Abstract
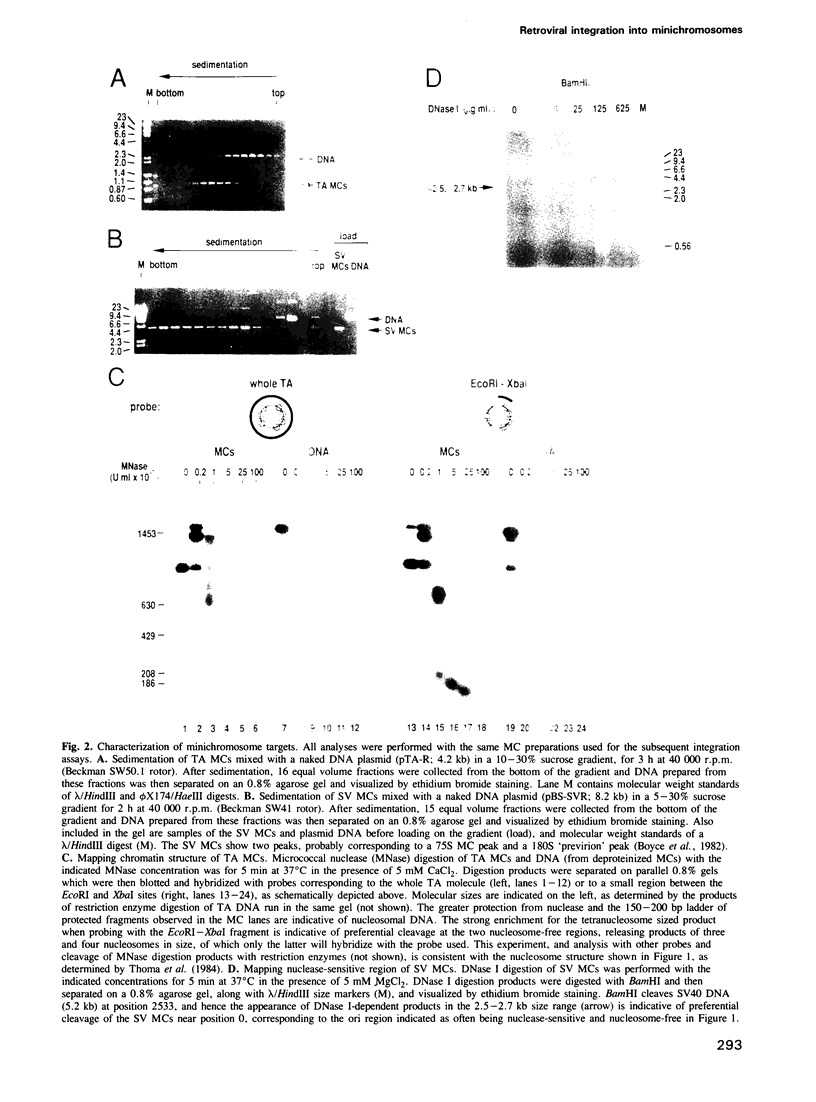
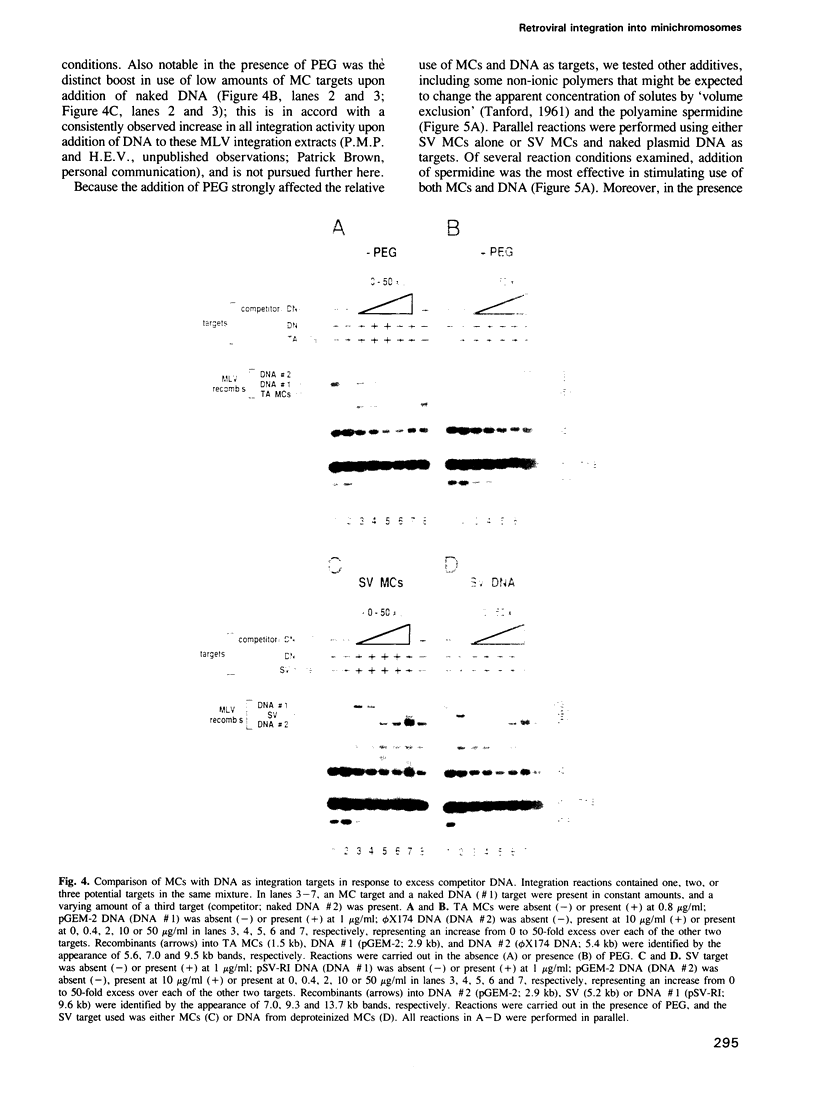
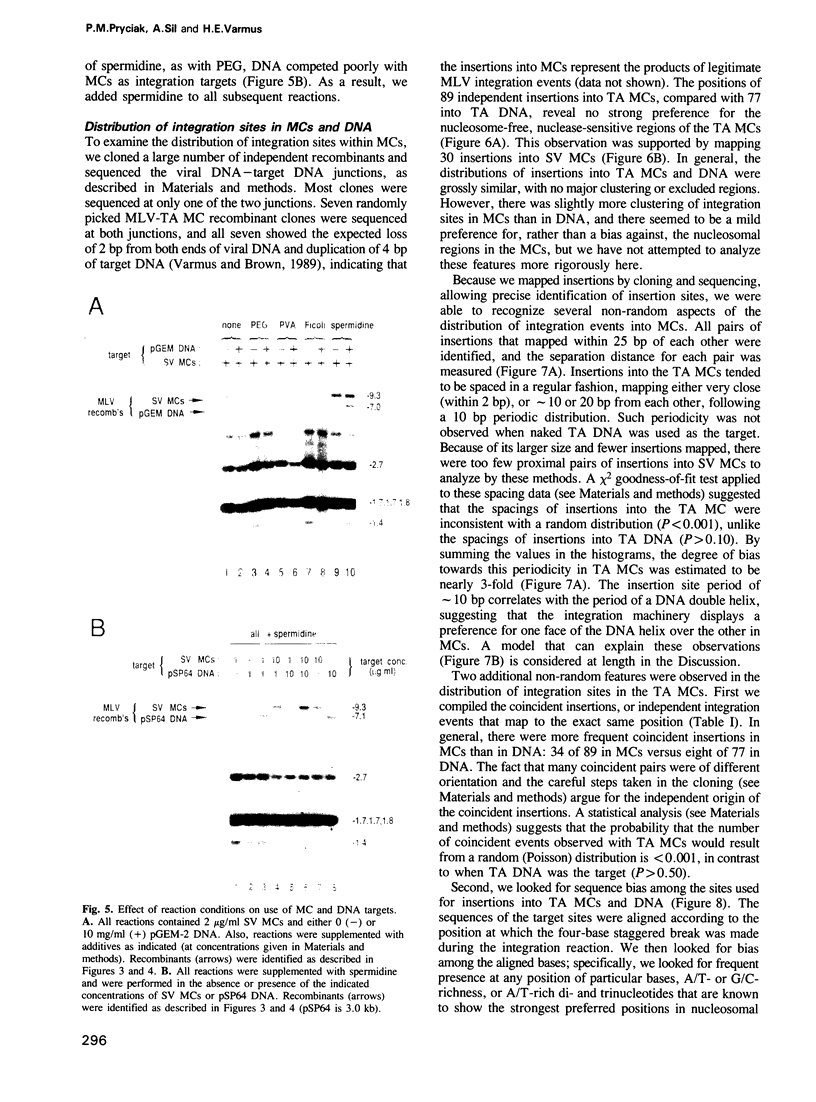
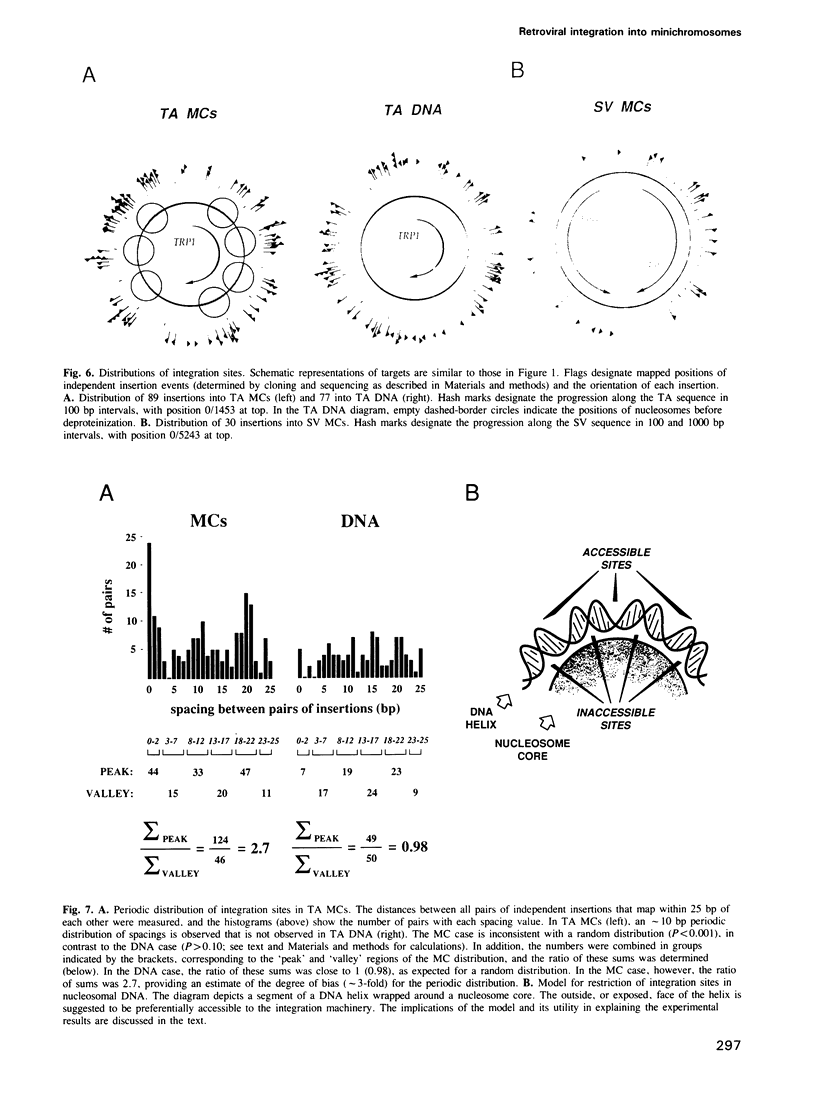
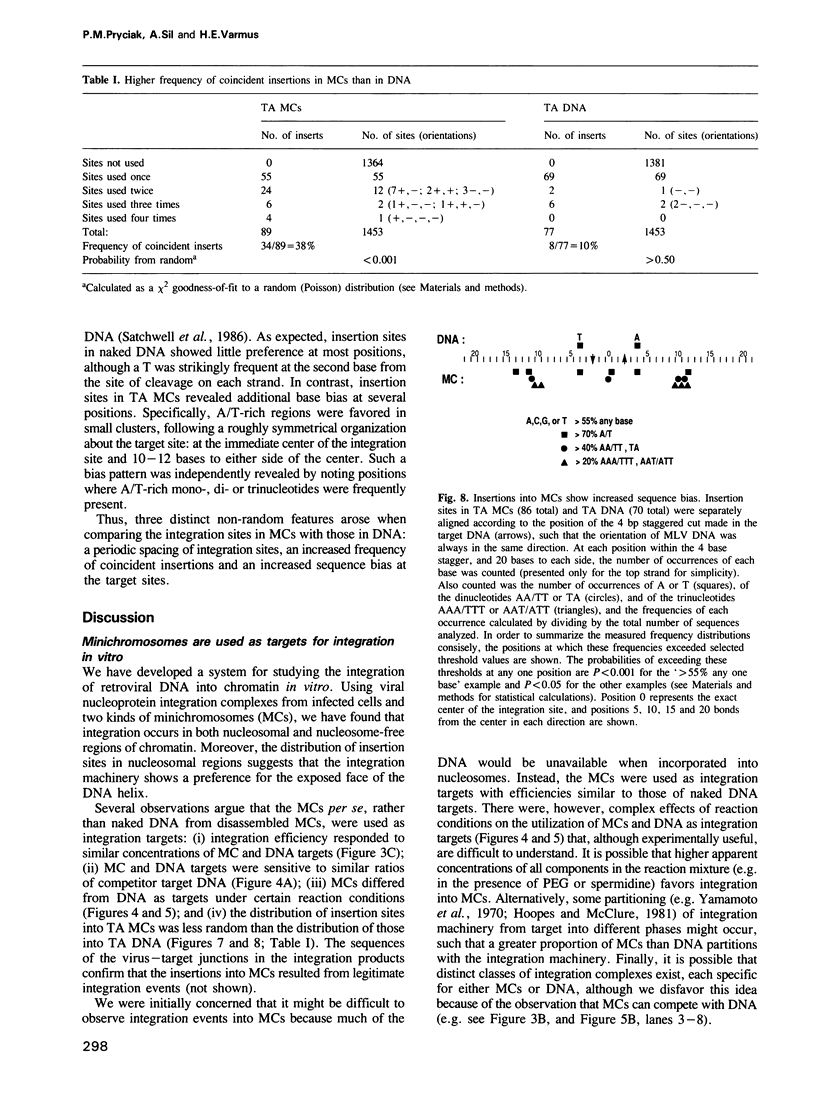
We describe here the use of chromatin as a target for retroviral integration in vitro. Extracts of cells newly infected with murine leukemia virus (MLV) provided the source of integration activity, and yeast TRP1ARS1 and SV40 minichromosomes served as simple models for chromatin. Both minichromosomes were used as targets for integration, with efficiencies comparable with that of naked DNA. In addition, under some reaction conditions the minichromosomes behaved as if they were used preferentially over naked DNAs in the same reaction. Mapping of integration sites by cloning and sequencing recombinants revealed that the integration machinery does not display a preference for nucleosome-free, nuclease-sensitive regions. The distributions of integration sites in TRP1ARS1 minichromosomes and a naked DNA counterpart were grossly similar, but in a detailed analysis the distribution in minichromosomes was found to be significantly more ordered: the sites displayed a periodic spacing of approximately 10 bp, many sites sustained multiple insertions and there was sequence bias at the target sites. These results are in accord with a model in which the integration machinery has preferential access to the exposed face of the nucleosomal DNA helix. The population of potential sites in chromatin therefore becomes more limited, in a manner dictated by the rotational orientation of the DNA sequence around the nucleosome core, and those sites are used more frequently than in naked DNA.