extracellular loop domains, matching sequence diversity of the unordered regions in CAP18-MPs. This is the first report describing that the signature of positive selection of cathelicidins is not associated with their direct bactericidal activity, but rather with the evolutionary variability of their endogenous receptors.
Introduction
Positive (Darwinian) selection promotes the fixation and spread of advantageous mutations in proteins throughout a population. Such selection can be detected by comparing the ratio of non-synonymous (dN) to synonymous (dS) substitutions in protein-coding loci. If the altered amino acids are selectively advantageous (that is, under positive selection), they will be fixed at a higher rate and the dN/dS-ratio (ω)) will be larger than one (Hill and Hastie, 1987; Hughes and Nei, 1988). On the basis of this criterion, a large number of protein-coding genes involved in diverse biological processes (for example, immunity, defense, reproduction and digestion and so on) have been recognized to evolve under positive selection (Yang, 2006). Among them, immune-related genes have been a main research focus because of their key roles in fighting against evolving pathogens. A paradigm in this aspect is the major histocompatibility complex (MHC), a set of cell surface proteins essential for binding pathogen-derived oligopeptides (antigens) and helping the adaptive immune system to recognize these foreign molecules in vertebrates (Hughes and Nei, 1988; Potts and Wakeland, 1990). During the host-pathogen co-evolutionary history, the antigen binding region of the MHC has undergone strong positive selection in response to the evolving pathogen-peptide diversity (Ejsmond and Radwan, 2015).
In addition to MHC, a series of multifunctional host defense peptides (HDPs) constitutes another model to study the evolution of the immune system. HDPs belong to a class of distinct antimicrobial peptides (AMPs) that are extensively distributed in vertebrates (Mansour et al., 2014). In comparison with non-vertebrate AMPs that primarily function as peptide antibiotics, HDPs have evolved immune modulatory functions via interacting with related receptors to boost the adaptive immune response (Mansour et al., 2014). In mammals, HDPs are generally grouped into two major families: β-structured defensins with three disulfide bonds, including α-, β- and θ-subfamilies (Schneider et al., 2005); and cathelicidins with highly variable folding types, ranging from linear α-helical peptides to disulfide bond-containing β-hairpins, and single residue-rich unstructured peptides (Zhu, 2008a).
Early efforts in identifying positive selection signatures of HDPs mainly concentrated on α- and β-defensins. In 2003, Morrison et al. (2003) firstly found evidence for signal sequence conservation but mature peptide divergence within subgroups of the murine β-defensin gene family. Subsequently, positive selection following gene duplication or speciation was widely identified in β-defensins from primates (Boniotto et al., 2003; Semple et al., 2003, 2005, 2006a; Radhakrishnan et al., 2005), bovines (Luenser and Ludwig, 2005), rodents (Semple et al., 2006b); caprine and ovine (Luenser et al., 2005) and chickens (Lynn et al., 2004; Higgs et al., 2007). Evidence for positive selection in mammalian α-defensins was also reported in 2004 (Lynn et al., 2004; Patil et al., 2004). Positive selection in cathelicidins was found in primate CAP18-MPs (see below; Zelezetsky et al., 2006) and the pro-region cathelin-like domain (CLD) in mammals (Zhu, 2008b)
In spite of the remarkable progress in recognizing signatures of positive selection during HDP evolution, a full understanding of their adaptive evolution cannot be achieved without considering the selective agents (the cause of evolution) responsible for such selection. External (for example, adaptation to different ecological niches) and internal (for example, co-evolution between receptor and ligand pairs) mechanisms have been considered as two major forces driving the evolution of proteins (MacColl, 2011; Morgan et al., 2012). To investigate the driving force in the evolution of cathelicidins, we analyzed the mature peptide of CAP18 (herein termed CAP18-MP). CAP18 (the abbreviation of cationic antimicrobial protein of 18 kD), originally isolated from rabbit granulocytes, belongs to a subfamily of peptides whose members have a mature C terminus of ~37 amino acids obtained by proteolytic cleavage (Larrick et al., 1993; Hirata et al., 1994). As the prototype of this subfamily, human LL-37 is among the most thoroughly studied CAP18-MP-type HDPs. It is an amphipathic α-helical peptide of 37 amino acids derived from the C-terminal end of the human CAP18 (Wang, 2008; Wang et al., 2014). As a multifunctional HDP of the innate immunity, LL-37 not only exhibits antibiotic activity via damaging bacterial membrane and/or DNA components but also immune-modulating properties that trigger intracellular signaling events via interacting with a diversity of receptors in host immune cells, such as membrane receptors—formyl peptide receptor 2 (FPR2), a member of the FPR family of G-protein-coupled receptors involved in host defense and inflammation, and P2X7, a member of the P2X family of ATP-gated cation channels (Xhindoli et al., 2016); and the intracellular receptor glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Mookherjee et al., 2009).
Apart from LL-37 in humans and the firstly characterized homolog from rabbits (called CAP18106–142; Larrick et al., 1993), CAP18-MPs have also been found and functionally characterized in other several mammalian species, such as rhesus monkey (RL-37), mouse (mCRAMP), rat (rCRAMP) and domestic cat (feCath; Travis et al., 2000; Leonard et al., 2011; Xhindoli et al., 2014). Although present as a single copy in mammalian genomes, functional diversification related to immune modulation has occurred among orthologues of CAP18-MPs. For example, human LL-37 potentiates the P2X7 activation (Elssner et al., 2004), but its ortholog in mice (mCRAMP) impairs murine P2X7 activation (Seil et al., 2010). In addition, LL-37 and mCRAMP both are agonists for FPR2, however, it is the former other than the latter can stimulate signaling by RNA through a FPR2-dependent pathway (Singh et al., 2013).
The CAP18-MP-type HDPs are a suitable model for resolving the relative importance of exogenous pathogens and endogenous receptors in driving HDP evolution because they function through different subdomains that target bacteria and receptors on host immune cells, respectively (Figure 1). For example, previous mutational assays have shown that the N and C termini of LL-37 are dispensable for its antibacterial activity but indispensable for its immunomodulatory activity (Gupta et al., 2015), suggesting a distinctive location of functional regions associated with these two immune properties. Its middle helical region is essentially responsible for antibacterial activity and the unordered C termini were found to interact with the receptor FPR2. Previously, an evolutionary analysis of the CAP18-MP-coding region implicated that 71% of codons were being subject to positive selection (Zelezetsky et al., 2006). However, this analysis was only limited to 21 highly similar primate sequences that might have skewed its implication. In this work, we have examined the evolution of the genes at the human cathelicidin locus and the orthologous loci in other mammals. The objectives were (i) to find positively selected sites (PSSs) in mammalian CAP18-MPs, (ii) to identify their evolutionary significance, and (iii) to test the driving forces of HDP evolution imposed by exogenous pathogens and endogenous receptors on the host immune cells, respectively (Figure 1). Our work highlights new PSSs occurring in mammals and suggests for the first time that signatures of positive selection in CAP18-MP is not associated with their direct bactericidal function. Instead, the evolutionary diversity of the receptors may be a major driving force for their evolution.
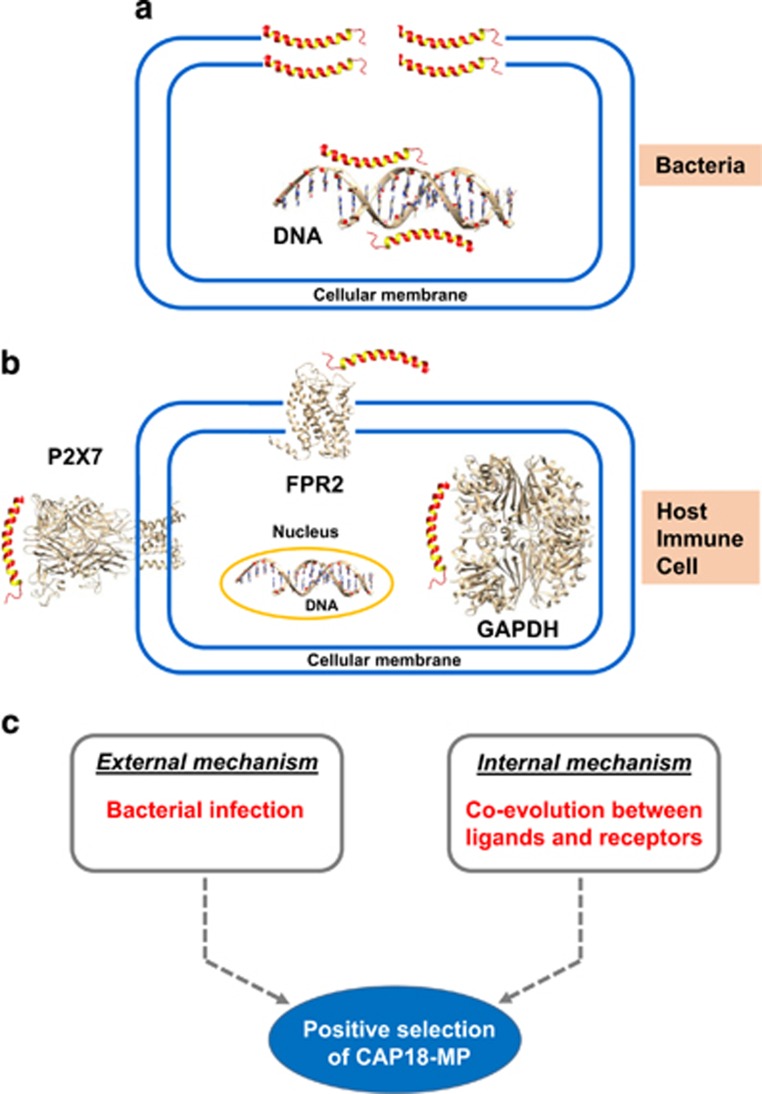
Figure 1.
