SUMMARY
Defects in DNA repair have been extensively linked to neurodegenerative diseases, but the exact mechanisms remain poorly understood. Here, we report that FUS, a RNA/DNA binding protein linked to amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD), plays a pivotal role in DNA damage response (DDR). We show that the function of FUS in DDR involves a direct interaction with histone deacetylase 1 (HDAC1), and that the recruitment of FUS to double stranded break (DSB) sites is important for proper DDR signaling. Remarkably, FUS proteins carrying familial ALS (fALS) mutations are defective in DDR and DNA repair, and show a diminished interaction with HDAC1. Moreover, increased DNA damage was also observed in human ALS patients harboring FUS mutations. Our findings suggest that an impaired DDR and DNA repair may contribute to the pathogenesis of neurodegenerative diseases linked to FUS mutations.
INTRODUCTION
Mutations in a large number of genes important for various DNA repair mechanisms have been linked to neurodegenerative diseases or to complex diseases with neurological components1, suggesting a role of DNA repair deficiency in the pathogenesis of neurodegeneration.For example, ataxia telangiectasia (AT) is caused by mutations in the ATM gene, which encodes a protein kinase that plays a crucial role in the cellular response to DNA double strand breaks (DSBs). Individuals with mutations in MRE11 and NBS1, components of the DNA damage sensor complex MRN (MRE11-RAD50-NBS1), present with severe neurological symptoms, along with manifestations including hypersensitivity to ionizing radiation and genome instability1,2. DNA damage and genome instability have also been linked to age-related neurodegenerative diseases such as Alzheimer’s and Parkinson’s diseases3–5. Furthermore, impairment of the DNA damage response (DDR) and DNA repair also contribute to motor neuron vulnerability. For example, mice lacking ERCC1, a protein involved in DNA excision repair, show age-related motor neuron degeneration6.
Fused-in-Sarcoma (FUS, also named TLS) is a multifunctional, multi-domain heterogeneous ribonucleoprotein (hnRNP) that belongs to the TET (TAF15, EWS, and TLS) family of RNA-binding proteins7. In the brain, FUS is predominantly expressed in the nucleus, but is able to shuttle between the nuclear and cytoplasmic compartments upon activity stimulation8,9. A number of previous studies have implicated FUS in the maintenance of genome stability and in the DNA repair. For example, inbred FUS−/− mice die perinatally and show genome instability, while embryonic fibroblasts derived from outbred FUS−/− mice display high chromosomal instability and radiation sensitivity10,11. FUS promotes the annealing of homologous DNA and the formation of DNA D-loops, an essential step in DNA repair by homologous recombination12,13. FUS is also shown to be phosphorylated by ATM following the induction of DSBs14, and to be involved in DNA damage-induced regulation of gene expression15. Recent studies have shown that mutations in FUS are causally linked to familial ALS (fALS-FUS), which is characterized by an aberrant accumulation of FUS in the cytoplasm of motor neurons and glia16,17. We therefore sought to determine whether a loss of FUS function affects the DDR and DNA repair, and to evaluate the impact of fALS FUS mutations upon the stability of the neuronal genome.
In this study, we show that FUS plays a pivotal role in the neuronal DDR and DNA repair. We further demonstrate that FUS interacts with HDAC1, a chromatin-modifying enzyme, to regulate DDR signaling and DNA repair. Remarkably, three out of the four fALS FUS mutants that we examined displayed severely impaired DNA repair efficiencies, whereas one mutant showed a mild impairment. The increased DNA damage was also observed in human ALS patients harboring FUS-R521C and FUS-P525L mutations. Our study provides a novel mechanism that is likely to contribute to the degeneration of motor neurons in fALS with FUS mutations.
RESULTS
FUS plays an important role in DDR and DNA repair
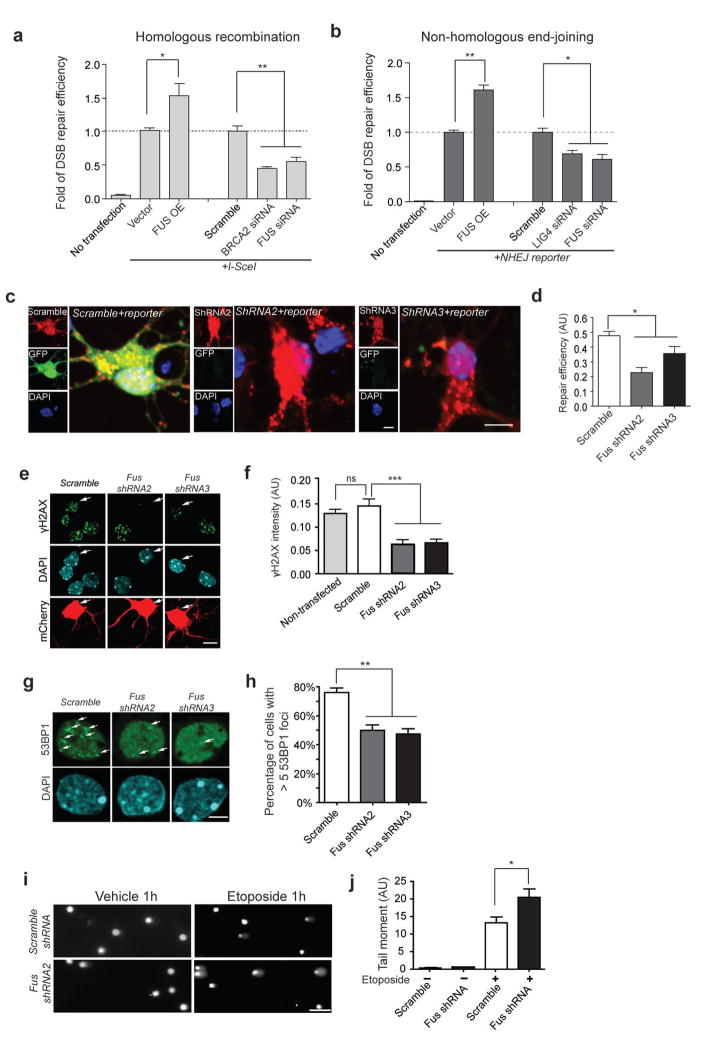
To characterize the role of FUS in repairing of double-stranded DNA breaks (DSBs), the most genotoxic type of DNA damage, we first used a modified U2OS reporter cell line (U2OS-GFP)18, which contains a non-functional eGFP (DR-eGFP) with a yeast endonuclease (I-SceI) cleavage site inserted into its coding sequence. In the absence of I-SceI, U2OS-GFP cells showed a very low background level of eGFP (No transfection, Fig. 1a). In contrast, the presence of I-SceI in these cells creates DSBs in the DR-eGFP DNA sequence, and successful repair of DSBs by homologous recombination (HR) results in the reconstitution of functional eGFP that can be quantified using Fluorescence Activated Cell Sorting (FACS) (assay illustrated in Supplemental Fig. 1). Using this approach, we observed that knockdown of BRCA2, a known component of the HR repair pathway19, impaired HR-mediated DSB repair (Fig 1a, Supplemental Fig. 2a). Knockdown of FUS with pooled FUS siRNAs (Supplemental Fig. 2a) led to a similar impairment, whereas overexpression of FUS significantly enhanced DSB repair efficiency in the U2OS-GFP reporter cell line. Thus, like BRCA2, FUS plays an important role in HR-mediated DSB repair.
Figure 1. FUS plays an important role in DNA damage response and repair in proliferative cells as well as in postmitotic neurons.
(a, b) Assays of DSB repair mediated by homologous recombination (HR) or non-homologous end-joining (NHEJ) in U2OS-GFP cells (illustrated in Supplemental Fig. 1). The number of FACS sorted GFP+ cells indicates DSB repair efficiency and is normalized to control I-SceI or NHEJ reporter-only transfected cells. OE: overexpression (mean ± SEM, *P<0.05, **P<0.01, unpaired t-test). (c, d) Primary cortical neurons were transfected with shRNAs together with pre-digested NHEJ reporter. The ratio of GFP+ cells to total mCherry+ cells indicates the repair efficiency. AU, arbitrary units. Scale bars: 8μm (mean ± SEM, n = 60–80, *P<0.05, unpaired t-test). (e, f) Neurons transfected with indicated shRNAs together with mCherry (white arrows) were labeled for γH2AX after 1 h etoposide treatment (5 μM) (mean ± SEM, n = 16–83, ***P<0.001, ns: no significant difference, unpaired t-test). Scale bar: 8 μm. (g, h) Primary cortical neurons transduced with lentivirus carrying indicated shRNAs were labeled with the anti-53BP1 antibody after 1 h etoposide treatment. White arrows, 53BP1 foci. The percentage of cells with more than five 53BP1 foci upon etoposide treatment was quantified (mean ± SEM, n = 402–619, **P<0.01, unpaired t-test). Scale bar: 4 μm. (I, j) Lentivirus-transduced neurons were treated with vehicle (DMSO) or etoposide for 1 h and harvested for comet assays (mean ± SEM, n = 13–18, *P<0.05, unpaired t-test). Scale bar: 100 μm.
