Abstract
Hepatocellular carcinoma (HCC) is a common malignancy of the liver. HCG11 is a member of long non-coding family, upregulation of which in HCC was proved by our previous study. In the present study, the role of HCG11 in the development of HCC was detected by focusing on the interaction between HCG11 and its target protein insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1). The expression status of HCG11 and IGF2BP1 was first investigated with clinical HCC samples. Then the expressions of HCG11 and IGF2BP1 were both inhibited in the human HCC cell line HepG2 and the cell viability, proliferation, apoptosis and metastasis potential of HepG2 cells were assessed. At molecular level, the expression levels of p-ERK, p-JNK, p-p38, p21 and cleaved caspase-3 were also determined to explain the pathways involved in the function of HCG11 in the progression of HCC. Expression of HCG11 and IGF2BP1 were significantly higher in HCC tissues than those in para-tumor tissues. Knockdown of both indicators led to decreased cell viability, proliferation, and migration ability in HepG2 cells while the cell apoptosis and G1 cell cycle arrest were induced after knockdown of HCG11 and IGF2BP1. In addition, suppressed activity of HCG11 and IGF2BP1 blocked the phosphorylation of anti-apoptosis factors, including ERK, JNK and p38 while the mitochondrial apoptosis in HCC cells was initiated by activation of p21 and cleaved caspase-3. HCG11 exerted its effect on HCC via interaction with IGF2BP1, leading to activation of MAPK signaling, which eventually promoted the progression of HCC.
Keywords: hepatocellular carcinoma, HCG11, IGF2BP1, apoptosis, mitogen-activated protein kinases
Introduction
Hepatocellular carcinoma (HCC) is a primary malignancy of the liver, now ranking the fifth of the most common cancers and third leading cause of cancer-related death worldwide (1,2). Curative treatment of HCC generally depends on surgical resection, but unfortunately, the residual liver tissue of HCC patients is still under high risk of relapse and the five-year recurrence rate of the primary tumor can be as high as 70% (3). It is confirmed that it is the high metastasis potential and recurrence rate that contribute to the poor outcome of HCC patients (4). Although plenty of effort has been made to improve the treatment efficiency of HCC, deficient understanding of the mechanism that drives the oncogenesis of the tumor has limited the development of effective therapies.
The molecular pathogenesis of HCC is complicated and associated by numerous events and genetic abnormalities which enable tumorigenesis of pre-neoplastic cells (5). Mutations of β-catenin gene, TP53 gene as well as deletion of the Axin1 and Axin2 genes are all common genetic abnormalities relevant to various etiologies of HCC (5). Except for the protein coding genes, in recent year, an emerging amount of studies show that non-coding RNAs (ncRNAs) also play important roles in onset of many cancer types, including HCC (1). Based on the length, ncRNAs can be classified into small ncRNAs (shorter than 200 nucleotides) and long ncRNAs (lncRNAs, longer than 200 nucleotides) (1). The latter can be further defined based on genome location and context as sense, antisense, bidirectional, intronic and intergenic lncRNA (6,7). As widely accepted, lncRNAs are involved in the regulation of tumor suppressors and oncogenes (8). In the study of Gupta et al (9), the authors indicate that lncRNA HOTAIR promotes metastasis of breast cancer by reprograming chromatin state. Additionally, overexpression of lncRNA H19 contributes to proliferation of gastric cancer cells (10). As for HCC, lncRNA members participate in progression of the cancer in various mechanisms, such as splicing regulation, lncRNA-miRNA interaction, lncRNA-protein interaction and genetic variation, which regulates cell proliferation, apoptosis and metastasis of HCC (11–13). Due to the easy detection of lncRNA in blood and urine and their close association with specific cancers, development of therapies targeting lncRNA may be a novel and effective treating option for HCC patients.
lncRNA HCG11 was previously reported to be a tumor suppressor of prostate cancer (PCa) (14) and also proved to be dys-regulated in breast cancer as well as gastric cancer (15,16). In our unpublished study, the expression status of HCG11 was validated in clinical HCC samples and the result showed an upregulated level of the indicator in HCC tumor tissues. Our data support the clinical relevance of HCG11 in carcinogenesis in multiple cancers. However, the underlying pathways associated with the function of HCG11 in cancers, especially HCC, remains unrevealed.
Therefore, in the present study, the expression of lncRNA HCG11 and its potential target protein insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) were quantified in clinical HCC and corresponding para-carcinoma tissues. Then the expressions of HCG11 and IGF2BP1 were both suppressed in the human HCC cell line HepG2. In addition to detection of proliferation, apoptosis and metastasis potential of HCC cells, the activities of molecules involved in MAPK signaling transduction were also determined to further explain the pathways through which HCG11 activates the progression of HCC.
Materials and methods
Chemicals and agents
Antibodies against IGF2BP1 (ab100999), phosphorylated p38 (p-p38) (ab31828), p-ERK (ab214362), p21 (p21), and cleaved caspase-3 (ab13847) were purchased from Abcam. IgG-HRP antibodies were purchased from Wuhan Boster Biological Technology, Ltd., (Wuhan, China) (#BA1054). Reverse Transcriptase kit (RT-PCR) was purchased from Takara. Total Protein Extraction kit was purchased from Thermo Fisher Scientific (#23227). Matrigel was obtained from Corning Inc., (Corning, NY, USA) (#354248). Hoechst staining kit was purchased from Sigma-Aldrich, St. Louis, MO, USA (#B2883). Cell-Light™ EdU Apollo® 488/567 In Vitro Imaging kit was purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China) (#C10327). PI/Annexin V-FITC apoptosis detection kit was purchased from MultiSciences Biotech Co., Ltd. (Hangzhou, China) (#CCS012).
Cell cultures
Human liver cell line L-02 and Human HCC cell lines Huh7, HepG2, SMMC-7721 and SK-HEP-1 were obtained from Cell Bank of the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% (v/v) mixed antibiotics.
Patients and collection of HCC specimens
HCC specimens and corresponding para-carcinoma tissues were collected from 20 patients in the Department of Liver Transplantation and Hepatobiliary Surgery (Shandong Provincial Hospital Affiliated to Shandong University) and preserved at −80°C. All the patients enrolled in the present study met the following criteria: i) diagnosed to be primary HCC; and ii) all the patients had complete information on clinicopathological and prognostic characteristics. The study was approved by the Ethics Committee of the Shandong Provincial Hospital Affiliated to Shandong University for related screening, inspection and data collection. All the subjects had signed a written informed consent form, and all the procedures were performed in accordance with the Declaration of Helsinki.
