Abstract
Gallbladder cancer (GBC) is one of the most unfavorable prognostic tumor, and immediate growth and distant metastasis are important factors associated with the poor prognosis of patients with this disease. Standard and variant isoforms of CD44 are associated with tumor growth, metastasis, and epithelial-mesenchymal transition (EMT), although their roles in GBC are unclear. We investigated the relationship between the CD44 isoforms with EMT, chemotaxis, and tumorigenicity. We analyzed CD44 expression in the GBC cell line NOZ and found that it comprises a major population that expressed CD44std+/CD44v9− (CD44s) and the minor population that expressed CD44std−/CD44v9+ (CD44v). CD44s cells exhibited increased chemotaxis and invasiveness compared with CD44v cells in in vitro cell migration and invasion assays. CD44s cells expressed higher and lower levels of mRNAs that encode vimentin and E-cadherin, respectively, compared with those of CD44v cells. CD44s cells expressed high levels of the transcription factors ZEB1 and ZEB2 that mediate EMT, and low levels of a splicing factor ESRP1 that controls the CD44 isoform switch. We performed in vivo mouse xenotransplantation analyses of CD44s and CD44v cells and found that CD44v cells exhibited relatively increased tumorigenicity. Immunohistochemical analysis of tissue microarrays revealed that high levels of CD44v9 and CD44std were associated with poorer prognosis. The expression of CD44std was also associated with poorly differentiated tumors and distant metastasis. In conclusion, CD44s was associated with a mesenchymal phenotype, increased chemotaxis and invasiveness, and decreased tumorigenicity. In contrast, CD44v cells exhibited an epithelial phenotype, decreased chemotaxis, decreased invasiveness, and increased tumorigenicity. These findings suggest that CD44v and CD44s cells play differently important roles in the progression and metastasis of GBC and the isoform switch triggers EMT.
Keywords: gallbladder cancer, CD44 standard isoform, CD44 variant 9 isoform, epithelial-mesenchymal transition, tumorigenicity, chemotaxis, flow cytometry
Introduction
Gallbladder cancer (GBC) is one of the intra-abdominal malignant tumors with the most unfavorable prognosis (5-year overall survival rate of <10%) (1,2). The only potentially curative therapy for GBC is surgical resection, including simple cholecystectomy, radical or extended cholecystectomy (sometimes accompanied by liver resection), bile duct resection, and pancreaticoduodenectomy. However, few patients undergo curative surgery because of distant metastasis or locally excessive invasion. Understanding the mechanisms of metastasis of GBC is, therefore, important to improve the survival of patients with GBC.
CD44 (also called H-CAM) is a type I transmembrane glycoprotein receptor that binds hyaluronic acid (3–5). The functions of CD44 include cellular adhesion, lymphocyte activation, homing, and migration. Further, CD44 can contribute to tumor progression, drug resistance, and cancer stem cell function (6). The gene encoding CD44 comprises 20 exons, with 10 in the variant region that generate isoforms through alternative splicing (6,7). The standard isoform is encoded by exons 1–5 and exons 16–20 and is the isoform with the smallest molecular weight. Seven of the standard exons encode the extracellular region, another exon is the transmembrane region, and other two domains comprise the cytoplasmic tail. Variant exons (exon 6–15, called as v1-v10) are alternatively spliced and inserted into the extracellular standard exons to generate variant isoforms. Accordingly, standard and variant isoforms harbor the same antigenic region.
CD44 variant 9 (CD44v9) is associated with the proliferation of cancer cells, inhibition of apoptosis, chemoresistance, EMT, and poor prognosis. CD44v9 expression is associated with the recurrence of primary early gastric cancer (8) as well as with the poor prognosis of patients with pancreatic cancer (9) or hepatocellular carcinoma (10). The CD44 variant isoform comprising exons v8–10 promotes the uptake of cystine for synthesis of the major antioxidant glutathione and contributes to the elimination of excess reactive oxygen species (11), which increases resistance to chemotherapy and radiotherapy. CD44v9 and CD44v6 interact with FAS, a programmed cell death receptor that inhibits apoptosis induced by its ligand FASL (12,13). Cancer cells that express CD44v9 resist chemotherapy by expressing the multidrug resistance protein 1 (14,15).
CD44 expression affects tumorigenicity or the EMT of GBC cells; however, there are few reports on whether CD44 isoforms are associated with these characteristics (16–18). To fill this gap in our knowledge, the aim of the present study was to investigate the relationship between the CD44 isoforms expressed by the GBC cells with EMT, chemotaxis, and tumorigenicity.
Materials and methods
Human GBC cell line and culture conditions
The human GBC cell line NOZ (19) was purchased from the Japanese Collection of Research Bioresources (Osaka, Japan). NOZ cells were cultured in a humidified atmosphere containing 5% CO2 at 37°C in DMEM/Ham's F-12 medium (Wako, Osaka, Japan) containing 10% fetal bovine serum (FBS) (Gibco/Thermo Fisher Scientific, Waltham, MA, USA) and 1% Antibiotic-Antimycotic 100× (Life Technologies, Carlsbad, CA, USA).
Flow cytometry and cell sorting
Cell surface proteins were detected using an allophycocyanin (APC)-conjugated anti-human CD44 monoclonal antibody (clone BD105, catalog number 130-095-177; Miltenyi Biotec GmbH, Bergisch Gladbach, Germany), a fluorescein isothiocyanate (FITC)-conjugated anti-human CD44 standard monoclonal antibody (CD44std; clone SFF-2, catalog number BMS113FI; Affymetrix Inc., Santa Clara, CA, USA), and an anti-human CD44v9 rat monoclonal primary antibody (clone RV3, catalog number LKG-M001; Cosmo Bio Co., Ltd., Tokyo, Japan). The secondary antibody used to detect CD44v9 was an APC-conjugated anti-rat IgG2a mouse monoclonal clone RG7/1.30 (catalog number 130-104-736; Miltenyi Biotec GmbH).
A FACS Aria II cell sorter and BD FACS Diva software (BD Biosciences, San Jose, CA, USA) were used for cell sorting. Monolayers of NOZ cells were dissociated using 0.025% Trypsin containing 0.01% EDTA (Life Technologies) and suspended in PBS containing 0.5% BSA. Cells were reacted with FITC or APC-conjugated antibodies according to the manufacturer's instructions. These cells were also stained with 7-aminoactinomycin D (7-AAD; Bio-Rad, Richmond, CA, USA) to exclude dead cells. All experiments were repeated at least five times. Cells were sorted to isolate populations of CD44std+-CD44v9− cells (CD44s) and CD44std−-CD44v9+ cells (CD44v), which were each cultured to confirm viability and proliferation. The sorted cells were used in the following study. To analyze sequential expression of cell surface markers, cells were reacted with primary antibodies and re-sorted after 1 or 2 weeks of culture, which was repeated three times using the same procedure.