We further assessed DSB repair efficiency mediated by the non-homologous end joining (NHEJ) mechanism using a previously described reporter construct in which the GFP is inactive due to the insertion of an adenoviral exon flanked by artificial introns in the GFP coding sequence20. DSBs can be generated within this insertion using a restriction enzyme and the pre-digested construct can be transfected into cells. Successful repair of DSBs using NHEJ restores the expression of GFP. Thus the percentage of GFP-positive cells is an indicator of successful repair by NHEJ mechanism (assay illustrated in Supplemental Fig. 1). We transfected the pre-digested reporter into U2OS cells together with siRNAs targeting either FUS or LIG4, a protein well known to be important in NHEJ-mediated DSB repair21. We found that, compared with cells infected with empty vector or scrambled siRNAs, overexpression of FUS enhanced, while the knockdown of FUS or LIG4 reduced, NHEJ-mediated DSB repair (Fig. 1b; Supplemental Fig. 2a). Together, these results suggest that FUS is important for both HR- and NHEJ-mediated DSB repair.
We next investigated whether FUS is also involved in the DDR and DNA repair in primary neurons. Since NHEJ is considered to be the primary mechanism for DNA DSB repair in postmitotic neurons22, we assessed whether FUS is important for NHEJ in neurons. We co-transfected primary mouse cortical neurons with the pre-digested NHEJ reporter and either scrambled shRNA, Fus shRNA2, or Fus shRNA3 (Supplemental Fig. 2b), and observed that the knockdown of FUS with either shRNA significantly reduced DSB repair efficiency compared to neurons transfected with scrambled shRNA (Fig. 1c, d), suggesting that FUS plays an important role in NHEJ-mediated DSB repair in neurons.
To further define the function of FUS in DDR and DNA repair, we treated mouse primary cortical neurons at 14 days in vitro (DIV) with etoposide to induce DNA damage23, and evaluated the DDR by measuring immunoreactivity for serine 139- phosphorylated histone H2AX (γH2AX), which is usually imperceptible in cells without DNA damage, but form immunoreactive foci proportional to the production of DSBs upon genotoxic reagent treatment24. Consistently, no γH2AX immunoreactivity was detected in vehicle-treated neurons (Supplemental Fig. 3a). Based on the results of of DNA repair assays (Fig. 1a–d), we expected to observe increased γH2AX immunoreactivity in neurons with FUS knockdown following etoposide treatment. Unexpectedly, while neurons expressing the scrambled shRNA exhibited robust γH2AX immunoreactivity in response to etoposide treatment, we observed diminished γH2AX signal in the etoposide-treated cortical neurons transfected with Fus shRNA2 or shRNA3 (Fig. 1e,f). To corroborate our observation, we examined immunoreactivity for 53BP1, a key mediator of the DDR primarily involved in NHEJ25. 53BP1 immunoreactivity was uniformly distributed in vehicle treated neurons (Supplemental Fig. 3b), but was rapidly recruited to form nuclear foci following etoposide treatment (Fig. 1g,h). Similar to the results observed with γH2AX labeling, etoposide-induced 53BP1 foci formation was markedly reduced by FUS knockdown in cortical neurons, indicated by a decrease in the number of cells containing more than five 53BP1 foci. Furthermore, the fluorescence intensity of phospho(p)-ChK2, a downstream component of the DNA repair pathway that has been shown to be involved in etoposide-induced neuronal death26, was also decreased in FUS knock-down neurons following etoposide treatment compared to control cells (Supplemental Fig. 4). Aberrant cell-cycle activity has been linked to DNA damage in many conditions involving neurodegeneration27. Therefore, we examined immunoreactivity for the cell-cycle marker Ki-6727. We did not observe any signal for Ki-67 following etoposide treatment in primary cortical neurons, indicating that cell cycle re-entry did not occur in these conditions (data not shown).
To directly assess the actual level of DNA strand breaks, we performed single cell gel electrophoresis assay (also known as comet assay), a sensitive method that assesses the integrity of DNA at the single cell level28. While vehicle-treated neurons showed few comet tails, indicative of few DNA strand breaks, etoposide-treated neurons expressing Fus shRNA2 showed significantly increased tail moments compared with those expressing scrambled shRNAs, indicating increased DNA strand breaks (Fig. 1i, j). Thus, despite the presence of increased DNA strand breaks, FUS knockdown resulted in the reduced DDR signaling shown by diminished γH2AX, 53BP1 and p-ChK2 foci. These results suggest that FUS is important for the induction of the DDR in response to DNA damage.
FUS recruitment to DSBs is an early event in DDR pathway
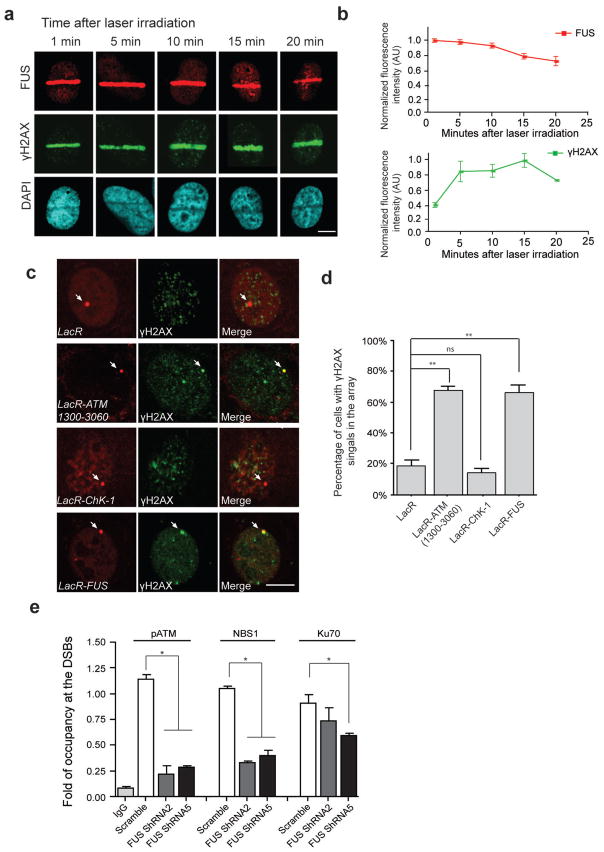
To gain further insight into the function of FUS in the DDR, we examined whether FUS could be recruited to the sites of DNA DSBs. To this end, we performed a micro-irradiation assay in U2OS cells, and investigated the dynamics of FUS recruitment to laser-induced DNA damage sites. The presence of FUS at the site of DNA damage appeared to already be at relatively high levels at 1 min following laser irradiation, the earliest time-point that could be measured with this assay, and was sustained up to 10 min, and gradually declining thereafter (Fig 2a,b). In contrast, immunoreactivity for γH2AX at the site of laser-induced DSBs increased steadily until reaching a maximum value 10–15 min following the laser damage, a time-course consistent with previous studies29. These findings indicate that the recruitment of FUS to sites of DNA damage appears to reach maximum levels while γH2AX is still accumulating.
Figure 2. FUS is rapidly recruited to DSBs and is one of the earliest proteins to respond to DNA damage.
(a, b) Immunolabeling for endogenous FUS and γH2AX at the indicated time following the induction of DSBs via laser micro-irradiation of U2OS cells. FUS and γH2AX immunoreactive intensities within the irradiated area were normalized to the signal from the whole nucleus. Signals from 5–10 cells at each time point were averaged and normalized to the highest intensity across the time period. Scale bar: 8μm. (c, d) Immunofluorescent images were taken of NIH2/4 cells transfected with FUS or the indicated repair factors fused to LacR-mCherry (Red) or LacR- mCherry vector only. DNA damage response (DDR) activation is indicated by the presence of γH2AX foci (green). Arrow pointed mCherry signal indicates the tethering of the mCherry-fused proteins to the genomic loci of LacO array. Scale bar: 8 μm. The percentage of cells with colocalized γH2AX and LacR-mCherry signals in total LacR-mCherry expressing cells were calculated (mean ± SEM, n = 60–70, **P<0.01, unpaired t-test). (e) The occupancy of pATM, NBS1, and Ku70 at DSB sites was assessed using chromatin immunoprecipitation (ChIP) assays in U2OS-GFP cells following FUS shRNA-mediated knock-down (mean ± SEM, *P<0.05, one-way ANOVA).
To further assess whether FUS responds to DNA damage in the early phases of the DNA repair pathway, we adapted a strategy that has been previously applied to demonstrate the hierarchy of proteins involved in the DDR30. It has been shown that even in the absence of actual DNA DSBs, stably tethering the transducer protein MDC1, ATM, or the MRN complex proteins to the chromatin is sufficient to initiate the DDR, as visualized by the presence of γH2AX immunoreactivity, while tethering Chk1 or Chk2, which participate in later stages of DSBs repair, is unable to elicit the DDR cascade30. To determine whether stably tethering FUS to the chromatin is sufficient to stimulate the formation of γH2AX foci, we cloned FUS into a Lac-repressor-mCherry construct (LacR-FUS-mCherry) and transfected it into the NIH-2/4 cell line, which harbors 256 Lac operator (LacO) repeats integrated into its genome. Consistent with previous reports, we observed a co-localization of γH2AX foci with the tethered ATM (1300–3060 aa), while tethering of Chk1, a downstream component of the DDR pathway, was unable to amplify the γH2AX signal (Fig. 2c,d). Importantly, we found that the immobilization of FUS to the chromatin is sufficient to initiate DDR, as indicated by the co-localization of γH2AX immunoreactivity with the LacR-FUS-mCherry signal. These results suggest that FUS is an integral component of the early phases of the DDR.