Vector construction and transfection
Specific siRNAs of HCG11 (5′-UUCUCCGAACGUGUCACGUTT-3′), negative control (5′-GCCAGAAUGUUCCUAUUUATT-3′) and IGF2BP1 (5′-GGCCAGUUCUUGGUCAAAUTT-3′) were synthesized by Shanghai GenePharma, Co., Ltd. (Shanghai, China). For transfection, HepG2 cells were transfected with siRNA or NC using Lipofectamine 2000 (Invitrogen, Carlsbad CA, USA). Each group was represented by three replicates.
Immunohistochemistry (IHC)
The HCC sections were incubated at 60°C for 2 h before incubation with dimethylbenzene for dewaxing. The sections were then hydrated with different concentrations of alcohol (95% for 2 min, 85% for 2 min, and 75% for 5 min) and washed with ddH2O for 2 min. Subsequently, the sections were fixed using 3% H2O2 for 15 min and washed with phosphate-buffered saline (PBS) three times, followed by incubation with a primary antibody IGF2BP1 (at 37°C for 30 min and then at 4°C overnight. After three washes with 0.01 M PBS (5 min each), a secondary antibody (HRP) was added to the sections for 30-min incubation at 37°C, followed by five cycles of PBS wash. Thereafter, the sections were incubated with HRP-labeled avidin for 30 min at 37°C, and reacted with DAB for 3–10 min before the reaction was stopped by ddH2O. The sections were re-stained with haematoxylin and dehydrated. The results were determined by scanning the sections using an Aperio ScanScope GL (Aperio Technologies, Vista, CA, USA) at ×40 magnification.
H&E staining
The histological changes in clinical tissues were observed using H&E staining: tissues were placed into Bouin solution (4% formaldehyde) for perfusion fixation. Then they were dehydrated using different concentration of alcohol and vitrified in dimethylbenzene. Samples were embedded in paraffin, sectioned and stained with H&E and the results were detected under a microscope at ×40 magnification.
Masson staining
To assess the collagen accumulation, Masson trichrome was utilized to demonstrate the change in tissue samples which were associated with formation of collagen following the method proposed by Flint and Lyons (17): briefly, sections were deparaffinized with toluene and rehydrated with graded alcohol. Three different dyes were used in order to differentiate between cells and extracellular matrix: i) celestine blue solution was used for cell staining (dark blue-black); ii) acid fuchsin solution was used for elastin (pink); and iii) methyl blue solution for collagen (blue). The results were detected under a microscope at ×40 magnification.
RT2-PCR
Total RNA was extracted using TRIzol method according to the manufacturer's instructions. cDNA templates were obtained by reverse transcription of RNA using Super M-MLV reverse transcriptase. The final reaction mix of 20 µl volume contained 10 µl Bestar® SYBR-Green qPCR Master mix, 0.5 µl of each primer (HCG11, forward, 5′-GCTCTATGCCATCCTGCTT-3′ and reverse, 5′-TCCCATCTCCATCAACCC-3′. IGF2BP1, forward, 5′-AAGACCTTACCCTTTACAACCC-3′ and reverse, 5′-GCAGCCACATCATTCTCATAG-3′. GAPDH, forward, 5′-TGTTCGTCATGGGTGTGAAC-3′ and reverse, 5′-ATGGCATGGACTGTGGTCAT-3′), 1 µl cDNA template, and 8 µl RNAse free H2O. Thermal cycling parameters for the amplification were set as follows: a denaturation step at 94°C for 2 min, followed by 40 cycles of amplification of 94°C for 20 sec, 58°C for 20 sec and 72°C for 20 sec. Melting curve was analyzed between 62 and 95°C and the relative expression levels of the target genes were calculated using the formula of 2−ΔΔct.
Western blotting
Total protein was extracted using the Total Protein Extraction kit according to the manufacturer's instructions. GAPDH was used as the internal reference protein. The concentrations of the protein samples were determined using the BCA method, and 40 µg protein from each sample was subjected to 10% sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE). After the proteins were transferred onto polyvinylidene difluoride (PVDF) membranes, the membranes were rinsed with TTBS and blocked with skimmed milk solution for 1 h. Thereafter, the membranes were incubated with the primary antibodies against IGF2BP1 (1:1,000), p-p38 (1:500), p-ERK (1:400), p-JNK (1:1,000), p21 (1:3,000), cleaved caspase-3 (1:1,500) or GAPDH (1:10,000) at 4°C overnight. After four washes with TTBS, the membranes were incubated with secondary HRP-conjugated IgG antibodies (1:20,000) for 45 min at 37°C. Following six washes with TTBS, the blots were developed using the Beyo ECL Plus reagent and the images were recorded in the Gel Imaging System. The relative expression levels of GREM1 and GLI3 were calculated by the Gel-Pro-Analyzer (Media Cybernetics, Inc., Rockville, MD, USA).
Cell viability and proliferation detection
Cell viability was detected using CCK-8 assay: exponentially growing HepG2 cells (2×105 cells/ml) were seeded into a well of a 96-well plate and incubated with different concentration of lead acetate (each concentration was represented by nine replicates) for 72 h. Every 24 h, CCK-8 solution (10 µl) was added to three randomly selected wells and the cultures were incubated at 37°C for 90 min. The OD values in different wells were detected with a mircoplate reader at 450 nm. Cell proliferation of HepG2 cells harvested from 48-h culture was assessed using EDU assay according to the manufacturer's instruction and the results was detected under an immunofluorescent microscope.
Colony formation assay
The capability of anchorage-independent growth of HepG2 cells was measured by the colony formation assay: cells were suspended in 10% serum media containing 0.35% agarose and inoculated into 35-mm plates at a density of 200 cells/plate. After 1 week of culture, the colonies were stained with Wright-Giemsa stain and the number of colonies in each plate was recorded. The colony formation rate was equal to the ratio of colony number to inoculated cell number per plate.