Transwell migration assay and invasion assay
The protocol used to analyze in vitro cell migration was based on the transwell migration assay (Boyden chamber assay) (20,21). Falcon cell culture inserts (Corning Inc., Corning, NY, USA) with a porous membrane (pore-size 8 μm) were placed into a 24-well plate (Corning Inc.). The attractant (medium containing 10% FBS) was added to the lower chamber, and sorted NOZ cells (50,000 cells) were suspended in serum-free medium and added to the upper chamber. The cells were incubated at 37°C in an atmosphere containing 5% CO2 for 24 h. The migrated cells attached to the lower side of the membrane were fixed and stained using a Differential Quik Stain kit (Polysciences, Warminster, PA, USA). Cells remaining on the upper side of membrane were removed with a cotton swab. The membranes containing the migrated cells were dried, and the cells were counted in three randomly selected fields (×200). The experiments were performed in triplicate, and each experiment was independently repeated three times.
Analysis of cell invasion was performed using a Matrigel Transwell invasion assay (22). Cell culture inserts with porous membranes (pore-size 8 μm) were coated overnight with 0.5 mg/ml Matrigel (Corning Inc.). The inserts were placed into a 24-well plate and the attractant (medium containing 10% FBS) was added to the lower chamber and sorted NOZ cells (50,000 cells) were suspended in serum-free medium and added to the upper chamber. The cells were incubated at 37°C in an atmosphere containing 5% CO2 for 24 h. The invaded cells attached to the lower side of the membrane were fixed and stained using the Differential Quik Stain kit (Polysciences). Cells remaining on the upper side of the membrane were removed with a cotton swab, and the membranes were dried. The number of invaded cells was counted in three randomly selected fields (×200). The experiments were performed in triplicate, and each experiment was independently repeated three times.
RNA extraction and quantitative real-time PCR
Total RNA was extracted using a NucleoSpin RNA column (Takara Bio Inc., Shiga, Japan), and cDNA was synthesized using a PrimeScript II 1st Strand cDNA Synthesis kit (Takara Bio) according to manufacturer's instructions. Quantitative real-time PCR was performed using SYBR Premix Ex Taq II (Takara Bio) and an Mx3000P real-time qPCR system (Agilent Technologies, Santa Clara, CA, USA). The ΔΔCt method was used to calculate mRNA levels (23). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was used as an internal control. Each experiment was performed in triplicate and independently repeated three times. The primer sequences were as follows: E-cadherin, 5′-GTCTGTCATGGAAGGTG CT-3′ (forward), 5′-TACGACGTTAGCCTCGTTC-3′ (reverse); Vimentin, 5′-AGTCCACTGAGTACCGGAGAC-3′ (forward), 5′-CATTTCACGCATCTGGCGTTC-3′ (reverse); ZEB1 (24), 5′-CAGCTTGATACCTGTGAATGGG-3′ (forward), 5′-TATCTGTGGTCGTGTGGGACT-3′ (reverse); ZEB2 (25), 5′-GCG ATGGTCATGCAGTCAG-3′ (forward), 5′-CAGGTGGCAG GTCATTTTCTT-3′ (reverse); ESRP1 (26), 5′-CAATATTG CCAAGGGAGGTG-3′ (forward), 5′-GTCCCCATGTGATG TTTGTG-3′ (reverse); GAPDH, 5′-AGCCTCAAGATCATCAGCAATGCC- 3′ (forward), 5′-TGTGGTCATGAGTCCTTCCACGAT-3′ (reverse); CD44 5′-AGCATCGGATTTGAGACCTG-3′ (forward), 5′-GTTGTTTGCTGCACAGATGG-3′ (reverse); CD44s (27), 5′-AAAGGAGCAGCACTTCAGGA-3′ (forward), 5′-TGTGTCTTGGTCTCTGGTAGC-3′ (reverse); CD44v9, 5′-ACCATCCAACAACTTCTACTCTGACA-3′ (forward), 5′-CCTTCAGAATGATTTGGGTCTCTT-3′ (reverse).
Tumorigenicity assay
Six-week-old female nude mice (BALB/c-nu, Charles River Laboratories Japan, Yokohama, Japan) were used for the tumorigenicity assay. Sorted NOZ cells were suspended in a 1:1 (v/v/) mixture of PBS and Matrigel (Corning Inc.). The mice were anesthetized using sevoflurane, and the suspensions of sorted cells (100–10,000 cells/0.1 ml) were injected subcutaneously into the right and left lumbar regions. The mice were maintained under standard conditions specified by institutional guidelines for animal care. The mice that were injected with 10,000 cells were sacrificed 4 weeks later, and tumor sizes and weights were measured. Tumor volumes were calculated as follows: Tumor volume = (long axis length in millimeters/2) × (short axis length/2)2 × 4π/3. The other mice were observed for 8 weeks when palpable tumors were observed. Tumors were histologically confirmed using hematoxylin and eosin (H&E) staining.
Tissue samples and tissue microarray (TMA) analysis
We collected a series of paraffin-embedded tissues specimens from 52 patients with GBC who underwent surgery between 1997 and 2010 at the Toyama University Hospital, Japan. The tumors were histologically diagnosed in the Department of Pathology. The final stage of GBC was confirmed pathologically according to the TNM Classification of Malignant Tumors, 7th edition. The ethics committee of the University of Toyama approved this study.
The TMA comprised 1.0-mm cores of tissues from the paraffin-embedded blocks of the surgical specimen described above. Paraffin blocks containing tumor tissue were selected, and the most representative areas encompassing the tumors were marked directly on the blocks according to the corresponding HE-stained slides. The array block was cut into sections that were placed onto glass slides for H&E staining and immunohistochemical analyses (28–30).
Immunohistochemical staining
The primary antibodies used for the immunohistochemical staining were as follows: anti-cytokeratin OSCAR polyclonal antibody (dilution 1:200, Abcam, Cambridge, UK) and anti-human CD44v9 monoclonal antibody (clone RV3; dilution 1:200, Cosmo Bio Co., Ltd.), CD44std (clone SFF-2, Affymetrix Inc.). The cancer cells were identified using the anti-cytokeratin OSCAR antibody, then CD44v9 and CD44std expression was detected in the membrane of cancer cells. The TMA score was calculated from staining intensity and the distribution score. The criteria for scoring were as follows: the staining distribution was scored as 0 (0–10%), 1 (11–50%), and 2 (51–100%) to indicate the percentage of tumor cells present in one tissue section. Staining intensity was scored as 0 (no staining of cancer cells), 1 (weak), 2 (moderate), and 3 (marked). We defined CD44v9-high/low and CD44std-high/low expression when the sum of the staining distribution and intensity score was ≥4/≤3 and ≥3/≤2, respectively. Two researchers uninformed of patient clinicopathological information independently scored the tissues, and when there was discordance, the investigators re-evaluated the data to assign a consensus score.