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambrose C., Blasquez V., Bina M. A block in initiation of simian virus 40 assembly results in the accumulation of minichromosomes containing an exposed regulatory region. Proc Natl Acad Sci U S A. 1986 May;83(10):3287–3291. doi: 10.1073/pnas.83.10.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose C., Lowman H., Rajadhyaksha A., Blasquez V., Bina M. Location of nucleosomes in simian virus 40 chromatin. J Mol Biol. 1990 Aug 20;214(4):875–884. doi: 10.1016/0022-2836(90)90342-J. [DOI] [PubMed] [Google Scholar]
- Ambrose C., Rajadhyaksha A., Lowman H., Bina M. Locations of nucleosomes on the regulatory region of simian virus 40 chromatin. J Mol Biol. 1989 Nov 20;210(2):255–263. doi: 10.1016/0022-2836(89)90328-8. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Bonne-Andrea C., Wong M. L., Alberts B. M. In vitro replication through nucleosomes without histone displacement. Nature. 1990 Feb 22;343(6260):719–726. doi: 10.1038/343719a0. [DOI] [PubMed] [Google Scholar]
- Bowerman B., Brown P. O., Bishop J. M., Varmus H. E. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989 Apr;3(4):469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- Boyce F. M., Sundin O., Barsoum J., Varshavsky A. New way to isolate simian virus 40 nucleoprotein complexes from infected cells: use of a thiol-specific reagent. J Virol. 1982 Apr;42(1):292–296. doi: 10.1128/jvi.42.1.292-296.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P. O., Bowerman B., Varmus H. E., Bishop J. M. Correct integration of retroviral DNA in vitro. Cell. 1987 May 8;49(3):347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- Brown P. O., Bowerman B., Varmus H. E., Bishop J. M. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker D. L., Sandmeyer S. B. Transfer RNA genes are genomic targets for de Novo transposition of the yeast retrotransposon Ty3. Genetics. 1990 Dec;126(4):837–850. doi: 10.1093/genetics/126.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R., Fujiwara T., Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990 Aug 24;62(4):829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- Dean A., Pederson D. S., Simpson R. T. Isolation of yeast plasmid chromatin. Methods Enzymol. 1989;170:26–41. doi: 10.1016/0076-6879(89)70041-0. [DOI] [PubMed] [Google Scholar]
- Drew H. R. Structural specificities of five commonly used DNA nucleases. J Mol Biol. 1984 Jul 15;176(4):535–557. doi: 10.1016/0022-2836(84)90176-1. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Eichinger D. J., Boeke J. D. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell. 1988 Sep 23;54(7):955–966. doi: 10.1016/0092-8674(88)90110-9. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gross D. S., Garrard W. T. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone function in transcription. Annu Rev Cell Biol. 1990;6:643–678. doi: 10.1146/annurev.cb.06.110190.003235. [DOI] [PubMed] [Google Scholar]
- Hoopes B. C., McClure W. R. Studies on the selectivity of DNA precipitation by spermine. Nucleic Acids Res. 1981 Oct 24;9(20):5493–5504. doi: 10.1093/nar/9.20.5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. W. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:428–453. doi: 10.1016/0076-6879(91)94034-a. [DOI] [PubMed] [Google Scholar]
- Katz R. A., Merkel G., Kulkosky J., Leis J., Skalka A. M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990 Oct 5;63(1):87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- King W., Patel M. D., Lobel L. I., Goff S. P., Nguyen-Huu M. C. Insertion mutagenesis of embryonal carcinoma cells by retroviruses. Science. 1985 May 3;228(4699):554–558. doi: 10.1126/science.3838595. [DOI] [PubMed] [Google Scholar]
