Abstract
Rationale: Difficulty of asthma ascertainment and its associated methodologic heterogeneity have created significant barriers to asthma care and research.
Objectives: We evaluated the validity of an existing natural language processing (NLP) algorithm for asthma criteria to enable an automated chart review using electronic medical records (EMRs).
Methods: The study was designed as a retrospective birth cohort study using a random sample of 500 subjects from the 1997–2007 Mayo Birth Cohort who were born at Mayo Clinic and enrolled in primary pediatric care at Mayo Clinic Rochester. Performance of NLP-based asthma ascertainment using predetermined asthma criteria was assessed by determining both criterion validity (chart review of EMRs by abstractor as a gold standard) and construct validity (association with known risk factors for asthma, such as allergic rhinitis).
Measurements and Main Results: After excluding three subjects whose respiratory symptoms could be attributed to other conditions (e.g., tracheomalacia), among the remaining eligible 497 subjects, 51% were male, 77% white persons, and the median age at last follow-up date was 11.5 years. The asthma prevalence was 31% in the study cohort. Sensitivity, specificity, positive predictive value, and negative predictive value for NLP algorithm in predicting asthma status were 97%, 95%, 90%, and 98%, respectively. The risk factors for asthma (e.g., allergic rhinitis) that were identified either by NLP or the abstractor were the same.
Conclusions: Asthma ascertainment through NLP should be considered in the era of EMRs because it can enable large-scale clinical studies in a more time-efficient manner and improve the recognition and care of childhood asthma in practice.
Keywords: informatics, retrospective study, electronic medical records
At a Glance Commentary
Scientific Knowledge on the Subject
Automated chart review using natural language processing for asthma ascertainment through electronic medical records reduces intrarater and interrater variability and inconsistency (i.e., methodologic heterogeneity) in asthma ascertainment.
What This Study Adds to the Field
In the era of electronic medical records, automated chart review using natural language processing helps clinicians and researchers identify children with recurrent asthma symptoms in a timely and consistent manner.
Asthma is the most common chronic illness in childhood, affecting 4–17% of children in the United States (1–5) and 2.8–37% of children worldwide, depending on the countries included in the estimate (6). Despite the availability of evidence-based guidelines for asthma management and effective asthma therapies, asthma continues to cause a significant morbidity and burden to our society (7).
Important concerns in current asthma care and research regarding identifying patients with asthma include the use of inconsistent asthma criteria (i.e., no standardized diagnostic criteria for asthma), asthma ascertainment process (e.g., physician-diagnosed asthma, symptom-based questionnaire, or billing codes), and sampling frame (e.g., different source population for cases [allergy clinic] and control subjects [primary care]). These challenges have led to inconsistent results of studies including genome-wide association studies, clinical trials, and biomarker studies (8–16). To more accurately assess clinically relevant information, such as symptoms and risk factors for asthma, manual chart review must be used because such information is typically embedded in text in electronic medical records (EMRs). To overcome the limitations of structured data (e.g., poor sensitivity [31%] of International Classification of Diseases [ICD] codes), manual chart review based on existing predetermined criteria has been widely used for asthma epidemiologic studies (17–21). However, this method is challenging, if not infeasible, especially for large-scale studies because applying asthma criteria to great volumes of medical records requires extraordinary effort and costs.
Growing deployments of EMR systems have established large practice-based longitudinal datasets, which allow for the identification of patient cohorts for epidemiologic investigations and population management. Natural language processing (NLP), which can extract information from narrative text automatically, has received a great deal of attention and has played a critical role in secondary use of the EMR for clinical and translational research (22–31). Previously, we developed an NLP algorithm, NLP-PAC, to automatically apply the Predetermined Asthma Criteria (PAC) for asthma ascertainment with significantly improved sensitivity (85%) when compared with structured data, such as ICD-9 codes (31%) (26). Using NLP-PAC has the potential to address some of the current challenges in asthma research and care because applying the NLP algorithm to large EMR datasets will enable large-scale clinical studies in asthma.
Herein, we applied NLP-PAC to an independent birth cohort with the purpose of further assessing both criterion and construct validity of the algorithm. Some of the results of our study have been previously reported in the form of an abstract (32).
Methods
Study Setting
Rochester, Minnesota, is centrally located in Olmsted County, and health care here is virtually self-contained within the community. Under the auspices of the Rochester Epidemiology Project, which links all inpatient and outpatient clinical diagnoses and information from every episode of care to each patient and health care provider, approximately 95% of Olmsted County residents’ medical records have been available for research purposes since 1966, allowing high fidelity of longitudinal studies (33). This resource has been electronically available at the Mayo Clinic since 1997 (i.e., the inception of EMR at Mayo Clinic).
Study Design and Subjects
This was a retrospective birth cohort study. The main aim of this study was to validate two aspects of our present NLP-PAC: criterion validity and construct validity (see Statistical Analysis). The accuracy of NLP-PAC (26) has been refined on the training cohort (n = 430), which is a random sample of the 2002–2006 birth cohort, Rochester, Minnesota, who had been enrolled in a previous asthma study (34). In this study, we evaluated the validity of the current version of NLP-PAC using an independent test cohort, by randomly sampling 500 subjects from the 1997–2007 Mayo Birth Cohort (n = 8,525) who were born at Mayo Clinic, enrolled in primary pediatric care at Mayo Clinic Rochester, and had EMR data available for applying the algorithm. The inclusion criteria were (1) a member of the Mayo Birth Cohort, (2) presence of research authorization for using medical record for research, (3) Olmsted County residency during the study period, (4) children who did not potentially have asthma-related medical records (e.g., diagnosis of asthma, bronchiolitis, pneumonia, wheezing) outside Mayo Clinic, and (5) children without any medical conditions fulfilling the exclusion criteria in Table 1. There was no requirement for a minimum follow-up to be included in the study.
