Abstract
Objective
Although driver mutation status is crucial to targeted therapy decision-making in non-small cell lung cancer (NSCLC), due to unavailable or inadequate biopsies, there are still many patients with unknown mutation status. A promising way to solve this problem is liquid biopsy, such as cell-free DNA (cfDNA) in peripheral blood. Additionally, due to the little amount of cfDNA, detecting methods with high sensitivity, specificity and economy are required in clinical practice. Here, we explored the feasibility of Competitive Allele-Specific TaqMan® PCR (CastPCR) detecting driver mutations in cfDNA from plasma in lung adenocarcinoma patients.
Results
Sensitivity, specificity, concordance, PPV and NPV of CastPCR detecting EGFR mutations in cfDNA was 56.4% (31/55), 94.2% (49/52), 74.8% (80/107), 91.2% (31/34) and 67.1% (49/73), respectively. Notably, specificity and PPV for p.T790M both reached 100.0%. For BRAF detection, it was 28.6% (2/7), 93.0% (93/100), 88.8% (95/107), 22.2% (2/9) and 94.9% (93/98), respectively.
Materials and Methods
Plasma specimens of 107 lung adenocarcinoma patients and their matched tumor formalin fixed paraffin embedded (FFPE) samples were analyzed. CastPCR was used to detect EGFR (c.2235_2249del, c.2236_2250del, c.2369C>T p.T790M, c.2573T>G p.L858R) and BRAF (c.1406G>C p.G469A, c.1799T>A p.V600E, c.1781A>G p.D594G) mutations. Mutation results of tumor tissue was set as gold standard, and the sensitivity, specificity, concordance, positive predictive value (PPV) and negative predictive value (NPV) were calculated for each mutation.
Conclusions
For patients whose tumor tissue is unavailable or inadequate, EGFR mutation detection in cfDNA with CastPCR could be first choice. Mutation positive results may provide reference for further clinical medication. While negative results indicate that detection in tissue should be considered as the following step. In this way, tumor tissue could be economized to the maximum extent and the risk of repeated percutaneous transthoracic lung biopsy could also be lowered to the maximum extent. For BRAF detection in cfDNA, CastPCR is a specific method while the sensitivity needs further exploration.
Keywords: NSCLC, cfDNA, CastPCR, driver mutation, EGFR
INTRODUCTION
Thanks to EGFR tyrosine kinase inhibitors (EGFR-TKIs), the treatment pattern of metastatic NSCLC has been revolutionized. Patients' progress free survival, overall response rate and quality of life have been ameliorated significantly [1–7]. However, in clinical practice, detection of driver mutations in tumor tissue is often not enough. First, sufficient tumor tissue is not readily available. For instance, only 35.9% (437/1217) and 20.3% (297/1466) of the advanced NSCLC patients had biopsied tissue that was suitable for testing in IPASS study [1] and INTEREST study [8], respectively. Second, due to heterogeneity, landscape of molecular information can't be covered with just one or two spots of biopsy [9]. Third, since tumor is evolving under drug therapy, dynamic and real-time monitoring of driver mutations during treatment is required. However, repeated biopsy is not practically feasible in clinical work for its invasiveness and risk. Fortunately, liquid biopsy such as cfDNA from plasma [10–12] has come into the picture. cfDNA originates from apoptosis, necrosis, phagocytosis, oncosis and active secretion of cells [13]. Studies have demonstrated that in cancer patients, cfDNA contains representation of the entire tumor genome [14, 15]. The mutation load was reported higher than 25.0% in one third of mutant plasma samples [13]. There have been multiple publications on cfDNA mutation detection in the last two decades [16–25]. According to Committee for Medicinal Products for Human Use (CHMP) of the European Medicines Agency (EMA), to predict therapeutic response of Iressa® (gefitinib), the use of circulating tumor DNA obtained from a blood sample has been allowed to assess EGFR mutation status in patients where a tumor sample is not an option[13]. Additionally, cfDNA was shown to be useful in dynamic monitoring of acquired resistance and prediction of relapse in various cancers, including NSCLC [26–30]. In clinical practice, methods with high sensitivity, specificity, convenience and economy are required. Development of molecular detection methods such as qPCR based methods, would allow for highly specific analysis of cfDNA [31]. Here, we evaluated the use of CastPCR detecting EGFR mutations (c.2235_2249del, c.2236_2250del, p.T790M, and p.L858R) in cfDNA form plasma in adenocarcinoma patients.
Reported incidence of BRAF mutations ranges from 0.5–9.0% in NSCLC [32]. Phase II clinical trial NCT01336634 has made its results public: BRAF targeted inhibitor dabrafenib has shown clinical activity in BRAF p.V600E-positive metastatic NSCLC. It suggested that for patients with limited therapeutic options, dabrafenib could represent a treatment option [33]. Detection of BRAF mutations in cfDNA hasn't been reported in NSCLC, thus we also evaluated CastPCR technology in detecting BRAF mutations (p.G469A, p.V600E and p.D594G).
