Abstract
Cancer is the leading cause of death in economically developed countries and the second leading cause of death in developing countries. The relationship between genetic polymorphisms and the risk of cancers has been widely researched. Excision repair cross-complementing group 2 (ERCC2) gene plays important roles in the nucleotide excision repair pathway. There is contrasting evidence on the association between the ERCC2 Asp312Asn polymorphism and the risk of cancer. We conducted a comprehensive meta-analysis in order to assess the correlation between these factors. We searched the PubMed, EMBASE, Science Direct, Web of Science, and CNKI databases for studies published from January 1, 2005 to January 1, 2016. Finally, 86 articles with 38,848 cases and 48,928 controls were included in the analysis. The overall analysis suggested a significant association between the ERCC2 Asp312Asn polymorphism and cancer risk. Furthermore, control source, ethnicity, genotyping method, and cancer type were used for subgroup analysis. The result of a trial sequential analysis indicated that the cumulative evidence is adequate; hence, further trials were unnecessary in the overall analysis for homozygote comparison. In summary, our results suggested that ERCC2 Asp312Asn polymorphism is associated with increased cancer risk. A significantly increased cancer risk was observed in Asian populations, but not in Caucasian populations. Furthermore, the ERCC2 Asp312Asn polymorphism is associated with bladder, esophageal, and gastric cancers, but not with breast, head and neck, lung, prostate, and skin cancers, and non-Hodgkin lymphoma. Further multi-center, well-designed studies are required to validate our results.
Keywords: ERCC2 Asp312Asn, polymorphism, cancer, meta-analysis, trial sequence analysis
INTRODUCTION
Cancer describes a group of diseases characterized by the uncontrolled growth and spread of abnormal cells [1]. It is the leading cause of death in economically developed countries and the second leading cause of death in developing countries [2]. According to statistics, a total of 1,658,370 new cancer cases and 589,430 cancer deaths were projected to occur in the United States in 2015 [3]. In general, cancer is the result of multiple environmental and genetic risk factors, as well as gene-environment interactions [4]. Among genetic factors, genetic and epigenetic mutations, such as aberrant DNA methylation, can lead to carcinogenesis [1].
Recently, the relationship between genetic polymorphisms and the risk of cancer has been widely researched. Among the polymorphic genes, excision repair cross-complementing group 2 (ERCC2), also called xeroderma pigmentosum group D (XPD), plays important roles in the nucleotide excision repair (NER) pathway [5]. The ERCC2 gene is located on chromosome 19q13.3, comprises 23 exons, and spans approximately 54,000 base pairs [6]. It encodes an evolutionarily conserved helicase, which has ATP-dependent helicase activity within its multi subunit core transcription factor IIH (TFIIH). The helicase participates in DNA unwinding as part of the NER pathway, and plays an important role in the recognition and repair of structurally unrelated DNA lesions containing bulky adducts and thymidine dimers [7, 8]. Some studies have shown that ERCC2 polymorphisms may be related to reduced DNA repair due to a possible reduction in its helicase activity [9, 10].
There are two important single nucleotide polymorphisms (SNPs) in the ERCC2 gene. One is the Lys751Gln polymorphism, which has been shown to be involved in genetic susceptibility to some cancer types. Another common ERCC2 polymorphism in the coding region is Asp312Asn (rs1799793) [11], which is characterized by a G to A transition at position 312 in exon 10 causing an aspartic acid (Asp) to asparagine amino acid (Asn) exchange [12]. This polymorphism has been widely studied for its association with susceptibility to cancer including brain [13], esophageal [14–16], head and neck [11], bladder [17–19], and breast cancers [20–22]. However, the results reported by these studies were inconsistent.
To provide a comprehensive assessment of and to clarify associations between the ERCC2 Asp312Asn polymorphisms and the risk of cancer, we performed a meta-analysis of all the eligible case-control studies.
RESULTS
Eligible studies
A total of 449 articles were reviewed, and eventually 86 articles with 38,848 cases and 48,928 controls met the inclusion criteria. Among these publications, there was 1 osteosarcoma [23], 1 hepatocellular cancer (HCC) [24], 3 oral cancer [25–27], 5 skin cancer [28–32], 5 colorectal cancer [23, 33–36], 6 head and neck cancer [37–42], 6 esophageal cancer [43–48], 6 non-Hodgkin lymphoma [49–54], 6 prostate cancer [55–60], 8 gastric cancer [61–67], 12 bladder cancer [68–79], 14 lung cancer [70, 80–92], and 15 breast cancer [23, 32, 93–105]. The detailed study selection process is shown in Figure 1. Table 1 presents the major characteristics of the 86 articles.
Figure 1. Flow chart showing the selection process for the included studies.
Table 1. Characteristics of the case–control studies included in the meta-analyses.
| First author | Year | Ethnicity | Countrya | Source of controls | Cancer site | Genotyping method | cases | controls | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Asp/Asp | Asp/Asn | Asn/Asn | Asp/Asp | Asp/Asn | Asn/Asn | |||||||
| Liu G | 2007 | Caucasian | USA | HB | esophageal cancer | PCR-RFLP | 75 | 92 | 16 | 144 | 160 | 32 |
| An | 2007 | Caucasian | USA | HB | head and neck cancera | PCR-RFLP | 330 | 395 | 104 | 370 | 386 | 98 |
| Harth | 2008 | Caucasian | Germany | HB | head and neck cancera | Real-time PCR | 113 | 158 | 40 | 101 | 145 | 52 |
| Abbasi | 2009 | Caucasian | Germany | PB | head and neck cancera | Real-time PCR | 93 | 119 | 34 | 258 | 304 | 82 |
| Ji | 2010 | Asian | Korea | HB | head and neck cancera | PCR | 235 | 29 | 0 | 309 | 30 | 3 |
| Gugatschka | 2011 | Caucasian | Austria | PB | head and neck cancera | TaqMan | 116 | 133 | 42 | 171 | 208 | 83 |
| Smedby | 2006 | Caucasian | Sweden | PB | non- Hodgkin lymphoma | PCR | 167 | 211 | 50 | 262 | 255 | 85 |
| Shen | 2006 | Caucasian | USA | PB | non- Hodgkin lymphoma | Real-time PCR | 199 | 189 | 57 | 226 | 238 | 70 |
| Song | 2008 | Asian | China | HB | non- Hodgkin lymphoma | PCR-RFLP | 256 | 47 | 4 | 265 | 35 | 3 |
| Baris | 2009 | Caucasian | Turkey | HB | non- Hodgkin lymphoma | PCR-RFLP | 13 | 16 | 4 | 15 | 27 | 10 |
| Worrillow | 2009 | Caucasian | England | PB | non- Hodgkin lymphoma | TaqMan | 270 | 265 | 79 | 316 | 335 | 79 |
| EI-Din | 2013 | Caucasian | Egypt | HB | non- Hodgkin lymphoma | PCR-RFLP | 30 | 37 | 14 | 38 | 44 | 18 |
| Capella G | 2008 | Mixed | Spain | PB | gastric cancer | PCR-RFLP | 110 | 96 | 38 | 444 | 532 | 159 |
| Zhou RM | 2007 | Asians | China | PB | gastric cancer | PCR-RFLP | 221 | 32 | 0 | 528 | 82 | 2 |
| Lou Y | 2006 | Asians | China | HB | gastric cancer | PCR-RFLP | 189 | 39 | 10 | 176 | 21 | 3 |
| Agalliu | 2010 | Caucasian | USA | PB | prostate cancer | PCR-RFLP | 545 | 575 | 120 | 527 | 528 | 166 |
| Agalliu | 2010 | African | USA | PB | prostate cancer | PCR-RFLP | 106 | 31 | 7 | 65 | 15 | 2 |
| Moreno V | 2006 | Caucasian | Spain | HB | colorectal cancer | PCR | 95 | 91 | 100 | 77 | 72 | 63 |
| Hansen RD | 2007 | Caucasian | Denmark | PB | colorectal cancer | TaqMan | 159 | 191 | 46 | 333 | 354 | 108 |
| De Ruyck | 2007 | Caucasian | Belgium | HB | Lung Cancer | PCR-RFLP | 44 | 53 | 13 | 49 | 46 | 14 |
| Zienolddiny | 2006 | Caucasian | Norway | PB | Lung Cancer | PCR | 119 | 102 | 54 | 120 | 121 | 49 |
| Matullo | 2006 | Caucasian | Europe | PB | Lung Cancer | PCR-RFLP | 49 | 48 | 19 | 418 | 506 | 170 |
| Hu | 2006 | Asian | China | HB | Lung Cancer | TaqMan | 850 | 116 | 4 | 874 | 111 | 1 |
