Abstract
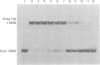
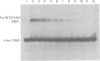
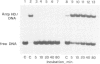
The mobility shift assay was used to study the competition of the minor groove binder distamycin A with either an Antennapedia homeodomain (Antp HD) peptide or derivatives of a fushi tarazu homeodomain (ftz HD) peptide for their AT-rich DNA binding site. The results show that distamycin and the homeodomain peptides compete under the conditions: (i) preincubation of DNA with distamycin and subsequent addition of HD peptide; (ii) simultaneous incubation of DNA with distamycin and HD peptide; and (iii) preincubation of DNA with HD peptide and subsequent addition of distamycin. There is also competition when using a peptide which lacks the N-terminal arm of ftz HD that is involved in contacts in the minor groove. It is proposed that the protein's binding affinity is diminished by distamycin-induced conformational changes of the DNA. The feasibility of the propagation of conformational changes upon binding in the minor groove is also shown for the inhibition of restriction endonucleases differing in the AT content of their recognition site and of their flanking DNA sequences. Thus, it is demonstrated that minor groove binders can compete with the binding of proteins in the major groove, providing an experimental indication for the influence of biological activities exerted by DNA ligands binding in the minor groove.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Affolter M., Percival-Smith A., Müller M., Leupin W., Gehring W. J. DNA binding properties of the purified Antennapedia homeodomain. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4093–4097. doi: 10.1073/pnas.87.11.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affolter M., Schier A., Gehring W. J. Homeodomain proteins and the regulation of gene expression. Curr Opin Cell Biol. 1990 Jun;2(3):485–495. doi: 10.1016/0955-0674(90)90132-x. [DOI] [PubMed] [Google Scholar]
- Barcelo F., Muzard G., Mendoza R., Révet B., Roques B. P., Le Pecq J. B. Removal of DNA curving by DNA ligands: gel electrophoresis study. Biochemistry. 1991 May 21;30(20):4863–4873. doi: 10.1021/bi00234a005. [DOI] [PubMed] [Google Scholar]
- Berleth T., Burri M., Thoma G., Bopp D., Richstein S., Frigerio G., Noll M., Nüsslein-Volhard C. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 1988 Jun;7(6):1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billeter M., Qian Y., Otting G., Müller M., Gehring W. J., Wüthrich K. Determination of the three-dimensional structure of the Antennapedia homeodomain from Drosophila in solution by 1H nuclear magnetic resonance spectroscopy. J Mol Biol. 1990 Jul 5;214(1):183–197. doi: 10.1016/0022-2836(90)90155-f. [DOI] [PubMed] [Google Scholar]
- Breslauer K. J., Remeta D. P., Chou W. Y., Ferrante R., Curry J., Zaunczkowski D., Snyder J. G., Marky L. A. Enthalpy-entropy compensations in drug-DNA binding studies. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8922–8926. doi: 10.1073/pnas.84.24.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broggini M., Ponti M., Ottolenghi S., D'Incalci M., Mongelli N., Mantovani R. Distamycins inhibit the binding of OTF-1 and NFE-1 transfactors to their conserved DNA elements. Nucleic Acids Res. 1989 Feb 11;17(3):1051–1059. doi: 10.1093/nar/17.3.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. G., Sanderson M. R., Skelly J. V., Jenkins T. C., Brown T., Garman E., Stuart D. I., Neidle S. Crystal structure of a berenil-dodecanucleotide complex: the role of water in sequence-specific ligand binding. EMBO J. 1990 Apr;9(4):1329–1334. doi: 10.1002/j.1460-2075.1990.tb08242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzik J. P., Auble D. T., deHaseth P. L. Specific activation of transcription initiation by the sequence-specific DNA-binding agents distamycin A and netropsin. Biochemistry. 1987 Feb 10;26(3):950–956. doi: 10.1021/bi00377a040. [DOI] [PubMed] [Google Scholar]