RESULTS
EGFR mutations
Of the 107 plasma specimens, 31.8% (34/107) cases were EGFR mutated, including 76.5% (26/34) sensitive mutations (c.2235_2249del/c.2236_2250del or/and p.L858R) and 26.5% (9/34) resistant mutations (p.T790M). A 19 + 20 double mutation (two different mutations found simultaneously in one patient) was found in one patient (0.9%, 1/107). In FFPE samples, 51.4% (55/107) cases were EGFR mutated: 72.3% (40/55) patients harbored sensitive mutations and 36.4%, (20/55) harbored resistant mutations (Tables 1, 2). In addition, 5.6% (6/107) mutation-positive patients were double-mutated: 3 with c.2235_2249del/c.2236_2250del + p.T790M, 1 with c.2235_2249del/c.2236_2250del + p.L858R and 2 with p.T790M + p.L858R. Sensitivity, specificity, concordance, PPV and NPV of CastPCR detecting EGFR mutations in cfDNA are listed in Tables 3, 4.
Table 1. EGFR sensitive mutations (c.2235_2249del/c.2236_2250del or/and p.L858R) in cfDNA and FFPE samples.
| FFPE | Mutation (−) | Mutation (+) | Total |
|---|---|---|---|
| cfDNA | |||
| Mutation (+) | 3 | 23 | 26 |
| Mutation (−) | 64 | 17 | 81 |
| Total | 67 | 40 | 107 |
Table 2. EGFR resistant mutations (p.T790M) in cfDNA and FFPE samples.
| FFPE | T790M (+) | T790M (−) | Total |
|---|---|---|---|
| cfDNA | |||
| T790M (+) | 9 | 0 | 9 |
| T790M (−) | 11 | 87 | 98 |
| Total | 20 | 87 | 107 |
Table 3. Parameters of CastPCR detecting EGFR and BRAF mutations in cfDNA#.
| Parameters | EGFR | BRAF |
|---|---|---|
| Sensitivity | 56.4%(31/55) | 28.6% (2/7) |
| Specificity | 94.2%(49/52) | 93.0% (93/100) |
| Concordance | 74.8%(80/107) | 88.8% (95/107) |
| PPV | 91.2% (31/34) | 22.2% (2/9) |
| NPV | 67.1% (49/73) | 94.9% (93/98) |
# Mutation results of FFPE was set the gold standard
PPV: Positive Predictive Value; NPV: Negative Predictive Value.
Table 4. Parameters of CastPCR detecting EGFR mutations in cfDNA#.
| Parameters | Del19 | L858R | Del19 L858R | T790M |
|---|---|---|---|---|
| Sensitivity | 66.7% (14/21) | 45.0% (9/20) | 57.5% (23/40) | 45% (9/20) |
| Specificity | 96.5% (83/86) | 100.0% (87/87) | 95.5% (64/67) | 100%(87/87) |
| Concordance | 90.7% (97/107) | 89.7% (96/107) | 81.3% (87/107) | 89.7%(96/107) |
| PPV | 82.4% (14/17) | 100.9% (9/9) | 88.5% (23/26) | 100.0%(9/9) |
| NPV | 92.2% (83/90) | 88.8% (87/98) | 79.0% (64/81) | 88.8%(87/98) |
#Mutation results of FFPE was set the gold standard
PPV: Positive Predictive Value; NPV: Negative Predictive Value.
BRAF mutations
In 107 specimens, 8.4% (9/107) were BRAF mutated. p.V600E accounted for 33.3% (3/9) and non-p.V600E accounted for 88.9% (8/9). Notably, 4 patients had complex mutations: 1 with p.D594G + p.V600E, 1 with p.G469A + p.D594G + p.V600E and 2 with p.G469A + p.D594G mutations. While in FFPE samples, 6.5% (7/107) patients were identified with BRAF mutations. 14.3% (1/7) was p.V600E and 85.7% (6/7) were non-p.V600E. Among the 6 non-p.V600E patients, p.D594G accounted for 83.3%(5/6) and p.G469A accounted for 16.7% (1/10). (Tables 4, 5) Sensitivity, specificity, concordance, PPV and NPV were 28.6% (2/7), 93.0% (93/100), 88.8% (95/107), 22.2% (2/9) and 94.9% (93/98), respectively.
Table 5. All the cases harboring BRAF mutations.
| Number | cfDNA | FFPE | Distant Metastasis |
|---|---|---|---|
| 1 | V600E | V600E | No |
| 2 | G469A | G469A | No |
| 8 | G469A | No | No |
| 9 | G469A | No | Pleura, Bone |
| 10 | G469A + D594G | No | Pleura, Lungs, Axillary Nodes |
| 11 | G469A + D594G | No | No |
| 12 | G469A + D594G + V600E | No | Lungs, Bone |
| 13 | D594G + V600E | No | No |
| 14 | D594G | No | Bone |
| 3 | No | D594G | No |
| 4 | No | D594G | No |
| 5 | No | D594G | Bone |
| 6 | No | D594G | Pleura |
| 7 | No | D594G | Pleura, Bone, Lung |
DISCUSSION
As the flourishing development of precision medicine in lung cancer, there is a pressing need for real-time information of clinical specimens, as well as the establishment of a detection platform with convenience, efficiency and economy. With its relative non-invasiveness, high repeatability and real-time dynamic, liquid biopsy is breaking new ground. For the first time, we evaluated the feasibility of CastPCR detecting EGFR and BRAF mutations in plasma cfDNA of a cohort lung adenocarcinoma patients.
Studies employing different methods detecting EGFR sensitive mutations in paired NSCLC tumor tissue and plasma were listed in Table 6. Though Amplification refractory mutation system (ARMS) is extensively adopted to detect tissue EGFR mutations in clinical practice, its sensitivity in cfDNA is not stable [34–36]. For denaturing high performance liquid chromatography (DHPLC), its complex detection procedures hinders its clinical promotion [10, 37, 38]. Several targeted or sequencing methods have shown good sensitivity (78.0–81.1%) and concordance (86.0–93.6%) on EGFR mutation detection, such as droplet digital PCR (ddPCR) and next-generation sequencing (NGS) [21]. Compared with their prohibitive cost, complex laboratory manipulations and high demand for operational platform, CastPCR is more feasible and economical.
