Abstract
Many studies have been conducted to investigate the association between miR-27 rs895819 A > G and miR-423 rs6505162 C > A and cancer risk; however, the results are not consistent. In order to acquire a more precise assessment of the correlation, we performed this meta-analysis. We searched PubMed, EMBASE and Web of Science databases to identify eligible studies. Pooled odds ratios (ORs) and 95% confidence intervals (CIs) were applied to evaluate the correlation of these two microRNA polymorphisms with cancer risk. Forty-five eligible studies from thirty-five articles were included in our analysis. The results showed that rs895819 was associated with a decreased cancer risk in Caucasians (AG vs. AA: OR = 0.87, 95% CI = 0.79-0.96; GG+AG vs. AA: OR = 0.89, 95% CI = 0.81-0.98). When grouped by ethnicity, an increased risk was observed in colorectal cancer (G vs. A: OR = 1.19, 95% CI = 1.08-1.32; GG vs. AA: OR = 1.58, 95% CI = 1.28-1.96; GG vs. AG+AA: OR = 1.58, 95% CI = 1.29-1.93), while a decreased risk was found in breast cancer (G vs. A: OR = 0.93, 95% CI = 0.87-0.99; GG+AG vs. AA: OR = 0.91, 95% CI = 0.83-0.99). For rs6505162, a significantly decreased cancer risk was observed in lung cancer under all five genetic models. To summarize, our results indicated that rs895819 was a protective factor for cancer in Caucasians and could increase colorectal cancer risk but decrease breast cancer risk. Moreover, rs6505162 was a protective factor for lung cancer.
Keywords: cancer, rs895819, rs6505162, meta-analysis
INTRODUCTION
MicroRNAs (miRNAs) are a class of small, endogenous, non-coding, single-stranded, highly conserved and tissue-specific RNA molecules which take part in the regulation of target mRNAs expression at the post-transcriptional level [1, 2]. By binding to the complementary sequence of the 3’ untranslated region of the specific mRNAs, miRNAs could increase or suppress the expression of multiple target genes, including cancer-associated genes, and thus are involved in many physiological and pathological process, such as cell proliferation, cell differentiation, cell apoptosis, carcinogenesis and development and so on [3, 4].
As the most common type of genetic variation in the human genome, single nucleotide polymorphisms (SNPs) occurring in miRNA genes have been found to be able to affect miRNA expression and function by interfering the interaction between miRNAs and their corresponding target mRNAs. A large number of studies have demonstrated that SNPs in the miRNA genes are associated with the occurrence of various diseases, such as cancer, so microRNA polymorphisms are considered to be a potentially important mechanism in the acquisition of cancer susceptibility [5, 6]. In recent years, some SNPs in miRNAs have been identified as oncogenes of many different types of cancer and found to play an important role in initiation and development of malignancies[7, 8].
However, for two common functional microRNA polymorphisms miR-27 rs895819 and miR-423 rs6505162, although many case-control studies involving various cancer types have been performed among different ethnic populations, there is currently no consensus on whether there exists an association between these two microRNA polymorphisms and cancer risk because of inconsistent results published studies reported. In addition, many relevant case-control studies on this theme have been published recently, which will expand the sample size and may have an impact on the evaluation of the association [9–13]. Therefore, we carried out an updated and systematic meta-analysis including those newly published articles to further assess the correlation of rs895819 and rs6505162 with cancer risk based on all available eligible studies at present.
RESULTS
Characteristics of included studies
The selection process of eligible studies is presented in Figure 1. A total of 1929 potentially relevant articles were preliminarily identified though a systematic publication search. After excluding duplicate literatures and further carefully reading titles and abstracts of the remaining studies, forty-one articles were performed full-text review for eligibility, among which six articles were excluded for the following reasons: three articles did not have sufficient data; three articles did not conform to HWE. Thus thirty-five articles were considered to meet our inclusion criteria [9–43]. Among them, seven articles involved not only rs895819 but also rs6505162 [9, 11, 14–15, 19, 29, 36]. Moreover, the study by Kontorovich et al. [36] was performed in both breast cancer and ovarian cancer, so we treated the study as two independent investigations. Ultimately, forty-five studies from thirty-five articles were included in quantitative synthesis. The main characteristics of the included studies are listed in Table 1. Of all the included studies, fifteen focused on breast cancer, eight on esophageal squamous cell carcinoma, six on gastric cancer, five on colorectal cancer, four on lung cancer, two on ovarian cancer, and one each on prostate cancer, gallbladder cancer, cervical cancer, renal cell cancer and hepatocellular carcinoma. In addition, thirty-five studies were carried out among Asians, and eight among Caucasians. A variety of genotyping methods including PCR-RFLP, MassARRAY, Taqman, Sequencing, PCR-LDR, SNaPshot, Allele-specific PCR, ARMS-PCR and HRM were applied in the included studies of this meta-analysis. Moreover, twenty studies were considered as moderate-quality studies (Newcastle-Ottawa Scale scores of these studies were 4-6), and other twenty-five studies were considered as high-quality studies (Newcastle-Ottawa Scale scores of these studies were 7 or above). Genotype distributions of all forty-five studies in the controls were in agreement with Hardy-Weinberg equilibrium (HWE).
Figure 1. Flow diagram of the selection process of included studies in this meta-analysis.
Table 1. The main characteristics of included studies in the meta-analysis.