- Cann J. R. Phenomenological theory of gel electrophoresis of protein-nucleic acid complexes. J Biol Chem. 1989 Oct 15;264(29):17032–17040. [PubMed] [Google Scholar]
- Celda B., Widmer H., Leupin W., Chazin W. J., Denny W. A., Wüthrich K. Conformational studies of d-(AAAAATTTTT)2 using constraints from nuclear overhauser effects and from quantitative analysis of the cross-peak fine structures in two-dimensional 1H nuclear magnetic resonance spectra. Biochemistry. 1989 Feb 21;28(4):1462–1471. doi: 10.1021/bi00430a006. [DOI] [PubMed] [Google Scholar]
- Coll M., Aymami J., van der Marel G. A., van Boom J. H., Rich A., Wang A. H. Molecular structure of the netropsin-d(CGCGATATCGCG) complex: DNA conformation in an alternating AT segment. Biochemistry. 1989 Jan 10;28(1):310–320. doi: 10.1021/bi00427a042. [DOI] [PubMed] [Google Scholar]
- Coll M., Frederick C. A., Wang A. H., Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn A., Bollekens J., Staub A., Benoist C., Mathis D. A multiplicity of CCAAT box-binding proteins. Cell. 1987 Sep 11;50(6):863–872. doi: 10.1016/0092-8674(87)90513-7. [DOI] [PubMed] [Google Scholar]
- Drew H. R., McCall M. J., Calladine C. R. Recent studies of DNA in the crystal. Annu Rev Cell Biol. 1988;4:1–20. doi: 10.1146/annurev.cb.04.110188.000245. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. Structural junctions in DNA: the influence of flanking sequence on nuclease digestion specificities. Nucleic Acids Res. 1985 Jun 25;13(12):4445–4467. doi: 10.1093/nar/13.12.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigon J., Denny W. A., Leupin W., Kearns D. R. Interactions of antitumor drugs with natural DNA: 1H NMR study of binding mode and kinetics. J Med Chem. 1984 Apr;27(4):450–465. doi: 10.1021/jm00370a007. [DOI] [PubMed] [Google Scholar]
- Fesen M., Pommier Y. Mammalian topoisomerase II activity is modulated by the DNA minor groove binder distamycin in simian virus 40 DNA. J Biol Chem. 1989 Jul 5;264(19):11354–11359. [PubMed] [Google Scholar]
- Finlay G. J., Baguley B. C. Potentiation by phenylbisbenzimidazoles of cytotoxicity of anticancer drugs directed against topoisomerase II. Eur J Cancer. 1990;26(5):586–589. doi: 10.1016/0277-5379(90)90083-6. [DOI] [PubMed] [Google Scholar]
- Fish E. L., Lane M. J., Vournakis J. N. Determination of equilibrium binding affinity of distamycin and netropsin to the synthetic deoxyoligonucleotide sequence d(GGTATACC)2 by quantitative DNase I footprinting. Biochemistry. 1988 Aug 9;27(16):6026–6032. doi: 10.1021/bi00416a030. [DOI] [PubMed] [Google Scholar]
- Forsblom S., Rigler R., Ehrenberg M., Philipson L. Kinetic studies on the cleavage of adenovirus DNA by restriction endonuclease Eco RI. Nucleic Acids Res. 1976 Dec;3(12):3255–3269. doi: 10.1093/nar/3.12.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring W. J., Müller M., Affolter M., Percival-Smith A., Billeter M., Qian Y. Q., Otting G., Wüthrich K. The structure of the homeodomain and its functional implications. Trends Genet. 1990 Oct;6(10):323–329. doi: 10.1016/0168-9525(90)90253-3. [DOI] [PubMed] [Google Scholar]