Scratch assay
Scratch assays were performed on the transfected cells to evaluate the migration ability of HepG2 cells. The cells were seeded in a 24-well plate at a density of 2×104 cells/well. Reference points were marked to guarantee the same area of image acquisition. The cells were allowed to grow into a confluent monolayer at 37°C for two days. Then the cell layer was scratched to generate a cell-free straight line. The cells were washed with PBS twice to remove debris at the edges of the scratch. Afterwards, cell migration towards the midline of the scratch was recorded in reference with the reference points. For each well, two images (at 0 and 48 h) were captured with a phase-contrast microscope and the gap distances were measured. Wound healing rate was defined as the percentage of gap closure, and the data were analyzed by Integrated Performance Primitives (IPP) software (6.0).
Transwell assay
Transwell assay was conducted to assess the invasiveness of HepG2 cells: 200 µl medium containing 2×104 cells were plated in the upper chamber of a Transwell system (BSA-coated porous polycarbonate membrane with a pore size of 8 µm; Corning Star Corp., Cambridge, MA, USA). Each polycarbonate membrane was pre-coated with 4 µl Matrigel (1.5 mg/ml; BD Biosciences, San Jose, CA, USA) and incubated at 37°C for 2 h to form a reconstituted basement membrane. The cells were allowed to migrate through the porous membrane at 37°C for 4 h. The cells in the upper surface of the membrane were completely removed, and the cells on the lower surfaces of the membrane were stained with 1% (w/v) crystal violet for 5 min. The cell number was determined using IPP software.
Hoechst 33258 staining
The morphological changes of cell nuclei of HepG2 cells harvested from 5 min after were detected using a Hoechst staining kit according to the manufacturer's instruction and the results were observed by fluorescence microscope.
Flow cytometry
Cell cycle was determined by flow cytometry. Briefly, propidium iodide (PI) was added to the cells to stain DNA in the dark. After 20 min of incubation at room temperature, the DNA contents were analyzed using a FACS flow cytometer (Accuri C6; BD Biosciences, San Jose, CA, USA).
Apoptotic process in HepG2 was detected using a PI/Annexin V-FITC Apoptosis Detection kit (Jingmei Biotech Co., Ltd., Beijing, China) was used according to the manufacturer's instructions. The apoptotic rates were analyzed using a FACScan flow cytometer (Accuri C6; BD Biosciences). The total apoptotic rate was equal to the sum of the late apoptotic rate (UR, upper right quadrant-advanced stage apoptosis) and the early apoptotic rate (LR, lower right quadrant-prophase apoptosis).
Statistical analysis
The data were expressed as mean ± SD (n=3). Difference between two groups was analyzed using t-test with a significant level of 0.05 (two-tailed P-value). All the statistical analyses were conducted using the SPSS version 19.0 (IBM, Armonk, NY, USA).
Results
The expression level of HCG11 in clinical samples is positively correlated with IGF2BP1
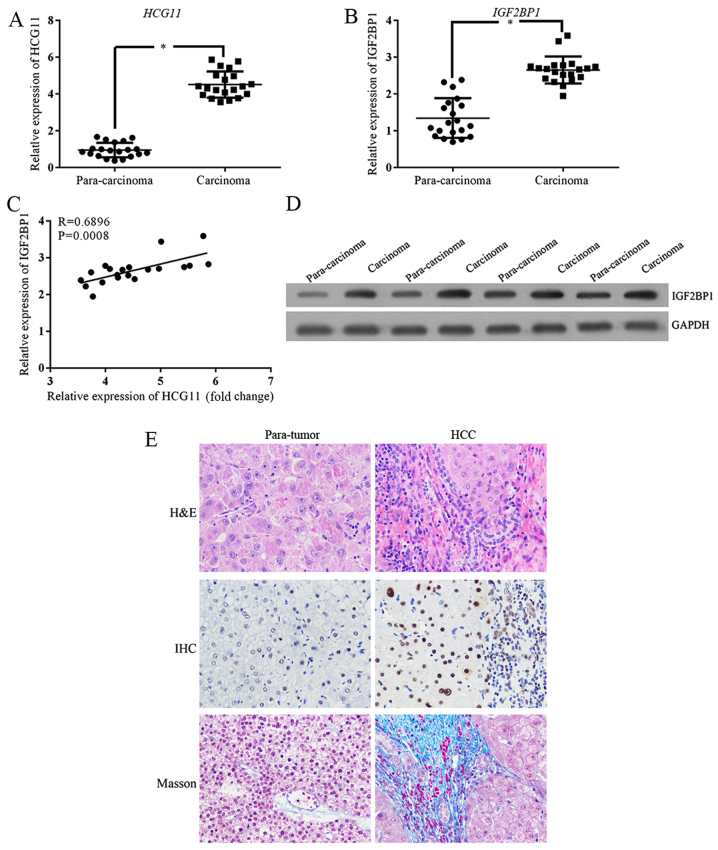
The expression levels of HCG11 and IGF2BP1 at mRNA level were quantified with RT2-PCR method. As shown in Fig. 1A and B, both indicators had a higher expression status in HCC samples. Based on the bioinformatics analysis, IGF2BP1 protein was a direct target of HCG11. Therefore, there should be some correlation between the expression levels of the two genes. By performing a correlation test, it was found that the level of IGF2BP1 was positively correlated with HCG11 (r=0.3556, P<0.000) (Fig. 1C). For IGF2BP1, the upregulated expression level was also confirmed at protein level (Fig. 1D).
Figure 1.
The expression of HCG11 and IGF2BP1 is upregulated in HCC samples. (A) RT2-PCR validation shows upregulated expression of HCG11 in carcinoma tissues. (B) RT2-PCR validation shows upregulated expression of IGF2BP1 in carcinoma tissues. (C) A significantly positive association was detected between expression levels of HCG11 and IGF2BP1. (D) Upregulation of IGF2BP1 at protein level was detected between carcinoma and para-carcinoma tissues. (E) Histological staining illustration of HCC samples: following H&E staining, more infiltration cells could be observed in carcinoma tissues. IGF2BP1 expression were confirmed by IHC analysis. Additionally, higher fibrosis was also observed in carcinoma tissue by Masson. *P<0.05 vs. para-carcinoma tissues. Magnification, ×40.
