Abstract
Single nucleotide polymorphisms (SNPs) of Excision repair cross-complementing group 2 (ERCC2) gene are suspected to affect the risk of pancreatic cancer. Many studies have reported the association between ERCC2 Lys751Gln polymorphism (rs13181) and the susceptibility to pancreatic cancer, but the outcomes remained controversial. To comprehensively determine this association, we conducted a meta-analysis based on a total of eight studies. Evidence for this association was obtained from the PubMed, EMBASE, Web of Science and Chinese National Knowledge Infrastructure (CNKI) databases. In general, a significant association was found between ERCC2 rs13181 polymorphism and the susceptibility to pancreatic cancer in four genetic models [CC vs. AA: OR = 1.56, (95% CI: 1.28-1.90), P = 0.470; AC/CC vs. AA: OR=1.20, (95% CI: 1.06-1.36), P = 0.396; CC vs. AC/CC: OR = 1.50; (95% CI: 1.24-1.81), P = 0.530; C vs. A: OR=1.22, (95%CI:1.11-1.34), P = 0.159]. Furthermore, stratified analyses by ethnicity indicated a significant association only in the Asian population. Our results indicate that the ERCC2 Lys751Gln polymorphism might be important in stimulating the development of pancreatic cancer, especially for Asians.
Keywords: pancreatic cancer, ERCC2, rs13181, polymorphism, meta-analysis
INTRODUCTION
As a highly lethal disease, Pancreatic cancer is correlated with a very poor prognosis, characterized by the close parallel between incidence and mortality [1]. In the United States, five-year survival rate in pancreatic cancer patients remains as low as 6% [2]. The low survival rate is attributed to several factors, of which perhaps the most important is the late stage at which most patients are diagnosed. However, there is still no standard program for screening patients at high risk of pancreatic cancer and the accurate genetic epidemiology of this cancer remains unknown. As a multi-factorial disease, many factors are known to play a key role in pancreatic cancer development, such as smoking, obesity, drinking, diabetes as well as environmental chemicals [3–5]. Nevertheless, even when individuals are exposed to similar risk factors, not all of them develop into pancreatic cancer, which indicate that hereditary factors might play an essential role in pancreatic carcinogenesis.
According to the genetic profiles of pancreatic cancer, genomic instability mediated by DNA repair deficiency is a vital event in development of pancreatic carcinoma. DNA repair machinery plays a crucial role in defending cells against environmental hazards like ionizing radiation, ultraviolet (UV) rays, diet and smoking. As a key DNA repair mechanism, Nucleotide excision repair (NER) can influence gene-gene rearrangement, deletion, translocation and amplification [6, 7]. Excision repair cross-complementation rodent repair deficiency group 2 (ERCC2), which locates on chromosome 19q13.3, is an important genetic complementation group encoding for proteins involved in the NER pathway and could reverse ionizing radiation-induced damage and DNA damage by chemotherapy [8, 9]. Polymorphism rs13181, located at position 751 in exon 23, is the most common polymorphism in the coding region of ERCC2 and characterized by an A > C substitution leading to a lysine (Lys) to glutamine (Gln) amino acid exchange [10].
There are many studies which focus on the relationship between this SNP and pancreatic cancer [11–18]. Jiao et al. (2007), Hocevar et al. (2014), Ying et al. (2015) and He et al. (2016) did not observe any significant association between rs13181 and the risk of pancreatic cancer [11–14]. However, McWilliams et al. (2008), Zhao et al. (2015), Yan et al. (2016) and Sileng et al. (2016) suggested that this polymorphism was associated with an increased susceptibility to pancreatic cancer [15–18]. Although these researches are all based on experiment results, their results are always inconsistent and the roles rs13181 plays in pancreatic cancer are still unclear. Therefore, there is a need to make it clear whether this polymorphism is associated with pancreatic cancer. In order to assess the real association, the latest and most convincing evidence was utilized in this meta-analysis. This is, to our knowledge, the first comprehensive meta-analysis concerning the association between ERCC2 rs13181 and the risk of pancreatic cancer.