- Lee Y. M., Coffin J. M. Efficient autointegration of avian retrovirus DNA in vitro. J Virol. 1990 Dec;64(12):5958–5965. doi: 10.1128/jvi.64.12.5958-5965.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linxweller W., Hörz W. Reconstitution experiments show that sequence-specific histone-DNA interactions are the basis for nucleosome phasing on mouse satellite DNA. Cell. 1985 Aug;42(1):281–290. doi: 10.1016/s0092-8674(85)80123-9. [DOI] [PubMed] [Google Scholar]
- Lorch Y., LaPointe J. W., Kornberg R. D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987 Apr 24;49(2):203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- Luchnik A. N., Bakayev V. V., Zbarsky I. B., Georgiev G. P. Elastic torsional strain in DNA within a fraction of SV40 minichromosomes: relation to transcriptionally active chromatin. EMBO J. 1982;1(11):1353–1358. doi: 10.1002/j.1460-2075.1982.tb01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter L. C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978 Sep 15;124(2):391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- Marck C. 'DNA Strider': a 'C' program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988 Mar 11;16(5):1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooslehner K., Karls U., Harbers K. Retroviral integration sites in transgenic Mov mice frequently map in the vicinity of transcribed DNA regions. J Virol. 1990 Jun;64(6):3056–3058. doi: 10.1128/jvi.64.6.3056-3058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsoulis G., Thomas W., Roghmann M. C., Winston F., Boeke J. D. Ty1 transposition in Saccharomyces cerevisiae is nonrandom. Genetics. 