Statistical analysis
Data are expressed as the mean ± standard deviation (SD). The Wilcoxon rank sum test and Fisher's exact test were used to compare the differences between groups. Overall survival (OS) was analyzed using the Kaplan-Meier method and the log-rank test. Statistical analysis was performed using JMP Pro 11.2 (SAS Institute Inc., Cary, NC, USA) and p-values <0.05 were considered significant.
Results
The NOZ cell line comprises two populations that express different CD44 isoforms
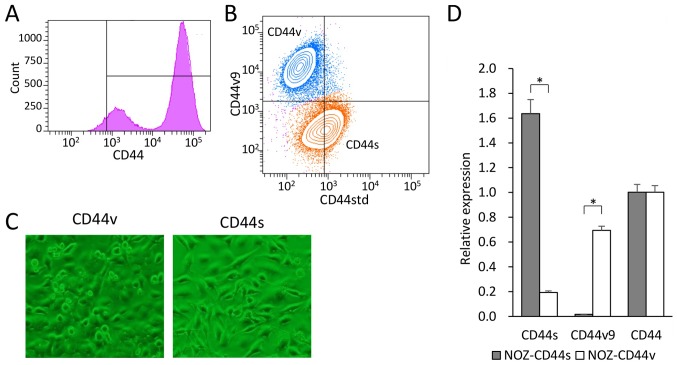
We used flow cytometry to analyze the cell surface expression of CD44 in the GBC cell line NOZ. CD44 expression was observed in 90% of the NOZ cells and showed a bimodal distribution (Fig. 1A). Double-staining analysis of CD44std and CD44v9 expression in the NOZ cell line revealed two populations (Fig. 1B). CD44-high cells accounted for approximately 70% of the NOZ cells, which expressed CD44std+ and CD44v9−. In contrast, CD44-low cells accounted for approximately 30% of the cells, which expressed CD44v9+ and CD44std−. We defined CD44s cells as CD44std+-CD44v9−, and CD44v cells as CD44std−-CD44v9+. Each group was sorted, collected, and used in the following experiments.
Figure 1.
(A) Histogram of cell surface CD44 expression in the human gallbladder cancer cell line NOZ. (B) Double-staining analysis of CD44std and CD44v9. CD44std+-CD44v9− (CD44s, orange) cells and CD44std−-CD44v9+ (CD44v, blue) cells were sorted. (C) Morphology of sorted NOZ-CD44v and CD44s cells cultured under normal conditions. (D) Reverse transcription-PCR analysis of mRNAs encoding CD44s, CD44v9, and CD44 (as well as pan-CD44) expressed by the sorted NOZ-CD44v and CD44s cells (*p<0.001).
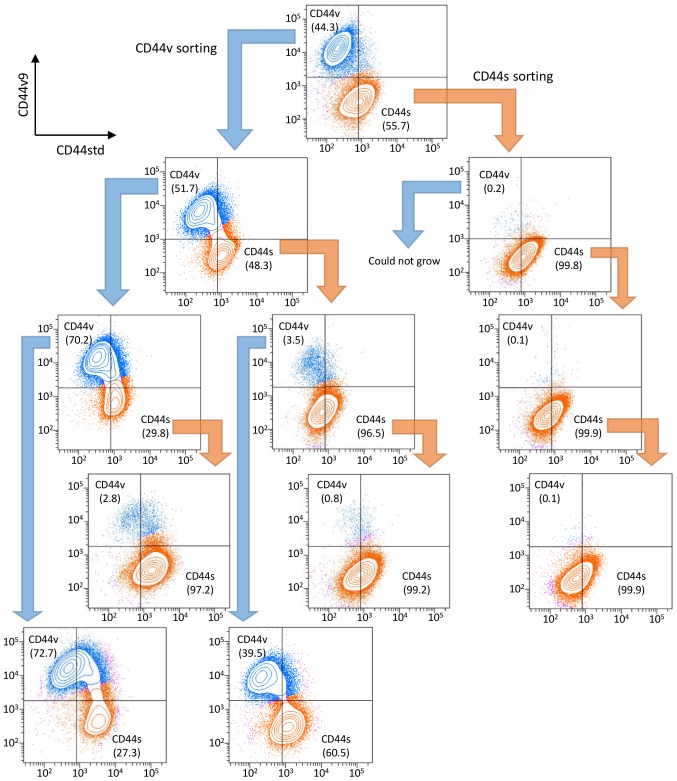
The CD44s and CD44v cells were recultured to confirm viability and proliferation. The NOZ-CD44s cells were spindle-shaped, and the NOZ-CD44v cells were rounded with a spindle or polygonal shape (Fig. 1C). The expressed CD44 isoforms of the sorted cells were genetically confirmed (Fig. 1D). The NOZ-CD44s sorted cells expressed significantly higher and lower levels of CD44s and CD44v9 mRNA compared with the NOZ-CD44v sorted cells. The whole CD44 mRNA levels did not differ between CD44s and CD44v sorted cells. We analyzed the sequential expression of CD44 isoforms of the sorted cells. One week after sorting, cells were restained and analyzed for CD44 isoform expression, and sequentially repeated three times (Fig. 2). The NOZ-CD44s cells were generated from CD44v cells (CD44v/CD44s range: 39.5–72.7/27.3–60.5), and NOZ-CD44v cells were not generated from CD44s cells (CD44v/CD44s range 0.1–3.5/96.5–99.9), suggesting that CD44s cells were derived from CD44v cells.
Figure 2.
Sequential expression analysis of CD44std and CD44v9. CD44std+/CD44v9− (CD44s) cells and CD44std−/CD44v9+ (CD44v) cells were recultured and analyzed 1 to 2 weeks after sorting. CD44std and CD44v9 expression in recultured NOZ-CD44v and NOZ-CD44s cells are shown below the former analysis. The populations of CD44s and CD44v are orange and blue, respectively, and the percentage of each population is shown. These procedures were repeated until the third sorting.