Table 6. Studies evaluating cfDNA EGFR sensitive mutations in patients with NSCLC.
| Study | Detection Method | Mutated patients (n) | Sensitivity (%) (n/total) | Specificity (%) (n/total) | Concordance (%) (n/total) |
|---|---|---|---|---|---|
| Bai et al. [10] | DHPLC | 77 | 81.8 (63/77) | 89.5 (137/153) | 74.0 (200/230) |
| Huang et al. [53] | DHPLC | 296 | 63.5 (188/296) | 84.6 (445/526) | 77.0 (633/822) |
| Liu et al. [35] | ARMS | 40 | 67.5 (27/40) | 100.0 (46/46) | 84.9 (73/86) |
| Kim et al. [54] | PNA-PCR | 35 | 17.1 (6/35) | 100.0 (5/5) | 27.5 (11/40) |
| Zhao et al. [55] | Mutant-enriched PCR | 45 | 35.6 (16/45) | 95.5 (63/66) | 71.2 (79/111) |
| Wang et al. [36] | ARMS | 68 | 22.1 (15/68) | 97.0 (64/66) | 59.0 (79/134) |
| Jing et al. [56] | HRMA | 45 | 66.4 (29/45) | 97.3 (73/75) | 85.0 (102/120) |
| Weber et al. [57] | Cobas EGFR test | 28 | 60.7 (17/28) | 96.4 (162/168) | 91.3 (179/196) |
| Yu et al. [58] | ddPCR | 93 | 61.3 (57/93) | 96.7 (118/122) | 81.4 (175/215) |
| Zhu et al. [59] | ddPCR | 37 | 81.1 (30/37) | 97.0 (131/135) | 93.6 (161/172) |
Abbreviations: DHPLC, denaturing high-performance liquid chromatography; ARMS, amplification refractory mutation system; PNA, peptide nucleic acid; PCR, polymerase chain reaction; HRMA, high resolution melting analysis.
EGFR T790M plays a key role in selecting the right patient for third-generation EGFR-TKI [39]. Up to date, U.S. Food and Drug Administration approved Cobas® as the only assay to detect EGFR specific mutations (i.e., exon 19 deletions and exon 21 L858R substitution mutations) in plasma specimens (www.fda.gov). In recent studies that adopted Cobas® testing platform for p. T790M mutation detection, the sensitivity was (51.0%) [40]. In our study, sensitivity detecting p.T790M was 45.0% (9/20). Notably, the specificity and concordance of p.T790M detection was 100.0% (87/87) and 89.7% (96/107), respectively. While in Cobas® the data presented as 77.0% and 61.0%, respectively [40]. Since the PPV and NPV for p.T790M was 100.0% (9/9) and 88.8% (87/98) in our study, for patients who acquire drug resistance to first generation EGFR-TKI, plasma detection of EGFR p.T790M by CastPCR could be first consideration. For those with cfDNA tested positive, third generation EGFR-TKI could be a treatment choice. Similarly, considering the PPV (91.2%, 31/34) and NPV (67.1%, 49/73) of EGFR detection with CastPCR in our study, for patients whose tumor tissue is not enough or unavailable, EGFR mutation detection with CastPCR in cfDNA could be first choice. For those with EGFR mutations in cfDNA, the result may be taken as reference for further clinical medication. While for those detected without EGFR mutations in cfDNA, further detection in tumor tissue is recommended. In this way, tumor tissue could be economized to the maximum extent and the risk of repeated percutaneous transthoracic lung biopsy could also be lowered to the maximum extent.
BRAF is another important drugable driver mutation in NSCLC. Compared with EGFR, the detection of BRAF mutation both in tumor tissue and plasma still has a long way to go. To the best of our knowledge, BRAF detection of cfDNA in NSCLC hasn't been reported yet. We performed BRAF mutation detection in 107 pairs of lung adenocarcinoma plasma cfDNA and matched FFPE DNA samples. CastPCR was applied to detect BRAF mutations in tumor tissues and 6.5% (7/107) patients were identified as mutation positive, consistent to the result concluded by NGS [32, 41]. Interestingly, p.V600E accounted for about half BRAF mutations in NSCLC in earlier studies [42–46], and recent studies employing NGS technology exhibited that non-p.V600E mutations is the majority (70.0–80.0%) of BRAF mutations in NSCLC [32, 41, 47]. In our study, the frequency of non-p.V600E was 85.7% (6/7), similar to that demonstrated by studies using NGS.
In BRAF mutation detection of cfDNA with CastPCR, the sensitivity, specificity and concordance was 28.6% (2/7), 93.0% (93/100) and 88.8% (95/107), respectively (Table 3). The low sensitivity may mainly related to the following reasons: Firstly, tumor heterogeneity. In the 12 patients whose mutation conditions were inconsistent, 58.3% (7/12) had distant metastasis. Thus their cfDNA released in plasma could probably carry different gene information compared with primary tumors. Secondly, detection of BRAF non-p.V600E mutations in exon 15 of cfDNA needs more investigation. In our study, sensitivity for p.V600E (locates on exon15) and p.G469A (locates on exon11) both reached 100.0%, while none of the five p.D594Gs (locate on exon15) in FFPE samples were detected in their matched plasma. Similarly, Couraud et al. [48] applied NGS to detect mutations of exon 11 and 15 in about 60 tumors and corresponding plasma, and better result was produced for mutations of exon 11 mutations than that of exon 15.
