Abstract
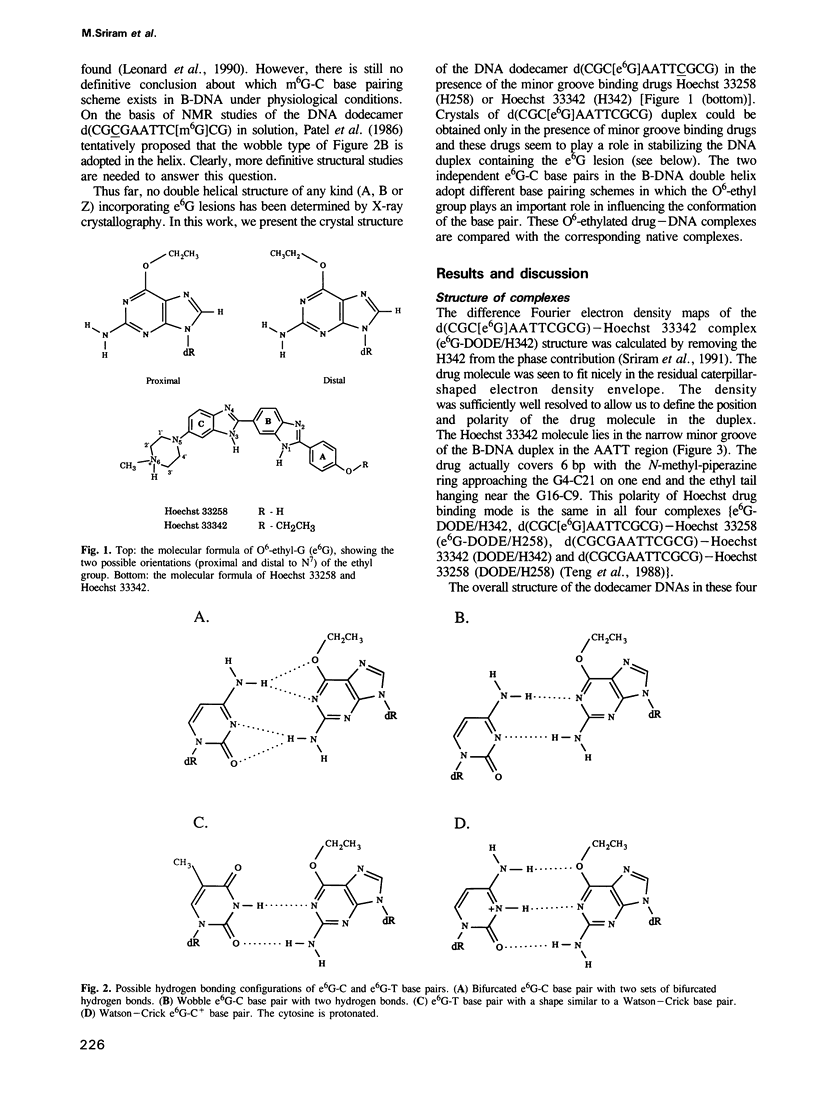
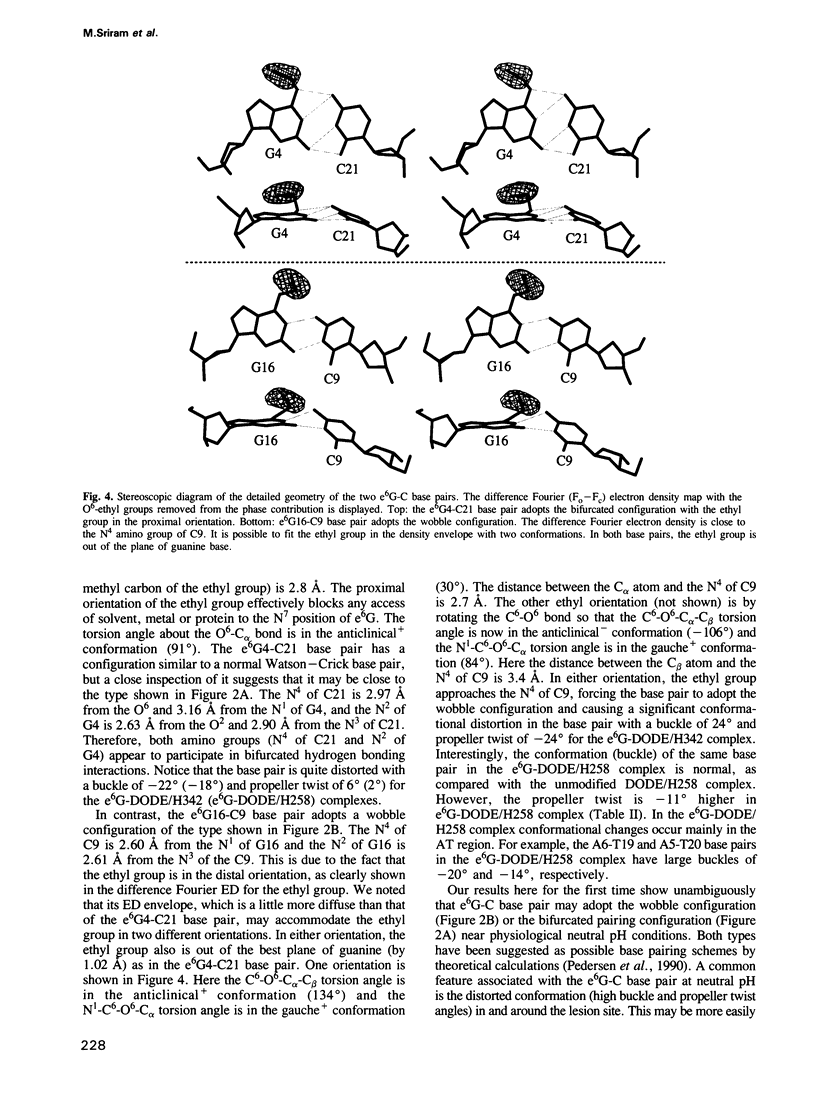
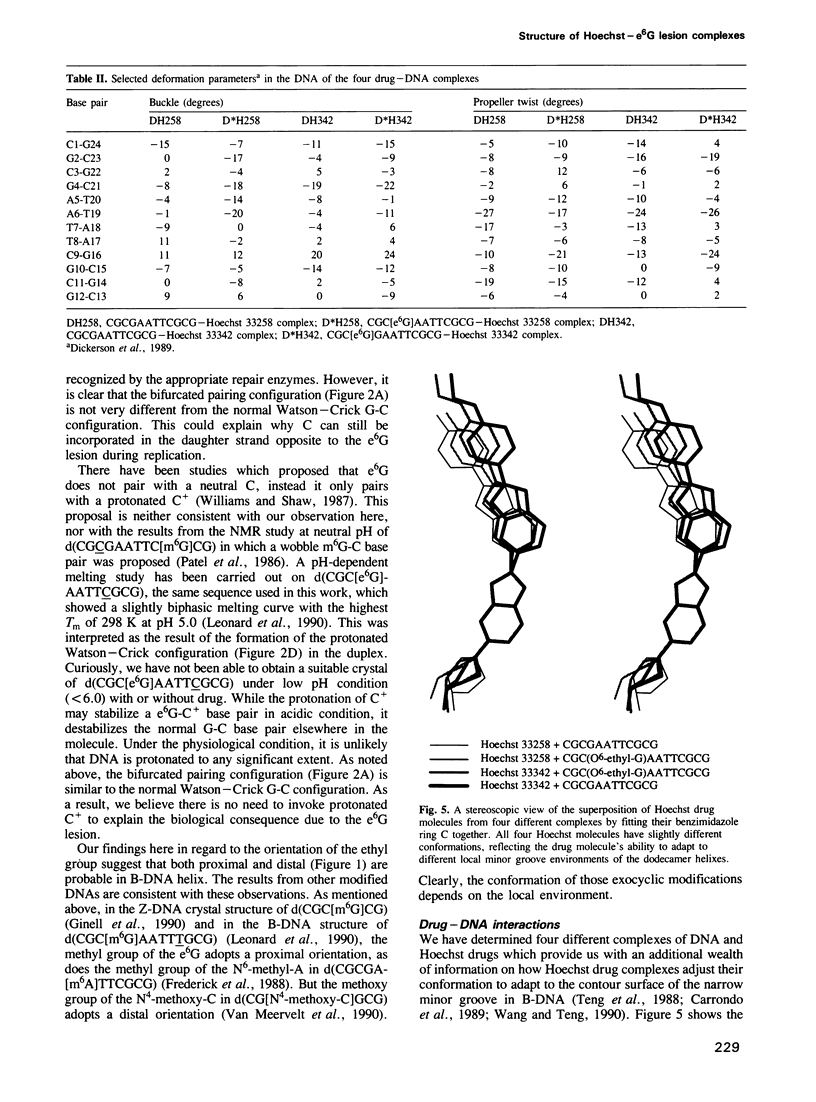
O6-ethyl-G (e6G) is an important DNA lesion, caused by the exposure of cells to alkylating agents such as N-ethyl-N-nitrosourea. A strong correlation exists between persistence of e6G lesion and subsequent carcinogenic conversion. We have determined the three-dimensional structure of a DNA molecule incorporating the e6G lesion by X-ray crystallography. The DNA dodecamer d(CGC[e6G]AATTCGCG), complexed to minor groove binding drugs Hoechst 33258 or Hoechst 33342, has been crystallized in the space group P212121, isomorphous to other related dodecamer DNA crystals. In addition, the native dodecamer d(CGCGAATTCGCG) was crystallized with Hoechst 33342. All three new structures were solved by the molecular replacement method and refined by the constrained least squares procedure to R-factors of approximately 16% at approximately 2.0 A resolution. In the structure of three Hoechst drug-dodecamer complexes in addition to the one published earlier [Teng et al. (1988) Nucleic Acids Res., 16, 2671-2690], the Hoechst molecule lies squarely at the central AATT site with the ends approaching the G4-C21 and the G16-C9 base pairs, consistent with other spectroscopic data, but not with another crystal structure reported [Pjura et al. (1987) J. Mol. Biol., 197, 257-271]. The two independent e6G-C base pairs in the DNA duplex adopt different base pairing schemes. The e6G4-C21 base pair has a configuration similar to a normal Watson-Crick base pair, except with bifurcated hydrogen bonds between e6G4 and C21, and the ethyl group is in the proximal orientation. In contrast, the e6G16-C9 base pair adopts a wobble configuration and the ethyl group is in the distal orientation.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aymami J., Coll M., van der Marel G. A., van Boom J. H., Wang A. H., Rich A. Molecular structure of nicked DNA: a substrate for DNA repair enzymes. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2526–2530. doi: 10.1073/pnas.87.7.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrondo M. A., Coll M., Aymami J., Wang A. H., van der Marel G. A., van Boom J. H., Rich A. Binding of a Hoechst dye to d(CGCGATATCGCG) and its influence on the conformation of the DNA fragment. Biochemistry. 1989 Sep 19;28(19):7849–7859. doi: 10.1021/bi00445a047. [DOI] [PubMed] [Google Scholar]