- Hanes S. D., Brent R. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell. 1989 Jun 30;57(7):1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- Harshman K. D., Dervan P. B. Molecular recognition of B-DNA by Hoechst 33258. Nucleic Acids Res. 1985 Jul 11;13(13):4825–4835. doi: 10.1093/nar/13.13.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., Scott M. P. What determines the specificity of action of Drosophila homeodomain proteins? Cell. 1990 Nov 30;63(5):883–894. doi: 10.1016/0092-8674(90)90492-w. [DOI] [PubMed] [Google Scholar]
- Hogan M., Dattagupta N., Crothers D. M. Transmission of allosteric effects in DNA. Nature. 1979 Apr 5;278(5704):521–524. doi: 10.1038/278521a0. [DOI] [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Kissinger C. R., Liu B. S., Martin-Blanco E., Kornberg T. B., Pabo C. O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990 Nov 2;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt C., Tovar K., Hillen W. Computer simulations and experimental studies of gel mobility patterns for weak and strong non-cooperative protein binding to two targets on the same DNA: application to binding of tet repressor variants to multiple and single tet operator sites. Nucleic Acids Res. 1991 Mar 11;19(5):1021–1028. doi: 10.1093/nar/19.5.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevit R. E., Wemmer D. E., Reid B. R. 1H NMR studies on the interaction between distamycin A and a symmetrical DNA dodecamer. Biochemistry. 1986 Jun 3;25(11):3296–3303. doi: 10.1021/bi00359a032. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Yoon C., Goodsell D., Pjura P., Dickerson R. E. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1376–1380. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyedov A. A., Grokhovsky S. L., Zhuze A. L., Nosikov V. V., Polyanovsky O. L. Distamycin A and its analogs as agents for blocking of endo R. EcoRI activity. Gene. 1977 Jul;1(5-6):389–395. doi: 10.1016/0378-1119(77)90043-9. [DOI] [PubMed] [Google Scholar]
- Käs E., Izaurralde E., Laemmli U. K. Specific inhibition of DNA binding to nuclear scaffolds and histone H1 by distamycin. The role of oligo(dA).oligo(dT) tracts. J Mol Biol. 1989 Dec 5;210(3):587–599. doi: 10.1016/0022-2836(89)90134-4. [DOI] [PubMed] [Google Scholar]
- Lane M. J., Dabrowiak J. C., Vournakis J. N. Sequence specificity of actinomycin D and Netropsin binding to pBR322 DNA analyzed by protection from DNase I. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3260–3264. doi: 10.1073/pnas.80.11.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughon A., Scott M. P. Sequence of a Drosophila segmentation gene: protein structure homology with DNA-binding proteins. Nature. 1984 Jul 5;310(5972):25–31. doi: 10.1038/310025a0. [DOI] [PubMed] [Google Scholar]
- Levy A., Weisman-Shomer P., Fry M. Distamycin paradoxically stimulates the copying of oligo(dA).poly(dT) by DNA polymerases. Biochemistry. 1989 Sep 5;28(18):7262–7267. doi: 10.1021/bi00444a018. [DOI] [PubMed] [Google Scholar]
- Malcolm A. D., Moffatt J. R. Differential reactivities at restriction enzyme sites. Biochim Biophys Acta. 1981 Sep 28;655(2):128–135. doi: 10.1016/0005-2787(81)90002-2. [DOI] [PubMed] [Google Scholar]
- Martello P. A., Bruzik J. P., deHaseth P., Youngquist R. S., Dervan P. B. Specific activation of open complex formation at an Escherichia coli promoter by oligo(N-methylpyrrolecarboxamide)s: effects of peptide length and identification of DNA target sites. Biochemistry. 1989 May 16;28(10):4455–4461. doi: 10.1021/bi00436a050. [DOI] [PubMed] [Google Scholar]