| Shen | 2005 | Asian | China | PB | Lung Cancer | PCR | 109 | 9 | 0 | 99 | 14 | 0 |
| Huang | 2006 | Mixed | USA | NA | Lung Cancer | PCR | 301 | 300 | 82 | 301 | 304 | 93 |
| Broberg | 2005 | Caucasian | Sweden | PB | bladder cancer | PCR | 16 | 29 | 12 | 61 | 71 | 13 |
| Matullo | 2005 | Caucasian | Italy | HB | bladder cancer | PCR-RFLP and TaqMan | 92 | 153 | 47 | 103 | 155 | 47 |
| Matullo | 2006 | Caucasian | European | PB | bladder cancer | TaqMan | 48 | 60 | 16 | 418 | 506 | 170 |
| Schabath | 2005 | Mixed | USA | HB | bladder cancer | PCR-RFLP | 225 | 215 | 57 | 248 | 179 | 50 |
| Andrew | 2006 | Mixed | USA | PB | bladder cancer | PCR-RFLP | 113 | 145 | 38 | 205 | 251 | 51 |
| Garcia-Closas | 2006 | Caucasian | Spain | HB | bladder cancer | PCR | 517 | 474 | 138 | 538 | 467 | 117 |
| Wu | 2006 | Caucasian | USA | HB | bladder cancer | PCR-RFLP | 264 | 283 | 78 | 283 | 243 | 65 |
| Fontana | 2008 | Caucasian | French | HB | bladder cancer | TaqMan | 25 | 19 | 7 | 21 | 18 | 6 |
| Chang | 2009 | Asian | China | HB | bladder cancer | PCR-RFLP | 153 | 98 | 57 | 199 | 67 | 42 |
| Gangwar | 2009 | Asian | India | HB | bladder cancer | PCR-RFLP | 72 | 100 | 34 | 128 | 104 | 18 |
| Mittal | 2012 | Asian | India | PB | bladder cancer | PCR | 78 | 100 | 34 | 128 | 104 | 18 |
| Ye | 2006 | Caucasian | Sweden | PB | esophageal cancer | PCR-RFLP | 61 | 92 | 24 | 176 | 237 | 57 |
| Tse | 2008 | Mixed | USA | HB | esophageal cancer | TaqMan | 117 | 150 | 43 | 199 | 206 | 49 |
| Pan | 2009 | Caucasian | USA | HB | esophageal cancer | TaqMan | 16 | 20 | 1 | 201 | 185 | 48 |
| Pan | 2009 | Caucasian | USA | HB | esophageal cancer | TaqMan | 137 | 163 | 43 | 201 | 185 | 48 |
| Huang | 2012 | Asian | China | HB | esophageal cancer | PCR-RFLP | 171 | 42 | 0 | 298 | 60 | 0 |
| Li | 2013 | Asian | China | HB | esophageal cancer | PCR-RFLP | 342 | 56 | 2 | 351 | 47 | 2 |
| Han | 2005 | Mixed | USA | PB | Skin Cancer | TaqMan | 88 | 99 | 19 | 342 | 373 | 121 |
| Wang LL | 2009 | Asian | China | HB | colorectal cancer | PCR-RFLP | 132 | 29 | 9 | 176 | 21 | 3 |
| Mahimkar MB | 2010 | Asian | India | NA | oral cancer | PCR-RFLP | 23 | 13 | 4 | 23 | 21 | 1 |
| Wang Y | 2007 | Caucasian | USA | HB | oral cancer | PCR and Taqman | 50 | 59 | 16 | 140 | 109 | 29 |
| Majumder M | 2007 | Asian | India | HB | oral cancer | PCR | 269 | 208 | 52 | 205 | 146 | 36 |
| Crew | 2007 | NA | USA | PB | breast cancer | Taqman | 415 | 478 | 138 | 490 | 454 | 139 |
| Jorgensen | 2007 | Caucasian | USA | PB | breast cancer | Taqman | 110 | 128 | 22 | 102 | 142 | 29 |
| Kuschel | 2005 | Australian | UK | PB | breast cancer | TaqMan | 1529 | 1530 | 497 | 1401 | 1437 | 430 |
| Lee | 2005 | Asian | Korea | HB | breast cancer | PCR | 475 | 50 | 3 | 401 | 41 | 3 |
| Bernard-Gallon | 2008 | NA | France | HB | breast cancer | Taqman | 403 | 383 | 118 | 458 | 418 | 118 |
| Debniak | 2006 | Polish | Poland | PB | breast cancer | PCR-RFLP | 672 | 785 | 269 | 180 | 252 | 79 |
| Jakubowska | 2010 | Polish | Poland | HB | breast cancer | PCR | 118 | 152 | 44 | 106 | 135 | 49 |
| Mechanic | 2006 | Caucasian | USA | PB | breast cancer | PCR-RFLP | 543 | 589 | 130 | 489 | 516 | 128 |
| Mechanic | 2006 | African-American | USA | PB | breast cancer | PCR-RFLP | 564 | 181 | 15 | 517 | 145 | 13 |
| Shen | 2006 | American | USA | PB | breast cancer | Taqman | 60 | 80 | 16 | 59 | 64 | 30 |
| Smith | 2008 | Caucasian | USA | HB | breast cancer | PCR | 126 | 137 | 41 | 161 | 188 | 42 |
| Smith | 2008 | African-American | USA | HB | breast cancer | PCR | 33 | 14 | 2 | 57 | 16 | 1 |
| Zhang | 2005 | Asian | China | PB | breast cancer | PCR-RFLP | 89 | 111 | 20 | 119 | 140 | 51 |
| Hussien | 2012 | Caucasian | Egypt | HB | breast cancer | PCR | 12 | 45 | 43 | 25 | 50 | 25 |
| Jelonek | 2010 | Mixed | Poland | PB | breast cancer | PCR-RFLP | 41 | 59 | 21 | 85 | 123 | 23 |
| Wang | 2010 | Asian | China | PB | breast cancer | PCR-RFLP | 624 | 388 | 220 | 925 | 315 | 193 |
| Zhou | 2012 | Asian | Asia | PB | Lung Cancer | PCR-RFLP | 85 | 18 | 0 | 85 | 17 | 1 |
| Sakoda | 2012 | Caucasian | USA | PB | Lung Cancer | TaqMan | 326 | 329 | 89 | 610 | 685 | 182 |
| Qian | 2011 | Asian | China | PB | Lung Cancer | PCR | 464 | 82 | 4 | 497 | 79 | 3 |
| Yin | 2009 | Asian | China | HB | Lung Cancer | PCR-RFLP | 246 | 38 | 1 | 255 | 30 | 0 |
| Raaschou-Nielsen | 2008 | Caucasian | Denmark | PB | Lung Cancer | PCR | 177 | 188 | 59 | 329 | 351 | 107 |
| Chang | 2008 | Latino-American | USA | PB | Lung Cancer | WGA | 60 | 40 | 8 | 192 | 93 | 12 |
| Chang | 2008 | African-American | USA | PB | Lung Cancer | WGA | 186 | 58 | 3 | 212 | 60 | 5 |
| Yin | 2007 | Asian | China | HB | Lung Cancer | PCR-RFLP | 200 | 1 | 0 | 170 | 0 | 1 |
| Lopez-Cima | 2007 | Caucasian | Spain | HB | Lung Cancer | PCR-RFLP | 240 | 221 | 55 | 260 | 230 | 43 |
| Han | 2005 | Mixed | USA | PB | Skin Cancer | TaqMan | 104 | 149 | 32 | 342 | 373 | 121 |
| Han | 2005 | Mixed | USA | PB | Skin Cancer | TaqMan | 128 | 115 | 37 | 342 | 373 | 121 |
| Lovatt | 2005 | Caucasian | UK | PB | Skin Cancer | PCR-RFLP | 224 | 219 | 66 | 151 | 163 | 65 |
| Li | 2006 | Mixed | USA | HB | Skin Cancer | PCR | 242 | 290 | 70 | 273 | 259 | 71 |
| Millikan | 2006 | Caucasian | USA | PB | Skin Cancer | PCR | 1039 | 1098 | 162 | 1039 | 1098 | 260 |
| Debniak | 2006 | Polish | Poland | mixed | Skin Cancer | PCR | 168 | 188 | 69 | 492 | 597 | 173 |
| Bau | 2007 | Asian | Taiwan | HB | prostate cancer | PCR | 62 | 39 | 22 | 310 | 106 | 63 |
| Mandal | 2010 | Asian | India | PB | prostate cancer | PCR | 76 | 56 | 39 | 99 | 81 | 20 |
| Lavende | 2010 | African | America | HB | prostate cancer | PCR and Taqman | 146 | 39 | 5 | 510 | 116 | 5 |
| Dhillon | 2011 | Caucasian | Australia | NA | prostate cancer | PCR-RFLP | 71 | 37 | 8 | 80 | 42 | 10 |
| Yuan T | 2011 | Asian | China | HB | gastric Cancer | PCR | 156 | 18 | 16 | 133 | 35 | 12 |
| Chen Z | 2011 | Asian | China | HB | gastric Cancer | PCR-RFLP | 75 | 118 | 15 | 220 | 111 | 8 |
| Zhang CZ | 2009 | Asian | China | HB | gastric Cancer | PCR-RFLP | 75 | 117 | 15 | 132 | 72 | 8 |
| Ruzzo A | 2007 | Caucasian | Italy | HB | gastric Cancer | PCR-RFLP | 23 | 26 | 20 | 41 | 67 | 13 |
| Deng Sl | 2010 | Asian | China | HB | gastric Cancer | PCR | 132 | 15 | 13 | 118 | 31 | 11 |
| Wu JS | 2014 | Asian | China | HB | HCC | PCR | 138 | 58 | 22 | 181 | 70 | 26 |
| Sambuddha | 2015 | Asian | Northeast India | NA | head and neck cancer | PCR | 32 | 40 | 8 | 57 | 31 | 4 |
| Benjamin | 2015 | Mexican | Mexica | HB | osteosarcoma | PCR | 21 | 3 | 4 | 68 | 8 | 21 |
| Benjamin | 2015 | Mexican | Mexica | HB | colorectal cancer | PCR | 74 | 26 | 8 | 81 | 23 | 15 |
| Benjamin | 2015 | Mexican | Mexica | HB | breast cancer | PCR | 54 | 9 | 8 | 54 | 1 | 19 |
| Min Ni | 2014 | Asian | China | HB | colorectal cancer | Real-time PCR | 182 | 26 | 5 | 210 | 27 | 3 |
| Volha P. Ramaniuk | 2014 | Belarusians | Belarus | HB | bladder cancer | PCR-RFLP | 99 | 178 | 56 | 128 | 169 | 71 |
| Aneta Mirecka | 2014 | Polish | Poland | PB | prostate cancer | real-time PCR | 199 | 249 | 124 | 377 | 218 | 32 |
a Country of first author.
Meta-analysis
Overall analysis
In the dominant model, increased cancer risk was found with an odds ratio (OR) of 1.110 (95% confidence interval [CI] 1.078-1.143, P<0.01). In the recessive model, significantly increased risk was determined with an OR of 1.059 (95% CI 1.013-1.108, P<0.01). Furthermore, when the homozygote and heterozygote comparisons were performed, increased risk was identified, with an OR of 1.103 (95% CI 1.052-1.157, P<0.01), and an OR of 1.106 (95% CI 1.072-1.141, P<0.01), respectively. Overall, the results of our meta-analysis showed a significant association between the ERCC2 polymorphism and cancer risk (Table 2).
Table 2. Results of overall and stratified meta-analyses.