The augmented expression levels of HCG11 and IGF2BP1 are associated with the progression of HCC
The upregulation of HCG11 and IGF2BP1 in carcinoma was associated with the progression of the tumor. As validated by H&E staining, more infiltration cells could be observed in carcinoma tissues when compared with para-carcinoma tissues (Fig. 1E). Additionally, higher fibrosis and IGF2BP1 (positive cells were stained brown) were also observed in carcinoma tissue by Masson and immunochemical staining, respectively (Fig. 1E). The association between upregulated levels of HCG11 and IGF2BP1 and progression of carcinoma implied the potential function of the two indicators in the oncogenesis of HCC.
Knockdown of HCG11 suppresses cell viability, proliferation and colony formation ability of HepG2 cells
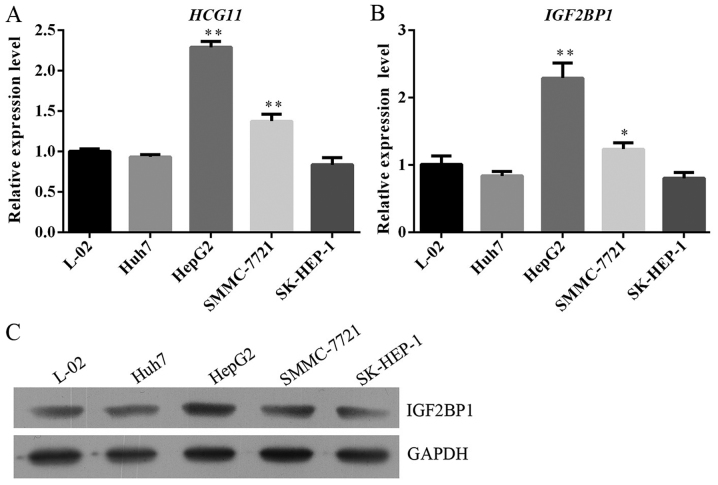
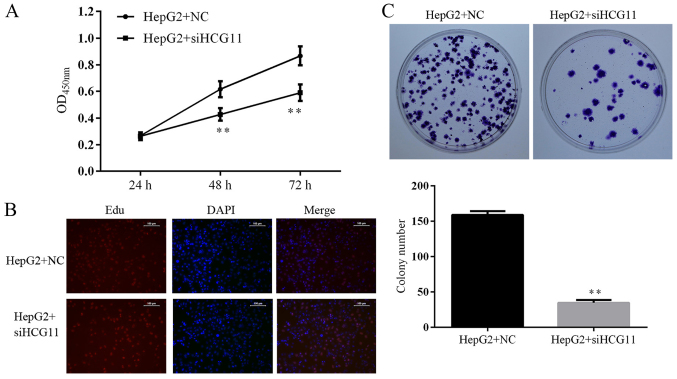
The expression levels of HCG11 and IGF2BP1 were quantified with reverse transcription real-time PCR (RT2-PCR) and the expression level of IGF2BP1 was quantified with western blotting as described. Based on both validations, cell line with highest expression levels of HCG11 and IGF2BP1 was employed for subsequent assays. As illustrated in Fig. 2, HepG2 had a dramatically higher level of both indicators when compared with other cell lines and was selected as the in vitro model of HCC in the present study. To determine the function of HCG11 in the progression of HCC, the expression of the gene was suppressed in HepG2 cells. Afterwards, cell viability, proliferation and colony formation ability of HepG2 cells in different groups were detected. Knockdown of HCG11 decreased the cell viability of HepG2 cells. For cells sampled from 48 and 72 h, the differences in OD450 between NC and siHCG11 groups were statistically significant (P<0.05) (Fig. 3A). Similar results were also observed in Edu assay which represented proliferation of HepG2 cells: as shown in Fig. 3B, less proliferating cells (stained red) were observed in HepG2+siHCG11 group when compared with HepG2+NC group. Colony formation ability is related to the anchorage-independent growth ability of tumor cells, which determines the tumorigenic feature of tumor cells. In the present study, knockdown of HCG11 significantly decreased the colony number in HepG2 cells (Fig. 3C), further confirming the key role of HCG11 in the progression of HCC.
Figure 2.
Selection of HCC cells with high expression of HCG11 and IGF2BP1. Human liver cell line L-02 was used as control. (A) RT2-PCR validation of HCG11 expression level in L-02, Hub7, HepG2, SMMC-7721 and SK-HEP-1 cells. (B) RT2-PCR validation of IGF2BP1 expression level in L-02, Hub7, HepG2, SMMC-7721 and SK-HEP-1 cells. (C) Western blot expression of IGF2BP1 level in L-02, Hub7, HepG2, SMMC-7721 and SK-HEP-1 cells. *P<0.05 vs. L-02 cells. **P<0.01 vs. L-02 cells.
Figure 3.
Knockdown of HCG11 decreases cell viability, inhibits cell proliferation and anchorage-independent growth in HepG2 cells. (A) Quantitative analysis of cell viability detected by CCK-8 assay. (B) Representative images of Edu assay, more proliferation cells were observed under immunofluoresecent detection in HepG2 cells with HCG11 knockdown. (C) Representative images and quantitative analysis of colony formation assay, positive cell number was decreased in HepG2 cells with HCG11 knockdown. **P<0.01 vs. HepG2+NC group.
Knockdown of HCG11 induces cell apoptosis and G1 cell cycle arrest in HepG2 cells
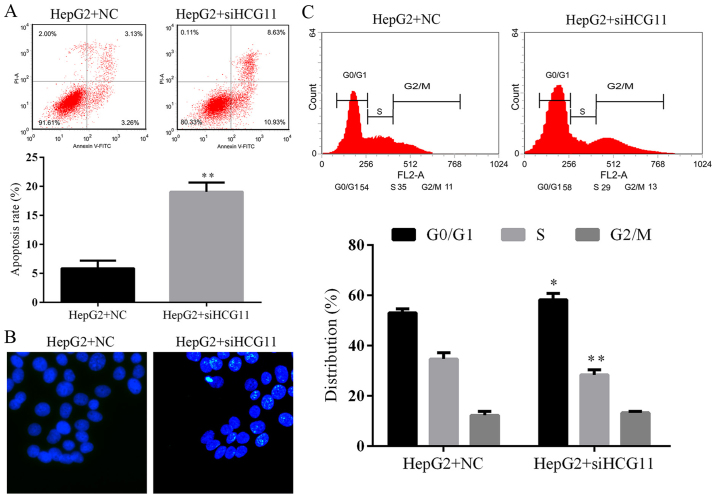
Suppression of HCG11 not only blocked the cell survival but also induced cell death in HepG2 cells. The apoptotic rate in HCG1 knockdown cells was significantly higher than that in normal HepG2 cells (P<0.05) (Fig. 4A). The results of flow cytometry was further validated by Hoechst staining, more Hoechst 33258-positive HepG2 cells were observed in HepG2+siHCG11 group in comparison to that in NC group (Fig. 4B). The increased apoptotic process in HCG11 knockdown cells was associated with G1 cell cycle arrest (Fig. 4C), indicating the cease of cell proliferation of HepG2 cells which was identical to the results of CCK-8 and Edu assays.