NOZ-CD44s cells have increased chemotaxis activity and invasiveness compared with NOZ-CD44v cells
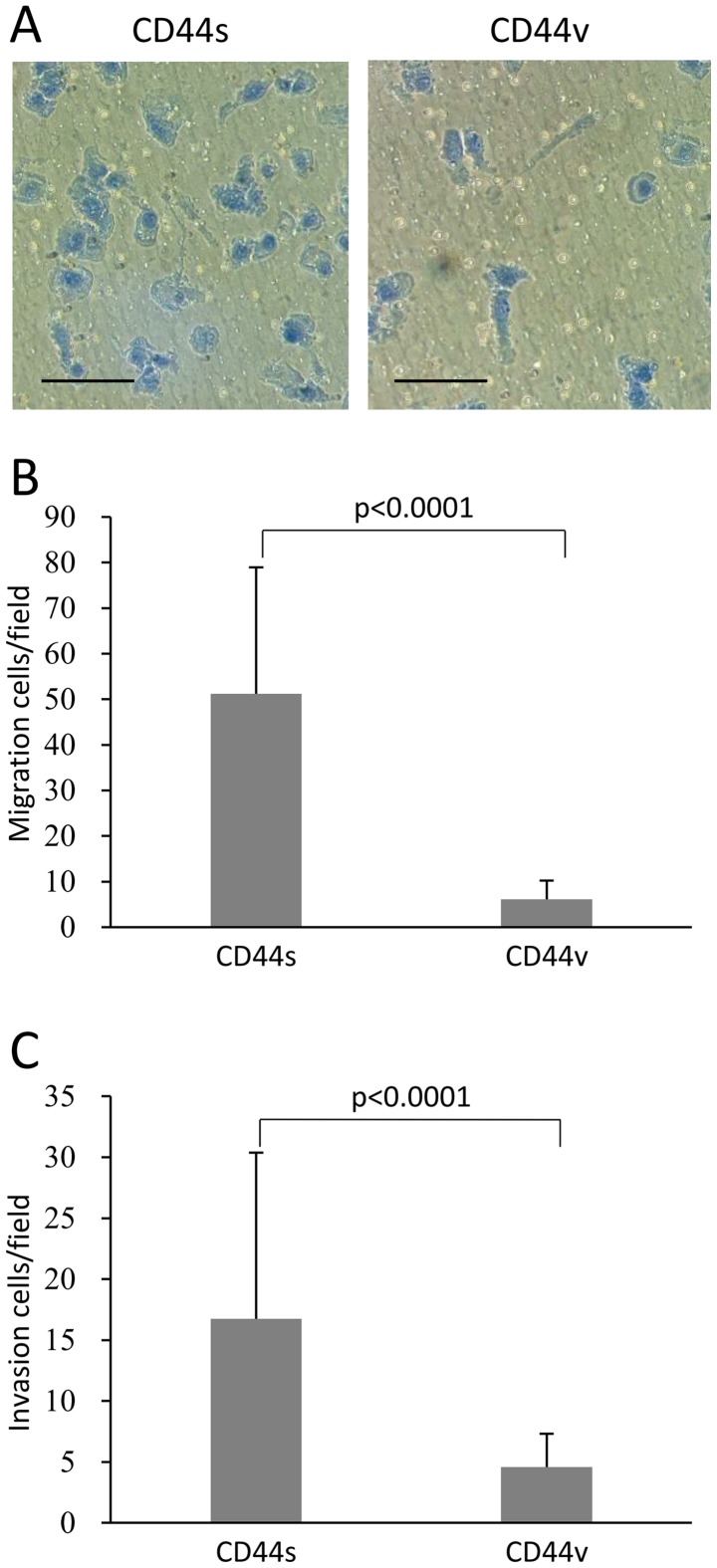
Analysis of chemotaxis in vitro revealed that significantly more NOZ-CD44s cells migrated compared with NOZ-CD44v cells (51.2±27.7 vs. 6.1±4.1 cells per field, p<0.0001) (Fig. 3A and B). Similarly, NOZ-CD44s cells were significantly more invasive compared with NOZ-CD44v cells (16.7±13.6 cells vs. 4.6±2.7 cells per field, p<0.0001) (Fig. 3C).
Figure 3.
(A) Morphology of migrating sorted fixed and stained NOZ-CD44v and CD44s cells. Scale bars, 100 μm. (B) Transwell migration assay of sorted NOZ-CD44v and CD44s cells. The bars represent the average number of migrated cells per field (×200) ± SD. (C) Transwell invasion assay of NOZ-CD44v and CD44s cells. The bars represent average number of invading cells per field (×200) ± SD.
NOZ-CD44s cells exhibit a mesenchymal phenotype
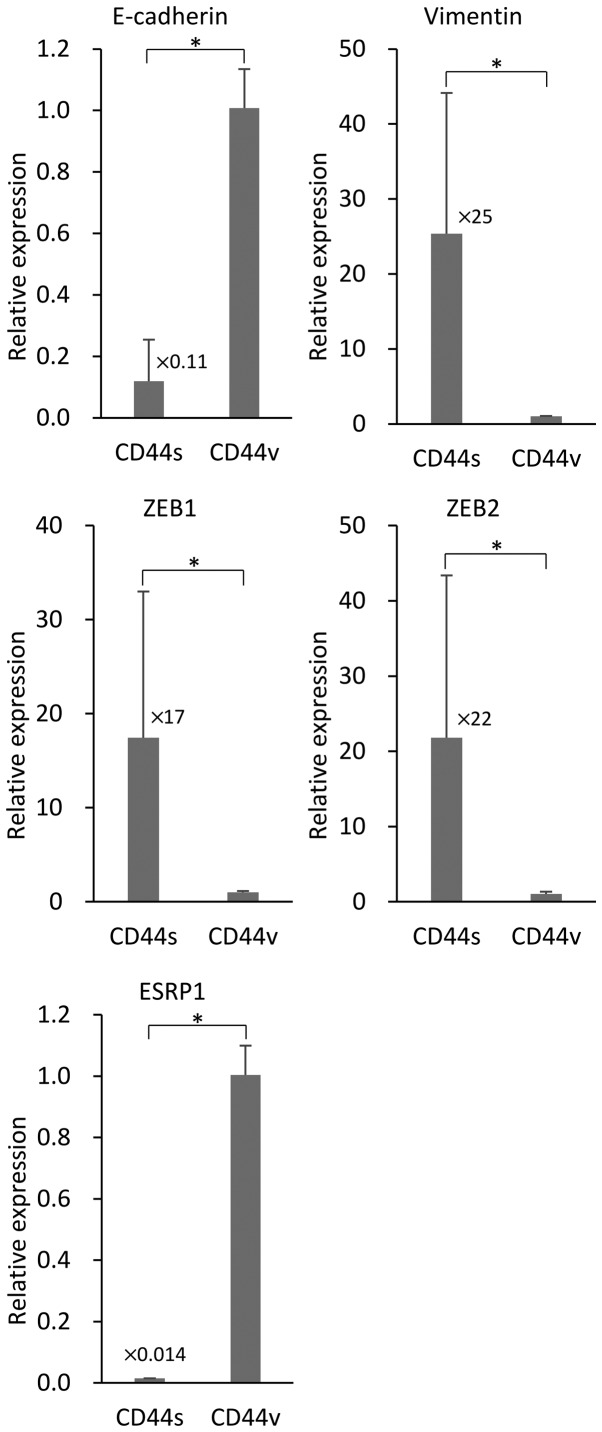
We investigated the association of the expression of the different CD44s and CD44v isoforms expressed by NOZ cells on EMT. We used RT-PCR to determine the levels of the mRNAs of the EMT markers E-cadherin and vimentin in the sorted NOZ cells. NOZ-CD44s expressed significantly higher levels of vimentin and lower levels of E-cadherin compared with CD44v cells (Fig. 4). The transcription factors ZEB1 (δEF1) and ZEB2 (SIP-1) that control EMT, were highly expressed in the NOZ-CD44s cells compared with NOZ-CD44v cells. The mRNA encoding the splicing factor ESRP1 that controls the CD44 isoform switch was expressed at lower levels in NOZ-CD44s cells compared with NOZ-CD44v cells.