The target’s diversity of LL-37 and proposed driving forces. (a) Membrane disruption and DNA binding mediating bacterial killing by LL-37; (b) Immune regulatory function of LL-37 on host immune cells via interacting with membrane receptors P2X7 and FPR2 and the intracellular receptor (glyceraldehydes 3-phosphate dehydrogenase, GAPDH) and DNA; (c) External or internal driving force for positive selection of CAP18-MP in mammals. PDB entries of the structures: 2K6O (LL-37); 2LNL (human CXCR1, the template for modeling human FPR2 structure); 4DW0 (zebrafish P2X4, the template for modeling human P2X7 structure); 1ZNQ (human GAPDH).
Materials and methods
Gene discovery, sequence alignment and phylogenetic tree construction
The amino acid sequence of human CAP18 (hCAP18) comprising a signal peptide, a CLD and the C-terminal mature LL-37 (Zhu, 2008b), was used as query to search for the whole-genome shotgun contigs (wgs) database by TBLASTN (http://blast.ncbi.nlm.nih.gov/Blast.cgi; Feb 3, 2015). Sequence alignment was carried out by ClustalX program (http://www.clustal.org). Phylogenetic trees were constructed on the basis of amino acid sequences by the neighbor-joining (NJ) algorithm, a distance-based method implemented in MEGA6 (http://www.megasoftware.net), with the Poisson substitution model and Gamma Distributed rates among sites. Gaps are treated by pairwise deletion. The multiple sequence alignment of CAP18 are provided in Supplementary Figure S1 as Supplementary Information.
Structural analysis
3D models reported here were built by comparative modeling (Ginalski, 2006). Except CAP18-MP(FV)(2-31), all the others were obtained by using human LL-37 (pdb entry 2K6O) as template at the Swiss-Model server (http://www.expasy.ch/swissmod/SWISS-MODEL.html). For modeling the structure of human FPR2, we chose human chemokine receptor CXCR1 (pdb entry 2LNL), a G-protein-coupled receptor (GPCR; Park et al., 2012), as template, which had a detectable sequence similarity to the target sequence. For all the models, initial backbone fitting and energy minimization steps were performed with the DeepView program (http://spdbv.vital-it.ch/) and further refined via submission to the Swiss-Model server. The model of CAP18-MP(FV; 2-31) was built based on the solution structure of the active domain of rabbit CAP18106-137 (pdb entry 1LYP) at the @TOME 2 server (http://atome.cbs.cnrs.fr/AT2B/meta.html).
Positive selection analysis
Positive selection is recognized when the non-synonymous to synonymous substitution rate ratio (ω=dN/dS) is more than 1. To estimate ω values among sites of mammalian CAP18-MP, we employed the CODEML program of the PAML software package (http://abacus.gene.ucl.ac.uk/software/paml.html) to analyze the data. Two pairs of codon-based likelihood models (M1a/M2a and M7/M8) were used to make two likelihood ratio tests (LRTs), in which the null models (M1a and M7) do not allow sites with ω>1 whereas their alternative models (M2a and M8) assume the existence of such sites (ω>1). Upon detection of positively selected signals, the calculation of posterior probabilities of ω classes for each site was completed using the Naive Empirical Bayes (NEB) and the Bays Empirical Bayes (BEB) analyses. Sites with a high probability (⩾99%) of coming from the class with ω>1 are likely to be under positive selection and were selected for experimental design.
Evolutionary conservation analysis
ConSurf, a bioinformatics tool for estimating the evolutionary conservation of amino positions in a protein molecule based on the phylogenetic relations among homologous sequences (Glaser et al., 2003), was used to analyze the FPR2 family. By using human FPR2 as query, we retrieved a total of 32 orthologs in mammals, all present as a sing copy (Supplementary Figure S2). A multiple sequence alignment and human FPR2 structure were used as input files for ConSurf analysis (http://consurf.tau.ac.il). The method used here was maximum likelihood (ML) with the JTT substitution model of amino acids.
Chemical synthesis and circular dichroism analysis
Peptides used here all were chemically synthesized by ChinaPeptides Co., Ltd. (Shanghai). Circular dichroism (CD) spectra of peptides were recorded on a Chirascan Plus spectropolarimeter (UK) at a protein concentration of 0.1 mg ml−1 dissolved in different solvents, including water, 50% trifluoroethanol (TFE) or phosphate-buffered saline (PBS, 135 mm NaCl, 4.7 mm KCl, 10 mm Na2HPO4, 2 mm NaH2PO4, pH 7.4). Spectra were measured at 20 °C from 260 to 185 nm with a quartz cell of 1.0 mm thickness. Data were collected at 1nm intervals with a scan rate of 60 nm/min. Data are expressed as mean residue molar ellipticity ([θ]), calculated as follows: [θ]=θ × (0.1 × MRW)/(L × C), where θ is the ellipticity (in millidegrees), C is the concentration (in mg ml−1), L is the path-length (in cm), and MRW is the mean residue weight (in Da).
Antibacterial assays
Antibacterial activity of peptides was assayed by the inhibition zone method (Hultmark, 1998). In brief, bacteria were incubated at 37 °C in LB medium (10 g tryptone, 5 g yeast extract, and 9 g NaCl in 1 litre of water) until the OD600 reached 0.5. 10 μl of bacterial culture were mixed in 6 ml of LB medium containing 0.8% agar and poured into Peri dishes of 9.0 cm diameter. Wells with a diameter of 2 mm were punched into the medium, filled with 2 μl of sample each well. For each peptide, three to five different doses by 2-fold dilution were used. After bacteria were incubated at 37 °C overnight, inhibition zones were measured and used to calculate lethal concentration (CL), a concentration just sufficient to inhibit bacterial growth (Hultmark, 1998; Gao and Zhu, 2010). CL values are calculated from a plot of d2 against log n, where d is the diameter (in cm) and n is the amount of sample applied in the well (in nmol). The plot is linear and thus CL can be calculated from the slope (k) and the intercept (m) of this plot. The formula used here is CL=2.93/ak10m/k, where a is the thickness of the bacterial plate and CL is in mm. This is a quantitative description of the qualitative inhibition zone assay since the days of Alexander Fleming and in most cases the calculated CL are comparable to the directly measured minimal inhibitory concentrations (MIC) via broth micro-dilution assay (Hultmark, 1998). Sources of bacteria used in this study are listed in Supplementary Table S3.
Membrane permeability assay
To assess the permeation ability of peptides on bacterial membrane, 5 × 105 B. megaterium or Micrococcus luteus cells in 500 μl of PBS were mixed with 1 μM propidium iodide (PI) for 5 min in the dark. After peptide was added, the increase in fluorescence was measured using an F-4500 FL spectrophotometer (Hitachi High- Technology Company). Once the basal fluorescence reached a constant value, peptides were added. Changes in fluorescence arbitrary were monitored (λexc=525 nm; λems=595 nm) and plotted as arbitrary units. Meucin-18 and vancomycin were used positive and negative controls, respectively (Zhu et al., 2012).
Scanning electron microscope
Salmonella typhimurium ATCC 14028 cells at exponential growth phase were treated with peptides at 10 × CL at 37 °C for 90 min. After centrifugation, bacterial pellets were fixed with 2.5% glutaraldehyde for 1 h, followed by washing three times with PBS. Dehydration was carried out with a series of graded ethanol solution. Cells were then dried using BAL-TEC CPD030 critical point dryer before being mounted on carbon tape, sputtered with platinum coating (BAL-TEC SCD005). Images were visualized in FEI QUANTA 200. Images were visualized in a Tecnai Spirit 120 KV microscope.
Hemolysis and serum stability
Hemolytic activity of peptides against fresh mouse erythrocytes was assayed according to the standard method (Gao et al., 2009). Melittin, a classical cytolytic peptide from bee venom (Raghuraman and Chattopadhyay, 2007), was used as control. To assess serum stability, peptides were incubated in water and fresh mouse serum for 0, 4, 24 and 44 h at 37 °C. Residual activities were measured by the inhibition zone assay with M. luteus.
