Abstract
Ovarian cancer is the first cause of death from gynaecological malignancy. Germline mutation in BRCA1 and 2, two genes involved in the mechanisms of reparation of DNA damage, are showed to be related with the incidence of breast and ovarian cancer, both sporadic and familiar. PARP is a family of enzymes involved in the base excision repair (BER) system. The introduction of inhibitors of PARP in patients with BRCA-mutated ovarian cancer is correlated with the concept of synthetic lethality. Among the PARP inhibitors introduced in clinical practice, niraparib showed interesting results in a phase III trial in the setting of maintenance treatment in ovarian cancer, after platinum-based chemotherapy. Interestingly, was niraparib showed to be efficacious not only in BRCA-mutated patients, but also in patients with other alterations of the homologous recombination (HR) system and in patients with unknown alterations. These results position niraparib as the first PARP-inhibitor with clinically and statistically significant results also in patients with no alterations in BRCA 1/2 and other genes involved in the DNA repair system. Even if the results are potentially practice-changing, the action of niraparib must be further studied and deepened.
Keywords: BRCA mutations, BRCAness, epithelial ovarian cancer, high-grade serous ovarian cancer, homologous recombination deficiency (HRD), niraparib, poly(ADP-ribose) polymerase inhibitors (PARPis), synthetic lethality concept, target therapy
Introduction
Epithelial ovarian cancer (EOC) accounts for 90% of all ovarian tumours and typically presents in post-menopausal women.1 It is the second most common malignant gynaecological disease and first cause of death from gynaecological malignancy.2,3
According to histopathological characteristics and to next-generation sequencing, EOC has been found to consist of a complex set of diseases. Several genetic or epigenetic alterations, strategic for tumorigenesis and progression, have been identified in heterogeneous subsets of patients.4
For example, BRCA mutations are most commonly associated with HGSOC (high-grade serous ovarian cancer).5 BRCA1 and BRCA2 are well known tumour suppressor genes, involved in the response to DNA damage. The prevalence of germline BRCA mutations (gBRCAm) in EOC has historically been estimated to be around 10–15%.6 Recent reports suggest that this may be an underestimated phenomenon, especially in women with HGSOC.7 The inhibition of PARP in the presence of homologous recombination deficiency (HRD) leads to cell death from gross genetic disarray due to a process called ‘synthetic lethality’.8,9
The emergence of the DNA repair pathway as a rational target in various cancers led to the development of the PARP inhibitors (PARPis).10
Such data support the use of routine evaluation of BRCA status in all patients with HGSOC, regardless of family history. This expansion in BRCA testing will require changes to the traditional genetic service indications, because today patients are screened for and referred according to family history. This diagnostic change will lead to tailored genetic testing, according to definite germline characteristics.
Determining the molecular events driving HGSOC progression and diffusion could advance the understanding of tumorigenesis and facilitate individualized treatment strategies for this lethal disease.
BRCA and PARP role in the DNA stability system
DNA continually undergoes structural alterations. These alterations can be divided into: base modifications; single-strand breaks (SSBs); double strand breaks (DSBs); and intrastrand or interstrand crosslinks.11 Fortunately, cells have evolved different mechanisms to maintain genomic integrity.12
There are at least five DNA repair mechanisms. Homologous recombination (HR) and non-homologous end joining (NHEJ) are responsible for DSB repair. The BRCA1 and BRCA2 proteins are important in maintaining genomic stability by promoting efficient and precise repair of DSB.13,14
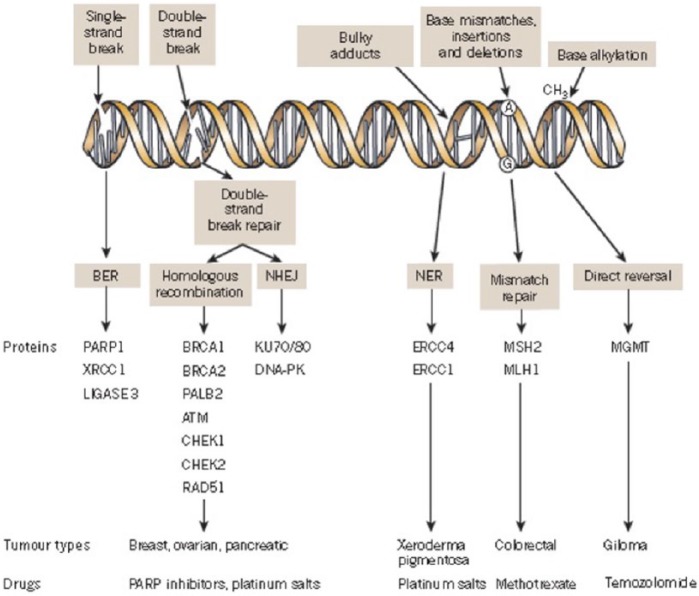
SSB repair mechanisms include base excision repair (BER), nucleotide excision repair (NER) and mismatch repair (MMR) pathways.15 The PARP family is involved in BER, which is the predominant pathway of the SSB repair system (Figure 1).16
Figure 1.
