Abstract
Different cortical areas are organized into distinct intra-cortical subnetworks. How descending pathways from the entire cortex interact subcortically as a network remains unclear. Here, we report an open-access comprehensive mesoscale cortico-striatal projectome—a detailed connectivity projection map from the entire cerebral cortex to the dorsal striatum or caudoputamen (CP) in rodents. Based on these projections, we use novel computational neuroanatomical tools to identify 29 distinct functional striatal domains. Further, we characterize different cortico-striatal networks and how they reconfigure across the rostral-caudal extent of the CP. The workflow was also applied to select cortico-striatal connections in two different mouse models of disconnection syndromes to demonstrate its utility in characterizing circuitry-specific connectopathies. Together, this work provides the structural basis for studying the functional diversity of the dorsal striatum and disruptions of cortico-basal ganglia networks across a broad range of disorders.
INTRODUCTION
Given its role in higher order functions like cognition and emotion, the cerebral cortex has been a main focus of neuroscience in the last century. Current advancements in microscopy and computational technologies have made it feasible to produce, collect, and analyze vast amounts of connectivity data to assemble neural networks of the neocortex in mice1,2 and rats3, which inform research about primate cortical networks4. Each cortical area sends descending projections that innervate subcortical structures through hierarchically organized, serially-ordered, multi-synaptic neural pathways. However, how descending pathways from cortex interact subcortically as a network largely remains unknown.
Projections from the cortex to dorsal striatum, or cortico-striatal projection pathways, serve as the initial input to the basal ganglia and therefore determine how cortical information is transposed, integrated, and processed within the basal ganglia to affect a range of sensorimotor, cognitive, and emotional functions5–10.Studies have demonstrated topographic cortico-striatal projection pathways in primates, cats, and rats5,11–17, 16–18,20–27 and to a lesser extent, in mice2,18.28 Current theory states that these pathways organize into a few functionally segregated, parallel cortico-basal ganglia-thalamo-cortical loops that convey sensorimotor, associative, or limbic information5,6,19,20. 5,6,30–33 This model has been influential in interpreting pathophysiology of disorders like Huntington’s and Parkinson’s diseases and neuropsychiatric diseases21–23.. 14,34,37–39 However, cortico-striatal connections suggest a more granular segregation of these parallel circuits, which could explain the variegated behaviors and symptoms associated with normal functions and dysfunctions of cortico-basal ganglia circuits21.
A hurdle to understanding the detailed organization of cortico-striatal projections at the network level and their role in disconnection syndromes like Huntington’s disease (HD) has been the inability to delineate striatal subdivisions. In rodent brain atlases, the entire dorsal striatum, or the caudoputamen (CP), remains one of the largest undivided structures24–26 predominantly because it does not lend itself well to subdivision by cyto- or chemoarchitectonic means. Fortunately, topographic projections from the cortex to the CP provide opportunity for identifying striatal subdivisions, as shown previously in rats13,16, and recently in mice2.
Extending upon this, we first created a comprehensive cortico-striatal projectome—a map of cortico-striatal projection pathways arising from virtually the entire mouse cerebral cortex. Computational neuroanatomic tools were developed to quantify each projection and graph theory identified 11 CP communities composed of 29 smaller domains based on their cortical afference. This information demonstrates how cortical information is integrated or segregated within the CP, which is important to understand the functional segregation of different cortico-basal ganglia circuits. All injection cases as well as the cortico-striatal projection map, are presented in a public-facing image database (http://www.mouseconnectome.org/iConnectome/page/search.jsp for injections; http://www.mouseconnectome.org/CorticalMap/page/menu.htm for projection map). Further, we characterized cortico-striatal networks based on how axonal projections from different cortical subnetworks converge or diverge within the CP. Finally, select cortico-striatal disconnections in two different mouse models of disconnection syndromes [zQ175 for HD and monoamine oxidase (MAO) A/B knockout for aggression and autism spectrum disorders] were identified and localized to unique CP domains. This demonstrates the utility of a parcellated CP in potentially explaining manifestations of broad cortico-basal ganglia disorder symptoms and provides insight into circuit-specific characterization of cortico-striatal dysfunctions in models of various diseases.
RESULTS
Constructing the cortico-striatal projectome
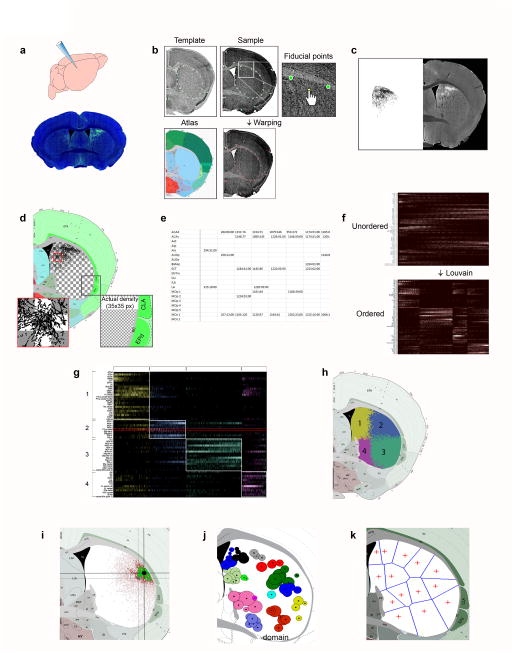
We employed a computational neuroanatomic approach to create a comprehensive mesoscopic mouse cortico-striatal projectome. This involved the production, collection, quantification, and analysis of ~150 anterograde tracer-labeled cortico-striatal pathways acquired from injections placed across the entire neocortex (Supplementary Fig. 1a), entorhinal and piriform areas and lateral and basolateral amygdalar nuclei. Small PHAL (Phaseolus vulgaris leucoagglutinin) anterograde tracer infusions confined to a single delineated structure were delivered iontophoretically to produce regional specific projection terminal patterns in the CP (Fig. 1a and Supplementary Fig. 1a). Double or triple anterograde tracer injections of PHAL, and adeno-associated viruses expressing green or red fluorescent protein (AAV-GFP, AAV-RFP), were also made to reveal direct spatial correlations of axonal terminals arising from different cortical areas (i.e., topography or interdigitation) (Supplementary Fig. 2a). Multiple retrograde tracer injections were made to validate the anterograde tracing data (Supplementary Fig. 1b). An informatics workflow was developed to reliably annotate, quantify, and graphically reconstruct 62 labeled pathways most representative of all cortical areas examined (Fig. 1b–e; see Online Methods for details regarding each step). First, each image to be analyzed was warped to its corresponding Allen Reference Atlas (ARA24) template (Fig 1b). Next, all cortico-striatal projection pathways were graphically reconstructed and plotted onto their corresponding levels of the ARA (Fig. 1c). Compilation of the segmented images was used to create a connectivity map (i.e., projectome), within which all pathways can be directly compared within a common neuroanatomic frame.
Fig. 1. Data production and informatics workflow.
Discrete anterograde tracer injections were made (a) and photomicrographs of the Nissl and tracer-labeled pathways were acquired. (b) Images were imported into an in-house software for warping. Fiducial markers were placed on the Nissl image of the sample to be warped, such that they matched the marker pattern of the corresponding atlas Nissl image. The sample was then deformably warped. (c) The warped PHAL channel was segmented and binarized, yielding a monochromatic image of axonal labeling. (d) The segmented image was superimposed onto its corresponding atlas level, within which the CP was divided into grid cells. Superimposed axons within each grid cell were quantified. (e) The quantified labeling for each injection was compiled into a spreadsheet and (f, top) the data expressed in a matrix with cortical areas along the y-axis and cells of the grid along the x-axis. (f, bottom) A community detection algorithm was utilized to group cortical areas projecting to a common set of cells. The matrix was reordered and color-coded (g) according to community structure. Cells boxed in red depict quantification of segmented image in c. (h) The grid cells were recolored to visualize, within the CP, the spatial arrangement of the communities to which they belong. (i) Communities were subdivided into domains using the centroid, or center of gravity, of each terminal field. The original labeling is shown in red and labeling after 95% cutoff, which was used for centroid generation, is shown in green. (j) Axonal fields within a community whose centroids were closely spaced defined a domain. (k) Using intensity weighted centroids as seed points, the CP was parcellated into Voronoi cells demonstrating the relative domain boundaries.
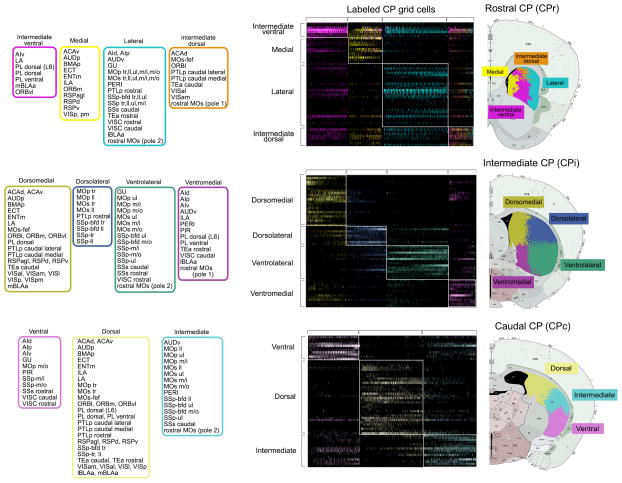
Second, to overcome the challenge of heterogeneous distribution patterns of cortico-striatal projections (Supplementary Fig. 1a), a grid-based approach was used to quantify the 62 pathways and index connectivity strength with anatomic specificity. The CP was divided into a grid of 35x35 pixel cells with a spatial dimension of 22.5 μm2 (~0.6 μm per pixel) (Fig. 1d). An overlap indexing process superimposed reconstructions onto the grid and computed an overlap value at each cell. These values were tabulated into a spreadsheet, which is graphically represented as a weighted matrix with cortical areas along the y-axis and CP grid cells along the x-axis (Fig. 1e–f). An implementation of the Louvain community detection algorithm27 was applied to cluster cortical injection sites with common CP termination fields across the rostral, intermediate, and caudal CP (Figs. 1f,g and 2). The Louvain algorithm employs a randomized, greedy optimization, which potentially reveals different community structures across multiple runs. To mitigate this, we ran the algorithm 1000 times and reported the community structure that emerged most often, or the community structure mode. For visualization, matrices were reordered and color coded according to community structure with communities of connections grouped close to the diagonal (Figs. 1g and 2). An accompanying color-coded CP illustrates the anatomic locations of the communities (Figs. 1h and 2).
Fig. 2. Visualization of CP communities and domains.
Matrices that visualize the CP communities across the CPr (top), CPi (middle), and CPc (bottom). Matrices show cortical injections along the y-axis and labeled CP cells along the x-axis. For visualization, matrices were reordered and color coded according to community structure. Corresponding color-coded CP illustrates the spatial arrangement of communities within dorsal striatum. Community color assignments across the CP were coordinated to reflect general projection trends of communities across the structure. For example, projections terminating in the dorsomedial community of the CPi (represented in mustard yellow) generally terminate in the medial community of the CPr (bright yellow) and the dorsal community of the CPc (pastel yellow). Constituents of each community are listed to the left in corresponding color-coded boxes. Cortical area abbreviations: ACAd, Anterior cingulate area, dorsal; ACAv, Anterior cingulate area, ventral; AId, Agranular insular area, dorsal; AIp, Agranular insular area, posterior; AIv, Agranular insular area, ventral; AUDd, Auditory area, dorsal; AUDp, Primary auditory area; AUDv, Primary auditory area, ventral; ECT, Ectorhinal area; ENTl, Entorhinal area, lateral; ENTm, Entorhinal area, medial; GU, Gustatory area; ILA, Infralimbic area; MOp, Primary motor area; MOp tr, MOp trunk; MOp ll, MOp lower limb; MOp ul, MOp upper limb; MOp m/i, MOp inner mouth; MOp m/o, MOp outer mouth; MOs, Secondary motor area; MOs tr, MOs trunk; MOs ll, MOs lower limb; MOs ul, MOs upper limb; MOs m/i, MOs inner mouth; MOs m/o, MOs outer mouth; MOs-fef, MOs frontal eye field; ORBl, Orbitofrontal area, lateral; ORBm, Orbitofrontal area, medial; ORBvl, Orbitofrontal area, ventrolateral; PERI, Perirhinal area; PIR, Piriform cortex; PL, Prelimbic area; PL dorsal (L6), Layer 6 of dorsal PL; PTLp, Posterior parietal association area; Rostral MOs (P1), rostral MOs pole 1; Rostral MOs (P2), rostral MOs pole 2; RSPagl, Retrosplenial area, agranular; RSPd, Retrosplenial area, dorsal; RSPv, Retrosplenial area, ventral; SUBd, Subiculum, dorsal; SSp, Primary somatosensory area; SSp-tr, SSp trunk; SSp-ll, SSp lower limb; SSp-ul, SSp upper limb; SSp-m/i, SSp inner mouth; SSp-m/o, SSp outer mouth; SSp-bfd, SSp barrel field; SSp-bfd tr, SSp-bfd trunk; SSp-bfd ll, SSp-bfd lower limb; SSp-bfd ul, SSp-bfd upper limb; SSp-bfd m/o, SSp-bfd outer mouth; TEa, Temporal association area; VISal, Visual area, anterolateral; VISam, Visual area, anteromedial; VISl, Visual area, lateral; VISp, Primary visual area; VISpl, Visual area, posterior lateral; VISpm, Visual area, posterior medial; VISC, Visceral area.