Results
Sequence and structural features of CAP18-MPs
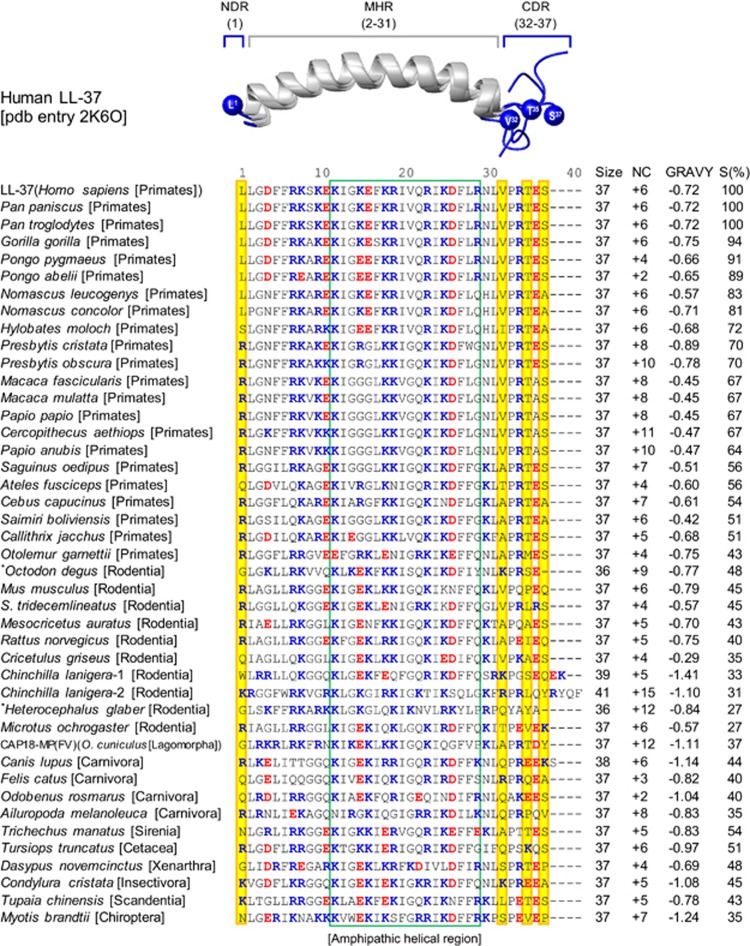
Our database search retrieved a total of 43 homologs of human CAP18, which are all distributed in mammalian species, including nine Orders (Primates, Rodentia, Lagomorpha, Carnivora, Sirenia, Insectivora, Chiroptera, Cingulata, and Scandentia). Their precursor organization comprises three parts, including an N-terminal signal peptide, an evolutionarily conserved cathelin-like domain (CLD) followed by a C-terminal CAP18-MP (Supplementary Figure S1). Except Chinchilla lanigera (Rodentia) that contains two duplicated CAP18 genes, others consist of a single copy. A phylogenetic tree based on the amino acid sequences of CAP18-MP is presented in Figure 2, according to which most members are clustered within their own Orders (Figure 2). Multiple sequence alignment revealed that these CAP18-MPs share 27–94% sequence similarity to LL-37 (Figure 3). They are composed of 36–41 amino acids with net positive charges of 2–12. All members, including those with low sequence similarity to LL-37, contain a conserved amphipathic architecture spanning residues 12 to 28 (Figure 3), as identified by their helical wheel projections (Figure 4). Grand average of hydropathicity (GRAVY) values of all these peptides were negative (ranging from −0.29 to −1.41; Figure 3), indicating their overall hydrophilicity (Kyte and Doolittle, 1982). Comparative modeling suggests that all these peptides have capacity to form a long α-helical segment, ranging from residues 2–31, with their N and C termini in an unordered conformation (data not shown).
Figure 2.
A Neighbor-Joining tree of the CAP18-MP subfamily from mammals. Branches in different colors represent different Orders of mammals (names on the left of the tree). The bootstrap values (50%) based on 1000 replications are indicated at each node. The scale bar represents amino acid substitutions of 0.10 per site.
Figure 3.
Multiple sequence alignment of the CAP18-MP family from mammals. Cationic and anionic amino acids are shown in blue and red, respectively, and sites identified to be under positive selection are shadowed in yellow. The NMR structure of human LL-37 (Wang, 2008) is used to map PSSs and define secondary structure element regions, which comprises three distinct parts: NDR (amino-terminal disordered region, residue 1), MHR (middle α-helical region, residues 2–31) and CDR (carboxyl-terminal disordered region, residues 32–37), in which regions forming a conserved amphipathic architecture is boxed in green. NC, net charge; GRAVY, grand average of hydropathicity, which was calculated by ProtParam as the sum of hydropathy values of all the amino acids, divided by the number of residues in the sequence (http://web.expasy.org/protparam/); S(%), sequence similarity percentage.
Figure 4.
Helical wheel projections of LL-37 and its orthologs. These images were generated at the server http://rzlab.ucr.edu/scripts/wheel/wheel.cgi, in which hydrophilic, hydrophobic, negatively charged, and positively charged residues are presented as circles, diamonds, triangles, and pentagons, respectively. The most hydrophobic residue is shown in green, and the amount of green is decreasing proportionally to the hydrophobicity. The most hydrophilic (uncharged) residues are coded red, and the amount of red is decreasing proportionally to the hydrophilicity. Charged residues are shown in light blue. These images illustrate a typical amphipathic architecture in residues 12–29 of the representative CAP18-MPs.
Positive selection in the unordered region of CAP18-MPs
Codon-based likelihood models that allow selection to vary across sites are a powerful statistical tool for detecting amino acid sites under positive selection. To identify positively selected sites (PSSs) in mammalian CAP18-MP-coding region, we employed two classical pairs of models (M1a and M2a; M7 and M8; Yang, 2006) to construct likelihood ratio tests (LRTs). The ML estimates (MLEs) under M0 predicts that all amino acid sites have a ω of 1.09, indicative of neutral selection. However, M0 fits the data worse than all other models (log likelihood values: −2119.85 vs −2105.98 to −2080.36; Table 1), suggesting the presence of heterogeneous selective pressure among sites. The MLEs under M2a and M8 both identified identical proportion (13%) of PSSs with ω values of 3.80–3.98 (Table 1). The LRT statistics are 50.84 for the comparison between M1a and M2a, and 49.42 for M7 and M8, both much greater than critical values from a χ2 distribution with degree of freedom=2 (P<0.001), indicating statistical significance for the presence of PSSs. M2a and M8 both identified four PSSs with P (posterior probabilities) ⩾0.99 under the Bayes Empirical Bayes (BEB) and Naive Empirical Bayes (NEB) methods, which were L1, V32, T35 and S37(numbered according to LL-37; Figure 3), showing some differences from the previous study based on a small data set from the primate lineage (Zelezetsky et al., 2006), with only two identical PSSs (L1 and V32). Mapping of the PSSs onto the LL-37 structure in sodium dodecyl sulfate micelles (Wang, 2008) revealed that they all are located on two unstructured terminal regions: L1 at the N terminus (named NDR), and V32, T35, and S37 at the C terminus (named CDR), without a PSS on the middle α-helical region (MHR; Figure 3). For the structure of LL-37 in dodecylphosphocholine micelles (Porcelli et al., 2008), the result is the same (data not show). When LL-37 was in complex with other micelles, such as lipopolysaccharides (LPS) and dioctanoylphosphatidylglycerol (D8PG), these regions are still disordered and mobile (Wang, 2008), in favor of structural location of the PSSs. The presence of PSSs in the mature peptide domain of mammalian cathelicidins is contrary to avian homologs that overall lack PSSs in the domain and their PSSs are mostly located within the signal peptide and the CLD (Cheng et al., 2015).
Table 1. Maximum likelihood estimates of parameters and sites inferred to be under positive selection in the CAP18/LL-37 family.
| Model | S | p | l | κ | Estimates of parameters | PSSs |
|---|---|---|---|---|---|---|
| M0 (one-ratio) | 16.74 | 1 | −2119.85 | 2.86 | ω=1.09 | None |
| M1a (Nearly Neutral) | 17.37 | 2 | −2105.98 | 2.70 | p0=0.27 (p1=0.73) | Not allowed |
| M2a (Positive Selection) | 18.54 | 4 | −2080.56 | 2.91 | ω0=0.35 (ω1=1) p0=0.07 (p1=0.80) | 1L**, 4D*, 32V**, 35T**, 37S** |
| p2=0.13 | ||||||
| ω0=0.14 (ω1=1) | ||||||
| ω2=3.98 | ||||||
| M7 (beta) | 17.18 | 2 | −12105.07 | 2.67 | p=0.79, q=0.23 | Not allowed |
| M8 (beta&ω>1) | 18.42 | 4 | −2080.36 | 2.89 | p=0.56, q=0.09 | 1L**, 4D*, 32V**, 35T**, 37S** |
| p0=0.87 (p1=0.13) | ||||||
| ωs=3.80 |
Abbreviations: κ, transition/transversion rate ratio; l, log likelihood; p, number of parameters in the ω distribution; S represents tree length.
Twice the log likelihood difference (2Δl) between null models (M1a and M7) and their alternative models (M2a and M8): M1a/M2a=50.84 (χ2 significant value: P<0.001); M7/M8=49.42 (P<0.001). PSSs identified by the Bayes Empirical Bayes (BEB) and Naive Empirical Bayes (NEB) methods under M2a and M8 with P (posterior probabilities) ⩾0.99 are indicated by **. Residues are numbered according to human LL-37. ω values in M2a and M8 as indicators of positive selection are boldfaced.
Molecular design of CAP18-MP(FV) mutants
Given that accumulated evidence of coming from the structure–activity relationship studies has indicated that the α-helix of LL-37 is its main antibacterial element (Wang, 2008), and the truncation of five amino acids from the C terminus of rabbit homolog (Larrick et al., 1993) did not impair its antibacterial activity, the discovery of the PSSs location onto the unstructured regions is unexpected. Because the nature of PSSs has implicated their functional significance in many protein families evolving under positive selection, and some constitute hot-spots of protein-protein interaction (Zhu et al., 2016), the absence of PSSs in the antibacterial region of CAP18-MPs raises a possibility that this class of HDPs might be not driven by mammalian pathogens. If confirmed, this might challenge the prevailing opinion that the co-evolution between hosts and pathogens serves as a force driving the evolution of HDPs.