| Model (Comparison) | Subgroup | No. of trials | I2(%) | Pa | Fixed | Random | P for bias |
|---|---|---|---|---|---|---|---|
| homozygote comparison (Asn/Asn vs. Asp/Asp) | Total | 95 | 68.3 | 0 | 1.103(1.052,1.157) | 1.170(1.060,1.293) | 0.079 |
| PB | 41 | 79.8 | 0 | 1.037(0.977,1.101) | 1.074(0.922,1.250) | 0.53 | |
| HB | 49 | 39 | 0.004 | 1.249(1.149,1.358) | 1.283(1.135,1.450) | 0.462 | |
| Asia | 30 | 48.3 | 0.003 | 1.664(1.461,1.894) | 1.734(1.371,2.192) | 0.961 | |
| Caucasian | 37 | 50.8 | 0 | 0.964(0.899,1.034) | 1.019(0.913,1.137) | 0.041 | |
| PCR | 29 | 65 | 0 | 1.041(0.951,1.140) | 1.175(0.983,1.404) | 0.054 | |
| PCR-RFLP | 38 | 62.5 | 0 | 1.160(1.068,1.260) | 1.238(1.053,1.455) | 0.054 | |
| Taqman | 18 | 24.8 | 0.163 | 1.003(0.921,1.093) | 0.983(0.878,1.100) | 0.16 | |
| Bladder cancer | 12 | 56.4 | 0.008 | 1.370(1.198,1.566) | 1.446(1.160,1.803) | 0.191 | |
| Breast cancer | 18 | 66.6 | 0 | 1.098(1.009,1.194) | 1.042(0.871,1.246) | 0.543 | |
| Esophageal cancer | 7 | 0 | 0.62 | 1.219(0.945,1.571) | 1.243(0.962,1.608) | 0.074 | |
| Gastric cancer | 8 | 65.3 | 0.005 | 1.517(1.167,1.972) | 1.876(1.105,3.186) | 0.258 | |
| Head and neck cancer | 6 | 52.4 | 0.062 | 0.993(0.814,1.212) | 0.989(0.707,1.384) | 0.909 | |
| Lung Cancer | 16 | 0 | 0.533 | 1.043(0.901,1.207) | 1.042(0.899,1.207) | 0.386 | |
| Prostate cancer | 7 | 93.5 | 0 | 1.570(1.314,1.874) | 2.038(0.848,4.894) | 0.419 | |
| Skin Cancer | 7 | 59.9 | 0.021 | 0.784(0.689,0.893) | 0.818(0.657,1.020) | 0.448 | |
| Non- Hodgkin lymphoma | 6 | 0 | 0.782 | 0.998(0.811,1.229) | 1.000(0.812,1.231) | 0.505 | |
| heterozygote comparison (Asp/Asn vs. Asp/Asp) | Total | 95 | 61.1 | 0 | 1.106(1.072,1.141) | 1.133(1.072,1.198) | 0.111 |
| PB | 41 | 64.7 | 0 | 1.061(1.020,1.104) | 1.064(0.988,1.146) | 0.889 | |
| HB | 49 | 53.9 | 0 | 1.205(1.143,1.270) | 1.229(1.128,1.339) | 0.329 | |
| Asia | 30 | 71.8 | 0 | 1.373(1.275,1.480) | 1.287(1.105,1.499) | 0.096 | |
| Caucasian | 37 | 0 | 0.801 | 1.034(0.988,1.083) | 1.034(0.987,1.082) | 0.526 | |
| PCR | 29 | 44.2 | 0.006 | 1.057(0.996,1.121) | 1.076(0.982,1.180) | 0.281 | |
| PCR-RFLP | 38 | 70 | 0 | 1.187(1.126,1.251) | 1.203(1.081,1.338) | 0.745 | |
| Taqman | 18 | 14.5 | 0.28 | 1.030(0.974,1.090) | 1.039(0.973,1.109) | 0.348 | |
| Bladder cancer | 12 | 31.2 | 0.142 | 1.235(1.128,1.353) | 1.265(1.125,1.423) | 0.231 | |
| Breast cancer | 18 | 70.7 | 0 | 1.086(1.025,1.149) | 1.101(0.972,1.248) | 0.42 | |
| Esophageal cancer | 7 | 0 | 0.994 | 1.213(1.051,1.401) | 1.213(1.051,1.401) | 0.932 | |
| Gastric cancer | 8 | 91.1 | 0 | 1.209(1.038,1.409) | 1.066(0.614,1.848) | 0.491 | |
| Head and neck cancer | 6 | 27.4 | 0.229 | 1.114(0.977,1.271) | 1.121(0.950,1.323) | 0.334 | |
| Lung Cancer | 16 | 0 | 0.808 | 1.000(0.918,1.090) | 1.001(0.918,1.091) | 0.294 | |
| Prostate cancer | 7 | 78.4 | 0 | 1.281(1.140,1.440) | 1.297(0.965,1.743) | 0.879 | |
| Skin Cancer | 7 | 36.5 | 0.15 | 1.018(0.938,1.105) | 1.023(0.913,1.146) | 0.868 | |
| Non- Hodgkin lymphoma | 6 | 27.7 | 0.227 | 1.038(0.907,1.187) | 1.047(0.881,1.244) | 0.938 | |
| dominant model((Asn/Asn+Asp/Asn) vs. Asp/Asp) | Total | 95 | 69.3 | 0 | 1.110(1.078,1.143) | 1.143(1.078,1.212) | 0.126 |
| PB | 41 | 75.9 | 0 | 1.060(1.021,1.101) | 1.067(0.981,1.160) | 0.754 | |
| HB | 49 | 56.6 | 0 | 1.217(1.158,1.278) | 1.237(1.139,1.343) | 0.587 | |
| Asia | 30 | 73.4 | 0 | 1.416(1.321,1.518) | 1.336(1.153,1.547) | 0.13 | |
| Caucasian | 37 | 3.2 | 0.414 | 1.020(0.976,1.065) | 1.021(0.976,1.068) | 0.102 | |
| PCR | 29 | 47.4 | 0.003 | 1.053(0.996,1.113) | 1.091(0.999,1.191) | 0.137 | |
| PCR-RFLP | 38 | 74.5 | 0 | 1.191(1.133,1.251) | 1.216(1.091,1.356) | 0.647 | |
| Taqman | 18 | 11.5 | 0.317 | 1.026(0.972,1.082) | 1.028(0.968,1.093) | 0.908 | |
| Bladder cancer | 12 | 50.2 | 0.024 | 1.266(1.162,1.379) | 1.309(1.148,1.494) | 0.242 | |
| Breast cancer | 17 | 73.4 | 0 | 1.091(1.034,1.151) | 1.083(0.958,1.223) | 0.962 | |
| Esophageal cancer | 7 | 0 | 0.989 | 1.214(1.057,1.394) | 1.214(1.057,1.394) | 0.236 | |
| Gastric cancer | 8 | 90.7 | 0 | 1.277(1.106,1.474) | 1.229(0.745,2.027) | 0.88 | |
| Head and neck cancer | 6 | 50.7 | 0.071 | 1.091(0.963,1.236) | 1.104(0.908,1.343) | 0.493 | |
| Lung Cancer | 15 | 0 | 0.763 | 1.010(0.931,1.097) | 1.010(0.931,1.097) | 0.474 | |
| Prostate cancer | 7 | 89.8 | 0 | 1.353(1.213,1.509) | 1.407(0.951,2.081) | 0.71 | |
| Skin Cancer | 7 | 37.6 | 0.142 | 0.968(0.895,1.046) | 0.978(0.877,1.090) | 0.682 | |
| Non- Hodgkin lymphoma | 6 | 9.4 | 0.356 | 1.033(0.909,1.173) | 1.035(0.901,1.189) | 0.932 | |
| recessive model (Asn/Asn vs. (Asp/Asp+Asp/Asn)) | Total | 95 | 62.7 | 0 | 1.059(1.013,1.108) | 1.108(1.016,1.208) | 0.098 |
| PB | 41 | 76.4 | 0 | 1.010(0.954,1.069) | 1.044(0.914,1.192) | 0.501 | |
| HB | 49 | 30.6 | 0.025 | 1.157(1.070,1.252) | 1.178(1.059,1.310) | 0.481 | |
| Asia | 30 | 35.8 | 0.032 | 1.445(1.275,1.637) | 1.515(1.240,1.852) | 0.668 | |
| Caucasian | 37 | 52.2 | 0 | 0.954(0.894,1.019) | 1.006(0.906,1.115) | 0.055 | |
| PCR | 29 | 64.2 | 0 | 1.022(0.939,1.113) | 1.131(0.959,1.335) | 0.107 | |
| PCR-RFLP | 38 | 53 | 0 | 1.087(1.006,1.175) | 1.147(1.002,1.314) | 0.152 | |
| Taqman | 18 | 28.8 | 0.123 | 0.987(0.911,1.609) | 0.958(0.859,1.069) | 0.082 | |
| Bladder cancer | 12 | 48.6 | 0.029 | 1.225(1.080,1.389) | 1.271(1.052,1.536) | 0.189 | |
| Breast cancer | 17 | 60.1 | 0.001 | 1.062(0.981,1.149) | 1.018(0.874,1.186) | 0.421 | |
| Esophageal cancer | 7 | 0 | 0.615 | 1.102(0.869,1.398) | 1.130(0.888,1.437) | 0.086 | |
| Gastric cancer | 8 | 39 | 0.119 | 1.563(1.215,2.011) | 1.739(1.190,2.541) | 0.341 | |
| Head and neck cancer | 6 | 35.4 | 0.171 | 0.951(0.790,1.144) | 0.944(0.729,1.223) | 0.815 | |
| Lung Cancer | 15 | 0 | 0.806 | 1.046(0.910,1.203) | 1.046(0.910,1.203) | 0.495 | |
| Prostate cancer | 7 | 92.4 | 0 | 1.406(1.186,1.667) | 1.851(0.846,4.050) | 0.357 | |
| Skin Cancer | 7 | 63.4 | 0.012 | 0.781(0.691,0.883) | 0.810(0.653,1.006) | 0.557 | |
| Non- Hodgkin lymphoma | 6 | 0 | 0.619 | 0.987(0.813,1.200) | 0.989(0.814,1.203) | 0.646 |
a P for heterogeneity.
Subgroup analysis
In order to evaluate the effects of specific study characteristics on the association between the ERCC2 polymorphism and cancer risk, we performed subgroup analysis if there were 6 or more studies. The ORs and 95% CIs were obtained from the subgroups of control source, ethnicity, genotyping method, and type of cancer. For control source subgroup, we found a significant association between the ERCC2 polymorphism and cancer risk when the source of the controls was hospital-based (HB). Meanwhile, when the studies recruited population-based (PB) control, no association was found. For ethnicity, no significant association was detected in Caucasians, but significant associations were observed in Asians. When stratified according to the genotyping method, significant associations were observed when the method was polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP). By comparison, no relationship was found when the methods used were PCR and TaqMan assay. According to the type of cancer, the ERCC2 polymorphism was associated with a significantly higher risk of bladder cancer. In contrast, we observed no association between this polymorphism and breast cancer. Similarly, the results of subgroups of other cancers indicated no association with the ERCC2 polymorphism, including head and neck, lung, prostate, and skin cancers and non-Hodgkin lymphoma. For the esophageal cancer group, a significant association was obtained in the heterozygote comparison, but not in the homozygote comparison and the recessive model. In the group with gastric cancer, the ERCC2 polymorphism was confirmed to increase the risk of cancer in the homozygote comparison and the recessive model, but not in the heterozygote comparison and the dominant model. The detailed results are shown in Table 2.
Test of heterogeneity
High heterogeneity was observed after the data were pooled (homozygote comparison: P for heterogeneity = 0, I2 = 68.3%). As shown in Table 2, when the subjects were stratified on the basis of the control source, high heterogeneity remained with PB controls (homozygote comparison: P for heterogeneity = 0, I2 = 79.8%). Additionally, in analyses of ethnicity, moderate heterogeneity was found in Asian studies (homozygote comparison: P for heterogeneity = 0.003, I2 = 48.3%), and high heterogeneity was found in Caucasian studies (homozygote comparison: P for heterogeneity = 0, I2 = 50.8%). Moreover, in analyses of genotyping methods, low heterogeneity was detected in the TaqMan group (homozygote comparison: P for heterogeneity = 0.163, I2 = 24.8%), but high heterogeneity was found in the PCR (homozygote comparison: P for heterogeneity = 0, I2 = 65%) and PCR-RFLP groups (homozygote comparison: P for heterogeneity = 0, I2 = 62.5%). Furthermore, heterogeneity was not detected in esophageal cancer studies (homozygote comparison: P for heterogeneity = 0.62, I2 = 0.0%), lung cancer studies (homozygote comparison: P for heterogeneity = 0.533, I2 = 0.0%), and non-Hodgkin lymphoma studies (homozygote comparison: P for heterogeneity = 0.782, I2 = 0.0%). Nonetheless, high heterogeneity was still present in studies of prostate cancer (homozygote comparison: P for heterogeneity = 0, I2 = 93.5%), bladder cancer (homozygote comparison: P for heterogeneity = 0.008, I2 = 56.4%), breast cancer (homozygote comparison: P for heterogeneity = 0, I2 = 66.6%), gastric cancer (homozygote comparison: P for heterogeneity = 0.005, I2 = 65.3%), head and neck cancer (homozygote comparison: P for heterogeneity = 0.062, I2 = 52.4%), and skin cancer (homozygote comparison: P for heterogeneity = 0.021, I2 = 59.9%).
Publication bias and sensitivity analysis
We used the Begg's funnel plot to estimate publication bias. There was no statistical evidence of publication bias in the overall analysis under each model (Figure 2). Table 2 shows the P details for bias. We also removed studies one by one to determine their effect on the test of heterogeneity, and evaluated the stability of the overall results; the results did not change in the overall analysis (Supplementary Table 1) neither in other analysis.
Figure 2.