Figure 4.
Knockdown of HCG11 induces apoptosis and G1 in HepG2 cells. (A) Representative images and quantitative analysis of cell apoptosis detected by flow cytometry. (B) Representative images of Hoechst staining, more apoptotic cells (stained bright light) were detected in HepG2 cells with HCG11 knockdown. (C) Representative images and quantitative analysis of cell cycle distribution detected by flow cytometry, proportion of cell distributed in G1 and S phases were increased in HepG2 cells with HCG11 knockdown. *P<0.05 vs. HepG2+NC group. **P<0.01 vs. HepG2+NC group.
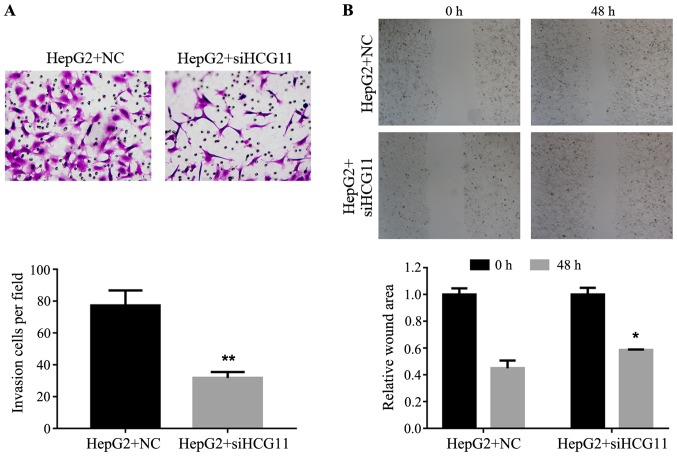
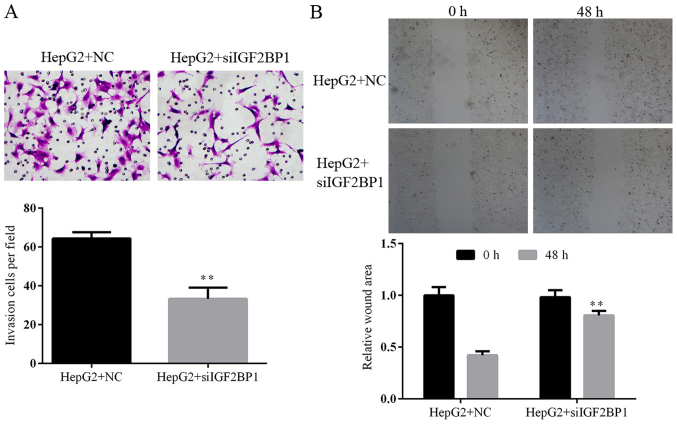
Knockdown of HCG11 suppresses the cell migration and invasion ability of HepG2 cells
Metastasis of a tumor depends on the migration and invasion ability of tumor cells. Effect of HCG11 knockdown on both features of HCC cells were detected in the present study. The invasion of HepG2 cells in Transwell assay was inhibited by HCG11 knockdown: due to the decreased expression level of the gene, less HepG2 cells were able to penetrate the membrane compared with normal HepG2 cells (Fig. 5A). In addition, as shown in Fig. 5B, lack of the function of HCG11 inhibited migration of HepG2 cells in the scratch assay, which was significantly represented by the delayed gap closure at 48 h (P<0.05).
Figure 5.
Knockdown of HCG11 decreases the invasion and migration potential of HepG2 cells. (A) Representative images and quantitative analysis of Transwell experiment, smaller number of cells penetrating membrane was detected in HepG2 cells with HCG11 knockdown. (B) Representative images and quantitative analysis of scratch assay, delayed closure was observed in HepG2 cells with HCG11 knockdown. *P<0.05 vs. HepG2+NC group. **P<0.01 vs. HepG2+NC group.
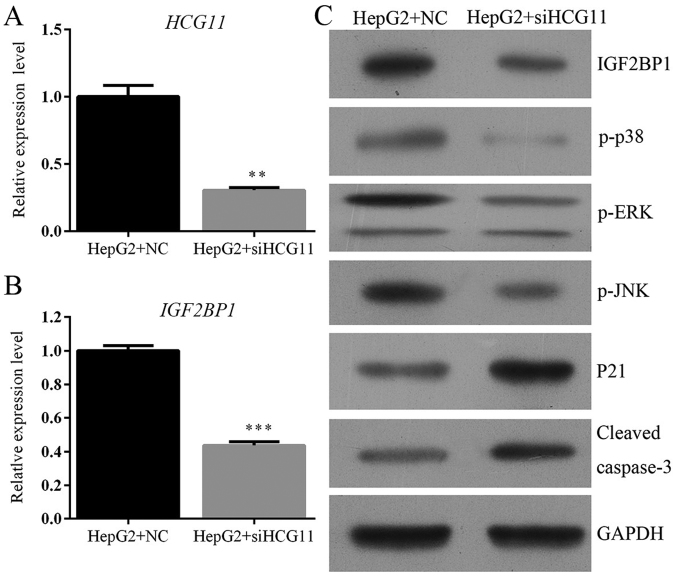
Knockdown of HCG11 antagonizes HCC via modulation of IGF2BP1 activity of MAPK pathways
Knockdown of HCG11 dramatically inhibited the expression of IGF2BP1 both at mRNA and protein levels (Fig. 6). IGF2BP1 is a key protumorigenic factor in HCC (18–20). The positive association between HCG11 and IGF2BP1 might explain the pathway through which HCG11 promoted the progression of HCC. To further explain the mechanism involved in the function of HCG11 in HCC, the activity of MAPK signaling was also detected in the present study. Results showed that inhibition of HCG11 resulted in the downregulation of anti-apoptosis indicators, including p-p38, p-ERK and p-JNK and upregulation of pro-apoptosis indicators, including p21 and cleaved caspase-3 (Fig. 6C), inferring that apoptosis induced by HCG11 knockdown was mediated via mitochondrial pathway (21).
