Abstract
The transcription factor, zinc-finger protein 545 (ZNF545), that belongs to the Kruppel-associated box zinc-finger protein (KRAB-ZFP) family, acts as a tumor suppressor and is inactivated by promoter methylation in cancers such as nasopharyngeal carcinoma, breast cancer, and gastric cancer, but its role in colorectal cancer (CRC) is unknown. The purpose of this study was to characterize the ZNF545 expression, methylation status, biological function, and related molecular mechanisms in CRC. The results showed that ZNF545 was expressed in adult normal colorectal tissues, but downregulated or silenced in CRC cell lines, and this mechanism was reversed by demethylation treatment with 5-aza-2′-deoxycytidine and trichostatin A. The results also showed that the expression of ZNF545 in primary CRC tissues was significantly downregulated compared to adjacent tissues (p<0.05). Overexpression of ZNF545 caused CRC cell cycle arrest and apoptosis, suppressed cell proliferation, and suppressed colony formation and migration in vitro, showing that ZNF545 can function as a tumor suppressor. This function was also shown in nude mice. Furthermore, Wnt/β-catenin, phosphatidylinositol 3 kinase/protein kinase B (PI3K/AKT), and mitogen-activated protein kinases/extracellular signal-regulated kinase (MAPK/ERK) signaling pathways participated in the regulation of ZNF545 in CRC cells. Together, the results suggested that ZNF545 functions as a tumor suppressor in CRC and is frequently inactivated by promoter methylation.
Keywords: zinc-finger protein 545, tumor suppressor, kruppel-associated box zinc-finger protein, methylation, colorectal cancer
Introduction
Colorectal cancer (CRC) is a common malignant tumor involving approximately 14 million new cases per year, and is the cause of approximately 690,000 deaths worldwide each year (1). The incidence of CRC increases with age; 79% of CRC patients are >60 years of age when diagnosed, and 34% are >80 years of age when diagnosed (2). Furthermore, it is predicted that the incidence of CRC in elderly patients will increase in the future. The pathogenesis of CRC involves activation of an oncogene and inactivation of a tumor suppressor gene (3), and the tumor suppressor gene inactivation is closely related to epigenetic changes. These epigenetic changes include DNA methylation, histone acetylation, and chromatin remodeling (4). DNA methylation plays an important role in the development and progression of different cancers such as CRC. Increased evidence has suggested that promoter methylation of tumor suppressor genes can be used as a tumor marker in the early diagnosis of multiple cancers (5,6). When CRC is diagnosed, it is often already in an advanced stage, so it is important to identify markers for early diagnosis of CRC.
Zinc finger proteins containing the Kruppel associated box (KRAB-ZFPs) belong to the zinc finger protein family, which is the largest family of transcription factors in mammals (7,8). KRAB-ZFPs regulate cell proliferation, differentiation, apoptosis, and formation of tumor cells, and play an important role in biological evolution (9). KRAB-ZFPs are transcription factors that bind to the promoter of target genes to inhibit or activate their expression (10), and to either inhibit or promote cancinogenesis. ZIC1, ZNF569 and ZFP932 have been reported to be tumor suppressor genes (11–13), and KLF5, SALL4 and ZNFEB have been reported to be oncogenes (14–16).
ZNF545, located on the 19q13 chromosome, is a member of the KRAB-ZFP family of tumor suppressor genes, and is downregulated and frequently methylated in multiple types of tumors (17–19). However, the effect of epigenetic inactivation of ZNF545 in colorectal carcinogenesis and its possible use as a tumor marker remain unclear. We therefore characterized the methylation status and expression of ZNF545 in CRC, together with its related functions and mechanism of action.
Materials and methods
Cell lines and tumor samples
Five CRC cell lines (HT-29, SW480, HCT116, CaCo-2, and LoVo) were used. The CRC cell lines, HT-29 and HCT116, were provided by Professor Q. Tao at the Chinese University of Hong Kong, and the SW480, CaCo-2, and LoVo cell lines were purchased from the Chinese Academy of Sciences. The cell lines were cultured with RPMI-1640 medium (Gibco-BRL, Karlsruhe, Germany) containing 10% fetal bovine serum (FBS) (ExCell Bio, Shanghai, China), and cultured in a 5% CO2 incubator at 37°C. The CRC tissues and paracarcinoma tissues, which were diagnosed by a pathologist, were obtained from patients during surgery at the First Affiliated Hospital of Chongqing Medical University (Chongqing, China). All patients signed an informed consent form, and the research protocol was approved by the Institutional Ethics Committee of the First Affiliated Hospital of Chongqing Medical University.
RNA, DNA, and protein extraction
Total RNA was extracted separately from 32 paired CRC tissues, surgical margin tissues, and five CRC cell lines using TRIzol® reagent (Life Technologies, Carlsbad, CA, USA). Genomic DNA was separately obtained from 24 CRC tissues, six normal colorectal tissues, and the five CRC cell lines using the QIAamp DNA Mini kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. The concentration of DNA and RNA were measured using a NanoDrop 2000 Spectrophotometer (Thermo Scientific, Rockford, IL, USA) and stored at −80°C. The experimental and control group of the HT-29 and SW480 cells were lysed using a protein extraction reagent (Thermo Scientific) that contained the protease inhibitor, phenylmethane sulfonyl fluoride, and a phosphatase inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA), and the lysate was then homogenized using a Ultrasonic Cell Grinder (Scientz, Ningbo). The supernatant was collected after centrifugation, and the concentration of protein in the supernatant was determined using the BCA protein kit (Thermo Scientific).
Semiquantitative polymerase chain reaction (PCR) and real-time PCR
ZNF545 expression in CRC cells and tissues was determined using semiquantitative PCR and quantitative PCR. The RNA (1 µg) was reverse transcribed to 20 µg of cDNA using the Reverse Transcription system (Promega, Madison, WI, USA). For semiquantitative PCR, the ZNF545 gene was amplified using GoTaq DNA polymerase (Promega) with 35 cycles, using GAPDH as an internal control. The primer sequences are listed in Table I. Quantitative PCR was performed with a SYBR mix green fluorescent reagent (Promega) using an ABI 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). β-actin was used as a control, and each sample was tested in triplicate.
Table I.