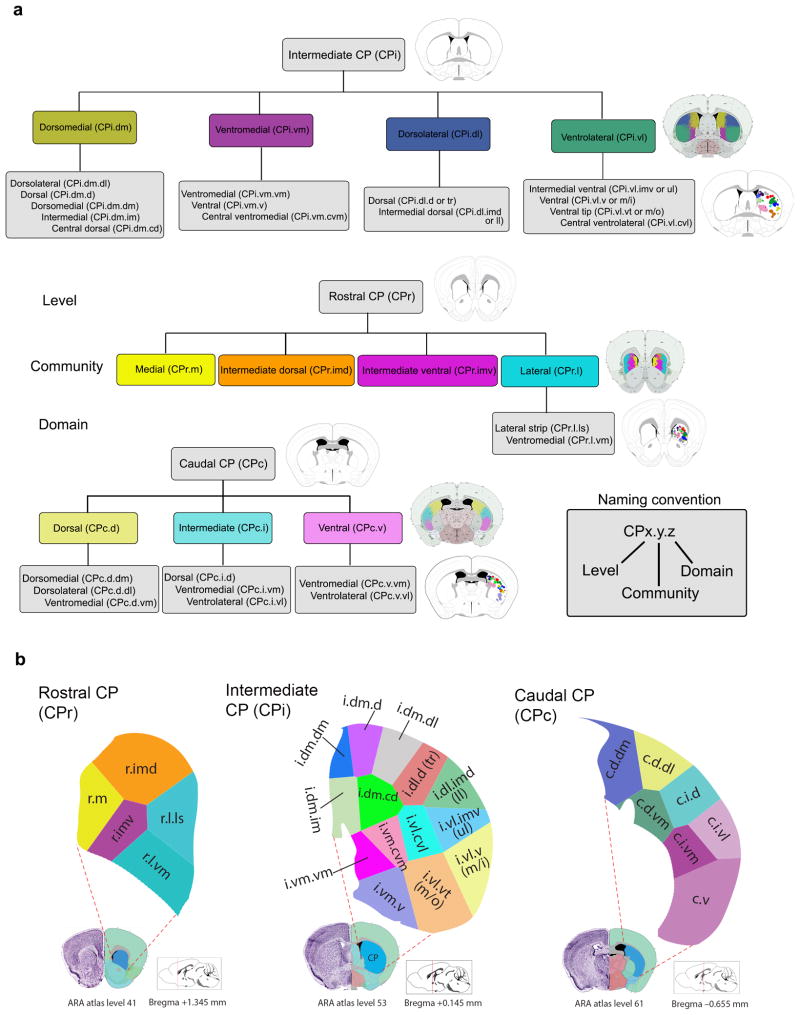
Subsequent analyses extracted smaller clusters of distinct projection fields, or domains, within the communities. Smaller domains were difficult to detect by the Louvain since it is not optimal for recognizing communities with members of unequal sizes. Accordingly, a measure of centrality for the CP projection fields, or the centroid, was used to identify the domains (Figs. 1i and 3a). Axonal fields within a community with closely spaced centroids were considered a domain of that community (Fig. 1j). By weighting the centroids by intensity to compute a set of Voronoi seed points, the CP was parcellated into Voronoi cells (Fig. 1k)44, which are recognized as functional domains based on their cortical input (Fig. 3b). Importantly, the organization of these Voronoi domains agreed closely with the raw data and thalamic injections provided confirmation of the domains.
Fig. 3. Summary of community and domain nomenclature and CP domain parcellations.
(a) Summary of communities and domains for the rostral, intermediate, and caudal CP are presented in a hierarchical tree. The nomenclature follows the CPx.y.z convention where x denotes the level (rostral, intermediate, or caudal), y the community, and z the domain. (b) Overview of connectivity-based CP domains defined with Voronoi parcellation. A total of 29 functional domains were identified in the rostral (CPr, left), intermediate (CPi, middle), and caudal (CPc, right) levels of the CP. Three domains that were identified based primarily from manual inspection of the data are not shown: the CPc.vl and CPc.v.vm, which are part of the CPc.v and the CPc.ext located at the tail end of the CP.
The density of total CP labeling from each cortical source was also quantified by computing complementary measures of intensity and span (Supplementary Fig. 3). While intensity measures the total pixel count of a terminal field (i.e., concentration of labeling), span is a measure of how many cells of the grid the labeling occupies (i.e., diffuseness of labeling). As such, intensity could be indicative of how strong a cortical area’s effect is within a specific region of the CP, while span indicates the breadth of its effect across the CP. These values were utilized to compute the Labeling Density Index (LDI), summarizing in a single value the labeling properties originating from an injection site (http://www.mouseconnectome.org/MCP/page/documents/tables?paperId=18 for tables containing these values).
Additionally, cortical projections to neighboring CP domains show varying degrees of overlap, which obscures hard boundary demarcation and suggests interaction across domains. Overlap values capturing this degree of CP convergence among individual cortical sources and domains were calculated and are available online (http://www.mouseconnectome.org/MCP/page/documents/tables?paperId=18).
Cortico-striatal projections to the intermediate CP (CPi)
Four communities were detected at the intermediate level of the CP (CPi): the dorsolateral (CPi.dl), ventrolateral (CPi.vl), dorsomedial (CPi.dm), and ventromedial (CPi.vm). Visualizations of this output show the four quadrant arrangement of the communities and their relative boundaries (Fig. 2). These communities were further subdivided into 14 distinct domains (Fig. 3b). The hierarchical organization of communities and domains, and all their cortical constituents are provided (Fig. 3a and Supplementary Fig. 4). Abbreviations of all cortical areas used throughout the report are available in the legend for Figure 2.
The somatic sensorimotor cortical-lateral striatal networks
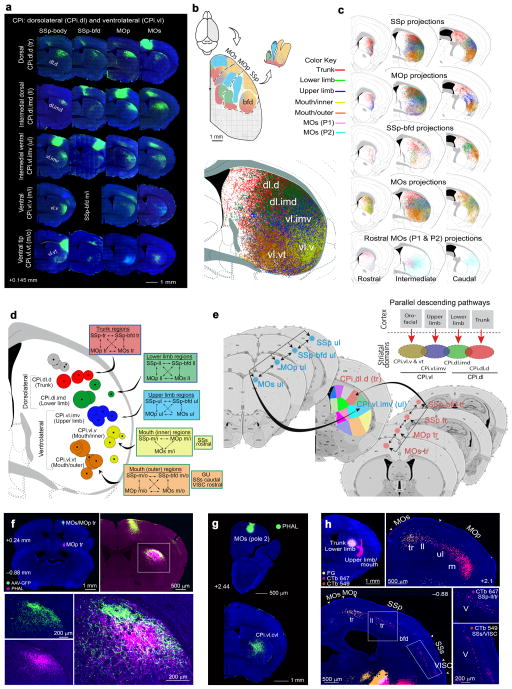
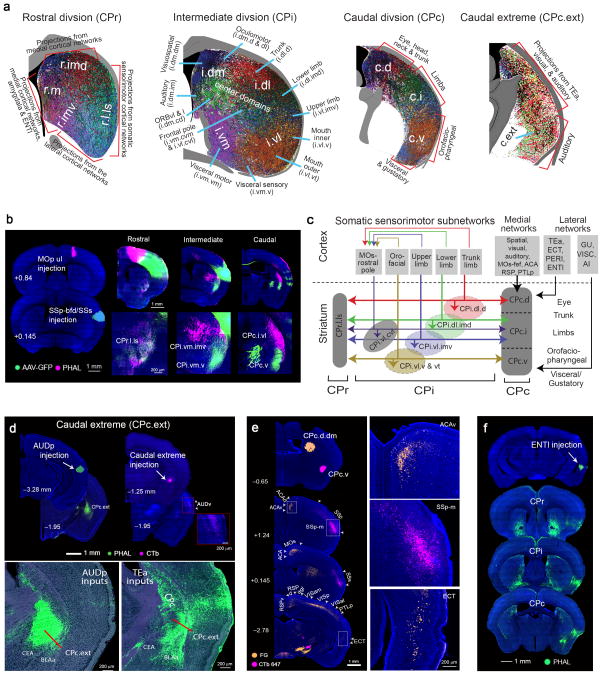
The CPi.dl and CPi.vl receive inputs from all somatic sensorimotor cortical areas (Fig. 4a–c; for injection sites and validation of projection specificities, see Supplementary Fig. 5a–d). At the cortical level, all of these areas organize into five distinct subnetworks that process trunk, lower limb, upper limb, and orofaciopharyngeal (mouth and nose) information1. Each subnetwork is comprised of domains within the primary somatosensory (SSp-body subfields), primary motor (MOp), and secondary motor (MOs) cortical areas that represent a specific body feature. Each node within the same somatic sensorimotor subnetwork is reciprocally interconnected1 (Supplementary Fig. 5e) and each sends projections that converge onto the same CP domain. Each sensorimotor subnetwork generates parallel descending projections that somatotopically innervate CPi lateral domains corresponding to their body region and subsequently form five parallel somatic sensorimotor cortical-lateral striatal networks (Figs. 4d,e and 7a).
Fig. 4. Somatotopic map of cortico-striatal projections to the CPi.
(a) Raw data showing somatic sensorimotor areas topographically projecting to different domains within the CPi.dl and CPi.vl communities. Each sensorimotor cortical area representing a unique body region sends converging input to their corresponding CP domain. (b) Dorsal view of the somatic sensorimotor cortical map color coded according to their domains (modified from 1) and maximum projection of their CP termination fields (bottom) showing distinct, partially overlapping domains. (c) Map depicting connections from body specific regions within SSp (row 1), MOp (row 2), SSp-bfd (row 3), and MOs (row 4) to CP, illustrating dorsal-ventral representation of trunk, lower limb, upper limb, inner mouth, and outer mouth observed in a. The map also demonstrates general trend of projections across the rostrocaudal CP. (d) Centroids of cortical areas grouped into CPi.dl and CPi.vl communities. Centroids of body specific SSp, SSp-bfd, MOp, and MOs topographically arrange and subdivide communities into domains. Importantly, centroids align with the raw data in a and reconstructions in c. PTLp rostral was grouped into the CPi.dl community. However, according to its centroid (denoted with *) and raw data (Supplementary Fig. 6d), it more appropriately grouped with structures in a dorsomedial domain. Cortical areas projecting to each domain are listed in boxes color coordinated with centroids (box stroke color denotes community). (e) Illustration of 5 parallel, relatively segregated somatic sensorimotor striatal networks. All SS and MO nodes within each subnetwork dedicated to a specific body region reciprocally connect (Supplementary Fig. 5e and 1) and send converging projections to CPi domain representing their corresponding body region. (f) Interdigitation of projection terminals within CPi trunk domain (CPi.dl.d) demonstrated by double anterograde injections MOs/MOp trunk regions. (g) Projections from rostral MOs (pole 2) arborize in the central CP, avoiding the periphery. (h) FG, CTb 647 and 549 injected in trunk, lower limb, and upper limb/mouth CP domains retrogradely label layer V CP-projecting cortical neurons in their corresponding SSp and MOp body subregions as well as SSs and VISC.
Fig. 7. Network reconfiguration across the CP.
(a) A hypothetical functional model of the CP domains based on all cortical afference to specific domains identified across the CP. (b) Double anterograde injections of AAV-GFP in SSp-bfd mouth/SSs and PHAL in MOp ul show segregation of somatic sensorimotor projection terminals in their corresponding domains in the CPi and CPc and intermixing of their axons in CPr, demonstrating reconfiguration of these subnetworks across the CP. (c) Schematic representation of subnetwork reorganization across the CP. Although relatively segregated in CPi, all sensorimotor subnetworks representing unique body information send converging inputs to a single domain in the CPr. At the CPc, inputs from the somatic sensorimotor nodes terminate in the same community as inputs from the medial (dorsal CPc) and lateral (ventral CPc) subnetworks providing greater opportunity for interaction across different subnetworks. (d) AUDp projections produce a unique dense terminal field in caudal parts of the CP, where projections from most cortical areas is absent (top panel, left). AUD projections to this domain, termed the caudal extreme (CPc.ext), were validated with a CTb injection, which retrogradely labeled neurons in AUDv (top panel, right). Bottom panels are magnified images of boxed region showing dense topographic projections from AUDp and TEa in the CPc.ext. CEA, central amygdalar nucleus; BLAa, basolateral amygdalar nucleus, anterior part. (e) FG and CTb 647 tracer injections in the dorsal and ventral CPc substantiate the cortical projections from ACA, MOs-fef, RSP, VIS, and ECT to CPc.d and from SSp-m to CPc.v. Right panels show magnified images of labeling. (f) Projections from ENTl to CPr, CPi, and CPc. Note the broad projections spanning across all CPi.dm and CPi.vm domains in CPi.