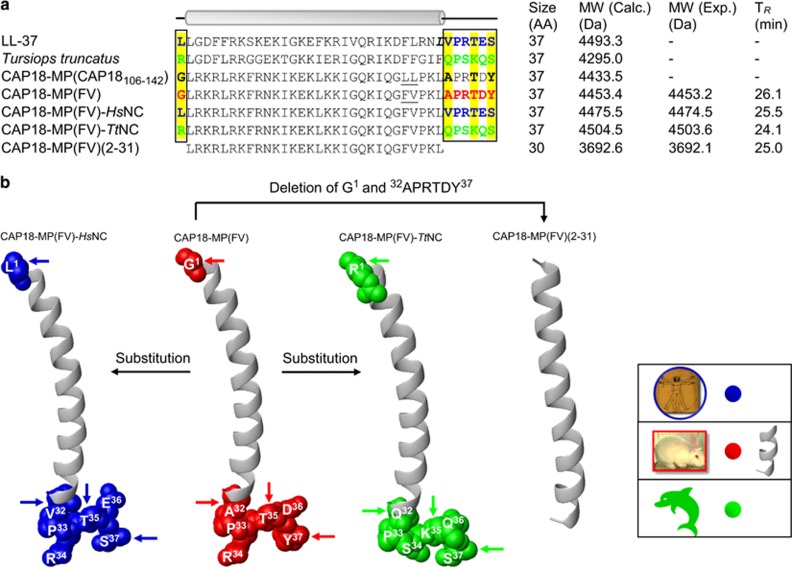
To test the above hypothesis, we used a chimeric approach to evaluate the antibacterial significance of PSSs in CAP18-MPs. This method has been successfully used in identification of a PSS-rich 13-aa ‘patch’ as retroviral restriction element of TRIM5α in primates (Sawyer et al., 2005). CAP18-MP(FV), a polymorphic variant of rabbit CAP18-MP (CAP18106-142; Travis et al., 2000) with two naturally mutated sites at 27 and 28 (from Leu-Leu to Phe-Val; GenBank No. NM_001082305; Figure 5a), was chosen as a scaffold to graft the PSS-containing unordered regions of whale (Tursiops truncatus) or human (Homo sapiens) homolog, including sites 1 and 32 to 37 (Figure 5a). Engineered chimeric peptides were termed CAP18-MP(FV)-TtNC and CAP18-MP(FV)-HsNC (Tt and Hs, the first letters of genus and species names; and NC, N and C termini). Meantime, these two regions in CAP18-MP(FV; G1 and 32APRTDY37) were also deleted and the mutant was named CAP18-MP(FV)(2-31; Figure 5a). Structural models of these peptides constructed by comparative modeling from templates of experimental structures (see Materials and Methods section) are shown in Figure 5b.
Figure 5.
Molecular design of chimeric and truncated CAP18-MP. (a) Sequence alignment of CAP18-MP and related peptides. Regions under positive selection are boxed and shown in different colors, blue for human LL-37, green for the whale T. truncatus LL-37 and red for rabbit CAP18-MP(FV). PSSs are shadowed in yellow. Two chimeric mutants possess a MHR of CAP18-MP(FV) but non-identical NDR and CDR, one from human and another from whale. The truncated peptide CAP18- MP(FV)(2-31) was designed to remove the NDR and CDR. Experimental molecular weights determined by MALDI-TOF and retention times at the C18 column of CAP18(FV) and its mutants are shown here; (b). Structural models illustrating the design strategy where substitution and deletion were involved. Color codes are the same to a. Arrows indicate residues under positive selection.
Structural features of CAP18(FV) and its mutants
Synthetic CAP18-MP(FV), CAP18-MP(FV)-HsNC, CAP18-MP(FV)-TtNC and CAP18-MP(FV)(2-31) had experimental molecular weights (MWs) of 4453.2 Da, 4474.5 Da, 4503.6 Da and 3692.1 Da, perfectly matching their calculated MWs of 4453.4 Da, 4475.5 Da, 4504.5 Da and 3692.6 Da, respectively (Figure 5). Circular dichroism (CD) spectral analysis indicated that CAP18-MP(FV) and its mutants behave similarly in water and saline buffer (PBS). In these two solvents, they remained a random coil structure, as identified by very low ellipticity above 210 nm and negative bands near 198 nm. However, in 50% trifluoroethanol (TFE), a solvent that mimics a membrane environment, these four peptides had negative bands at 222 and 208 nm, and a positive band at 193 nm. These characteristic spectral signatures indicated that they adopted a predominant α-helical structure (Figure 6). In addition, we found that the [θ]222/[θ]208 ratios were <1 for all the peptides in 50% TFE, suggesting a monomeric helical form, different from LL-37 that exists as an aggregative helical state in both saline buffer and in the presence of membrane components (Morgera et al., 2009). Structural features of these peptides elucidated from their CD data are highly similar to those of the murine CAP18-MP (mCRAMP; Gallo et al., 1997; Yu et al., 2002).
Figure 6.
Circular dichroism spectra of CAP18-MP(FV) and its mutants. Three solvents were used: H2O, 50% TFE and PBS, and peptide concentrations used were 0.1 mg ml−1. The spectra in the far UV were obtained as described in Methods. The α-helical concentration of peptides was calculated according to the CDSSTR method implemented in the DICHROWEB online server for protein secondary structure analyses from CD data (http://dichroweb.cryst.bbk.ac.uk).
Functional features of CAP18(FV) and its mutants
To compare whether there is activity differential among CAP18-MP(FV) and its mutants, we used a wide range of bacterial strains to assay their sensitivity to the peptides, including both Gram-positive and Gram-negative bacteria (Supplementary Table S3). The LB agar-plates used here contain a physiologically-relevant salt component (154 mm NaCl). As shown in Table 2, deletion or substitution of the positively selected regions led to no significant alterations in activity and antibacterial spectrum. Similar to CAP18-MP(FV), the mutants exhibited activity on an array of Gram-positive bacteria, including Micrococcus luteus, Bacillus (B. megaterium and B. subtilis, rather than B. cereus); Streptomyces (S. griseus and S. scabiei); Streptococcus (S. mutans, S. salivarius and S. sanguinis). For Staphylococcus, these four peptides displayed activity dependent on different species or isolates. For example, they were active on methicillin-resistant coagulase negative Staphylococci (MRCNS), penicillin-resistant S. epidermidis (PRSE), S. warneri and a series of clinic isolates of S. aureus, but no activity on other isolates, such as S. aureus strains (CGMCC 1.89, J685, J698, J700, J706, J708, and J710), penicillin-sensitive Staphylococcus epidermidis (PSSE) P1111 and penicillin-resistant Staphylococcus aureus (MRSA) P1386 (Table 2). Relative to Gram-positive bacteria, CAP18-MP(FV) and its mutants exhibited a wider spectrum of activity towards a variety of Gram-negative bacteria, including Alcaligenes faecalis, Xanthomonas oryzae pv. oryzae, Stenotrophomonas maltophilia, Pseudomonas solanacearum, and multiple isolates from Escherichia coli, Pseudomonas aeruginosa and Salmonella. In this assay, the only one Gram-negative bacterium resistant to all these peptides was Serratia marcescens ATCC 14041 (Table 3).
Table 2. Lethal concentrations (C L , μm) of CAP18(FV) and its mutants against Gram-positive bacteria.
| CAP18(FV) | CAP18(FV)(2-31) | CAP18(FV)-HsNC | CAP18(FV)-TtNC | |
|---|---|---|---|---|
| BC CGMCC 1.1846 | NA | NA | NA | NA |
| BM CGMCC 1.0459 | 1.40 | 1.46 | 1.64 | 1.92 |
| BS CGMCC 1.2428 | 1.10 | 1.45 | 1.80 | 2.36 |
| ML CGMCC 1.0290 | 0.94 | 0.90 | 1.18 | 1.50 |
| SA CGMCC 1.89 | NA | NA | NA | NA |
| SA (J685,J698,J700 J706,J708,J710) | NA | NA | NA | NA |
| PSSE P1111 | NA | NA | NA | NA |
| MRCNS P1369 | 1.57 | 1.44 | 1.48 | 2.09 |
| MRSA P1374 | 1.20 | 1.46 | 2.08 | 2.13 |
| MRSA P1381 | 1.73 | 1.46 | 1.85 | 2.17 |
| PRSA P1383 | 1.19 | 1.35 | 1.74 | 2.29 |
| PRSE P1386 | NA | NA | NA | NA |
| PRSE P1389 | 1.00 | 1.15 | 1.30 | 1.87 |
| SW ATCC 1.2824 | 1.38 | 0.78 | 1.00 | 2.17 |
| SG NBRC 13350 | 6.36 | 8.84 | 5.68 | ND |
| SSCA CGMCC 4.1765 | 1.84 | 3.10 | 8.93 | 8.66 |
| SM ATCC 1.2499 | 1.67 | 0.73 | 2.02 | 4.97 |
| SSAL ATCC 1.2498 | 2.25 | 2.66 | 2.79 | 4.13 |
| SSAN ATCC 1.2497 | 1.16 | 1.75 | 2.81 | 3.83 |
Abbreviations: BC, Bacillus cereus; BM, Bacillus megaterium; BS, Bacillus subtilis; ML, Micrococcus luteus; MRCNS, Methicillin-resistant coagulase negative Staphylococci; MRSA, Methicillin-resistant Staphylococcus aureus; PRSA, Penicillin-resistant Staphylococcus aureus; PRSE, Penicillin-resistant Staphylococcus epidermidis; PSSE, Penicillin- sensitive Staphylococcus epidermidis; SA, Staphylococcus aureus; SG, Streptomyces griseus; SM, Streptococcus mutans; SSCA, Streptomyces scabiei; SSAL, Streptococcus salivarius; SSAN, Streptococcus sanguinis; SW, Staphylococcus warneri. NA means ‘no activity’, as identified by no inhibition zone observed at 0.8 nmol peptide each well. ND means ‘activity not determined’.