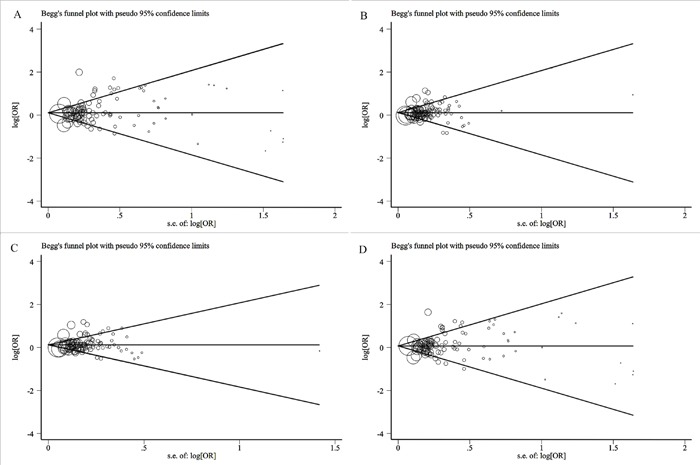
(A) Begg's funnel plot for the publication bias test in the overall analysis under homozygote comparison. (B) Begg's funnel plot for the publication bias test in the overall analysis under heterozygote comparison. (C) Begg's funnel plot for the publication bias test in the overall analysis under dominant model. (D) Begg's funnel plot for the publication bias test in the overall analysis under recessive model.
Trial sequential analysis (TSA)
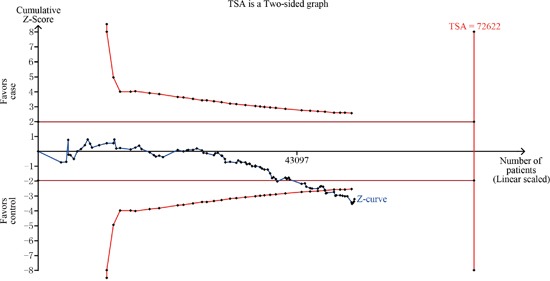
In the overall analysis for homozygote comparison, the required information size was 72,622 patients to demonstrate the issue (Figure 3), and the result showed that the Z-curve had crossed the trial monitoring boundary before reaching the required information size, indicating that the cumulative evidence is adequate and further trials are unnecessary.
Figure 3. TSA for overall analysis under homozygote comparison.
DISCUSSION
Nowadays, cancer is one of the most important global public health problems [106]. Personalized analysis and improved methods of cancer diagnoses can be provided, based on an understanding of the association between genetic polymorphisms and cancer risk [107]. In the relationship between gene polymorphisms and cancer risk, the ERCC2 Asp312Asn polymorphism is an important risk factor. Impaired DNA repair capacity is a risk factor for the development of cancer. The ERCC2 Asp312Asn polymorphism influences DNA repair through the NER pathway. To date, many publications have shown an association between the ERCC2 Asp312Asn polymorphism and risk of cancer. However, the results remain controversial. In order to resolve this conflict, we performed a meta-analysis that evaluates the relationship between the ERCC2 Asp312Asn polymorphism and risk of cancer.
In our meta-analysis, the association of the ERCC2 Asp312Asn polymorphism with the risk of cancer was evaluated in 38,848 cases and 48,928 controls. A significant association was observed between the ERCC2 Asp312Asn polymorphism and overall cancer risk in all genetic models. To the best of our knowledge, this is the most comprehensive meta-analysis on this topic until now. Moreover, the result of the TSA indicated that the cumulative evidence is adequate and further trials are unnecessary in the overall analysis for homozygote comparison.
In the subgroup analysis based on ethnicity, a significantly increased cancer risk was observed in Asian populations, but not in Caucasian populations. One possible reason for these discrepancies is that different ethnicities may have distinct genetic backgrounds, and therefore, tumor susceptibility can be influenced by ethnicity [108]. Moreover, this may indicate that these groups have distinct environmental or genetic cancer co-etiologies [109]. In subgroup analysis based on the control source, we found that a significantly increased cancer risk was observed in HB studies, but not in PB studies. The former may have certain biases for such controls and may only represent a sample of an ill-defined reference population. Furthermore, HB controls may not be representative of the general population or it may be that numerous subjects in the PB controls were individuals susceptible to cancer [110]. In the subgroup analysis based on the genotyping method, a significantly increased cancer risk was found in the PCR-RFLP studies, but not in the PCR or TaqMan studies. A possible reason for this may be that the different genotyping methods are specialized for different aspects, and the results would be more accurate and reliable if the same genotyping method was applied in different studies [111].
In the subgroup analysis according to the cancer site, a significant association with the ERCC2 Asp312Asn polymorphism was observed for bladder, esophageal, and gastric cancers; however, no significant association was observed for breast, head and neck, lung, prostate, and skin cancers, and non- Hodgkin lymphoma. Some previous meta-analyses assessed the effect of the ERCC2 Asp312Asn polymorphism on the risk of these cancers and reached conclusions consistent with those of our study. For example, Li et al. [19] and Wen et al. [14] suggested that the ERCC2 Asp312Asn polymorphism might be associated with an increased risk of bladder cancer and esophageal cancer, respectively. Yin et al. [48] showed that this polymorphism might be a potential biomarker of gastric cancer susceptibility in the overall population. In contrast, Yan et al. [21], Hu et al. [11], and Zhu et al. [112] suggested that the ERCC2 Asp312Asn polymorphism was not associated with breast cancer, head and neck cancer, and skin cancer, respectively. Moreover, Chen et al. [113], Feng et al. [12], and Ma et al. [114] suggested that the ERCC2 Asp312Asn polymorphism contributed to the risk of non-Hodgkin lymphoma, lung cancer, and prostate cancer, respectively. Because we only included studies published from 2005 to 2016, we drew different conclusions in lung cancer and prostate cancer studies. Therefore, more research should be undertaken in the future. Moreover, the exact mechanism for the associations between different cancer sites and the ERCC2 Asp312Asn polymorphism is not clear; the mechanism of carcinogenesis may differ between different cancer sites and the ERCC2 genetic variants may exert varying effects in different cancers [115].
Notably, HCC, osteosarcoma, oral cancer, and colorectal cancer were not included for further analysis as there were fewer than 6 studies available for analysis for such cancers. Wu et al. indicated that the ERCC2 Asp312Asn polymorphism was not associated with the development of HCC [24]. Gomez-Diaz et al. demonstrated no relationship between ERCC2 Asp312Asn polymorphism and osteosarcoma [23]. Interestingly, based on a study by Mahimkar et al. this polymorphism was associated with an overall increase in chromosomal damage in oral cancer [25]. Wang et al. [35] observed a slightly lower statistical significance between the ERCC2 Asp312Asn polymorphism and colorectal cancer. In fact, this polymorphism has also been shown to be related to other diseases; previous studies have indicated that it may have a role in the development of ultraviolet-related diseases, such as maturity onset cataract. [116]. However, no significant association of this polymorphism was found with either idiopathic azoospermia [117] or arsenic-related skin lesions [118]. Therefore, the equivocal association between the ERCC2 Asp312Asn polymorphism and some diseases remains to be confirmed.
Heterogeneity is a major concern for meta-analysis [119]. In our overall analysis, high heterogeneity was observed for all genetic models. However, when data were pooled in to subgroups according the control source, ethnicity, genotyping method, and cancer type, the heterogeneity decreased. Sensitivity analysis showed that the results have sufficient statistical power. There are some limitations of our meta-analysis that should be addressed. First, subgroup analysis cannot be conducted based on sex, age, lifestyle, and other factors owing to insufficient data. Second, some cancers, such as oral cancer and colorectal cancer, were not suitable for further analysis because of the small sample sizes. Thus, more studies on these cancers should be conducted in the future. Third, a single gene has only a moderate effect on cancer development; hence, the ERCC2 gene may influence susceptibility of cancer along with other genes. However, enough data for further analysis is not available. Finally, only published articles were included in the analysis; therefore, unpublished data may modify our conclusions.
In summary, our meta-analysis suggested that the ERCC2 Asp312Asn polymorphism is associated with increased cancer risk. A significantly increased cancer risk was observed in Asian populations, but not in Caucasian populations. Moreover, our results indicated that this polymorphism is associated with bladder, esophageal, and gastric cancers, but not with breast, head and neck, lung, prostate, and skin cancers, and non-Hodgkin lymphoma. In addition, stratification analyses based on the control source also indicated that this polymorphism was associated with cancer risk in the HB populations, but not in the PB populations. In subgroup analysis according to the genotyping method, a significantly increased cancer risk was found in the PCR-RFLP studies, but not in the PCR and TaqMan studies. Considering the limitations of this study, further multi-center, well-designed research should be undertaken in the future.
MATERIALS AND METHODS
Literature search
A systematic search of articles relating to the ERCC2 Asp312Asn polymorphism and cancer was conducted by 2 researchers, using the PubMed, EMBASE, Science Direct, Web of Science and the China National Knowledge Infrastructure (CNKI) databases. The search included studies published between January 1, 2005 and January 1, 2016. The search strategy was based on various combinations of the following terms: “xeroderma pigmentosum group d protein “[MeSH Terms] OR “xeroderma pigmentosum group d protein” [All Fields] OR “ercc2” [All Fields]) AND Asp312Asn [All Fields] AND (“neoplasms” [MeSH Terms] OR “neoplasms” [All Fields] OR “cancer” [All Fields]. In addition, the reference lists of the publications identified were searched for further relevant studies. The PRISMA Checklist was used for this meta-analysis (Supplementary Table 2).
Selection criteria
The following inclusion criteria were set and reviewed by two independent investigators: (I) case-control study; (II) evaluation of the ERCC2 Asp312Asn polymorphism and cancer; and (III) detailed data available for calculating ORs and the corresponding 95% CIs. Studies were excluded if they: (I) had no control population; (II) were review articles or previous meta-analyses; (III) contained insufficient or duplicate data; or (IV) had no full text available.
Data extraction
Two authors performed data extraction independently. For all publications, the following data were extracted: first author, year of publication, ethnicity of the population, country, source of cases and controls, cancer site, genotyping method, and number of cases and controls.
Trial sequential analysis
To evaluate whether our meta-analysis had sufficient sample size to reach firm conclusions about the effect of interventions [120], TSA was used in this meta-analysis. If the cumulative Z curve in results exceeds the TSA boundary, a sufficient level of evidence for the anticipated intervention effect may have been reached and no further trials are needed. However, when the Z curve does not exceed the TSA boundaries and the required information size has not been reached, evidence to draw a conclusion is insufficient [121]. We used two-sided tests, type I error set at 5%, and power set at 80%. The required information size was calculated based on a relative risk reduction of 10%. Trials ignored in interim appear to be due to too low use of information (<1.0%) by the software. TSA was performed using the TSA software (version 0.9.5.5).
Statistical analysis
The primary objective of our meta-analysis was to calculate ORs and their 95% CIs to evaluate the association between ERCC2 Asp312Asn and cancer risks. In our included studies, no clear models had been chosen; thus, the following genetic models were used: homozygote comparison (Asn/Asn vs. Asp/Asp), heterozygote comparison (Asp/Asn vs. Asp/Asp), recessive model (Asn/Asn vs. Asp/Asp+Asp/Asn), and dominant model (Asn/Asn+Asp/Asn vs. Asp/Asp). The statistical heterogeneity assumption was evaluated using I2 statistics to quantify any inconsistency arising from inter-research variability that was derived from heterogeneity instead of random chance [107]. An I2 value from 0-25% indicates low heterogeneity, 25-50% moderate heterogeneity and ≥50% high heterogeneity [122]. Two models (fixed-effect model and random-effect model) were used for analysis [123]. When I2< 50%, we used a fixed effect model and when I2 ≥50%, we performed a random effect model [124, 125]. We used sensitivity analyses by omitting each study in turn to determine the effect of heterogeneity on the test, and evaluated the stability of the overall results [107]. Potential publication bias was assessed using the Begg's linear regression test [126]. Notably, subgroup analysis was not performed when there were fewer than 6 studies available, because the small number may have resulted in insufficient power [107]. All statistical analyses were performed using the STATA statistical software package (version 12.0; StataCorp, College Station, TX).