Figure 4.
Reverse transcription-PCR analysis of mRNAs expressed by sorted NOZ-CD44v and CD44s cells. The bars represent average ratios compared with NOZ-CD44v cells ± SD. The numbers next to the bars represent the fold-change in CD44s mRNA compared with CD44v mRNA (*p<0.001).
NOZ-CD44v cells are highly tumorigenic in vivo
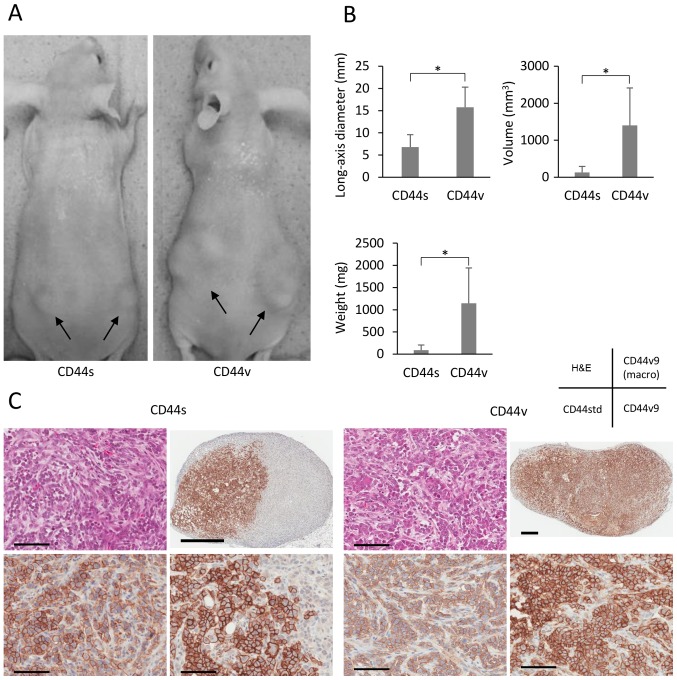
When we investigated the tumorigenicity of CD44s and CD44v cells in nude mice, we found that the former were less tumorigenic (Fig. 5A). Although each cell population formed tumors at all sites where 10,000 cells were injected, the respective long-axis diameters, tumor volumes, and weights of the tumors formed by CD44s and CD44v cells were 6.8±2.8 and 15.7±4.5 mm, 131±160 and 1401±1013 mm3, and 0.09±0.12 and 1.15±0.80 g (all p<0.001). NOZ-CD44v cells (100 per site) formed tumors at 10 out of 12 sites, whereas NOZ-CD44s cells did not generate tumors (Table I). The tumors were pathologically confirmed using H&E staining, and immunohistochemical analysis of CD44v9 expression revealed that CD44s cells generated partly CD44v9-rich tumors (Fig. 5C).
Figure 5.
(A) Representative mouse xenotransplantation experiments using sorted NOZ-CD44v and CD44s cells. CD44v and CD44s cells were subcutaneously injected into bilateral lumber legions. Arrows indicate tumors. (B) Comparisons of long-axis diameters, calculated volumes, and weights of the tumors from 10,000 cells 4 weeks after injection (*p<0.001). (C) Hematoxylin and eosin (H&E) staining and immunohistochemical analysis of CD44std and CD44v9 (with macro image) expression in tumors generated from NOZ-CD44s and CD44v sorted cells. Scale bars, 100 μm (micro) and 1 mm (macro).
Table I.
In vivo tumorigenicity analysis in which NOZ-CD44s and CD44v cells were injected into nude mice subcutaneously.
| Number of cells injected | Tumor incidence
|
||
|---|---|---|---|
| 100 | 1000 | 10000 | |
| CD44s | 0/12 | 4/12 | 12/12 |
| CD44v | 10/12 | 11/12 | 12/12 |
CD44v9 expression associates with poor prognosis
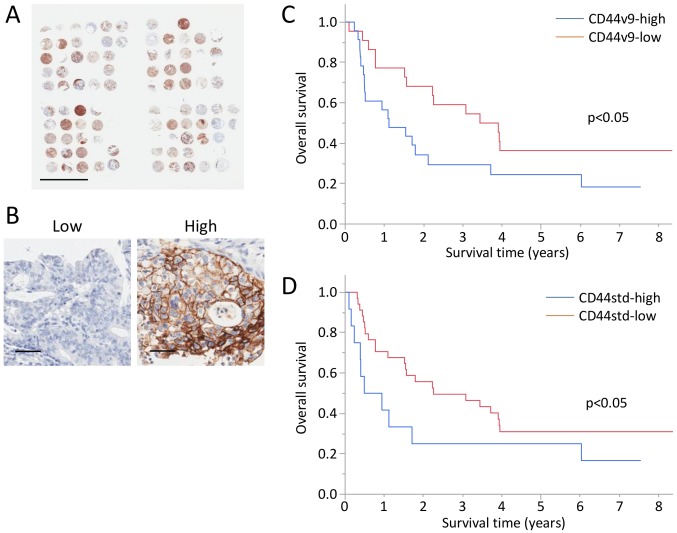
A TMA of GBC was performed, and CD44v9 and CD44std expression was analyzed in 45 and 47 tumors from patients with GBC (Fig. 6A and Table II). Immunohistochemical analysis detected high levels of CD44v9 and CD44std expression in 23 and 12 tumors (Fig. 6B). No significant differences were detected in age, sex, serum CA19-9 levels, and pathological T, N, ly, and v factors associated with the expression of CD44v9 and CD44std. CD44v9 expression was significantly associated with serum CEA levels. Whereas, CD44std expression was, significantly associated with poorly differentiation and distant metastasis.