| First author | Year | Country | Ethnicity | Cancer type | Genotyping methods | Source of controls | Case (n) | Control (n) | HWE | NOS | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AB | BB | AA | AB | BB | |||||||||
| rs895819 | ||||||||||||||
| Jiang | 2016 | China | Asian | GC | MassARRAY | HB | 480 | 356 | 59 | 537 | 389 | 62 | 0.447 | 7 |
| Jiang | 2016 | China | Asian | CRC | TaqMan | HB | 245 | 176 | 87 | 275 | 222 | 65 | 0.053 | 7 |
| Yin | 2016 | China | Asian | LC | TaqMan | HB | 321 | 217 | 37 | 318 | 252 | 38 | 0.199 | 7 |
| Bian | 2015 | China | Asian | CRC | TaqMan | HB | 199 | 143 | 70 | 205 | 166 | 41 | 0.389 | 7 |
| He | 2015 | China | Asian | BC | MassARRAY | PB | 251 | 165 | 34 | 232 | 181 | 37 | 0.839 | 7 |
| Gupta | 2015 | India | Asian | GBC | TaqMan | PB | 75 | 81 | 20 | 26 | 24 | 3 | 0.399 | 6 |
| Nikolic | 2015 | Serbia | Caucasian | PC | ASPCR | PB | 151 | 172 | 30 | 152 | 137 | 19 | 0.101 | 6 |
| Qi | 2015 | China | Asian | BC | TaqMan | PB | 101 | 159 | 61 | 95 | 139 | 56 | 0.686 | 7 |
| Zhang | 2015 | China | Asian | BC | MassARRAY | PB | 196 | 150 | 30 | 106 | 70 | 14 | 0.605 | 7 |
| Yin | 2015 | China | Asian | LC | TaqMan | HB | 138 | 103 | 17 | 167 | 125 | 18 | 0.391 | 6 |
| Cao | 2014 | China | Asian | CRC | PCR-RFLP | PB | 92 | 113 | 49 | 114 | 93 | 31 | 0.089 | 7 |
| Kupcinskas | 2014 | Countriesa | Caucasian | CRC | TaqMan | HB | 87 | 79 | 25 | 203 | 191 | 34 | 0.235 | 4 |
| Kupcinskas | 2014 | Countriesb | Caucasian | GC | TaqMan | HB | 181 | 144 | 38 | 156 | 164 | 30 | 0.151 | 6 |
| Song | 2014 | China | Asian | GC | Sequencing | HB | 105 | 124 | 49 | 131 | 111 | 36 | 0.110 | 7 |
| Xiong | 2014 | China | Asian | CC | PCR-LDR | HB | 48 | 40 | 15 | 223 | 170 | 24 | 0.255 | 7 |
| Zhang | 2014 | China | Asian | ESCC | SNaPshot | PB | 613 | 414 | 82 | 719 | 466 | 90 | 0.226 | 7 |
| Ma | 2013 | China | Asian | BC | MassARRAY | HB | 97 | 76 | 16 | 106 | 70 | 14 | 0.605 | 6 |
| Wei | 2013 | China | Asian | ESCC | MassARRAY | HB | 216 | 143 | 20 | 208 | 139 | 30 | 0.322 | 6 |
| Zhang | 2013 | China | Asian | BC | Sequencing | PB | 152 | 96 | 16 | 137 | 103 | 15 | 0.447 | 7 |
| Catucci | 2012 | Italy | Caucasian | BC | TaqMan | HB | 547 | 388 | 90 | 803 | 633 | 157 | 0.051 | 5 |
| Hezova | 2012 | Czech | Caucasian | CRC | TaqMan | HB | 88 | 86 | 23 | 93 | 94 | 25 | 0.867 | 6 |
| Shi | 2012 | China | Asian | RCC | TaqMan | HB | 334 | 213 | 47 | 288 | 262 | 50 | 0.373 | 7 |
| Zhang | 2012 | China | Asian | BC | PCR-RFLP | PB | 60 | 144 | 41 | 75 | 109 | 59 | 0.123 | 7 |
| Zhou | 2012 | China | Asian | GC | MassARRAY | HB | 166 | 122 | 7 | 214 | 167 | 32 | 0.941 | 5 |
| Sun | 2010 | China | Asian | GC | PCR-RFLP | HB | 115 | 135 | 54 | 145 | 119 | 40 | 0.053 | 6 |
| Kontorovich | 2010 | Israel | Asian | BC | MassARRAY | HB | 98 | 78 | 11 | 101 | 82 | 15 | 0.769 | 5 |
| Kontorovich | 2010 | Israel | Asian | OC | MassARRAY | HB | 43 | 34 | 3 | 101 | 82 | 15 | 0.769 | 5 |
| Yang | 2010 | Germany | Caucasian | BC | Sequencing | PB | 576 | 486 | 127 | 605 | 660 | 151 | 0.142 | 7 |
| rs6505162 | ||||||||||||||
| Yin | 2016 | China | Asian | LC | TaqMan | HB | 389 | 166 | 20 | 368 | 205 | 35 | 0.366 | 7 |
| Morales | 2016 | Chile | Caucasian | BC | TaqMan | HB | 125 | 229 | 86 | 284 | 385 | 138 | 0.700 | 5 |
| Jiang | 2016 | China | Asian | GC | MassARRAY | HB | 593 | 255 | 32 | 656 | 288 | 41 | 0.192 | 7 |
| Shen | 2016 | China | Asian | ESCC | SNaPshot | PB | 920 | 421 | 59 | 1421 | 680 | 84 | 0.814 | 7 |
| Zhang | 2015 | China | Asian | BC | MassARRAY | PB | 231 | 131 | 20 | 110 | 69 | 10 | 0.847 | 7 |
| Zhu | 2015 | China | Asian | ESCC | MassARRAY | PB | 99 | 122 | 21 | 109 | 140 | 31 | 0.159 | 7 |
| Yin | 2015 | China | Asian | LC | TaqMan | HB | 177 | 74 | 7 | 190 | 106 | 14 | 0.872 | 6 |
| He | 2015 | China | Asian | BC | MassARRAY | PB | 292 | 142 | 16 | 299 | 129 | 22 | 0.103 | 7 |
| Ma | 2014 | China | Asian | HCC | MassARRAY | HB | 652 | 297 | 42 | 643 | 313 | 30 | 0.273 | 7 |
| Ma | 2013 | China | Asian | BC | MassARRAY | HB | 127 | 57 | 8 | 110 | 69 | 10 | 0.847 | 6 |
| Umar | 2013 | India | Asian | ESCC | ARMS-PCR | HB | 90 | 132 | 67 | 96 | 143 | 70 | 0.233 | 7 |
| Yin | 2013 | China | Asian | ESCC | PCR-LDR | HB | 374 | 197 | 29 | 425 | 207 | 19 | 0.299 | 7 |
| Wang | 2013 | Countriesc | Black | ESCC | TaqMan | PB | 207 | 128 | 16 | 376 | 184 | 12 | 0.052 | 6 |
| Wang | 2013 | Countriesc | Mixed | ESCC | TaqMan | PB | 89 | 84 | 14 | 198 | 188 | 34 | 0.249 | 5 |
| Smith | 2012 | Australia | Caucasian | BC | HRM | HB | 60 | 95 | 24 | 52 | 80 | 42 | 0.307 | 7 |
| Kontorovich | 2010 | Israel | Caucasian | BC | iPLEX | PB | 68 | 88 | 34 | 55 | 102 | 49 | 0.899 | 5 |
| Kontorovich | 2010 | Israel | Caucasian | OC | iPLEX | PB | 31 | 26 | 22 | 55 | 102 | 49 | 0.899 | 5 |
A: the major allele; B: the minor allele; ASPCR: allele-specific PCR; GC: gastric cancer; CRC: colorectal cancer; LC: lung cancer; BC: breast cancer; GBC: gallbladder cancer; PC: prostate cancer; CC: cervical cancer; ESCC: esophageal squamous cell carcinoma; RCC: renal cell cancer; OC: ovarian cancer; HCC: hepatocellular carcinoma; PB: population based; HB: hospital based; a: Lithuania and Latvia; b: German, Lithuanian and Latvian; c: South Africa.