RESULTS
Literature search and study characteristics
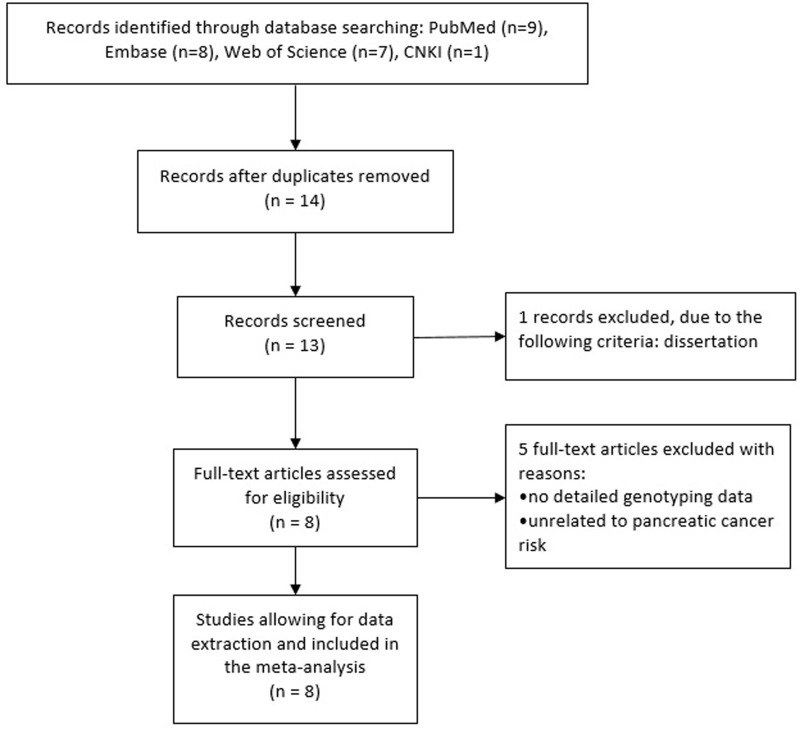
The flow diagram of study exclusion and inclusion with specific reasons is shown in Figure 1. We identified 14 records, among which 8 papers appeared to be eligible for inclusion and were retrieved in full texts [11–18]. Among the six excluded articles, one was dissertation, four were not association studies on the risk of pancreatic cancer and one lacked sufficient data for estimating an odds ratio (OR) with 95% CI. Eventually, a total of eight case-control studies (1,980 cases and 2,317 controls) were ultimately included in the meta-analysis, and the details of each study were recorded in Table 1. As a result, each group of them was considered separately for pooling stratified analysis. These studies were divided into two groups based on the ethnicity of study participants, i.e., studies involving Caucasian population (3 studies) and those involving Asian population (5 studies).
Figure 1. The flow diagram of retrieval for this study.
Table 1. Characteristics of the 8 studies included in the meta-analysis.
| Author | Year | Ethnicity | Source of controla | Case | Control | Susceptibilityb | P value of HWEc | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AC | CC | AA | AC | CC | ||||||
| He MG | 2016 | Asian | HB | 119 | 78 | 20 | 143 | 86 | 15 | N | 0.668 |
| Sileng A | 2016 | Asian | HB | 116 | 103 | 35 | 138 | 121 | 18 | Y | 0.209 |
| Yan D | 2016 | Asian | HB | 118 | 70 | 38 | 167 | 65 | 31 | Y | 0.000 |
| Ying MF | 2015 | Asian | PB | 113 | 56 | 26 | 159 | 70 | 25 | N | 0.000 |
| Zhao FL | 2015 | Asian | HB | 131 | 72 | 43 | 159 | 64 | 23 | Y | 0.000 |
| Hocevar AB | 2014 | Caucasian | HB | 15 | 11 | 5 | 21 | 16 | 3 | N | 0.984 |
| Mcwilliams RR | 2008 | Caucasian | PB | 186 | 211 | 76 | 241 | 291 | 79 | Y | 0.544 |
| Jiao L | 2007 | Caucasian | PB | 124 | 184 | 30 | 147 | 203 | 32 | N | 0.001 |
a HB, hospital-based studies; PB, population-based studies.
b “Y” indicates an association between the rs13181 polymorphism and risk of pancreatic cancer; “N” means no association between rs13181 and the risk of pancreatic cancer.
c HWE, Hardy–Weinberg equilibrium; P > 0.05 indicates that the participants in the control group met the HWE.
Association between the rs13181 polymorphism and risk of pancreatic cancer
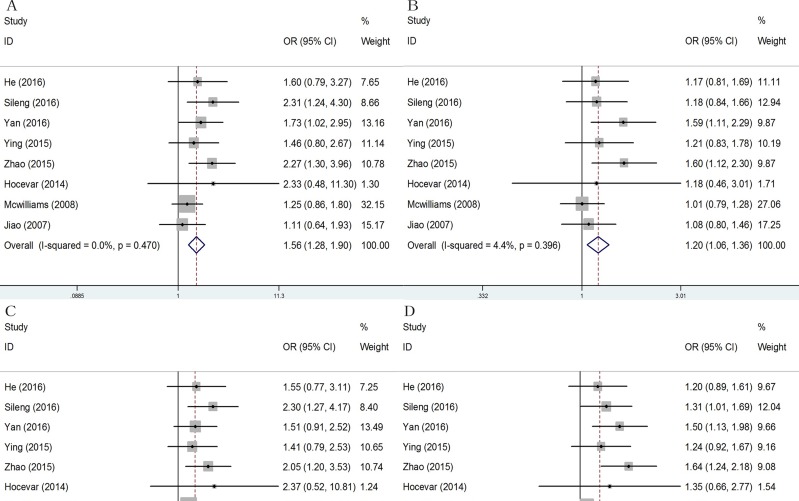
Forest plots of overall analyses with different models on the association between the rs13181 polymorphism and risk of pancreatic cancer are shown in Figure 2. All the results of overall and subgroup analyses are listed in Table 2.