Figure 6.
(A) Tissue microarray analysis of the tumors of 52 patients with gallbladder cancer. Scale bars, 5 mm. (B) Representative CD44v9-low and high tumors. Scale bars, 50 μm. Kaplan-Meier curves of overall survivals of patients with GBC based on the expression of CD44v9 (C) and CD44std (D).
Table II.
CD44v9 and CD44std expression in tumors from patients with GBC.
| Factors | CD44v9
|
p-value | CD44std
|
p-value | ||
|---|---|---|---|---|---|---|
| Low (N=22) |
High (N=23) |
Low (N=22) |
High (N=23) |
|||
| Age | 71.1±10.7 | 69.2±11.5 | 0.566 | 70.9±12.0 | 68.0±8.2 | 0.450 |
| Sex | 0.928 | 0.702 | ||||
| Male | 6 (27%) | 6 (26%) | 10 (29%) | 2 (17%) | ||
| Female | 16 (73%) | 17 (74%) | 25 (71%) | 10 (83%) | ||
| CEA (ng/ml) | 4.1±3.1 | 12.9±20.9 | 0.032 | 7.2±11.9 | 13.2±22.7 | 0.243 |
| CA19-9 (U/ml) | 249.4±609.2 | 161.4±327.3 | 0.549 | 237.5±515.3 | 133.8±346.4 | 0.521 |
| Differentiation | 0.588 | 0.024 | ||||
| Well/Mod | 21 (95%) | 17 (89%) | 33 (97%) | 6 (67%) | ||
| Poor | 1 (5%) | 2 (11%) | 1 (3%) | 3 (33%) | ||
| pT | 0.850 | 0.261 | ||||
| 1–2 | 13 (62%) | 13 (59%) | 22 (65%) | 5 (45%) | ||
| 3–4 | 8 (38%) | 9 (41%) | 12 (35%) | 6 (55%) | ||
| pN | 0.790 | 0.733 | ||||
| Negative | 11 (55%) | 13 (59%) | 18 (56%) | 8 (67%) | ||
| Positive | 9 (45%) | 9 (41%) | 14 (44%) | 4 (33%) | ||
| M | 0.153 | 0.045 | ||||
| Negative | 16 (73%) | 12 (52%) | 24 (69%) | 4 (33%) | ||
| Positive | 6 (27%) | 11 (48%) | 11 (31%) | 8 (67%) | ||
| ly | 0.208 | 0.161 | ||||
| Negative | 11 (52%) | 6 (29%) | 16 (47%) | 2 (20%) | ||
| Positive | 10 (48%) | 15 (71%) | 18 (53%) | 8 (80%) | ||
| v | 0.059 | 0.074 | ||||
| Negative | 15 (71%) | 9 (43%) | 22 (65%) | 3 (30%) | ||
| Positive | 6 (29%) | 12 (57%) | 12 (35%) | 7 (70%) | ||
The 3-year OS rates of the CD44v9-high, CD44v9-low, CD44std-high, and CD44std-low groups were 29.4, 59.1, 25.0, and 49.5%, respectively (Fig. 6C and D).
Discussion
In the present study, we showed that the GBC cell line NOZ comprises two populations that expressed different CD44 isoforms. The major population (CD44s) expressed CD44std+/CD44v9−. The CD44s cells showed the mesenchymal phenotype, and they showed strong chemotactic and invasive abilities. On the other hand, they showed weakly tumorigenic ability. The minor population (CD44v) expressed CD44std−/CD44v9+. This population showed the epithelial phenotype, and it showed weakly chemotactic and invasive abilities. However, it showed highly tumorigenic ability. These isoforms of CD44 in the NOZ sorted cells were also genetically identified. CD44v cells generated CD44s cells in vitro, although the xenotransplantation study indicated that CD44v cells were generated from CD44s cells in vivo. In patients with GBC, we demonstrated that both phenotypes CD44v9high and CD44stdhigh were significantly associated with poor prognosis. CD44v9high was significantly associated with high CEA levels, and CD44stdhigh was significantly associated with poor differentiation and distant metastasis.
It is important to clarify whether CD44 represents particular CD44 isoform or pan-CD44. Each variant isoform possesses specific epitopes, although most epitopes of the CD44 standard isoform are included in the variant isoforms. Numerous anti-CD44 antibodies react with CD44 variant isoforms as well as the CD44 standard isoform, although these isoforms are associated with different phenotypes. Thus, the isoforms of CD44 (or pan-CD44) should be identified when studies using CD44-specific antibodies are planned. We show here that the GBC cell line NOZ comprised populations of CD44 standard and variant 9 isoforms that were associated with different phenotypes, although flow cytometry using the anti-CD44 antibody detected both isoforms. We then used isoform-specific antibodies CD44std and CD44v9 to show that the CD44s and CD44v cells expressed the respective cognate isoforms.
CD44 isoforms are associated with EMT. It is reported that pancreatic cancer cells that express CD44v9 showed epithelial phenotype (14), and hepatocellular carcinomas that express CD44 standard isoform showed the mesenchymal phenotype (7,31). Further, breast cancer cells are reported to show isoform switch from variant isoform to the standard isoform through EMT (32), ESRP1 is critical for regulating the isoform switch (32,33), and signaling through AKT, which is activated by the CD44 standard isoform, is essential for driving EMT (32). Moreover, CD44 isoform switch occurs in colorectal cancer cells (27). When ESRP1 is silenced, the CD44 variant isoform is replaced by the standard isoform that promotes EMT. Further, the CD44 standard isoform is essential for EMT, because CD44 knockdown inhibits vimentin expression (32). In cancers of the biliary tract, expression of the CD44 standard isoform is associated with well-differentiated cancers (16), and CD44v6 isoforms are associated with moderate-poorly differentiated cancers and Stage IV tumors (17,34). Our TMA analysis, however, showed that CD44std expression was associated with poorly differentiated cancers and distant metastasis, which was compatible with the mesenchymal phenotype of the CD44s showed in the in vitro studies.