Meta-analysis results
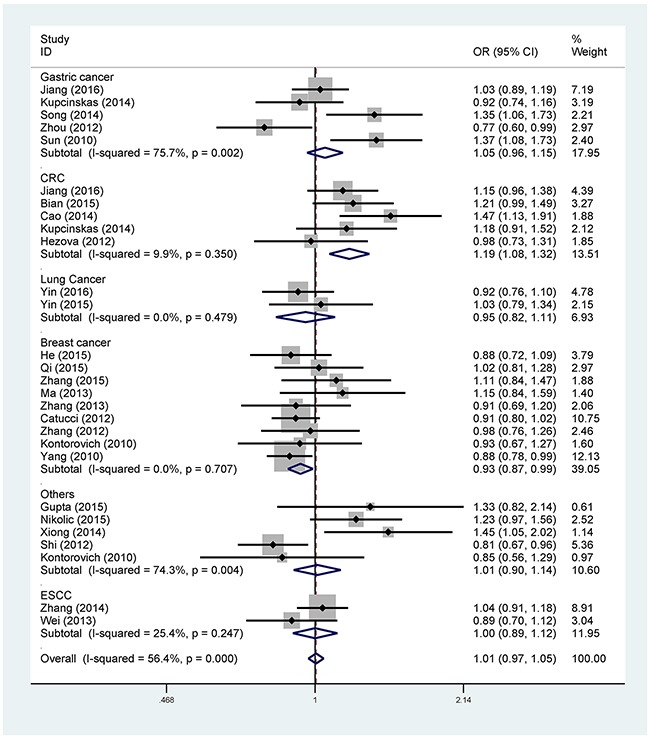
The main meta-analysis results of the correlation between rs895819 A > G and cancer susceptibility are listed in Table 2. In the overall analysis, we couldn't detect an association of the rs895819 polymorphism with cancer risk in all five genetic models. However, when analysis was stratified by ethnicity, a decreased cancer risk was observed in the Caucasian population (AG vs. AA: OR = 0.87, 95% CI = 0.79-0.96; GG+AG vs. AA: OR = 0.89, 95% CI = 0.81-0.98), while no significant correlation was found among Asians in all the genetic models (Table 2) (Figure 2). Moreover, the stratified analysis by cancer type revealed that rs895819 was associated with an increased risk of colorectal cancer (G vs. A: OR = 1.19, 95% CI = 1.08-1.32; GG vs. AA: OR = 1.58, 95% CI = 1.28-1.96; GG vs. AG+AA: OR = 1.58, 95% CI = 1.29-1.93), and a decreased risk of breast cancer (G vs. A: OR = 0.93, 95% CI = 0.87-0.99; GG+AG vs. AA: OR = 0.91, 95% CI = 0.83-0.99) (Table 2) (Figure 3). Nevertheless, we didn't find an association of the rs895819 polymorphism with risks of lung cancer, gastric cancer, prostate cancer, gallbladder cancer, cervical cancer, renal cell cancer, ovarian cancer and esophageal squamous cell carcinoma. We also performed the subgroup analysis by source of controls and the result indicated that there was no association between rs895819 and cancer risk in either population-based subgroup or hospital-based subgroup.
Table 2. Meta-analysis results of the association between rs895819 and cancer risk.
| Variables | Na | G vs. A | GG vs. AA | AG vs. AA | GG+AG vs. AA | GG vs.AG+AA | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | I2 (%) | OR (95% CI) | I2 (%) | OR (95% CI) | I2(%) | OR (95% CI) | I2 (%) | OR (95% CI) | I2(%) | ||
| Overall | 28 | 1.03 (0.97,1.10)b | 56.4 | 1.12 (0.97,1.29)b | 52.2 | 0.99 (0.91,1.06)b | 42.6 | 1.01 (0.94,1.09)b | 49.7 | 1.11 (0.97,1.26)b | 49.6 |
| Ethnicity | |||||||||||
| Asians | 22 | 1.05 (0.97,1.13)b | 55.9 | 1.13 (0.95,1.34)b | 54.2 | 1.02 (0.93,1.11)b | 36.6 | 1.04 (0.95,1.13)b | 44.9 | 1.10 (0.93,1.30)b | 54.9 |
| Caucasians | 6 | 0.94 (0.88,1.01) | 48.7 | 0.98 (0.83,1.15) | 36.8 | 0.87 (0.79,0.96) | 43.0 | 0.89 (0.81,0.98) | 49.0 | 1.05 (0.90,1.22) | 23.1 |
| Cancer type | |||||||||||
| BC | 9 | 0.93 (0.87,0.99) | 0.0 | 0.90 (0.78,1.04) | 0.0 | 0.97 (0.84,1.11)b | 50.3 | 0.91 (0.83,0.99) | 33.7 | 0.92 (0.80,1.05) | 0.0 |
| CRC | 5 | 1.19 (1.08,1.32) | 9.9 | 1.58 (1.28,1.96) | 0.0 | 0.98 (0.85,1.14) | 29.3 | 1.11 (0.97,1.27) | 23.0 | 1.58 (1.29,1.93) | 0.0 |
| LC | 2 | 0.95 (0.82,1.11) | 0.0 | 1.02 (0.69,1.51) | 0.0 | 0.90 (0.74,1.09) | 0.0 | 0.91 (0.76,1.10) | 0.0 | 1.07 (0.73,1.57) | 0.0 |
| GC | 5 | 1.06 (0.87,1.29)b | 75.7 | 1.08 (0.68,1.70)b | 74.7 | 1.06 (0.86,1.31)b | 60.8 | 1.08 (0.85,1.36)b | 70.9 | 1.05 (0.71,1.56)b | 68.6 |
| ESCC | 2 | 1.00 (0.89,1.12) | 25.4 | 0.95 (0.72,1.26) | 54.1 | 1.03 (0.89,1.19) | 0.0 | 1.02 (0.88,1.17) | 0.0 | 0.94 (0.72,1.24) | 52.4 |
| Others | 5 | 1.09 (0.84,1.41)b | 74.3 | 1.35 (0.73,2.49)b | 69.0 | 0.99 (0.75,1.31)b | 59.5 | 1.05 (0.77,1.43)b | 69.4 | 1.35 (0.80,2.28)b | 60.5 |
| Source of controls | |||||||||||
| PB | 10 | 1.04 (0.94,1.15)b | 55.9 | 1.05 (0.91,1.21) | 22.2 | 1.06 (0.90,1.24)b | 63.7 | 1.07 (0.92,1.24)b | 63.8 | 1.03 (0.90,1.18) | 15.2 |
| HB | 18 | 1.03 (0.94,1.11)b | 59.0 | 1.12 (0.91,1.37)b | 61.7 | 0.94 (0.88,1.01) | 21.4 | 0.99 (0.90,1.08)b | 40.5 | 1.14 (0.94,1.37)b | 58.9 |
a: number of studies; b: calculating based on random-effects model; BC: breast cancer; CRC: colorectal cancer; LC: lung cancer; GC: gastric cancer; ESCC: esophageal squamous cell carcinoma.
Figure 2. Forest plot of the association between rs895819 and cancer risk in subgroup analysis by ethnicity under heterozygote model.

Figure 3. Forest plot of the association between rs895819 and cancer risk in subgroup analysis by cancer type under allele contrast model.