Table 1.
Predetermined Asthma Criteria
| Patients were considered to have definite asthma if a physician had made a diagnosis of asthma and/or if each of the following three conditions were present, and they were considered to have probable asthma if only the first two conditions were present: |
| 1. History of cough with wheezing, and/or dyspnea; or history of cough and/or dyspnea plus wheezing on examination |
| 2. Substantial variability in symptoms from time to time or periods of weeks or more when symptoms were absent |
| 3. Two or more of the following: |
| • Sleep disturbance by nocturnal cough and wheeze |
| • Nonsmoker (14 yr or older) |
| • Nasal polyps |
| • Blood eosinophilia higher than 300/μl |
| • Positive wheal and flare skin tests; or elevated serum IgE |
| • History of hay fever or infantile eczema; or cough, dyspnea, and wheezing regularly on exposure to an antigen |
| • Pulmonary function tests showing one FEV1 or FVC less than 70% predicted and another with at least 20% improvement to an FEV1 of higher than 70% predicted; or methacholine challenge test showing 20% or greater decrease in FEV1 |
| • Favorable clinical response to bronchodilator |
| Patients were excluded from our previous study if any of these conditions were present: |
| • Pulmonary function tests that showed FEV1 to be consistently below 50% predicted or diminished diffusion capacity |
| • Tracheobronchial foreign body at or about the incidence date |
| • Hypogammaglobulinemia (IgG <2.0 mg/ml) or other immunodeficiency disorder |
| • Wheezing occurring only in response to anesthesia or medications |
| • Bullous emphysema or pulmonary fibrosis on chest radiograph |
| • PiZZ alpha-1 antitrypsin |
| • Cystic fibrosis |
| • Other major chest disease, such as juvenile kyphoscoliosis or bronchiectasis |
Predetermined Asthma Criteria
Drs. John Yunginger and Charles Reed, renowned researchers and clinicians for asthma, developed and validated the original PAC for retrospective studies among children and adults based on chart review (Table 1) (35). To our knowledge, these are the only existing predetermined criteria for asthma that determines asthma status and the index date of incident asthma retrospectively based on medical records. As defined by PAC, most cases of probable asthma (85%) became definite asthma over time (35, 36). PAC was found to have high reliability, and extensive epidemiologic work for asthma has used PAC showing the excellent construct validity in identifying known risk factors for asthma and asthma-related adverse outcomes (e.g., microbial infections) (35–45). Index date was defined as the date when the PAC was met for the first time.
NLP-PAC for PAC
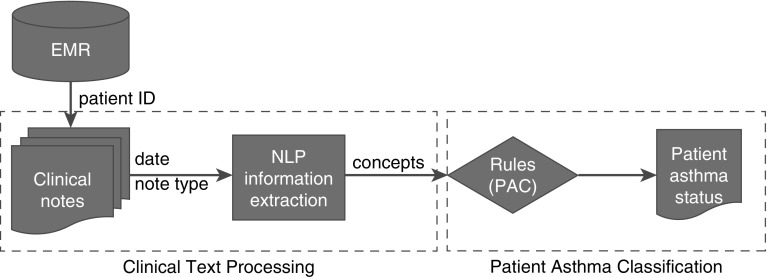
The development of NLP algorithm for PAC was previously described in detail (26, 27), and a high-level diagram of the system is depicted in Figure 1. Briefly, there are two components in NLP-PAC: the text processing component, which extracts concepts in PAC (delineated in Table 1) from medical records; and the asthma status ascertainment component, which classifies asthma status at a patient level using pattern-based rules, assertion status (e.g., nonnegated [had wheezing vs. denied wheezing], associated with patient [not family history]), and section constraints (e.g., diagnosis). Some primary concepts were combined into secondary concepts to meet the criteria (e.g., “wheezing” and “coughing”). The algorithm was implemented using the open-source NLP pipeline MedTagger (http://ohnlp.org/index.php/MedTagger) developed by Mayo Clinic (46).
Figure 1.
A high-level diagram of the system (i.e., NLP-PAC). There are two components in NLP-PAC: the text processing component extracts concepts in PAC delineated in Table 1 from medical records, and the asthma status ascertainment component classifies asthma status at a patient level using pattern-based rules, assertion status (e.g., nonnegated, associated with patient), and section constraints (e.g., diagnosis). EMR = electronic medical records; NLP = natural language processing; PAC = predetermined asthma criteria.
Asthma Ascertainment by NLP-PAC and Chart Review of EMR by Abstractor
For each patient, we retrieved medical records that were available up to September 1, 2015. Only EMR narratives were used by both NLP-PAC and abstractor to ascertain asthma status. After completing asthma ascertainment by NLP-PAC and the abstractor using the training cohort (34), we performed an error analysis for false positives (i.e., NLP indicates “yes” for asthma, but abstractor indicates “no”) and false negatives (i.e., vice versa) to revise and refine NLP-PAC through a reiterative process. Any discrepancies (i.e., false positives and false negatives) were adjudicated by an independent reviewer (M.A.P., an allergy specialist). We applied this NLP-PAC to the test cohort for validation. Chart review of EMR by an abstractor was performed independently. For chart review of the test cohort’s EMR, only one abstractor (M.C.R.) ascertained asthma status by PAC after checking interrater agreement with an independent rater (physician, C.-I.W.) showing 100% agreement using five random samples. The data abstractor was blinded to the asthma status by the NLP algorithm throughout the data collection phase. Once they finished asthma ascertainment, an independent reviewer (C.-I.W.) reviewed charts of subjects with discrepancy between gold standard (i.e., abstractor’s review of EMR) and NLP-PAC, and reconciled the discrepancy and confirmed it with an allergist (M.A.P.).