Our findings described above that the CD44 standard and variant 9 isoforms were associated with mesenchymal and epithelial phenotypes, respectively, are consistent with a switch from the CD44 variant to the standard isoform through EMT. The levels of the mRNAs encoding the transcription factors ZEB1 and ZEB2 were increased in cells that expressed the CD44 standard isoform that was associated with promoting EMT. Further, the levels of the mRNA encoding the splicing factor ESRP1 were lower in cells that expressed the CD44 standard isoform, which triggered the CD44-isoform switch and then promoted EMT. Further, we showed that the CD44s cells were generated from CD44v cells under the normal culture condition. There may, therefore be an auto-trigger which induces the CD44 isoform switch through NOZ cell progression.
CD44 is associated with tumorigenicity of many cancers such as the breast (35), colon, head and neck, and pancreas (36). In cholangiocarcinoma, CD44+CD24+EpCAMhigh cells exhibit high tumorigenic potential (37). However, these studies did not analyze CD44 isoforms. In contrast, analyses using xenotransplantation of mice reveal that colon (15) and breast cancer (33) cells that express CD44v9 are highly tumorigenic. Further, pancreatic cancer cells express CD44v9 when they re-enter mitosis (14). Clinical studies reveal that CD44v9 expression is associated with the recurrence of early gastric cancer (8) and with poor OS and recurrence-free survival of patients with hepatocellular carcinoma (10). Moreover, RT-PCR analyses show that increased CD44v9 expression in pancreatic cancer tissues correlates with lymph node metastasis, liver metastasis, TNM stage progression, and decreased overall survival rate (9). Although the relationship between CD44v9 expression and tumorigenicity is unknown, CD44v9 was proposed to activate the Ras/Rac1/RhoA pathway, leading to the migration, growth, invasion, and survival of tumor cells (37–39). In the present study, CD44v9 cells had higher tumorigenicity in terms of tumor burden and tumor incidence compared with CD44s cells. Further, both tumors generated from CD44v9 and CD44s cells expressed CD44v9 immunohistochemically. Although the mechanism by which the CD44s generated CD44v9-rich tumors is unclear, it is suggesting that a switch from the standard to variant isoform is induced through engraftment or the progression of tumor formation.
In conclusion, expression of the CD44 standard isoform was associated with a mesenchymal phenotype, increased chemotaxis, increased invasiveness, and lower tumorigenicity. In contrast, expression of the CD44 variant 9 isoform was associated with an epithelial phenotype, decreased chemotaxis, decreased invasiveness, and increased tumorigenicity. These findings suggest that these CD44 isoforms play different roles in cancer progression and metastasis. Therefore, investigating the potential role and mechanism of the isoform switch may lead to the development of novel therapeutic interventions or diagnostic agents that target splicing or specific isoforms that will improve the treatment of patients with GBC.
Acknowledgments
We thank Mr. Masahiko Kawahara for his technical assistance.
References
- 1.Misra S, Chaturvedi A, Misra NC, Sharma ID. Carcinoma of the gallbladder. Lancet Oncol. 2003;4:167–176. doi: 10.1016/S1470-2045(03)01021-0. [DOI] [PubMed] [Google Scholar]
- 2.Gourgiotis S, Kocher HM, Solaini L, Yarollahi A, Tsiambas E, Salemis NS. Gallbladder cancer. Am J Surg. 2008;196:252–264. doi: 10.1016/j.amjsurg.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 3.Hofmann M, Rudy W, Zöller M, Tölg C, Ponta H, Herrlich P, Günthert U. CD44 splice variants confer metastatic behavior in rats: Homologous sequences are expressed in human tumor cell lines. Cancer Res. 1991;51:5292–5297. [PubMed] [Google Scholar]
- 4.Sneath RJS, Mangham DC. The normal structure and function of CD44 and its role in neoplasia. Mol Pathol. 1998;51:191–200. doi: 10.1136/mp.51.4.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodison S, Urquidi V, Tarin D. CD44 cell adhesion molecules. Mol Pathol. 1999;52:189–196. doi: 10.1136/mp.52.4.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zöller M. CD44: Can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11:254–267. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- 7.Okabe H, Ishimoto T, Mima K, Nakagawa S, Hayashi H, Kuroki H, Imai K, Nitta H, Saito S, Hashimoto D, et al. CD44s signals the acquisition of the mesenchymal phenotype required for anchorage-independent cell survival in hepatocellular carcinoma. Br J Cancer. 2014;110:958–966. doi: 10.1038/bjc.2013.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirata K, Suzuki H, Imaeda H, Matsuzaki J, Tsugawa H, Nagano O, Asakura K, Saya H, Hibi T. CD44 variant 9 expression in primary early gastric cancer as a predictive marker for recurrence. Br J Cancer. 2013;109:379–386. doi: 10.1038/bjc.2013.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Chen K, Jiang P, Zhang X, Li X, Li Z. CD44v/CD44s expression patterns are associated with the survival of pancreatic carcinoma patients. Diagn Pathol. 2014;9:79. doi: 10.1186/1746-1596-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kakehashi A, Ishii N, Sugihara E, Gi M, Saya H, Wanibuchi H. CD44 variant 9 is a potential biomarker of tumor initiating cells predicting survival outcome in hepatitis C virus-positive patients with resected hepatocellular carcinoma. Cancer Sci. 2016;107:609–618. doi: 10.1111/cas.12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishimoto T, Nagano O, Yae T, Tamada M, Motohara T, Oshima H, Oshima M, Ikeda T, Asaba R, Yagi H, et al. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell. 2011;19:387–400. doi: 10.1016/j.ccr.2011.01.038. [DOI] [PubMed] [Google Scholar]
- 12.Mielgo A, van Driel M, Bloem A, Landmann L, Günthert U. A novel antiapoptotic mechanism based on interference of Fas signaling by CD44 variant isoforms. Cell Death Differ. 2006;13:465–477. doi: 10.1038/sj.cdd.4401763. [DOI] [PubMed] [Google Scholar]
- 13.Nagano O, Okazaki S, Saya H. Redox regulation in stem-like cancer cells by CD44 variant isoforms. Oncogene. 2013;32:5191–5198. doi: 10.1038/onc.2012.638. [DOI] [PubMed] [Google Scholar]