1989 Oct;123(2):269–279. doi: 10.1093/genetics/123.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J., Masson J. M., Kleina L. G., Abelson J., Miller J. H. Construction of two Escherichia coli amber suppressor genes: tRNAPheCUA and tRNACysCUA. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6548–6552. doi: 10.1073/pnas.83.17.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudet P., Weiss E., Regnier E. Preparation of simian virus 40 minichromosomes. Methods Enzymol. 1989;170:14–25. doi: 10.1016/0076-6879(89)70040-9. [DOI] [PubMed] [Google Scholar]
- Pederson D. S., Thoma F., Simpson R. T. Core particle, fiber, and transcriptionally active chromatin structure. Annu Rev Cell Biol. 1986;2:117–147. doi: 10.1146/annurev.cb.02.110186.001001. [DOI] [PubMed] [Google Scholar]
- Pederson D. S., Venkatesan M., Thoma F., Simpson R. T. Isolation of an episomal yeast gene and replication origin as chromatin. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7206–7210. doi: 10.1073/pnas.83.19.7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmann T., Wrange O. Specific glucocorticoid receptor binding to DNA reconstituted in a nucleosome. EMBO J. 1988 Oct;7(10):3073–3079. doi: 10.1002/j.1460-2075.1988.tb03172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piña B., Brüggemeier U., Beato M. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell. 1990 Mar 9;60(5):719–731. doi: 10.1016/0092-8674(90)90087-u. [DOI] [PubMed] [Google Scholar]