Input from body region-specific areas of the SSp-body subfields subdivide the CPi.dl and CPi.vl into five smaller domains, which reveal a clear somatotopic map of the mouse body. This map has a dorsal to ventral orientation, with the dorsal domain (CPi.dl.d) receiving densest input from the SSp-tr (trunk), the intermedial dorsal (CPi.dl.imd) from the SSp-ll (lower limb), the intermedial ventral (CPi.vl.imv) from the SSp-ul (upper limb), and the ventral (CPi.vl.v) and ventral tip (CPi.vl.vt) domains from rostral-caudal positions within the SSp-m (mouth) (Fig. 4a–d). Downstream projections suggest the rostral likely represents a feature of the inner mouth (SSp-m/i; i.e., tongue, buccal wall), while the caudal represents a feature of the outer mouth (SSp-m/o; i.e., mandible, snout) (Supplementary Fig. 5c).
Each of these five body domains also receives input from regions within the barrel cortex (SSp-bfd), MOp, and MOs that presumably correspond to trunk, lower limb, upper limb, inner mouth, and outer mouth (Fig. 4a–d). The exception is that no projections from the SSp-bfd to the m/i domain were detected. Notably, within a domain, axonal terminals arising from different cortical areas also display interdigitation rather than complete overlap (Fig. 4f). The CPi.vl receives additional input from the secondary somatosensory cortex (SSs) and from extra-somatic sensorimotor cortices. The rostral SSs projects to the inner mouth domain, while fibers from the caudal SSs, the gustatory area (GU) and visceral area (VISC) rostral arborize in the outer mouth domain (Fig. 4d,h).
Additionally, the rostral MOs (pole 2) forms part of the CPi.vl community and given its unique labeling pattern is designated its own domain: the ventrolateral central (CPi.vl.cvl) (Fig. 4g and Supplementary Fig. 6a,b). The central domains are so named since terminal fields within these domains are restricted to the center of the CP rather than to its peripheral edges.
Multi-fluorescent retrograde tracer injections into different domains of CPi.dl and CPi.vl, reveal the distinct spatial distribution patterns of these striatal projecting neurons in the different regions of somatic sensorimotor cortical areas, which agree with the anterograde labeling. Injections in the trunk, lower limb, and upper limb/mouth domains of the CP retrogradely label CP-projecting cortical neurons in their corresponding body regions within SSp, SSp-bfd, MOs, and MOp. Injections in upper limb/mouth domains also label cells in the SSs and VISC validating these projections (Fig. 4h and Supplementary Fig. 5f).
The medial cortical-dorsomedial striatal subnetworks
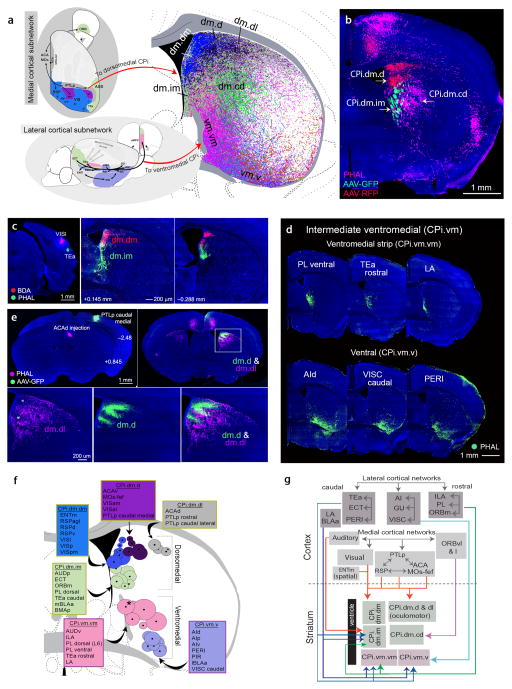
The dorsomedial CPi (CPi.dm) receives inputs primarily from cortical areas within the so-called medial cortical subnetwork (Fig. 5a,g). This subnetwork is composed of the visual (VIS) and auditory (AUD) areas, as well as a number of higher-order association areas along the medial cerebral cortex, including the anterior cingulate (ACA), retroplenial (RSP), and posterior parietal association (PTLp) areas1. This subnetwork is primarily involved in processing and transferring visual, spatial, auditory, and somatic sensory (trunk/lower limb) information to the ventromedial prefrontal cortex [i.e., ventrolateral orbital (ORBvl), prelimbic (PL), and infralimbic (ILA) cortical areas] Descending projections arising from these cortical areas form several parallel medial cortical-dorsomedial striatal networks that innervate 5 different domains in the CPi.dm (Fig. 5a,g).
Fig. 5. Cortical projections to the dorsomedial and ventromedial CPi communities.
(a) Dorsal and lateral views of cortical regions within the medial and lateral cortical subnetworks, respectively (modified from1). Areas within the medial cortical subnetwork project to domains within dorsomedial CPi (CPi.dm), while those in the lateral cortical subnetwork project to ventromedial CPi (CPi.vm) domains. Colors of cortical areas denote domain assignments. Overlap of reconstructed pathways of areas projecting CPi.dm and CPi.vm highlights unique domains. (b) Triple anterograde tracer injections in VISam (AAV-RFP), ORBvl (PHAL), and PL (AAV-GFP) show relative boundaries of CPi.dm.d, CPi.dm.im, and CPi.dm.cd. (c) Double injections of BDA and PHAL in VISl and TEa show segregation of CPi.dm.dm and CPi.dm.im. (d) Raw data from representative cases of cortical areas projecting to CPi.vm illustrates the CPi.vm.vm and CPi.vm.v domains. ic, internal capsule. (e) Double injections of AAV-GFP (PTLp caudal medial) and PHAL (ACAd) highlight interdigitating boundary between CPi.dm.d and CPi.dm.dl. (f) Centroids of cortical areas projecting to CPi.dm and CPi.vm display high topography and identify domains. Cortical areas projecting to each domain are listed in a box that is color coordinated with the centroids (stroke color of box denotes community). The algorithm grouped LA with cortical structures projecting to the CPi.dm; however, its centroid (denoted with an *) placed it outside of the 5 domains of this community and into CPi.vm.vm. (g) Wiring schema depicts connections among cortical areas within the medial cortical subnetwork dedicated to transferring visual, auditory, and some somatosensory (trunk/lower limb) information to the ORBvl through interconnected nodes involving the VIS, ACA, RSP, PTLp, and AUD1. Nodes within this subnetwork project to domains in the CPi.dm, forming a medial cortical-dorsomedial striatal network. Diagram also depicts the anterolateral cortical-ventromedial striatal subnetwork. The anterolateral cortical subnetwork (i.e., AI, GU, VISC, PIR) is heavily connected with PL and ILA. Downstream projections of each of these nodes generally target the domains of the CPi.vm.
The first is the dorsomedial strip (CPi.dm.dm) (Fig. 5c), a vertical narrow strip running adjacent to the dorsal half of the lateral ventricle that receives information from the primary (VISp), posteromedial (VISpm), and lateral (VISl) visual cortices, the medial entorhinal cortex (ENTm), and from the dorsal (RSPd), ventral (RSPv), and agranular retrosplenial areas (RSPagl) (Fig. 5a,f and Supplementary Fig. 6d,f,i). The ENTm contains grid cells and is critical in processing spatial information28. All divisions of the RSP share reciprocal connections with all visual cortices 1 and RSPv is the only neocortical recipient of inputs from the dorsal subiculum (SUBd), which has head direction cells that provide spatial information for navigation29. Convergence of neural inputs from these areas suggests the CPi.dm.dm may be a visuospatial convergence zone for integrating visual and spatial information (Fig. 7a).
The intermedial strip (CPi.dm.im) is immediately ventral to the dorsomedial strip and neighbors the ventral half of the lateral ventricle (Fig. 5a–c and Supplementary Fig. 6d). The CPi.dm.im is characterized by inputs from the primary auditory area (AUDp) and the anterior basolateral amygdalar nucleus (BLAa) (Figs. 5f and 7a), which plays an essential role in Pavlovian fear conditioning30. The CPi.dm.im further receives input from the dorsal prelimbic cortex (PL dorsal), medial orbital cortex (ORBm), caudal parts of the temporal association areas (TEa caudal), the ectorhinal cortex (ECT), and posterior basomedial amygdala (BMAp) (Fig. 5f and Supplementary Fig. 6d).
Two other domains of the CPi.dm are the dorsolateral (CPi.dm.dl) and dorsal (CPi.dm.d) (Fig. 3a,b,e). The densest fibers to these domains organize like horizontal stripes that stack underneath the corpus callosum (Fig. 5b,e and Supplementary Fig. 2c). The CPi.dm.d begins from the dorsal-most edge of the lateral ventricle and fans out laterally. In addition to direct inputs from the anterolateral and anteromedial VIS areas (VISal, VISam), this domain receives inputs from the MOs frontal eye field (MOs-fef) (as shown previously in rats31), the ventral anterior cingulate area (ACAv), and the posterior parietal association area (PTLp) caudal medial (Fig. 5f and Supplementary Figs. 2 and 6d,e,h). Efferents to the CPi.dm.dl project from the dorsal ACA (ACAd), PTLp caudal lateral, and the PTLp rostral (Fig. 5e,f and Supplementary Fig. 6d). All of these cortical areas are involved in processing visual information, eye movement, spatial attention, navigation, and spatially-guided motor planning1,32, suggesting that these two adjacent domains, CPi.dm.d and CPi.dm.dl, correspond to a specific oculomotor zone (Fig. 7a) identified in rats and primates that involves reactive saccadic eye control31,33. Projections to the CPi.dm.dl were validated with retrograde tracers that back-labeled neurons in ACAd and PTLp caudal lateral regions (Supplementary Fig. 6k).
The final domain of the CPi.dm is the central dorsal (CPi.dm.cd), which is ventral to the CPi.dm.d and CPi.dm.dl and lateral to the CPi.dm.im (Fig. 5a,b). Inputs to this domain are provided by the lateral and ventrolateral parts of the orbital area (ORBl, ORBvl) and as is characteristic of central domains, they largely avoid the periphery of the CP (Fig. 5f and Supplementary Fig. 6b,c).
The lateral cortical-ventromedial striatal network
Cortical structures in the fourth CPi community send topographically organized projections to three distinct, partially overlapping domains in the ventromedial CPi (CPi.vm). The ventromedial strip (CPi.vm.vm) is ventral to the intermedial strip and juxtaposed to the internal capsule (Fig. 5a and Supplementary Fig. 6g). Fibers projecting to the CPi.vm.vm appear diagonal, running along the dorsoventral extent of the internal capsule and hence are distinguishable from fibers projecting to its dorsally adjacent intermedial strip domain (Fig. 5d) and its ventrolaterally adjacent ventral domain (introduced below). The CPi.vm.vm is characterized by heavy inputs from layer 6 of the dorsal PL [PL dorsal (L6)], PL ventral, and infralimbic cortical area (ILA) (Figs. 5f and 7a). These medial prefrontal areas regulate autonomic and neuroendocrine activities through their projections to the central amygdalar nucleus (CEA), bed nuclei of the stria terminalis, hypothalamus, and lower brainstem7. Further, the CPi.vm.vm collects input from the ventral auditory cortex (AUDv) and rostral temporal association area (TEa rostral) (Fig. 5f).
The second CPi.vm domain is the ventral (CPi.vm.v), located along the ventral rim of the CP (Fig. 5a,d and Supplementary Fig. 6j). It starts from the ventral-most end of the internal capsule and lies between the CPi.vm.vm and the outer mouth domain. It compiles input from the caudal parts of the visceral area (VISC caudal), gustatory (GU), and the dorsal (AId), posterior (AIp), and ventral (AIv) agranular insular areas (Fig. 5f ). These cortical areas are within the anterolateral insular cortical subnetwork1 and are presumably involved in the control of visceral, gustatory, and pain information processing34,35 (Figs. 5g and 7a). Nodes within this cortical subnetwork also connect with the ILA and PL1, which primarily innervate the CPi.vm.vm. Together, these structures form the lateral cortical-ventromedial striatal network (Fig. 5g). The CPi.vm.v receives additional input from the piriform cortex (PIR), perirhinal area (PERI), and lateral part of the BLAa (lBLAa) (Fig. 5f). Projections to the CPi.vm.vm and CPi.vm.v were validated with retrograde tracing (data not shown).
The central ventromedial (CPi.vm.cvm) is the final domain of the CPi.vm community, which receives input from the rostral-most part of the MOs [rostral MOs, pole 1 (P1)] whose inputs again are confined to the central CP (Supplementary Fig. 6b,c).