The primers used in this study.
| Primer | Sequences (5′–3′) | Product sizes (bp) | Annealing temperature (°C) | Cycles | |
|---|---|---|---|---|---|
| RT-PCR | ZNF545-F | GAGCCTTGGAAAGTTGTGAG | 245 | 55 | 35 |
| q-PCR | ZNF545-R | GGCATTTTCACACTACTGAAG | |||
| MSP | ZNF545-M1 | TTTTTTTTAGGTTTTGTCGCGTC | 177 | 60 | 40 |
| ZNF545-M2 | CTACTAAAAAAACCGAACGCG | ||||
| ZNF545-U1 | TTTTTTTTTAGGTTTTGTTGTGTT | 164 | 58 | 40 | |
| ZNF545-U2 | CCAAACACACTCACAAAATACA |
Methylation-specific PCR analyses of ZNF545
As previously described (20), methylation-specific PCR (MSP) was used to detect the ZNF545 promoter methylation status. bisulfite-treated DNA (50 ng) was mixed with AmpliTaq Gold polymerase (Applied Biosystems), mgCl2, and deoxynucleotide triphosphates for the MSP amplification reaction. The methylation-specific primers are listed in Table I. The PCR amplification was performed for a total of 40 cycles with an annealing temperature of 60°C or 58°C for 30 sec. The final products were analyzed on a 2% agarose gel, and then recorded using a Molecular Imager (Bio-Rad, Hercules, CA, USA).
5-Aza-2′-deoxycytidine (Aza) and trichostatin A (TSA) treatment
Aza, a DNA methyltransferase (DNMT) inhibitor, makes DNMT inactivation through DNMT covalent bonding with thiol on cysteine residues, causing reactivation genes silenced by promoter methylation. TSA, a histone deacetylase inhibitor, plays an important role in controlling the tightness of DNA around histone. Combination treatment of Aza and TSA leads the synergistic activation of methylated genes.
CRC cell lines, SW480 and HT-29, were cultured and demethylated. The cells were treated with Aza (Sigma-Aldrich) in the dark at a final concentration of 10 mM/l for 3 days or TSA at a final concentration of 100 mM/l for 1 day, and further treated with or without 100 mM/l TSA (Cayman Chemical, Ann Arbor, MI, USA) for another 1 day.
The establishment of stable cell lines
A ZNF545-expressing plasmid was provided by Professor Q. Tao at the Chinese University of Hong Kong. After SW480 and HT-29 cells were plated into 6-well plates, the cells were transfected with pcDNA3.1-ZNF545 or pcDNA3.1 (+) vectors using Lipofectamine® 2000 reagent (Invitrogen) and cultured with serum-free RPMI-1640 medium. After 4–6 h, the medium containing 10% FBS was changed to a selection medium containing 400 µg/ml G418 (Invitrogen/Gibco) and incubated for 48 h. Approximately 14 days later, the HT-29 and SW480 cells were stably expressing pcDNA3.1 and pcDNA3.1-ZNF545.
The cell proliferation and colony formation assays
SW480 and HT-29 cells stably expressing ZNF545 or pcDNA 3.1 were seeded into 96-well plates at 2,000 cells/well, and the cell viability was measured using MTS (Promega) at 0, 24, 48 and 72 h using a Multiskan Spectrum™ (Tecan, Switzerland). The stably expressing cells were counted and seeded into 6-cell plates at 500 cells/well. Visible cell clones appeared in ~2 weeks, then they were fixed with 4% paraformaldehyde for 30 min and then stained with crystal violet solution for 30 min. Finally, the cells were scanned using a CanoScan 8800F (Canon, Tokyo, Japan).
Flow cytometry of the cell cycle and apoptosis
The cell cycle and apoptosis were measured using flow cytometry as previously described (20). For the cell cycle measurements, SW480 and HT-29 cells stably expressing ZNF545 or pcDNA 3.1 were digested and washed twice using precooled phosphate-buffered saline (PBS), then fixed with ice-cold 70% ethanol, stored at 4°C overnight, and then stained with propidium iodide (PI), followed by analysis using CellQuest software (BD Biosciences, San Jose, CA, USA). After the stably transfected cells were collected and washed twice with PBS, they were stained with Annexin V-FITC/PI in the dark, and the cell apoptosis was measured using CellQuest software, followed by data analyses.
The analyses of wound healing
Stably ZNF545- or vector-transfected SW480 or HT-29 cells were seeded into 6-well plates, and when the cells reached 95% confluency, a sterile P-20 pipette was used to scratch wounds across each well, followed by removal of cell debris using PBS. The cells were then cultured with medium without FBS to minimize cell proliferation. The images of the wound closure areas of SW480 cells were observed at 0, 48, and 60 h, and the images of the wound closure areas of HT-29 cells were observed at 0, 24, 48, and 84 h using a light microscope (CTR4000; Leica, Germany).
Western blot analyses
A total of 40 mg of protein lysate was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to a polyvinylidene fluoride (PVDF) membrane (Bio-Rad). The primary antibodies were against cleaved caspase-3 (12742; Cell Signaling Technology, Danvers, MA, USA), cleaved poly(ADP-ribose) polymerase (PARP) (9541; Cell Signaling Technology), BAX (5023; Cell Signaling Technology), ZNF545 (sc-102235; Santa Cruz Biotechnology, Santa Cruz, CA, USA), PI3K (ab125571; Abcam, Cambridge, UK), phospho-PI3K (Abcam), Akt (4691; Cell Signaling Technology), phospho-Akt (ser473) (4060; Cell Signaling Technology), phospho-GSK3β (ser9) (9323; Cell Signaling Technology), β-catenin (2677; Cell Signaling), active β-catenin (4270; Cell Signaling), c-Myc (1472-1; Epitomic), cyclin D1 (2261; Epitomics), β-actin (LK-ab008-100; Liankebio, China), and Flag (F3165; Sigma-Aldrich).
In vivo tumorigenicity
Six groups of 4-week-old nude mice were used. Empty vector, stably transfected SW480 cells (3×106 cells suspended in 200 µl of PBS) were injected subcutaneously into the left dorsal flank of nude mice, and ZNF545-transfected SW480 cells (3×106 cells suspended in 200 µl of PBS) were injected subcutaneously into the right dorsal flank. After the tumor mass appeared, its length and width were measured every 3 days using a microcaliper. As previously described (23), the tumor volume (mm3) was calculated using the equation: volume = length × width2/2. The mice were euthanized and the tumors were removed when the tumor length reached 1.5 cm. All tumor tissues were weighed before fixation in 4% paraformaldehyde and embedded in paraffin, followed by preparation of the sections for immunohistochemical analyses. All experimental procedures were approved by the Animal Ethics Committee of the Experimental Animal Center of the Chongqing Medical University, Chongqing, China.
Immunohistochemical staining
The samples of excised tumor tissues from nude mice were fixed in 4% paraformaldehyde before being embedded in paraffin, followed by sectioning into 4-µm slices. After the sections were dewaxed for 2 h in a 60°C incubator, dewaxing and alcohol dehydration were performed in the following order: xylene I, xylene II, xylene III, xylene IV, absolute ethyl alcohol, 95% ethyl alcohol, 80% ethyl alcohol, and 70% ethyl alcohol. Antigen retrieval was performed by boiling the slides in citrate buffer solution for 20 min, followed by cooling at room temperature. The sections were then incubated in 3% hydrogen peroxide for 20 min to prevent endogenous peroxidase activity and washed three times with PBS, then incubated in sheep serum for 20 min. The sections were incubated at 4°C overnight with rabbit anti-PCNA monoclonal antibody. The next day, the sections were warmed to room temperature for 2 h, then washed three times with PBS and incubated with rabbit secondary antibody for 20 min. The slices were washed three times with PBS and incubated with horseradish peroxidase-labeled streptomycin anti-biotin antibody for 20 min. The sections were washed three times, followed by color development with DAB. The sections were washed with water, and the cell nuclei were dyed using hematoxylin, followed by observation using light microscopy.