The phosphorylation of H2AX has been proposed to regulate the formation of repair foci containing proteins such as MDC1 and NBS1 by acting as a docking site on the chromatin, and functionally, to stabilize repair protein complexes at sites of DNA damage24,29. Based on the observation that the recruitment of FUS precedes the appearance of γH2AX foci at the sites of damage, and that FUS depletion results in impaired H2AX phosphorylation in neurons, we speculated that the accumulation of DNA repair complex components might be impaired following FUS knockdown. To test this idea, we transfected our previously described U2OS-GFP cells with the I-SceI endonuclease, after which the cells were harvested and chromatin immunoprecipitation (ChIP) was performed using antibodies against phosphorylated (p)ATM, NBS1, and the NHEJ pathway protein Ku70, followed by quantitative (q)PCR using primers flanking the I-SceI cleavage site in the DR-GFP sequence31. This method allows us to quantitatively analyze the presence of these proteins at the programmed DSB sites created by I-SceI cleavage. We observed that knockdown of FUS resulted in a decreased retention of NBS1, pATM, and Ku70 at DSB sites (Fig. 2e). The impairment of Ku70 accumulation was less severe than that of pATM or NBS1, reflecting the fact that HR is the predominant DNA repair mechanism in proliferative cells1. Nevertheless, these results are in agreement with our observation that γH2AX levels were diminished following FUS knock-down, and support the notion that FUS functions as one of the earliest proteins in the cellular DDR, and that the loss of FUS results in a failure of DDR activation.
FUS and HDAC1 interact, and are required for DNA repair
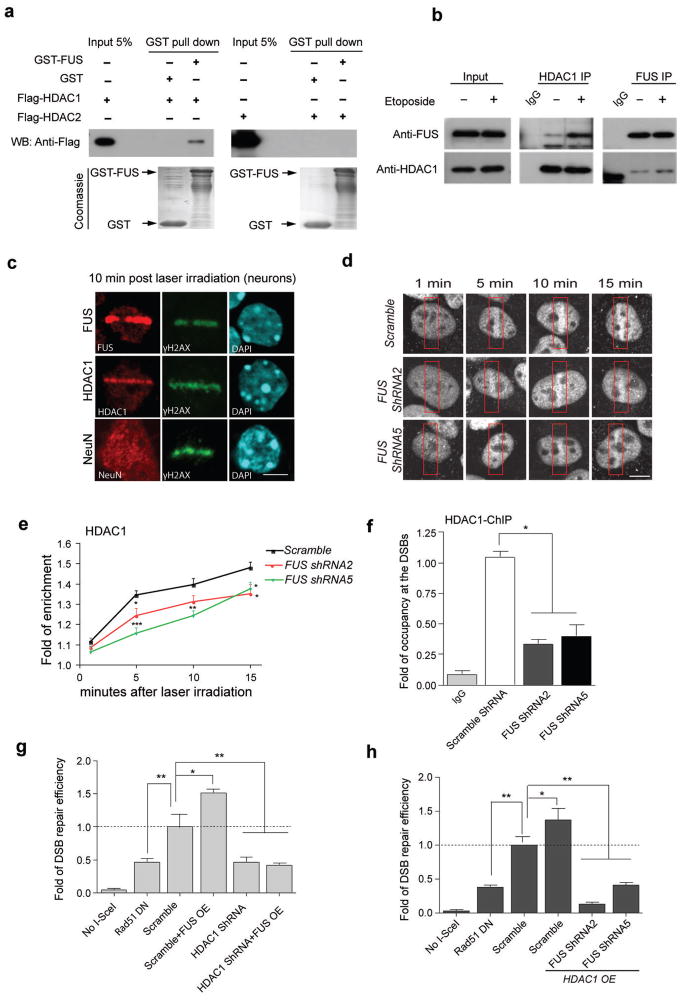
A critical feature of the DDR is the rapid activation and loading of a series of sensor, mediator, and repair proteins into complexes on the chromatin1. To gain mechanistic insight into the function of FUS in this response, it is necessary to determine what proteins it interacts with following DNA damage. Recently, we found that HDAC1 plays a prominent role in DNA repair in postmitotic neurons32. Notably, in mouse brain lysates, FUS readily co-immunoprecipitated with HDAC1 and HDAC2 (Supplemental Fig. 5). Using an in vitro protein-binding assay, we found that HDAC1, but not HDAC2, directly interacts with FUS (Fig. 3a). More importantly, while the interaction of FUS and HDAC1 is detectable under physiological conditions in cortical neurons, the induction of DNA damage by etoposide treatment markedly enhances this interaction (Fig. 3b), suggesting that FUS and HDAC1 form a complex in response to DNA damage.
Figure 3. FUS directly interacts with HDAC1, and both proteins are necessary for successful DNA repair.
(a) Purified GST-FUS protein was incubated with recombinant Flag-HDAC1 or Flag-HDAC2, precipitated with anti-GST beads, and blotted with anti-Flag antibody. A duplicate SDS-PAGE gel was stained with Coomassie blue as input controls. (b) Immunoprecipitation and blotting with antibodies against HDAC1 and FUS in cultured primary neurons with 1 h etoposide or vehicle treatment. Full-length blots are presented in Supplemental Fig. 10. (c) Immunoreactivity for FUS, HDAC1, NeuN and γH2AX in micro-irradiated primary cortical neurons. Scale bar, 4μm. (d, e) HDAC1 immunolabeling of U2OS cells that were infected with indicated shRNA-expressing lentiviruses, and were subjected to laser micro-irradiation. Red boxes indicate damaged area. Cells were fixed and stained for endogenous HDAC1 at indicated time following irradiation. Scale bar = 4μm. Fold enrichment of HDAC1 signal intensity at the lesioned area compared to the whole nucleus was quantified as a function of time following irradiation (mean ± SEM, n = 6–11, *P<0.05, **P<0.01, ***P<0.001, unpaired t-test). (f) The U2OS-GFP cell line, transduced with indicated lentivirus, was subjected to ChIP followed by qPCR to evaluate the occupancy of HDAC1 to the DSBs created by I-SceI (mean ± SEM, *P<0.05, one-way ANOVA). (g,h) DSB repair efficiency was evaluated in U2OS-GFP cells by co-transfecting the indicated constructs together with I-SceI. Percentage of GFP+ cells was analyzed by FACS and fold changes were normalized to cells transduced with lentivirus carrying scrambled shRNA (mean ± SEM, *P<0.05, **P<0.01, unpaired t-test).
Consistent with these results, we found that both FUS and HDAC1 were recruited to laser-induced DNA damage sites in primary cortical neurons. Conversely, the distribution of NeuN was not affected by laser-induced DNA damage (Fig. 3c). The recruitment of HDAC1 to the sites of DNA DSBs appeared to exhibit similar kinetics as that of γH2AX (Supplemental Fig. 6) suggesting that it is delayed compared to FUS (Fig. 2a,b). To determine whether FUS is necessary for the recruitment of HDAC1 to the sites of DSBs, we analyzed HDAC1 immunoreactivity at the laser micro-irradiated areas in FUS knockdown U2OS cells or in control cells (Fig. 3d,e). We show that the enrichment of HDAC1 to the sites of DNA damage was reduced following FUS knockdown. We then expressed I-SceI in U2OS-GFP cells and performed ChIP-qPCR with an antibody against HDAC1 and primers amplifying the DNA region surrounding the I-SceI site. These experiments showed that the knockdown of FUS significantly reduced the presence of HDAC1 at DSBs compared to control (Fig. 3f). Taken together, these results suggest that FUS is important for the recruitment and stable retention of HDAC1 at DNA DSB sites.
To gain insight into how the interaction between FUS and HDAC1 regulates DNA repair, we again used HR repair assay illustrated in Supplemental Fig. 1a. As with FUS, we found that the overexpression of HDAC1 promotes, whereas knockdown of HDAC1 impairs, DNA repair (Fig. 3g). Moreover, while overexpression of FUS and HDAC1 can both enhance DSB repair, overexpression of one component cannot rescue DNA repair deficit following knockdown of the other (Fig. 3g,h). As a positive control for impaired DSB repair, we included the dominant-negative RAD51 (Rad51DN) in these experiments, which has previously been demonstrated to reduce HR mediated DNA repair in the same assay33. These data indicate that FUS and HDAC1 depend upon each other to promote DNA repair.