Figure 2. Forest plots of pancreatic cancer risk associated with ERCC2 rs13181 A > C polymorphism.
Four models showed statistical significance between ERCC2 rs13181 A > C and pancreatic cancer risk and the specific values were as follows. A. Homozygote model (CC vs. AA): OR = 1.56, 95% CI: 1.28-1.90; B. Dominant model (AC/CC vs. AA): OR = 1.20, 95% CI: 1.06-1.36; C. Recessive model (CC vs. AC/CC): OR = 1.50; 95% CI: 1.24-1.81; D. Allele model (C vs. A): OR = 1.22, 95% CI:1.11-1.34.
Table 2. Meta-analysis results of association between rs13181 A > C polymorphism and pancreatic cancer risk.
| AC vs AA | CC vs AA | AC/CC vs AA | CC vs AC/AA | C vs A | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Variables | Na | OR(95%CI) | Pb/I2(%) | OR(95%CI) | Pb/I2(%) | OR(95%CI) | Pb/I2(%) | OR(95%CI) | Pb/I2(%) | OR(95%CI) | Pb/I2(%) |
| Total | 8 | 1.10(0.96-1.25) | 0.636/0.0 | 1.56(1.28-1.90) | 0.470/0.0 | 1.20(1.06-1.36) | 0.396/4.4 | 1.50(1.24-1.81) | 0.530/0.0 | 1.22(1.11-1.34) | 0.159/33.7 |
| Ethnicity | |||||||||||
| Asian | 5 | 1.20(1.00-1.43) | 0.584/0.0 | 1.87(1.43-2.43) | 0.770/0.0 | 1.34(1.14-1.57) | 0.534/0.0 | 1.74(1.35-2.25) | 0.718/0.0 | 1.37(1.21-1.56) | 0.507/0.0 |
| Caucasian | 3 | 0.99(0.82-1.21) | 0.809/0.0 | 1.23(0.91-1.67) | 0.681/0.0 | 1.04(0.86-1.25) | 0.904/0.0 | 1.25(0.94-1.65) | 0.583/0.0 | 1.07(0.94-1.23) | 0.808/0.0 |
| Source of controlc | |||||||||||
| PB | 3 | 1.02(0.85-1.22) | 0.707/0.0 | 1.25(0.95-1.65) | 0.803/0.0 | 1.07(0.90-1.27) | 0.713/0.0 | 1.25(0.97-1.62) | 0.758/0.0 | 1.09(0.96-1.24) | 0.661/0.0 |
| HB | 5 | 1.20(0.99-1.46) | 0.567/0.0 | 1.99(1.49-2.66) | 0.895/0.0 | 1.36(1.14-1.62) | 0.568/0.0 | 1.85(1.39-2.45) | 0.809/0.0 | 1.40(1.22-1.61) | 0.593/0.0 |
| HWEd | |||||||||||
| Yes | 4 | 0.99(0.82-1.19) | 0.939/0.0 | 1.52(1.14-2.01) | 0.365/5.7 | 1.09(0.92-1.29) | 0.853/0.0 | 1.53(1.17-1.99) | 0.379/2.8 | 1.16(1.02-1.32) | 0.618/0.0 |
| No | 4 | 1.23(1.02-1.49) | 0.535/0.0 | 1.60(1.21-2.11) | 0.339/10.7 | 1.33(1.11-1.58) | 0.259/25.5 | 1.47(1.12-1.92) | 0.397/0.0 | 1.29(1.13-1.47) | 0.061/59.4 |
a Number of studies;
b P value of Q test for heterogeneity;
c PB, population-based; HB, hospital-based;
d HWE, Hardy-Weinberg equilibrium.
Of the eight studies included, four reported an association between the rs13181 polymorphism and risk of pancreatic cancer while the others did not. A significant association was observed between ERCC2 Lys751Gln polymorphism and susceptibility to pancreatic cancer in four genetic models [CC vs. AA: OR = 1.56, (95% CI: 1.28-1.90), P = 0.470, I2 = 0.0%; AC/CC vs. AA: OR = 1.20, (95% CI: 1.06-1.36), P = 0.396, I2 = 4.4%; CC vs. AC/CC: OR = 1.50; (95% CI: 1.24-1.81), P = 0.530, I2 = 0.0%; C vs. A: OR = 1.22, (95% CI:1.11-1.34), P = 0.159, I2 = 33.7%] (Figure 2A-2D, Table 2).