Table 3. Lethal concentrations (C L , μM) of CAP18(FV) and its mutants against Gram-negative bacteria.
| CAP18(FV) | CAP18(FV)(2-31) | CAP18(FV)-HsNC | CAP18(FV)-TtNC | |
|---|---|---|---|---|
| AF CGMCC 1.1837 | 1.80 | 1.92 | 2.31 | 2.65 |
| EC ATCC 25922 | 1.46 | 1.63 | 2.07 | 2.75 |
| EC DH5α | 1.60 | 1.46 | 2.21 | 2.51 |
| EC JM109 | 1.52 | 1.50 | 2.11 | 3.00 |
| EC TOP10 | 1.62 | 1.33 | 1.93 | 2.85 |
| EC Am. J16c | 1.67 | 1.01 | 1.84 | 2.89 |
| EC Am. J23a | 1.44 | 1.33 | 1.95 | 2.89 |
| EC CIP. J14b | 2.19 | 1.77 | 2.67 | 2.17 |
| EC D. G2b | 2.19 | 1.33 | 3.49 | 3.85 |
| EC D. J45b | 2.19 | 1.41 | 2.67 | 5.23 |
| SMAR ATCC 14041 | NA | NA | NA | NA |
| PA O1 | 1.40 | 2.10 | 1.95 | 3.00 |
| PA 14 | 2.04 | 2.67 | 3.22 | 3.75 |
| PA FRD1 | 3.03 | 3.33 | 2.96 | 4.46 |
| PA 374 | 1.79 | 4.42 | 4.77 | 3.89 |
| PA 11082603 | 2.19 | 2.66 | 2.89 | 5.77 |
| PA 11082616 | 2.91 | 3.54 | 1.47 | 7.20 |
| PA 11092304 | 3.13 | 3.54 | 2.67 | 5.23 |
| PA 11092618 | 2.91 | 2.66 | 1.84 | 4.34 |
| PA DH | 2.19 | 1.77 | 2.89 | 4.34 |
| PA QT1 | 2.91 | 2.66 | 2.89 | 4.34 |
| PS | 1.46 | 2.19 | 1.78 | 2.31 |
| SE ATCC 14028 | 1.21 | 1.47 | 2.65 | 1.08 |
| SM DZ. 3G | 1.94 | 2.92 | 3.70 | 2.17 |
| SM WFH. 9N | 1.67 | 1.41 | 1.95 | 2.05 |
| SM WFM. 5N | 1.67 | 3.54 | 2.67 | 2.17 |
| SM ZQ. 17N | 1.67 | 3.54 | 1.95 | 2.89 |
| SM ZQ. 18X | 1.67 | 3.54 | 2.89 | 2.89 |
| SM ZQ. 21X | 1.94 | 2.36 | 2.89 | 2.89 |
| SMAL CGMCC 1.1788 | 3.93 | 2.99 | 3.21 | 10.67 |
| Xanthomonas oryzae pv. oryzae | 5.67 | 4.86 | 2.98 | 10.19 |
Abbreviations: AF, Alcaligenes faecalis; EC, Escherichia coli; PA, Pseudomonas aeruginosa; PS, Pseudomonas solanacearum; SE, Salmonella enterica; SM, Salmonella; SMAL, Stenotrophomonas maltophilia; SMAR, Serratia marcescens. NA means 'no activity', as identified by no inhibition zone observed at 0.8 nmol peptide each well.
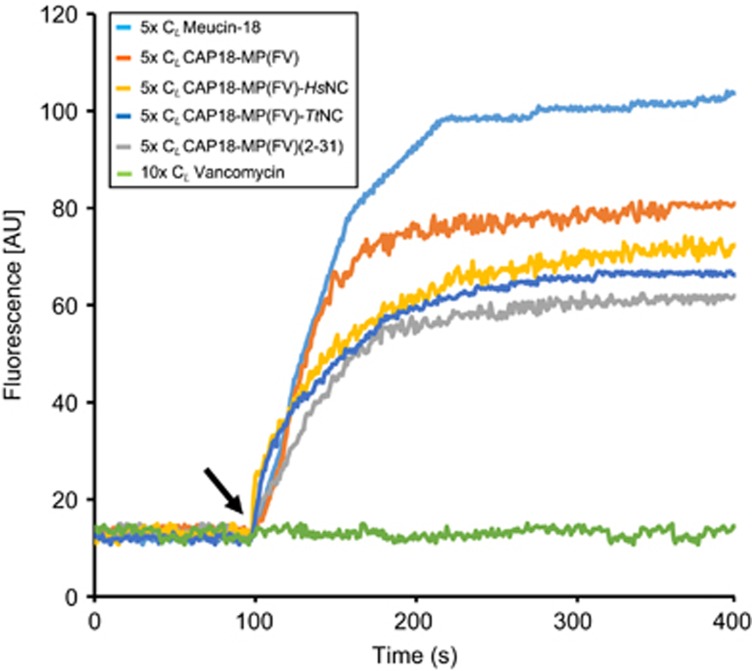
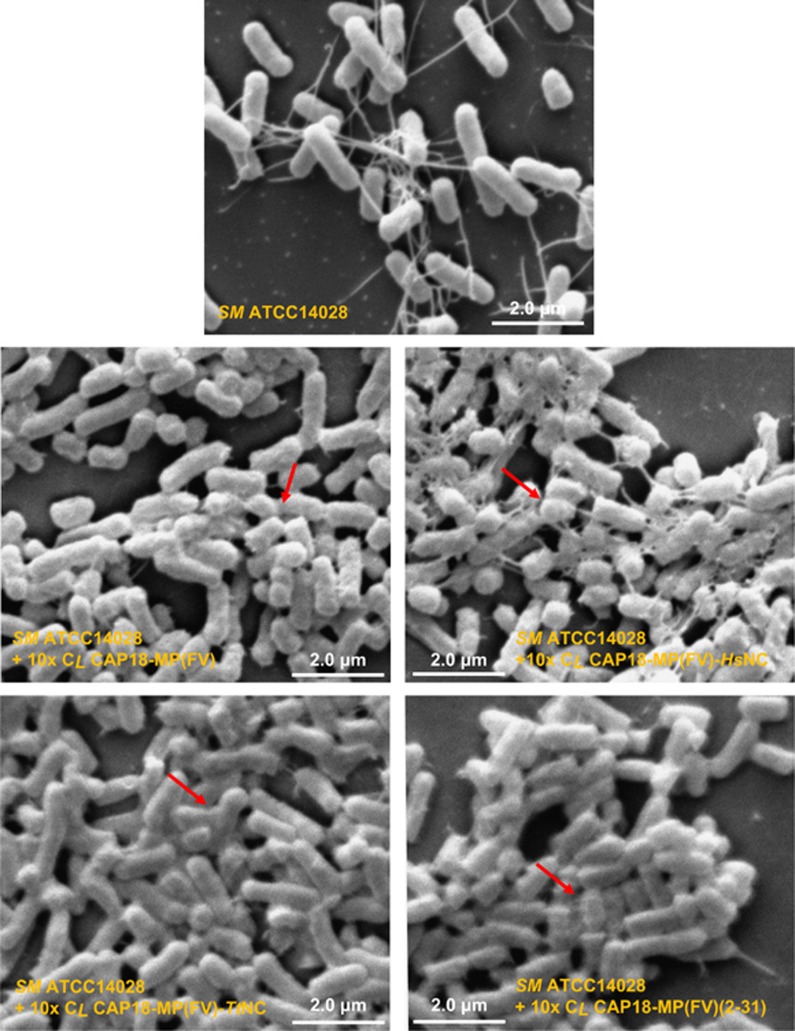
Membrane permeability assay and scanning electron microscopy were used to investigate the action modes of CAP18-MP(FV) and its mutants. Similar to LL-37 (Burton and Steel, 2009), these peptides all exhibited similar membrane-disruptive capacity on bacterial cells, as identified by rapid increase in fluorescence when they were added to B. megaterium cells preincubated with PI, a nucleic acid binding dye, indicating that immediate membrane damage occurred, leading to DNA release (Figure 7). The strain Micrococcus luteus gave a similar result while its fluorescence was slightly lower than B. megaterium (data not shown). Electron microscopic observation revealed morphological changes and cell aggregation in Salmonella enterica treated by the peptides, likely due to the membrane damage leading to the release of cellular contents (Figure 8).
Figure 7.
Effect of peptides on the membrane integrity of B. megaterium cells. In each run, peptides at 5-fold CL were added when the basal fluorescence remained constant 100 s. Vancomycin and meucin-18 were used as negative and positive control, respectively.
Figure 8.