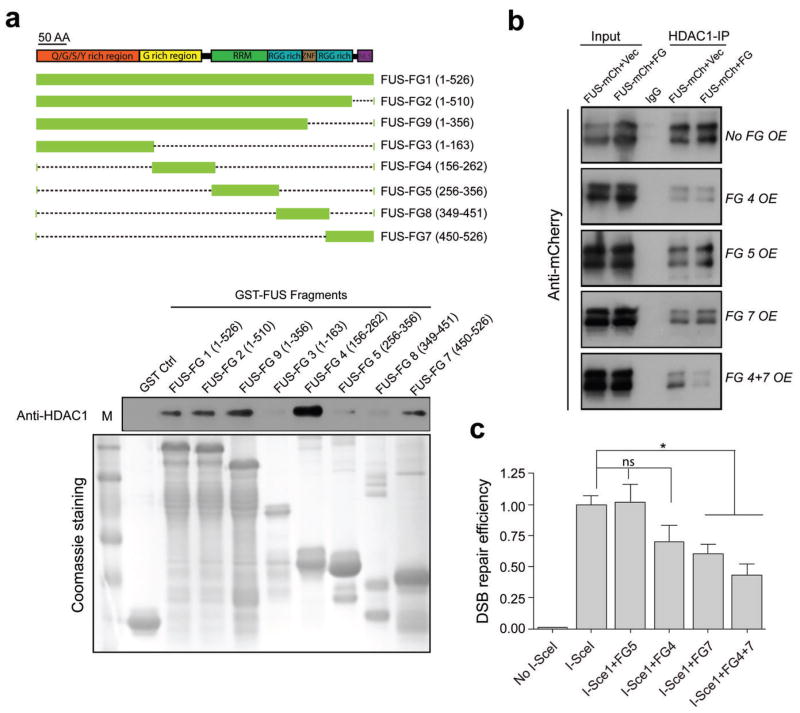
To characterize the domain(s) of FUS responsible for the FUS/HDAC1 interaction, we generated a battery of GST-tagged FUS fragments representing the various putative functional domains of FUS (Fig. 4a). Using GST pull-down assays with recombinant HDAC1, we found that two independent fragments of FUS, the G-rich (FG4) and the C-terminal (FG7) domains, co-immunoprecipitated with HDAC1. Notably, these two regions of FUS are the resident domains for the majority of the fALS mutations identified in FUS to date (Fig. 5a).
Figure 4. The G-rich and C-terminal domains of FUS directly interact with HDAC1, and this interaction is important for successful DSB repair.
(a) Top, schematic of various GST-FUS fragments (FG) that were generated corresponding to functional domains of FUS. RRM: RNA recognition motif; ZNF: zinc finger domain; RGG: the arginine, glycine rich domain; NLS: nuclear localization signal. Bottom, in vitro GST-pull down assay for mapping the functional domains of FUS that directly interact with HDAC1. GST tagged FUS fragments were incubated with recombinant HDAC1, pulled down using GST beads and blotted with anti-HDAC1 antibody. Protein inputs were shown by Coomassie blue staining of a duplicate gel. M: protein marker. (b) FUS fragments 4, 5, and 7 were co-transfected with full length FUS fused with mCherry (FUS-mCherry) in 293T cell as indicated and processed for immunoprecipitation with anti-HDAC1 antibody, and then blotted with anti-mCherry antibody. OE: overexpression. Vec: vector. Full-length blots are presented in Supplemental Fig. 10. (c) FUS fragments 4, 5, and 7 were over-expressed, together with I-SceI in U2OS-GFP cell line to evaluate DSB repair efficiency. GFP+ cells were analyzed by FACS. Repair efficiency was normalized to cells transfected with I-SceI alone (mean ± SEM, *P<0.05. unpaired t-test).
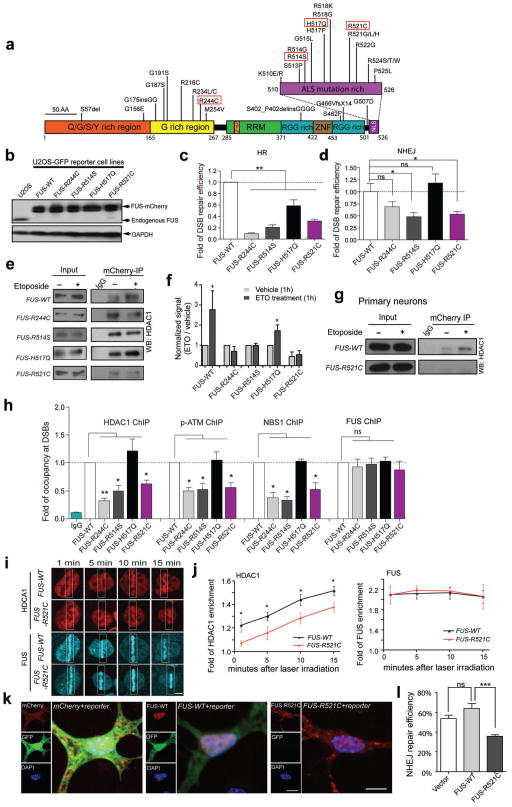
Figure 5. Cells carrying human fALS FUS mutations exhibit impaired DNA repair efficiency and a diminished FUS/HDAC1 interaction.
(a) FUS mutations identified in fALS patients. Red rectangles indicate those investigated in this study. (b) Anti-FUS blot of stable U2OS-GFP cell lines created by replacing endogenous FUS with mCherry tagged wild type or mutated FUS. (c, d) DNA repair assays of U2OS-GFP FUS cell lines (mean ± SEM, *P<0.05, **P< 0.01, unpaired t-test). (e, f) The interaction between FUS and HDAC1 in U2OS FUS cell lines. The signal intensity of etoposide-treated samples was compared to that of vehicle treated samples (mean ± SEM, *P<0.05, unpaired t-test). (g) The interaction between HDAC1 and FUS-WT or FUS-R521C in primary neurons. Full-length blots are presented in Supplemental Fig. 10. (h) ChIP-qPCR for analysis of the retention of HDAC1, pATM, NBS1 and FUS to the I-SceI created DSBs in U2OS-GFP FUS cell lines (mean ± SEM, *P< 0.05, **P< 01, unpaired t-test). (i, j) Representative images of endogenous HDAC1 (stained with anti-HDAC1 antibody) and overexpressed FUS-mCherry at the indicated time point following laser irradiation of FUS-WT or FUS-R521C U2OS cells. White boxes, lesioned area. Scale bar: 4 μm. Fluorescence intensities within lesioned area were normalized to those of the whole nucleus (mean ± SEM, n = 6–10, *P<0.05, unpaired test.). (k, l) DNA repair efficiency mediated by NHEJ was measured by the percentage of GFP+ cells in mCherry expressing cells (mean ± SEM, n = 60–80, ***P<0.001, unpaired t-test). Scale bar: 10 μm.
We next sought to determine whether overexpression of the HDAC1-interacting FUS fragments could interfere with the interaction of HDAC1 with FUS. We observed that whereas transfection with FG4, FG5, or FG7 alone did not affect the binding of FUS to HDAC1, transfection with FG4 and FG7 together (FG4+7) significantly reduced the FUS/HDAC1 interaction (Fig. 4b; Supplemental Fig. 7). To investigate whether the disruption of the endogenous FUS/HDAC1 interaction using overexpression of FUS fragments 4 and 7 leads to impaired DNA repair, we transfected U2OS-GFP cells with FUS fragments FG4, FG5, FG7, and FG4+7 twenty-four hours prior to the introduction of I-SceI. FACS analysis of GFP-positive cells following DSB induction revealed that FG5 overexpression did not affect the DNA repair efficiency, while overexpression of FG4 showed a trend (although not statistically significant) towards a reduction in DSB repair. Overexpression of FG7 showed a stronger impairment of DSB repair (Fig. 4c), while co-expression of FG4 and FG7 caused the most severe impairment in DSB repair. These findings indicate that the G-rich and C-terminal domains of FUS are crucial for DSB repair, and that efficient DNA damage repair may involve the direct interaction of FUS with HDAC1.
fALS FUS mutants are defective in DNA repair and HDAC1 interaction
To test the impact of fALS FUS mutations upon DDR and DNA repair, we selected 4 different fALS FUS mutants including R244C, R514S, H517Q and R521C for this study (Fig. 5a) 16,17. Transfection of WT and mutant FUS into primary cultured neurons demonstrated that WT, FUS-R244C, and H517Q are predominantly expressed only in the nucleus, whereas approximately 80%–90% of FUS-R514S and FUS-R521C transfected cells showed both nuclear and cytoplasmic expression, with the majority of FUS protein still located in the nucleus (Supplemental Fig. 8).