Complex BRAF mutations have been reported in different solid tumors. For example, c.1801A>G (p.K601E) + c.1796C>G (p.T599I) in thyroid carcinoma [50], p.T599I + c.1798_1799delinsAA (p.V600K) and c.1798A>T (p.T599T) + p.V600E + c.1803A>T (p.K601N) in melanoma [50, 51]. And we firstly found complex BRAF mutations in plasma cfDNA of lung adenocarcinoma: two with p.G469A + p.D594G, one with p.D594G + p.V600E and one with p.G469A + p.D594G + p.V600E. The meaning of these complex mutations still needs to be excavated in the future.
MATERIALS AND METHODS
Materials
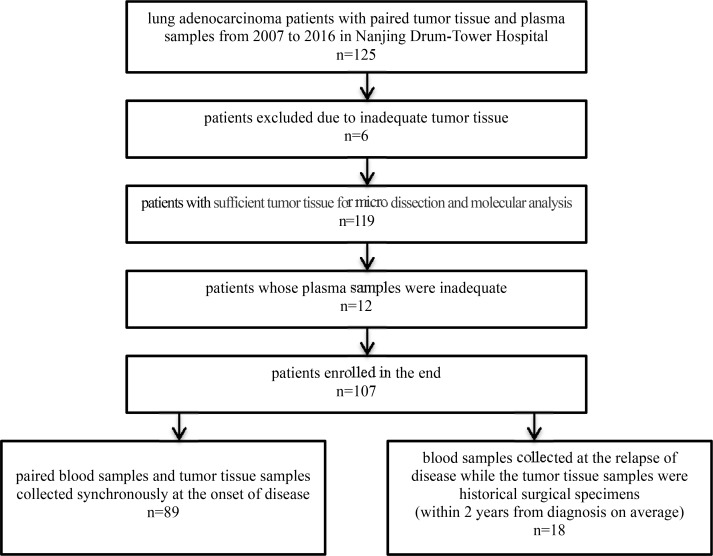
All samples used in this study were obtained from Nanjing Drum-Tower Hospital form 2007 to 2016. 125 lung adenocarcinoma patients were enrolled, and data of 107 paired samples was analyzed in the end (Figure 1). Patients' clinicopathological features are provided in Table 7. 83.2% (89/107) blood samples was collected when patients were diagnosed, together with their paired tumor tissue. 16.8% (18/107) blood samples was collected right after the recurrence, while tumor samples came from the historical surgical specimens (duration of diagnosis to recurrence ranges from 1 to 46 months, medium 18 months), and 61.1% (11/18) of them had experienced chemotherapy after surgery.
Figure 1. Workflow of patients enrollment.
Table 7. Clinicopathological features of the 107 lung adenocarcinoma patients.
| Clinicopathological Features | Number of Cases | % |
|---|---|---|
| Gender | ||
| Male | 60 | 56.1 |
| Female | 47 | 43.9 |
| Age (years) | ||
| ≥ 60 60 | 53 | 49.5 |
| < 60 | 54 | 50.5 |
| Smoking History | ||
| Smokers# | 47 | 43.9 |
| Non-smokers | 60 | 56.1 |
| Distant Metastasis | ||
| Yes | 62 | 57.9 |
| No | 45 | 42.1 |
| Differentiation Degree | ||
| Well | 9 | 8.4 |
| Medium-Poor | 98 | 91.6 |
| Clinical Stages | ||
| I–III | 42 | 39.3 |
| IV | 65 | 60.7 |
| Tumor Size (cm)* | ||
| ≤ 3 | 16 | 38.1 |
| ≥ 3 | 26 | 61.9 |
| Lymph Node Metastasis* | ||
| Yes | 31 | 73.8 |
| No | 11 | 26.2 |
#including 6 former smokers (defined as those had quit smoking ≥ 1 year) [45].
*42 patients that underwent surgical resection were analyzed.
FFPE samples were cut into slices and each slice was 4μm thick. For every FFPE sample, a random slice was performed with the HE staining, then two pathologists identified and marked the tumor tissue through the observation of microscope. 1–2 mL peripheral blood was collected into an EDTA-containing tube, centrifuged at 3000 rpm/min for 15 min at room temperature, then the plasma portion was pipetted carefully, aliquoted to 0.5 mL Eppendorf tubes and stored at −80°C. All patients enrolled in the study provided written informed consent for the use of their resected/biopsied tumor tissue and peripheral blood. The study was approved by the Ethics Committee of Nanjing Drum-Tower Hospital.
DNA extraction
DNA of FFPE samples was extracted using TIANamp FFPE DNA Kit (Tiangen Biotech, Beijing) according to manufacturer's instructions. For DNA extraction of cfDNA, after comparing the reference Ct value of DNA extracted with MinElute Virus Spin Kit (QIAamp, Germany) and phenol-chloroform method in our preliminary experiment, we adopted the latter method (data not shown). NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA) was used to quantify cfDNA.