- McCarthy J. G., Williams L. D., Rich A. Chemical reactivity of potassium permanganate and diethyl pyrocarbonate with B DNA: specific reactivity with short A-tracts. Biochemistry. 1990 Jun 26;29(25):6071–6081. doi: 10.1021/bi00477a027. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Garber R. L., Wirz J., Kuroiwa A., Gehring W. J. A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell. 1984 Jun;37(2):403–408. doi: 10.1016/0092-8674(84)90370-2. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Levine M. S., Hafen E., Kuroiwa A., Gehring W. J. A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. 1984 Mar 29-Apr 4Nature. 308(5958):428–433. doi: 10.1038/308428a0. [DOI] [PubMed] [Google Scholar]
- McHugh M. M., Woynarowski J. M., Sigmund R. D., Beerman T. A. Effect of minor groove binding drugs on mammalian topoisomerase I activity. Biochem Pharmacol. 1989 Jul 15;38(14):2323–2328. doi: 10.1016/0006-2952(89)90472-3. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Mortensen U. H., Stevnsner T., Krogh S., Olesen K., Westergaard O., Bonven B. J. Distamycin inhibition of topoisomerase I-DNA interaction: a mechanistic analysis. Nucleic Acids Res. 1990 Apr 25;18(8):1983–1989. doi: 10.1093/nar/18.8.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Affolter M., Leupin W., Otting G., Wüthrich K., Gehring W. J. Isolation and sequence-specific DNA binding of the Antennapedia homeodomain. EMBO J. 1988 Dec 20;7(13):4299–4304. doi: 10.1002/j.1460-2075.1988.tb03328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W., Crothers D. M. Studies of the binding of actinomycin and related compounds to DNA. J Mol Biol. 1968 Jul 28;35(2):251–290. doi: 10.1016/s0022-2836(68)80024-5. [DOI] [PubMed] [Google Scholar]
- Nilsson M. G., Skarped C., Magnusson G. Structure at restriction endonuclease MboI cleavage sites protected by actinomycin D or distamycin A. FEBS Lett. 1982 Aug 23;145(2):360–364. doi: 10.1016/0014-5793(82)80200-7. [DOI] [PubMed] [Google Scholar]
- Nosikov V. V., Braga E. A., Karlishev A. V., Zhuze A. L., Polyanovsky O. L. Protection of particular cleavage sites of restriction endonucleases by distamycin A and actinomycin D. Nucleic Acids Res. 1976 Sep;3(9):2293–2301. doi: 10.1093/nar/3.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otting G., Qian Y. Q., Billeter M., Müller M., Affolter M., Gehring W. J., Wüthrich K. Protein--DNA contacts in the structure of a homeodomain--DNA complex determined by nuclear magnetic resonance spectroscopy in solution. EMBO J. 1990 Oct;9(10):3085–3092. doi: 10.1002/j.1460-2075.1990.tb07505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Schevitz R. W., Zhang R. G., Lawson C. L., Joachimiak A., Marmorstein R. Q., Luisi B. F., Sigler P. B. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988 Sep 22;335(6188):321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Pelton J. G., Wemmer D. E. Structural characterization of a 2:1 distamycin A.d(CGCAAATTGGC) complex by two-dimensional NMR. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5723–5727. doi: 10.1073/pnas.86.15.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival-Smith A., Müller M., Affolter M., Gehring W. J. The interaction with DNA of wild-type and mutant fushi tarazu homeodomains. EMBO J. 1990 Dec;9(12):3967–3974. doi: 10.1002/j.1460-2075.1990.tb07617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y. Q., Billeter M., Otting G., Müller M., Gehring W. J., Wüthrich K. The structure of the Antennapedia homeodomain determined by NMR spectroscopy in solution: comparison with prokaryotic repressors. Cell. 1989 Nov 3;59(3):573–580. doi: 10.1016/0092-8674(89)90040-8. [DOI] [PubMed] [Google Scholar]
- Reeves R., Nissen M. S. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J Biol Chem. 1990 May 25;265(15):8573–8582. [PubMed] [Google Scholar]
