Abstract
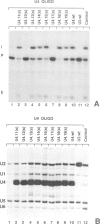
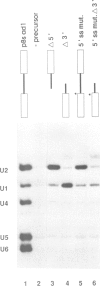
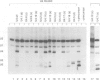
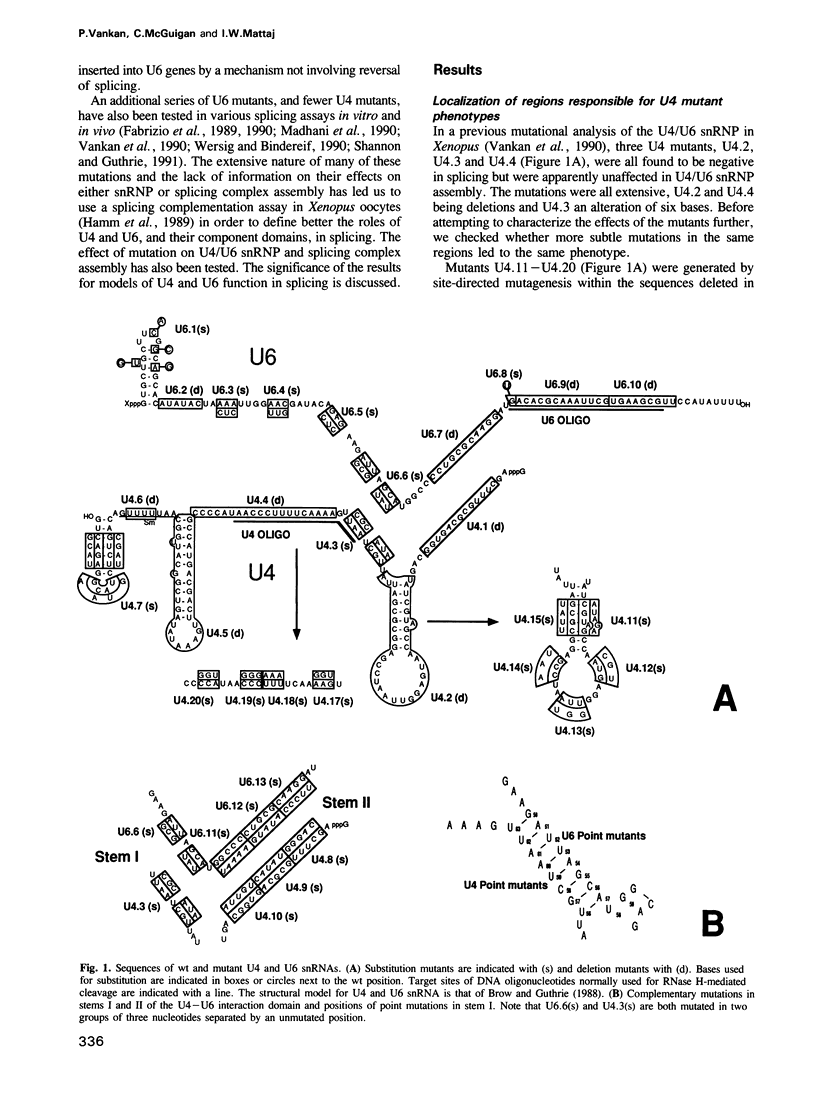
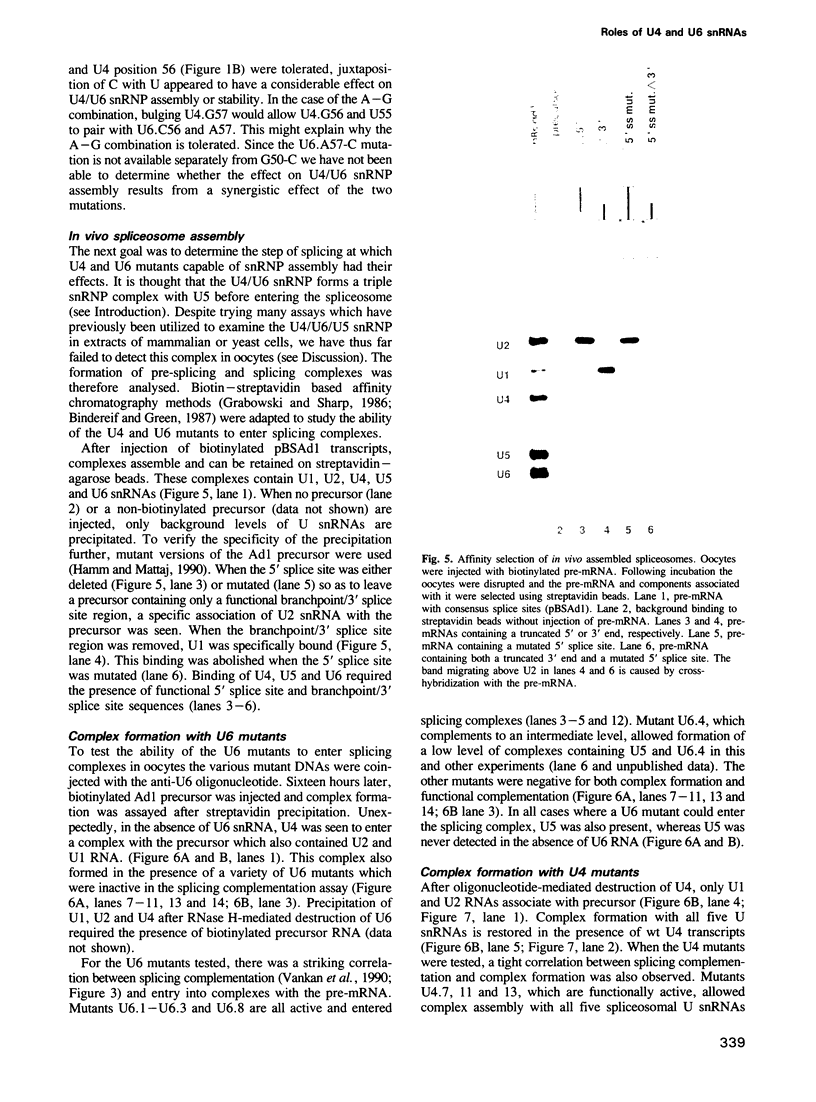
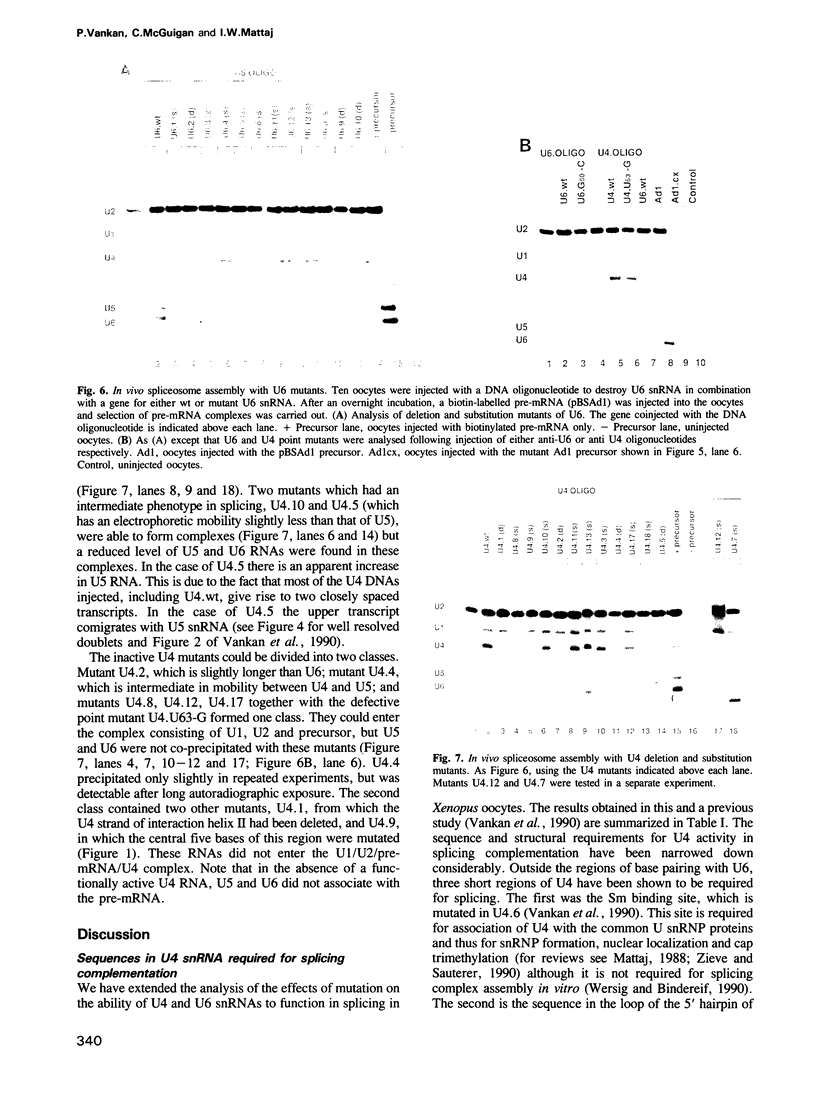
A series of U4 and U6 snRNA mutants was analysed in Xenopus oocytes to determine whether they block splicing complex assembly or splicing itself. All the U4 and U6 mutants found to be inactive in splicing complementation resulted in defects in assembly of either U4/U6 snRNP or of splicing complexes. No mutants were found to separate the entry of U5 and U6 snRNAs into splicing complexes and neither of these RNAs was able to associate with the pre-mRNA in the absence of U4. In the absence of U6 snRNA, however, U4 entered a complex containing pre-mRNA as well as the U1 and U2 snRNAs. U6 nucleotides whose mutation resulted in specific blockage of the second step of splicing in Saccharomyces cerevisiae are shown not to be essential for splicing in the oocyte assay. The results are discussed in terms of the roles of U4 and U6 in the assembly and catalytic steps of the splicing process.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barabino S. M., Blencowe B. J., Ryder U., Sproat B. S., Lamond A. I. Targeted snRNP depletion reveals an additional role for mammalian U1 snRNP in spliceosome assembly. Cell. 1990 Oct 19;63(2):293–302. doi: 10.1016/0092-8674(90)90162-8. [DOI] [PubMed] [Google Scholar]
- Behrens S. E., Lührmann R. Immunoaffinity purification of a [U4/U6.U5] tri-snRNP from human cells. Genes Dev. 1991 Aug;5(8):1439–1452. doi: 10.1101/gad.5.8.1439. [DOI] [PubMed] [Google Scholar]
- Bindereif A., Green M. R. An ordered pathway of snRNP binding during mammalian pre-mRNA splicing complex assembly. EMBO J. 1987 Aug;6(8):2415–2424. doi: 10.1002/j.1460-2075.1987.tb02520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindereif A., Wolff T., Green M. R. Discrete domains of human U6 snRNA required for the assembly of U4/U6 snRNP and splicing complexes. EMBO J. 1990 Jan;9(1):251–255. doi: 10.1002/j.1460-2075.1990.tb08102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D. L., Pinto A. L. U5 small nuclear ribonucleoprotein: RNA structure analysis and ATP-dependent interaction with U4/U6. Mol Cell Biol. 1989 Aug;9(8):3350–3359. doi: 10.1128/mcb.9.8.