CastPCR
CastPCR technology combines allele specific TaqMan® qPCR with allele-specific blocker (ASB) oligonucleotides that effectively suppress nonspecific amplification from the off-target allele. In our preliminary experiment, we assessed the limit of detection (LOD, minimum percentage of mutant alleles in a wild type background required for reliable mutation detection) of some of the assays with corresponding mutated cell lines. It turned out to be that the sensitivity was consistent to that provided by the manufacturer: 0.1% for EGFR c.2235_2249del, p.L858R and BRAF p.V600E, 1.0% for EGFR p.T790M. Due to unavailable corresponding cell lines, LOD of EGFR c.2236_2250del, BRAF p.G469A and p.D594G was not assessed. Concentration of extracted DNA was determined by a NanoDrop spectrophotometer (NanoDrop Technologies, Inc., Thermo Fisher Scientific, Wilmington, DE, USA), and adjusted to a concentration of 10 ng/μL. PCR mutation detection assays were then conducted with 2μL of each DNA sample. Every DNA sample was analyzed by CastPCR using the EGFR_6223_mu, EGFR_6225_mu, EGFR_6224_mu, EGFR_6240_mu, BRAF_460_mu, BRAF_467_mu and BRAF_460_mu assays for the detection of c.2235_2249del, c.2236_2250del, p.L858R, p.T790M p.G469A, p.D594G and p.V600E, respectively (Life Technologies, USA). The threshold cycle was set as 0.2 according to the handbook of Taqman Mutation Detection Assay by CastPCR. CastPCR was run in a final volume of 10μL in a 96 well plate including 5μL TaqMan® Genotyping Master Mix (2X), 2μL Prepared gDNA sample, 2μL Nuclease-free water and 1μL TaqMan® Mutation Detection Assay(10X) (mutant allele or gene reference assay). Positive control was set for each sample detecting EGFR c.2235_2249del, p.T790M, p.L858R and BRAF p.V600E. Negative and blank control was set for every assay. Three replicates were set for each assay. PCR were performed on Stratagene MX3000P real-time PCR system (Stratagene, USA). PCR conditions: 95°C for 10 minutes, followed by 5 cycles of 92°C for 15 seconds and 58°C for 1 minute and then 40 cycles of 92°C for 15 seconds and 60°C for 1 minute. According to the handbook of Taqman Mutation Detection Assay by CastPCR, reference Ct value was required between 17 and 27, and samples with a ΔCt of less than 9.96 (9.61 for EGFR p.T790M) were considered positive for mutation, where ΔCt = Ct mut - Ct ref. Otherwise the sample was defined as mutation negative. Detection results of tumor tissue was set the gold standard.
Statistical analysis
Sensitivity, specificity, concordance, PPV and NPV were calculated as follows [52]: Sensitivity = number of true positives/(number of true positives + number of false negatives); Specificity = number of true negatives/ (number of true negatives + number of false positives); Concordance = (number of true positives + number of true negatives) / (number of total cases); PPV = number of true positives/(number of true positives + number of false positives); NPV = number of true negatives/(number of true negatives + number of false negatives).
CONCLUSIONS
For the first time, we evaluated the possibility of CastPCR detecting EGFR and BRAF mutations in cfDNA of plasma from 107 lung adenocarcinoma patients. For patients whose tumor tissue is not enough or unavailable, EGFR mutation detection with CastPCR in cfDNA could be first choice. For those with EGFR mutations in cfDNA, the result may be taken as reference for further clinical medication. While for those detected without EGFR mutations in cfDNA, further detection in tumor tissue is recommended. Additionally, CastPCR technology was firstly employed to detect BRAF mutations in plasma specimens of lung adenocarcinoma and their matched FFPE samples. We found that for BRAF mutation detection, CastPCR is a specific method while its sensitivity, especially for BRAF non-p.V600E mutations on exon 15, needs further exploration.
ACKNOWLEDGMENTS AND FUNDING
We would like to thank all the stuff of the comprehensive cancer center of Drum Tower Hospital.
Footnotes
CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
REFERENCES
- 1.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang JJ, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 2.Kwong A, Shin VY, Au CH, Law FB, Ho DN, Ip BK, Wong AT, Lau SS, To RM, Choy G, Ford JM, Ma ES, Chan TL. Abstract P2-09-20: Evaluation on the mutation screening by next-generation sequencing in hereditary breast and ovarian cancer: Implementation of recurrent mutation panel. Cancer Res. 2016 doi: 10.1016/j.jmoldx.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, Fujita Y, Okinaga S, Hirano H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 4.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R, Pallares C, Sanchez JM, Porta R, Cobo M, Garrido P, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 5.Chung C. Tyrosine kinase inhibitors for epidermal growth factor receptor gene mutation-positive non-small cell lung cancers: an update for recent advances in therapeutics. J Oncol Pharm Pract. 2016;22:461–476. doi: 10.1177/1078155215577810. [DOI] [PubMed] [Google Scholar]