Other Variables
To assess construct validity, one abstractor (A.S.) collected all pertinent variables known to be risk factors for asthma. After checking interrater agreement with another rater (C.-I.W.), both raters showed 100% agreement for each variable for a random sample of five subjects such as a family history of asthma; a history of other atopic diseases, such as allergic rhinitis or eczema; maternal smoking during pregnancy and household smoking exposure after birth; cesarean section; breastfeeding; and birth weight. The data sources for these variables included birth certificates, well-child visit notes, and the clinical note sections of family history and final diagnosis.
Statistical Analysis
In our present study, we summarized the characteristics of each study subject. Performance of NLP-PAC was assessed for criterion and construct validity. For criterion validity, performance of the algorithm was assessed by using agreement rate, kappa index, sensitivity, specificity, positive predictive value, and negative predictive value for concordance in asthma status between NLP-PAC and manual chart review as a gold standard. Construct validity was tested using logistic regression models by assessing the association of NLP-PAC results with the known risk factors for asthma, because asthma status ascertained by NLP-PAC is expected to be correlated with the known risk factors for asthma if it captures the underlying construct (i.e., asthma). The construct validity of NLP-PAC was compared with that of EMR chart review by an abstractor. Odds ratios and their corresponding 95% confidence intervals were presented. All statistical analyses were performed using JMP statistical software package version 10 (SAS Institute, Inc., Cary, NC).
Results
Characteristics of Study Subjects
The characteristics of study subjects are summarized in Table 2. Out of 500 children in the test cohort, three were excluded because of conditions that may make it hard to differentiate asthma-related wheezing from others, such as mild pectus excavatum with respiratory symptoms, tracheomalacia, or paradoxical vocal cord motion. Among the 497 eligible study subjects, 255 (51%) were males, 383 (77%) white persons, and the median age at last follow-up date was 11.5 years (interquartile range, 9.0–14; range, 4.7–17.9). All study subjects had more than 4 years of follow-up, and each subject had multiple EMR notes documented by multiple health care providers (e.g., nurse note, physician note) during study follow-up period. The median (interquartile range) numbers of EMR notes per subject were 100 (73–140) and 63 (44–92) among subjects with and without asthma, respectively (P < 0.001).
Table 2.
Characteristics of Study Subjects
| Variables | n = 497 |
|---|---|
| Age at last follow-up date, yr, median (IQR) | 11.5 (9.0–14.5) |
| Male | 255 (51%) |
| White | 383 (77%) |
| Allergic rhinitis | 84 (16%) |
| Eczema | 147 (29%) |
| Family history of asthma | 131 (26%) |
| Smoking during pregnancy | 31 (6%) |
| Smoking exposure after birth | 32 (6%) |
| Breastfeeding | 361 (78%) |
| Cesarean section | 113 (22%) |
| Birth weight, kg, median (IQR) | 3.3 (3.0–3.7) |
Definition of abbreviation: IQR = interquartile range.
Concordance in Asthma Status between NLP-PAC and Chart Review of EMR by Abstractor (Criterion Validity)
The results are summarized in Table 3. NLP-PAC identified 158 subjects meeting asthma criteria, whereas manual chart review identified 147 subjects with 131 by both approaches. Kappa index and agreement for asthma status between NLP-PAC and chart review by abstractor were 0.91 and 0.96, respectively, suggesting excellent agreement. Sensitivity, specificity, positive predictive value, and negative predictive value for NLP-PAC in asthma ascertainment using chart review by abstractor as the gold standard showed high predictive values (97%, 95%, 90%, and 98%, respectively). These results were similar between children with and without lung function test among those aged 6 years or older (Table 3).
Table 3.
Agreement for Asthma Status between NLP Algorithm and Manual Chart Review (Gold Standard) (n = 497)*
| Criterion Validity | Unweighted Cohen’s Kappa | Overall Agreement | Sensitivity | Specificity | Positive Predictive Value | Negative Predictive Value |
|---|---|---|---|---|---|---|
| All subjects (n = 497) | 0.91 | 0.96 | 143/147 (97%) | 335/350 (95%) | 143/158 (90%) | 335/339 (98%) |
| Children ≥6 yr (n = 491) | 0.90 | 0.96 | 143/147 (97%) | 329/344 (95%) | 143/158 (90%) | 329/333 (98%) |
| Lung function test, yes (n = 51) | 0.92 | 0.98 | 43/43 (100%) | 7/8 (87%) | 43/44 (97%) | 7/7 (100%) |
| Lung function test, no (n = 440) | 0.89 | 0.95 | 100/104 (96%) | 322/336 (95%) | 100/114 (87%) | 322/326 (98%) |
Definition of abbreviation: NLP = natural language processing.
Sensitivity, specificity, positive predictive value, and negative predictive value were calculated as performance of NLP with manual chart review as a gold standard. For example, for the sensitivity among all subjects, 147 was the number of patients whose asthma status was ascertained as “yes” by manual chart review, and 143 was the number of patients whose asthma status was ascertained as “yes” by the NLP among 147 subjects ascertained by manual chart review.
Asthma prevalence of 31%.