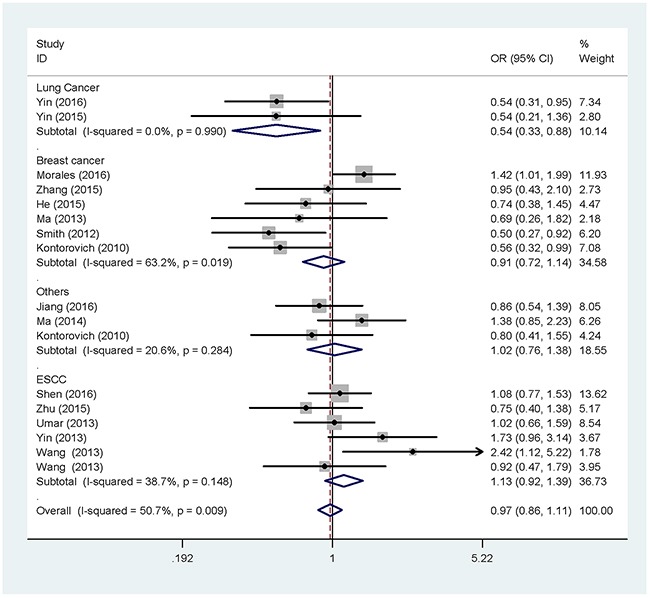
For rs6505162 C > A, no significant risk association was observed in the overall analysis. Similarly, when grouped by ethnicity, we couldn't detect a correlation of the rs6505162 polymorphism with cancer risk in either Caucasians or Asians. However, after subgroup analysis by cancer type, the rs6505162 polymorphism was found to be associated with a decreased risk of lung cancer in all five genetic models (A vs. C: OR = 0.75, 95% CI = 0.63-0.88; AA vs. CC: OR = 0.54, 95% CI = 0.33-0.88; AC vs. CC: OR = 0.76, 95% CI = 0.62-0.93; AA+AC vs. CC: OR = 0.73, 95% CI = 0.60-0.89; AA vs. AC+CC: OR = 0.59, 95% CI = 0.37-0.95) (Table 3) (Figure 4). Besides, subgroup analysis by source of controls revealed that there was no correlation between rs6505162 and cancer susceptibility in either population-based subgroup or hospital-based subgroup.
Table 3. Meta-analysis results of the association between rs6505162 and cancer risk.
| Variables | Na | A vs. C | AA vs. CC | AC vs. CC | AA+AC vs. CC | AA vs. AC+CC | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | I2(%) | OR (95% CI) | I2(%) | OR (95% CI) | I2(%) | OR (95% CI) | I2(%) | OR (95% CI) | I2(%) | ||
| Overall | 17 | 0.96 (0.88,1.04)b | 56.1 | 0.93 (0.77,1.14)b | 50.7 | 0.96 (0.87,1.05)b | 40.2 | 0.95 (0.86,1.05)b | 50.6 | 0.96 (0.81,1.14)b | 40.6 |
| Ethnicity | |||||||||||
| Asians | 11 | 0.96 (0.90,1.02) | 35.3 | 0.96 (0.82,1.13) | 27.7 | 0.94 (0.87,1.01) | 0.0 | 0.94 (0.88,1.01) | 16.9 | 0.98 (0.84,1.15) | 19.2 |
| Caucasians | 4 | 0.88 (0.66,1.17)b | 78.3 | 0.7 7(0.44,1.35)b | 77.0 | 0.85 (0.54,1.36)b | 78.0 | 0.84 (0.54,1.30)b | 79.3 | 0.86 (0.56,1.31)b | 69.7 |
| Cancer type | |||||||||||
| LC | 2 | 0.75 (0.63,0.88) | 0.0 | 0.54 (0.33,0.88) | 0.0 | 0.76 (0.62,0.93) | 0.0 | 0.73 (0.60,0.89) | 0.0 | 0.59 (0.37,0.95) | 0.0 |
| BC | 6 | 0.91 (0.75,1.09)b | 67.3 | 0.79 (0.52,1.19)b | 63.2 | 1.03 (0.90,1.19) | 51.7 | 0.93 (0.74,1.18)b | 62.5 | 0.87 (0.71,1.07)b | 47.1 |
| ESCC | 6 | 1.04 (0.96,1.13) | 33.9 | 1.13 (0.92,1.39) | 38.7 | 1.02 (0.92,1.13) | 0.0 | 1.04 (0.94,1.14) | 5.6 | 1.12 (0.92,1.37) | 30.8 |
| Others | 3 | 0.98 (0.88,1.09) | 0.0 | 1.02 (0.76,1.38) | 20.6 | 0.92 (0.81,1.06) | 63.7 | 0.94 (0.83,1.07) | 43.9 | 1.14 (0.85,1.52) | 5.3 |
| Source of controls | |||||||||||
| PB | 8 | 0.98 (0.91,1.06) | 40.0 | 0.93 (0.76,1.13) | 36.2 | 0.97 (0.88,1.08) | 44.1 | 0.98 (0.89,1.08) | 45.9 | 0.98 (0.81,1.19) | 22.0 |
| HB | 9 | 0.94 (0.83,1.06)b | 67.7 | 0.93 (0.69,1.26)b | 61.9 | 0.96 (0.88,1.05) | 43.7 | 0.94 (0.82,1.09)b | 58.7 | 0.9 3(0.72,1.21)b | 55.5 |
a: number of studies; b: calculating based on random-effects model; LC: lung cancer; BC: breast cancer; ESCC: esophageal squamous cell carcinoma.
Figure 4. Forest plot of the association between rs6505162 and cancer risk in subgroup analysis by cancer type under homozygote model.
Sensitivity analysis
In order to evaluate the stability of our results, we performed the sensitivity analysis to assess the effect of each individual study on the pooled ORs. After sequentially excluding each eligible studies, the corresponding ORs were not substantially changed, showing that the results of this meta-analysis are statistically stable.
Heterogeneity analysis
Q test and I2 test were used to assess the heterogeneity among studies. When obvious between-study heterogeneity (P value of Q test was less than 0.1 or I2 value was greater than 75%) was observed in the overall pooled analysis or subgroup analysis, the random-effects model would be applied because it could generate wider confidence intervals. Otherwise, we would select fixed-effects model to conduct related data analysis.
Publication bias
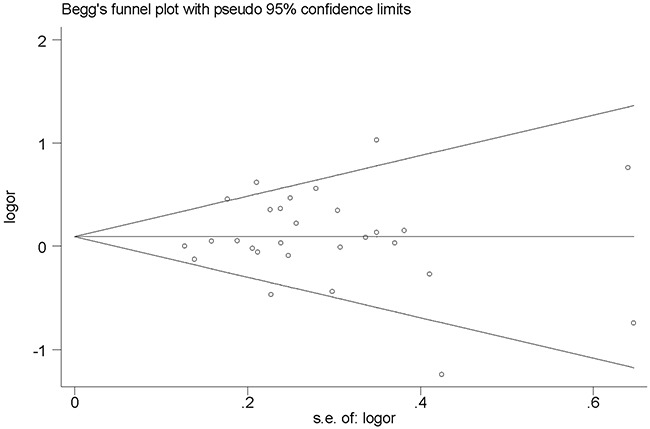
The Begg's funnel plot and Egger's test were performed to examine the publication bias in this meta-analysis. For rs895819, no publication bias was found in allele contrast model, homozygote model and recessive model, and the shape of the funnel plots appeared to relatively symmetrical (Figure 5). However, there was evidence of significant publication bias in the other two genetic models (P values of Begg's test and Egger's test were less than 0.05). Similarly, we could not observe a publication bias for the rs6505162 polymorphism in three of five comparison models including homozygote model, heterozygote model and recessive model. Interestingly, for both allele contrast model and recessive model, P values of Egger's test in these two genetic models were all > 0.05, while that of Begg's test were all < 0.05, which might be due to small sample size and limited number of studies included in this meta-analysis. Therefore, there might exist publication bias in our study.