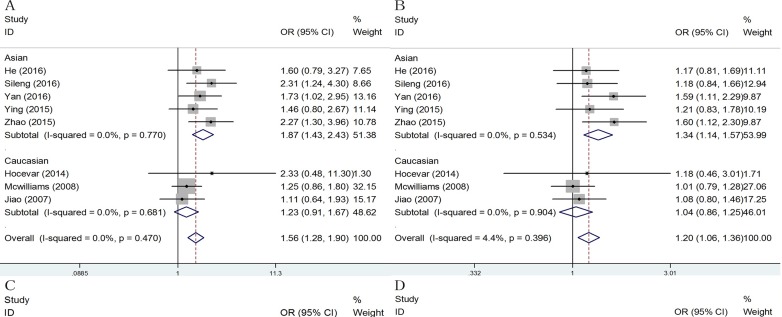
Subgroup analysis by ethnicity showed that a significant association was identified in the Asian population [CC vs. AA: OR = 1.87, (95% CI: 1.43-2.43), P = 0.770, I2 = 0.0%; AC/CC vs. AA: OR = 1.34, (95%CI: 1.14-1.57), P = 0.534, I2 = 0.0%; CC vs. AC/CC: OR = 1.74; (95% CI: 1.35-2.25), P = 0.718, I2 = 0.0%; C vs. A: OR = 1.37, (95% CI:1.21-1.56), P = 0.507, I2 = 0.0%], but not in the Caucasian population (Figure 3A-3D, Table 2). In addition, stratified analysis by design of study showed a significant relationship in hospital-based studies (Supplementary Figure 1A-1D). Next, the Hardy-Weinberg equilibrium (HWE) of each study was taken into consideration. After eliminating studies whose distribution of genotype in controls deviated from HWE, the outcome remained statistically significant. (Supplementary Figure 2A-2D)
Figure 3. Subgroup analysis of ethnicity for ERCC2 rs13181 polymorphism and pancreatic cancer.
Statistical significance was observed in Asian population under four genetic models. A. Homozygote model: OR = 1.87, 95% CI: 1.43-2.43; B. Dominant model: OR = 1.34, 95% CI: 1.14-1.57;C. Recessive model: OR = 1.74; 95% CI: 1.35-2.25; D. Allele model: OR = 1.37, 95% CI:1.21-1.56.
Test of heterogeneity
The genetic heterogeneity between studies was evaluated based on all the five models and the data from the selected studies. By the Chi-squared-based Q-test, heterogeneity between studies was not identified in overall genetic models (P > 0.1). (Table 2)
Sensitivity and publication bias analysis
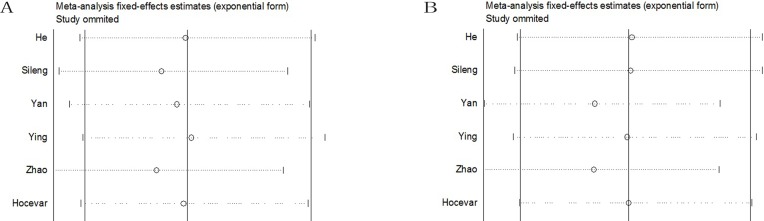
To further validate the robustness of the outcomes, we conducted sensitivity analyses by repeating the meta-analysis while sequentially omitting the studies included (one omitted each time) for every genotype model. The pooled ORs were not influenced significantly by removal of each single study under four genetic models (Figure 4A-4D), suggesting that the results of this meta-analysis were stable.
Figure 4. The sensitivity analysis of pancreatic cancer risk associated with ERCC2 rs13181 A > C polymorphism.
The pooled ORs were not influenced significantly by removal of each single study under four genetic models. A. Homozygote model; B. Dominant model; C. Recessive model; D. Allele model.
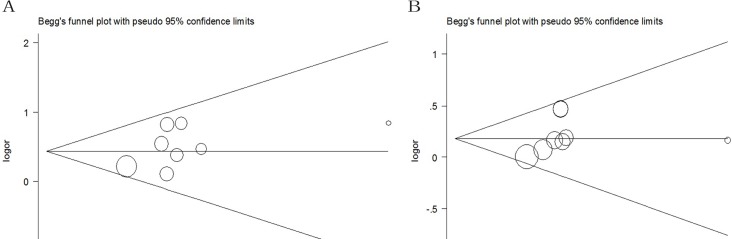
To determine the possible publication bias of the literature, Begg's test and Egger's test were performed. The results of both Begg's and Egger's test showed no evidence of publication bias for ERCC2 Lys751Gln polymorphisms (Table 3). In addition, the funnel plots of the homozygous, dominant, recessive and allele models were symmetrical inverted funnels (Figure 5A-5D), which suggested no significant publication bias. The outcomes above indicated that the conclusions of our meta-analysis were stable and credible.