- Raines M. A., Lewis W. G., Crittenden L. B., Kung H. J. c-erbB activation in avian leukosis virus-induced erythroblastosis: clustered integration sites and the arrangement of provirus in the c-erbB alleles. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2287–2291. doi: 10.1073/pnas.82.8.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S., DeGregori J. V., von Melchner H., Ruley H. E. Retrovirus promoter-trap vector to induce lacZ gene fusions in mammalian cells. J Virol. 1991 Mar;65(3):1507–1515. doi: 10.1128/jvi.65.3.1507-1515.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohdewohld H., Weiher H., Reik W., Jaenisch R., Breindl M. Retrovirus integration and chromatin structure: Moloney murine leukemia proviral integration sites map near DNase I-hypersensitive sites. J Virol. 1987 Feb;61(2):336–343. doi: 10.1128/jvi.61.2.336-343.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeyer S. B., Hansen L. J., Chalker D. L. Integration specificity of retrotransposons and retroviruses. Annu Rev Genet. 1990;24:491–518. doi: 10.1146/annurev.ge.24.120190.002423. [DOI] [PubMed] [Google Scholar]
- Saragosti S., Moyne G., Yaniv M. Absence of nucleosomes in a fraction of SV40 chromatin between the origin of replication and the region coding for the late leader RNA. Cell. 1980 May;20(1):65–73. doi: 10.1016/0092-8674(80)90235-4. [DOI] [PubMed] [Google Scholar]
- Satchwell S. C., Drew H. R., Travers A. A. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986 Oct 20;191(4):659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Scherdin U., Rhodes K., Breindl M. Transcriptionally active genome regions are preferred targets for retrovirus integration. J Virol. 1990 Feb;64(2):907–912. doi: 10.1128/jvi.64.2.907-912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzberg P., Colicelli J., Goff S. P. Deletion mutants of Moloney murine leukemia virus which lack glycosylated gag protein are replication competent. J Virol. 