- Coll M., Aymami J., van der Marel G. A., van Boom J. H., Rich A., Wang A. H. Molecular structure of the netropsin-d(CGCGATATCGCG) complex: DNA conformation in an alternating AT segment. Biochemistry. 1989 Jan 10;28(1):310–320. doi: 10.1021/bi00427a042. [DOI] [PubMed] [Google Scholar]
- Coll M., Frederick C. A., Wang A. H., Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll M., Sherman S. E., Gibson D., Lippard S. J., Wang A. H. Molecular structure of the complex formed between the anticancer drug cisplatin and d(pGpG): C222(1) crystal form. J Biomol Struct Dyn. 1990 Oct;8(2):315–330. doi: 10.1080/07391102.1990.10507808. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Haran T. E., Nadeau J. G. Intrinsically bent DNA. J Biol Chem. 1990 May 5;265(13):7093–7096. [PubMed] [Google Scholar]
- Dickerson R. E. Definitions and nomenclature of nucleic acid structure components. Nucleic Acids Res. 1989 Mar 11;17(5):1797–1803. doi: 10.1093/nar/17.5.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Dickerson R. E. Structure of a B-DNA dodecamer. III. Geometry of hydration. J Mol Biol. 1981 Sep 25;151(3):535–556. doi: 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- Frederick C. A., Quigley G. J., van der Marel G. A., van Boom J. H., Wang A. H., Rich A. Methylation of the EcoRI recognition site does not alter DNA conformation: the crystal structure of d(CGCGAm6ATTCGCG) at 2.0-A resolution. J Biol Chem. 1988 Nov 25;263(33):17872–17879. doi: 10.2210/pdb4dnb/pdb. [DOI] [PubMed] [Google Scholar]
- Ginell S. L., Kuzmich S., Jones R. A., Berman H. M. Crystal and molecular structure of a DNA duplex containing the carcinogenic lesion O6-methylguanine. Biochemistry. 1990 Nov 20;29(46):10461–10465. doi: 10.1021/bi00498a005. [DOI] [PubMed] [Google Scholar]