Table 3. The result of Begg and Egger's tests.
| Risk model | Egger' s test | Begg' s test | ||
|---|---|---|---|---|
| T statistic | P value | Z statistic | P value | |
| Homozygous (CC vs. AA) | 1.32 | 0.235 | 1.11 | 0.266 |
| Dominant (AC/CC vs. AA) | 1.00 | 0.355 | 1.11 | 0.266 |
| Recessive (CC vs.AC/AA) | 1.35 | 0.226 | 1.36 | 0.174 |
| Allele (C vs. A) | 1.50 | 0.185 | 1.11 | 0.266 |
Figure 5. Begg's funnel plot of pancreatic cancer risk associated with ERCC2 rs13181 A > C polymorphism.
The funnel plots of A. homozygous, B. dominant, C. recessive and D. allele models are symmetrical inverted funnels, which suggests no significant publication bias.
DISCUSSION
Pancreatic cancer is a highly lethal disease, for which mortality closely parallels incidence [2]. As a multi-factor disease, genetic mutation has been found to play a key role in its development and progression. Understanding the etiology and genetic background of pancreatic cancer is important for screening high-risk populations and promoting the development of molecular-targeted therapy.
Many previous genetic association studies on pancreatic cancer risk have focused on the effects of single nucleotide polymorphisms in ERCC2 gene. ERCC2, also known as Xeroderma pigmentosum D (XPD), is a key DNA repair gene in nucleotide excision repair (NER) pathway which could repair a wide variety of structurally DNA lesions, including cross-links, bulky adducts [19], thymidine dimers, oxidative DNA damage [20] and alkylating damage [21]. SNPs in exons of DNA repair genes can affect their protein activity, leading to differences of individual NER and DNA repair capacity (DRC) that may influence the susceptibility to pancreatic cancer. As the most frequently assessed variant, the ERCC2 Lys751Gln polymorphism is thought to be associated with many cancers [22–27]. In 2012, Sobti et al. [22] showed that ERCC2 mutations are associated with an increased risk of urinary bladder cancer in North Indian population. Meanwhile, Samson et al. [23] identified that polymorphisms of the ERCC2 gene might contribute to tumorigenesis in breast cancer among south Indian population. In addition, the ERCC2 gene could also increase the risk of hepatocellular carcinoma [24], acute lymphoblastic leukemia [25], lung cancer [26] and melanoma [27]. Recently, more studies have shown that the ERCC2 gene polymorphisms plays a key role in the tumorigenesis of pancreatic cancer [11–18]. However, the results remain inconsistent. In addition, there were no publically available GWAS databases and GWAS analysis which had evaluated this SNP and pancreatic cancer before. In order to elucidate the real association, this analysis was performed.
The current study is the first meta-analysis of the association between ERCC2 rs13181 and the risk of pancreatic cancer. Analysis among all subjects suggested a significant increase in the risk of pancreatic cancer associated with Gln/Gln or Lys/Gln genotype. All studies included were found homogeneous without any study disproportionately driving the combined estimates. In our meta-analysis, the genetic heterogeneity between the selected studies was evaluated, and no significant heterogeneity was observed in the homozygous, dominant, recessive and allele models.
Interestingly, the association remained statistically significant in subgroups (Asians, HB and the studies consistent with HWE) analyses. In subgroup analysis of ethnicity, our results showed that the C allele of rs13181 had a 1.22-fold risk of pancreatic cancer in overall populations, a 1.37-fold risk (95% CI 1.21-1.56) in Asian populations. Compared to the dominant and recessive models, the homozygous model showed the highest odds ratio in all populations (OR = 1.56, 95% CI 1.28-1.90) and in the Asian population (OR = 1.87, 95% CI 1.43-2.43). The differences between Asians and other races may be partly due to the different genetic backgrounds and environments or lifestyles.
Meta-analysis is a very powerful tool for analyzing cumulative data of studies where the individual sample sizes are small and the statistical power is low. However, there are still some limitations in the current meta-analysis. To begin with, our analysis was based on ORs estimated without adjustment for several potential confounding variables, because of a lack of information about cigarette smoking [28], chronic pancreatitis [29], diabetes [30] and a family history of pancreatic cancer [31], which are known to have significant effects on the development of pancreatic cancer. Secondly, more studies from all over the world should be performed to make our conclusions more persuasive, because our meta-analysis lacks studies in African populations. Finally, as a multi-factorial disease, pancreatic cancer results from complex interactions including a variety of genetic and environmental factors, suggesting pancreatic cancer susceptibility could not be influenced by any single gene. More researches exploring the influencing factors are required in the future.