1983 May;46(2):538–546. doi: 10.1128/jvi.46.2.538-546.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selten G., Cuypers H. T., Zijlstra M., Melief C., Berns A. Involvement of c-myc in MuLV-induced T cell lymphomas in mice: frequency and mechanisms of activation. EMBO J. 1984 Dec 20;3(13):3215–3222. doi: 10.1002/j.1460-2075.1984.tb02281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton E. R., Wassarman P. M., DePamphilis M. L. Structure, spacing, and phasing of nucleosomes on isolated forms of mature simian virus 40 chromosomes. J Biol Chem. 1980 Jan 25;255(2):771–782. [PubMed] [Google Scholar]
- Shih C. C., Stoye J. P., Coffin J. M. Highly preferred targets for retrovirus integration. Cell. 1988 May 20;53(4):531–537. doi: 10.1016/0092-8674(88)90569-7. [DOI] [PubMed] [Google Scholar]
- Shih C. K., Linial M., Goodenow M. M., Hayward W. S. Nucleotide sequence 5' of the chicken c-myc coding region: localization of a noncoding exon that is absent from myc transcripts in most avian leukosis virus-induced lymphomas. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4697–4701. doi: 10.1073/pnas.81.15.4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno K., Temin H. M. No apparent nucleotide sequence specificity in cellular DNA juxtaposed to retrovirus proviruses. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7357–7361. doi: 10.1073/pnas.77.12.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C., Goff S., Gilboa E., Paskind M., Mitra S. W., Baltimore D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: implications for retrovirus integration. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3932–3936. doi: 10.1073/pnas.77.7.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C., Hoffman J., Goff S. P., Baltimore D. Intramolecular integration within Moloney murine leukemia virus DNA. J Virol. 