Figure 5. Begg's funnel plot of rs895819 in recessive model.
DISCUSSION
In the current meta-analysis, a total of forty-five eligible case-control studies from thirty-five articles were included and their pooled results were used to assess the association of microRNA polymorphisms including miR-27 rs895819 and miR-423 rs6505162 with cancer susceptibility. We found that the rs895819 polymorphism was associated with a decreased cancer risk in Caucasians and breast cancer, while a significantly increased risk for developing cancer could be detected in colorectal cancer under allele contrast model, homozygote model and recessive model. In addition, for the rs6505162 polymorphism, a significantly decreased risk for cancer could be observed in lung cancer when data were stratified by cancer type.
As an important member of the microRNAs family, miR-27 is located in the intergenic region of chromosome 19 and is involved in the occurrence, invasion and metastasis of many types of cancer by acting as an oncogene. The miR-27 displays its oncogenic property through regulating the expression of target genes including FOXO1 [44], BTG2 [45], PHB [46] and other cancer-related genes [47]. Studies have reported that genetic mutations in miRNA precursors (pre-miRNA) can alter miRNA expression levels and their binding with several nuclear factors during miRNA processing [48]. Consequently, rs895819, an A to G nucleotide substitution occurring in pre-miR-27a, can down-regulate miR-27 by influencing its expression and/or maturation, and thus may lead to a reduction of individual susceptibility to cancer. Our results support this inference. In our study, we found that rs895819 could decrease the risk of breast cancer in allele contrast model and dominant model.
In addition to being able to function as an oncogene, many studies have demonstrated that miR-27a can also play an essential role as a tumor-suppressor gene in the development and progression of various cancer. For example, Bao and colleagues found that miR-27a could act as a tumor suppressor for colorectal cancer through inhibiting colon cancer cell proliferation and migration, and inducing colon cancer cell apoptosis [49]. As described in the previous paragraph, the rs895819 polymorphism can down-regulate the expression of miR-27, which will affect the tumor suppressor function of miR-27, so rs895819 may be able to promote the occurrence and development of colorectal cancer by attenuating miR-27's tumor suppressive effect. Therefore, rs895819 may be a risk factor for colorectal cancer, which is consistent with the results of our study. In our meta-analysis, the rs895819 polymorphism was found to be associated with an increased risk of colorectal cancer in allele contrast model, homozygote model and recessive model. In addition, three latest publications reported similar results [50–52], which will make our conclusions more credible. Moreover, when analysis was stratified by ethnicity, a decreased cancer risk could be detected among Caucasians, indicating that the distinction in genetic backgrounds among different ethnic groups might have an impact on cancer susceptibility, which was in accordance with the results of five previously published studies [52–56] and further demonstrated the reliability of our results.
For rs6505162, our results showed that there was no association between rs6505162 and cancer risk in the overall pooled analysis. When grouped by race, similar results could still be observed among both Asian populations and Caucasian populations. However, in the subgroup analysis by cancer type, a significantly decreased cancer risk was found in lung cancer, demonstrating that rs6505162 polymorphism might be a protective factor for lung cancer. But considering that only two studies were included in the analysis, we should treat the result with caution. Our results were different from a previously published study by Hu et al. [57], which included eight studies and found that rs6505162 was a risk factor for cancer, especially for bladder cancer. This discrepancy may be due to differences in sample sizes and ethnic groups: compared with his study, our study included seventeen eligible studies, which would expand the sample size more than twice and thus get a more credible assessment of the association.
However, some limitations of this meta-analysis should be addressed. Firstly, several important individual information such as age, sex, environmental factor was not available for most of studies included in our meta-analysis, so we couldn't carry out a more detailed analysis grouped by other non-genetic risk factors of cancer. Secondly, there existed a moderate degree of heterogeneity among the included studies, however, when analysis stratified by cancer type or race, between-study heterogeneity obviously reduced, demonstrating that heterogeneity might partly stem from differences in cancer types and ethnicities. Thirdly, many kinds of genotyping methods were applied in the included studies of our meta-analysis, which might have a potential impact on the results. Fourthly, publication bias existed for both rs895819 and rs6505162 because only studies collected into PubMed, EMBASE and Web of Science databases and published in English were selected into our meta-analysis. Those literatures not collected into these three databases or published not in English were not included in our analysis, which might contain other eligible studies and thus cause publication bias. Therefore, in view of these limitations, we should cautiously treat the results of our study.
In conclusion, our results indicate that miR-27 rs895819 is a protective factor for developing cancer in Caucasian populations and could increase colorectal cancer risk but decrease breast cancer risk. For miR-423 rs6505162, it could decrease the risk of lung cancer. More multi-center large-scale case-control studies with larger sample sizes are needed to confirm our results.
MATERIALS AND METHODS
Literature and search strategy
A comprehensive literature search of all eligible studies published before October 21, 2016 was conducted in PubMed, EMBASE and Web of Science electronic databases. During our searching process, the following retrieval items were applied: (“microRNA 27” OR “microRNA-27” OR “miR-27” OR “rs895819” OR “microRNA 423” OR “microRNA-423” OR “miR-423” OR “rs6505162”) AND (“polymorphism” OR “SNP” OR “variation” OR “locus” OR “mutation”) AND (“cancer” OR “tumor” OR “malignancy” OR “carcinoma” OR “neoplasm”).
Criterion for study selection
All eligible studies included in this meta-analysis must meet the following criteria: (1) evaluating the association of rs895819/rs6505162 with cancer risk; (2) case-control design; (3) providing sufficient genotyping data for estimating odds ratios (ORs) and 95% confidence intervals (CIs); (4) genotype frequencies of subjects in controls were in accordance with Hard-Weinberg equilibrium (HWE); (5) all cancer cases were confirmed by pathology. The exclusion criteria were as follows: (1) not case-control studies; (2) case reports, comments or review articles; (3) duplicate articles; (4) the control groups did not conform to HWE; (5) lack of enough genotyping data.
Data extraction and quality assessment
Two independent investigators carefully collected the following information from each study included in this meta-analysis: first author, year, country, ethnicity, cancer type, genotyping method, source of controls, number of cases and controls, genotype distribution of cases and controls, and P value of Hardy-Weinberg equilibrium (HWE) in controls. Any inconsistency was resolved through discussion until a consensus was reached. Moreover, we also evaluated the methodological quality of included studies based on Newcastle-Ottawa Scale (NOS), which scored studies according to three aspects: selection, comparability, and exposure. Therefore, all studies could be divided into three categories: “low quality” studies (score 0-3); “moderate quality” studies (score 4-6); “high quality” studies (score 7-9).