Scanning electron microscopy (SEM) observation of the effects of peptides on sensitive bacteria. The Gram-negative Salmonella enterica ATCC 14028 was selected as a representative. 10x CL peptide doses were used to treat the bacteria for 90 min at 37 °C. Arrows indicate cells with a visible morphological change.
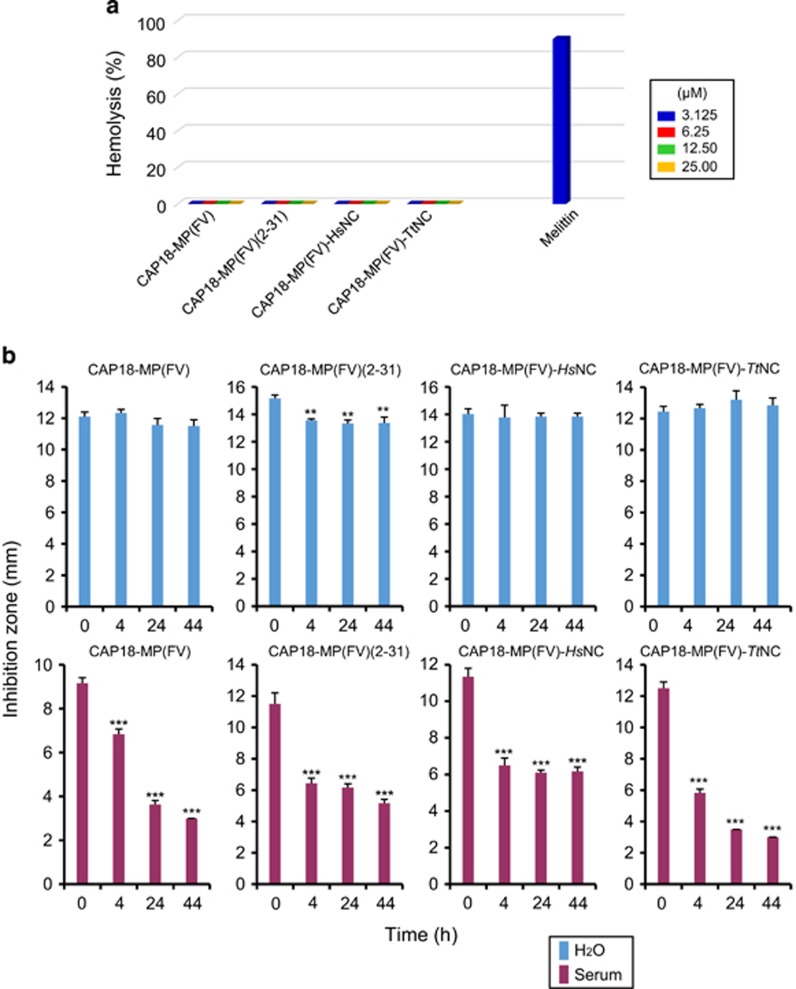
In addition to the similarities in antibacterial activity and action mode, these peptides all were not toxic on mouse blood cells even at 25 μM (Figure 9a), a concentration that exceeded several-folds of their lethal doses to bacteria. After incubated in mouse serum for only 4 hr, these peptides significantly reduced their antibacterial activity, however, incubation in water even up to 44 h resulted in no significant loss of activity (Figure 9b), suggesting their sensitivity on serum proteases. This observation also hints that accelerated substitutions in the unordered regions does not confer the peptide’s stability.
Figure 9.
Hemolysis and serum stability. (a) Hemolysis of CAP18-MP(FV) and its mutants on mouse blood cells. Melittin was used as positive control; (b) Serum stability of CAP18-MP(FV) and its mutants. For comparison, H2O stability of the peptides was also evaluated in parallel. Data are expressed as mean±s.d. (n=3). Student’s unpaired t-test was used to compare means between the group treated with the serum or H2O and the group without treatment. **P<0.005; ***P<0.001.
Evolutionary variability of LL-37 receptors
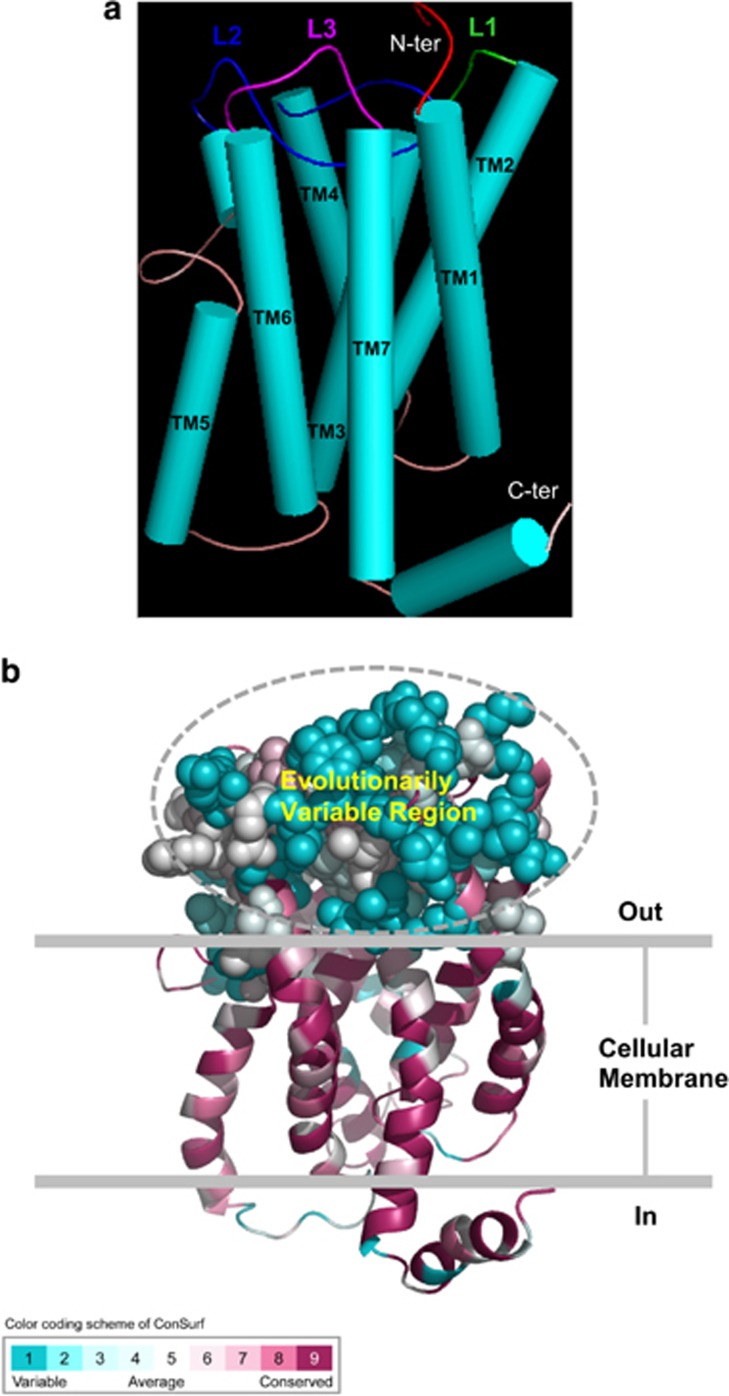
It is known that LL-37 regulates the adaptive immune response via binding to different receptors on host immune cells, such as P2X7, MrgX2 (Mas-related gene X2, a novel G-protein-coupled receptor in human mast cells), and FPR2 that belongs to a class of Gi protein-coupled receptors (Xhindoli et al., 2016). Analyses of the evolutionary conservation of these receptors in mammals might help provide clues on driving forces of the evolution of CAP18-MPs in a cooperative co-evolutionary manner. In particular, the finding that a selective antagonist (WRW4) for FPR2 signaling that directly binds to the formyl peptide binding pocket is capable of blocking the LL-37 functions (Verjans et al., 2016) provides evidence for direct interaction between LL-37 and FPR2. Among these receptors, FPR2 is also the only one identified to bind to the C-terminal, PSS-rich region of LL-37 (Singh et al., 2014). We therefore put our focus on this receptor. To investigate putative co-evolution between CAP18-MP and FPR2, simple Mantel test implemented in the zt software (Bonnet and Van de Peer, 2002) was used to compare their distance matrices, from which their evolutionary trees were constructed (Supplementary Figure S3). This analysis gave a significant correlation of 0.54 (P=0.0002) and thus provided a support for their co- evolutionary relationship. To observe structural location of evolutionarily conserved and variable sites, we firstly modeled the three-dimensional (3D) models of human FPR2 (Figure 10a). It is a typical GPCR containing seven transmembrane helices (TM1–TM7) associated with three extracellular loops (L1–L3). Then we used ConSurf to define the conserved and variable sites in the 3D structure. As shown in Figure 10b, the extracellular loop regions of FPR2 show high evolutionary variability, in contrast to the helices that are overall conserved (Figure 10b). It was found that both terminal regions of LL-37 are required for wild-type level of interaction with RNA to activate TLR3 signaling through FPR2 (Singh et al., 2014), in line with our opinion that these positively selected regions are involved in immune regulation via interacting with the receptors.
Figure 10.
Evolutionary conservation of the FPR2 family in mammals. (a) The cartoon model of human FPR2 structure (residues 16 - 321, Supplementary Figure S2). The structure is displayed by PYMOL (http://www.pymol.org/). Seven transmembrane helices and three extracellular loops are shown; (b) The ConSurf result showing the evolutionarily variable extracellular region, displayed as cyan sphere and circled in dotted line. The multiple sequence alignment for conservation analysis is provided in Supplementary Figure S2.