Association of Asthma Status of NLP-PAC and Chart Review of EMR by Abstractor with the Known Risk Factors (Construct Validity)
The known risk factors for asthma identified by NLP were the same as the ones identified by the abstractor. Children with asthma determined by NLP-PAC had higher odds of having a family history of asthma, a history of allergic rhinitis and eczema, maternal smoking during pregnancy, smoking exposure after birth, and no breastfeeding history compared with those without asthma (P < 0.05 in each), but not for cesarean section, or low birth weight (Table 4). Asthma status by manual chart review showed similar results in terms of the association with known risk factors for asthma, except eczema with marginal significance.
Table 4.
Associations of Asthma Status Determined by NLP and Abstractor with Known Risk Factors for Asthma (n = 497)
| By NLP |
By Manual Chart Review |
|||||||
|---|---|---|---|---|---|---|---|---|
| No Asthma (n = 339) | Asthma (n = 158) | OR (95% CI) | P Value | No asthma (n = 350) | Asthma (n = 147) | OR (95% CI) | P Value | |
| Age, yr, median (IQR)* | 11.2 (8.6–13.9) | 12.4 (9.5–15.4) | 1.0 (1.0–1.1) | 0.01 | 11.1 (8.6–13.9) | 12.6 (9.8–15.5) | 1.0 (1.0–1.1) | 0.001 |
| Male, n (%) | 165 (48) | 90 (56) | 1.3 (0.9–2.0) | 0.08 | 172 (49) | 83 (56) | 1.3 (0.9–1.9) | 0.13 |
| White, n (%) | 263 (77) | 120 (75) | 0.9 (0.5–1.4) | 0.68 | 271 (77) | 112 (76) | 0.9 (0.5–1.4) | 0.76 |
| Allergic rhinitis, n (%) | 38 (11) | 46 (29) | 3.2 (2.0–5.2) | <0.001 | 40 (11) | 44 (29) | 3.3 (2.0–5.3) | <0.001 |
| Eczema, n (%) | 90 (26) | 57 (36) | 1.5 (1.0–2.3) | 0.03 | 96 (27) | 51 (34) | 1.4 (0.9–2.1) | 0.07 |
| Family history of asthma, n (%) | 69 (20) | 62 (39) | 2.5 (1.6–3.8) | <0.001 | 72 (20) | 59 (40) | 2.5 (1.7–3.9) | <0.001 |
| Smoking during pregnancy (missing: 10),† n (%) | 13 (3) | 18 (11) | 3.1 (1.5–6.6) | 0.001 | 15 (4) | 16 (11) | 2.7 (1.2–5.6) | 0.006 |
| Smoking exposure after birth (missing: 36),‡ n (%) | 15 (4) | 17 (10) | 2.3 (1.1–4.8) | 0.01 | 14 (4) | 18 (12) | 3.0 (1.4–6.2) | 0.001 |
| Breastfeeding (missing: 38), n (%) | 255 (81) | 106 (72) | 0.5 (0.3–0.9) | 0.01 | 263 (81) | 98 (72) | 0.5 (0.3–0.9) | 0.02 |
| Cesarean section, n (%) | 78 (23) | 35 (22) | 0.9 (0.6–1.4) | 0.83 | 81 (23) | 32 (21) | 0.9 (0.5–1.4) | 0.73 |
| Birth weight, kg, median (IQR) | 3.3 (3.0–3.7) | 3.3 (3.0–3.7) | 0.9 (0.7–1.3) | 0.89 | 3.3 (2.9–3.7) | 3.3 (3.0–3.7) | 1.0 (0.7–1.3) | 0.92 |
Definition of abbreviations: CI = confidence interval; IQR = interquartile range; NLP = natural language processing; OR = odds ratio.
Age at the last follow-up date.
Maternal smoking status during pregnancy.
Household smoking exposure status between ages 0 and 6 years.
Comparison of Time Efficiency between NLP-PAC and Abstractor’s Chart Review–based Asthma Ascertainment
We compared time spent ascertaining asthma status between NLP-PAC versus chart review of EMR by abstractor to assess time efficiency. It took 384 hours for data abstractors to complete chart review of EMR for asthma ascertainment of 430 study subjects, whereas it took 22 minutes (2.3-GHz single laboratory top) to run NLP-PAC for the same subjects. The time spent running NLP-PAC was calculated only for running the algorithm, not including time spent developing NLP-PAC. These findings demonstrate that after development, the NLP algorithm can enable large scale clinical studies in a highly time-efficient manner.
Discussion
To our knowledge, this is the first study that demonstrates that asthma status ascertained by NLP algorithm using EMR has excellent concordance with chart review of EMR by abstractor (i.e., criterion validity) and is associated with the known risk factors for asthma (i.e., construct validity). Our study results suggest feasibility of determining asthma status by an NLP algorithm. Although our findings need to be replicated by future studies with a larger sample size using different EMR systems, they suggest the huge potential of leveraging NLP for asthma care and research in the EMR and big data era.