We created five stably-transfected U2OS-GFP cell lines in which endogenous FUS expression was knocked down by shRNA5 targeting the 3′UTR of human FUS, and replaced with wild type (WT) FUS, or with FUS harboring the 4 selected fALS mutations, all tagged with mCherry. Western blot analysis verified the efficient knockdown of endogenous FUS in each cell line, and all five lines showed comparable expression of mCherry-tagged WT or mutant FUS (Fig. 5b). We then expressed I-SceI in these five cell lines to induce programmed DSBs, and assessed the percentage of GFP-positive cells 48 hours post-transfection using FACS. Compared to cells expressing FUS-WT, cells expressing fALS FUS mutants showed decreased percentage of GFP-positive cells, indicating deficient DSB repair (Fig. 5c). FUS-R244C and FUS-R514S presented the most severe deficiency in HR-mediated DSB repair, while the FUS-R521C cell line demonstrated a more modest deficiency, and FUS-H517Q had only a marginal defect. Nevertheless, these data indicate that fALS-associated mutations in FUS interfere with the DNA repair pathway. We further transfected the same stable cell lines with the NHEJ reporter construct to evaluate the DSB repair through the NHEJ mechanism. We observed that, similar to HR, the FUS-R514S and R521C mutants were also impaired in NHEJ-mediated DSB repair (Fig. 5d). However, no DSB repair deficiency was observed with the FUS-H517Q mutant, and the FUS-R244C mutant only moderately impaired NHEJ-mediated repair. Thus, all of the fALS mutations studied impacted HR-mediated DNA DSB repair, while R244C, R514S, and R521C negatively affected NHEJ-mediated DNA DSB repair in the U2OS-GFP cell line.
We next examined whether the FUS/HDAC1 interaction in response to DNA damage was altered with the introduction of fALS associated FUS mutations. The interaction of HDAC1 with FUS was compared in the absence or presence of etoposide treatment using immunoprecipitation with antibodies against mCherry in the five stably-transfected FUS cell lines. Consistent with our findings in cultured cortical neurons, the interaction of FUS-WT with HDAC1 was enhanced following etoposide treatment (Fig. 5e, f). However, while the baseline interaction of HDAC1 with FUS-R244C and FUS-R514S remained detectable, no significant increase was observed following etoposide treatment in cells expressing these two FUS mutants. This suggests that FUS carrying R244C or R514S loses its DNA damage-stimulated interaction with HDAC1. We found that the presence of H517Q did not alter the baseline interaction of FUS with HDAC1, and that this interaction was further enhanced upon etoposide treatment. Thus, the intact interaction of FUS-H517Q with HDAC1 parallels its modest effect on DNA DSB repair. In contrast, we detected little interaction of FUS-R521C with HDAC1 prior to etoposide treatment, and no significant increase in HDAC1 binding following DNA damage, suggesting that the R521C mutation of FUS not only interferes with the interaction of FUS with HDAC1 under physiological conditions, but also disrupts the DNA damage-induced enhancement of the FUS/HDAC1 interaction. Consistently, FUS-R521C failed to interact with HDAC1 following DNA damage in cultured primary cortical neurons (Fig. 5g). Thus, in both proliferating cells and primary neurons, fALS-linked FUS mutations alter the ability of FUS to associate with HDAC1 in response to DNA damage.
To explore whether fALS FUS mutations interfere with the recruitment of DNA damage sensor and transducer factors following DNA damage, we investigated the accumulation of pATM, NBS1, and HDAC1 to the sites of DSBs in U2OS-GFP cells using a ChIP- qPCR assay. Each of the five cell lines was transfected with I-SceI to introduce programmed DSBs in the GFP reporter gene, and ChIP was carried out with antibodies against HDAC1, pATM, NBS1, and FUS. We found that the accumulation of these proteins to the DNA damage sites was impaired in cells expressing the R244C, R514G, and R521C FUS mutants, but not in H517Q-expressing cells (Fig. 5h). And we were surprised to find that the recruitment of the mutant FUS proteins to sites of DNA damage appeared to be normal, and was comparable to that of wild type FUS (Fig. 5h). This observation was confirmed using laser micro-irradiation experiments (Fig. 5i, j and Supplemental Fig. 9). In contrast, the recruitment of HDAC1 to the laser irradiated sites was impaired in FUS-R521C cells (Fig. 5i, j). Together, these results indicate that FUS proteins harboring the fALS mutations are capable of responding to DNA damage, and are recruited to the sites of DNA damage. However, these mutants either interfere with or are impaired in the formation or stabilization of DNA repair complex at the sites of DSBs.
Since our results indicate that FUS-R521C mutant is the most defective in the HDAC1 interaction, we set out to investigate its role in DNA repair in primary cortical neurons via the NHEJ mechanism. To this end, we transfected cultured mouse cortical neurons with the NHEJ reporter construct together with mCherry-tagged FUS-WT, FUS-R521C, or mCherry alone. The proportion of mCherry-expressing cells that were also GFP-positive was used as an indicator of successful NHEJ-mediated DNA DSB repair. We found that the overexpression of FUS-WT in primary neurons only slightly increased DSB repair efficiency, and this effect was not statistically significant. In contrast, overexpression of FUS-R521C caused a marked reduction in NHEJ-mediated DSB repair (Fig. 5k, l).
fALS-FUS patients show increased DNA damage
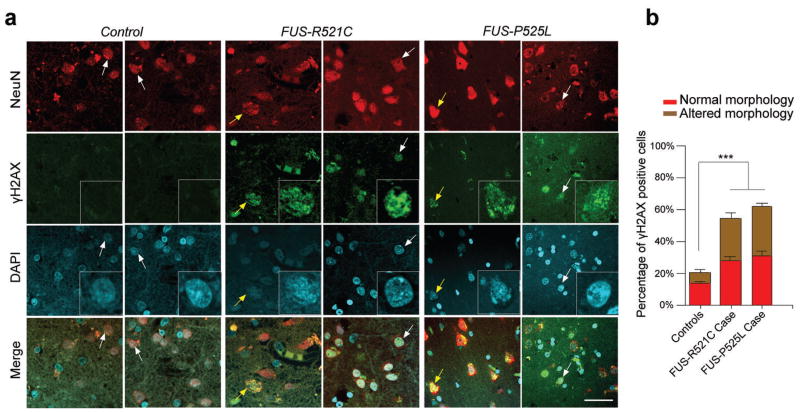
To further assess the significance of fALS FUS mutants in the DNA repair and its disease relevance, we examined DNA damage in brain sections from the motor cortex of three control individuals and two familial ALS patients, harboring the R521C16 or the P525L34 FUS mutation, respectively. We observed increased levels of DNA damage, indicated by γH2AX immunoreactivity, in NeuN-positive neurons of both of these patients (53% to 61%), compared to the controls (20%) (Fig. 6a, b; Supplemental Table 1). It is worth noting that approximately 52% and 46% of the NeuN/γH2AX double positive cells in FUS-R521C or -P525L brain sections, respectively, exhibit abnormal morphology, such as anomalous nuclear labeling (DAPI), while the remaining 48% (R521C) and 54% (P525L) of the NeuN/γH2AX double positive cells appear indistinguishable from surrounding NeuN positive cells, suggesting that the accumulation of DNA damage does not simply represent the “wear and tear” of dying cells, but is perhaps one of the early events that can predispose neurons to deterioration and death. Nevertheless, these data are consistent with our observations in the U2OS cells and primary cultured neurons, and suggest that, like FUS-R521C, the FUS-P525L fALS mutant may also exhibits deficits in DNA repair.
Figure 6. fALS patients harboring FUS mutations exhibit increased DNA damage.
(a, b) Representative images of γH2AX staining in human postmortem motor cortex sections of controls and ALS patients harboring FUS-R521C or FUS-P525L mutation. Higher magnificent image of arrow pointed cells was shown in the γH2AX and DAPI panels. Note that 48% and 54% of the NeuN+, γH2AX+ cells (white arrow) are morphologically normal and indistinguishable from surrounding NeuN+, γH2AX− cells in FUS-R521C and FUS-P525L brain sections, respectively. While the remaining cells exhibit altered morphology (yellow arrow). Error bar represents SD between slides. 5–6 images per slide, 5 slides each patient (mean ± SEM, ***P<0.001, one-way ANOVA). Scale bar: 20 μm.
DISCUSSION
In this study, we demonstrate that FUS plays an important role in mediating the DDR and DNA repair in postmitotic neurons and present the first evidence that fALS mutations in FUS lead to accumulated DNA damage in neurons. Furthermore, we recapitulated our in vitro findings in postmortem brain sections from human patients harboring fALS FUS mutations. Our data suggest a novel mechanism by which impaired DDR and DNA repair may significantly influence the pathogenesis of neurodegenerative diseases caused by FUS mutations.
Recruitment of FUS to DSB is required for efficient repair
FUS has been suggested to be involved in the formation of D-loops, an essential step in homologous recombination, and normally presents in chromosome pairing, DNA repair and telomeres12,13. Accordingly, FUS−/− mice present high levels of genome instability, enhanced ionizing radiation sensitivity, and increased numbers of unpaired and mispaired chromosomal axes in pre-meiotic spermatocytes10,11. In the current study, we find that FUS loss-of-function also results in increased DNA damage in neurons.
Immunoreactivity for γH2AX, a modification that is a prerequisite for DSB repair, is one of the earliest markers of DNA damage29. Upon DNA damage, H2AX is rapidly phosphorylated to γH2AX by ATM, which facilitates the retention of a number of proteins including NBS1, MDC1, and 53BP1 to the vicinity of the DNA breaks24. We observed, in primary neurons, that FUS depletion led to a dampening of the DDR, reflected by decreased H2AX phosphorylation, and an increase in the amount of DNA damage. This finding indicates that FUS is important for neurons to appropriately signal the presence of DNA damage, and subsequently activate repair pathways. In support of this notion, we demonstrate that the recruitment of FUS to DNA damage sites precedes the accumulation of γH2AX, and that stably tethering FUS to chromatin is sufficient to elicit the DDR (Fig. 2a–d). This notion is also supported by previous observations that FUS−/− mice exhibit some striking similarities with ATM−/− and H2AX−/− mice10,11,35,36, including enhanced radiation sensitivity, growth retardation, immunodeficiency, and increased genomic instability. Together, these data suggest that depletion of FUS results in an impaired ATM/γH2AX signaling.