Statistical method
The association of rs895819 and rs6505162 with cancer susceptibility was estimated by pooled odds ratios (ORs) and 95% confidence intervals (CIs) under five different genetic models including allele contrast model, homozygote model, heterozygote model, dominant model and recessive model. Both Q-statistical test and I2 test were applied to assess the between-study heterogeneity in this meta-analysis. If there was significant heterogeneity among included studies (P value of Q statistic was < 0.1 or I2 value was > 75%), ORs with corresponding 95% CIs would be calculated using the random-effects model; otherwise, the fixed-effects model would be selected. The subgroup analysis was undertaken according to ethnicity, cancer type (investigations consisting of only one study were merged into the “others” group) and source of controls. We conducted sensitivity analysis by sequentially omitting a single study each time to assess the influence of individual study on the pooled ORs. Moreover, we employed Begg's funnel plot and Egger's test (P < 0.05 was considered significant) to assess whether there existed publication bias. Besides, chi-square test was applied to calculate P value of Hardy-Weinberg equilibrium (HWE) in controls. All data analyses performed in the present analysis were carried out using STATA 12.0 software (StataCorp LP, College Station, TX, USA). All statistical tests were two sided, and P < 0.05 indicated statistical significance.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
GRANT SUPPORT
This work was supported by the National Natural Science Foundation, China (No. 81274136), and the Program for New Century Excellent Talents in Universities of China (No. NCET-110439).
REFERENCES
- 1.Shivdasani RA. MicroRNAs: regulators of gene expression and cell differentiation. BLOOD. 2006;108:3646–3653. doi: 10.1182/blood-2006-01-030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Massirer KB, Pasquinelli AE. The evolving role of microRNAs in animal gene expression. BIOESSAYS. 2006;28:449–452. doi: 10.1002/bies.20406. [DOI] [PubMed] [Google Scholar]
- 3.Li X, Chen W, Zeng W, Wan C, Duan S, Jiang S. microRNA-137 promotes apoptosis in ovarian cancer cells via the regulation of XIAP. Br J Cancer. 2017;116:66–76. doi: 10.1038/bjc.2016.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie L, Sadovsky Y. The function of miR-519d in cell migration, invasion, and proliferation suggests a role in early placentation. PLACENTA. 2016;48:34–37. doi: 10.1016/j.placenta.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qian F, Feng Y, Zheng Y, Ogundiran TO, Ojengbede O, Zheng W, Blot W, Ambrosone CB, John EM, Bernstein L, Hu JJ, Ziegler RG, Nyante S, Bandera EV, Ingles SA, Press MF, et al. Genetic variants in microRNA and microRNA biogenesis pathway genes and breast cancer risk among women of African ancestry. HUM GENET. 2016;135:1145–1159. doi: 10.1007/s00439-016-1707-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee AR, Park J, Jung KJ, Jee SH, Kim-Yoon S. Genetic variation rs7930 in the miR-4273-5p target site is associated with a risk of colorectal cancer. Onco Targets Ther. 2016;9:6885–6895. doi: 10.2147/OTT.S108787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang S, He S, Qiu G, Lu J, Wang J, Liu J, Fan L, Zhao W, Che X. MicroRNA-125b promotes invasion and metastasis of gastric cancer by targeting STARD13 and NEU1. Tumour Biol. 2016;37:12141–12151. doi: 10.1007/s13277-016-5094-y. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Chen Y, Zhao P, Zang L, Zhang Z, Wang X. MicroRNA-19a functions as an oncogene by regulating PTEN/AKT/pAKT pathway in myeloma. Leuk Lymphoma. 2016:1–9. doi: 10.1080/10428194.2016.1213827. [DOI] [PubMed] [Google Scholar]
- 9.Jiang J, Jia ZF, Cao DH, Wu YH, Sun ZW, Cao XY. Association of the miR-146a rs2910164 polymorphism with gastric cancer susceptibility and prognosis. FUTURE ONCOL. 2016;12:2215–2226. doi: 10.2217/fon-2016-0224. [DOI] [PubMed] [Google Scholar]
- 10.Jiang Y, Lin DH, Xu JP, Chen WX, Zheng SJ, Song L. Genotype GG of rs895819 Functional Polymorphism Within miR-27a Might Increase Genetic Susceptibility to Colorectal Cancer in Han Chinese Population. J CLIN LAB ANAL. 2016;30:351–355. doi: 10.1002/jcla.21862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin Z, Cui Z, Ren Y, Xia L, Wang Q, Zhang Y, He Q, Zhou B. Association between polymorphisms in pre-miRNA genes and risk of lung cancer in a Chinese non-smoking female population. LUNG CANCER. 2016;94:15–21. doi: 10.1016/j.lungcan.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Morales S, Gulppi F, Gonzalez-Hormazabal P, Fernandez-Ramires R, Bravo T, Reyes JM, Gomez F, Waugh E, Jara L. Association of single nucleotide polymorphisms in Pre-miR-27a, Pre-miR-196a2, Pre-miR-423, miR-608 and Pre-miR-618 with breast cancer susceptibility in a South American population. BMC GENET. 2016;17:109. doi: 10.1186/s12863-016-0415-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen F, Chen J, Guo S, Zhou Y, Zheng Y, Yang Y, Zhang J, Wang X, Wang C, Zhao D, Wang M, Zhu M, Fan L, Xiang J, Xia Y, Wei Q, et al. Genetic variants in miR-196a2 and miR-499 are associated with susceptibility to esophageal squamous cell carcinoma in Chinese Han population. Tumour Biol. 2016;37:4777–4784. doi: 10.1007/s13277-015-4268-3. [DOI] [PubMed] [Google Scholar]
- 14.Yin Z, Cui Z, Guan P, Li X, Wu W, Ren Y, He Q, Zhou B. Interaction between Polymorphisms in Pre-MiRNA Genes and Cooking Oil Fume Exposure on the Risk of Lung Cancer in Chinese Non-Smoking Female Population. PLOS ONE. 2015;10:e128572. doi: 10.1371/journal.pone.0128572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang P, Ma F, Xu B. [Polymorphisms of microRNA genes and their association with breast cancer risk] Zhonghua Zhong Liu Za Zhi. 2015;37:501–507. [Article in Chinese] [PubMed] [Google Scholar]
- 16.Qi P, Wang L, Zhou B, Yao WJ, Xu S, Zhou Y, Xie ZB. Associations of miRNA polymorphisms and expression levels with breast cancer risk in the Chinese population. GENET MOL RES. 2015;14:6289–6296. doi: 10.4238/2015.June.11.2. [DOI] [PubMed] [Google Scholar]