Because literature on determination of asthma status by NLP does not currently exist, it is difficult to compare our study findings with others. We previously reported the performance of the original NLP algorithm (26), in which the results showed that sensitivity, specificity, positive predictive value, and negative predictive value were 81%, 95%, 84%, and 94%, respectively (asthma prevalence is 31% in this cohort). Although the original study showed feasibility and reasonable performance of the NLP algorithm, the original NLP algorithm for asthma ascertainment was based on a small convenience sample (n = 112) and did not assess construct validity. The typical approach for assessing performance of NLP in the medical informatics literature was determining criterion validity against chart review of EMR by abstractor (e.g., sensitivity, specificity, positive and negative predictive value, area under the curve, and F-measure). This approach, however, is used for outcomes with clear-cut laboratory definitions or imaging/biopsy definitions, such as diabetes, hypercholesterolemia, or cancer (29–31). In this present study, we examined construct validity by assessing the association of asthma status determined by NLP versus chart review of EMR by abstractor for those known risk factors for asthma. We not only significantly improved criterion validity, but we also successfully demonstrated the association of asthma status determined by our NLP algorithm with the known risk factors for asthma, such as allergic rhinitis or eczema. Some risk factors for asthma were not associated with asthma status by NLP in this study, such as cesarean section and low birth weight, which showed inconsistent association with asthma in the literature, likely caused by the weaker effect of these risk factors on asthma or measurement error of these variables rather than poor performance of NLP (44, 47, 48).
In the present validation study, we improved the way that the NLP algorithm captured and interpreted key words and sentences of PAC in a way to reduce false positives of NLP. For example, one of the main sources for false positives of NLP is a negated sentence, such as “Patient had wheezing” and “Patient denied wheezing,” which cannot be differentiated with a simple key search function, whereas NLP was designed and improved to rule out those negated sentences. Furthermore, hypothetical sentences (e.g., as part of patient instruction, such as “cold medicine in case of cough”) and nonpatient issues (e.g., sister had wheezing) have been trained using abstractors’ annotations, and thus, NLP performance was improved.
There is one noteworthy finding in our study. When we performed an error analysis and disconcordant cases between the NLP algorithm and data abstractor were adjudicated by an independent reviewer, we discovered that the NLP algorithm (computer) was correct for some false-positive cases. It suggests that data abstractors might potentially miss some important asthma-related symptom events embedded in texts of medical records. Although this error by data abstractors is likely to be nondifferential, it could be escalated if the volume of medical records to be reviewed was larger. Another advantage of NLP algorithm for asthma ascertainment is its capability of identifying the asthma index date (the date when PAC was met for the first time), which allows temporality assessment in epidemiologic studies. In the present study, among the 147 subjects with asthma ascertained by chart review of EMR by abstractor, 118 asthma index dates by NLP (80%) were within 4 weeks of those defined by chart review of EMR by abstractor, including 102 (69%) with exact same index date, supporting the utility of the NLP algorithm for determining a time component of events of interest, which is essential to determine the association between exposures and outcomes in observational studies (17–19).
There have been prior attempts at applying NLP in asthma research. Himes and coworkers (22) identified risk factors for asthma exacerbation, such as race and body mass index, using a logistic regression model, and Zeng and coworkers (23) developed an algorithm to extract asthma as a principal diagnosis from clinical notes (22, 23). Even 20 years ago, there were studies using NLP for monitoring patient care by extracting asthma-related information from discharge summaries or outpatient progress notes (24, 25). However, these studies focused on text searching capabilities of NLP (i.e., record level) but did not apply the text processing and classification components of NLP at a patient level. Therefore, our presented work is the first report for applying an NLP approach to text extraction, processing, and classification of patients for asthma status based on a predetermined criteria. This NLP approach will help clinicians and researchers address the limitations of using structured data (e.g., poor sensitivity of ICD codes in identifying asthma), labor-intensive manual chart review, self-reported data or symptoms (recall bias), and biomarkers or laboratory tests (e.g., impracticality for large-scale studies). This NLP-based approach for identifying patients with asthma will help maximize clinical benefits of NLP for asthma care in practice along with other NLP algorithms for monitoring asthma care, minimizing the chance of a delayed diagnosis of asthma (23–25, 49).
The main strength of our study is the epidemiologic advantages of our study setting and design in conducting retrospective studies, which enabled us to capture all inpatient and outpatient asthma-related events for this present study. Also, our study is a birth cohort study, which allowed us to follow subjects longitudinally since birth. Additional strengths of this study include evolution of NLP algorithm through incorporating free text data (e.g., asthma symptom). Our NLP algorithm has the capability to determine a time component of events of interest, such as index date of asthma, which are important for assessing relationships of asthma and other events or for determining temporal trends of asthma. In addition, our study highlights the potential use of NLP algorithms for ascertaining asthma and other chronic conditions, such as rheumatoid arthritis, as a clinical decision support tool (50, 51).
The main limitation was that we limited our analysis to study subjects who were born at and received medical care from Mayo Clinic for validation purposes. Future studies need to address whether our NLP algorithm for PAC can be applied to different EMR systems with similar validity, which our group is currently working on. Another limitation of this study is a potential misclassification of asthma status by PAC, because the presence of prebronchodilator airflow obstruction or airway hyperresponsiveness is not required to be considered asthmatic by PAC (which are not routinely performed in asthma care among children) (52). When we examined the criterion validity among children with and without lung function tests, it did not significantly affect the results (98% with lung function tests vs. 95% without). Although we acknowledge the potential misclassification of asthma because of the lack of lung function tests in our study, NLP algorithm for PAC may still be more suitable for large-scale clinical research for asthma than commonly used methods for asthma ascertainment in the current literature, such as self-report and ICD codes. Despite the small number of subjects, currently, the identification of children meeting the exclusion criteria in Table 1 relies solely on abstractors. Developing an NLP algorithm for exclusion criteria might be necessary for consistency in the future. Finally, our study results need to be replicated in a different study setting to ascertain the accuracy of the NLP algorithm. After completing development, validation, and optimization, we intend to explore various options to share and disseminate this algorithm with the research community.