In conclusion, our meta-analysis suggests that ERCC2 Lys751Gln polymorphism is a risk factor of pancreatic cancer for all of the ethnicities, and presence of this polymorphism in Asian population will increase their susceptibility to pancreatic cancer. However, additional larger and ethnically diverse studies are needed to further clarify the role of this polymorphism in the development of pancreatic cancer.
MATERIALS AND METHODS
Primary search strategy
We searched for relevant studies up to July 18, 2016 in both English and Chinese through PubMed, Web of Science, EMBSE and the China National Knowledge Infrastructure (CNKI) platforms database with the following terms and their combinations: “ERCC2 or XPD”, “polymorphism or variant”, “Lys751Gln”, “rs13181”. “K751Q” and “pancreatic cancer”, “pancreatic ductal adenocarcinoma”. To prevent the loss of any important data, we also identified additional investigations by screening the reference lists of key studies and reviews.
Inclusion criteria and exclusion criteria
Studies involved had to satisfy the inclusion criteria: (a) case-control design was utilized; (b) researches focused on the association of ERCC2 Lys751Gln (rs13181) polymorphisms with the risk of pancreatic cancer; (c) sufficient data for estimating an odds ratio (OR) with 95% CI were available. The major exclusion criteria were as follows: (a) no obtainable genotype frequency data; (b) unpublished papers, dissertations, conference articles, reviews and duplication of publications (select the study in the latest and largest sample size); (c) studies designed as a case-case or case-only study.
Data extraction
All the following information was extracted separately by two investigators (Y Wu, Z Lu) and recorded in a standardized form, including: first author's name, year of publication, ethnicity of each study population, source of controls, sample size, genotyping method, number of pancreatic cancer cases and controls, allele frequencies and genotype distributions of ERCC2 rs13181 in pancreatic cancer cases and controls respectively, and results of the Hardy-Weinberg equilibrium (HWE) test, as shown in Table 1. Inconsistencies were resolved by a discussion involving a senior investigator (K Jiang).
Genetic model
The rs13181 polymorphism includes the two alleles A and C, of which C is the minor allele. C is assumed to be the high-risk allele and A is the low-risk allele. We selected the homozygous model (C/C vs. A/A), heterozygous model (A/C vs. A/A), dominant model (C/C + A/C vs. A/A), recessive model (C/C vs. A/C + A/A) and allele model (C vs A) for further meta-analysis.
Statistical analysis
Stata software (version 12.0; StataCorp LP, College Station, TX) was applied in the whole statistical analyses. P values were all two-sided and regarded as statistically significant if less than 0.05.
To evaluate the strength of association between rs13181 and the risk of pancreatic cancer, the pooled odds ratios (ORs) with 95% CIs were calculated in the five genetic models. Meanwhile, between-study heterogeneity was evaluated using two methods including Cochran's Q-statistic and I2 = (Q-(k-1))/Q*100% statistic. Cochran's Q statistic approximately follows a χ2 distribution with k-1 degrees of freedom (k stands for the number of studies for analysis). A significant Q-statistic (P < 0.1) indicates heterogeneity among selected studies. I2 is a measure of heterogeneity and a statistic that indicates the percentage of variance in a meta-analysis that is attributable to study heterogeneity. The intervals including 0-25%, 25-50%, 50-75% and 75-100%, represent the low, moderate, large and extreme heterogeneity. The interval I2 > 50% indicates statistically significant heterogeneity. If heterogeneity P value was lower than 0.10, we considered the heterogeneity to be significant and random-effects model (The DerSimonian-Laird method) was used [32]. Otherwise, the fixed effects model (the Mantel-Haenszel method) was used [33]. Then, we conducted the subgroup analyses by collecting similar characteristics from the eligible studies, such as ethnicity (Asian and Caucasian), source of controls (population-based and hospital-based), and HWE (yes and no).
Sensitivity analysis was performed to assess the influence of individual studies on the pooled ORs, with the method of calculating the outcomes again by omitting one single study each time. Publication bias was evaluated with Begg's funnel plots and Egger's linear regression method and a P < 0.05 was set as the significance threshold [34]. HWE was checked by the goodness-of-fit chi-square test and a P < 0.05 was considered as a significantly selective bias [35].