Previous studies have defined peptide ligand-binding domains of the formyl peptide receptor family of receptors, which include extracellular loops and their adjacent transmembrane domains (Quehenberger et al., 1993). Our work suggests that the extracellular loop regions could also constitute a putative binding site for LL-37 via its C terminus, namely PSSs of CAP18-MPs binding to the evolutionarily variable regions of FPR2s, a reflection of their co-evolution. This also provides a reasonable explanation for the previous proposal that sequence difference between human and murine FPR2 proteins might affect the ability to interact with LL-37 and mCRAMP (Singh et al., 2013).
Discussion
The finding that the PSSs of mammalian CAP18-MPs are located onto two unordered termini of their structures is remarkable because these regions are dispensable for their antibacterial activity. We found that deletion of the unordered and PSS-rich N- and C-terminal regions in rabbit CAP18-MP led to no significant changes in antibacterial spectrum, potency and action mode against multiple Gram- positive and Gram-negative bacteria. Further exchanges of these regions resulted in similar results. These observations suggest that accelerated substitutions in the structurally unordered regions might not be driven by their pathogens, in line with the opinion that the antimicrobial properties of cathelicidins in mammals may not be the main function because they enhance host defense against infection primarily through modulatory mechanisms in vivo (Beaumont et al., 2014).
This appears to be different from the defensin-type HDPs. For example, modification of chicken β-defensin-8 at PSSs can enhance specific antimicrobial activity, suggesting that positive selection at particular sites might be implicated in directing the antimicrobial response against specific pathogens (Higgs et al., 2007). In addition, evolutionary analyses of a primate β-defensin gene cluster suggest that the evolutionary forces driving the proliferation and diversification of the primate β- defensins may be related to reproductive specialization and/or the host-parasite co-evolutionary process (Radhakrishnan et al., 2005). A similar conclusion was also made with regard to the evolution of β-defensin 2 in primates potentially driven, at least in part, by different environmental pressures so as to modulate antimicrobial activity (Antcheva et al., 2004). For mammalian α-defensins, it is proposed that their positive selection is a response to potentially infectious challenges by fast-evolving microbes (Lynn et al., 2004).
In our opinion, the differences in 3D structures and action modes between CAP18-MPs and defensins could explain their differential driving forces. It is known that the defensins kill bacteria by binding to lipid II via residues in a relatively rigid scaffold to inhibit cell wall synthesis (Oppedijk et al., 2016), and thus small variations might lead to a large antibacterial activity alteration, as observed in a fungal defensin (Wu et al., 2016). In this case, functional divergence driven by pathogens occurs more easily. On the contrary, CAP18-MPs kill bacteria via a membrane-disruptive mechanism, in which mutations leading to functional changes should occur rarely because an amphiphilic α-helical structure accommodating more sequence mutations can retain bactericidal activity. In other words, positive selection could exert minor impact on the antibacterial function of this class of α-helical peptides. It is also worth mentioning that previous study based on a small data set from primates identified several PSSs in the α-helical region and elucidated a correlation between structural variations and antimicrobial activity (Zelezetsky et al., 2006). However, when a larger data set that extends to mammals was used, the PSSs located in the helical region disappeared, suggesting that at a large genetic scale, positive selection primarily acts on the unstructured receptor-binding regions.
Alternatively, based on the role of the positively selected regions in interacting with host cell receptors, we proposed that positive selection of CAP18-MPs might be driven by their receptors’ evolutionary variability (Figure 10b), as previously observed in scorpion toxins affecting sodium channels. In this example, toxins bind to the highly variable channel receptor sites via their hot PSSs (Zhang et al., 2015; Zhu et al., 2016). Our proposal is further strengthened by functional characterization of the C terminus of LL-37 in interacting with FPR2 (Singh et al., 2014). Apart from FPR2, P2X7 might also act as a driving force for the evolution of CAP18-MPs because similar evolutionary conservation is also observed in this receptor family, in which the transmembrane and the cytoplasmic domains are highly conserved, while the highest divergence occurred in the extracellular loops involved in ligand binding (Wiley et al., 2011). However, to draw a decisive conclusion, it is necessary to determine the structure of the complex between the CAP18-MP and its receptor.
Data archiving
All sequence data are available from GenBank (https://www.ncbi.nlm.nih.gov/) and their accession numbers are provided in Supplementary Table S1 and S2 as Supplementary Information.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31570773) and the State Key Laboratory of Integrated Management of Pest Insects and Rodents (Grant No. ChineseIPM1512).
Author contributions
SZ conceived and designed this study. BG performed the experiments. SZ wrote and reviewed the manuscript. The two authors have seen the manuscript and agreed to submit it to Heredity.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
The authors declare no conflict of interest.
Supplementary Material
References
- Antcheva N, Boniotto M, Zelezetsky I, Pacor S, Verga Falzacappa MV, Crovella S et al. (2004). Effects of positively selected sequence variations in human and Macaca fascicularis β-defensins 2 on antimicrobial activity. Antimicrob Agents Chemother 48: 685–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont PE, McHugh B, Gwyer Findlay E, Mackellar A, Mackenzie KJ, Gallo RL et al. (2014). Cathelicidin host defence peptide augments clearance of pulmonary Pseudomonas aeruginosa infection by its influence on neutrophil function in vivo. PLoS One 9: e99029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniotto M, Tossi A, DelPero M, Sgubin S, Antcheva N, Santon D et al. (2003). Evolution of the β defensin 2 gene in primates. Genes Immun 4: 251–257. [DOI] [PubMed] [Google Scholar]
- Bonnet E, Van de Peer Y. (2002). zt: a software tool for simple and partial Mantel tests. J Stat Softw 7: 1–12. [Google Scholar]
- Burton MF, Steel PG. (2009). The chemistry and biology of LL-37. Nat Prod Rep 26: 1572–1584. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Prickett MD, Gutowska W, Kuo R, Belov K, Burt DW. (2015). Evolution of the avian β-defensin and cathelicidin genes. BMC Evol Biol 15: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejsmond MJ, Radwan J. (2015). Red queen processes drive positive selection on major histocompatibility complex (MHC) genes. PLoS Comput Biol 11: e1004627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elssner A, Duncan M, Gavrilin M, Wewers MD. (2004). A novel P2X7 receptor activator, the human cathelicidin-derived peptide LL37, induces IL-1b processing and release. J Immunol 172: 4987–4994. [DOI] [PubMed] [Google Scholar]
- Gallo RL, Kim KJ, Bernfield M, Kozak CA, Zanetti M, Merluzzi L et al. (1997). Identification of CRAMP, a cathelin-related antimicrobial peptide expressed in the embryonic and adult mouse. J Biol Chem 272: 13088–13093. [DOI] [PubMed] [Google Scholar]
- Gao B, Sherman P, Luo L, Bowie J, Zhu S. (2009). Structural and functional characterization of two genetically related meucin peptides highlights evolutionary divergence and convergence in antimicrobial peptides. FASEB J 23: 1230–1245. [DOI] [PubMed] [Google Scholar]
- Gao B, Zhu S. (2010). Characterization of a hymenoptaecin-like antimicrobial peptide in the parasitic wasp Nasonia vitripennis. Process Biochem 45: 139–146. [Google Scholar]
- Ginalski K. (2006). Comparative modeling for protein structure prediction. Curr Opin Struct Biol 16: 172–177. [DOI] [PubMed] [Google Scholar]
- Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E et al. (2003). ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 19: 163–164. [DOI] [PubMed] [Google Scholar]
- Gupta K, Subramanian H, Ali H. (2015). Modulation of host defense peptide-mediated human mast cell activation by LPS. Innate Immun 22: 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs R, Lynn DJ, Cahalane S, Alaña I, Hewage CM, James T et al. (2007). Modification of chicken avian β-defensin-8 at positively selected amino acid sites enhances specific antimicrobial activity. Immunogenetics 59: 573–580. [DOI] [PubMed] [Google Scholar]
- Hill RE, Hastie ND. (1987). Accelerated evolution in the reactive centre regions of serine protease inhibitors. Nature 326: 96–99. [DOI] [PubMed] [Google Scholar]
- Hirata M, Shimomura Y, Yoshida M, Morgan JG, Palings I, Wilson D et al. (1994). Characterization of a rabbit cationic protein (CAP18) with lipopolysaccharide- inhibitory activity. Infect Immun 62: 1421–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Nei M. (1988). Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335: 167–170. [DOI] [PubMed] [Google Scholar]