Cortical projections to rostral (CPr) and caudal CP (CPc)
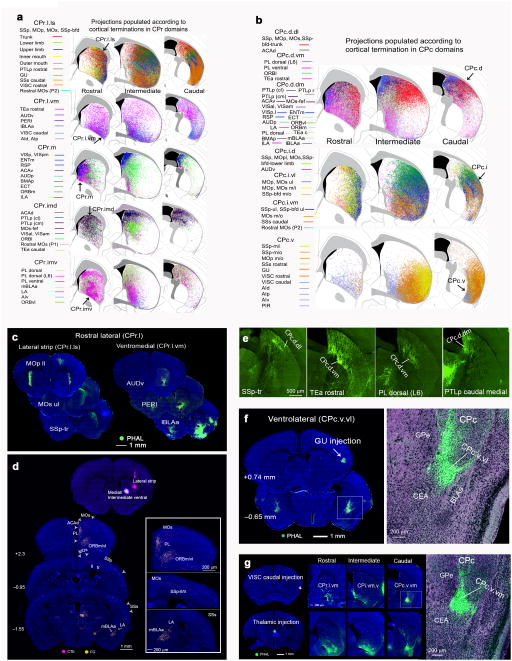
Compared to the intermediate CP, cortical inputs to the rostral (CPr) and caudal (CPc) exhibit two major features: (1) high integration, especially among inputs from cortical areas within different cortical subnetworks for cross modality integration; and (2) extensive reconfiguration, meaning cortical areas sending convergent inputs to the same CPi domain may innervate different domains in the CPr and CPc, while projections to different CPi domains may converge into the same CPr or CPc domain (Supplementary Fig. 4b,c for CPr and CPc communities and domains and their cortical constituents and Fig. 3b for relative domain boundaries). For example, moving rostrally from the CPi to the CPr, the relatively segregated projections from cortical areas within different somatic sensorimotor subnetworks converge into a newly identified narrow strip along the lateral edge, the CPr.l.ls (Figs. 6a,c and 7a–c and Supplementary Fig. 7a), suggesting its potential integrative role in synchronizing and coordinating motor actions. Moving caudally from CPi to CPc, inputs from the different somatic sensorimotor subnetworks maintain a rough topography. For instance, inputs from cortical areas within the trunk somatic subnetwork (i.e., SSp-tr, MOp tr, MOs tr, and SSp-bfd tr) terminate in the dorsal part (CPc.d; primarily in the dorsolateral domain, CPc.d.dl), which provides a potential interface for integrating somatic inputs with cortical inputs of different modalities (see below) (Figs. 6b,e and 7a,c and Supplementary Fig. 7d,e).
Fig. 6. Convergence and reconfiguration of cortico-striatal projections in CPr and CPc.
(a) Map showing reconstructions of projections populated according to CPr domains (i.e., row 1 shows projections from all cortical areas that send input to CPr.l.ls; row 2 depicts projections from cortical areas to CPr.l.vm). Reconstructions demonstrate CPr domains (column 1) and show general trend of projections through CPi and CPc. For example, projections to lateral CPi tend to terminate in lateral CPr, while projections terminating in the medial CPi terminate in the medial CPr. (b) Map showing reconstructions of projections according to CPc communities and delineating the dorsal, intermediate, and ventral regions of CPc (column 3). The termination pattern of cortical areas clustered into these caudal communities across CPi and CPr are also depicted (i.e., projections terminating in dorsal CPi terminate in dorsal CPc, while those terminating in lateral CPi terminate in intermediate parts of CPc. (c) Raw data from representative cases showing projections to CPr.l.ls and CPr.l.vm. (d) FG and CTb 647 injections into the lateral strip (CPr.l.ls) and medial/intermediate ventral CP (CPr.m/CPr.imv), respectively validate cortical projections to those domains and communities. FG retrogradely labeled neurons in PL, ORBvl, LA, mBLAa, while CTb labeled neurons in MOs, SSp, SSs. Right panels show magnification of retrograde labeling. (e) Representative cases demonstrating raw labeling to domains within CPc.d. (f) GU injection reveals the ventrolateral domain of CPc.v (CPc.v.vl). Labeling in boxed region magnified at right. CEA, central amygdalar nucleus; GPe, globus pallidus external part; BLA, basolateral amygdalar nucleus, anterior part. (g) Injections in VISC caudal produce unique labeling in CPc.v.vm (magnified on right). An injection in the thalamus validates this CPc.v.vm domain. Thalamic injection validates the general projection trends observed across the rostral-caudal extent of the CP for ventral domains (i.e., tendency of projections to CPi.vm.v to terminate in ventral domains of CPr and CPc). Labeling in inset magnified at right.
The intermediate part of the CPc (CPc.i) receives predominant inputs from cortical areas within the lower and upper limb somatic subnetworks, as well as MOs frontal pole and remaining SSp-bfd domains (Fig. 6b and Supplementary Fig. 7d–e). Here, these axonal terminals from different cortical areas still retain topography, but also display extensive intermingling or interdigitation. Convergence of these cortical inputs suggests that the CPc.i may coordinate movements of the limbs (Fig. 7a). Finally, most cortical areas within the mouth subnetworks (i.e., SSp-m/i, SSp-m/o, MOp m/o) generate projections that primarily terminate in CPc.v (Figs. 6b and 7a and Supplementary Fig. 7d–e).
Similarly, axonal projections arising from cortical areas innervating the relatively segregated CPi.dm.dm (i.e., VISp, VISpm, ENTm, RSP, ACAv) and CPi.dm.im (i.e., AUDp, ECT, amygdala) extend rostrally to converge in the CPr.m (near the ventricle) (Fig. 6a and Supplementary Fig. 7a,b), and caudally to terminate in a CPc.d domain, CPc.d.dm (Fig. 6b and Supplementary Fig. 7d,e). Although little is known about its functional role, this narrow strip along the wall of lateral ventricle across the entire CP specifically integrates multi-modality information, like visual, spatial, and auditory inputs with inputs from association areas like RSP, ACAv, ORBm, amygdala (BLAa, LA, BMAp), and temporal association areas associated with perception (TEa, ECT, ENTl) (Fig. 7a,c).
Axonal projections from cortical areas that primarily innervate the proposed oculomotor zone of the CPi.dm.d and dl (i.e., MOs-fef, ACAd, PTLp caudomedial, VISam, VISal) extend rostrally to target the CPr.imd (Fig. 6a and Supplementary Fig. 7a–c). Caudally, these axons heavily terminate in the CPc.d.vm, but also intermingle with cortical inputs in two adjacent domains, the CPc.d.dm and CPc.d.dl (Fig. 6b and Supplementary Fig. 7d). Interestingly, axonal projections from cortical areas that innervate CPi.vm.vm (i.e., ILA, PL, TEa, LA) also converge in CPc.d (Fig. 6b). Taken together, the CPc.d appears to be an important convergence zone that integrates cortical inputs associated with body trunk (including head and neck, SSp-tr, MOp tr, MOs tr 1), eye movements (i.e., MOs-fef, ACAd, RSP), attention and decision making (i.e., ACA, ORB, PTLp), and fear memory (i.e., ILA, PL, BLAa, LA) (Fig. 7a).
As previously described, all areas within the anterolateral insular subnetworks1, including VISC and all AI divisions generate dense projections in CPi.vm.v at the intermediate CP (Fig. 5f). These axons extend rostrally into the ventrolateral edge of CPr (defined here as CPr.l.vm) (Fig. 6c), and caudally into CPc.v (Fig. 6a,b,g). Dense projections arising from the visceral (VISC) or gustatory (GU) cortical areas generate two densely packed adjacent terminal balls in CPc.v, identified here as the CPc.v.m and CPc.v.vl, respectively (Fig. 6f,g). Additionally, the entire CPc.v receives dense inputs from all divisions of the AI (Supplementary Fig. 7e). Classically, these areas within the anterolateral insular subnetworks (i.e., VISC, GU, AId, AIv, AIp) are known to process visceral and inner state information and to regulate autonomic function through their inputs to the CEA—the visceral motor striatum35,36. Little is known about the functional relevance of cortico-striatal projections from these classic limbic areas to CP. However, the ventral edge comprised of the CPi.vm.v, CPi.vl.vt, and CPc.v receives somatomotor information pertaining to mouth, and input from rostral VISC and GU. Thus, this ventral edge is in a strategic position to integrate visceral, gustatory, olfactory, and somatic information to regulate orofaciopharyngeal movements associated with feeding behavior or stereotyped motor sequences for predatory eating37,38 (Fig. 7a,c). This functional role is complementary to that of the ventrally adjacent CEA—a striatal-like structure which receives similar cortical inputs, but primarily regulates autonomic function36.
Axonal terminals in CPc.v disappear at the caudal tail of the CP. Instead, extremely dense terminals from the AUDp cluster in a region adjacent to the LA and CEA, earning it a separate domain termed the (CPc.ext) (Fig. 7a,d). Additionally, dense projections from the TEa fill a gap between the AUDp terminals and external capsule in this caudal extreme domain (Fig. 7d), which also receives inputs from VIS, PTLp, ORBvl, ORBm, and SSs caudal.
Cortical afference to CPr and CPc domains also were validated with retrograde tracing. FG and CTb 647 infused into the lateral strip (CPr.l.ls) and medial/intermediate ventral CPr (CPr.m/CPr.imv) labeled neurons in the PL, ORBvl, LA, mBLAa and in the MOs, SSp, and SSs, respectively (Fig. 6d). Further, FG and CTb 647 tracer injections in the CPc.d and CPc.v labeled neurons in ACA, RSP, VIS, PTLp, ECT, and SSp-m and SSs, respectively, validating the cortical projections to these domains (Fig. 7e).
The lateral entorhinal (ENTl) projections to the CP
The ENTl strongly connects with almost the entire cortex and is therefore suggested to be a critical hub of interaction for cortical information1. It was excluded from the analysis due to its broad, dispersed projections to the rim of CPi (level 53), which spanned across six domains (Fig. 7f). These broad CP connections mimic its broad characteristic connections within the cortex, where it is connected with the medial prefrontal cortex, all cortical areas in the lateral cortical subnetworks, the hippocampus, olfactory cortical areas, and amygdala1.
Cortico-striatal disconnections in HD and MAO A/B KO mice
A detailed murine cortico-striatal projectome can provide a structural frame for studying disconnection pathogenesis of disorders recaptured in mouse models21–23,39. Accordingly, we applied our computational neuroanatomical workflow and newly delineated CP domains to quantify select cortico-striatal projection pathways in two mouse models that each represent a different category of disconnection syndrome. The first was the heterozygote zQ715 mouse, a full length knock-in model of Huntington’s disease (HD)40 — an inherited neurodegenerative disorder with motor, psychiatric, and cognitive deficits. Heterozygotes were chosen because unlike homozygotes, they do not exhibit cortical and striatal cell loss and striatal shrinkage as measured stereologically in aged animals40, which could result in a general disruption of all cortico-striatal circuits rather than circuit-specific compromises. The second model was the MAO A/B knockout41, recognized primarily for its aggressive behavior, but also for the neuropathological features it shares with models of autism spectrum disorders (ASD), including reduced thickness of the corpus callosum and increased dendritic arborization of pyramidal neurons in the prefrontal cortex41, both typical phenotypes of developmental disconnection syndromes42.
Comparisons of select cortico-striatal connections were made in zQ175 (at 12 months old) and MAO A/B knockout mice (at 12–14 months old), with their age matched wild type (WT) littermates. Specifically, projections from the upper limb motor regions (MOs ul and MOp ul) were studied for zQ175 subjects, while orbitofrontal projections were the target of investigation in MAO mice. A standardized injection protocol was instituted to accurately assess quantitative differences of connectivity strength between groups. This controlled individual variability of injection sizes and ensured the reproducibility of injection locations (Supplementary Fig. 8a,c,d)1. Signal intensity was normalized by injection size to control for label variability produced by injections of different sizes (Supplementary Fig. 8b).