Interaction of FUS and HDAC1 is required for DNA repair
In the current work, we show that FUS directly interacts with HDAC1 both in vivo and in vitro, and that this interaction is important for the DDR and efficient DNA repair. These observations are consistent with the recently elucidated roles of HDAC1 in DNA repair and the maintenance of genomic stability, and provide novel mechanistic insights into the function of FUS in DDR and DNA repair37.
As a key component of the NuRD complex, the reduced binding of HDAC1 to the chromatin is considered as one potential factor underlying defects in chromatin structure and accumulation of persistent DNA damage in Hutchinson-Gilford Progeria Syndrome38,39. Reduced HDAC1 binding may also result in dysregulation of histone acetylation upon DNA damage37, abnormalities in heterochromatin formation, as well as aberrant expansion and re-condensing of the chromatin structure in DNA repair process39,40. We previously demonstrated that the dysregulation of HDAC1 sensitizes neurons to DNA damage and induces aberrant cell cycle re-entry, while the overexpression of HDAC1 protects neurons from genotoxic agents27. Additionally, we found that HDAC1−/− neurons exhibit increased DNA damage following ETO treatment32, which phenocopies, at least partially, our observation in FUS mutants in vitro and in vivo. Taken together, our current study indicates that FUS, in conjunction with HDAC1, plays an important role in maintaining genome stability and integrity in the neuron, and that the impairment of this interaction may contribute to accumulated DNA damage and, eventually, the pathogenesis of fALS.
Our current study indicates the direct interaction of FUS with HDAC1, but not HDAC2, in response to DNA damage, and underscores the functional importance of this interaction for DNA repair and cell survival in neurons. In contrast, we previously showed that HDAC2 negatively regulates synaptic plasticity, learning, and memory, and that its expression is elevated in Alzheimer’s disease41,42. These studies collectively suggest that, despite their structural homology, HDAC1 and HDAC2 may play distinct roles in adult neurons.
It is of particular interest that the G-rich and C-terminal domains of FUS, which harbor most of the fALS mutations, are the two domains necessary for FUS to interact with HDAC1. FUS has been shown to promote homologous DNA pairing, a key step in HR, whereas the oncogenic fusion protein FUS-CHOP, in which the C-terminal domain of FUS was replaced by the DNA-binding and leucine zipper dimerization domain of CHOP7,43, is unable to promote DNA pairing. Since the G-rich domain is also present in the FUS-CHOP protein, these data together suggest that the interaction of the C-terminal domain of FUS with HDAC1 may be more relevant to DNA repair. Notably, it has been shown that FUS directly interacts with CBP/p300, an acetyltransferase, through its N-terminal domain, and leads to the inhibition of CCND1 transcription following DNA damage15, suggesting that FUS may play multiple roles in response to DNA damage.
DNA damage in the pathogenesis of FUS linked ALS
The accumulation of DNA damage has been widely implicated in neurodegenerative conditions such as Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, and premature aging diseases such as Progeroid syndromes. In addition, increased levels of 8-hydroxydeoxyguanosine (8-OHdG) residues, a marker of oxidative DNA damage, have been identified in the spinal cord of both sporadic and familial ALS patients44. Age-related motor neuron degeneration has been observed in mice lacking the DNA repair protein Ercc1, suggesting that the accumulation of DNA damage contributes to the motor neuron vulnerability6. Our observation of increased DNA damage in postmortem brain samples from fALS patients supports the relevance of FUS dysfunction to human disease.
Intriguingly, FUS mutations are associated with earlier disease onset compared to SOD1, TARDBP, or C9ORF72 mutations in patients 45,46. Thus, impaired DNA repair may render motor neurons more vulnerable to intrinsic and/or extrinsic genotoxicity. Also of note is that these fALS FUS mutations may not cause a complete loss of function of FUS protein. This is supported by our study showing that FUS mutant proteins are still recruited to sites of DNA damage, but are impaired in the later steps of assembly or stabilization of the repair complex (Fig 5. h–j, Supplemental Fig. 9). Combined with earlier work, these studies show that the role of FUS in the human brain is multifaceted and warrants a great deal of further study.
Online Methods
Expression constructs
All constructs were assembled using standard cloning methods and confirmed by DNA sequencing. All shRNAs used in this study were cloned into a lentiviral backbone derived from FUGW47 with dual promoter cassettes in which an H1 promoter drives shRNA expression and a ubiquitin promoter drives mCherry expression. The sequences are FUS shRNA2 = CAGAGTTACAGTGGTTATG; FUS shRNA3 = GCTACGGACAACAACAAAG; FUS 3′UTR shRNA5 = CTATACCTCTGGTTCCCAT. For overexpression constructs, point mutations were generated using Quickchange Kit from Promega, and then cloned into the lentiviral vector Lv-PGK-mCherry48. For GST-FUS and GST-FUS fragment constructs, full length FUS and FUS fragments were PCR-amplified and cloned into the pGEX vector. For LacR constructs, FUS and HDAC1 were cloned into Cherry-LacRep (Addgene plasmid 18985). FUS overexpression constructs were generated by fusing mCherry to the N-terminal of FUS and subcloning this into the Lv-CAG lentiviral vector.
siRNAs
siRNAs against FUS (sc-40563), BRCA2 (sc-29825) and LIG4 (sc-37394) were purchased from Santa Cruz biotech. These consisted of three to five 19–25 nt siRNAs designed to knockdown the specific target gene. For detailed information, please view the Santa Cruz website at: http://www.scbt.com/research/sirna_and_shrna_plasmid_and_lentivirus.html
Antibodies
The antibodies used in this study include: mouse monoclonal anti-FUS/TLS (Santa Cruz biotech, sc-47711, 1:500) for western blot, rabbit polyclonal antibody anti-TLS/FUS (Abcam, ab70381, ab84078) for immunohistochemistry (IHC, 1:500) and immunoprecipitation (IP, 1μg). Rabbit polyclonal anti-HDAC1 (Abcam ab-7028), mouse monoclonal anti-HDAC1 (Abcam, ab-31236) for chromatin immunoprecipitation (ChIP, 1μg) and western blot (1:1,000). Rabbit polyclonal anti-NBS1 (Novus Biologicals, NB100-143, NB-100-60654), mouse monoclonal anti-Ku70 (Abcam, ab3108), rabbit anti-pATM (phospho S1891; Abcam, ab81292) for ChIP, rabbit anti-RFP (mCherry; Abcam, ab62341) for IP, mouse monoclonal anti-γH2AX (phosphor S139; Millipore, 05-636,1:1,000), rabbit monoclonal anti-γH2AX (Epitomics, 2212-1,1:1,000) for immunolabeling and western blot, rabbit anti-53BP1 (Bethyl, A300-272A, 1:500) for immunolabeling, rabbit anti-Histone H2A antibody (Abcam, ab13923, 1:1,000), anti-Histone H3 (Abcam, ab1791, 1:50,000) for western blotting, mouse monoclonal anti-BRCA2 (Millipore 05-666); rabbit anti-LIG4 (Abcam, ab26039, 1:1,000), rabbit anti-Phospho-Chk2 (#2661, Cell Signaling Technology, 1:1,000), mouse monoclonal anti-NeuN (Millipore, MAB377, 1:1,000), rabbit anti-Chk2 (#2662, Cell Signaling Technology, 1:1,000), and rabbit anti-Phospho-ATM (Abcam, ab36810, 1: 500) for wester blotting and immunostaining.
Cell culture and transfection/transduction
HEK293T and U2OS cell lines were cultured and maintained with Dulbecco’s modified Eagle’s medium (DMEM, Mediatech or Invitrogen) supplemented with 10% FBS. For primary neuron culture, cortices from E15 Swiss-Webster mice were dissected and neurons were dissociated in 1 x HBSS (Invitrogen) containing papain (Worthington) and DNase at 37°C. Neurons were plated on poly-D-lysine- and laminin- (BD Biosciences) coated glass coverslips (for imaging) or 10 cm plates (for biochemistry). Neurons were maintained in Neurobasal/B27 medium (Invitrogen), supplemented with Glutamax and penicillin/streptomycin. Lentivirus was directly added to the media at DIV 4–6 for transduction. For transient transfection, DIV5 neurons were exposed to the Lipofectamine 2000 reagent (Invitrogen; 11668) and DNA constructs for less than 1 hr in antibiotic-free media; cells were then washed and maintained for 24 hr for overexpression and 48 hr for knockdown experiments.