- Hultmark D. (1998) Quantification of antimicrobial activity, using the inhibition-zone assay. In: Wiesner A, Dumpy AG, Marmaras VJ, Morishima I, Sugumaran M, Yamakawa M (eds). Techniques in insect immunology. SOS Publications: Fair Haven, NJ, USA, pp 103–107. [Google Scholar]
- Kyte J, Doolittle RF. (1982). A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132. [DOI] [PubMed] [Google Scholar]
- Larrick JW, Hirata M, Shimomoura Y, Yoshida M, Zheng H, Zhong J et al. (1993). Antimicrobial activity of rabbit CAP18-derived peptides. Antimicrob Agents Chemother 37: 2534–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard BC, Chu H, Johns JL, Gallo RL, Moore PF, Marks SL et al. (2011). Expression and activity of a novel cathelicidin from domestic cats. PLoS One 6: e18756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luenser K, Fickel J, Ludwig A. (2005). Evolution of caprine and ovine β-defensin genes. Immunogenetics 57: 487–498. [DOI] [PubMed] [Google Scholar]
- Luenser K, Ludwig A. (2005). Variability and evolution of bovine β-defensin genes. Genes Immun 6: 115–122. [DOI] [PubMed] [Google Scholar]
- Lynn DJ, Higgs R, Gaines S, Tierney J, James T, Lloyd AT et al. (2004). Bioinformatic discovery and initial characterisation of nine novel antimicrobial peptide genes in the chicken. Immunogenetics 56: 170–177. [DOI] [PubMed] [Google Scholar]
- MacColl ADC. (2011). The ecological causes of evolution. Trends Ecol Evol 26: 514–522. [DOI] [PubMed] [Google Scholar]
- Mansour SC, Pena OM, Hancock REW. (2014). Host defense peptides: front-line immunomodulators. Trends Immunol 35: 443–450. [DOI] [PubMed] [Google Scholar]
- Mookherjee N, Lippert DN, Hamill P, Falsafi R, Nijnik A, Kindrachuk J et al. (2009). Intracellular receptor for human host defense peptide LL-37 in monocytes. J Immunol 183: 2688–2696. [DOI] [PubMed] [Google Scholar]
- Morgan CC, Shakya K, Webb A, Walsh TA, Lynch M, Loscher CE et al. (2012). Colon cancer associated genes exhibit signatures of positive selection at functionally significant positions. BMC Evol Biol 12: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgera F, Vaccari L, Antcheva N, Scaini D, Pacor S, Tossi A. (2009). Primate cathelicidin orthologues display different structures and membrane interactions. Biochem J 417: 727–735. [DOI] [PubMed] [Google Scholar]
- Morrison GM, Semple CA, Kilanowski FM, Hill RE, Dorin JR. (2003). Signal sequence conservation and mature peptide divergence within subgroups of the murine β-defensin gene family. Mol Biol Evol 20: 460–470. [DOI] [PubMed] [Google Scholar]
- Oppedijk SF, Martin NI, Breukink E. (2016). Hit 'em where it hurts: The growing and structurally diverse family of peptides that target lipid-II. Biochim Biophys Acta 1858: 947–957. [DOI] [PubMed] [Google Scholar]
- Park SH, Das BB, Casagrande F, Tian Y, Nothnagel HJ, Chu M et al. (2012). Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature 491: 779–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil A, Hughes A, Zhang G. (2004). Rapid evolution and diversification of mammalian α-defensins as revealed by comparative analysis of rodent and primate genes. Physiol Genomics 20: 1–11. [DOI] [PubMed] [Google Scholar]
- Porcelli F, Verardi R, Shi L, Henzler-Wildman KA, Ramamoorthy A, Veglia G et al. (2008). NMR structure of the cathelicidin-derived human antimicrobial peptide LL-37 in dodecylphosphocholine micelles. Biochemistry 47: 5565–5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts W, Wakeland E. (1990). Evolution of diversity at the major histocompatibility complex. Trends Ecol Evol 5: 181–186. [DOI] [PubMed] [Google Scholar]
- Quehenberger O, Prossnitz ER, Cavanagh SL, Cochrane CG, Ye RD. (1993). Multiple domains of the N-formyl peptide receptor are required for high-affinity ligand binding. Construction and analysis of chimeric N-formyl peptide receptors. J Biol Chem 268: 18167–18175. [PubMed] [Google Scholar]
- Radhakrishnan Y, Hamil KG, Yenugu S, Young SL, French FS, Hall SH et al. (2005). Identification, characterization, and evolution of a primate β-defensin cluster. Genes Immun 6: 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman H, Chattopadhyay A. (2007). Melittin: a membrane-active peptide with diverse functions. Biosci Rep 27: 189–223. [DOI] [PubMed] [Google Scholar]
- Sawyer SL, Wu LI, Emerman M, Malik HS. (2005). Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci USA 102: 2832–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JJ, Unholzer A, Schaller M, Schäfer-Korting M, Korting HC. (2005). Human defensins. J Mol Med (Berl) 83: 587–595. [DOI] [PubMed] [Google Scholar]
- Seil M, Kabré E, Nagant C, Vandenbranden M, Fontanils U, Marino A et al. (2010). Regulation by CRAMP of the responses of murine peritoneal macrophages to extracellular ATP. Biochim Biophys Acta 1798: 569–578. [DOI] [PubMed] [Google Scholar]
- Semple CA, Gautier P, Taylor K, Dorin JR. (2006. a). The changing of the guard: Molecular diversity and rapid evolution of β-defensins. Mol Divers 10: 575–584. [DOI] [PubMed] [Google Scholar]
- Semple CA, Maxwell A, Gautier P, Kilanowski FM, Eastwood H, Barran PE et al. (2005). The complexity of selection at the major primate β-defensin locus. BMC Evol Biol 5: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple CA, Rolfe M, Dorin JR. (2003). Duplication and selection in the evolution of primate β-defensin genes. Genome Biol 4: R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple CA, Taylor K, Eastwood H, Barran PE, Dorin JR. (2006. b). β-defensin evolution: selection complexity and clues for residues of functional importance. Biochem Soc Trans 34: 257–262. [DOI] [PubMed] [Google Scholar]
- Singh D, Qi R, Jordan JL, Mateo LS, Kao CC. (2013). The human antimicrobial peptide LL-37, but not the mouse ortholog, mCRAMP, can stimulate signaling by poly(I:C) through a FPRL1-dependent pathway. J Biol Chem 288: 8258–8268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D, Vaughhan R, Kao CC. (2014). LL-37 peptide enhancement of signal transduction by toll-like receptor 3 is regulated by pH. J Biol Chem 289: 27614–27624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis SM, Anderson NN, Forsyth WR, Espiritu C, Conway BD, Greenberg EP et al. (2000). Bactericidal activity of mammalian cathelicidin-derived peptides. Infect Immun 68: 2748–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verjans ET, Zels S, Luyten W, Landuyt B, Schoofs L. (2016). Molecular mechanisms of LL-37-induced receptor activation: An overview. Peptides 85: 16–26. [DOI] [PubMed] [Google Scholar]
- Wang G. (2008). Structure of human host defense cathelicidin LL-37 and its smallest antimicrobial peptide KR-12 in lipid micelles. J Biol Chem 283: 32637–32643. [DOI] [PubMed] [Google Scholar]
- Wang G, Mishra B, Epand RF, Epand RM. (2014). High-quality 3D structure shine light on antibacterial, anti-biofilm and antiviral activities of human cathelicidin LL-37 and its fragments. Biochim Biophys Acta 1838: 2160–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley JS, Sluyter R, Gu BJ, Stokes L, Fuller SJ. (2011). The human P2X7 receptor and its role in innate immunity. Tissue Antigens 78: 321–332. [DOI] [PubMed] [Google Scholar]
- Wu J, Gao B, Zhu S. (2016). Single-point mutation-mediated local amphipathic adjustment dramatically enhances antibacterial activity of a fungal defensin. FASEB J 30: 2602–2614. [DOI] [PubMed] [Google Scholar]
- Xhindoli D, Pacor S, Benincasa M, Scocchi M, Gennaro R, Tossi A et al. (2016). The human cathelicidin LL-37 - A pore-forming antibacterial peptide and host-cell modulator. Biochim Biophys Acta 1858: 546–566. [DOI] [PubMed] [Google Scholar]
- Xhindoli D, Pacor S, Guida F, Antcheva N, Tossi A. (2014). Native oligomerization determines the mode of action and biological activities of human cathelicidin LL-37. Biochem J 457: 263–275. [DOI] [PubMed] [Google Scholar]
- Yang Z. (2006) Computational Molecular Evolution. Oxford University Press: Oxford, England. [Google Scholar]
- Yu K, Park K, Kang SW, Shin SY, Hahm KS, Kim Y et al. (2002). Solution structure of a cathelicidin-derived antimicrobial peptide, CRAMP as determined by NMR spectroscopy. J Pept Res 60: 1–9. [DOI] [PubMed] [Google Scholar]
- Zelezetsky I, Pontillo A, Puzzi L, Antcheva N, Segat L, Pacor S et al. (2006). Evolution of the primate cathelicidin. J Biol Chem 281: 19861–19871. [DOI] [PubMed] [Google Scholar]
- Zhang S, Gao B, Zhu S. (2015). Target-driven evolution of scorpion toxins. Sci Rep 5: 14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Peigneur S, Gao B, Zhang S, Tytgat J, Zhu S. (2016). Target-driven positive selection at hot spots of scorpion toxins uncovers their potential in design of insecticides. Mol Biol Evol 33: 1907–1920. [DOI] [PubMed] [Google Scholar]
- Zhu S. (2008. a). Did cathelicidins, a family of multifunctional host-defense peptides, arise from a cysteine protease inhibitor? Trends Microbiol 16: 353–360. [DOI] [PubMed] [Google Scholar]
- Zhu S. (2008. b). Positive selection targeting the cathelin-like domain of the antimicrobial cathelicidin family. Cell Mol Life Sci 65: 1285–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Gao B, Harvey P, Craik D. (2012). Dermatophytic defensin with antiinfective potential. Proc Natl Acad Sci USA 109: 8495–8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.