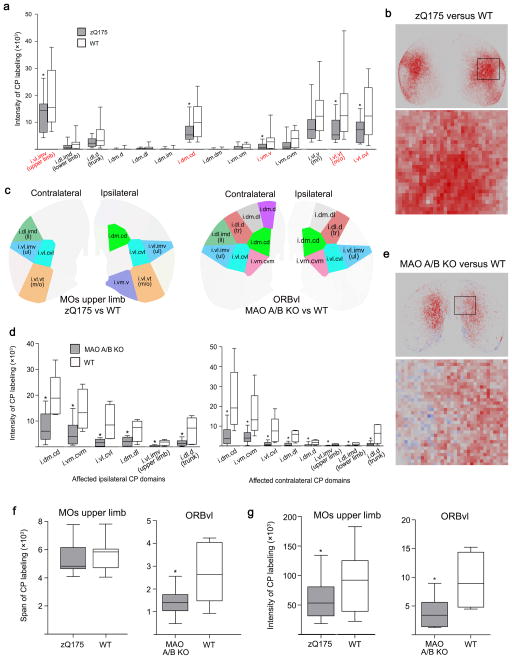
In zQ175 animals, projections from MOs ul to CPi showed a significant reduction in overall label intensity when the CPi was quantified as a single structure (Fig. 8g). Post-hoc domain level assessments showed that ipsilateral reductions were specifically in the CPi.dl.imv, CPi.dm.cd, CPi.vm.v, CPi.vl.vt, and CPi.vl.cvl domains (Fig. 8a–c and Supplementary Fig. 9a). Contralaterally, significant reductions were observed in mostly the same domains: the CPi.dl.imv, CPi.dl.imd, CPi.vl.vt, and CPi.vl.cvl (Fig. 8c and Supplementary Fig. 9c). The reduction in signal within these domains ranged from ~40–60% in both hemispheres (P<.05 for all). These reductions in signal intensity did not affect the span of axonal terminals in the CPi as a whole (ipsilateral, P=.43; contralateral, P=.42; Fig. 8f and Supplementary Fig. 9f). Importantly, projections from the MOp ul remained unaffected, suggesting pathway specific loss rather than generalized perturbation of all cortico-striatal connections (Supplementary Fig. 9d,e).
Fig. 8. Domain-specific pathological connections in zQ175 and MAO A/B KO mice.
(a) Boxplot showing domain specific loss of signal intensity from the MOs upper limb in zQ175 mice compared to wild type (WT) littermates in ipsilateral CP (zQ175, n=13; WT, n=15). Boxes depict the range between the upper and lower quartiles, whiskers depict the maximum and minimum values, and the bar indicates the median value. Red denotes domains within which connections were affected. [Domain, Mean/SEM zQ175 or MAO group, Mean/SEM WT group, P value, t value, degrees of freedom: i.vl.imv (ul), 12740/1926, 20560/2822, 0.031, 2.289, 24; i.dl.imd (ll), 1205/379, 2269/653, 0.173, 1.410, 22; i.dl.d (tr), 2370/453, 4494/1067, 0.084, 1.832, 18; i.dm.d, 65.16/38.17, 181.7/98.30, 0.2846, 1.103, 18; i.dm.im, 156.9/79.37, 147.5/93.66, 0.940, 0.0767, 25; i.dm.cd, 6427/1087, 10990/1700, 0.034, 2.260, 23; i.dm.dm, 46.73,/42.48, 79.25/56.45, 0.649, 0.460, 24; i.vm.vm, 538.4/227.4, 889.1/225.8, 0.284, 1.094, 25; i.vm.v, 1114/336, 10990/1700, 0.048, 2.122, 18; i.vm.cvm, 1804/671.4, 4036/869.8, 0.0504, 2.056, 25; i.vl.v, 9269/1850, 14390/2295, 0.095, 1.738, 25; i.vl.vt (m/o), 6964/1192, 14890/2787, 0.018, 2.615, 18; i.vl.cvl, 6770/1216, 13890/2376, 0.015, 2.668, 20]. (b,e) Pixelgrams illustrating differences in signal intensity detected between zQ175 and MAO A/B KO mice and their WT littermates within each 35x35 pixel cell. Red denotes a reduction in signal compared to WT, while blue indicates an increase. Saturation of color scales with percent change. (c) Visualization of the bar graphs showing domain-specific signal intensity reductions from MOs upper limb and ORBvl in zQ175 (left) and MAO A/B KO (right) mice, respectively. (d) Boxplots showing domain specific loss of signal intensity from the ORBvl in MAO A/B KO mice compared to WT in ipsilateral (left) and contralateral (right) CP (MAO A/B, n=10; WT, n=6). Only domains within which significant changes were detected are shown. [Ipsilateral CPi: i.vl.imv (ul), 22.11,/10.43, 164.2/37.29, 0.014, 3.670, 5; i.dl.imd (ll), 730.9/182.6, 3771/1576, 0.114, 1.916, 5; i.dl.d (tr), 149/35.35, 649.9/190.5, 0.049, 2.585, 5; i.dm.d, 3460/1297, 7171/2416, 0.218, 1.353, 7; i.dm.dl, 199.6/50.78, 652.6/154.6, 0.032, 2.780, 6; i.dm.im, 10650/2879, 23640/13100, 0.377, 0.9683, 5; i.dm.cd, 736.3/175.3, 1953/313.6, 0.010, 3.386, 8; i.dm.dm, 1336/551.3, 3036/1590, 0.351, 1.011, 6; i.vm.vm, 17010/5028, 30220/11120, 0.315, 1.083, 7; i.vm.v, 10400/2930, 24090/6629, 0.108, 1.888, 6; i.vm.cvm, 513.4/144.1, 1425/285.6, 0.025, 2.848, 7; i.vl.v (m/i), 1933/728, 3535/1214, 0.290, 1.132, 8; i.vl.vt (m/o), 4556/1233, 8541/1663, 0.083, 1.925, 10; i.vl.cvl, 158.7/42.76, 935.6/255.8, 0.030, 2.996, 5; Contralateral CPi: i.vl.imv (ul), 9.934/4.762, 66.22/14.83, 0.011, 3.614, 6; i.dl.imd (ll), 9.880/4.902, 98.34/24.94, 0.018, 3.481, 5; i.dl.d (tr), 79.11/26.30, 617.5/175.1, 0.029, 2.935, 7; i.dm.dl, 68.73/25.66, 520.9/109.8, 0.010, 4.012, 5; i.dm.im, 9358/4310, 19100/7382, 0.287, 1.140, 8; i.dm.cd, 502.6/148.4, 2242/587.3, 0.035, 2.871, 5; i.dm.dm, 271.7/184.3, 1686/890.8, 0.181, 1.555, 5; i.vm.vm, 11210/3949, 26130/5758, 0.061, 2.138, 9; i.vm.v, 7857/2429, 9863/2260, 0.556, 0.6049, 13; i.vm.cvm, 444.8/94.5, 1549/429.5, 0.049, 2.512, 5; i.vl.v (m/i), 272.2/145.1, 1145/371, 0.071, 2.192, 6; i.vl.vt (m/o), 2499/944.1, 3170/759.2, 0.589, 0.5539, 13; i.vl.cvl, 110.6/29.52, 782.3/246.2, 0.042, 2.708, 5]. (f) Boxplots of differences in the span of MOs upper limb and ORBvl projection fields within the CP in zQ175 and MAO A/B KO mice compared to WT. [MOs ul: 5348/288.9, 5654/250.8, 0.432, 0.7991, 24; ORBvl: 1425/177.1, 2715/494, 0.049, 2.459, 6]. (g) Boxplots showing the reduction in signal intensity in the entire ipsilateral CP from the MOs upper limb (left) and ORBvl (right) in zQ175 and MAO A/B mice, respectively. [MOs ul: 57120/9124, 90420/12830, 0.045, 2.115, 24; ORBvl: 3799/842.1, 9372/1764, 0.006, 3.221, 14]. * denotes significant difference (P<0.05) detected by two-tailed t test with Welch’s correction. Intensity is measured as total number of labeled pixels. Span is measured as total number of labeled cells.
MAO A/B knockouts displayed significantly reduced normalized signal intensity values compared to WT littermates, in both ipsi- and contralaterally projecting axons in the entire CPi from ORBvl (P<.05 for both) (Fig. 8g and Supplementary Fig. 9b). The span of the projections was also significantly reduced both ipsi- and contralaterally in the MAO A/B knockouts (P<.05 for both; Fig. 8f and Supplementary Fig. 9f). Domain-level analysis revealed reductions in the ipsilateral CPi.dm.cd, CPi.vm.cvm, CPi.vl.cvl, CPi.dl.imv, CPi.dm.dl, and CPi.dl.d (P<.05; Fig. 8d,e). Contralaterally, reductions were observed in the CPi.dm.cd, CPi.vm.cvm, CPi.vl.cvl, CPi.dm.dl, CPi.dl.d, CPi.dm.d, CPi.dl.imd, and CPi.dl.imv (P<.05 for all; Fig. 8c–e). The reductions in signal intensity within these domains ranged from ~60–90%.
DISCUSSION
Cortical projections to striatum have been examined across different species5,11–17. However, compiling a comprehensive cortico-striatal map from studies that use different species, strains, and methodologies is challenging and potentially leads to inconsistent conclusions. A comprehensive, reliable connectivity map requires systematic data collection and analysis workflows. Here, we systematically collected and analyzed striatal projections from all cortical areas that project to the dorsal striatum and created the most comprehensive cortico-striatal connectivity map that is available for any mammalian species. For example, partial somatotopic maps of the CP have been described for primates, cats, and rats11,12,14. However, in this study, we identified CP projections from nearly all somatic sensorimotor cortical areas representing five body subregions for a complete somatotopic map across the entire CP. A thorough investigation of CP projections of intra-cortical regions was conducted which showed that different areas within a single cortical area project to different CP domains, i.e., rostral versus caudal regions of SSs, VISC, PTLp, TEa, dorsal versus ventral parts of the PL, and medial and lateral parts of BLAa. Importantly, comprehensive assessment of the convergence and divergence of all cortico-striatal projections was conducted, which to our knowledge has not been conducted for any species. This data was used to objectively subdivide the CP into 29 domains whose anatomic locations, relative boundaries, and cortical afference are clearly delineated across the CP (Fig. 3b and Supplementary Fig. 4). The functional relevance of these domains are implicated by their cortical afference and can be directly tested using different methodological approaches (Fig. 7a).
Further, the current understanding of the brain is that it functions as a system of interacting networks rather than a collection of independent circuits or structures43, highlighting the importance of approaching connectomics from the network perspective. We previously assembled the global neural networks of the mouse neocortex1, which revealed that all cortical areas are organized into several functionally specific subnetworks: three somatic sensorimotor, two medial, and two lateral. Continuing on this trajectory, we used the cortico-striatal projectome to construct the cortico-striatal networks based on our previously identified cortical subnetworks. We found that descending projections from cortical subnetworks extensively reconfigure across the rostral, intermediate, and caudal divisions of the CP. This extensive convergence, divergence, and reconfiguration suggests that the dorsal striatum provides an important subcortical interface to mediate segregation, integration, and interaction of information from different cortical networks.
Projections from cortical hubs to center CPi domains
At the cortical level, several regions of the cortex potentially serve as hubs that bridge the communication between different subnetworks1. The cortical connectivity architecture of the frontal MOs poles and ORBvl warrants their identification as potential hubs1. The ORBvl receives convergent inputs from visual, auditory, and somatosensory cortices and also pre-integrated information from association cortices like PTLp and ACA. Similarly, the MOs rostral poles receive massive converged inputs from all other somatic sensorimotor networks1. These three structures generate unique localized projections to the three center domains of CPi, namely CPi.dm.cd, CPi.vl.cvl, and CPi.vl.cvm. Their terminal fields in the center domains are mostly confined to the center of the CPi and do not encroach into the periphery. Unlike the peripheral CPi domains that receive input from multiple cortical areas, the number of cortical areas that project to each central domain is limited to these single structures. This suggests that these central domains may not be as integrative in nature as their peripheral counterparts. It may be because the central domains receive highly processed information, which obviates the need for further integration. This physical and connectional difference of the peripheral and central domains leaves the impression of a center/periphery-like arrangement within the intermediate CP (CPi) reminiscent of the core (inner region) and shell (outer region) organization of the nucleus accumbens (ACB) in the ventral striatum44,45.
Relevance of CP communities and domains
A large majority of studies examining the functional relevance of the dorsal striatum have approached the CP as a quadrant structure, often assigning functions to its dorsolateral, dorsomedial, ventrolateral, and to a much lesser extent the ventromedial divisions. The lack of a standard structural framework regarding the relative boundaries of these quadrants makes comparisons and interpretation across studies difficult. This report offers the structural frame (Fig. 2). Further, the functions of the specific domains can partially be inferred from their cortical input. From this perspective, our cortico-striatal projectome aligns nicely with striatal functions proposed in the literature. For example, lesions of a region of the dorsolateral CP that corresponds roughly to the trunk region in the current report, disrupt the implementation of a stereotyped grooming sequence46. This is congruent with studies that demonstrate the role of the dorsolateral CP in habitual behaviors8,9. Lesions of the ventrolateral CP, which corresponds roughly to the mouth region in this report, disrupt implementation of stereotyped action sequences exhibited during predatory hunting including handling and biting38. Together, these data suggest that the subregions of the lateral CP are involved in the implementation of stereotyped behaviors involving their corresponding body regions (i.e., trunk for grooming, mouth and upper limb for handling and eating).