Statistical analysis
All statistical analyses were performed using SPSS statistical software for Windows, version 16. Student's t-test was used to compare the expression of ZNF545 in CRC tissues and adjacent normal tissues. The four table Chi-square test or the t-test were used to analyze categorical variables. A value of p<0.05 was considered statistically significant.
Results
ZNF545 is downregulated in CRC tissues and cell lines
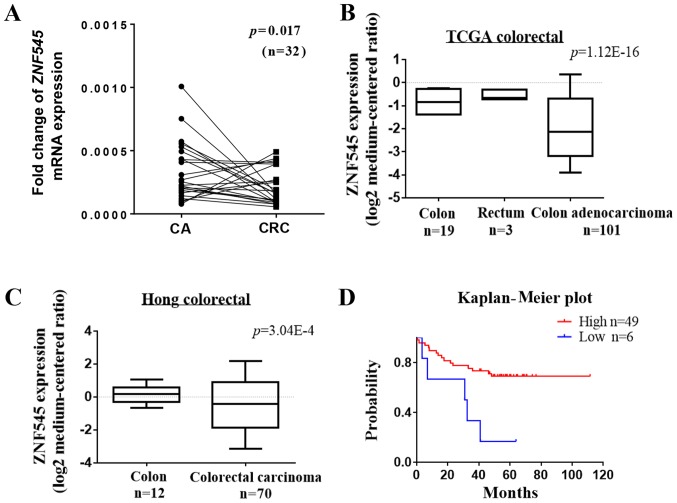
The messenger RNA (mRNA) expression of ZNF545 in CRC and its corresponding adjacent tissues was detected by quantitative PCR. The results showed that the expression of ZNF545 was downregulated in tumor tissues compared with the adjacent tissue (p<0.05, Fig. 1A). RT-PCR was then used to examine the expression of ZNF545 in the HT-29, SW480, HCT116, CoCo-2, and LoVo colorectal cell lines. ZNF545 expression was downregulated or silenced in all cell lines compared with the vector (Fig. 2A). In addition, analyses of an online microarray database (www.oncomine.org, Compendia Bioscience, Ann Arbor, MI, USA) showed that ZNF545 was downregulated in CRC tissues but not in normal adult colorectal tissues (p<0.001; Fig. 1B and C). Prognostic analyses from the PrognoScan database (http://www.prognoscan.org/) showed that patients with higher expression of ZNF545 mRNA had significantly increased relapse-free survival compared with those with lower ZNF545 mRNA levels (Fig. 1D). Taken together, these results suggested that ZNF545 was a cancer suppressor gene.
Figure 1.
The expression and prognosis of ZNF545 in CRC tissues. (A) Expression of ZNF545 mRNA in human CRC and its corresponding adjacent tissues detected by qRT-PCR. (B and C) The expression of ZNF545 (median of expression intensity) in CRC and normal colorectal tissues from Oncomine database. (D) The correlation between ZNF545 expression and CRC patient prognosis. The Prognoscan database was used to compared the ZNF545 expression and patient prognosis.
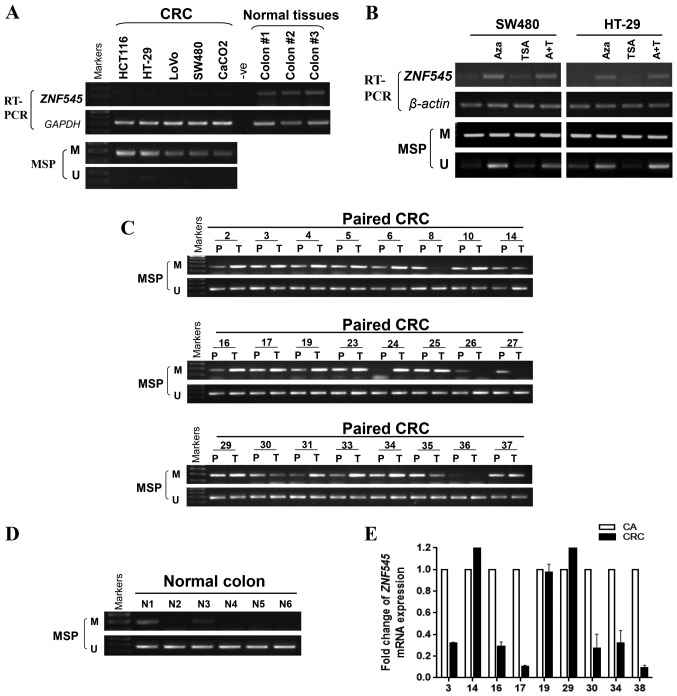
Figure 2.
The expression and methylation status of ZNF545 in CRC cell lines and colorectal tissues. (A) Expression of ZNF545 mRNA in CRC cell lines and normal colorectal tissues, and the methylation status of ZNF545 in CRC cell lines. (B) Pharmacological demethylation of ZNF545 by Aza (A) or TSA (T) restored its expression. ZNF545 expression before and after drug treatment was determined by RT-PCR, and demethylation was confirmed by MSP. (C) The methylation status of the ZNF545 promoter in CRC tissues, CRC adjacent tissues, and (D) normal colon tissues. M, methylated; U, unmethylated. (E) ZNF545 expression levels in 9 paired CRC tissues and adjacent tissues were detected by qPCR. CA, colorectal tumor adjacent tissues; CRC, colorectal cancer tissues.
The ZNF545 promoter was frequently methylated during CRC
Aberrant promoter methylation is a common mechanism involved in tumor suppressor gene silencing in cancers (21,22). To identify whether the decreased expression of ZNF545 was due to promoter methylation, we determined the methylation status of the ZNF545 promoter in five CRC cell lines using MSP. The results showed that ZNF545 was methylated in all cell lines that were associated with silenced or reduced ZNF545 expression, while normal colorectal tissues expressed significantly higher levels of ZNF545 mRNA (Fig. 2A). We then treated SW480 and HT-29 cells which have low-expression of ZNF545 with Aza, TSA and combination, results showed that its expression was restored together with increasing unmethylated alleles of the ZNF545 promoter (Fig. 2B). The results suggested that promoter methylation is directly responsible for ZNF545 downregulation in colorectal cancer cells.