- 17.Nikolic Z, Savic PD, Vucic N, Cidilko S, Filipovic N, Cerovic S, Vukotic V, Romac S, Brajuskovic G. Assessment of association between genetic variants in microRNA genes hsa-miR-499, hsa-miR-196a2 and hsa-miR-27a and prostate cancer risk in Serbian population. EXP MOL PATHOL. 2015;99:145–150. doi: 10.1016/j.yexmp.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Gupta A, Sharma A, Yadav A, Rastogi N, Agrawal S, Kumar A, Kumar V, Misra S, Mittal B. Evaluation of miR-27a, miR-181a, and miR-570 genetic variants with gallbladder cancer susceptibility and treatment outcome in a North Indian population. MOL DIAGN THER. 2015;19:317–327. doi: 10.1007/s40291-015-0159-y. [DOI] [PubMed] [Google Scholar]
- 19.He B, Pan Y, Xu Y, Deng Q, Sun H, Gao T, Wang S. Associations of polymorphisms in microRNAs with female breast cancer risk in Chinese population. Tumour Biol. 2015;36:4575–4582. doi: 10.1007/s13277-015-3102-2. [DOI] [PubMed] [Google Scholar]
- 20.Bian Q, Chen JJ, Gu JP, Xu J. Association between pre-miR-27a functional polymorphism and risk of colorectal cancer in north Chinese Han population. Onco Targets Ther. 2015;8:3003–3007. doi: 10.2147/OTT.S89754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Huang X, Xiao J, Yang Y, Zhou Y, Wang X, Liu Q, Yang J, Wang M, Qiu L, Zheng Y, Zhang P, Li J, Wang Y, Wei Q, Jin L, et al. Pri-miR-124 rs531564 and pri-miR-34b/c rs4938723 polymorphisms are associated with decreased risk of esophageal squamous cell carcinoma in Chinese populations. PLOS ONE. 2014;9:e100055. doi: 10.1371/journal.pone.0100055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiong XD, Luo XP, Cheng J, Liu X, Li EM, Zeng LQ. A genetic variant in pre-miR-27a is associated with a reduced cervical cancer risk in southern Chinese women. GYNECOL ONCOL. 2014;132:450–454. doi: 10.1016/j.ygyno.2013.12.030. [DOI] [PubMed] [Google Scholar]
- 23.Song B, Yan G, Hao H, Yang B. rs11671784 G/A and rs895819 A/G polymorphisms inversely affect gastric cancer susceptibility and miR-27a expression in a Chinese population. Med Sci Monit. 2014;20:2318–2326. doi: 10.12659/MSM.892499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kupcinskas J, Bruzaite I, Juzenas S, Gyvyte U, Jonaitis L, Kiudelis G, Skieceviciene J, Leja M, Pauzas H, Tamelis A, Pavalkis D, Kupcinskas L. Lack of association between miR-27a, miR-146a, miR-196a-2, miR-492 and miR-608 gene polymorphisms and colorectal cancer. Sci Rep. 2014;4:5993. doi: 10.1038/srep05993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kupcinskas J, Wex T, Link A, Leja M, Bruzaite I, Steponaitiene R, Juzenas S, Gyvyte U, Ivanauskas A, Ancans G, Petrenkiene V, Skieceviciene J, Kupcinskas L, Malfertheiner P. Gene polymorphisms of micrornas in Helicobacter pylori-induced high risk atrophic gastritis and gastric cancer. PLOS ONE. 2014;9:e87467. doi: 10.1371/journal.pone.0087467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao Y, Hu J, Fang Y, Chen Q, Li H. Association between a functional variant in microRNA-27a and susceptibility to colorectal cancer in a Chinese Han population. GENET MOL RES. 2014;13:7420–7427. doi: 10.4238/2014.September.12.8. [DOI] [PubMed] [Google Scholar]
- 27.Zhang N, Huo Q, Wang X, Chen X, Long L, Jiang L, Ma T, Yang Q. A genetic variant in pre-miR-27a is associated with a reduced breast cancer risk in younger Chinese population. GENE. 2013;529:125–130. doi: 10.1016/j.gene.2013.07.041. [DOI] [PubMed] [Google Scholar]
- 28.Wei J, Zheng L, Liu S, Yin J, Wang L, Wang X, Shi Y, Shao A, Tang W, Ding G, Liu C, Chen S, Gu H. MiR-196a2 rs11614913 T > C polymorphism and risk of esophageal cancer in a Chinese population. HUM IMMUNOL. 2013;74:1199–1205. doi: 10.1016/j.humimm.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 29.Ma F, Zhang P, Lin D, Yu D, Yuan P, Wang J, Fan Y, Xu B. There is no association between microRNA gene polymorphisms and risk of triple negative breast cancer in a Chinese Han population. PLOS ONE. 2013;8:e60195. doi: 10.1371/journal.pone.0060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y, Du WD, Chen G, Ruan J, Xu S, Zhou FS, Zuo XB, Lv ZJ, Zhang XJ. Association analysis of genetic variants in microRNA networks and gastric cancer risk in a Chinese Han population. J Cancer Res Clin Oncol. 2012;138:939–945. doi: 10.1007/s00432-012-1164-8. [DOI] [PubMed] [Google Scholar]
- 31.Zhang M, Jin M, Yu Y, Zhang S, Wu Y, Liu H, Liu H, Chen B, Li Q, Ma X, Chen K. Associations of miRNA polymorphisms and female physiological characteristics with breast cancer risk in Chinese population. Eur J Cancer Care (Engl) 2012;21:274–280. doi: 10.1111/j.1365-2354.2011.01308.x. [DOI] [PubMed] [Google Scholar]
- 32.Shi D, Li P, Ma L, Zhong D, Chu H, Yan F, Lv Q, Qin C, Wang W, Wang M, Tong N, Zhang Z, Yin C. A genetic variant in pre-miR-27a is associated with a reduced renal cell cancer risk in a Chinese population. PLOS ONE. 2012;7:e46566. doi: 10.1371/journal.pone.0046566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hezova R, Kovarikova A, Bienertova-Vasku J, Sachlova M, Redova M, Vasku A, Svoboda M, Radova L, Kiss I, Vyzula R, Slaby O. Evaluation of SNPs in miR-196-a2, miR-27a and miR-146a as risk factors of colorectal cancer. World J Gastroenterol. 2012;18:2827–2831. doi: 10.3748/wjg.v18.i22.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Catucci I, Verderio P, Pizzamiglio S, Bernard L, Dall’Olio V, Sardella D, Ravagnani F, Galastri L, Barile M, Peissel B, Zaffaroni D, Manoukian S, Radice P, Peterlongo P. The SNP rs895819 in miR-27a is not associated with familial breast cancer risk in Italians. Breast Cancer Res Treat. 2012;133:805–807. doi: 10.1007/s10549-012-2011-y. [DOI] [PubMed] [Google Scholar]
- 35.Yang R, Schlehe B, Hemminki K, Sutter C, Bugert P, Wappenschmidt B, Volkmann J, Varon R, Weber BH, Niederacher D, Arnold N, Meindl A, Bartram CR, Schmutzler RK, Burwinkel B. A genetic variant in the pre-miR-27a oncogene is associated with a reduced familial breast cancer risk. Breast Cancer Res Treat. 2010;121:693–702. doi: 10.1007/s10549-009-0633-5. [DOI] [PubMed] [Google Scholar]