In conclusion, the NLP approach for asthma ascertainment is a useful tool for asthma research and care in the era of the EMR and big data because it enables large-scale clinical studies and population management. It significantly improves cost and time efficiency in identifying children with asthma symptoms, while also improving methodologic heterogeneity in asthma ascertainment.
Acknowledgments
Acknowledgment
The authors thank the original natural language processing project staff and Mrs. Kelly Okeson for her administrative assistance.
Footnotes
Supported by National Institutes of Health grants R01 HL126667 and R21AI116839-01 and T. Denny Sanford Pediatric Collaborative Research Fund. This work was made possible by Rochester Epidemiology project R01-AG34676 from the National Institute on Aging and Clinical and Translational Science Award grant UL1 TR000135 from the National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of National Institutes of Health.
Author Contributions: Y.J.J. and H.L. had full access to all of the data in the study, take responsibility for the integrity of the data and the accuracy of the data analysis, and had authority over manuscript preparation and the decision to submit the manuscript for publication. Study concept and design, C.-I.W., S.S., H.L., E.R., M.A.P., H.K., I.T.C., and Y.J.J. Acquisition, analysis, or interpretation of data, C.-I.W., S.S., M.C.R., A.S., H.L., E.R., G.V., M.A.P., and Y.J.J. Drafting of the manuscript, C.-I.W., S.S., and Y.J.J. Critical revision of the manuscript for important intellectual content, C.-I.W., S.S., M.C.R., A.S., E.R., G.V., K.A.B., M.A.P., H.K., I.T.C., H.L., and Y.J.J. Statistical analysis, C.-I.W., S.S., and E.R. Study supervision, C.-I.W., S.S., M.C.R., A.S., E.R., G.V., K.A.B., M.A.P., H.K., I.T.C., H.L., and Y.J.J.
Originally Published in Press as DOI: 10.1164/rccm.201610-2006OC on April 4, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Azad MB, Coneys JG, Kozyrskyj AL, Field CJ, Ramsey CD, Becker AB, Friesen C, Abou-Setta AM, Zarychanski R. Probiotic supplementation during pregnancy or infancy for the prevention of asthma and wheeze: systematic review and meta-analysis. BMJ. 2013;347:f6471. doi: 10.1136/bmj.f6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention (CDC) Vital signs: asthma prevalence, disease characteristics, and self-management education: United States, 2001--2009. MMWR Morb Mortal Wkly Rep. 2011;60:547–552. [PubMed] [Google Scholar]
- 3.Lethbridge-Cejku M, Vickerie J. Summary of health statistics for U.S. adults: national health interview survey, 2003. Vital Health Stat 10. 2005;(225):1–161. [PubMed] [Google Scholar]
- 4.Stanton MW.The high concentration of U.S. health care expenditures. Research in Action 2006 (Issue 19) [accessed 2017 Jul 17]. Available from: https://meps.ahrq.gov/data_files/publications/ra19/ra19.pdf
- 5.Schiller JS, Lucas JW, Peregoy JA. Summary health statistics for U.S. adults: national health interview survey, 2011. Vital Health Stat 10. 2012;(256):1–218. [PubMed] [Google Scholar]
- 6.Asher MI, Montefort S, Björkstén B, Lai CK, Strachan DP, Weiland SK, Williams H ISAAC Phase Three Study Group. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood: ISAAC Phases One and Three repeat multicountry cross-sectional surveys. Lancet. 2006;368:733–743. doi: 10.1016/S0140-6736(06)69283-0. [DOI] [PubMed] [Google Scholar]
- 7.Murray CJ, Atkinson C, Bhalla K, Birbeck G, Burstein R, Chou D, Dellavalle R, Danaei G, Ezzati M, Fahimi A, et al. U.S. Burden of Disease Collaborators The state of US health, 1990-2010: burden of diseases, injuries, and risk factors JAMA 2013310591–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Howard TD, Zheng SL, Haselkorn T, Peters SP, Meyers DA, Bleecker ER. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol. 2010;125:328–335. doi: 10.1016/j.jaci.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferreira MA, Matheson MC, Duffy DL, Marks GB, Hui J, Le Souëf P, Danoy P, Baltic S, Nyholt DR, Jenkins M, et al. Australian Asthma Genetics Consortium. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet. 2011;378:1006–1014. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyers DA. Genetics of asthma and allergy: what have we learned? J Allergy Clin Immunol. 2010;126:439–446, quiz 447–448. doi: 10.1016/j.jaci.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ducharme FM, Lemire C, Noya FJD, Davis GM, Alos N, Leblond H, Savdie C, Collet J-P, Khomenko L, Rivard G, et al. Preemptive use of high-dose fluticasone for virus-induced wheezing in young children. N Engl J Med. 2009;360:339–353. doi: 10.1056/NEJMoa0808907. [DOI] [PubMed] [Google Scholar]
- 12.Panickar J, Lakhanpaul M, Lambert PC, Kenia P, Stephenson T, Smyth A, Grigg J. Oral prednisolone for preschool children with acute virus-induced wheezing. N Engl J Med. 2009;360:329–338. doi: 10.1056/NEJMoa0804897. [DOI] [PubMed] [Google Scholar]