To investigate whether ZNF545 promoter methylation existed in CRC tissues, MSP was used to examine 32 primary colorectal carcinomas and six normal colorectal tissues. The clinical information of colorectal carcinoma tissues are shown in Table II. We observed that ZNF545 methylation was detected in 28 out of 32 (87.5%) CRC tissues, but in only two out of six (33.33%) normal colorectal tissues (Fig. 2C and D and Table III p=0.0116). Nine pairs of primary CRC tissues and para-carcinoma tissues which were randomly selected in the 32 CRC tissues, were also tested for ZNF545 expression by qPCR. Most tumors have lower ZNF545 mRNA expression compared with their paired adjacent tissues (Fig. 2E), while displaying a higher level of promoter methylation.
Table II.
The clinical information of colorectal carcinoma tissues.
| No. | Age | Size (cm) | Sex (1M2F) | Grade | Lymph metastasis | Distant metastasis | Localization | T-stage | N-status | Phase | Pathological type |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CRC2 | 42 | 3,2 | 2 | 3 | 0 | 0 | Middle, down | 3 | N0 | IIA | Ad |
| CRC3 | 78 | 2 | 2 | 2 | 1 | 0 | Middle | 3 | N1 | IIIB | Ad |
| CRC4 | 65 | 2 | 1 | 2 | 0 | 0 | Left | 3 | N1c | IIIB | Ad |
| CRC5 | 63 | 3 | 2 | 3 | 1 | 0 | Left | 4b | N1 | IIIC | Ad |
| CRC6 | 82 | 3,2 | 1 | 2 | 0 | 0 | Right, down | 3 | N0 | IIA | Ad |
| CRC8 | 72 | 2 | 1 | 2–3 | 0 | 0 | Middle | 3 | N0 | IIA | Ad |
| CRC10 | 76 | 2 | 2 | 2 | 0 | 0 | Right | 3 | N0 | IIA | Ad |
| CRC14 | 67 | 2 | 1 | 2–3 | 0 | 0 | Left | 1 | N0 | I | Ad |
| CRC16 | 76 | 2 | 1 | 2 | 0 | 0 | Left | 3 | N0 | IIA | Ad |
| CRC17 | 79 | 2 | 2 | 2 | 4 | 0 | Left | 3 | N2a | IIIB | Ad |
| CRC19 | 55 | 2 | 1 | 2 | 0 | 0 | Left | 3 | N0 | IIA | Ad |
| CRC23 | 76 | 2 | 2 | 2 | 2 | 0 | Left | 3 | N1 | IIIB | Ad |
| CRC24 | 44 | 2 | 2 | X | 0 | 0 | Left | 3 | N0 | IIA | Ad |
| CRC25 | 73 | 2 | 2 | 2 | 0 | 0 | Left | 3 | N0 | IIA | Ad |
| CRC26 | 62 | 3.2 | 1 | 2–3 | 7 | 0 | Left | 3 | N2b | IIIC | Ad |
| CRC27 | 66 | 2 | 1 | X | 1 | 0 | Left | 4b | N1 | IIIC | MN |
| CRC29 | 65 | 2 | 2 | 2 | 5 | 0 | Right | 3 | N2a | IIIB | Ad |
| CRC30 | 78 | 3 | 1 | 2 | 1 | 0 | Left | 3 | N1 | IIIB | Ad |
| CRC31 | 66 | 2 | 1 | 2 | 0 | 1 | Left | 3 | N1c | IVA | Ad |
| CRC33 | 78 | 2 | 2 | 2 | 0 | 0 | Right | 3 | N1 | IIIB | Ad |
| CRC34 | 70 | 2 | 1 | 2–3 | 0 | 0 | Right | 2 | N0 | I | Ad |
| CRC35 | 59 | 2 | 2 | 2 | 0 | 0 | Left | 3 | N0 | IIA | Ad |
| CRC36 | 45 | 2 | 1 | 2 | 0 | 0 | Left | 3 | N0 | IIA | Ad |
| CRC37 | 67 | 2 | 2 | 2 | 0 | 0 | Right | 3 | N0 | IIA | Ad |
| CRC38 | 76 | 2 | 2 | 2 | 1 | 0 | Right | 3 | N1 | IIIB | Ad |
| CRC39 | 64 | 2 | 1 | 2 | 0 | 0 | Left | 3 | N0 | IIA | Ad |
| CRC40 | 43 | 3 | 1 | 2 | 5 | 0 | Right | 3 | N2a | IIIB | Ad |
| CRC41 | 72 | 3 | 1 | 2 | 0 | 0 | Right | 3 | N0 | IIA | Ad |
| CRC42 | 68 | 2 | 1 | 2 | 0 | 0 | Left | 2 | N0 | I | Ad |
| CRC43 | 57 | 3 | 1 | 2 | 0 | 0 | Right | 3 | N0 | IIA | Ad |
| CRC44 | 80 | 2,2,2 | 1 | 2 | 2 | 0 | Left, down | 3 | N1 | IIIB | Ad |
| CRC45 | 54 | 2 | 2 | 2 | 0 | 0 | Right | 3 | N0 | IIA | Ad |
Ad, adenocarcinoma; MN, mixed neurosecretion.
Table III.
Methylation status of the ZNF545 promoter in colorectal cancer.
| Samples |
ZNF545 promoter
|
Frequency of methylation | |
|---|---|---|---|
| Methylation | Unmethylation | ||
| CRC (n=32) | 28 | 4 | 87.5% |
| CN (n=6) | 2 | 4 | 33.3% |
CRC, colorectal cancer; CN, colorectal normal tissues.