- 6.Solomon BJ, Mok T, Kim DW, Wu YL, Nakagawa K, Mekhail T, Felip E, Cappuzzo F, Paolini J, Usari T, Iyer S, Reisman A, Wilner KD, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014;371:2167–2177. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 7.Shaw AT, Kim DW, Nakagawa K, Seto T, Crino L, Ahn MJ, De Pas T, Besse B, Solomon BJ, Blackhall F, Wu YL, Thomas M, O'Byrne KJ, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 8.Kim ES, Hirsh V, Mok T, Socinski MA, Gervais R, Wu YL, Li LY, Watkins CL, Sellers MV, Lowe ES, Sun Y, Liao ML, Osterlind K, et al. Gefitinib versus docetaxel in previously treated non-small-cell lung cancer (INTEREST): a randomised phase III trial. Lancet. 2008;372:1809–1818. doi: 10.1016/S0140-6736(08)61758-4. [DOI] [PubMed] [Google Scholar]
- 9.de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, Jamal-Hanjani M, Shafi S, Murugaesu N, Rowan AJ, Gronroos E, Muhammad MA, Horswell S, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bai H, Mao L, Wang HS, Zhao J, Yang L, An TT, Wang X, Duan CJ, Wu NM, Guo ZQ, Liu YX, Liu HN, Wang YY, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol. 2009;27:2653–2659. doi: 10.1200/JCO.2008.17.3930. [DOI] [PubMed] [Google Scholar]
- 11.Yung TK, Chan KC, Mok TS, Tong J, To KF, Lo YM. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res. 2009;15:2076–2084. doi: 10.1158/1078-0432.CCR-08-2622. [DOI] [PubMed] [Google Scholar]
- 12.Brevet M, Johnson ML, Azzoli CG, Ladanyi M. Detection of EGFR mutations in plasma DNA from lung cancer patients by mass spectrometry genotyping is predictive of tumor EGFR status and response to EGFR inhibitors. Lung Cancer. 2011;73:96–102. doi: 10.1016/j.lungcan.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016;35:347–376. doi: 10.1007/s10555-016-9629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan KC, Jiang P, Zheng YW, Liao GJ, Sun H, Wong J, Siu SS, Chan WC, Chan SL, Chan AT. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem. 2013;59:211–224. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- 15.Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, O'shaughnessy J, Kinzler KW, Parmigiani G, Vogelstein B. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012;4:162ra154. doi: 10.1126/scitranslmed.3004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taly V, Pekin D, Benhaim L, Kotsopoulos SK, Le CD, Li X, Atochin I, Link DR, Griffiths AD, Pallier K. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem. 2013;59:1722. doi: 10.1373/clinchem.2013.206359. [DOI] [PubMed] [Google Scholar]
- 17.Higgins MJ, Jelovac D, Barnathan E, Blair B, Slater S, Powers P, Zorzi J, Jeter SC, Oliver GR, Fetting J. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin Cancer Res. 2012;18:3462–3469. doi: 10.1158/1078-0432.CCR-11-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spindler KL, Pallisgaard N, Vogelius I, Jakobsen A. Quantitative cell-free DNA, KRAS, and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin Cancer Res. 2012;18:1177–1185. doi: 10.1158/1078-0432.CCR-11-0564. [DOI] [PubMed] [Google Scholar]
- 19.Dawson SJ, Rosenfeld N, Caldas C. Circulating tumor DNA to monitor metastatic breast cancer. Breast Diseases A Year Book Quarterly. 2013;24:350–352. doi: 10.1056/NEJMc1306040. [DOI] [PubMed] [Google Scholar]
- 20.Dowler NA, Spindler KL, Pallisgaard N, Andersen RF, Jakobsen A. Levels of cell-free DNA and plasma KRAS during treatment of advanced NSCLC. Oncol Rep. 2014;31:969–974. doi: 10.3892/or.2013.2906. [DOI] [PubMed] [Google Scholar]
- 21.Kukita Y, Uchida J, Oba S, Nishino K, Kumagai T, Taniguchi K, Okuyama T, Imamura F, Kato K. Quantitative identification of mutant alleles derived from lung cancer in plasma cell-free DNA via anomaly detection using deep sequencing data. Plos One. 2013;8:e81468. doi: 10.1371/journal.pone.0081468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mouliere F, El Massoudi S, Gongora C, Guedj AS, Robert B, Del Rio M, Molina F, Lamy PJ, Lopez-Crapez E, Mathonnet M, Ychou M, Pezet D, Thierry AR. Circulating Cell-Free DNA from Colorectal Cancer Patients May Reveal High KRAS or BRAF Mutation Load. Transl Oncol. 2013;6:319. doi: 10.1593/tlo.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perkins G, Yap TA, Pope L, Cassidy AM, Dukes JP, Riisnaes R, Massard C, Cassier PA, Miranda S, Clark J. Multi-purpose utility of circulating plasma DNA testing in patients with advanced cancers. Plos One. 2012;7:e47020. doi: 10.1371/journal.pone.0047020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spindler KG, Appelt AL, Pallisgaard N, Andersen RF, Jakobsen A. KRAS-mutated plasma DNA as predictor of outcome from irinotecan monotherapy in metastatic colorectal cancer. Brit J Cancer. 2013;109:3067–3072. doi: 10.1038/bjc.2013.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spindler KL, Sorensen MM, Pallisgaard N, Andersen RF, Havelund BM, Ploen J, Lassen U, Jakobsen AK. Phase II trial of temsirolimus alone and in combination with irinotecan for KRAS mutant metastatic colorectal cancer: outcome and results of KRAS mutational analysis in plasma. Acta Oncologica. 2013;52:963. doi: 10.3109/0284186X.2013.776175. [DOI] [PubMed] [Google Scholar]