SUPPLEMENTARY MATERIALS TABLES
Acknowledgments
The authors gratefully acknowledge the National Natural Science Foundation of China (Grants number: 81160097; 21463006); the Guangxi Natural Science Foundation (Grants number: 2011GXNSFA018175, 2013GXNSFGA019005, 2016GXNSFDA380010); the Guangxi scientific research and technology development project (Grant number: Guikegong1355005-5-7); Youth Science Foundation of Guangxi Medical University (Grant number: GXMUYSF2014014); Guangxi medical and healthcare technology research and development project contract (Grant number: S201303-06); Students’ platform for innovation and entrepreneurship training program (Grants Number: 201510598003, 201610598112). The authors thank Editage English service for language editing services.
Abbreviations
- NER
nucleotide excision repair
- ERCC2
excision repair cross-complementing group 2
- XPD
Xeroderma pigmentosum group D
- TFIIH
transcription factor IIH
- SNPs
single nucleotide polymorphisms
- Asn
asparagine amino acid
- HB
hospital-based
- PB
population-based
- HCC
hepatocellular cancer
- CNKI
China National Knowledge Infrastructure
- TSA
trial sequential analysis
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Castro J, Ribo M, Benito A, Vilanova M. Mini-review: nucleus-targeted ribonucleases as antitumor drugs. Curr Med Chem. 2013;20:1225–31. doi: 10.2174/0929867311320100003. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 4.Huang X, Gao Y, He J, Cai J, Ta N, Jiang H, Zhu J, Zheng J. The association between RFC1 G80A polymorphism and cancer susceptibility: Evidence from 33 studies. J Cancer. 2016;7:144–52. doi: 10.7150/jca.13303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qixing M, Gaochao D, Wenjie X, Rong Y, Feng J, Lin X, Mantang Q, Qiang C. Predictive Value of Ercc1 and Xpd Polymorphisms for Clinical Outcomes of Patients Receiving Neoadjuvant Therapy: A Prisma-Compliant Meta-Analysis. Medicine (Baltimore) 2015;94:e1593. doi: 10.1097/md.0000000000001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benhamou S, Sarasin A. ERCC2 /XPD gene polymorphisms and lung cancer: a HuGE review. Am J Epidemiol. 2005;161:1–14. doi: 10.1093/aje/kwi018. [DOI] [PubMed] [Google Scholar]
- 7.Alanazi M, Pathan AA, Ajaj SA, Khan W, Shaik JP, Al Tassan N, Parine NR. DNA Repair Genes XRCC1, XRCC3, XPD, and OGG1 Polymorphisms among the Central Region Population of Saudi Arabia. Biol Res. 2013;46:161–7. doi: 10.4067/s0716-97602013000200007. [DOI] [PubMed] [Google Scholar]
- 8.Ouyang FD, Yang FL, Chen HC, Khan MA, Huang FM, Wan XX, Xu AH, Huang X, Zhou MJ, Fang Q, Zhang DZ. Polymorphisms of DNA repair genes XPD, XRCC1, and OGG1, and lung adenocarcinoma susceptibility in Chinese population. Tumour Biol. 2013;34:2843–8. doi: 10.1007/s13277-013-0844-6. [DOI] [PubMed] [Google Scholar]
- 9.Clarkson SG, Wood RD. Polymorphisms in the human XPD (ERCC2) gene, DNA repair capacity and cancer susceptibility: an appraisal. DNA Repair (Amst) 2005;4:1068–74. doi: 10.1016/j.dnarep.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Lunn RM, Helzlsouer KJ, Parshad R, Umbach DM, Harris EL, Sanford KK, Bell DA. XPD polymorphisms: effects on DNA repair proficiency. Carcinogenesis. 2000;21:551–5. doi: 10.1093/carcin/21.4.551. [DOI] [PubMed] [Google Scholar]
- 11.Hu YY, Yuan H, Jiang GB, Chen N, Wen L, Leng WD, Zeng XT, Niu YM. Associations between XPD Asp312Asn polymorphism and risk of head and neck cancer: a meta-analysis based on 7,122 subjects. PLoS One. 2012;7:e35220. doi: 10.1371/journal.pone.0035220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Z, Ni Y, Dong W, Shen H, Du J. Association of ERCC2/XPD polymorphisms and interaction with tobacco smoking in lung cancer susceptibility: a systemic review and meta-analysis. Mol Biol Rep. 2012;39:57–69. doi: 10.1007/s11033-011-0710-9. [DOI] [PubMed] [Google Scholar]
- 13.Geng P, Ou J, Li J, Liao Y, Wang N, Xie G, Sa R, Liu C, Xiang L, Liang H. A Comprehensive Analysis of Influence ERCC Polymorphisms Confer on the Development of Brain Tumors. Mol Neurobiol. 2015;53:2705–14. doi: 10.1007/s12035-015-9371-3. [DOI] [PubMed] [Google Scholar]
- 14.Wen F, Zhao Z, Liu C, Yin Q, Weng J, Wang Y, Ma Y. A pooled analysis of the ERCC2 Asp312Asn polymorphism and esophageal cancer susceptibility. Tumour Biol. 2014;35:2959–65. doi: 10.1007/s13277-013-1380-0. [DOI] [PubMed] [Google Scholar]
- 15.Zhu ML, He J, Wang M, Sun MH, Jin L, Wang X, Yang YJ, Wang JC, Zheng L, Xiang JQ, Wei QY. Potentially functional polymorphisms in the ERCC2 gene and risk of esophageal squamous cell carcinoma in Chinese populations. Sci Rep. 2014;4:6281. doi: 10.1038/srep06281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang R, Zhang C, Malik A, Shen ZD, Hu J, Wu YH. Xeroderma pigmentosum group D polymorphisms and esophageal cancer susceptibility: a meta-analysis based on case-control studies. World J Gastroenterol. 2014;20:16765–73. doi: 10.3748/wjg.v20.i44.16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C, Jiang Z, Liu X. XPD Lys (751) Gln and Asp (312) Asn polymorphisms and bladder cancer risk: a meta-analysis. Mol Biol Rep. 2010;37:301–9. doi: 10.1007/s11033-009-9693-1. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y, Yang Y. Complex association between ERCC2 gene polymorphisms, gender, smoking and the susceptibility to bladder cancer: a meta-analysis. Tumour Biol. 2014;35:5245–57. doi: 10.1007/s13277-014-1682-x. [DOI] [PubMed] [Google Scholar]
- 19.Li SX, Dai QS, Chen SX, Zhang SD, Liao XY, Deng X, Chi HB, Li FJ, Zhu JH, Jiang YY. Xeroderma pigmentosum complementation group D (XPD) gene polymorphisms contribute to bladder cancer risk: a meta-analysis. Tumour Biol. 2014;35:3905–15. doi: 10.1007/s13277-013-1519-z. [DOI] [PubMed] [Google Scholar]
- 20.Jiang Z, Li C, Xu Y, Cai S, Wang X. Associations between XPD polymorphisms and risk of breast cancer: a meta-analysis. Breast Cancer Res Treat. 2010;123:203–12. doi: 10.1007/s10549-010-0751-0. [DOI] [PubMed] [Google Scholar]
- 21.Yan Y, Liang H, Light M, Li T, Deng Y, Li M, Li S, Qin X. XPD Asp312Asn and Lys751Gln polymorphisms and breast cancer susceptibility: a meta-analysis. Tumour Biol. 2014;35:1907–15. doi: 10.1007/s13277-013-1256-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao L, Qiu LX, Yu L, Yang Z, Yu XJ, Zhong Y, Hu XC, Yu L. The association between ERCC2 Asp312Asn polymorphism and breast cancer risk: a meta-analysis involving 22,766 subjects. Breast Cancer Res Treat. 2010;123:227–31. doi: 10.1007/s10549-010-0754-x. [DOI] [PubMed] [Google Scholar]
- 23.Gómez-Díaz B, de la Luz Ayala-Madrigal M, Gutiérrez-Angulo M, Valle-Solis AE, Linares-González LM, González-Guzmán R, Cruz-Guillén D, Cedeño-Garcidueñas AL, Canto P, López-Hernández LB. Analysis of ERCC1 and ERCC2 gene variants in osteosarcoma, colorectal and breast cancer. Oncol Lett. 2015;9:1657–61. doi: 10.3892/ol.2015.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu JS, Chen YP, Wang LC, Yang YJ, Deng CW, Hou BX, He ZL, Chen JX. Implication of polymorphisms in DNA repair genes with an increased risk of hepatocellular carcinoma. Genet Mol Res. 2014;13:3812–8. doi: 10.4238/2014.May.16.5. [DOI] [PubMed] [Google Scholar]
- 25.Mahimkar MB, Samant TA, Kannan S, Patil T. Influence of genetic polymorphisms on frequency of micronucleated buccal epithelial cells in leukoplakia patients. Oral Oncol. 2010;46:761–6. doi: 10.1016/j.oraloncology.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Spitz MR, Lee JJ, Huang M, Lippman SM, Wu X. Nucleotide excision repair pathway genes and oral premalignant lesions. Clin Cancer Res. 2007;13:3753–8. doi: 10.1158/1078-0432.ccr-06-1911. [DOI] [PubMed] [Google Scholar]
- 27.Majumder M, Sikdar N, Ghosh S, Roy B. Polymorphisms at XPD and XRCC1 DNA repair loci and increased risk of oral leukoplakia and cancer among NAT2 slow acetylators. Int J Cancer. 2007;120:2148–56. doi: 10.1002/ijc.22547. [DOI] [PubMed] [Google Scholar]
- 28.Han J, Colditz GA, Liu JS, Hunter DJ. Genetic variation in XPD, sun exposure, and risk of skin cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:1539–44. doi: 10.1158/1055-9965.epi-04-0846. [DOI] [PubMed] [Google Scholar]
- 29.Lovatt T, Alldersea J, Lear JT, Hoban PR, Ramachandran S, Fryer AA, Smith AG, Strange RC. Polymorphism in the nuclear excision repair gene ERCC2/XPD: association between an exon 6-exon 10 haplotype and susceptibility to cutaneous basal cell carcinoma. Hum Mutat. 2005;25:353–9. doi: 10.1002/humu.20158. [DOI] [PubMed] [Google Scholar]
- 30.Li C, Hu Z, Liu Z, Wang LE, Strom SS, Gershenwald JE, Lee JE, Ross MI, Mansfield PF, Cormier JN, Prieto VG, Duvic M, Grimm EA, et al. Polymorphisms in the DNA repair genes XPC, XPD, and XPG and risk of cutaneous melanoma: a case-control analysis. Cancer Epidemiol Biomarkers Prev. 2006;15:2526–32. doi: 10.1158/1055-9965.epi-06-0672. [DOI] [PubMed] [Google Scholar]
- 31.Millikan RC, Hummer A, Begg C, Player J, de Cotret AR, Winkel S, Mohrenweiser H, Thomas N, Armstrong B, Kricker A, Marrett LD, Gruber SB, Culver HA, et al. Polymorphisms in nucleotide excision repair genes and risk of multiple primary melanoma: the Genes Environment and Melanoma Study. Carcinogenesis. 2006;27:610–8. doi: 10.1093/carcin/bgi252. [DOI] [PubMed] [Google Scholar]