A panoply of DNA repair mechanisms maintains genomic stability. DNA is continually exposed to a series of insults that cause a range of lesions, from single-strand breaks (SSBs) to base alkylation events. The choice of repair mechanism is largely defined by the type of lesion, but factors such as the stage of the cell cycle also have a role. Key proteins involved in each DDR mechanism, the tumour types usually characterized by DDR defects and the drugs that target these defects are shown. Figure modified, with permission, from Lord and colleagues 2012.
BER, base excision repair; DDR, DNA damage response; NER, nucleotide excision repair; NHEJ, non-homologous end joining.
Poly(ADP-ribose) polymerase (PARP) is a family of nuclear proteins that sense and bind to DNA SSB and subsequently activate the BER pathway by recruiting additional repair factors, modifying target proteins with ADP-ribose units. Of the 17 known members of the PARP superfamily in humans, PARP-1 accounts for >90% of cellular DNA repair activity and remains the most studied.17,18
When an SSB occurs, PARP-1 is recruited, undergoing a conformational change inducing the C-terminal catalytic domain to transfer ADP-ribose moieties from cellular (NAD+) to protein acceptors. This activation leads to addition of PAR chains, that results in a relaxation of chromatin and subsequent recruitment of DNA repair factors, such as XRCC1. XRCC1 is crucial for DNA repair, initially assembling and activating the BER machinery through the modification of several proteins such as histones and topoisomerases, but subsequently ‘switching off’ the BER machinery by decreasing the affinity of both histones and PARP-1 to DNA.19
In cases of small to moderate damage, PARP-1 allows restoration of the genomic integrity and the return to normal cellular function. However, emerging evidence has implicated PARP-1 over-activation in unregulated PAR synthesis, depleting NAD, and consequently ATP, eventually leading to widespread cell death.20
PARP inhibitors in BRCA-mutated EOC and synthetic lethality concept
PARPis mediate their anti-cancer effects as catalytic inhibitors able to block repair of DNA SSBs by the BER/SSBR pathway. The initial clinical development of PARPis focused on their role as chemo-sensitizers, while their single-agent activity was unknown. Ten years ago, two articles published in Nature reported that BRCA1/2 heterozygote or wild-type cell lines were 100- to 1000-fold less sensitive to PARP inhibitors than cells deficient in BRCA1 and 2.8,9 The conclusion of both studies was that the BRCA-deficient cells were selectively sensitive to PARP inhibition by a mechanism of ‘synthetic lethality’: cancer cells are selectively sensitive to the inactivation of two genes or pathways when inactivation of either gene or pathway alone is non-lethal.21
If the cell cannot initiate HR, as is the case with BRCA1/2-mutant tumours, it resorts to more error-prone pathways, such as NHEJ or single-strand annealing, which can cause gross chromosomal mutations, growth inhibition and eventual cell death.22
Then, patients with EOC with germline mutations in either BRCA1 or BRCA2 genes exhibit impaired ability to repair DNA DSB via HR, and show a heightened sensitivity to inhibitors of the BER pathway.23,24
Sometimes, sporadic EOC share pathological and clinical traits of BRCA mutation-associated cases, in the absence of a gBRCAm. This condition is termed ‘BRCAness’.23,25 Genes possibly involved in HR include PALB2, RAD51C, RAD51D, BRIP1, BARD1, Chk2Mre11A, MSH6, NBN and RAD50.25,26
Niraparib in ovarian cancer treatment
Niraparib (MK-4827) is a potent PARP-1 and PARP-2 inhibitor, with in vitro IC50 = 3.8 and 2.1 nm. Niraparib was showed to selectively inhibit proliferation of BRCA1 and BRCA2 deficient cell lines. Like for other PARPis, also niraparib induces cell cycle arrest in the G2/M phase, followed by apoptosis and mitosis dysregulation. Correlations between niraparib, BRCA mutation status and DNA damage was evaluated in HGSOC patient-derived xenograft models. These xenograft model showed interesting results, both in niraparib monotherapy and niraparib maintenance after combination of niraparib and chemotherapy. Other genes, involved in HR mechanisms, seemed to be related with niraparib action.27 Niraparib showed activity also in a CAPAN-1 pancreatic cancer xenograft model. In preclinical models, PARP-1/2 showed efficacy in combination with platinum, alkylating and methylation agents, radiation therapy and topoisomerase inhibitors.28 Niraparib enhanced efficacy of radiation therapy in breast and lung cancer models.29