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe B. J., Sproat B. S., Ryder U., Barabino S., Lamond A. I. Antisense probing of the human U4/U6 snRNP with biotinylated 2'-OMe RNA oligonucleotides. Cell. 1989 Nov 3;59(3):531–539. doi: 10.1016/0092-8674(89)90036-6. [DOI] [PubMed] [Google Scholar]
- Bordonné R., Banroques J., Abelson J., Guthrie C. Domains of yeast U4 spliceosomal RNA required for PRP4 protein binding, snRNP-snRNP interactions, and pre-mRNA splicing in vivo. Genes Dev. 1990 Jul;4(7):1185–1196. doi: 10.1101/gad.4.7.1185. [DOI] [PubMed] [Google Scholar]
- Brow D. A., Guthrie C. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988 Jul 21;334(6179):213–218. doi: 10.1038/334213a0. [DOI] [PubMed] [Google Scholar]
- Brow D. A., Guthrie C. Splicing a spliceosomal RNA. Nature. 1989 Jan 5;337(6202):14–15. doi: 10.1038/337014a0. [DOI] [PubMed] [Google Scholar]
- Cech T. R. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell. 1986 Jan 31;44(2):207–210. doi: 10.1016/0092-8674(86)90751-8. [DOI] [PubMed] [Google Scholar]
- Chabot B., Black D. L., LeMaster D. M., Steitz J. A. The 3' splice site of pre-messenger RNA is recognized by a small nuclear ribonucleoprotein. Science. 1985 Dec 20;230(4732):1344–1349. doi: 10.1126/science.2933810. [DOI] [PubMed] [Google Scholar]
- Cheng S. C., Abelson J. Spliceosome assembly in yeast. Genes Dev. 1987 Nov;1(9):1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- Datta B., Weiner A. M. Genetic evidence for base pairing between U2 and U6 snRNA in mammalian mRNA splicing. Nature. 1991 Aug 29;352(6338):821–824. doi: 10.1038/352821a0. [DOI] [PubMed] [Google Scholar]
- Fabrizio P., Abelson J. Two domains of yeast U6 small nuclear RNA required for both steps of nuclear precursor messenger RNA splicing. Science. 1990 Oct 19;250(4979):404–409. doi: 10.1126/science.2145630. [DOI] [PubMed] [Google Scholar]
- Fabrizio P., McPheeters D. S., Abelson J. In vitro assembly of yeast U6 snRNP: a functional assay. Genes Dev. 1989 Dec;3(12B):2137–2150. doi: 10.1101/gad.3.12b.2137. [DOI] [PubMed] [Google Scholar]