- 26.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, Rajan S, Humphray S, Becq J, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 28.Rago C, Huso DL, Diehl F, Karim B, Liu G, Papadopoulos N, Samuels Y, Velculescu VE, Vogelstein B, Kinzler KW. Serial assessment of human tumor burdens in mice by the analysis of circulating DNA. Cancer Res. 2007;67:9364–9370. doi: 10.1158/0008-5472.CAN-07-0605. [DOI] [PubMed] [Google Scholar]
- 29.Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong AS. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 30.Zheng D, Ye X, Sun Y, Zhang M, Wang J, Zhang J, Ni J, Zhang L, Zhang H, Luo J. Noninvasive monitoring of acquired EGFR-T790M mutation and discovery of its heterogeneity in patients with advanced NSCLC treated with EGFR-TKI. Asco Meeting Abstract. 2014 [Google Scholar]
- 31.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carter J, Tseng LH, Zheng G, Dudley J, Illei P, Gocke CD, Eshleman JR, Lin MT. Non-p.V600E BRAF Mutations Are Common Using a More Sensitive and Broad Detection Tool. Am J Clin Pathol. 2015;144:620–628. doi: 10.1309/AJCP85ATMJOZOUDJ. [DOI] [PubMed] [Google Scholar]
- 33.Planchard D, Kim TM, Mazieres J, Quoix E, Riely G, Barlesi F, Souquet PJ, Smit EF, Groen HJ, Kelly RJ, Cho BC, Socinski MA, Pandite L, et al. Dabrafenib in patients with BRAF-positive advanced non-small-cell lung cancer: a single-arm, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016 doi: 10.1016/S1470-2045(16)00077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goto K, Ichinose Y, Ohe Y, Yamamoto N, Negoro S, Nishio K, Itoh Y, Jiang H, Duffield E, McCormack R, Saijo N, Mok T, Fukuoka M. Epidermal growth factor receptor mutation status in circulating free DNA in serum: from IPASS, a phase III study of gefitinib or carboplatin/paclitaxel in non-small cell lung cancer. J Thorac Oncol. 2012;7:115–121. doi: 10.1097/JTO.0b013e3182307f98. [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Lu Y, Zhu G, Lei Y, Zheng L, Qin H, Tang C, Ellison G, Mccormack R, Ji Q. The diagnostic accuracy of pleural effusion and plasma samples versus tumour tissue for detection of EGFR mutation in patients with advanced non-small cell lung cancer: comparison of methodologies. J Clin Pathol. 2013;66:1065–1069. doi: 10.1136/jclinpath-2013-201728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang S, Han X, Hu X, Wang X, Zhao L, Tang L, Feng Y, Wu D, Sun Y, Shi Y. Clinical significance of pretreatment plasma biomarkers in advanced non-small cell lung cancer patients. Clinica chimica acta. 2014;430:63. doi: 10.1016/j.cca.2013.12.026. [DOI] [PubMed] [Google Scholar]
- 37.Bai H, Wang Z, Wang Y, Zhuo M, Zhou Q, Duan J, Yang L, Wu M, An T, Zhao J, Wang J. Detection and clinical significance of intratumoral EGFR mutational heterogeneity in Chinese patients with advanced non-small cell lung cancer. PLoS One. 2013;8:e54170. doi: 10.1371/journal.pone.0054170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Z, Wang Z, Bai H, Wu M, An T, Zhao J, Yang L, Duan J, Zhuo M, Wang Y. The detection of EGFR mutation status in plasma is reproducible and can dynamically predict the efficacy of EGFR-TKI. Thoracic Cancer. 2012;3:334–340. doi: 10.1111/j.1759-7714.2012.00133.x. [DOI] [PubMed] [Google Scholar]
- 39.McCoach CE, Jimeno A. Osimertinib, a third-generation tyrosine kinase inhibitor targeting non-small cell lung cancer with EGFR T790M mutations. Drugs Today (Barc) 2016;52:561–568. doi: 10.1358/dot.2016.52.10.2541343. [DOI] [PubMed] [Google Scholar]
- 40.Wu YL, Jenkins S, Ramalingam S, Han JY, Delmonte A, Hsia TC, Laskin J, Kim SW, He Y, Patel S, Hodge R, Marotti M, Papadimitrakopoulou V, et al. MA08.03 Osimertinib vs Platinum-Pemetrexed for T790M-Mutation Positive Advanced NSCLC (AURA3): Plasma ctDNA Analysis. J Thorac Oncol. 12:S386. [Google Scholar]
- 41.Zheng G, Tseng LH, Chen G, Haley L, Illei P, Gocke CD, Eshleman JR, Lin MT. Clinical detection and categorization of uncommon and concomitant mutations involving BRAF. BMC cancer. 2015;15:779. doi: 10.1186/s12885-015-1811-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cardarella S, Ogino A, Nishino M, Butaney M, Shen J, Lydon C, Yeap BY, Sholl LM, Johnson BE, Janne PA. Clinical, pathologic, and biologic features associated with BRAF mutations in non-small cell lung cancer. Clin Cancer Res. 2013;19:4532–4540. doi: 10.1158/1078-0432.CCR-13-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmid K, Oehl N, Wrba F, Pirker R, Pirker C, Filipits M. EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin Cancer Res. 2009;15:4554–4560. doi: 10.1158/1078-0432.CCR-09-0089. [DOI] [PubMed] [Google Scholar]