- 13.Haldar P, Pavord ID, Shaw DE, Berry MA, Thomas M, Brightling CE, Wardlaw AJ, Green RH. Cluster analysis and clinical asthma phenotypes. Am J Respir Crit Care Med. 2008;178:218–224. doi: 10.1164/rccm.200711-1754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore WC, Meyers DA, Wenzel SE, Teague WG, Li H, Li X, D’Agostino R, Jr, Castro M, Curran-Everett D, Fitzpatrick AM, et al. National Heart, Lung, and Blood Institute’s Severe Asthma Research Program. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med. 2010;181:315–323. doi: 10.1164/rccm.200906-0896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzpatrick AM, Teague WG, Meyers DA, Peters SP, Li X, Li H, Wenzel SE, Aujla S, Castro M, Bacharier LB, et al. National Institutes of Health/National Heart, Lung, and Blood Institute Severe Asthma Research Program. Heterogeneity of severe asthma in childhood: confirmation by cluster analysis of children in the National Institutes of Health/National Heart, Lung, and Blood Institute severe asthma research program. J Allergy Clin Immunol. 2011;127:382–389. doi: 10.1016/j.jaci.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lazic N, Roberts G, Custovic A, Belgrave D, Bishop CM, Winn J, Curtin JA, Hasan Arshad S, Simpson A. Multiple atopy phenotypes and their associations with asthma: similar findings from two birth cohorts. Allergy. 2013;68:764–770. doi: 10.1111/all.12134. [DOI] [PubMed] [Google Scholar]
- 17.Juhn YJ, Kita H, Yawn BP, Boyce TG, Yoo KH, McGree ME, Weaver AL, Wollan P, Jacobson RM. Increased risk of serious pneumococcal disease in patients with asthma. J Allergy Clin Immunol. 2008;122:719–723. doi: 10.1016/j.jaci.2008.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Capili CR, Hettinger A, Rigelman-Hedberg N, Fink L, Boyce T, Lahr B, Juhn YJ. Increased risk of pertussis in patients with asthma. J Allergy Clin Immunol. 2012;129:957–963. doi: 10.1016/j.jaci.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bjur KA, Lynch RL, Fenta YA, Yoo KH, Jacobson RM, Li X, Juhn YJ. Assessment of the association between atopic conditions and tympanostomy tube placement in children. Allergy Asthma Proc. 2012;33:289–296. doi: 10.2500/aap.2012.33.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wi CI, Park MA, Juhn YJ. Development and initial testing of Asthma Predictive Index for a retrospective study: an exploratory study. J Asthma. 2015;52:183–190. doi: 10.3109/02770903.2014.952438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wi CI, Kim BS, Mehra S, Yawn BP, Park MA, Juhn YJ. Risk of herpes zoster in children with asthma. Allergy Asthma Proc. 2015;36:372–378. doi: 10.2500/aap.2015.36.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Himes BE, Kohane IS, Ramoni MF, Weiss ST. Characterization of patients who suffer asthma exacerbations using data extracted from electronic medical records. AMIA Annu Symp Proc. 2008:308–312. [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng QT, Goryachev S, Weiss S, Sordo M, Murphy SN, Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak. 2006;6:30. doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sager N, Lyman M, Bucknall C, Nhan N, Tick LJ. Natural language processing and the representation of clinical data. J Am Med Inform Assoc. 1994;1:142–160. doi: 10.1136/jamia.1994.95236145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ertle AR, Campbell EM, Hersh WR. Automated application of clinical practice guidelines for asthma management. Proc AMIA Annu Fall Symp. 1996:552–556. [PMC free article] [PubMed] [Google Scholar]
- 26.Wu ST, Sohn S, Ravikumar KE, Wagholikar K, Jonnalagadda SR, Liu H, Juhn YJ. Automated chart review for asthma cohort identification using natural language processing: an exploratory study. Ann Allergy Asthma Immunol. 2013;111:364–369. doi: 10.1016/j.anai.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu ST, Juhn YJ, Sohn S, Liu H. Patient-level temporal aggregation for text-based asthma status ascertainment. J Am Med Inform Assoc. 2014;21:876–884. doi: 10.1136/amiajnl-2013-002463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ford E, Carroll J, Smith H, Davies K, Koeling R, Petersen I, Rait G, Cassell J. What evidence is there for a delay in diagnostic coding of RA in UK general practice records? An observational study of free text. BMJ Open. 2016;6:e010393. doi: 10.1136/bmjopen-2015-010393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Safarova MS, Liu H, Kullo IJ. Rapid identification of familial hypercholesterolemia from electronic health records: The SEARCH study. J Clin Lipidol. 2016;10:1230–1239. doi: 10.1016/j.jacl.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng L, Wang Y, Hao S, Shin AY, Jin B, Ngo AD, Jackson-Browne MS, Feller DJ, Fu T, Zhang K, et al. Web-based real-time case finding for the population health management of patients with diabetes mellitus: a prospective validation of the natural language processing-based algorithm with statewide electronic medical records. JMIR Med Inform. 2016;4:e37. doi: 10.2196/medinform.6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eide MJ, Tuthill JM, Krajenta RJ, Jacobsen GR, Levine M, Johnson CC. Validation of claims data algorithms to identify nonmelanoma skin cancer. J Invest Dermatol. 2012;132:2005–2009. doi: 10.1038/jid.2012.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wi CI, Sohn S, Ryu E, Liu H, Park MA, Juhn YJ. Automated chart review for asthma ascertainment: an innovative approach for asthma care and research in the era of electronic medical record [abstract] J Allergy Clin Immunol. 2016;137(Suppl 2):AB196. [Google Scholar]