- 32.Debniak T, Scott RJ, Huzarski T, Byrski T, Masojc B, van de Wetering T, Serrano-Fernandez P, Gorski B, Cybulski C, Gronwald J, Debniak B, Maleszka R, Kladny J, et al. XPD common variants and their association with melanoma and breast cancer risk. Breast Cancer Res Treat. 2006;98:209–15. doi: 10.1007/s10549-005-9151-2. [DOI] [PubMed] [Google Scholar]
- 33.Moreno V, Gemignani F, Landi S, Gioia-Patricola L, Chabrier A, Blanco I, Gonzalez S, Guino E, Capella G, Canzian F. Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res. 2006;12:2101–8. doi: 10.1158/1078-0432.ccr-05-1363. [DOI] [PubMed] [Google Scholar]
- 34.Hansen RD, Sorensen M, Tjonneland A, Overvad K, Wallin H, Raaschou-Nielsen O, Vogel U. XPA A23G, XPC Lys939Gln, XPD Lys751Gln and XPD Asp312Asn polymorphisms, interactions with smoking, alcohol and dietary factors, and risk of colorectal cancer. Mutat Res. 2007;619:68–80. doi: 10.1016/j.mrfmmm.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 35.Joshi AD, Corral R, Siegmund KD, Haile RW, Le Marchand L, Martinez ME, Ahnen DJ, Sandler RS, Lance P, Stern MC. Red meat and poultry intake, polymorphisms in the nucleotide excision repair and mismatch repair pathways and colorectal cancer risk. Carcinogenesis. 2009;30:472–9. doi: 10.1093/carcin/bgn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ni M, Zhang WZ, Qiu JR, Liu F, Li M, Zhang YJ, Liu Q, Bai J. Association of ERCC1 and ERCC2 polymorphisms with colorectal cancer risk in a Chinese population. Sci Rep. 2014;4:4112. doi: 10.1038/srep04112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.An J, Liu Z, Hu Z, Li G, Wang LE, Sturgis EM, El-Naggar AK, Spitz MR, Wei Q. Potentially functional single nucleotide polymorphisms in the core nucleotide excision repair genes and risk of squamous cell carcinoma of the head and neck. Cancer Epidemiol Biomarkers Prev. 2007;16:1633–8. doi: 10.1158/1055-9965.epi-07-0252. [DOI] [PubMed] [Google Scholar]
- 38.Harth V, Schafer M, Abel J, Maintz L, Neuhaus T, Besuden M, Primke R, Wilkesmann A, Thier R, Vetter H, Ko YD, Bruning T, Bolt HM, et al. Head and neck squamous-cell cancer and its association with polymorphic enzymes of xenobiotic metabolism and repair. J Toxicol Environ Health A. 2008;71:887–97. doi: 10.1080/15287390801988160. [DOI] [PubMed] [Google Scholar]
- 39.Abbasi R, Ramroth H, Becher H, Dietz A, Schmezer P, Popanda O. Laryngeal cancer risk associated with smoking and alcohol consumption is modified by genetic polymorphisms in ERCC5, ERCC6 and RAD23B but not by polymorphisms in five other nucleotide excision repair genes. Int J Cancer. 2009;125:1431–9. doi: 10.1002/ijc.24442. [DOI] [PubMed] [Google Scholar]
- 40.Ji YB, Tae K, Lee YS, Lee SH, Kim KR, Park CW, Park BL, Shin HD. XPD Polymorphisms and Risk of Squamous Cell Carcinoma of the Head and Neck in a Korean Sample. Clin Exp Otorhinolaryngol. 2010;3:42–7. doi: 10.3342/ceo.2010.3.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gugatschka M, Dehchamani D, Wascher TC, Friedrich G, Renner W. DNA repair gene ERCC2 polymorphisms and risk of squamous cell carcinoma of the head and neck. Exp Mol Pathol. 2011;91:331–4. doi: 10.1016/j.yexmp.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Das S, Bhowmik A, Bhattacharjee A, Choudhury B, Naiding M, Laskar AK, Ghosh SK, Choudhury Y. XPD, APE1, and MUTYH polymorphisms increase head and neck cancer risk: effect of gene-gene and gene-environment interactions. Tumour Biol. 2015;36:7569–79. doi: 10.1007/s13277-015-3472-5. [DOI] [PubMed] [Google Scholar]
- 43.Liu G, Zhou W, Yeap BY, Su L, Wain JC, Poneros JM, Nishioka NS, Lynch TJ, Christiani DC. XRCC1 and XPD polymorphisms and esophageal adenocarcinoma risk. Carcinogenesis. 2007;28:1254–8. doi: 10.1093/carcin/bgm020. [DOI] [PubMed] [Google Scholar]
- 44.Ye W, Kumar R, Bacova G, Lagergren J, Hemminki K, Nyren O. The XPD 751Gln allele is associated with an increased risk for esophageal adenocarcinoma: a population-based case-control study in Sweden. Carcinogenesis. 2006;27:1835–41. doi: 10.1093/carcin/bgl017. [DOI] [PubMed] [Google Scholar]
- 45.Tse D, Zhai R, Zhou W, Heist RS, Asomaning K, Su L, Lynch TJ, Wain JC, Christiani DC, Liu G. Polymorphisms of the NER pathway genes, ERCC1 and XPD are associated with esophageal adenocarcinoma risk. Cancer Causes Control. 2008;19:1077–83. doi: 10.1007/s10552-008-9171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pan J, Lin J, Izzo JG, Liu Y, Xing J, Huang M, Ajani JA, Wu X. Genetic susceptibility to esophageal cancer: the role of the nucleotide excision repair pathway. Carcinogenesis. 2009;30:785–92. doi: 10.1093/carcin/bgp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang CG, Liu T, Lv GD, Liu Q, Feng JG, Lu XM. Analysis of XPD genetic polymorphisms of esophageal squamous cell carcinoma in a population of Yili Prefecture, in Xinjiang, China. Mol Biol Rep. 2012;39:709–14. doi: 10.1007/s11033-011-0789-z. [DOI] [PubMed] [Google Scholar]
- 48.Yin QH, Liu C, Hu JB, Meng RR, Li L, Wang YJ. XPD Lys751Gln and Asp312Asn polymorphisms and gastric cancer susceptibility: a meta-analysis of case-control studies. Asian Pac J Cancer Prev. 2013;14:231–6. doi: 10.7314/apjcp.2013.14.1.231. [DOI] [PubMed] [Google Scholar]
- 49.Smedby KE, Lindgren CM, Hjalgrim H, Humphreys K, Schollkopf C, Chang ET, Roos G, Ryder LP, Falk KI, Palmgren J, Kere J, Melbye M, Glimelius B, et al. Variation in DNA repair genes ERCC2, XRCC1, and XRCC3 and risk of follicular lymphoma. Cancer Epidemiol Biomarkers Prev. 2006;15:258–65. doi: 10.1158/1055-9965.epi-05-0583. [DOI] [PubMed] [Google Scholar]
- 50.Shen M, Zheng T, Lan Q, Zhang Y, Zahm SH, Wang SS, Holford TR, Leaderer B, Yeager M, Welch R, Kang D, Boyle P, Zhang B, et al. Polymorphisms in DNA repair genes and risk of non-Hodgkin lymphoma among women in Connecticut. Hum Genet. 2006;119:659–68. doi: 10.1007/s00439-006-0177-2. [DOI] [PubMed] [Google Scholar]
- 51.Song B, Zhu JY, Liu J, Wang ZH, Shi Y, Lü LY, Zheng Y. [Association of Gene Polymorphisms in the DNA Repair Gene XPD with Risk of Non-Hodgkin's Lymphoma]. [Article in Chinese] Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2008;16:97–100. [PubMed] [Google Scholar]
- 52.Baris S, Celkan T, Batar B, Guven M, Ozdil M, Ozkan A, Apak H, Yildiz I. Association between genetic polymorphism in DNA repair genes and risk of B-cell lymphoma. Pediatr Hematol Oncol. 2009;26:467–72. doi: 10.3109/08880010903096201. [DOI] [PubMed] [Google Scholar]
- 53.Worrillow L, Roman E, Adamson PJ, Kane E, Allan JM, Lightfoot TJ. Polymorphisms in the nucleotide excision repair gene ERCC2/XPD and risk of non-Hodgkin lymphoma. Cancer Epidemiol. 2009;33:257–60. doi: 10.1016/j.canep.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 54.El-Din MA, Khorshied MM, El-Saadany ZA, El-Banna MA, Reda Khorshid OM. Excision repair cross-complementing group 2/Xeroderma pigmentousm complementation group D (ERCC2/XPD) genetic variations and susceptibility to diffuse large B cell lymphoma in Egypt. Int J Hematol. 2013;98:681–6. doi: 10.1007/s12185-013-1462-1. [DOI] [PubMed] [Google Scholar]
- 55.Agalliu I, Kwon EM, Salinas CA, Koopmeiners JS, Ostrander EA, Stanford JL. Genetic variation in DNA repair genes and prostate cancer risk: results from a population-based study. Cancer Causes Control. 2010;21:289–300. doi: 10.1007/s10552-009-9461-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mirecka A, Paszkowska-Szczur K, Scott RJ, Gorski B, van de Wetering T, Wokolorczyk D, Gromowski T, Serrano-Fernandez P, Cybulski C, Kashyap A, Gupta S, Golab A, Slojewski M, et al. Common variants of xeroderma pigmentosum genes and prostate cancer risk. Gene. 2014;546:156–61. doi: 10.1016/j.gene.2014.06.026. [DOI] [PubMed] [Google Scholar]
- 57.Bau DT, Wu HC, Chiu CF, Lin CC, Hsu CM, Wang CL, Wang RF, Tsai FJ. Association of XPD polymorphisms with prostate cancer in Taiwanese patients. Anticancer Res. 2007;27:2893–6. [PubMed] [Google Scholar]
- 58.Mandal RK, Gangwar R, Mandhani A, Mittal RD. DNA repair gene X-ray repair cross-complementing group 1 and xeroderma pigmentosum group D polymorphisms and risk of prostate cancer: a study from North India. DNA Cell Biol. 2010;29:183–90. doi: 10.1089/dna.2009.0956. [DOI] [PubMed] [Google Scholar]
- 59.Lavender NA, Komolafe OO, Benford M, Brock G, Moore JH, Vancleave TT, States JC, Kittles RA, Kidd LC. No association between variant DNA repair genes and prostate cancer risk among men of African descent. Prostate. 2010;70:113–9. doi: 10.1002/pros.21048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dhillon VS, Yeoh E, Fenech M. DNA repair gene polymorphisms and prostate cancer risk in South Australia—results of a pilot study. Urol Oncol. 2011;29:641–6. doi: 10.1016/j.urolonc.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 61.Yuan T, Deng S, Chen M, Chen W, Lu W, Huang H, Xia J. Association of DNA repair gene XRCC1 and XPD polymorphisms with genetic susceptibility to gastric cancer in a Chinese population. Cancer Epidemiol. 2011;35:170–4. doi: 10.1016/j.canep.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 62.Chen Z, Zhang C, Xu C, Li K, Hou R, Li D, Cheng X. Effects of selected genetic polymorphisms in xeroderma pigmentosum complementary group D on gastric cancer. Mol Biol Rep. 2011;38:1507–13. doi: 10.1007/s11033-010-0258-0. [DOI] [PubMed] [Google Scholar]