Overexpression of ZNF545 inhibits cell proliferation and clonogenicity of CRC cells in vitro
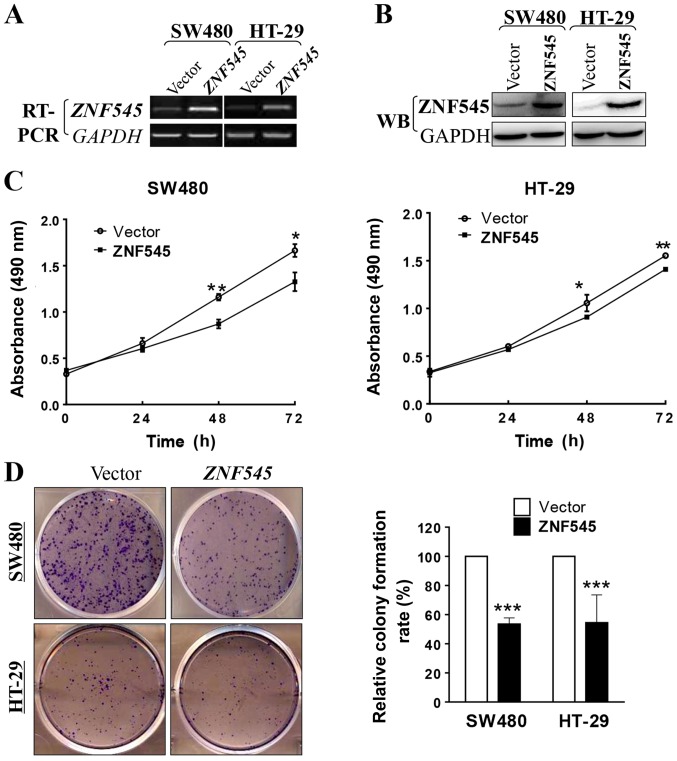
To determine whether ZNF545 is a functional tumor suppressor gene in CRC we used the MTS assay and the colony formation assay to measure the growth-inhibiting effects of overexpressed ZNF545 in SW480 and HT-29 cells. The transfection efficiencies of our ZNF545 constructs were confirmed by RT-PCR and western blotting of tumor cell lines and stable cell lines that were selected, respectively (Fig. 3A and B). The MTS cell proliferation assay showed that the cell viability of the ZNF545-overexpressing cells significantly decreased at 48 and 72 h compared with cells transfected with the empty vector (p<0.05; Fig. 3C). The colony formation assay showed that the colony number of cells transfected with ZNF545 was reduced by ~45% when compared with that transfected with the empty vector in SW480 and HT-29 cells (p<0.001; Fig. 3D), indicating that expression of ZNF545 decreased the cloning efficiency. Together, these results showed that ZNF545 inhibited cell proliferation in CRC.
Figure 3.
Overexpression of ZNF545 inhibits the cell proliferation and colony formation of CRC cells. (A and B) Expression of ZNF545 by RT-PCR and western blot analysis in vector- and ZNF545-transfected SW480 and HT-29 cells. (C) MTS cell proliferation assay for vector- and ZNF545-transfected SW480 and HT-29 cells. Asterisks indicate a significant level of proliferation compared to controls (*p<0.05; **p<0.01). (D) Representative colony formation assay in vector- and ZNF545-transfected SW480 and HT-29 cells. The numbers of the colonies in vector-transfected controls were set to 100%. The values are expressed as mean ± SD from three experiments (***p<0.001).
Overexpression of ZNF545 induces cell cycle arrest and apoptosis of CRC cells
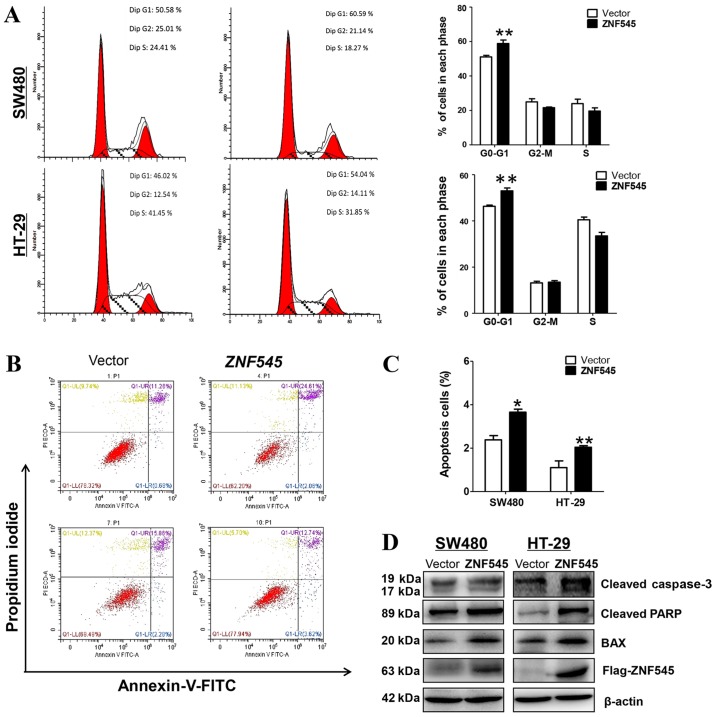
Further studies characterized the effects of ZNF545 on apoptosis and the cell cycle of CRC cells. Fig. 4A shows that the stably-transfected SW480 and HT-29 cells showed a significantly increased number of cells accumulating in the G0/G1 phase when compared with control cells (p<0.01), suggesting that the inhibition of cell proliferation by ZNF545 was likely mediated by a cell cycle arrest at the G0/G1 interface. In addition, flow cytometry showed that the percentage of apoptotic Annexin V-PI-positive cells increased with the overexpression of ZNF545 (Fig. 4B and C).
Figure 4.
Re-expression of ZNF545 induces cell cycle arrest and apoptosis of SW480 and HT-29 cells. (A) The effect of cell cycle distribution of vector- and ZNF545-transfected SW480 and HT-29 cells was detected by flow cytometry analysis. Left, representative flow cytometry plots. Right, histograms of cell cycle alterations. (B) Induction of apoptosis detected by flow cytometric analysis with Annexin V-FITC and PI-staining. (C) Quantity analysis of apoptotic cells. Statistical signifcance was assessed by Student's t-test. The graph displays the mean ± SD of three independent experiments. *p<0.05; **p<0.01. (D) Western blot analysis of apoptosis-related proteins in vector-, ZNF545-transfected SW480 and HT-29 cells. β-actin was used as a control.
We also determined the apoptosis of CRC cell lines by western blot analyses of Bax, the active form of caspase-3, and PARP. Fig. 4D shows the increased expression of Bax, cleaved-caspase-3, and cleaved-PARP in both cell lines when compared with cells transfected with the control vector. Together, these results suggested that ZNF545 inhibited cell proliferation by apoptosis and by mediating the cell cycle arrest at the G0/G1 interface.
ZNF545 inhibits CRC cell migration
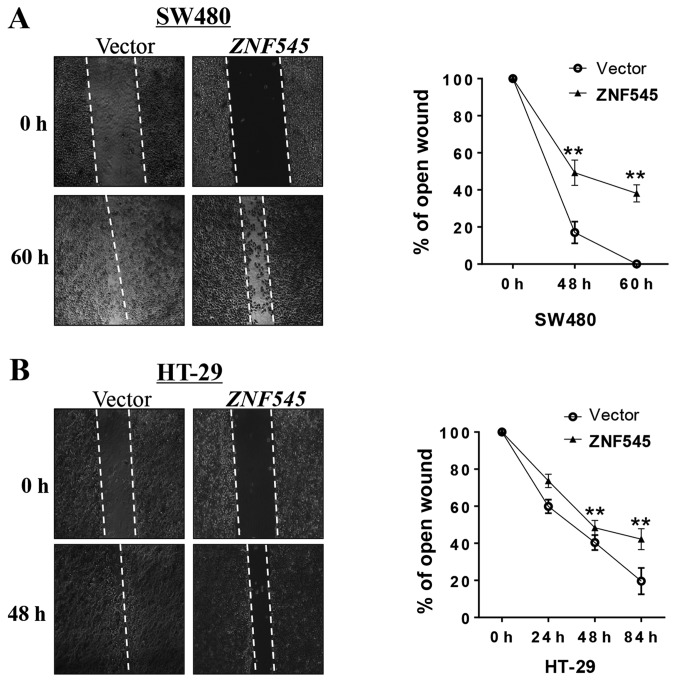
A wound-healing assay was performed to test the effect of ZNF545 on CRC cell migration. Cells expressing ZNF545 spread along the wound edges significantly more slowly than the control cells (Fig. 5), suggesting that ZNF545 suppressed the migration of SW480 and HT-29 cells.