- 36.Kontorovich T, Levy A, Korostishevsky M, Nir U, Friedman E. Single nucleotide polymorphisms in miRNA binding sites and miRNA genes as breast/ovarian cancer risk modifiers in Jewish high-risk women. INT J CANCER. 2010;127:589–597. doi: 10.1002/ijc.25065. [DOI] [PubMed] [Google Scholar]
- 37.Sun Q, Gu H, Zeng Y, Xia Y, Wang Y, Jing Y, Yang L, Wang B. Hsa-mir-27a genetic variant contributes to gastric cancer susceptibility through affecting miR-27a and target gene expression. CANCER SCI. 2010;101:2241–2247. doi: 10.1111/j.1349-7006.2010.01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu J, Yang L, You W, Cui X, Chen Y, Hu J, Liu W, Li S, Song X, Wei Y, Zhang W, Li F. Genetic variation in miR-100 rs1834306 is associated with decreased risk for esophageal squamous cell carcinoma in Kazakh patients in northwest China. Int J Clin Exp Pathol. 2015;8:7332–7340. [PMC free article] [PubMed] [Google Scholar]
- 39.Ma Y, Wang R, Zhang J, Li W, Gao C, Liu J, Wang J. Identification of miR-423 and miR-499 polymorphisms on affecting the risk of hepatocellular carcinoma in a large-scale population. Genet Test Mol Biomarkers. 2014;18:516–524. doi: 10.1089/gtmb.2013.0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Umar M, Upadhyay R, Prakash G, Kumar S, Ghoshal UC, Mittal B. Evaluation of common genetic variants in pre-microRNA in susceptibility and prognosis of esophageal cancer. Mol Carcinog. 2013;52:E10–E18. doi: 10.1002/mc.21931. [DOI] [PubMed] [Google Scholar]
- 41.Yin J, Wang X, Zheng L, Shi Y, Wang L, Shao A, Tang W, Ding G, Liu C, Liu R, Chen S, Gu H. Hsa-miR-34b/c rs4938723 T>C and hsa-miR-423 rs6505162 C>A polymorphisms are associated with the risk of esophageal cancer in a Chinese population. PLOS ONE. 2013;8:e80570. doi: 10.1371/journal.pone.0080570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Vogelsang M, Schafer G, Matejcic M, Parker MI. MicroRNA polymorphisms and environmental smoke exposure as risk factors for oesophageal squamous cell carcinoma. PLOS ONE. 2013;8:e78520. doi: 10.1371/journal.pone.0078520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith RA, Jedlinski DJ, Gabrovska PN, Weinstein SR, Haupt L, Griffiths LR. A genetic variant located in miR-423 is associated with reduced breast cancer risk. Cancer Genomics Proteomics. 2012;9:115–118. [PubMed] [Google Scholar]
- 44.Myatt SS, Wang J, Monteiro LJ, Christian M, Ho KK, Fusi L, Dina RE, Brosens JJ, Ghaem-Maghami S, Lam EW. Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. CANCER RES. 2010;70:367–377. doi: 10.1158/0008-5472.CAN-09-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou L, Liang X, Zhang L, Yang L, Nagao N, Wu H, Liu C, Lin S, Cai G, Liu J. MiR-27a-3p functions as an oncogene in gastric cancer by targeting BTG2. ONCOTARGET. 2016 doi: 10.18632/oncotarget.10460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang T, Lu W, Xu W, Anderson L, Bacanamwo M, Thompson W, Chen YE, Liu D. MicroRNA-27 (miR-27) targets prohibitin and impairs adipocyte differentiation and mitochondrial function in human adipose-derived stem cells. J BIOL CHEM. 2013;288:34394–34402. doi: 10.1074/jbc.M113.514372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X, Mertens-Talcott SU, Zhang S, Kim K, Ball J, Safe S. MicroRNA-27a Indirectly Regulates Estrogen Receptor {alpha} Expression and Hormone Responsiveness in MCF-7 Breast Cancer Cells. ENDOCRINOLOGY. 2010;151:2462–2473. doi: 10.1210/en.2009-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parpart S, Wang XW. microRNA Regulation and Its Consequences in Cancer. Curr Pathobiol Rep. 2013;1:71–79. doi: 10.1007/s40139-012-0002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bao Y, Chen Z, Guo Y, Feng Y, Li Z, Han W, Wang J, Zhao W, Jiao Y, Li K, Wang Q, Wang J, Zhang H, Wang L, Yang W. Tumor suppressor microRNA-27a in colorectal carcinogenesis and progression by targeting SGPP1 and Smad2. PLOS ONE. 2014;9:e105991. doi: 10.1371/journal.pone.0105991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu F, Dear K, Huang L, Liu L, Shi Y, Nie S, Liu Y, Lu Y, Xiang H. Association between microRNA-27a rs895819 polymorphism and risk of colorectal cancer: A meta-analysis. Cancer Genet. 2016;209:388–394. doi: 10.1016/j.cancergen.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Pan XM, Xiao X, Qin HJ, Zhang Z, Li ZH, Gao LB, Jia J. MicroRNA variants and colorectal cancer risk: a meta-analysis. GENET MOL RES. 2016:15. doi: 10.4238/gmr.15038478. [DOI] [PubMed] [Google Scholar]
- 52.Feng Y, Duan F, Song C, Zhao X, Dai L, Cui S. Systematic evaluation of cancer risk associated with rs2292832 in miR149 and rs895819 in miR27a: a comprehensive and updated metaanalysis. ONCOTARGET. 2016;7:22368–22384. doi: 10.18632/oncotarget.8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bai RP, Weng Y, Su LL, Jin MJ, Xu ZP, Lu LQ, Chen GD. Association of a pre-miR-27a polymorphism with cancer risk: an updated meta-analysis. Asian Pac J Cancer Prev. 2014;15:10107–10114. doi: 10.7314/apjcp.2014.15.23.10107. [DOI] [PubMed] [Google Scholar]
- 54.Zhong S, Chen Z, Xu J, Li W, Zhao J. Pre-mir-27a rs895819 polymorphism and cancer risk: a meta-analysis. MOL BIOL REP. 2013;40:3181–3186. doi: 10.1007/s11033-012-2392-3. [DOI] [PubMed] [Google Scholar]
- 55.Xu Q, He CY, Liu JW, Yuan Y. Pre-miR-27a rs895819A/G polymorphisms in cancer: a meta-analysis. PLOS ONE. 2013;8:e65208. doi: 10.1371/journal.pone.0065208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Z, Lai J, Wang Y, Nie W, Guan X. The Hsa-miR-27a rs895819 (A>G) polymorphism and cancer susceptibility. GENE. 2013;521:87–90. doi: 10.1016/j.gene.2013.02.042. [DOI] [PubMed] [Google Scholar]
- 57.Hu Y, Yu CY, Wang JL, Guan J, Chen HY, Fang JY. MicroRNA sequence polymorphisms and the risk of different types of cancer. Sci Rep. 2014;4:3648. doi: 10.1038/srep03648. [DOI] [PMC free article] [PubMed] [Google Scholar]