1981 Oct;40(1):164–172. doi: 10.1128/jvi.40.1.164-172.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrader T. E., Crothers D. M. Artificial nucleosome positioning sequences. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990 Jan 25;343(6256):387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Stafford D. W. Structural features of a phased nucleosome core particle. Proc Natl Acad Sci U S A. 1983 Jan;80(1):51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo J. M., Stahl H., Koller T., Knippers R. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J Mol Biol. 1986 May 5;189(1):189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
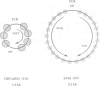
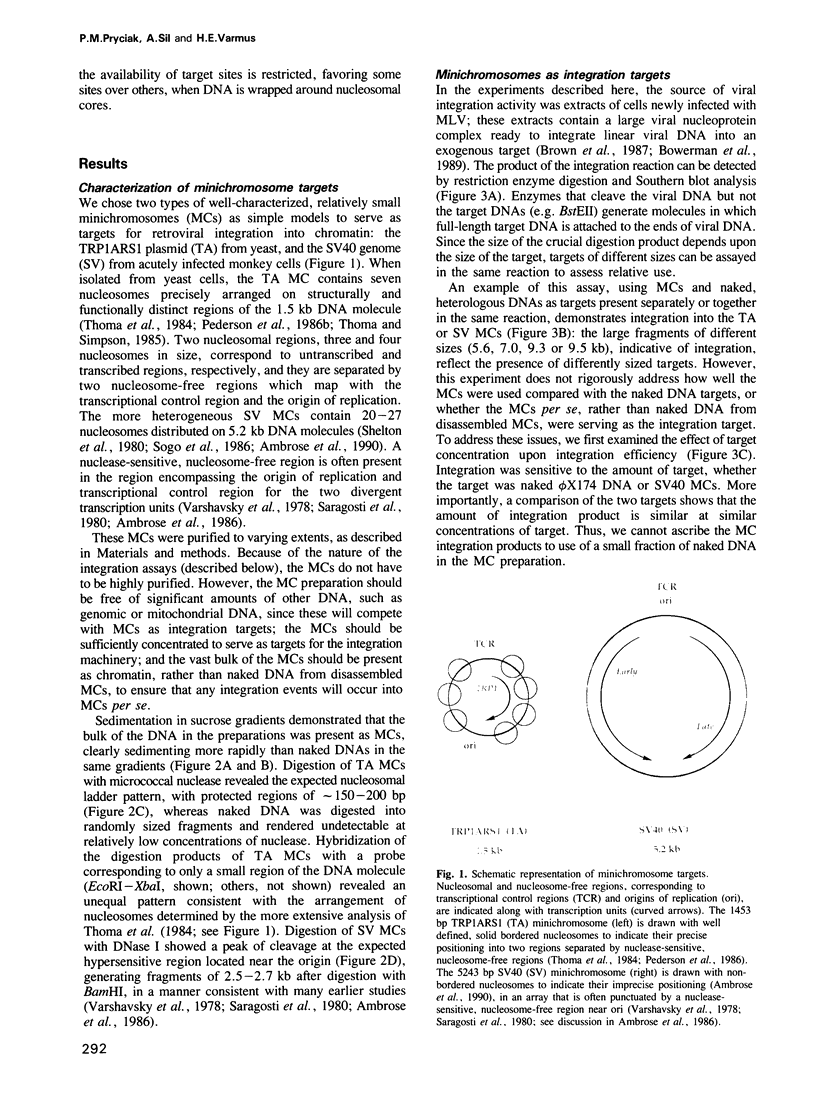
- Thoma F., Bergman L. W., Simpson R. T. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J Mol Biol. 1984 Aug 25;177(4):715–733. doi: 10.1016/0022-2836(84)90046-9. [DOI] [PubMed] [Google Scholar]
- Thoma F., Simpson R. T. Local protein-DNA interactions may determine nucleosome positions on yeast plasmids. Nature. 1985 May 16;315(6016):250–252. doi: 10.1038/315250a0. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Sundin O. H., Bohn M. J. SV40 viral minichromosome: preferential exposure of the origin of replication as probed by restriction endonucleases. Nucleic Acids Res. 1978 Oct;5(10):3469–3477. doi: 10.1093/nar/5.10.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaya S., Steffen D. L., Robinson H. L. Acceptor sites for retroviral integrations map near DNase I-hypersensitive sites in chromatin. J Virol. 1986 Nov;60(2):683–692. doi: 10.1128/jvi.60.2.683-692.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]