- Larsen T. A., Goodsell D. S., Cascio D., Grzeskowiak K., Dickerson R. E. The structure of DAPI bound to DNA. J Biomol Struct Dyn. 1989 Dec;7(3):477–491. doi: 10.1080/07391102.1989.10508505. [DOI] [PubMed] [Google Scholar]
- Leonard G. A., Thomson J., Watson W. P., Brown T. High-resolution structure of a mutagenic lesion in DNA. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9573–9576. doi: 10.1073/pnas.87.24.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Sedgwick B., Sekiguchi M., Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- Loechler E. L., Green C. L., Essigmann J. M. In vivo mutagenesis by O6-methylguanine built into a unique site in a viral genome. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6271–6275. doi: 10.1073/pnas.81.20.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loontiens F. G., McLaughlin L. W., Diekmann S., Clegg R. M. Binding of Hoechst 33258 and 4',6'-diamidino-2-phenylindole to self-complementary decadeoxynucleotides with modified exocyclic base substituents. Biochemistry. 1991 Jan 8;30(1):182–189. doi: 10.1021/bi00215a027. [DOI] [PubMed] [Google Scholar]
- Nelson H. C., Finch J. T., Luisi B. F., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987 Nov 19;330(6145):221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- Parkinson J. A., Barber J., Douglas K. T., Rosamond J., Sharples D. Minor-groove recognition of the self-complementary duplex d(CGCGAATTCGCG)2 by Hoechst 33258: a high-field NMR study. Biochemistry. 1990 Nov 6;29(44):10181–10190. doi: 10.1021/bi00496a005. [DOI] [PubMed] [Google Scholar]
- Parthasarathy R., Fridey S. M. Conformation of O6-alkylguanosines: molecular mechanism of mutagenesis. Carcinogenesis. 1986 Feb;7(2):221–227. doi: 10.1093/carcin/7.2.221. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Shapiro L., Kozlowski S. A., Gaffney B. L., Jones R. A. Structural studies of the O6meG.C interaction in the d(C-G-C-G-A-A-T-T-C-O6meG-C-G) duplex. Biochemistry. 1986 Mar 11;25(5):1027–1036. doi: 10.1021/bi00353a012. [DOI] [PubMed] [Google Scholar]
- Pedersen L. G., Darden T. A., Deerfield D. W., 2nd, Anderson M. W., Hoel D. G. A theoretical study of the effect of methylation or ethylation at O6-guanine in the structure and energy of DNA double strands. Carcinogenesis. 1988 Sep;9(9):1553–1562. doi: 10.1093/carcin/9.9.1553. [DOI] [PubMed] [Google Scholar]