- Grabowski P. J., Sharp P. A. Affinity chromatography of splicing complexes: U2, U5, and U4 + U6 small nuclear ribonucleoprotein particles in the spliceosome. Science. 1986 Sep 19;233(4770):1294–1299. doi: 10.1126/science.3638792. [DOI] [PubMed] [Google Scholar]
- Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991 Jul 12;253(5016):157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Hamm J., Dathan N. A., Mattaj I. W. Functional analysis of mutant Xenopus U2 snRNAs. Cell. 1989 Oct 6;59(1):159–169. doi: 10.1016/0092-8674(89)90878-7. [DOI] [PubMed] [Google Scholar]
- Hamm J., Mattaj I. W. An abundant U6 snRNP found in germ cells and embryos of Xenopus laevis. EMBO J. 1989 Dec 20;8(13):4179–4187. doi: 10.1002/j.1460-2075.1989.tb08603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm J., Mattaj I. W. Monomethylated cap structures facilitate RNA export from the nucleus. Cell. 1990 Oct 5;63(1):109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- Hausner T. P., Giglio L. M., Weiner A. M. Evidence for base-pairing between mammalian U2 and U6 small nuclear ribonucleoprotein particles. Genes Dev. 1990 Dec;4(12A):2146–2156. doi: 10.1101/gad.4.12a.2146. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986 Sep 12;46(6):845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987 Jun 19;49(6):763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- Lamm G. M., Blencowe B. J., Sproat B. S., Iribarren A. M., Ryder U., Lamond A. I. Antisense probes containing 2-aminoadenosine allow efficient depletion of U5 snRNP from HeLa splicing extracts. Nucleic Acids Res. 1991 Jun 25;19(12):3193–3198. doi: 10.1093/nar/19.12.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A. I., Konarska M. M., Grabowski P. J., Sharp P. A. Spliceosome assembly involves the binding and release of U4 small nuclear ribonucleoprotein. Proc Natl Acad Sci U S A. 1988 Jan;85(2):411–415. doi: 10.1073/pnas.85.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrain P., Seraphin B., Rosbash M. Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol Cell Biol. 1988 Sep;8(9):3755–3760. doi: 10.1128/mcb.8.9.