Figure 5.
ZNF545 suppresses the cell motility in SW480 and HT-29 cells. (A and B) Left, representative images of wound healing assay. Right, the percentage of wound healing compared with that of controls at each time-point as indicated, asterisks indicate a significant level of cell motility compared to controls, Mean ± SD of three independent experiments (**p<0.01).
ZNF545 inhibits the growth of tumor xenografts in nude mice
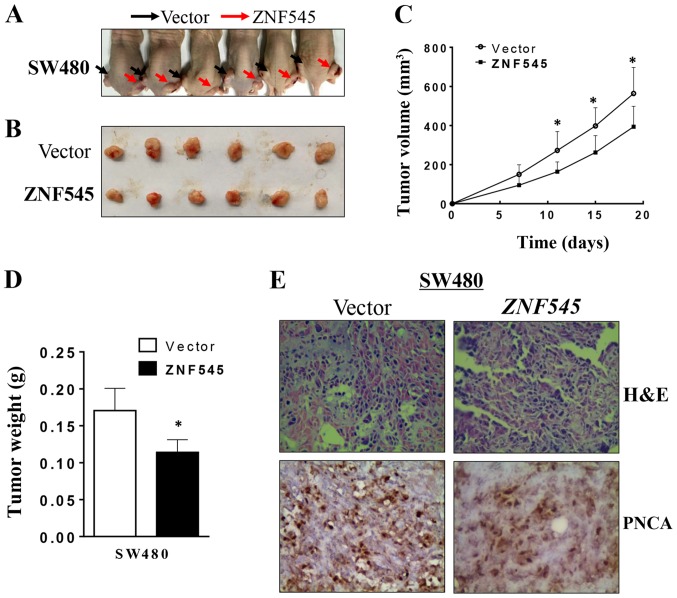
We also determined if ZNF545 could suppress the growth of CRC cells in vivo. SW480 cells stably transfected with ZNF545 or an empty vector formed tumors in nude mice (Fig. 6A). The subcutaneous tumor growth curve of SW480 cells stably-transfected with ZNF545 or with empty vector showed a significant difference (p<0.05; Fig. 6B), and the tumor volumes from the ZNF545-treated animals were significantly smaller than that of the control group (p<0.05; Fig. 6C). Moreover, the mean weight of the tumors formed by ZNF545-transfected cells was significantly lower than that of tumors formed from the empty vector control cells (p<0.05; Fig. 6D), suggesting that ZNF545 inhibited the growth of CRC cells in vivo. After using PCNA to stain the tissue sections from nude mice, immunohistochemical analyses showed that cell proliferation in the tumors from empty vector-treated cells was greater than that of tumors from the ZNF545-treated animals (Fig. 6E).
Figure 6.
ZNF545 inhibits tumorigenicity of CRC in vivo. (A and B) Representative images of tumor growth in nude mice. Left dorsal flank of nude mice is the vector group, right dorsal flank of nude mice is overexpression ZNF545 group. (C) Growth curve for the ZNF545-expressing tumor compared to the control in nude mice. (D) Tumor weight of ZNF545 expressing cells in nude mice compared with control tumors, *p<0.05. (E) Representative images of H&E staining and IHC analyses of the expression of PCNA in tumors from nude mice.
The tumor suppressive property of ZNF545 was mediated by the Wnt/β-catenin, PI3K/AKT, and MAPK/ERK signaling pathways
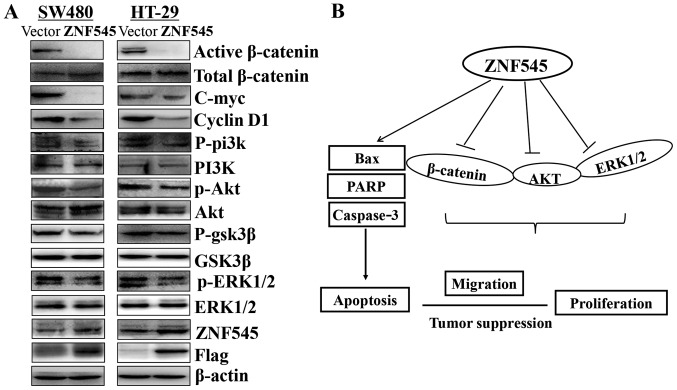
Multiple zinc finger family members have been reported to interact the β-catenin activity, and we investigated whether ZNF545 functions as a tumor suppressor through Wnt/β-catenin signaling pathway. The results showed that the expression of active-catenin was downregulated. Moreover, we characterized the effects of ZNF545 on the PI3K/Akt and ERK signaling pathways. The results showed that phospho-AKT and phospho-ERK1/2 were downregulated, indicating that the two pathways also participated in the carcinogenic process. GSK3β is a downstream target gene of the PI3K/AKT pathway and ZNF545 resulted in a profound reduction of GSK3β (Fig. 7A).
Figure 7.
The mechanism of ZNF545 acts on CRC cell lines. (A) Overexpression of ZNF545 in SW480 and HT-29 cells disrupted Wnt/β-catenin, PI3K/AKT, and MAPK/ERK1/2 signaling pathways. Western blot analysis of β-catenin, AKT, ERK1/2, and the related downstream targets in SW480 and HT-29 cells. β-actin was used as an internal control. (B) The schematic diagram of antitumorigenesis function mechanisms of ZNF545 deriving from the western blot analysis data.
Discussion
Increasing evidence has shown that ZNF545 plays an important role in the development of various cancers. ZNF545 is abnormally expressed, and acts as a suppressor gene in some tumors. For example, ZNF545 was poorly expressed in breast cancer (19) and gastric cancer (23), and suppressed breast cancer and gastric cancer proliferation. Furthermore, the Oncomine database showed that ZNF545 was significantly downregulated in multiple types of CRC tissues when compared to normal adult colorectal tissues. In the present study, we found that the expression of ZNF545 was frequently downregulated in CRC cells, as well as in primary colorectal tumor tissues when compared with surgical margin tissues, suggesting that ZNF545 may be a tumor suppressor in CRC.
It has been reported that the decreased expression of ZNF545 is related to its methylation, and it is generally accepted that promoter methylation is a major mechanism involved in gene changes such as gene silencing or gene downregulation that have been found in human malignant tumors (21). Using MSP and a demethylation treatment, we found that the reduced expression of ZNF545 was associated with methylation of the promoter, suggesting that DNA methylation is a regulatory mechanism of ZNF545 inactivation in CRC.