Figure 6.
Knockdown of HCG11 inhibits expression of IGF2BP1 and MAPK signaling, which induced the activation of p21/caspase-3 dependent mitochondrial apoptosis process in HepG2 cells. (A) RT2-PCR validation showed downregulation of HCG11 after HCG11 inhibition. (B) RT2-PCR validation showed downregulation of IGF2BP1 after HCG11 inhibition in HepG2 cells. (C) Western blotting validation showed downregulation of IGF2BP1, p-p38, p-ERK, p-JNK and upregulation of p21 and cleaved caspase-3 after HCG11 inhibition in HepG2 cells. **P<0.01 vs. HepG2+NC group. ***P<0.001 vs. HepG2+NC group.
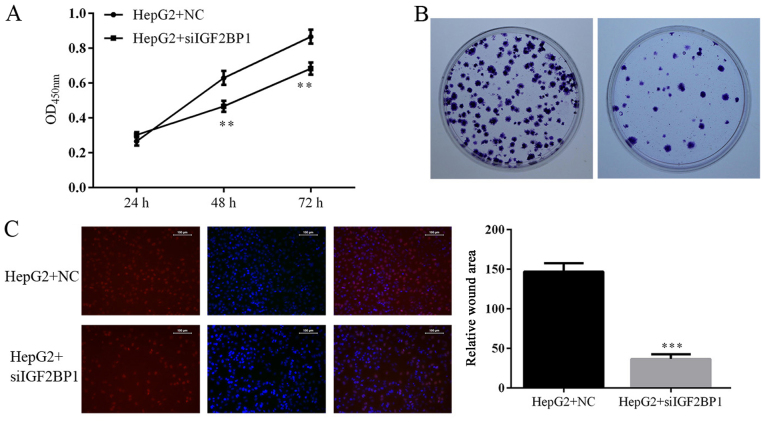
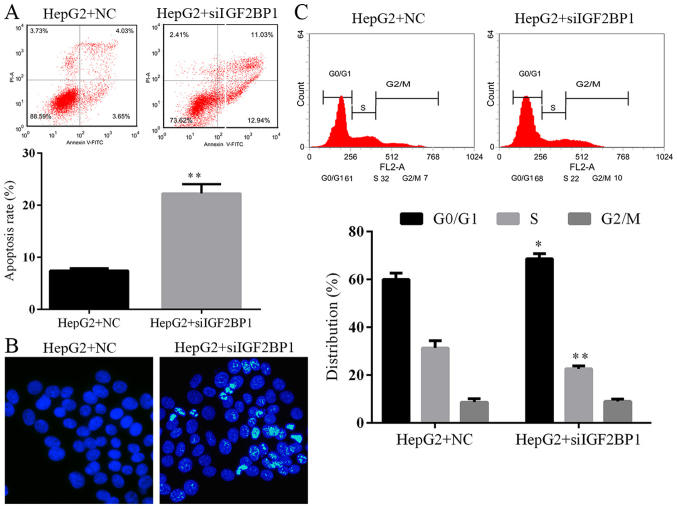
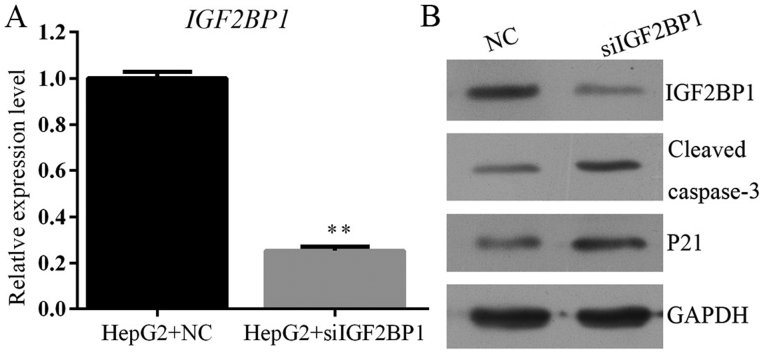
Suppression of IGF2BP1 antagonizes HCC by inducing activity of p21 and caspase-3 signaling
The function of IGF2BP1 on HCC was further confirmed by knocking down the activity of the gene in HepG2 cells. Identical to the effect of IGF2BP1 suppression due to HCG11 knockdown, inhibition of IGF2BP1 by shRNA also decreased cell viability, proliferation, and colony formation ability while induced cell apoptosis and G1 cell cycle arrest in HCC cells (Figs. 7 and 8). The metastasis potential of HepG2 cells was blocked by IGF2BP1 knockdown as well (Fig. 9). At molecular level, knockdown of IGF2BP1 induced the expression of p21 and cleaved caspase-3 (Fig. 10). These results inferred that the modulation effect of HCG11 on progression of HCC and p21/caspase-3 signaling was dependent on functioning of IGF2BP1. Taken together, in the oncogenesis of HCC, HCG11 modulated the activity of IGF2BP1 and its downstream signaling, such as MAPK and p21/capase-3, which then influenced the growth and metastasis of the tumor.
Figure 7.
Knockdown of IGF2BP1 decreases cell viability, inhibited cell proliferation and anchorage-independent growth in HepG2 cells. (A) Quantitative analysis of cell viability as detected by CCK-8 assay. (B) Representative images of Edu assay, more proliferation cells could be observed under immunofluoresecent detection in HepG2 cells with IGF2BP1 knockdown. (C) Representative images and quantitative analysis of colony formation assay, positive cell number was decreased in HepG2 cells with IGF2BP1 knockdown. **P<0.01 vs. HepG2+NC group. ***P<0.001 vs. HepG2+NC group.
Figure 8.