- 33.Yawn BP, Yawn RA, Geier GR, Xia Z, Jacobsen SJ. The impact of requiring patient authorization for use of data in medical records research. J Fam Pract. 1998;47:361–365. [PubMed] [Google Scholar]
- 34.Voge GA, Carey WA, Ryu E, King KS, Wi CI, Juhn YJ. What accounts for the association between late preterm births and risk of asthma? Allergy Asthma Proc. 2017;38:152–156. doi: 10.2500/aap.2017.38.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yunginger JW, Reed CE, O’Connell EJ, Melton LJ, III, O’Fallon WM, Silverstein MD. A community-based study of the epidemiology of asthma. Incidence rates, 1964-1983. Am Rev Respir Dis. 1992;146:888–894. doi: 10.1164/ajrccm/146.4.888. [DOI] [PubMed] [Google Scholar]
- 36.Juhn YJ, Kita H, Lee LA, Swanson RJ, Smith R, Bagniewski SM, Weaver AL, Pankratz VS, Jacobson RM, Poland GA. Childhood asthma and measles vaccine response. Ann Allergy Asthma Immunol. 2006;97:469–476. doi: 10.1016/S1081-1206(10)60937-4. [DOI] [PubMed] [Google Scholar]
- 37.Beard CM, Yunginger JW, Reed CE, O’Connell EJ, Silverstein MD. Interobserver variability in medical record review: an epidemiological study of asthma. J Clin Epidemiol. 1992;45:1013–1020. doi: 10.1016/0895-4356(92)90117-6. [DOI] [PubMed] [Google Scholar]
- 38.Hunt LW, Jr, Silverstein MD, Reed CE, O’Connell EJ, O’Fallon WM, Yunginger JW. Accuracy of the death certificate in a population-based study of asthmatic patients. JAMA. 1993;269:1947–1952. [PubMed] [Google Scholar]
- 39.Silverstein MD, Reed CE, O’Connell EJ, Melton LJ, III, O’Fallon WM, Yunginger JW. Long-term survival of a cohort of community residents with asthma. N Engl J Med. 1994;331:1537–1541. doi: 10.1056/NEJM199412083312301. [DOI] [PubMed] [Google Scholar]
- 40.Bauer BA, Reed CE, Yunginger JW, Wollan PC, Silverstein MD. Incidence and outcomes of asthma in the elderly. A population-based study in Rochester, Minnesota. Chest. 1997;111:303–310. doi: 10.1378/chest.111.2.303. [DOI] [PubMed] [Google Scholar]
- 41.Silverstein MD, Yunginger JW, Reed CE, Petterson T, Zimmerman D, Li JT, O’Fallon WM. Attained adult height after childhood asthma: effect of glucocorticoid therapy. J Allergy Clin Immunol. 1997;99:466–474. doi: 10.1016/s0091-6749(97)70072-1. [DOI] [PubMed] [Google Scholar]
- 42.Juhn YJ, Qin R, Urm S, Katusic S, Vargas-Chanes D. The influence of neighborhood environment on the incidence of childhood asthma: a propensity score approach. J Allergy Clin Immunol. 2010;125:838–843. doi: 10.1016/j.jaci.2009.12.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Juhn YJ, Sauver JS, Katusic S, Vargas D, Weaver A, Yunginger J. The influence of neighborhood environment on the incidence of childhood asthma: a multilevel approach. Soc Sci Med. 2005;60:2453–2464. doi: 10.1016/j.socscimed.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 44.Juhn YJ, Weaver A, Katusic S, Yunginger J. Mode of delivery at birth and development of asthma: a population-based cohort study. J Allergy Clin Immunol. 2005;116:510–516. doi: 10.1016/j.jaci.2005.05.043. [DOI] [PubMed] [Google Scholar]
- 45.Yawn BP, Yunginger JW, Wollan PC, Reed CE, Silverstein MD, Harris AG. Allergic rhinitis in Rochester, Minnesota residents with asthma: frequency and impact on health care charges. J Allergy Clin Immunol. 1999;103:54–59. doi: 10.1016/s0091-6749(99)70525-7. [DOI] [PubMed] [Google Scholar]
- 46.Liu H, Bielinski S, Sohn S, Murphy S, Wagholikar K, Jonnalagadda S, Ravikumar KE, Wu S, Kullo I, Chute C. An information extraction framework for cohort identification using electronic health records. AMIA Summits Transl Sci Proc. 2013;2013:149–153. [PMC free article] [PubMed] [Google Scholar]
- 47.Bager P, Wohlfahrt J, Westergaard T. Caesarean delivery and risk of atopy and allergic disease: meta-analyses. Clin Exp Allergy. 2008;38:634–642. doi: 10.1111/j.1365-2222.2008.02939.x. [DOI] [PubMed] [Google Scholar]
- 48.Salam MT, Margolis HG, McConnell R, McGregor JA, Avol EL, Gilliland FD. Mode of delivery is associated with asthma and allergy occurrences in children. Ann Epidemiol. 2006;16:341–346. doi: 10.1016/j.annepidem.2005.06.054. [DOI] [PubMed] [Google Scholar]
- 49.Childhood Asthma Management Program Research Group. The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Control Clin Trials. 1999;20:91–120. [PubMed] [Google Scholar]
- 50.Liao KP, Cai T, Gainer V, Goryachev S, Zeng-treitler Q, Raychaudhuri S, Szolovits P, Churchill S, Murphy S, Kohane I, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res (Hoboken) 2010;62:1120–1127. doi: 10.1002/acr.20184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO, III, Birnbaum NS, Burmester GR, Bykerk VP, Cohen MD, et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010;62:2569–2581. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
- 52.Yawn BP, Rank MA, Cabana MD, Wollan PC, Juhn YJ. Adherence to asthma guidelines in children, tweens, and adults in primary care settings: a practice-based network assessment. Mayo Clin Proc. 2016;91:411–421. doi: 10.1016/j.mayocp.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]