It has been reported that the methylation of ZNF545 can be used as a prognostic marker for early stage hepatocellular carcinoma (HCC) after thermal ablation (24), and low expression of ZNF545 was determined positively correlated with the poor prognosis in CRC patients through analyzing the PrognoScan database. However, there was an insufficient number of clinical samples of CRC in our study, so we could not determine the possible relationship between ZNF545 methylation and the pathological features and survival prognoses of CRCs. Further analyses of more clinical samples is therefore necessary to determine if ZNF545 is indeed a prognostic marker for CRC.
The biological functions of ZNF545 in CRC were also studied by in vitro and in vivo overexpression of ZNF545. Previous studies reported that expression of ZNF545 inhibited clone formation and induced apoptosis in HCT116 CRC cells (18). The results of the present study showed that ZNF545 affected apoptosis and G0/G1 cell cycle arrest to inhibit cell proliferation and clone formation in vitro. Furthermore, using the wound healing assay, we showed that overexpression of ZNF545 markedly inhibited cell migration. The inhibition of tumor growth by overexpression of ZNF545 was also tested in vivo, by monitoring the formation of tumor xenografts in nude mice. Flow cytometry and western blot analyses were used to show the induction of apoptosis by expression of ZNF545 in CRC cells. Flow cytometry showed that expression of ZNF545 resulted in early apoptosis in CRC cells, and western blot analyses showed increased expression of Bax, cleaved caspase-3, and cleaved PARP. Taken together, the results suggested that ZNF545 functions as a tumor suppressor in CRC.
The members of KRAB-ZFPs family have N-terminal KRAB domain and C-terminal C2H2 domain, and the C2H2 domain can function as structural motif to bind to the promoter of DNA or RNA. Previous studies have reported that many ZFPs influence cell biological function through Wnt/β-catenin pathway, such as ZNF488, ZNF191, KLF4 and ZIC2. ZNF488 acts as an oncogene via the Wnt/β-catenin pathway to induce EMT in nasopharyngeal carcinoma (25). ZNF191 can directly bind to the β-catenin promoter and invoke the expression of β-catenin to promote cell proliferation in hepatoma cell lines (26). Recent report showed that the KLF4 binds to the β-catenin through its C-terminus which contains three zinc-finger domains, inhibits Wnt/β-catenin signaling pathway and suppresses cell proliferation in colorectal cancer (27). The zinc finger domain of ZIC2 is required for the interaction with TCF4, and ZIC2 can modulate Wnt pathway in 293T cells (28). Whether ZNF545 performs a similar mechanism that acts on Wnt/β-catenin pathway in colorectal cancer remains unclear.
Wnt signaling plays an important role in the formation and maintenance of human intestinal epithelium (29). The deregulated Wnt signaling is an early event in colon carcinogenesis (30). Our data showed that re-expression of ZNF545 inhibited the activated β-catenin and its downstream target genes in colorectal cancer cell lines.
It has been reported that the PI3K/AKT and MAPK/ERK pathways can promote the biological function of proliferation, and migration in multiple cancer cells (31–33). AKT is a key component in the PI3K pathway, where it suppresses cell survival and growth (34). Our results showed that when compared to the empty vector group, the expression level of phosphorylated AKT and phosphorylated GSK-3β were decreased in the ZNF545 group. The total amount of ERK1/2 protein was unchanged in the ZNF545- and vector-transfected cells, but phosphorylated ERK1/2 levels decreased in the ZNF545-transfected cells. Thus, ZNF545 may inhibit the PI3K/Akt/GSK3β and MAPK/ERK pathways to reduce the cell proliferation and migration ability in colorectal cancer cells. Identification of the target gene of ZNF545 (RNA sequence or Chip sequence) will therefore assist in identifying its role in the progression of CRC.
In conclusion, these results suggested that ZNF545 functions as a tumor suppressor in CRC, and that its inactivation results from promoter methylation. ZNF545 may be an epigenetic biomarker for colorectal tumors. Overall, ZNF545 can regulate cell proliferation, apoptosis, and metastasis by affecting the Wnt/β-catenin, PI3K/AKT, and MAPK/ERK signaling pathways.
Acknowledgments
The authors thank Professor Qian Tao (the Chinese University of Hong Kong, Hong Kong, China) for generously providing CRC cell lines, plasmid and primers, technical assistance, and help with the whole design of the experiment. This study was supported by National Natural Science Foundation of China (no. 31420103915) and special research funds from The Chinese University of Hong Kong. This study was also supported by National Key Clinical Specialties Construction Program of China (no. [2013] 544).
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics. CA Cancer J Clin. 2012;2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2016;2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dehan P, Kustermans G, Guenin S, Horion J, Boniver J, Delvenne P. DNA methylation and cancer diagnosis: New methods and applications. Expert Rev Mol Diagn. 2009;9:651–657. doi: 10.1586/erm.09.53. [DOI] [PubMed] [Google Scholar]
- 6.Shivapurkar N, Gazdar AF. DNA methylation based biomarkers in non-invasive cancer screening. Curr Mol Med. 2010;10:123–132. doi: 10.2174/156652410790963303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4:231. doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lupo A, Cesaro E, Montano G, Zurlo D, Izzo P, Costanzo P. KRAB-zinc finger proteins: A repressor family displaying multiple biological functions. Curr Genomics. 2013;14:268–278. doi: 10.2174/13892029113149990002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huntley S, Baggott DM, Hamilton AT, Tran-Gyamfi M, Yang S, Kim J, Gordon L, Branscomb E, Stubbs L. A comprehensive catalog of human KRAB-associated zinc finger genes: Insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006;16:669–677. doi: 10.1101/gr.4842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cowger JJ, Zhao Q, Isovic M, Torchia J. Biochemical characterization of the zinc-finger protein 217 transcriptional repressor complex: Identification of a ZNF217 consensus recognition sequence. Oncogene. 2007;26:3378–3386. doi: 10.1038/sj.onc.1210126. [DOI] [PubMed] [Google Scholar]