- 44.An SJ, Chen ZH, Su J, Zhang XC, Zhong WZ, Yang JJ, Zhou Q, Yang XN, Huang L, Guan JL, Nie Q, Yan HH, Mok TS, et al. Identification of enriched driver gene alterations in subgroups of non-small cell lung cancer patients based on histology and smoking status. PLoS One. 2012;7:e40109. doi: 10.1371/journal.pone.0040109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marchetti A, Felicioni L, Malatesta S, Grazia Sciarrotta M, Guetti L, Chella A, Viola P, Pullara C, Mucilli F, Buttitta F. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol. 2011;29:3574–3579. doi: 10.1200/JCO.2011.35.9638. [DOI] [PubMed] [Google Scholar]
- 46.Sasaki H, Shitara M, Yokota K, Okuda K, Hikosaka Y, Moriyama S, Yano M, Fujii Y. Braf and erbB2 mutations correlate with smoking status in lung cancer patients. Exp Ther Med. 2012;3:771–775. doi: 10.3892/etm.2012.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kinno T, Tsuta K, Shiraishi K, Mizukami T, Suzuki M, Yoshida A, Suzuki K, Asamura H, Furuta K, Kohno T, Kushima R. Clinicopathological features of nonsmall cell lung carcinomas with BRAF mutations. Annals of oncology. 2014;25:138–142. doi: 10.1093/annonc/mdt495. [DOI] [PubMed] [Google Scholar]
- 48.Couraud S, Vaca-Paniagua F, Villar S, Oliver J, Schuster T, Blanche H, Girard N, Tredaniel J, Guilleminault L, Gervais R, Prim N, Vincent M, Margery J, et al. Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: a proof-of-concept study from BioCAST/IFCT-1002. Clin Cancer Res. 2014;20:4613–4624. doi: 10.1158/1078-0432.CCR-13-3063. [DOI] [PubMed] [Google Scholar]
- 49.Afkhami M, Karunamurthy A, Chiosea S, Nikiforova MN, Seethala R, Nikiforov YE, Coyne C. Histopathologic and Clinical Characterization of Thyroid Tumors Carrying the BRAF(K601E) Mutation. Thyroid. 2016;26:242–247. doi: 10.1089/thy.2015.0227. [DOI] [PubMed] [Google Scholar]
- 50.Zalaudek I, Ciarrocchi A, Piana S, Argenziano G, Torricelli F, Sancisi V, Gandolfi G, Longo C, Moscarella E, Banzi C, Nicoli D. A novel BRAF mutation in association with primary amelanotic melanoma with oral metastases. J Eur Acad Dermatol. 2015;29:387–390. doi: 10.1111/jdv.12358. [DOI] [PubMed] [Google Scholar]
- 51.Wilson MA, Morrissette JJ, McGettigan S, Roth D, Elder D, Schuchter LM, Daber RD. What you are missing could matter: a rare, complex BRAF mutation affecting codons 599, 600, and 601 uncovered by next generation sequencing. Cancer Genet. 2014;207:272–275. doi: 10.1016/j.cancergen.2014.06.022. [DOI] [PubMed] [Google Scholar]
- 52.Yang H, Cai L, Zhang Y, Tan H, Deng Q, Zhao M, Xu X. Sensitive detection of EGFR mutations in cerebrospinal fluid from lung adenocarcinoma patients with brain metastases. J Mol Diagn. 2014;16:558–563. doi: 10.1016/j.jmoldx.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 53.Huang Z, Wang Z, Bai H, Wu M, An T, Zhao J, Yang L, Duan J, Zhuo M, Wang Y. The detection of EGFR mutation status in plasma is reproducible and can dynamically predict the efficacy of EGFR-TKI. Thoracic Cancer. 2012;3:334–340. doi: 10.1111/j.1759-7714.2012.00133.x. [DOI] [PubMed] [Google Scholar]
- 54.Kim HR, Lee SY, Hyun DS, Lee MK, Lee HK, Choi CM, Yang SH, Kim YC, Lee YC, Kim SY. Detection of EGFR mutations in circulating free DNA by PNA-mediated PCR clamping. J Exp Clin Canc Res. 2013;32:50. doi: 10.1186/1756-9966-32-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao X, Han RB, Zhao J, Wang J, Yang F, Zhong W, Zhang L, Li LY, Wang MZ. Comparison of epidermal growth factor receptor mutation statuses in tissue and plasma in stage I–IV non-small cell lung cancer patients. Respiration. 2013;85:119. doi: 10.1159/000338790. [DOI] [PubMed] [Google Scholar]
- 56.Jing CW, Wang Z, Cao HX, Ma R, Wu JZ. High resolution melting analysis for epidermal growth factor receptor mutations in formalin-fixed paraffin-embedded tissue and plasma free DNA from non-small cell lung cancer patients. Asian Pac J Cancer P. 2014;14:6619. doi: 10.7314/apjcp.2013.14.11.6619. [DOI] [PubMed] [Google Scholar]
- 57.Weber B, Meldgaard P, Hager H, Wu L, Wei W, Tsai J, Khalil A, Nexo E, Sorensen BS. Detection of EGFR mutations in plasma and biopsies from non-small cell lung cancer patients by allele-specific PCR assays. BMC cancer. 2014;14:294. doi: 10.1186/1471-2407-14-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, Xu Y, Zhong W, Zhao J, Chen M, Zhang L, Li L, Wang M. Total DNA input is a crucial determinant of the sensitivity of plasma cell-free DNA EGFR mutation detection using droplet digital PCR. Oncotarget. 2017;8:5861–5873. doi: 10.18632/oncotarget.14390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu G, Ye X, Dong Z, Lu YC, Sun Y, Liu Y, McCormack R, Gu Y, Liu X. Highly Sensitive Droplet Digital PCR Method for Detection of EGFR-Activating Mutations in Plasma Cell-Free DNA from Patients with Advanced Non-Small Cell Lung Cancer. J Mol Diagn. 2015;17:265–272. doi: 10.1016/j.jmoldx.2015.01.004. [DOI] [PubMed] [Google Scholar]