While the dorsolateral CP is involved in automatized motor sequences or habits8–10, the dorsomedial is involved in goal-directed behaviors8,10, spatial learning47, and orienting saccadic eye movements based on reward expectancy48. The cortical afference to the dorsomedial quadrant reported here also agrees with these behavioral designations. For example, the PTLp is involved in the computational aspects of goal-directed actions including movement planning49. Functionally distinct regions of the PTLp code movement plans associated with saccadic eye movements49. It does so by transforming sensory information into a common reference frame that is modulated by information from the eyes and head49. We identified three distinct regions of the PTLp that project primarily to the dorsomedial CP where they potentially get integrated with all the necessary sensory information. For instance, within CPi.dm.d, PTLp input converges with projections from the MOs frontal eye field and secondary visual areas (VISam, VISal). This domain also highly overlaps with, and hence communicates with, the CPi.dm.dm, the primary striatal target of visual and spatial information. Consequently, the proposed roles of goal-directed behaviors, spatial learning, and saccadic movements assigned to the dorsomedial CP are aligned with the cortical afference to this region.
Characterization of circuit-specific connectopathies
We used our mesoscale connectomics approach to characterize specific pathological cortico-striatal connections in HD zQ175 mice and MAO A/B knockouts. Our data showed that in HD, MOs ul projections to specific lateral CP domains is affected. Such characterization of circuit-specific connectopathies in animal models is important to constrain future experimentation to specific cortical and striatal regions and will offer strategies for more targeted therapeutics50. For instance, reductions in the MOs ul projection to CPi was limited to 5 of the 14 domains in the HD model. Therefore, testing of novel therapeutics to prevent cortico-striatal axonal loss in HD should focus on these five domains of the CPi to assess for improvement. Similarly, physiological investigations of responses in this cortico-striatal pathway would likely find differences in medium spiny neuron responses postsynaptic to the CPi domains showing the greatest disease-related changes.
ONLINE METHODS
The data were produced as part of the Mouse Connectome Project (MCP, www.MouseConnectome.org) at the University of Southern California (USC) (project formerly at the University of California, Los Angeles). Data collection for the zQ175 and MAO A/B knockout mice were conducted at USC.
Subjects
Overall, data from 8-week old male C57BL/6J mice (n=91; Jackson Laboratories) were used to trace all cortical pathways. Sixty-two pathways most representative of all the cortical areas examined were reconstructed and used for the analysis. For the HD and MAO model experiments, 12-month old male heterozygous zQ175 HD mice (n=19) and wild type controls (n=21; Jackson Laboratories), and 12–14-month old male MAO A/B knockout mice (n=10) and wild types (n=6), were used. The background strain of the zQ175 animals is the C57Bl/6J. The MAO A/B knockout were originally generated in a C57Bl/6J/129Sv strain, whose males were subsequently backcrossed over 25 generations with 129/SvEv females, then backcrossed over 25 generations with C57 B1/6J females. No randomization was used in the experiments reported in this paper. Although no formal a priori power analysis was conducted, group sizes were comparable to those utilized in previous reports40,41,51.
Animals were housed in pairs in a room that was temperature (21–22 °C), humidity (51%), and light controlled (12-hr light:12-hr dark cycle). The mice were allowed at least 1 week to adapt to their living conditions prior to stereotaxic surgeries for the delivery of tracers. Subjects had ad libitum access to tap water and mouse chow throughout the experiments. All experiments were conducted according to the regulatory standards set by the National Institutes of Health Guide for the Care and Use of Laboratory Animals and by the institutional guidelines set by the Institutional Animal Care and Use Committee at USC and the Animal Research Committee at UCLA.
Tracers and surgeries
Experimental procedures for tracer injections have been described previously52. Briefly, double coinjections of anterograde and retrograde tracers were delivered to virtually all anatomically delineated regions of the cortex and into select regions of the amygdala and thalamus. Phaseolus vulgaris leucoagglutinin (PHAL; 2.5%; Vector Laboratories) and cholera toxin subunit b (AlexaFluor 647 conjugate, 0.25%; Invitrogen) were coinjected, while biotinylated dextran amine (BDA; FluoroRuby, 5%; Invitrogen) was injected in combination with Fluorogold (FG; 1%; Fluorochrome, LLC). Small localized injections (~200–500 μm) were confined within domains of cortical areas and produced consistent, specific, and highly topographic patterns across the rostral-caudal extent of the CP (Supplementary Fig. 1a). The labeling from PHAL injections was primarily used for automated quantification (see below). Multiple retrograde tracers were injected into different CP domains within a single animal to validate the anterograde tracing data (Supplementary Fig. 1b). Retrograde tracers included FG and CTb 647, 488, and 549 conjugates (0.25%; Invitrogen). Adeno-associated viruses encoding enhanced green fluorescent protein (AAV-GFP; AAV2/1.hSynapsin.EGFP.WPRE.bGH; Penn Vector Core) and tdTomato (AAV1.CAG.tdtomato.WPRE.SV40; Penn Vector Core) were used in cases in which multiple anterograde tracer injections were used to reveal direct spatial correlations of axonal terminals arising from different cortical areas (i.e., topography or interdigitation) (Supplementary Fig. 2a). Although the images in the paper are unique exemplars, the majority of injections were successfully repeated anywhere from 1–17 times (see Supplementary Table 1). For zQ175 and MAO A/B knockout mice, only PHAL tracer injections and labeling were used for quantification. Either one (PHAL) or three weeks (for AAV-GFP) was allowed for tracer transport after which animals were perfused and their brains were extracted.
Surgeries for tracer infusions were performed under isoflurane anesthesia (Hospira, Inc.). Mice were initially anesthetized in an induction chamber primed with isoflurane and were subsequently mounted to the stereotaxic apparatus where they were maintained under anesthetic state via a vaporizer (Datex-Ohmeda). The isoflurane was vaporized and mixed with oxygen (0.5 L/min) and nitrogen (1 L/min). The percent of isoflurane in the gas mixture was maintained between 2 and 2.5 throughout the surgery. Tracers were delivered iontophoretically using glass micropipettes whose outside tip diameters measured approximately 10–30 μm. A positive 5 μAmp, 7-second alternating injection current was delivered for 10 minutes (Stoelting Co.).
Tissue preparation
Following an overdose injection of sodium pentobarbital, each animal was transcardially perfused with approximately 50 ml of 0.9% NaCl followed by 50 ml of 4% paraformaldehyde solution (PFA; pH 9.5). The brains were post-fixed in 4% PFA for 24–48 hours at 4°C after which they were embedded in 3% Type I-B agarose (Sigma-Aldrich) prior to sectioning. Four series of coronal sections were sliced at 50-μm thickness with a Compresstome (VF-700, Precisionary Instruments, Greenville, NC) and prepared for processing.
Immunofluorescence staining
One series of sections was stained for PHAL using the free-floating method for immunofluorescence. Briefly, sections were transferred to a blocking solution containing normal donkey serum (Vector Laboratories) and Triton X-100 (VWR) for 1 hour. Following three 5-minute rinses, sections were incubated in a KPBS solution comprised of donkey serum, Triton, and a 1:1000 concentration of rabbit anti-PHAL antibody for 48–72 hours at 4°C (Vector Laboratories, #AS-2300; see our previous work53 for validation of this antibody). Sections were rinsed three times in KPBS and then soaked for 3 hours in the secondary antibody solution, which contained donkey serum, Triton, and a 1:500 concentration of anti-rabbit IgG conjugated with Alexa Fluor® 488 or 647 (Invitrogen, 488: #A-21206, 647: #A-31573). Following three KBS rinses, the sections were counterstained with a fluorescent Nissl stain, NeuroTrace® 435/455 (NT; 1:500; Invitrogen, #N21479). The sections were then mounted and coverslipped using 65% glycerol.
Imaging and post-acquisition processing for publication to iConnectome
Initially, all sections were scanned as high-resolution virtual slide image (VSI) files using an Olympus VS110 high-throughput microscope using identical exposure parameters for all cases. Each image, comprised of five channels (PHAL, CTb, BDA, FG, and NT), was flipped to the correct left-right orientation, matched to the nearest ARA level, and converted to tiff format prior to being registered. Following registration, the NeuroTrace® fluorescent Nissl stain was converted to a bright-field image. Next, each channel for every image was adjusted for brightness and contrast to maximize labeling visibility and quality in iConnectome. Importantly, these adjustments were selective for files published to the iConnectome viewer. Thresholded images, which were used for the quantification of the data (discussed below), were not affected by changes in image contrast and brightness. Following final modifications (i.e., skewness, angles) and JPEG2000 file format conversions, images were published to iConnectome (http://www.mouseconnectome.org/iConnectome/page/search.jsp).
Informatics workflow
An informatics workflow was specifically designed to reliably warp, reconstruct, annotate, and analyze the labeled pathways in a high-throughput fashion (Fig. 1). The workflow was applied toward analyzing projections from 62 representative cortical injection cases to sections with characteristic labeling across the rostral (CPr), intermediate (CPi), and caudal CP (CPc). The labeling at level 53 (+0.145 mm from bregma) was the target of rich projections from all cortical areas, representative of all cortico-striatal projections, and exhibited the most segregation of labeling in terms of unique domains. Therefore, it was selected as the representative section for the intermediate CP. The CP at levels 41 (+1.345 mm from bregma) and 61 (–0.655 mm from bregma) displayed distinguishable labeling with a higher degree of integration of cortical inputs and far fewer discrete termination fields compared to the intermediate CP and were therefore selected to represent the rostral and caudal CP, respectively.
For analysis of data collected from zQ175 and MAO A/B knockout mice, researchers were blind to the genotype of the animals across the entire pipeline of image processing including image acquisition, segmentation, and quantification (steps discussed below).
The first step of our informatics workflow was a streamlined image registration pipeline. Facilitated by semi-automated human interaction, the registration system warped each individual image onto its corresponding level of the ARA with a level of detail such that even small nuclei were reliably registered.
After warping, a segmentation algorithm was applied to each image. Segmentation employed pixel-wise global and local thresholding to produce binary images with labeling (positive signal; 1) and background (negative signal; 0). The global threshold value was set to six standard deviations above the mean of a 700x700 pixel sample array within the CP that had been user-designated as without labeling. Individual pixels greater than this global threshold were accordingly designated as positive. Local threshold values were set dynamically. Pixels were examined element-wise against their surrounding 101x101 array. Any pixel value greater than the mean intensity value of neighborhood pixels (plus a small additional constant) was also assigned as labeling. Conversely, any pixel jointly below global and local threshold values was designated background. These reconstructions represented all labeling, including axonal pathways, terminal boutons, and varicosities.
An important concern when employing automated analysis of connectivity is the issue of fibers of passage getting annotated as functional connections when in fact no synapses exist. Pathways devoid of synapses can produce bright labeling that get annotated as positive pixels. This is especially relevant with regard to the fascicles indigenous to the dorsal striatum. However, the degree to which these axons rather than terminals are labeled is tracer dependent. PHAL labels far less CP fibers of passage than AAV tracers. Triple anterograde tracer injections of PHAL, AAV-GFP, and AAV-RFP in the MOs upper limb demonstrate this notion. Fascicles are intensely labeled by the AAV tracers, but not by the PHAL (Supplementary Fig. 2b). Further, higher resolution confocal imaging at 40x magnification clearly shows the abundance of PHAL-labeled varicosities and boutons, both of which are indicative of synaptic contacts54 (Supplementary Fig. 2c), and both of which are far greater in number than axons of passage. This suggests that positive pixels detected using our analysis were most likely representative of connections rather than fibers of passage since PHAL labeled pathways were primarily used for quantitative analysis.
The registration output also served as source images for both a comprehensive cortico-striatal connectivity map of the CP, as well as input to our automated annotation system. The cortico-striatal connectivity map contains a collection of 62 reconstructed cortico-striatal pathways within the ARA neuroanatomic frame (http://www.mouseconnectome.org/CorticalMap/page/map/protected.htm?projectId=5). Reconstruction of each pathway was rendered into an individual layer, allowing patterns of efference from multiple cortical source regions to be composed and compared by activation and deactivation of layers (Fig. 6a for example).
To facilitate annotation, the CP at atlas levels 41 (+1.345 mm), 53 (+0.145 mm), and 61 (–0.655) was divided into a grid of 35x35pixel cells with a spatial dimension of 22.5 μm2 (~0.6 μm per pixel). An overlap value at each cell was computed by projecting the segmented labeling output onto the grid. We employed this overlap process to annotate the 62 cortical injection cases. We further analyzed the annotation data to objectively identify groups of cortical injection sites that send converging input within different CP regions. To perform this final stage of the informatics pipeline, we first built an adjacency matrix out of our annotation data. The graph structure of the data is relatively simple: nodes and connections are organized as a multi-tree with two levels: the cortex and the CP. We therefore were able to apply a Louvain community detection algorithm27 to the data and identify clusters of injection sites with similar CP termination fields. The algorithm implementing the Louvain analysis was obtained from the Brain Connectivity Toolbox and executed in MATLAB55. Given the modularity of the data (i.e., highly topographic labeling), a modularity optimization algorithm like the Louvain was well suited. The Louvain algorithm has a lower asymptotic complexity than other clustering algorithms, employing a randomized, greedy optimization. However, the element of randomness makes it probable that the algorithm will reveal a different community structure over multiple runs. To mitigate this issue, the algorithm was run 1000 times. The community structure that emerged most often, which we defined as the community structure mode (borrowing from statistics) is reported. A mean and standard deviation for the number of communities that was detected across the 1000 runs was also computed. The community structure mode for the CPi occurred 23/1000 times with a q value of 0.4963. The mean and standard deviation for the number of communities for this division were 3.99 ± 0.35. For the rostral CP, the community structure mode reported occurred 32/1000 runs with a q value of 0.325 and a mean of 4.265± 0.578 for number of communities. Finally, the structure mode for the CPc occurred 23/1000 times with a q value of 0.442 and a mean of 3.07± 0.299. Community structure analysis executed on randomized graph models corresponding to each CP level (i.e., rostral, intermediate, caudal) returned 1000 unique communities. These different, essentially opposite, results demonstrate that the communities detected by the Louvain with the non-randomized data were statistically significant.