Single cell electrophoresis (Comet) assays
DIV10–14 primary cortical neurons were treated with etoposide at 5 μM final concentration for 1 hr before being collected for the assay. Single-cell gel electrophoresis under alkaline condition was performed using a Comet assay kit (Trevigen, 4250-050-K). Samples stained with SYBR–green were observed using a Zeiss LSM 510 confocal microscope. Images were analyzed using CometScore software (TriTek).
Western blotting and immunoprecipitation
Cells or brain tissues were collected and lysed in RIPA buffer. The lysates were incubated for 30 min on ice, centrifuged for 15 min at 4,000 g at 4°C, and the supernatant was collected. The lysates were subjected to SDS–PAGE followed by immunoblotting. For histone extraction, primary cultured neurons or brain tissues were homogenized in 1X TX buffer (50 mM Tris HCl, 150 mM NaCl, 2 mM EDTA, 1% Triton-100), rotated at 4°C for 15 min, and then spun down at 4,000 rpm for 10 min. Pellets were resuspended in 250 μl TX-HCl (0.2 N HCl) buffer, incubated on ice for 30 min, spun down at 14,000 rpm for 10 min, and the supernatant, which contains the histones, was collected for western blotting. Immunoprecipitation was performed as described before49
Chromatin Immunoprecipitation
ChIP was performed as described49. Briefly, cells were chemically crosslinked by the addition of one-tenth volume of fresh 11% formaldehyde solution for 15 min at room temperature, homogenized, resuspended, lysed in lysis buffer, and sonicated to solubilize and shear crosslinked DNA. We used a Misonix Sonicator 3000 and sonicated at power 6 for 20 s pulses (90 s pause between pulses) at 4°C while samples were immersed in an ice bath. The resulting whole-cell extract was incubated overnight at 4°C with 100 μl Dynal Protein G magnetic beads that had been pre-incubated with 10 μg antibody. Beads were washed five times with RIPA buffer and once with TE buffer containing 50 mM NaCl. Bound complexes were eluted from the beads by heating at 65°C with occasional vortexes and crosslinking was reversed by overnight incubation at 65°C. Whole-cell DNA extract (reserved from the sonication step) was also treated for crosslink reversal. Immunoprecipitated DNA and whole-cell DNA were then purified by treatment with RNase A, proteinase K, and multiple phenol/chloroform/isoamyl alcohol extractions. The recovered chromatin fragments were subjected to quantitative real-time PCR for 39 cycles using primer pairs specific for 250–500 bp segments corresponding to targeted genomic sequence.
Fluorescence activated cell sorting (FACS)
U2OS cells were transfected with plasmids using Lipofectamine 2000 according to the manufacturer’s protocol (Invitrogen). Cells were collected 60 hr following transfection, washed twice, and resuspended in 1 ml of PBS containing 0.05 mg/ml propidium iodide and 10% FBS. GFP-positive cells were analyzed using a FACScan instrument (Becton & Dickinson).
Micro-irradiation and fluorescence microscopy
Micro-irradiation was generally performed as described previously50. Briefly, U2OS cells and primary cortical neurons were cultured on gridded glass coverslips (MatTek) and were pre-sensitized with 100 nM Hoechst 33242 (Sigma) for 30 min. A Zeiss LSM510 upright laser scanning confocal microscope equipped with a 63X, 1.4NA Plan-Apo water-immersion objective was used for all micro-irradiation experiments. At 3X digital magnification, a ROI of 1μm width (or 0.7μm width for neurons) was selected within each cell. A 405 nm diode laser set to 100% transmission was used to scan the selected ROI for 25 iterations. These conditions generated a detectable DDR confined to the ROI without detectable cytotoxic effects. Following irradiation, cells were maintained for designated time periods before fixing with 4% PFA and immunolabeling with anti-FUS, anti-HDAC1 and anti-γH2AX antibodies to detect endogenous protein recruitment or anti-RFP to detect overexpressed wild-type or mutant FUS. For each time point, data were collected from 5–15 cells and the signal intensity within the irradiated area was measured by Image J and normalized to the signal of the whole nucleus.
Real-time qPCR
Real-time qPCR was carried out using SsoFastTM Evagreen Supermix (Bio-Rad) and a CFX96 real-time PCR Detection system (Bio-Rad). For mRNA analysis, data were normalized to β-tubulin signals. For ChIP-qPCR in I-SceI-transfected cells, data was normalized to GAPDH in input DNA 4.
Immunolabeling, confocal imaging, and quantification
Neurons or transfected cells cultured on glass coverslips were fixed in 4% PFA for 10 min and washed and blocked with 10% donkey serum in PBST(0.3% Triton-X). Primary and secondary antibodies were diluted in blocking buffer and incubated with cells overnight at 4°C and 1 hr at RT, respectively. Images were taken with a Zeiss LSM510 confocal microscope. CellProfiler automated image analysis software51 was used to measure whole-cell γH2AX signal in an unbiased manner.
GST protein purification and in vitro GST pulldown assay
GST fusion proteins were prepared using the E. coli BL21 strain. Transformed BL21 cells were cultured overnight and protein expression was induced for 3 hours with 0.2 mM IPTG. The cell pellets were collected and sonicated in TNT buffer with 4 × 30 second bursts (5/50 setting, no bubbles), then spun down at 10,000 rpm for 30 min at 4°C. The supernatant was collected for GST protein purification with GST beads (Amershan Biosci #27-4574-01), and Bio-Rad’s disposable chromatography columns (polyprep #731-1550). A more detailed protocol is available upon request. For in vitro protein-protein binding assay, 30 μg of GST-fusion proteins were incubated in 500 μl ELB/glycerol buffer (10% glycerol, 150 mM NaCl, 0.1% NP-40, 50 mM Tris pH 8.0, 5 mM EDTA) with 30 μl GST beads slurry overnight. The GST beads were washed 3 times and resuspended in 500 μl ELB buffer and incubated with 5 μg Flag-HDAC1 protein for at least 4 hr. Co-immunoprecipitated protein was eluted in 50 μl elution buffer, from which 20 μl was used for western blotting.
RNA extraction and reverse transcription
RNA was extracted with TRIzol (Invitrogen) using a homogenizer according to the manufacturer’s protocol. RNA quality was assessed using the Nanodrop spectrophotometer (ND-1000). cDNA was synthesized using SuperScript III (Invitrogen) according to the manufacturer’s protocol.
HR and NHEJ reporter assay
The I-SceI construct and the U2OS-GFP cell line were kindly provided by Dr. David Sinclair, and the NHEJ reporter construct was kindly provided by Dr. Vera Gorbunova20. The schematic of these constructs and the procedures of the report assays were illustrated in Supplemental Figure 1. For HR reporter assay, the I-SceI construct was co-transfected with either shRNA, siRNA or overexpression constructs into U2OS-GFP cell line in 24-well plates and FACS analysis were performed 60 hours after transfection. For NHEJ reporter assay, the NHEJ reporter construct was digested overnight with HindIII at 37°C and gel-purified. The pre-digested construct was co-transfected with either shRNA, siRNA or overexpression constructs into U2OS cells or cultured primary neurons using Amaxa™ Mouse Neuron Nucleofector™ Kit (Lonza, VPG-1001) (Amaxa Program No. O-05). FUS and NHEJ reporter construct ratio was 2:1. Three days following transfection, cells were harvested for FACS analysis or fixed for staining. The percentage of GFP-positive cells (successful NHEJ repair) in the total mCherry-positive cells was calculated as an indicator of the NHEJ repair efficiency.
Human materials and immunolabeling
Human brain sections were kindly provided by Ian Mackenzie at the University of British Columbia and by Eric Huang at the University of California San Francisco. The protocol was approved by the UCSF IRB. Detailed information was included in Supplemental Table 1. The brain tissues were fixed in paraformaldehyde, paraffin-embedded, and sectioned at 10 μm thickness. For immunolabeling, following deparaffinization (60°C for 30 min, Xylene 10min, 100% EtOH 20 dips, 95% EtOH 10 dips, 75% EtOH 10 dips, H2O 1 min), brain sections were processed for antigen retrieval with a modified buffer (Tris-HCl, 0.1 mM, pH 9.5, 2.5% Urea, 0,05% Tween-20), blocked with 5% milk in TBS (pH 7.4) at room temperature and incubated at 4°C overnight in TBS containing 5% milk with the following antibodies: rabbit monoclonal anti-γH2AX (Epitomics, 2212-1, 1: 500); anti-NeuN (Millipore, MAB377 1: 300). The next day, they were washed in PBS (pH 7.4) for three times, 5 min each time, and incubated in secondory antibodies (1: 400) for 1 hour at room temperature. All images were obtained using a Zeiss LSM 510 confocal microscope.
Statistical analysis
No statistical methods were used to pre-determine sample sizes, but our sample sizes are similar to those reported in previous publications32. Data were collected randomly and assessed blindly. Data distribution was assumed to be normal but was not formally tested. Statistic methods used in each figures were specified in the corresponding figure legends. Every statistical test was based on at least three biological replicates unless specified in the legends.