Knockdown of IGF2BP1 induces apoptosis and G1 in HepG2 cells. (A) Representative images and quantitative analysis of cell apoptosis as detected by flow cytometry. (B) Representative images of Hoechst staining, more apoptotic cells (stained bright light) were detected in HepG2 cells with IGF2BP1 knockdown. (C) Representative images and quantitative analysis of cell cycle distribution as detected by flow cytometry, proportion of cell distributed in G1 and S phased were increased in HepG2 cells with IGF2BP1 knockdown. **P<0.01 vs. HepG2+NC group.
Figure 9.
Knockdown of IGF2BP1 decreased the invasion and migration potential of HepG2 cells. (A) Representative images and quantitative analysis of Transwell experiment, smaller number of cells penetrating membrane was detected in in HepG2 cells with IGF2BP1 knockdown. (B) Representative images and quantitative analysis of scratch assay, delayed closure was observed in HepG2 cells with IGF2BP1 knockdown. **P<0.01 vs. HepG2+NC group.
Figure 10.
Knockdown of IGF2BP1 inhibits expression of MAPK signaling, inducing the activation of p21/caspase-3 dependent mitochondrial apoptosis process in HepG2 cells. (A) RT2-PCR validation showed downregulation of HCG11 after IGF2BP1 inhibition. (B) Western blotting validation showed downregulation of IGF2BP1, p-p38, p-ERK, p-JNK and upregulation of p21 and cleaved caspase-3 after IGF2BP1 inhibition in HepG2 cells. **P<0.01 vs. HepG2+NC group.
Discussion
The lncRNA HCG11 was previously reported to be dysregulated in multiple types of cancers (15) and has been proposed to be a promising indicator for prognosis of cancers. In our previous study, it was found that the HCG11 was upregulated in clinical HCC samples, which made us explore the exact role of HCG11 in the oncogenesis and development of HCC. In this study, the expression levels of HCG11 and its target protein IGF2BP1 were quantified in tumor and corresponding para-tumor tissues. The results showed that both indicators were overexpressed in HCC tumors and there was a positive correlation between the expression status of the two indicators. Subsequently, a series of in vitro assays demonstrated that HCG11 was a protumorigenic factor for HCC and the function of the lncRNA was dependent on IGF2BP1, which could modulate the function of downstream MAPK and mitochondrial apoptosis pathways.
Abnormal expressions of multiple lncRNA have been validated to be associated with hepatocarcinogenesis and play key roles in the metastasis and prognosis of the HCC (11,22,23). In the study of Deng et al (24), the authors indicated that lncRNA colon cancer associated transcript-1 (CCAT1) determines the tumor size, microvascular invasion, and prognosis of HCC patients via let-7/HMGA2 signaling transduction. In addition, in the study of Quagliata et al (25), the results showed a promoting effect of lncRNA HOTTIP on the metastasis of HCC cells through modulation on WDR5/MLL pathway. In this study, the key role of HCG11 in the progression of HCC was first validated. Knockdown of the lncRNA not only suppressed the cell viability and growth of HCC cells but also led to inhibition on metastasis potential of HCC cells, which represented the pro-HCC effect of HCG11. Involvement of HCG11 was also proved in PCa, but as reported by Zhang and his colleagues (14), HCG11 in PCa was downregulated and associated with a poor prognosis of PCa patients. Although few studies regarding the role of HCG11 in carcinogenesis have been conducted, it seemed that function of this lncRNA member was tumor type-specific.
To explicitly uncover the function and associated mechanism of HCG11 in the progression of HCC, the potential target of HCG11 was analyzed with bioinformatics tools. Among multiple targets of HCG11, the function of IGF2BP1 in promoting HCC development has been long confirmed (18–20), and was thus selected for subsequent analysis regarding the mechanism of HCG11 in HCC. There are three members in IGF2BP family, including IGF2BP1, IGF2BP2 and IGF2BP3, all of which are bona fide oncofetal proteins involved in various cancers (18,20,26). For IGF2BP1, several targets of the protein are known to play exceptional roles in developmental processes and neoplastic transformation. As previously reported, molecules contributing to the oncogenesis of HCC, including IGF2, ACTB, or c-Myc have downstream effect on the function of IGF2BP1 (27–30). Moreover, IGF2BP1 can also induce the phosphorylation of ERK in HCC cells (19), which was also confirmed in the present study. Suppression of IGF2BP1 by HCG11 knockdown and IGF2BP1 specific shRNA both resulted in inhibition of activity of MAPK members and induced HCC cell apoptosis through mitochondrial pathways.
MAPK pathways are crucial signaling components in converting extracellular stimuli into a wide range of cellular responses (31). In the onset of various tumors, three major MAPKs, ERK, JNK and p38 are activated by mitogen or environmental stress (31). Although the function of MAPKs in the development cancers are complicated, it is confirmed that MAPKs control cell differentiation, proliferation, survival and migration of specific cancer cells (31). Based on the results of western blotting, knockdown of HCG11 and IGF2BP1 both resulted a decreased activity of ERK, JNK and p38. The changed pattern of the three MAPKs explained apoptosis initiated in HCG11 and IGF2BP1 knockdown HCC cells in that suppression of MAPK pathways initiated p12/caspase-3 mediated apoptosis (21,31). Moreover, induced production of metalloproteinases and inflammatory cytokines by MAPKs is critical for the metastasis potential of cancers and inhibited activity of MAPK pathway certainly led to decreased migration ability of cancer cells. The above mentioned conclusion was clearly validated in this study, inhibition of HCG11 and IGF2BP1 augmented the production of p12 and caspase-3, which was synchronized with the induced apoptosis and decreased migration of HCC cells.
In conclusion, findings outlined in the present study for the first time verified the involvement of lncRNA HCG11 in the survival and metastasis of HCC. By interacting with IGF2BP1, HCG11 promoted cell proliferation and invasion of human HCC cells. Moreover, with detection at molecular levels, it was found that modulation of IGF2BP1 by HCG11 exerted its effect via MAPK pathways. Knockdown of either indicator would lead to induced apoptosis and suppressed migration ability in HCC cells. Further characterization of pathways associated with function of HCG1 will facilitate the development of therapeutic strategies for HCC.
Acknowledgments
This study is supported by the National Natural Science Foundation of China (no. 81602550).