SUPPLEMENTARY MATERIALS FIGURES
ACKNOWLEDGMENTS AND FUNDING
Funded by the National Natural Science Foundation of China (No. 81272382 to Kuirong Jiang, No. 81300351 to Zipeng Lu, No. 81572337 to Jingjing Zhang), the Natural Science Foundation of Jiangsu Province (No. BK2012881) and the “Six Talent Peaks” Project of Jiangsu Province (2014-WSW-006).
Footnotes
CONFLICTS OF INTEREST
The authors have declared that no competing interests exist.
REFERENCES
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Gillen S, Schuster T, Meyer Zum Büschenfelde C, Friess H, Kleeff J. Preoperative/neoadjuvant therapy in pancreatic cancer: a systematic review and meta-analysis of response and resection percentages. PLoS Med. 2010;7:e1000267. doi: 10.1371/journal.pmed.1000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antwi SO, Eckert EC, Sabaque CV, Leof ER, Hawthorne KM, Bamlet WR, Chaffee KG, Oberg AL, Petersen GM. Exposure to environmental chemicals and heavy metals, and risk of pancreatic cancer. Cancer Causes Control. 2015;26:1583–1591. doi: 10.1007/s10552-015-0652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim VM, Ahuja N. Early detection of pancreatic cancer. Chin J Cancer Res. 2015;27:321–331. doi: 10.3978/j.issn.1000-9604.2015.07.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng Z, Zheng R, He Y, Sun X, Wang N, Chen T, Chen W. Risk Factors for Pancreatic Cancer in China: A Multicenter Case-Control Study. J Epidemiol. 2016;26:64–70. doi: 10.2188/jea.JE20140148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berwick M, Vineis P. Markers of DNA repair and susceptibility to cancer in humans: an epidemiologic review. J Natl Cancer Inst. 2000;92:874–897. doi: 10.1093/jnci/92.11.874. [DOI] [PubMed] [Google Scholar]
- 7.Shields PG, Harris CC. Cancer risk and low-penetrance susceptibility genes in gene-environment interactions. J. Clin. Oncol. 2000;18:2309–2315. doi: 10.1200/JCO.2000.18.11.2309. [DOI] [PubMed] [Google Scholar]
- 8.Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2002;11:1513–1530. [PubMed] [Google Scholar]
- 9.Alanazi M, Pathan AA, Ajaj SA, Khan W, Shaik JP, Al Tassan N, Parine NR. DNA repair genes XRCC1, XRCC3, XPD, and OGG1 polymorphisms among the central region population of Saudi Arabia. Biol Res. 2013;46:161–167. doi: 10.4067/S0716-97602013000200007. [DOI] [PubMed] [Google Scholar]
- 10.Pabalan N, Francisco-Pabalan O, Sung L, Jarjanazi H, Ozcelik H. Meta-analysis of two ERCC2 (XPD) polymorphisms, Asp312Asn and Lys751Gln, in breast cancer. Breast Cancer Res Treat. 2010(124):531–541. doi: 10.1007/s10549-010-0863-6. [DOI] [PubMed] [Google Scholar]
- 11.Jiao L, Hassan MM, Bondy ML, Abbruzzese JL, Evans DB, Li D. The XPD Asp312Asn and Lys751Gln polymorphisms, corresponding haplotype, and pancreatic cancer risk. Cancer Lett. 2007;245:61–68. doi: 10.1016/j.canlet.2005.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hocevar BA, Kamendulis LM, Pu X, Perkins SM, Wang ZY, Johnston EL, DeWitt JM, Li L, Loehrer PJ, Klaunig JE, Chiorean EG. PLoS One. 2014;9:e90052. doi: 10.1371/journal.pone.0090052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ying MF, Zhao R. Role of single nucleotide polymorphisms of DNA repair genes in susceptibility to pancreatic cancer in Chinese population. Genet Mol Res. 2016:15. doi: 10.4238/gmr.15017273. [DOI] [PubMed] [Google Scholar]
- 14.He MG, Zheng K, Tan D, Wang ZX. Association between ERCC1 and ERCC2 gene polymorphisms and susceptibility to pancreatic cancer. Genet Mol Res. 2016:15. doi: 10.4238/gmr.15017879. [DOI] [PubMed] [Google Scholar]
- 15.McWilliams RR, Bamlet WR, Cunningham JM, Goode EL, de Andrade M, Boardman LA, Petersen GM. Polymorphisms in DNA repair genes, smoking, and pancreatic adenocarcinoma risk. Cancer Res. 2008;68:4928–4935. doi: 10.1158/0008-5472.CAN-07-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao F, Shang Y, Zeng C, Gao D, Li K. Association of single nucleotide polymorphisms of DNA repair genes in NER pathway and susceptibility to pancreatic cancer. Int J Clin Exp Pathol. 2015;8:11579–11586. [PMC free article] [PubMed] [Google Scholar]