Subsequently, to aid visualization we employed the community structure to re-order and color code an adjacency matrix such that connections were placed close to the diagonal. An accompanying color-coded CP illustrates the spatial arrangement of the communities.
The raw data and connectivity map showed the existence of topographically organized projection fields within these individual communities, suggesting the existence of relatively smaller CP domains with a more restricted number of cortical sources. Since modularity optimization algorithms like the Louvain are not optimal for detecting smaller communities and communities with members of unequal size, subsequent analyses were performed to extract the smaller clusters of projection fields embedded within the superordinate communities. Accordingly, a measure of centrality for the CP projection fields of each cortical area was computed. The centroid, or the arithmetic average coordinate of a reconstructed terminal field (i.e., center of gravity), was used as this measure. Generally, projections from the cortex terminated most densely within a circumscribed CP domain and more diffusely to regions adjacent to the condensed terminal field, providing some degree of overlap between neighboring regions and obscuring clear boundary demarcation. However, the segregation among the dense cores of the terminal fields from different cortical regions was clear. To better represent the dense cores of these projections, only pixels in the densest subregions of the field were analyzed with the centroid method. This was achieved by making the entry of a pixel of positive labeling into the centroid analysis conditional on that pixel being contained within a cell of the grid that had a density percentile, with respect to all cells within the grid, equal to or greater than 95%, that is, only pixels within the top 5% of grid cells, based on labeling density were utilized for the centroid analysis.
To further subdivide the CP into smaller domains objectively, we constructed a Voronoi diagram56 that designates in the CP a number of Voronoi cells (or domains), built upon the centroid analysis previously conducted. To create these regions, for each domain a single intensity-weighted average location of its constituent centroids was computed. Specifically, at each domain with N centroids let
be the coordinates of each centroid in the domain. Let
be the intensity (in [0, 1]) of each centroid. Finally, let
be the weighted location of each centroid. Per domain then, the Voronoi point was calculated as the element-wise sum of WL divided by the element-wise sum of C:
The resulting average location at each domain was used as a Voronoi seed point. Executing a standard euclidean Voronoi algorithm on the seed points, the resulting tessellation is shown (Figs. 1k and 3b). As evidenced by the high degree of terminal field overlap between adjacent domains, hard borders between domains do not exist. Therefore, borders between Voronoi cells simply localize zones of strongest terminal convergence among members of a subdomain (i.e., where interaction between convergent sources of cortical efference is most prominent).
In this work we also quantified density of labeling terminating in the CP, defining the complementary measures intensity and span for this purpose. Intensity is simply a sum of the labeled pixels across the CP. Span complements the intensity value, and is a count of the cells of the grid containing labeling. They were calculated using the following equations:
Intensity IR and Span SR:
where P, C are the number of pixels and cells respectively with positive labeling for region R.
Normalized Intensity IR and Span SR:
where i...N is all injection sites for the given level.
To summarize label density, the intensity and span values were further utilized to compute the Labeling Density Index (LDI). The LDI, computed by taking the quotient of the (sum-to-unity normalized) intensity and span, summarized in a single value the labeling properties originating from an injection site. We designated LDI values greater than 1.0 as concentrated, with lesser LDI values defined as diffuse. The equation used to calculate LDI is as follows:
Labeling Density Index LDIR:
Tables containing the calculated intensity, span, and LDI values for each cortico-striatal projection are available online (http://www.mouseconnectome.org/MCP/page/documents/tables?paperId=18).
Overlap refers to a convergence of terminal fields within the CP between cortical source areas. The overlap value between a source and target ROI is the ratio of common labeling among the two ROIs to total source labeling. To corroborate the domains derived from visual analysis of injection site centroids, higher order overlap values, denoting overlap between domains, were computed from the injection site-scale overlap values. Whereas domains were constituted by multiple cortical areas, overlap values between any two domains were simply the mean of injection site-scale overlaps from one cortical area with another. Overlap was calculated using the following equation:
Common labeling (COMR, S) between regions R, S:
where L is the number of labeled pixels in common locations across injection sites R, S
Overlap OVRR, S between regions R, S:
Tables containing overlap values between individual injection sites and between domains are also available (http://www.mouseconnectome.org/MCP/page/documents/tables?paperId=18).
Code availability
We employed the Brain Connectivity Toolbox (BCT) to analyze the connectivity data, taking particular advantage of the Louvain algorithm implementation55. The BCT is freely available at https://sites.google.com/site/bctnet/. The other computer codes used to generate the findings of this study are available from the corresponding author upon request.
Assessment of injection sites
All cortical injection cases included in this work are, in our judgment, prototypical representatives of each cortical area. We have previously demonstrated our targeting accuracy with respect to injection placement, our attention to injection location, and the fidelity of labeling patterns derived from injections to the same cortical location (Supplementary Fig. 2 in 1). In the current report, we also demonstrate our injection placement accuracy and the consistent labeling resulting from injections placed in the same cortical areas. In one case, we applied three different anterograde tracers in the MOs upper limb, which resulted in similar labeling patterns, showing both consistency of injection placement and fidelity of labeling across the injections (Supplementary Fig. 2b). Repeated injections also were applied in double or triple anterograde tracing experiments to show topography and interdigitation. These injections also validated the striatal projection patterns of the cortical areas involved. For example, injections into the VISam (AAV-RFP), PL dorsal (AAV-GFP), and ORBvl (PHAL) showed the topographic positions of unique CP domains (Fig. 5b), but also validated the striatal projections of these cortical areas demonstrated in different cases (Supplementary Fig. 6c,d).
The somatosensory cortices representing each body subfield and barrel fields lay adjacent to one another (Supplementary Fig. 5a) and therefore cross contamination of injection sites is a legitimate concern when striatal convergence from these cortical areas is considered. First, injection sites were re-imaged using lower exposure parameters than those used to capture labeled neurons and axons. This ensured proper identification of injection locations. Next, the specificity of the injection sites were validated by examining their thalamic and brainstem projections (Supplementary Fig. 5c–d). Injections into the SSp-tr, ll, ul, and m regions showed clear topographic projections to their corresponding terminals in the ventral posteromedial (VPM) and ventral posterolateral (VPL) nuclei of the thalamus. Trunk representations were in dorsal regions, followed by lower limb which were lateral to upper limb representations, with orofacial representations in the most ventral and medial parvocellular regions57–60. Consistent with brainstem body representations, SSp-ll labeled the gracile nucleus, the SSp-ul the cuneate, and the SSp-m the dorsal parts of the spinal trigeminal (SPV) nucleus with SSp-m/o represented dorsal and lateral to the SSp-m/i regions57,61–63. Specificity of SSp-bfd injections were validated by observing their topographic projections to the medial part of the posterior thalamus (POm), to the VPM, and to approximately the ventral third of the spinal trigeminal, precisely where whisker representations reside (Supplementary Fig. 5d)57,58,64.
For the HD and MAO experiments, only injections located within cortical layer V of MOs ul (for the HD experiment; Supplementary Fig. 8a) and ORBvl (for the MAO experiment; Supplementary Fig. 8c) were included in the analysis. Examination of the relationship between injection site size and cortico-striatal terminal field size was conducted. Injection sites were rescanned using lower exposure settings to clearly visualize neurons that had absorbed the tracer (Supplementary Fig. 8a,c). The number of PHAL-labeled cells in the section closest to the center of the injection site, i.e., containing the greatest number of labeled cells, was manually counted. PHAL injections typically result in a dense cluster of labeled neurons; injections resulting in a diffuse labeling pattern, or injections in which the background staining was too high to discern the number of PHAL-labeled neurons, were excluded from the analysis. Pooling the remaining subjects, linear regression of injection site size to cortico-striatal terminal intensity revealed a significant correlation between the two variables: the greater the injection site size, the greater the terminal intensity. This relationship was found in both for MOs ul injections (P < 0.05, r = 0.415) and the ORBvl injections (Supplementary Fig. 8b; P < 0.05, r = 0.615). This relationship approached significance for the MOp ul injections (Supplementary Fig. 8b). Therefore, most analyses were conducted using normalized terminal intensity values, in which the raw intensity value for each subject was divided by its injection site size. Further, injection sizes of zQ175 and MAO mice did not differ from their respective WT littermates (Supplementary Fig. 8d; P=.214 for zQ175 and 0.776 for MAO). Exclusion of erroneous injections and counting of labeled cells was performed by experimenters blind to group affiliation.
Statistical analyses
Differences in normalized intensity and span values between mutants and their wild type littermates were statistically compared using a two-sided t-test with Welch’s correction for heteroscedasticity. Correlations between injection site size and intensity of CP labeling were investigated using Pearson’s r. Normality of the data distributions was assumed, but not formally tested. A supplementary methods and statistics checklist is available where all group sizes, means, standard errors, p values, t values, and degrees of freedom are available for all statistical tests conducted in this work (URL). Additionally, the raw data upon which all boxplots and scatter plots are based are also available.
Data availability
All image data comprising the projectome, cortico-striatal projection maps, and tables of intensity, span, labeling density indices, and overlap values, are freely available at www.MouseConnectome.org.
Supplementary Material
Acknowledgments
This work was supported by NIH/NIMH MH094360-01A1 (HWD), NIH/NCI U01CA198932-01 (HWD), the CHDI Foundation (HWD) and LONIR P41 EB015922 (AWT). The authors are grateful to Drs. Larry W. Swanson and Harvey Karten to serve on Advisory Council for the Mouse Connectome Project (www.MouseConnectome.org).