- 63.Ruzzo A, Canestrari E, Maltese P, Pizzagalli F, Graziano F, Santini D, Catalano V, Ficarelli R, Mari D, Bisonni R, Giordani P, Giustini L, Lippe P, et al. Polymorphisms in genes involved in DNA repair and metabolism of xenobiotics in individual susceptibility to sporadic diffuse gastric cancer. Clin Chem Lab Med. 2007;45:822–8. doi: 10.1515/cclm.2007.143. [DOI] [PubMed] [Google Scholar]
- 64.Yuan T, Deng S, Chen M, Chen W, Lu W, Huang H, Xia J. Association of DNA repair gene polymorphisms with genetic susceptibility to gastric cancer. J Mol Diagn Ther. 2010;2:371–4. doi: 10.1016/j.canep.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 65.Zhang CZ, Chen ZP, Xu CQ, Ning T, Li DP, Hou RP. [Correlation of XPD gene with susceptibility to gastric cancer]. [Article in Chinese] Ai Zheng. 2009;28:1163–7. doi: 10.5732/cjc.009.10340. [DOI] [PubMed] [Google Scholar]
- 66.Yi L, Song BQ, He XM. [Association of single nucleotide polymorphism in DNA repair gene XPD with gastric cancer in Han population from northeast region of China]. [Article in Chinese] Shijie Huaren Xiaohua Zazhi. 2006;14:3143–6. [Google Scholar]
- 67.Zhou R, Li Y, Wand N, Dong X, Zhang X, Guo W. Correlation between single nucleotide polymorphism of DNA repair gene XPD and the risks of esophageal squamous cell carcinoma and gastric cardiacadeno carcinoma. Epidemiological Research. 2007;27:118–33. doi: 10.1155/2016/7495135. [DOI] [Google Scholar]
- 68.Broberg K, Bjork J, Paulsson K, Hoglund M, Albin M. Constitutional short telomeres are strong genetic susceptibility markers for bladder cancer. Carcinogenesis. 2005;26:1263–71. doi: 10.1093/carcin/bgi063. [DOI] [PubMed] [Google Scholar]
- 69.Matullo G, Guarrera S, Sacerdote C, Polidoro S, Davico L, Gamberini S, Karagas M, Casetta G, Rolle L, Piazza A, Vineis P. Polymorphisms/haplotypes in DNA repair genes and smoking: a bladder cancer case-control study. Cancer Epidemiol Biomarkers Prev. 2005;14:2569–78. doi: 10.1158/1055-9965.epi-05-0189. [DOI] [PubMed] [Google Scholar]
- 70.Matullo G, Dunning AM, Guarrera S, Baynes C, Polidoro S, Garte S, Autrup H, Malaveille C, Peluso M, Airoldi L, Veglia F, Gormally E, Hoek G, et al. DNA repair polymorphisms and cancer risk in non-smokers in a cohort study. Carcinogenesis. 2006;27:997–1007. doi: 10.1093/carcin/bgi280. [DOI] [PubMed] [Google Scholar]
- 71.Schabath MB, Delclos GL, Grossman HB, Wang Y, Lerner SP, Chamberlain RM, Spitz MR, Wu X. Polymorphisms in XPD exons 10 and 23 and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2005;14:878–84. doi: 10.1158/1055-9965.epi-04-0235. [DOI] [PubMed] [Google Scholar]
- 72.Andrew AS, Nelson HH, Kelsey KT, Moore JH, Meng AC, Casella DP, Tosteson TD, Schned AR, Karagas MR. Concordance of multiple analytical approaches demonstrates a complex relationship between DNA repair gene SNPs, smoking and bladder cancer susceptibility. Carcinogenesis. 2006;27:1030–7. doi: 10.1093/carcin/bgi284. [DOI] [PubMed] [Google Scholar]
- 73.Garcia-Closas M, Malats N, Real FX, Welch R, Kogevinas M, Chatterjee N, Pfeiffer R, Silverman D, Dosemeci M, Tardon A, Serra C, Carrato A, Garcia-Closas R, et al. Genetic variation in the nucleotide excision repair pathway and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2006;15:536–42. doi: 10.1158/1055-9965.epi-05-0749. [DOI] [PubMed] [Google Scholar]
- 74.Wu X, Gu J, Grossman HB, Amos CI, Etzel C, Huang M, Zhang Q, Millikan RE, Lerner S, Dinney CP, Spitz MR. Bladder cancer predisposition: a multigenic approach to DNA-repair and cell-cycle-control genes. Am J Hum Genet. 2006;78:464–79. doi: 10.1086/500848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fontana L, Bosviel R, Delort L, Guy L, Chalabi N, Kwiatkowski F, Satih S, Rabiau N, Boiteux JP, Chamoux A, Bignon YJ, Bernard-Gallon DJ. DNA repair gene ERCC2, XPC, XRCC1, XRCC3 polymorphisms and associations with bladder cancer risk in a French cohort. Anticancer Res. 2008;28:1853–6. [PubMed] [Google Scholar]
- 76.Chang CH, Wang RF, Tsai RY, Wu HC, Wang CH, Tsai CW, Chang CL, Tsou YA, Liu CS, Bau DT. Significant association of XPD codon 312 single nucleotide polymorphism with bladder cancer susceptibility in Taiwan. Anticancer Res. 2009;29:3903–7. [PubMed] [Google Scholar]
- 77.Gangwar R, Ahirwar D, Mandhani A, Mittal RD. Influence of XPD and APE1 DNA repair gene polymorphism on bladder cancer susceptibility in north India. Urology. 2009;73:675–80. doi: 10.1016/j.urology.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 78.Mittal RD, Mandal RK. Genetic variation in nucleotide excision repair pathway genes influence prostate and bladder cancer susceptibility in North Indian population. Indian J Hum Genet. 2012;18:47–55. doi: 10.4103/0971-6866.96648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramaniuk VP, Nikitchenko NV, Savina NV, Kuzhir TD, Rolevich AI, Krasny SA, Sushinsky VE, Goncharova RI. Polymorphism of DNA repair genes OGG1, XRCC1, XPD and ERCC6 in bladder cancer in Belarus. Biomarkers. 2014;19:509–16. doi: 10.3109/1354750x.2014.943291. [DOI] [PubMed] [Google Scholar]
- 80.Zhou M, Wan HY, Gao BL, Ding YJ, Jun RX. Genetic polymorphisms of XPD and CDA and lung cancer risk. Oncol Lett. 2012;4:247–51. doi: 10.3892/ol.2012.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sakoda LC, Loomis MM, Doherty JA, Julianto L, Barnett MJ, Neuhouser ML, Thornquist MD, Weiss NS, Goodman GE, Chen C. Germ line variation in nucleotide excision repair genes and lung cancer risk in smokers. Int J Mol Epidemiol Genet. 2012;3:1–17. [PMC free article] [PubMed] [Google Scholar]
- 82.Qian B, Zhang H, Zhang L, Zhou X, Yu H, Chen K. Association of genetic polymorphisms in DNA repair pathway genes with non-small cell lung cancer risk. Lung Cancer. 2011;73:138–46. doi: 10.1016/j.lungcan.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 83.Yin Z, Su M, Li X, Li M, Ma R, He Q, Zhou B. ERCC2, ERCC1 polymorphisms and haplotypes, cooking oil fume and lung adenocarcinoma risk in Chinese non-smoking females. J Exp Clin Cancer Res. 2009;28:153. doi: 10.1186/1756-9966-28-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raaschou-Nielsen O, Sorensen M, Overvad K, Tjonneland A, Vogel U. Polymorphisms in nucleotide excision repair genes, smoking and intake of fruit and vegetables in relation to lung cancer. Lung Cancer. 2008;59:171–9. doi: 10.1016/j.lungcan.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 85.Chang JS, Wrensch MR, Hansen HM, Sison JD, Aldrich MC, Quesenberry CP, Jr, Seldin MF, Kelsey KT, Kittles RA, Silva G, Wiencke JK. Nucleotide excision repair genes and risk of lung cancer among San Francisco Bay Area Latinos and African Americans. Int J Cancer. 2008;123:2095–104. doi: 10.1002/ijc.23801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yin J, Vogel U, Ma Y, Qi R, Sun Z, Wang H. A haplotype encompassing the variant allele of DNA repair gene polymorphism ERCC2/XPD Lys751Gln but not the variant allele of Asp312Asn is associated with risk of lung cancer in a northeastern Chinese population. Cancer Genet Cytogenet. 2007;175:47–51. doi: 10.1016/j.cancergencyto.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 87.Lopez-Cima MF, Gonzalez-Arriaga P, Garcia-Castro L, Pascual T, Marron MG, Puente XS, Tardon A. Polymorphisms in XPC, XPD, XRCC1, and XRCC3 DNA repair genes and lung cancer risk in a population of northern Spain. BMC Cancer. 2007;7:162. doi: 10.1186/1471-2407-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V, Stangeland L, Phillips DH, Canzian F, Haugen A. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis. 2006;27:560–7. doi: 10.1093/carcin/bgi232. [DOI] [PubMed] [Google Scholar]
- 89.Hu Z, Xu L, Shao M, Yuan J, Wang Y, Wang F, Yuan W, Qian J, Ma H, Wang Y, Liu H, Chen W, Yang L, et al. Polymorphisms in the two helicases ERCC2/XPD and ERCC3/XPB of the transcription factor IIH complex and risk of lung cancer: a case-control analysis in a Chinese population. Cancer Epidemiol Biomarkers Prev. 2006;15:1336–40. doi: 10.1158/1055-9965.epi-06-0194. [DOI] [PubMed] [Google Scholar]
- 90.Shen M, Berndt SI, Rothman N, Demarini DM, Mumford JL, He X, Bonner MR, Tian L, Yeager M, Welch R, Chanock S, Zheng T, Caporaso N, et al. Polymorphisms in the DNA nucleotide excision repair genes and lung cancer risk in Xuan Wei, China. Int J Cancer. 2005;116:768–73. doi: 10.1002/ijc.21117. [DOI] [PubMed] [Google Scholar]
- 91.Huang WY, Berndt SI, Kang D, Chatterjee N, Chanock SJ, Yeager M, Welch R, Bresalier RS, Weissfeld JL, Hayes RB. Nucleotide excision repair gene polymorphisms and risk of advanced colorectal adenoma: XPC polymorphisms modify smoking-related risk. Cancer Epidemiol Biomarkers Prev. 2006;15:306–11. doi: 10.1158/1055-9965.epi-05-0751. [DOI] [PubMed] [Google Scholar]
- 92.De Ruyck K, Szaumkessel M, De Rudder I, Dehoorne A, Vral A, Claes K, Velghe A, Van Meerbeeck J, Thierens H. Polymorphisms in base-excision repair and nucleotide-excision repair genes in relation to lung cancer risk. Mutat Res. 2007;631:101–10. doi: 10.1016/j.mrgentox.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 93.Crew KD, Gammon MD, Terry MB, Zhang FF, Zablotska LB, Agrawal M, Shen J, Long CM, Eng SM, Sagiv SK, Teitelbaum SL, Neugut AI, Santella RM. Polymorphisms in nucleotide excision repair genes, polycyclic aromatic hydrocarbon-DNA adducts, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2007;16:2033–41. doi: 10.1158/1055-9965.epi-07-0096. [DOI] [PubMed] [Google Scholar]