Supplementary Material
Acknowledgments
We thank Dr. Tom Misteli for kindly providing the NIH2/4 cell line and Lac-R constructs, Matthew Dobbin for technical assistance on microirradiation experiments, Drs. Alison Mungenast, Ram Madabhushi, Adam Bero, Jay Penney and Alexi Nott for critical reading of the manuscript, and other members of Tsai and Huang labs for helpful discussions. We also thank Drs. Robert H. Brown Jr and Lawrence J. Hayward for insightful discussions. WY.W. was supported by a Postdoctoral Fellowship from the Simons Foundation; J.J. was supported by HHMI EXROP (exceptional research opportunities program). This work is also supported by NS078839 (to L.-H.T.), the Dept. of Veterans Affairs BX001108 (to E.J.H.), the Muscular Dystrophy Association MDA217592 (to E.J.H.). L.-H. T. is a member of the Neurodegeneration Consortium. E.J.H. is a member of the Consortium for Frontotemporal Dementia Research. L.-H.T. is an investigator of the Howard Hughes Medical Institute.
Footnotes
ATHOUR CONTRIBUTIONS
W.Y.W. performed DNA repair assay, in vitro protein interaction, domain mapping assay, created mutant cell lines, and completed immunoprecipitation, chromatin IP and qPCR assay. L.P. performed micro-irradiation assay, comet assays, and evaluated DNA damage response in cultured neurons. S.S. purified full-length and fragment GST tagged proteins. E.J.Q. and M.S. helped on neuronal culture. J.J. helped on in vitro GST pull down assay. L.-H.T. and E.J.H. supervised the project, and W.Y.W. and L.-H.T. wrote the manuscript.
References
- 1.Rass U, Ahel I, West SC. Defective DNA repair and neurodegenerative disease. Cell. 2007;130:991–1004. doi: 10.1016/j.cell.2007.08.043. [DOI] [PubMed] [Google Scholar]
- 2.McKinnon PJ. DNA repair deficiency and neurological disease. Nat Rev Neurosci. 2009;10:100–112. doi: 10.1038/nrn2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender A, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 4.Lu T, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 5.Anderson AJ, Su JH, Cotman CW. DNA damage and apoptosis in Alzheimer’s disease: colocalization with c-Jun immunoreactivity, relationship to brain area, and effect of postmortem delay. J Neurosci. 1996;16:1710–1719. doi: 10.1523/JNEUROSCI.16-05-01710.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Waard MC, et al. Age-related motor neuron degeneration in DNA repair-deficient Ercc1 mice. Acta Neuropathol. 2010;120:461–475. doi: 10.1007/s00401-010-0715-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan AY, Manley JL. The TET family of proteins: functions and roles in disease. J Mol Cell Biol. 2009;1:82–92. doi: 10.1093/jmcb/mjp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujii R, et al. The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr Biol. 2005;15:587–593. doi: 10.1016/j.cub.2005.01.058. [DOI] [PubMed] [Google Scholar]
- 9.Fujii R, Takumi T. TLS facilitates transport of mRNA encoding an actin-stabilizing protein to dendritic spines. J Cell Sci. 2005;118:5755–5765. doi: 10.1242/jcs.02692. [DOI] [PubMed] [Google Scholar]
- 10.Kuroda M, et al. Male sterility and enhanced radiation sensitivity in TLS(−/−) mice. Embo J. 2000;19:453–462. doi: 10.1093/emboj/19.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hicks GG, et al. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat Genet. 2000;24:175–179. doi: 10.1038/72842. [DOI] [PubMed] [Google Scholar]
- 12.Bertrand P, Akhmedov AT, Delacote F, Durrbach A, Lopez BS. Human POMp75 is identified as the pro-oncoprotein TLS/FUS: both POMp75 and POMp100 DNA homologous pairing activities are associated to cell proliferation. Oncogene. 1999;18:4515–4521. doi: 10.1038/sj.onc.1203048. [DOI] [PubMed] [Google Scholar]
- 13.Baechtold H, et al. Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLS/FUS and is able to promote D-loop formation. J Biol Chem. 1999;274:34337–34342. doi: 10.1074/jbc.274.48.34337. [DOI] [PubMed] [Google Scholar]
- 14.Gardiner MTR, Vandermoere F, Morrice NA, Rouse J. Identification and characterization of FUS/TLS as a new target of ATM. Biochem J. 2008;415 doi: 10.1042/BJ20081135. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwiatkowski TJ, Jr, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 18.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel KJ, et al. Involvement of Brca2 in DNA repair. Mol Cell. 1998;1:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 20.Seluanov A, Mittelman D, Pereira-Smith OM, Wilson JH, Gorbunova V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc Natl Acad Sci U S A. 2004;101:7624–7629. doi: 10.1073/pnas.0400726101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferguson DO, et al. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc Natl Acad Sci U S A. 2000;97:6630–6633. doi: 10.1073/pnas.110152897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma S. Age-related nonhomologous end joining activity in rat neurons. Brain Res Bull. 2007;73:48–54. doi: 10.1016/j.brainresbull.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Hande KR. Etoposide: four decades of development of a topoisomerase II inhibitor. Eur J Cancer. 1998;34:1514–1521. doi: 10.1016/s0959-8049(98)00228-7. [DOI] [PubMed] [Google Scholar]
- 24.Fillingham J, Keogh MC, Krogan NJ. GammaH2AX and its role in DNA double-strand break repair. Biochem Cell Biol. 2006;84:568–577. doi: 10.1139/o06-072. [DOI] [PubMed] [Google Scholar]
- 25.Anderson L, Henderson C, Adachi Y. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol Cell Biol. 2001;21:1719–1729. doi: 10.1128/MCB.21.5.1719-1729.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keramaris E, Hirao A, Slack RS, Mak TW, Park DS. Ataxia telangiectasia-mutated protein can regulate p53 and neuronal death independent of Chk2 in response to DNA damage. J Biol Chem. 2003;278:37782–37789. doi: 10.1074/jbc.M304049200. [DOI] [PubMed] [Google Scholar]
- 27.Kim D, et al. Deregulation of HDAC1 by p25/Cdk5 in neurotoxicity. Neuron. 2008;60:803–817. doi: 10.1016/j.neuron.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collins AR. The comet assay for DNA damage and repair: principles, applications, and limitations. Mol Biotechnol. 2004;26:249–261. doi: 10.1385/MB:26:3:249. [DOI] [PubMed] [Google Scholar]
- 29.Pilch DR, et al. Characteristics of gamma-H2AX foci at DNA double-strand breaks sites. Biochem Cell Biol. 2003;81:123–129. doi: 10.1139/o03-042. [DOI] [PubMed] [Google Scholar]
- 30.Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 2008;320:1507–1510. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinstock DM, Nakanishi K, Helgadottir HR, Jasin M. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 2006;409:524–540. doi: 10.1016/S0076-6879(05)09031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dobbin MM, et al. SIRT1 collaborates with ATM and HDAC1 to maintain genomic stability in neurons. Nat Neurosci. 2013;16:1008–1015. doi: 10.1038/nn.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stark JM, et al. ATP hydrolysis by mammalian RAD51 has a key role during homology-directed DNA repair. J Biol Chem. 2002;277:20185–20194. doi: 10.1074/jbc.M112132200. [DOI] [PubMed] [Google Scholar]
- 34.Huang EJ, et al. Extensive FUS-immunoreactive pathology in juvenile amyotrophic lateral sclerosis with basophilic inclusions. Brain Pathol. 2010;20:1069–1076. doi: 10.1111/j.1750-3639.2010.00413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Y, et al. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 1996;10:2411–2422. doi: 10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]
- 36.Celeste A, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller KM, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pegoraro G, et al. Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol. 2009;11:1261–1267. doi: 10.1038/ncb1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol. 2011;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- 40.Dinant C, Houtsmuller AB, Vermeulen W. Chromatin structure and DNA damage repair. Epigenetics Chromatin. 2008;1:9. doi: 10.1186/1756-8935-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guan JS, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graff J, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crozat A, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 44.Ferrante RJ, et al. Evidence of increased oxidative damage in both sporadic and familial amyotrophic lateral sclerosis. J Neurochem. 1997;69:2064–2074. doi: 10.1046/j.1471-4159.1997.69052064.x. [DOI] [PubMed] [Google Scholar]
- 45.Millecamps S, et al. Phenotype difference between ALS patients with expanded repeats in C9ORF72 and patients with mutations in other ALS-related genes. J Med Genet. 2012;49:258–263. doi: 10.1136/jmedgenet-2011-100699. [DOI] [PubMed] [Google Scholar]
- 46.Huang EJ, et al. Extensive FUS-immunoreactive pathology in juvenile amyotrophic lateral sclerosis with basophilic inclusions. Brain Pathol. 2010;20:1069–1076. doi: 10.1111/j.1750-3639.2010.00413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 48.Wang W, et al. Genetically encoding unnatural amino acids for cellular and neuronal studies. Nat Neurosci. 2007;10:1063–1072. doi: 10.1038/nn1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao J, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kruhlak MJ, Celeste A, Nussenzweig A. Monitoring DNA breaks in optically highlighted chromatin in living cells by laser scanning confocal microscopy. Methods Mol Biol. 2009;523:125–140. doi: 10.1007/978-1-59745-190-1_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. gb-2006-7-10-r100 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.