References
- 1.Shi L, Peng F, Tao Y, Fan X, Li N. Roles of long noncoding RNAs in hepatocellular carcinoma. Virus Res. 2016;223:131–139. doi: 10.1016/j.virusres.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Al Salloom AA. An update of biochemical markers of hepatocellular carcinoma. Int J Health Sci (Qassim) 2016;10:121–136. [PMC free article] [PubMed] [Google Scholar]
- 3.Yang JD, Roberts LR. Hepatocellular carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 2010;7:448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Budhu A, Forgues M, Ye QH, Jia HL, He P, Zanetti KA, Kammula US, Chen Y, Qin LX, Tang ZY, et al. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10:99–111. doi: 10.1016/j.ccr.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 5.Tejeda-Maldonado J, García-Juárez I, Aguirre-Valadez J, González-Aguirre A, Vilatobá-Chapa M, Armengol-Alonso A, Escobar-Penagos F, Torre A, Sánchez-Ávila JF, Carrillo-Pérez DL. Diagnosis and treatment of hepatocellular carcinoma: An update. World J Hepatol. 2015;7:362–376. doi: 10.4254/wjh.v7.i3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Ma L, Bajic VB, Zhang Z. On the classification of long non- coding RNAs. RNA Biol. 2013;10:925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: Regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 9.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- 11.Huang JL, Zheng L, Hu YW, Wang Q. Characteristics of long non-coding RNA and its relation to hepatocellular carcinoma. Carcinogenesis. 2014;35:507–514. doi: 10.1093/carcin/bgt405. [DOI] [PubMed] [Google Scholar]
- 12.Yang X, Xie X, Xiao YF, Xie R, Hu CJ, Tang B, Li BS, Yang SM. The emergence of long non-coding RNAs in the tumorigenesis of hepatocellular carcinoma. Cancer Lett. 2015;360:119–124. doi: 10.1016/j.canlet.2015.02.035. [DOI] [PubMed] [Google Scholar]
- 13.He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, Li J. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344:20–27. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Zhang P, Wan X, Su X, Kong Z, Zhai Q, Xiang X, Li L, Li Y. Downregulation of long non-coding RNA HCG11 predicts a poor prognosis in prostate cancer. Biomed Pharmacother. 2016;83:936–941. doi: 10.1016/j.biopha.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 15.Liu H, Li J, Koirala P, Ding X, Chen B, Wang Y, Wang Z, Wang C, Zhang X, Mo YY. Long non-coding RNAs as prognostic markers in human breast cancer. Oncotarget. 2016;7:20584–20596. doi: 10.18632/oncotarget.7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu Y, Chen T, Li G, Yu X, Lu Y, Wang H, Teng L. LncRNAs: Emerging biomarkers in gastric cancer. Future Oncol. 2015;11:2427–2441. doi: 10.2217/fon.15.175. [DOI] [PubMed] [Google Scholar]
- 17.Wu Z, Yang L, Cai L, Zhang M, Cheng X, Yang X, Xu J. Detection of epithelial to mesenchymal transition in airways of a bleomycin induced pulmonary fibrosis model derived from an α-smooth muscle actin-Cre transgenic mouse. Respir Res. 2007;8:1. doi: 10.1186/1465-9921-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutschner T, Hämmerle M, Pazaitis N, Bley N, Fiskin E, Uckelmann H, Heim A, Groβ M, Hofmann N, Geffers R, et al. Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) is an important protumorigenic factor in hepatocellular carcinoma. Hepatology. 2014;59:1900–1911. doi: 10.1002/hep.26997. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Cheng J, Zeng Z, Wang Y, Li X, Xie Q, Jia J, Yan Y, Guo Z, Gao J, et al. Comprehensive profiling of novel microRNA-9 targets and a tumor suppressor role of microRNA-9 via targeting IGF2BP1 in hepatocellular carcinoma. Oncotarget. 2015;6:42040–42052. doi: 10.18632/oncotarget.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou X, Zhang CZ, Lu SX, Chen GG, Li LZ, Liu LL, Yi C, Fu J, Hu W, Wen JM, et al. miR-625 suppresses tumour migration and invasion by targeting IGF2BP1 in hepatocellular carcinoma. Oncogene. 2015;34:965–977. doi: 10.1038/onc.2014.35. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki A, Tsutomi Y, Yamamoto N, Shibutani T, Akahane K. Mitochondrial regulation of cell death: Mitochondria are essential for procaspase 3-p21 complex formation to resist Fas-mediated cell death. Mol Cell Biol. 1999;19:3842–3847. doi: 10.1128/MCB.19.5.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shibata C, Otsuka M, Kishikawa T, Ohno M, Yoshikawa T, Takata A, Koike K. Diagnostic and therapeutic application of noncoding RNAs for hepatocellular carcinoma. World J Hepatol. 2015;7:1–6. doi: 10.4254/wjh.v7.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng L, Yang SB, Xu FF, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18. doi: 10.1186/s13046-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z, Boldanova T, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–923. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bell JL, Wächter K, Mühleck B, Pazaitis N, Köhn M, Lederer M, Hüttelmaier S. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): Post-transcriptional drivers of cancer progression? Cell Mol Life Sci. 2013;70:2657–2675. doi: 10.1007/s00018-012-1186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farina KL, Hüttelmaier S, Musunuru K, Darnell R, Singer RH. Two ZBP1 KH domains facilitate β-actin mRNA localization, granule formation, and cytoskeletal attachment. J Cell Biol. 2003;160:77–87. doi: 10.1083/jcb.200206003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hüttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 29.Bernstein PL, Herrick DJ, Prokipcak RD, Ross J. Control of c-myc mRNA half-life in vitro by a protein capable of binding to a coding region stability determinant. Genes Dev. 1992;6:642–654. doi: 10.1101/gad.6.4.642. [DOI] [PubMed] [Google Scholar]
- 30.Ioannidis P, Mahaira LG, Perez SA, Gritzapis AD, Sotiropoulou PA, Kavalakis GJ, Antsaklis AI, Baxevanis CN, Papamichail M. CRD-BP/IMP1 expression characterizes cord blood CD34+ stem cells and affects c-myc and IGF-II expression in MCF-7 cancer cells. J Biol Chem. 2005;280:20086–20093. doi: 10.1074/jbc.M410036200. [DOI] [PubMed] [Google Scholar]
- 31.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]