- 14.Kiuchi S, Ikeshita S, Miyatake Y, Kasahara M. Pancreatic cancer cells express CD44 variant 9 and multidrug resistance protein 1 during mitosis. Exp Mol Pathol. 2015;98:41–46. doi: 10.1016/j.yexmp.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Kimura Y, Goi T, Nakazawa T, Hirono Y, Katayama K, Urano T, Yamaguchi A. CD44 variant exon 9 plays an important role in colon cancer initiating cells. Oncotarget. 2013;4:785–791. doi: 10.18632/oncotarget.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalekou H, Miliaras D. Immunohistochemical study of microvessel density, CD44 (standard form), p53 protein and c-erbB2 in gallbladder carcinoma. J Gastroenterol Hepatol. 2004;19:812–818. doi: 10.1111/j.1440-1746.2004.03357.x. [DOI] [PubMed] [Google Scholar]
- 17.Yanagisawa N, Mikami T, Mitomi H, Saegusa M, Koike M, Okayasu I. CD44 variant overexpression in gallbladder carcinoma associated with tumor dedifferentiation. Cancer. 2001;91:408–416. doi: 10.1002/1097-0142(20010115)91:2<408::AID-CNCR1015>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 18.Yamaguchi A, Zhang M, Goi T, Fujita T, Niimoto S, Katayama K, Hirose K. Expression of variant CD44 containing variant exon v8-10 in gallbladder cancer. Oncol Rep. 2000;7:541–544. doi: 10.3892/or.7.3.541. [DOI] [PubMed] [Google Scholar]
- 19.Homma S, Hasumura S, Nagamori S, Kameda H. Establishment and characterization of a human gall bladder carcinoma cell line NOZ. Hum Cell. 1988;1:95–97. In Japanese. [PubMed] [Google Scholar]
- 20.Kramer N, Walzl A, Unger C, Rosner M, Krupitza G, Hengstschläger M, Dolznig H. In vitro cell migration and invasion assays. Mutat Res. 2013;752:10–24. doi: 10.1016/j.mrrev.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Valster A, Tran NL, Nakada M, Berens ME, Chan AY, Symons M. Cell migration and invasion assays. Methods. 2005;37:208–215. doi: 10.1016/j.ymeth.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Long J, Luo G, Liu C, Cui X, Satoh K, Xiao Z, Zhang B, Xu J, Ni Q, Li M, et al. Development of a unique mouse model for pancreatic cancer lymphatic metastasis. Int J Oncol. 2012;41:1662–1668. doi: 10.3892/ijo.2012.1613. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, Berx G, Cano A, Beug H, Foisner R. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24:2375–2385. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- 25.Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D, van Roy F. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7:1267–1278. doi: 10.1016/S1097-2765(01)00260-X. [DOI] [PubMed] [Google Scholar]
- 26.Ishii H, Saitoh M, Sakamoto K, Kondo T, Katoh R, Tanaka S, Motizuki M, Masuyama K, Miyazawa K. Epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) suppress cancer cell motility via different mechanisms. J Biol Chem. 2014;289:27386–27399. doi: 10.1074/jbc.M114.589432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mashita N, Yamada S, Nakayama G, Tanaka C, Iwata N, Kanda M, Kobayashi D, Fujii T, Sugimoto H, Koike M, et al. Epithelial to mesenchymal transition might be induced via CD44 isoform switching in colorectal cancer. J Surg Oncol. 2014;110:745–751. doi: 10.1002/jso.23705. [DOI] [PubMed] [Google Scholar]
- 28.Sekine S, Shimada Y, Nagata T, Moriyama M, Omura T, Watanabe T, Hori R, Yoshioka I, Okumura T, Sawada S, et al. Prognostic significance of aquaporins in human biliary tract carcinoma. Oncol Rep. 2012;27:1741–1747. doi: 10.3892/or.2012.1747. [DOI] [PubMed] [Google Scholar]
- 29.Sekine S, Shimada Y, Nagata T, Sawada S, Yoshioka I, Matsui K, Moriyama M, Omura T, Osawa S, Shibuya K, et al. Role of aquaporin-5 in gallbladder carcinoma. Eur Surg Res. 2013;51:108–117. doi: 10.1159/000355675. [DOI] [PubMed] [Google Scholar]
- 30.Fukuoka J, Fujii T, Shih JH, Dracheva T, Meerzaman D, Player A, Hong K, Settnek S, Gupta A, Buetow K, et al. Chromatin remodeling factors and BRM/BRG1 expression as prognostic indicators in non-small cell lung cancer. Clin Cancer Res. 2004;10:4314–4324. doi: 10.1158/1078-0432.CCR-03-0489. [DOI] [PubMed] [Google Scholar]
- 31.Mima K, Okabe H, Ishimoto T, Hayashi H, Nakagawa S, Kuroki H, Watanabe M, Beppu T, Tamada M, Nagano O, et al. CD44s regulates the TGF-β-mediated mesenchymal phenotype and is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Res. 2012;72:3414–3423. doi: 10.1158/0008-5472.CAN-12-0299. [DOI] [PubMed] [Google Scholar]
- 32.Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, Cheng C. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest. 2011;121:1064–1074. doi: 10.1172/JCI44540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yae T, Tsuchihashi K, Ishimoto T, Motohara T, Yoshikawa M, Yoshida GJ, Wada T, Masuko T, Mogushi K, Tanaka H, et al. Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat Commun. 2012;3:883. doi: 10.1038/ncomms1892. [DOI] [PubMed] [Google Scholar]
- 34.Gu MJ, Jang BI. Clinicopathologic significance of Sox2, CD44 and CD44v6 expression in intrahepatic cholangiocarcinoma. Pathol Oncol Res. 2014;20:655–660. doi: 10.1007/s12253-014-9745-2. [DOI] [PubMed] [Google Scholar]
- 35.Han Z, Chen Z, Zheng R, Cheng Z, Gong X, Wang D. Clinicopathological significance of CD133 and CD44 expression in infiltrating ductal carcinoma and their relationship to angiogenesis. World J Surg Oncol. 2015;13:56. doi: 10.1186/s12957-015-0486-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 37.Wang M, Xiao J, Shen M, Yahong Y, Tian R, Zhu F, Jiang J, Du Z, Hu J, Liu W, et al. Isolation and characterization of tumorigenic extrahepatic cholangiocarcinoma cells with stem cell-like properties. Int J Cancer. 2011;128:72–81. doi: 10.1002/ijc.25317. [DOI] [PubMed] [Google Scholar]
- 38.Wang SJ, Bourguignon LYW. Role of hyaluronan-mediated CD44 signaling in head and neck squamous cell carcinoma progression and chemoresistance. Am J Pathol. 2011;178:956–963. doi: 10.1016/j.ajpath.2010.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akasaka J, Uekuri C, Shigetomi H, Koike M, Kobayashi H. Hepatocyte nuclear factor (HNF)-1β and its physiological importance in endometriosis. Biomed Rep. 2013;1:13–17. doi: 10.3892/br.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]