- 17.Yan D, He TY, Ding W, Xu XJ, Wang XY. Relationship between the interaction of xeroderma pigmentosum group D polymorphism with smoking and the susceptibility to pancreatic cancer. Zhonghua Shi Yong Wai Ke Za Zhi. 2016;33:12–15. [Google Scholar]
- 18.Sileng A, Pan R, Li G, Wang W, Li J, Zhang M, Li M, Zhang Z, Lin J, Liao C. ERCC1 rs3212986 and ERCC2 rs13181 gene polymorphisms contributes to the susceptibility to pancreatic cancer in a Chinese population. International Journal of Clinical and Experimental Pathology. 2016;9:5686–5693. [Google Scholar]
- 19.De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol. 2000;20:7980–7990. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braithwaite E, Wu X, Wang Z. Repair of DNA lesions: mechanisms and relative repair efficiencies. Mutat Res. 1999;424:207–219. doi: 10.1016/s0027-5107(99)00020-2. [DOI] [PubMed] [Google Scholar]
- 21.Chen ZP, Malapetsa A, McQuillan A, Marcantonio D, Bello V, Mohr G, Remack J, Brent TP, Panasci LC. Evidence for nucleotide excision repair as a modifying factor of O6-methylguanine-DNA methyltransferase-mediated innate chloroethylnitrosourea resistance in human tumor cell lines. Mol Pharmacol. 1997;52:815–820. doi: 10.1124/mol.52.5.815. [DOI] [PubMed] [Google Scholar]
- 22.Sobti RC, Kaur S, Sharma VL, Singh SK, Hosseini SA, Kler R. Susceptibility of XPD and RAD51 genetic variants to carcinoma of urinary bladder in North Indian population. DNA Cell Biol. 2012;31:199–210. doi: 10.1089/dna.2011.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samson M, Singh SS, Rama R, Sridevi V, Rajkumar T. XPD Lys751Gln increases the risk of breast cancer. Oncol Lett. 2011;2:155–159. doi: 10.3892/ol.2010.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long XD, Ma Y, Zhou YF, Yao JG, Ban FZ, Huang YZ, Huang BC. XPD codon 312 and 751 polymorphisms, and AFB1 exposure, and hepatocellular carcinoma risk. BMC Cancer. 2009;9:400. doi: 10.1186/1471-2407-9-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Batar B, Güven M, Bariś S, Celkan T, Yildiz I. DNA repair gene XPD and XRCC1 polymorphisms and the risk of childhood acute lymphoblastic leukemia. Leuk Res. 2009;33:759–763. doi: 10.1016/j.leukres.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Chang JS, Wrensch MR, Hansen HM, Sison JD, Aldrich MC, Quesenberry CP, Jr, Seldin MF, Kelsey KT, Kittles RA, Silva G, Wiencke JK. Nucleotide excision repair genes and risk of lung cancer among San Francisco Bay Area Latinos and African Americans. Int J Cancer. 2008;123:2095–2104. doi: 10.1002/ijc.23801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kertat K, Rosdahl I, Sun XF, Synnerstad I, Zhang H. The Gln/Gln genotype of XPD codon 751 as a genetic marker for melanoma risk and Lys/Gln as an important predictor for melanoma progression: a case-control study in the Swedish population. Oncol Rep. 2008;20:179–183. [PubMed] [Google Scholar]
- 28.Iodice S, Gandini S, Maisonneuve P, Lowenfels AB. Tobacco and the risk of pancreatic cancer: a review and meta-analysis. Langenbecks Arch Surg. 2008;393:535–545. doi: 10.1007/s00423-007-0266-2. [DOI] [PubMed] [Google Scholar]
- 29.Raimondi S, Lowenfels AB, Morselli-Labate AM, Maisonneuve P, Pezzilli R. Pancreatic cancer in chronic pancreatitis; aetiology, incidence, and early detection. Best Pract Res Clin Gastroenterol. 2010;24:349–358. doi: 10.1016/j.bpg.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 30.Silverman DT, Hoover RN, Brown LM, Swanson GM, Schiffman M, Greenberg RS, Hayes RB, Lillemoe KD, Schoenberg JB, Schwartz AG, Liff J, Pottern LM, Fraumeni JF., Jr Why do black Americans have a higher risk of pancreatic cancer than white Americans? Epidemiology. 2003;14:45–54. doi: 10.1097/00001648-200301000-00013. [DOI] [PubMed] [Google Scholar]
- 31.Hruban RH, Canto MI, Goggins M, Schulick R, Klein AP. Update on familial pancreatic cancer. Adv Surg. 2010;44:293–311. doi: 10.1016/j.yasu.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 33.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 34.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 35.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.