Footnotes
AUTHOR CONTRIBUTIONS
H.-W. and H.H. conceived, designed, and managed the entire project, conducted manual analysis of the raw data, and along with N.N.F. prepared the manuscript and the figures. N.N.F. along with H.H., H.-W., I.B., M.B., and M.S.B. refined and finalized the applied methods of analyses. I.B. led the informatics team and along with M.B. created and executed all aspects of the informatics workflow and wrote the corresponding methods. B.Z. and L.G. made tracer injections and conducted subsequent tissue processing and imaging, M.Y.S. processed raw images in preparation for uploading to iConnectome, and S.Y. managed iConnectome for both raw image uploads and connectivity map displays. N.N.F. managed and conducted experiments pertaining to zQ175 and MAO knockout mice. X.W.Y. contributed his expertise regarding discussion of striatal function under normal and diseased conditions. J.C.S. provided the MAO A/B knockout mice and contributed to the interpretation of the data resulting from those subjects. A.W.T. served as project advisor. H.-W. conceived and led the Mouse Connectome Project. All authors offered constructive guidance for the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Zingg B, et al. Neural networks of the mouse neocortex. Cell. 2014;156:1096–1111. doi: 10.1016/j.cell.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oh SW, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bota M, Sporns O, Swanson LW. Architecture of the cerebral cortical association connectome underlying cognition. Proc Natl Acad Sci U S A. 2015;112:E2093–2101. doi: 10.1073/pnas.1504394112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song HF, Kennedy H, Wang XJ. Spatial embedding of structural similarity in the cerebral cortex. Proc Natl Acad Sci U S A. 2014;111:16580–16585. doi: 10.1073/pnas.1414153111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexander GE, DeLong MR, Strick PL. Parallel organization of functionally segregated circuits linking basal ganglia and cortex. Annu Rev Neurosci. 1986;9:357–381. doi: 10.1146/annurev.ne.09.030186.002041. [DOI] [PubMed] [Google Scholar]
- 6.Parent A, Hazrati LN. Functional anatomy of the basal ganglia. I The cortico-basal ganglia-thalamo-cortical loop. Brain Res Brain Res Rev. 1995;20:91–127. doi: 10.1016/0165-0173(94)00007-c. [DOI] [PubMed] [Google Scholar]
- 7.Swanson LW. Cerebral hemisphere regulation of motivated behavior. Brain Res. 2000;886:113–164. doi: 10.1016/s0006-8993(00)02905-x. [DOI] [PubMed] [Google Scholar]
- 8.Yin HH, Knowlton BJ. The role of the basal ganglia in habit formation. Nat Rev Neurosci. 2006;7:464–476. doi: 10.1038/nrn1919. [DOI] [PubMed] [Google Scholar]
- 9.Graybiel AM. Habits, rituals, and the evaluative brain. Annu Rev Neurosci. 2008;31:359–387. doi: 10.1146/annurev.neuro.29.051605.112851. [DOI] [PubMed] [Google Scholar]
- 10.Balleine BW, O’Doherty JP. Human and rodent homologies in action control: corticostriatal determinants of goal-directed and habitual action. Neuropsychopharmacology. 2010;35:48–69. doi: 10.1038/npp.2009.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malach R, Graybiel AM. Mosaic architecture of the somatic sensory-recipient sector of the cat’s striatum. J Neurosci. 1986;6:3436–3458. doi: 10.1523/JNEUROSCI.06-12-03436.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flaherty AW, Graybiel AM. Corticostriatal transformations in the primate somatosensory system. Projections from physiologically mapped body-part representations. J Neurophysiol. 1991;66:1249–1263. doi: 10.1152/jn.1991.66.4.1249. [DOI] [PubMed] [Google Scholar]
- 13.Berendse HW, Galis-de Graaf Y, Groenewegen HJ. Topographical organization and relationship with ventral striatal compartments of prefrontal corticostriatal projections in the rat. J Comp Neurol. 1992;316:314–347. doi: 10.1002/cne.903160305. [DOI] [PubMed] [Google Scholar]
- 14.Brown LL, Smith DM, Goldbloom LM. Organizing principles of cortical integration in the rat neostriatum: corticostriate map of the body surface is an ordered lattice of curved laminae and radial points. J Comp Neurol. 1998;392:468–488. [PubMed] [Google Scholar]
- 15.Alloway KD, Crist J, Mutic JJ, Roy SA. Corticostriatal projections from rat barrel cortex have an anisotropic organization that correlates with vibrissal whisking behavior. J Neurosci. 1999;19:10908–10922. doi: 10.1523/JNEUROSCI.19-24-10908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mailly P, Aliane V, Groenewegen HJ, Haber SN, Deniau JM. The rat prefrontostriatal system analyzed in 3D: evidence for multiple interacting functional units. J Neurosci. 2013;33:5718–5727. doi: 10.1523/JNEUROSCI.5248-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Averbeck BB, Lehman J, Jacobson M, Haber SN. Estimates of projection overlap and zones of convergence within frontal-striatal circuits. J Neurosci. 2014;34:9497–9505. doi: 10.1523/JNEUROSCI.5806-12.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan WX, Mao T, Dudman JT. Inputs to the dorsal striatum of the mouse reflect the parallel circuit architecture of the forebrain. Front Neuroanat. 2010;4:147. doi: 10.3389/fnana.2010.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haber SN. The primate basal ganglia: parallel and integrative networks. J Chem Neuroanat. 2003;26:317–330. doi: 10.1016/j.jchemneu.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Voorn P, Vanderschuren LJ, Groenewegen HJ, Robbins TW, Pennartz CM. Putting a spin on the dorsal-ventral divide of the striatum. Trends Neurosci. 2004;27:468–474. doi: 10.1016/j.tins.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Joel D. Open interconnected model of basal ganglia-thalamocortical circuitry and its relevance to the clinical syndrome of Huntington’s disease. Mov Disord. 2001;16:407–423. doi: 10.1002/mds.1096. [DOI] [PubMed] [Google Scholar]
- 22.Nelson AB, Kreitzer AC. Reassessing models of basal ganglia function and dysfunction. Annu Rev Neurosci. 2014;37:117–135. doi: 10.1146/annurev-neuro-071013-013916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shepherd GM. Corticostriatal connectivity and its role in disease. Nat Rev Neurosci. 2013;14:278–291. doi: 10.1038/nrn3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong HW. The Allen Reference Atlas: A digital color brain atlas of the C57BL/6J male mouse. John Wiley and Sons, Inc; Hoboken, NJ, USA: 2007. [Google Scholar]
- 25.Paxinos G, Watson C. The rat brain in stereotaxic coordinates. 7. Academic Press; San Diego, CA, USA: 2013. [Google Scholar]
- 26.Swanson LW. Brain maps: structure of the rat brain. 3. Academic Press; San Diego, CA, USA: 2004. [Google Scholar]
- 27.Blondel VD, Guillaume JL, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech. 2008:P10008. [Google Scholar]
- 28.Moser EI, Kropff E, Moser MB. Place cells, grid cells, and the brain’s spatial representation system. Annu Rev Neurosci. 2008;31:69–89. doi: 10.1146/annurev.neuro.31.061307.090723. [DOI] [PubMed] [Google Scholar]
- 29.Taube JS. Head direction cells and the neurophysiological basis for a sense of direction. Prog Neurobiol. 1998;55:225–256. doi: 10.1016/s0301-0082(98)00004-5. [DOI] [PubMed] [Google Scholar]
- 30.Fanselow MS, Poulos AM. The neuroscience of mammalian associative learning. Annu Rev Psychol. 2005;56:207–234. doi: 10.1146/annurev.psych.56.091103.070213. [DOI] [PubMed] [Google Scholar]
- 31.Reep RL, Cheatwood JL, Corwin JV. The associative striatum: organization of cortical projections to the dorsocentral striatum in rats. J Comp Neurol. 2003;467:271–292. doi: 10.1002/cne.10868. [DOI] [PubMed] [Google Scholar]
- 32.Reep RL, Corwin JV. Posterior parietal cortex as part of a neural network for directed attention in rats. Neurobiol Learn Mem. 2009;91:104–113. doi: 10.1016/j.nlm.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 33.Thakkar KN, van den Heiligenberg FM, Kahn RS, Neggers SF. Frontal-subcortical circuits involved in reactive control and monitoring of gaze. J Neurosci. 2014;34:8918–8929. doi: 10.1523/JNEUROSCI.0732-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allen GV, Saper CB, Hurley KM, Cechetto DF. Organization of visceral and limbic connections in the insular cortex of the rat. J Comp Neurol. 1991;311:1–16. doi: 10.1002/cne.903110102. [DOI] [PubMed] [Google Scholar]
- 35.Craig AD. How do you feel--now? The anterior insula and human awareness. Nat Rev Neurosci. 2009;10:59–70. doi: 10.1038/nrn2555. [DOI] [PubMed] [Google Scholar]
- 36.Swanson LW, Petrovich GD. What is the amygdala? Trends Neurosci. 1998;21:323–331. doi: 10.1016/s0166-2236(98)01265-x. [DOI] [PubMed] [Google Scholar]
- 37.Pisa M. Motor somatotopy in the striatum of rat: manipulation, biting and gait. Behav Brain Res. 1988;27:21–35. doi: 10.1016/0166-4328(88)90106-4. [DOI] [PubMed] [Google Scholar]
- 38.dos Santos LM, et al. The role of the ventrolateral caudoputamen in predatory hunting. Physiol Behav. 2012;105:893–898. doi: 10.1016/j.physbeh.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 39.Griffiths KR, Morris RW, Balleine BW. Translational studies of goal-directed action as a framework for classifying deficits across psychiatric disorders. Front Syst Neurosci. 2014;8:101. doi: 10.3389/fnsys.2014.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heikkinen T, et al. Characterization of neurophysiological and behavioral changes, MRI brain volumetry and 1H MRS in zQ175 knock-in mouse model of Huntington’s disease. PLoS One. 2012;7:e50717. doi: 10.1371/journal.pone.0050717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bortolato M, et al. Monoamine oxidase A and A/B knockout mice display autistic-like features. Int J Neuropsychopharmacol. 2013;16:869–888. doi: 10.1017/S1461145712000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geschwind DH, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Curr Opin Neurobiol. 2007;17:103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 43.Sporns O. Contributions and challenges for network models in cognitive neuroscience. Nat Neurosci. 2014;17:652–660. doi: 10.1038/nn.3690. [DOI] [PubMed] [Google Scholar]
- 44.Zahm DS. An integrative neuroanatomical perspective on some subcortical substrates of adaptive responding with emphasis on the nucleus accumbens. Neurosci Biobehav Rev. 2000;24:85–105. doi: 10.1016/s0149-7634(99)00065-2. [DOI] [PubMed] [Google Scholar]
- 45.Groenewegen HJ, Wright CI, Beijer AV, Voorn P. Convergence and segregation of ventral striatal inputs and outputs. Ann N Y Acad Sci. 1999;877:49–63. doi: 10.1111/j.1749-6632.1999.tb09260.x. [DOI] [PubMed] [Google Scholar]
- 46.Cromwell HC, Berridge KC. Implementation of action sequences by a neostriatal site: a lesion mapping study of grooming syntax. J Neurosci. 1996;16:3444–3458. doi: 10.1523/JNEUROSCI.16-10-03444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Devan BD, White NM. Parallel information processing in the dorsal striatum: relation to hippocampal function. J Neurosci. 1999;19:2789–2798. doi: 10.1523/JNEUROSCI.19-07-02789.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hikosaka O, Nakamura K, Nakahara H. Basal ganglia orient eyes to reward. J Neurophysiol. 2006;95:567–584. doi: 10.1152/jn.00458.2005. [DOI] [PubMed] [Google Scholar]
- 49.Cohen YE, Andersen RA. A common reference frame for movement plans in the posterior parietal cortex. Nat Rev Neurosci. 2002;3:553–562. doi: 10.1038/nrn873. [DOI] [PubMed] [Google Scholar]
- 50.Steinberg EE, Christoffel DJ, Deisseroth K, Malenka RC. Illuminating circuitry relevant to psychiatric disorders with optogenetics. Curr Opin Neurobiol. 2015;30:9–16. doi: 10.1016/j.conb.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith KS, Graybiel AM. A dual operator view of habitual behavior reflecting cortical and striatal dynamics. Neuron. 2013;79:361–374. doi: 10.1016/j.neuron.2013.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hintiryan H, et al. Comprehensive connectivity of the mouse main olfactory bulb: analysis and online digital atlas. Front Neuroanat. 2012;6:30. doi: 10.3389/fnana.2012.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biag J, et al. Cyto- and chemoarchitecture of the hypothalamic paraventricular nucleus in the C57BL/6J male mouse: a study of immunostaining and multiple fluorescent tract tracing. J Comp Neurol. 2012;520:6–33. doi: 10.1002/cne.22698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gerfen CR, Sawchenko PE. An anterograde neuroanatomical tracing method that shows the detailed morphology of neurons, their axons and terminals: immu nohistochemical localization of an axonally transported plant lectin, Phaseolus vulgaris leucoagglutinin (PHA-L) Brain Res. 1984;290:219–238. doi: 10.1016/0006-8993(84)90940-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. NeuroImage. 2010;52:1059–69. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 56.Voronoï G. Nouvelles applications des paramètres continus à la théorie des formes quadratiques Deuxième mémoire Recherches sur les parallélloèdres primitifs. Journal für die reine und angewandte Mathematik. 1908;134:198–287. [Google Scholar]
- 57.Fabri M, Burton H. Topography of connections between primary somatosensory cortex and posterior complex in rat: a multiple fluorescent tracer study. Brain Res. 1991;538:351–357. doi: 10.1016/0006-8993(91)90455-5. [DOI] [PubMed] [Google Scholar]
- 58.Hoogland PV, Welker E, Van der Loos H. Organization of the projections from barrel cortex to thalamus in mice studied with Phaseolus vulgaris-leucoagglutinin and HRP. Exp Brain Res. 1987;68:73–87. doi: 10.1007/BF00255235. [DOI] [PubMed] [Google Scholar]
- 59.Liao CC, Chen RF, Lai WS, Lin RC, Yen CT. Distribution of large terminal inputs from the primary and secondary somatosensory cortices to the dorsal thalamus in the rodent. J Comp Neurol. 2010;518:2592–2611. doi: 10.1002/cne.22354. [DOI] [PubMed] [Google Scholar]
- 60.Nambu A. Somatotopic organization of the primate Basal Ganglia. Front Neuroanat. 2011;5:26. doi: 10.3389/fnana.2011.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li XG, Florence SL, Kaas JH. Areal distributions of cortical neurons projecting to different levels of the caudal brain stem and spinal cord in rats. Somatosens Mot Res. 1990;7:315–335. doi: 10.3109/08990229009144711. [DOI] [PubMed] [Google Scholar]
- 62.Remple MS, Henry EC, Catania KC. Organization of somatosensory cortex in the laboratory rat (Rattus norvegicus): Evidence for two lateral areas joined at the representation of the teeth. J Comp Neurol. 2003;467:105–118. doi: 10.1002/cne.10909. [DOI] [PubMed] [Google Scholar]
- 63.Cerkevich CM, Qi HX, Kaas JH. Corticocortical projections to representations of the teeth, tongue, and face in somatosensory area 3b of macaques. J Comp Neurol. 2014;522:546–572. doi: 10.1002/cne.23426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Welker E, Hoogland PV, Van der Loos H. Organization of feedback and feedforward projections of the barrel cortex: a PHA-L study in the mouse. Exp Brain Res. 1988;73:411–435. doi: 10.1007/BF00248234. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All image data comprising the projectome, cortico-striatal projection maps, and tables of intensity, span, labeling density indices, and overlap values, are freely available at www.MouseConnectome.org.