- 94.Kuschel B, Chenevix-Trench G, Spurdle AB, Chen X, Hopper JL, Giles GG, McCredie M, Chang-Claude J, Gregory CS, Day NE, Easton DF, Ponder BA, Dunning AM, et al. Common polymorphisms in ERCC2 (Xeroderma pigmentosum D) are not associated with breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2005;14:1828–31. doi: 10.1158/1055-9965.epi-04-0807. [DOI] [PubMed] [Google Scholar]
- 95.Jorgensen TJ, Visvanathan K, Ruczinski I, Thuita L, Hoffman S, Helzlsouer KJ. Breast cancer risk is not associated with polymorphic forms of xeroderma pigmentosum genes in a cohort of women from Washington County, Maryland. Breast Cancer Res Treat. 2007;101:65–71. doi: 10.1007/s10549-006-9263-3. [DOI] [PubMed] [Google Scholar]
- 96.Lee SA, Lee KM, Park WY, Kim B, Nam J, Yoo KY, Noh DY, Ahn SH, Hirvonen A, Kang D. Obesity and genetic polymorphism of ERCC2 and ERCC4 as modifiers of risk of breast cancer. Exp Mol Med. 2005;37:86–90. doi: 10.1038/emm.2005.12. [DOI] [PubMed] [Google Scholar]
- 97.Bernard-Gallon D, Bosviel R, Delort L, Fontana L, Chamoux A, Rabiau N, Kwiatkowski F, Chalabi N, Satih S, Bignon YJ. DNA repair gene ERCC2 polymorphisms and associations with breast and ovarian cancer risk. Mol Cancer. 2008;7:36. doi: 10.1186/1476-4598-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jakubowska A, Gronwald J, Menkiszak J, Gorski B, Huzarski T, Byrski T, Toloczko-Grabarek A, Gilbert M, Edler L, Zapatka M, Eils R, Lubinski J, Scott RJ, et al. BRCA1-associated breast and ovarian cancer risks in Poland: no association with commonly studied polymorphisms. Breast Cancer Res Treat. 2010;119:201–11. doi: 10.1007/s10549-009-0390-5. [DOI] [PubMed] [Google Scholar]
- 99.Mechanic LE, Millikan RC, Player J, de Cotret AR, Winkel S, Worley K, Heard K, Heard K, Tse CK, Keku T. Polymorphisms in nucleotide excision repair genes, smoking and breast cancer in African Americans and whites: a population-based case-control study. Carcinogenesis. 2006;27:1377–85. doi: 10.1093/carcin/bgi330. [DOI] [PubMed] [Google Scholar]
- 100.Shen J, Desai M, Agrawal M, Kennedy DO, Senie RT, Santella RM, Terry MB. Polymorphisms in nucleotide excision repair genes and DNA repair capacity phenotype in sisters discordant for breast cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:1614–9. doi: 10.1158/1055-9965.epi-06-0218. [DOI] [PubMed] [Google Scholar]
- 101.Smith TR, Levine EA, Freimanis RI, Akman SA, Allen GO, Hoang KN, Liu-Mares W, Hu JJ. Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis. 2008;29:2132–8. doi: 10.1093/carcin/bgn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang L, Zhang Z, Yan W. Single nucleotide polymorphisms for DNA repair genes in breast cancer patients. Clin Chim Acta. 2005;359:150–5. doi: 10.1016/j.cccn.2005.03.047. [DOI] [PubMed] [Google Scholar]
- 103.Hussien YM, Gharib AF, Awad HA, Karam RA, Elsawy WH. Impact of DNA repair genes polymorphism (XPD and XRCC1) on the risk of breast cancer in Egyptian female patients. Mol Biol Rep. 2012;39:1895–901. doi: 10.1007/s11033-011-0935-7. [DOI] [PubMed] [Google Scholar]
- 104.Jelonek K, Gdowicz-Klosok A, Pietrowska M, Borkowska M, Korfanty J, Rzeszowska-Wolny J, Widlak P. Association between single-nucleotide polymorphisms of selected genes involved in the response to DNA damage and risk of colon, head and neck, and breast cancers in a Polish population. J Appl Genet. 2010;51:343–52. doi: 10.1007/bf03208865. [DOI] [PubMed] [Google Scholar]
- 105.Wang HC, Liu CS, Wang CH, Tsai RY, Tsai CW, Wang RF, Chang CH, Chen YS, Chiu CF, Bau DT, Huang CY. Significant association of XPD Asp312Asn polymorphism with breast cancer in Taiwanese patients. Chin J Physiol. 2010;53:130–5. doi: 10.4077/cjp.2010.amk005. [DOI] [PubMed] [Google Scholar]
- 106.Lin QJ, Yang F, Jin C, Fu DL. Current status and progress of pancreatic cancer in China. World J Gastroenterol. 2015;21:7988–8003. doi: 10.3748/wjg.v21.i26.7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang M, Wang Y, Fang T, Cai Y, Xu Y, Yan C, Zhang L, Liang C. Common polymorphisms in CD44 gene and susceptibility to cancer: A systematic review and meta-analysis of 45 studies. Oncotarget. 2016;7:76021–76035. doi: 10.18632/oncotarget.12580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang CD, Li HT, Liu K, Lin ZD, Peng QL, Qin X, He M, Wu H, Mo ZN, Yang XL. Impact of caspase-8 (CASP8) -652 6N del and D302H polymorphisms on prostate cancer in different ethnic groups. Asian Pac J Cancer Prev. 2014;15:7713–8. doi: 10.7314/apjcp.2014.15.18.7713. [DOI] [PubMed] [Google Scholar]
- 109.Fu W, Zhuo ZJ, Chen YC, Zhu J, Zhao Z, Jia W, Hu JH, Fu K, Zhu SB, He J, Liu GC. NFKB1-94insertion/deletion ATTG polymorphism and cancer risk: Evidence from 50 case-control studies. Oncotarget. 2016;8:9806–9822. doi: 10.18632/oncotarget.14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xie SZ, Liu ZZ, Yu JH, Liu L, Wang W, Xie DL, Qin JB. Association between the MTHFR C677T polymorphism and risk of cancer: evidence from 446 case-control studies. Tumour Biol. 2015;36:8953–72. doi: 10.1007/s13277-015-3648-z. [DOI] [PubMed] [Google Scholar]
- 111.Tang J, Li X, Jiang X, Xu W, Xu Z, Wang W, Liu B, Lv Q, Zhang W. The association between rs9642880 gene polymorphism and bladder cancer risk: a meta-analysis. Int J Clin Exp Med. 2015;8:20236–44. [PMC free article] [PubMed] [Google Scholar]
- 112.Zhu HL, Bao JM, Lin PX, Li WX, Zou ZN, Huang YE, Chen Q, Shen H. XPD Lys751Gln and Asp312Asn polymorphisms and susceptibility to skin cancer: a meta-analysis of 17 case-control studies. Asian Pac J Cancer Prev. 2014;15:6619–25. doi: 10.7314/apjcp.2014.15.16.6619. [DOI] [PubMed] [Google Scholar]
- 113.Chen S, Zhu JH, Wang F, Huang SY, Xue WQ, Cui Z, He J, Jia WH. Association of the Asp312Asn and Lys751Gln polymorphisms in the XPD gene with the risk of non-Hodgkin's lymphoma: evidence from a meta-analysis. Chin J Cancer. 2015;34:108–14. doi: 10.1186/s40880-015-0001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ma Q, Qi C, Tie C, Guo Z. Genetic polymorphisms of xeroderma pigmentosum group D gene Asp312Asn and Lys751Gln and susceptibility to prostate cancer: a systematic review and meta-analysis. Gene. 2013;530:309–14. doi: 10.1016/j.gene.2013.08.053. [DOI] [PubMed] [Google Scholar]
- 115.Wu KG, He XF, Li YH, Xie WB, Huang X. Association between the XPD/ERCC2 Lys751Gln polymorphism and risk of cancer: evidence from 224 case-control studies. Tumour Biol. 2014;35:11243–59. doi: 10.1007/s13277-014-2379-x. [DOI] [PubMed] [Google Scholar]
- 116.Padma G, Mamata M, Reddy KR, Padma T. Polymorphisms in two DNA repair genes (XPD and XRCC1)—association with age related cataracts. Mol Vis. 2011;17:127–33. [PMC free article] [PubMed] [Google Scholar]
- 117.Ji G, Gu A, Xia Y, Lu C, Liang J, Wang S, Ma J, Peng Y, Wang X. ERCC1 and ERCC2 polymorphisms and risk of idiopathic azoospermia in a Chinese population. Reprod Biomed Online. 2008;17:36–41. doi: 10.1016/s1472-6483(10)60290-8. [DOI] [PubMed] [Google Scholar]
- 118.McCarty KM, Smith TJ, Zhou W, Gonzalez E, Quamruzzaman Q, Rahman M, Mahiuddin G, Ryan L, Su L, Christiani DC. Polymorphisms in XPD (Asp312Asn and Lys751Gln) genes, sunburn and arsenic-related skin lesions. Carcinogenesis. 2007;28:1697–702. doi: 10.1093/carcin/bgm099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xiao F, Lan A, Lin Z, Song J, Zhang Y, Li J, Gu K, Lv B, Zhao D, Zeng S, Zhang R, Zhao W, Pan Z, et al. Impact of CAG repeat length in the androgen receptor gene on male infertility - a meta-analysis. Reprod Biomed Online. 2016;33:39–49. doi: 10.1016/j.rbmo.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 120.Varvaki Rados D, Catani Pinto L, Reck Remonti L, Bauermann Leitao C, Gross JL. The Association between Sulfonylurea Use and All-Cause and Cardiovascular Mortality: A Meta-Analysis with Trial Sequential Analysis of Randomized Clinical Trials. PLoS Med. 2016;13:e1001992. doi: 10.1371/journal.pmed.1001992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang G, Zhang L, Lou S, Chen Y, Cao Y, Wang R, Zhang L, Tang P. Effect of Dexmedetomidine in Preventing Postoperative Side Effects for Laparoscopic Surgery: A Meta-Analysis of Randomized Controlled Trials and Trial Sequential Analysis (PRISMA) Medicine (Baltimore) 2016;95:e2927. doi: 10.1097/md.0000000000002927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang Y, Li M, Xiao F, Teng R, Zhang C, Lan A, Gu K, Li J, Wang D, Li H, Jiang L, Zeng S, He M, et al. Impact of partial DAZ1/2 deletion and partial DAZ3/4 deletion on male infertility. Gene. 2015;571:9–16. doi: 10.1016/j.gene.2015.07.083. [DOI] [PubMed] [Google Scholar]
- 123.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhai Y, Dai Z, He H, Gao F, Yang L, Dong Y, Lu J. A PRISMA-compliant meta-analysis of MDM4 genetic variants and cancer susceptibility. Oncotarget. 2016;7:73935–73944. doi: 10.18632/oncotarget.12558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Luo M, Yang Y, Luo D, Liu L, Zhang Y, Xiao F, Yang J, Zhang C, Fu S, Luo Z. Tumor necrosis factor-alpha promoter polymorphism 308 G/A is not significantly associated with esophageal cancer risk: a meta-analysis. Oncotarget. 2016;7:79901–79913. doi: 10.18632/oncotarget.13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L. Comparison of two methods to detect publication bias in meta-analysis. Jama. 2006;295:676–80. doi: 10.1001/jama.295.6.676. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