- Pjura P. E., Grzeskowiak K., Dickerson R. E. Binding of Hoechst 33258 to the minor groove of B-DNA. J Mol Biol. 1987 Sep 20;197(2):257–271. doi: 10.1016/0022-2836(87)90123-9. [DOI] [PubMed] [Google Scholar]
- Rich A., Nordheim A., Wang A. H. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- Singer B., Bodell W. J., Cleaver J. E., Thomas G. H., Rajewsky M. F., Thon W. Oxygens in DNA are main targets for ethylnitrosourea in normal and xeroderma pigmentosum fibroblasts and fetal rat brain cells. Nature. 1978 Nov 2;276(5683):85–88. doi: 10.1038/276085a0. [DOI] [PubMed] [Google Scholar]
- Teng M. K., Usman N., Frederick C. A., Wang A. H. The molecular structure of the complex of Hoechst 33258 and the DNA dodecamer d(CGCGAATTCGCG). Nucleic Acids Res. 1988 Mar 25;16(6):2671–2690. doi: 10.1093/nar/16.6.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomale J., Huh N. H., Nehls P., Eberle G., Rajewsky M. F. Repair of O6-ethylguanine in DNA protects rat 208F cells from tumorigenic conversion by N-ethyl-N-nitrosourea. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9883–9887. doi: 10.1073/pnas.87.24.9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomale J., Huh N. H., Nehls P., Eberle G., Rajewsky M. F. Repair of O6-ethylguanine in DNA protects rat 208F cells from tumorigenic conversion by N-ethyl-N-nitrosourea. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9883–9887. doi: 10.1073/pnas.87.24.9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meervelt L., Moore M. H., Lin P. K., Brown D. M., Kennard O. Molecular and crystal structure of d(CGCGmo4CG): N4-methoxycytosine.guanine base-pairs in Z-DNA. J Mol Biol. 1990 Dec 5;216(3):773–781. doi: 10.1016/0022-2836(90)90398-6. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Williams L. D., Shaw B. R. Protonated base pairs explain the ambiguous pairing properties of O6-methylguanine. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1779–1783. doi: 10.1073/pnas.84.7.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]