- Schröter H., Maier G., Ponstingl H., Nordheim A. DNA intercalators induce specific release of HMG 14, HMG 17 and other DNA-binding proteins from chicken erythrocyte chromatin. EMBO J. 1985 Dec 30;4(13B):3867–3872. doi: 10.1002/j.1460-2075.1985.tb04159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz P. G., Dervan P. B. Distamycin and penta-N-methylpyrrolecarboxamide binding sites on native DNA. A comparison of methidiumpropyl-EDTA-Fe(II) footprinting and DNA affinity cleaving. J Biomol Struct Dyn. 1984 Mar;1(5):1133–1147. doi: 10.1080/07391102.1984.10507508. [DOI] [PubMed] [Google Scholar]
- Scott M. P., Tamkun J. W., Hartzell G. W., 3rd The structure and function of the homeodomain. Biochim Biophys Acta. 1989 Jul 28;989(1):25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- Selby C. P., Sancar A. Noncovalent drug-DNA binding interactions that inhibit and stimulate (A)BC excinuclease. Biochemistry. 1991 Apr 23;30(16):3841–3849. doi: 10.1021/bi00230a006. [DOI] [PubMed] [Google Scholar]
- Skorobogaty A., Brownlee R. T., Chandler C. J., Kyratzis I., Phillips D. R., Reiss J. A., Trist H. The DNA-association and biological activity of a new bis(14-thiadaunomycin). Anticancer Drug Des. 1988 Jun;3(1):41–56. [PubMed] [Google Scholar]
- Struhl K. Molecular mechanisms of transcriptional regulation in yeast. Annu Rev Biochem. 1989;58:1051–1077. doi: 10.1146/annurev.bi.58.070189.005155. [DOI] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Travers A. A. DNA conformation and protein binding. Annu Rev Biochem. 1989;58:427–452. doi: 10.1146/annurev.bi.58.070189.002235. [DOI] [PubMed] [Google Scholar]
- Treisman J., Gönczy P., Vashishtha M., Harris E., Desplan C. A single amino acid can determine the DNA binding specificity of homeodomain proteins. Cell. 1989 Nov 3;59(3):553–562. doi: 10.1016/0092-8674(89)90038-x. [DOI] [PubMed] [Google Scholar]
- Ushay H. M., Tullius T. D., Lippard S. J. Inhibition of the BamHI cleavage and unwinding of pBR322 deoxyribonucleic acid by the antitumor drug cis-dichlorodiammineplatinum(II). Biochemistry. 1981 Jun 23;20(13):3744–3748. doi: 10.1021/bi00516a012. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Hertzberg R. P., Dervan P. B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II). Proc Natl Acad Sci U S A. 1982 Sep;79(18):5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton R. P., Ptashne M. A new-specificity mutant of 434 repressor that defines an amino acid-base pair contact. 1987 Apr 30-May 6Nature. 326(6116):888–891. doi: 10.1038/326888a0. [DOI] [PubMed] [Google Scholar]
- Woynarowski J. M., McHugh M., Sigmund R. D., Beerman T. A. Modulation of topoisomerase II catalytic activity by DNA minor groove binding agents distamycin, Hoechst 33258, and 4',6-diamidine-2-phenylindole. Mol Pharmacol. 1989 Feb;35(2):177–182. [PubMed] [Google Scholar]
- Woynarowski J. M., Sigmund R. D., Beerman T. A. DNA minor groove binding agents interfere with topoisomerase II mediated lesions induced by epipodophyllotoxin derivative VM-26 and acridine derivative m-AMSA in nuclei from L1210 cells. Biochemistry. 1989 May 2;28(9):3850–3855. doi: 10.1021/bi00435a034. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zimmer C. Effects of the antibiotics netropsin and distamycin A on the structure and function of nucleic acids. Prog Nucleic Acid Res Mol Biol. 1975;15(0):285–318. doi: 10.1016/s0079-6603(08)60122-1. [DOI] [PubMed] [Google Scholar]
- Zimmer C., Wähnert U. Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Prog Biophys Mol Biol. 1986;47(1):31–112. doi: 10.1016/0079-6107(86)90005-2. [DOI] [PubMed] [Google Scholar]