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner E. A., Lerner M. R., Janeway C. A., Jr, Steitz J. A. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci U S A. 1981 May;78(5):2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lührmann R., Kastner B., Bach M. Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim Biophys Acta. 1990 Nov 30;1087(3):265–292. doi: 10.1016/0167-4781(90)90001-i. [DOI] [PubMed] [Google Scholar]
- Madhani H. D., Bordonné R., Guthrie C. Multiple roles for U6 snRNA in the splicing pathway. Genes Dev. 1990 Dec;4(12B):2264–2277. doi: 10.1101/gad.4.12b.2264. [DOI] [PubMed] [Google Scholar]
- Newman A., Norman C. Mutations in yeast U5 snRNA alter the specificity of 5' splice-site cleavage. Cell. 1991 Apr 5;65(1):115–123. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- Okano Y., Medsger T. A., Jr Newly identified U4/U6 snRNP-binding proteins by serum autoantibodies from a patient with systemic sclerosis. J Immunol. 1991 Jan 15;146(2):535–542. [PubMed] [Google Scholar]
- Pikielny C. W., Rymond B. C., Rosbash M. Electrophoresis of ribonucleoproteins reveals an ordered assembly pathway of yeast splicing complexes. 1986 Nov 27-Dec 3Nature. 324(6095):341–345. doi: 10.1038/324341a0. [DOI] [PubMed] [Google Scholar]
- Ruby S. W., Abelson J. An early hierarchic role of U1 small nuclear ribonucleoprotein in spliceosome assembly. Science. 1988 Nov 18;242(4881):1028–1035. doi: 10.1126/science.2973660. [DOI] [PubMed] [Google Scholar]
- Ruby S. W., Abelson J. Pre-mRNA splicing in yeast. Trends Genet. 1991 Mar;7(3):79–85. doi: 10.1016/0168-9525(91)90276-V. [DOI] [PubMed] [Google Scholar]
- Scherly D., Boelens W., van Venrooij W. J., Dathan N. A., Hamm J., Mattaj I. W. Identification of the RNA binding segment of human U1 A protein and definition of its binding site on U1 snRNA. EMBO J. 1989 Dec 20;8(13):4163–4170. doi: 10.1002/j.1460-2075.1989.tb08601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin B., Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989 Oct 20;59(2):349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- Shannon K. W., Guthrie C. Suppressors of a U4 snRNA mutation define a novel U6 snRNP protein with RNA-binding motifs. Genes Dev. 1991 May;5(5):773–785. doi: 10.1101/gad.5.5.773. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Abovich N., Rosbash M. Genetic depletion indicates a late role for U5 snRNP during in vitro spliceosome assembly. Nucleic Acids Res. 1991 Jul 25;19(14):3857–3860. doi: 10.1093/nar/19.14.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Rosbash M. The yeast branchpoint sequence is not required for the formation of a stable U1 snRNA-pre-mRNA complex and is recognized in the absence of U2 snRNA. EMBO J. 1991 May;10(5):1209–1216. doi: 10.1002/j.1460-2075.1991.tb08062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani T., Ohshima Y. The gene for the U6 small nuclear RNA in fission yeast has an intron. Nature. 1989 Jan 5;337(6202):87–90. doi: 10.1038/337087a0. [DOI] [PubMed] [Google Scholar]
- Tani T., Ohshima Y. mRNA-type introns in U6 small nuclear RNA genes: implications for the catalysis in pre-mRNA splicing. Genes Dev. 1991 Jun;5(6):1022–1031. doi: 10.1101/gad.5.6.1022. [DOI] [PubMed] [Google Scholar]
- Vankan P., McGuigan C., Mattaj I. W. Domains of U4 and U6 snRNAs required for snRNP assembly and splicing complementation in Xenopus oocytes. EMBO J. 1990 Oct;9(10):3397–3404. doi: 10.1002/j.1460-2075.1990.tb07541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wersig C., Bindereif A. Conserved domains of human U4 snRNA required for snRNP and spliceosome assembly. Nucleic Acids Res. 1990 Nov 11;18(21):6223–6229. doi: 10.1093/nar/18.21.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. A., Manley J. L. Base pairing between U2 and U6 snRNAs is necessary for splicing of a mammalian pre-mRNA. Nature. 1991 Aug 29;352(6338):818–821. doi: 10.1038/352818a0. [DOI] [PubMed] [Google Scholar]
- Xu Y., Petersen-Bjørn S., Friesen J. D. The PRP4 (RNA4) protein of Saccharomyces cerevisiae is associated with the 5' portion of the U4 small nuclear RNA. Mol Cell Biol. 1990 Mar;10(3):1217–1225. doi: 10.1128/mcb.10.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieve G. W., Sauterer R. A. Cell biology of the snRNP particles. Crit Rev Biochem Mol Biol. 1990;25(1):1–46. doi: 10.3109/10409239009090604. [DOI] [PubMed] [Google Scholar]