- 11.Qiang W, Zhao Y, Yang Q, Liu W, Guan H, Lv S, Ji M, Shi B, Hou P. ZIC1 is a putative tumor suppressor in thyroid cancer by modulating major signaling pathways and transcription factor FOXO3a. J Clin Endocrinol Metab. 2014;99:E1163–E1172. doi: 10.1210/jc.2013-3729. [DOI] [PubMed] [Google Scholar]
- 12.Huang X, Yuan W, Huang W, Bai Y, Deng Y, Zhu C, Liang P, Li Y, Du X, Liu M, et al. ZNF569, a novel KRAB-containing zinc finger protein, suppresses MAPK signaling pathway. Biochem Biophys Res Commun. 2006;346:621–628. doi: 10.1016/j.bbrc.2006.05.109. [DOI] [PubMed] [Google Scholar]
- 13.Huang GJ, He Z, Ma L. ZFP932 suppresses cellular Hedgehog response and Patched1 transcription. Vitam Horm. 2012;88:309–332. doi: 10.1016/B978-0-12-394622-5.00014-6. [DOI] [PubMed] [Google Scholar]
- 14.Li A, Jiao Y, Yong KJ, Wang F, Gao C, Yan B, Srivastava S, Lim GS, Tang P, Yang H, et al. SALL4 is a new target in endometrial cancer. Oncogene. 2015;34:63–72. doi: 10.1038/onc.2013.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tune CE, Pilon M, Saiki Y, Dosch HM. Sustained expression of the novel EBV-induced zinc finger gene, ZNFEB, is critical for the transition of B lymphocyte activation to oncogenic growth transformation. J Immunol. 2002;168:680–688. doi: 10.4049/jimmunol.168.2.680. [DOI] [PubMed] [Google Scholar]
- 16.Nakaya T, Ogawa S, Manabe I, Tanaka M, Sanada M, Sato T, Taketo MM, Nakao K, Clevers H, Fukayama M, et al. KLF5 regulates the integrity and oncogenicity of intestinal stem cells. Cancer Res. 2014;74:2882–2891. doi: 10.1158/0008-5472.CAN-13-2574. [DOI] [PubMed] [Google Scholar]
- 17.Wang S, Cheng Y, Du W, Lu L, Zhou L, Wang H, Kang W, Li X, Tao Q, Sung JJ, et al. Zinc-finger protein 545 is a novel tumour suppressor that acts by inhibiting ribosomal RNA transcription in gastric cancer. Gut. 2013;62:833–841. doi: 10.1136/gutjnl-2011-301776. [DOI] [PubMed] [Google Scholar]
- 18.Cheng Y, Liang P, Geng H, Wang Z, Li L, Cheng SH, Ying J, Su X, Ng KM, Ng MH, et al. A novel 19q13 nucleolar zinc finger protein suppresses tumor cell growth through inhibiting ribosome biogenesis and inducing apoptosis but is frequently silenced in multiple carcinomas. Mol Cancer Res. 2012;10:925–936. doi: 10.1158/1541-7786.MCR-11-0594. [DOI] [PubMed] [Google Scholar]
- 19.Xiao Y, Xiang T, Luo X, Li C, Li Q, Peng W, Li L, Li S, Wang Z, Tang L, et al. Zinc-finger protein 545 inhibits cell proliferation as a tumor suppressor through inducing apoptosis and is disrupted by promoter methylation in breast cancer. PLoS One. 2014;9:e110990. doi: 10.1371/journal.pone.0110990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Li J, Cui Y, Li T, Ng KM, Geng H, Li H, Shu XS, Li H, Liu W, et al. CMTM3, located at the critical tumor suppressor locus 16q22.1, is silenced by CpG methylation in carcinomas and inhibits tumor cell growth through inducing apoptosis. Cancer Res. 2009;69:5194–5201. doi: 10.1158/0008-5472.CAN-08-3694. [DOI] [PubMed] [Google Scholar]
- 21.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 22.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 23.Deng J, Liang H, Ying G, Dong Q, Zhang R, Yu J, Fan D, Hao X. Poor survival is associated with the methylated degree of zinc-finger protein 545 (ZNF545) DNA promoter in gastric cancer. Oncotarget. 2015;6:4482–4495. doi: 10.18632/oncotarget.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu J, Li X, Tao Q, Yu XL, Cheng ZG, Han ZY, Guo M, Liang P. Hypermethylation of ZNF545 is associated with poor prognosis in patients with early-stage hepatocellular carcinoma after thermal ablation. Gut. 2015;64:1836–1837. doi: 10.1136/gutjnl-2015-310258. [DOI] [PubMed] [Google Scholar]
- 25.Zong D, Yin L, Zhong Q, Guo WJ, Xu JH, Jiang N, Lin ZR, Li MZ, Han P, Xu L, et al. ZNF488 enhances the invasion and tumorigenesis in nasopharyngeal carcinoma via the Wnt signaling pathway involving epithelial mesenchymal transition. Cancer Res Treat. 2016;48:334–344. doi: 10.4143/crt.2014.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu G, Jiang S, Wang C, Jiang W, Liu Z, Liu C, Saiyin H, Yang X, Shen S, Jiang D, et al. Zinc finger transcription factor 191, directly binding to β-catenin promoter, promotes cell proliferation of hepatocellular carcinoma. Hepatology. 2012;55:1830–1839. doi: 10.1002/hep.25564. [DOI] [PubMed] [Google Scholar]
- 27.Zhang W, Chen X, Kato Y, Evans PM, Yuan S, Yang J, Rychahou PG, Yang VW, He X, Evers BM, et al. Novel cross talk of Kruppel-like factor 4 and beta-catenin regulates normal intestinal homeostasis and tumor repression. Mol Cell Biol. 2006;26:2055–2064. doi: 10.1128/MCB.26.6.2055-2064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pourebrahim R, Houtmeyers R, Ghogomu S, Janssens S, Thelie A, Tran HT, Langenberg T, Vleminckx K, Bellefroid E, Cassiman JJ, et al. Transcription factor Zic2 inhibits Wnt/β-catenin protein signaling. J Biol Chem. 2011;286:37732–37740. doi: 10.1074/jbc.M111.242826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doerks T, Copley RR, Schultz J, Ponting CP, Bork P. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 2002;12:47–56. doi: 10.1101/gr.203201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 31.Datta SR, Brunet A, Greenberg ME. Cellular survival: A play in three Akts. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 32.Qing H, Gong W, Che Y, Wang X, Peng L, Liang Y, Wang W, Deng Q, Zhang H, Jiang B. PAK1-dependent MAPK pathway activation is required for colorectal cancer cell proliferation. Tumour Biol. 2012;33:985–994. doi: 10.1007/s13277-012-0327-1. [DOI] [PubMed] [Google Scholar]
- 33.Levidou G, Saetta AA, Gigelou F, Karlou M, Papanastasiou P, Stamatelli A, Kavantzas N, Michalopoulos NV, Agrogiannis G, Patsouris E, et al. ERK/pERK expression and B-raf mutations in colon adenocarcinomas: Correlation with clinicopathological characteristics. World J Surg Oncol. 2012;10:47. doi: 10.1186/1477-7819-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khor TO, Gul YA, Ithnin H, Seow HF. Positive correlation between overexpression of phospho-BAD with phosphorylated Akt at serine 473 but not threonine 308 in colorectal carcinoma. Cancer Lett. 2004;210:139–150. doi: 10.1016/j.canlet.2004.01.017. [DOI] [PubMed] [Google Scholar]