The genomic analysis by the Cancer Genome Atlas evaluated the rate of mutations in 489 patients with HGSOC, finding 9% germline mutations in BRCA1, 8% germline mutations in BRCA2 and 3% somatic mutations in both genes. Other significant mutations were highlighted in other HR genes: EMSY (8%), PTEN (7%), RAD51 (3%), ATM or ATR (3%) and Fanconi anaemia genes (5%). Overall, HRD could be present in about half of HGSOC cases.30 Other data from Pennington and colleagues showed the presence of germline and somatic loss-of-function mutations in 30 genes, involved in the HR pathway, in 390 cases of ovarian carcinoma. In this case, 367 individuals and 390 carcinomas were evaluated: 87 (24%) had a germline HR mutation; 32 (9%) had a somatic mutation. Four patients (1.1%) had both a somatic and germline mutation. In particular, 68 mutations occurred in BRCA1, 23 in BRCA2 and 32 in another 11 genes (ATM, BARD1, BRIP1, CHEK1, CHEK2, FAM175A, MRE11A, NBN, PALB2, RAD51C, and RAD51D). Mutations of HR-related genes were present also in non-squamous ovarian cancer.31
Niraparib has been studied in a phase I dose-escalation trial.32 In part A of the trial, eligible patients had several kinds of malignancies, not suitable for standard treatments. Enrichment for germline BRCA1 and BRCA2 mutations was conducted prospectively, to detect whether administration of niraparib could be more effective in patients with defective HR function. Mutations were established using Myriad Genetics BRCA mutation testing. In part B of the trial only patients with sporadic platinum-resistant EOC and castration-resistant prostate cancer were enrolled. At the end of part A, 400 mg was found to be the maximum tolerated dose (MTD) due to grade 4 reversible thrombocytopenia; 300 mg was defined as the recommended dose in phase II trials, evaluated in an additional 10 patients. All toxicities were manageable and reversible.
Overall, 77 patients showed a response according to RECIST criteria. Among the patients in part A, 29 showed mutations in BRCA1 or BRCA2. Twenty-two patients had ovarian cancer. Twenty of these 22 patients had measurable disease; partial response (PR) was observed in eight patients (40%), using doses between 80 mg and 400 mg. Three of nine patients with platinum-resistant EOC showed RECIST and Ca125 responses; another patient had stable disease (SD) for 120 days. Five patients of the 22 with BRCA wild-type serous ovarian cancer achieved a durable PR. Two of these responding patients were platinum-resistant, three were platinum-sensitive.
Combination of niraparib with temozolomide was evaluated in an open-label phase I trial in 19 patients [ClinicalTrials.gov identifier: NCT01294735]. MTD was found to be 40 mg/day plus temozolomide 150 mg/m2/die. The most frequent adverse events were thrombocytopenia, leukopenia, nausea and fatigue. A patient with glioblastoma achieved PR after four cycles, while SD was achieved in two patients (a malignant melanoma and a serous ovarian cancer patient).
Other combination attempts were evaluated in three trials: a phase I trial in which niraparib was associated with carboplatin or carboplatin/paclitaxel or carboplatin/liposomal doxorubicin [ClinicalTrials.gov identifier: NCT01110603]; a phase Ib trial evaluated niraparib in combination with pegylated liposomal doxorubicin [ClinicalTrials.gov identifier: NCT01227941]; and a phase I study, started in Japan, in patients with advanced solid tumours [ClinicalTrials.gov identifier: NCT01226901]. None of these trials are yet completed.
NOVA trial33 is a phase III randomized double-blind trial, in which niraparib was evaluated versus placebo in patients with platinum-sensitive ovarian cancer expressing BRCA mutation or high-grade histology (Table 1).
Table 1.
Published trials with niraparib.
| Authors | Drug | Ph | Pts | Lines of therapy | BRCA status |
Pl. Sens. | ORR (%) |
mPFS (m) |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mt n (%) | Wt n (%) | Mt | Wt | HRD+ | |||||||
| Sandhu and colleagues32 | NIR | I | 100 | II–IV | 29 (29) | 71(71) | S | 40 (in gBRCAm) | |||
| Mirza and colleagues33 | NIR | III | 372 | III | 136 | 231 | S | Nr | 21 | 9.3 | 12.9 |
| PLA | 181 | 65 () | 114 | S | Nr | 5.5 | 3.9 | 3.8 | |||
gBRCAm, germline BRCA mutated; HRD+, homologous recombination deficiency positive; mPFS, median progression-free survival; Mt, mutated; NIR, niraparib; ORR, overall response rate; Ph, phase; PLA, placebo; Pts, patients; Wt, wild-type; Pl. Sens, Platinum sensitivity; S, sensitive.
In the NOVA trial all patients enrolled had platinum-sensitive disease, with at least two platinum-based chemotherapy regimens in their clinical history. Patients, divided in two cohorts on the basis of the BRCA mutation, determined with BRAC analysis testing (Myriad Genetics, Salt Lake City, USA), were assigned a 2:1 ratio of niraparib 300 mg or placebo once daily in 28-day cycles. Before the database lock, tissue samples in the non-BRCA cohort were analysed using the myChoice HRD test (Myriad Genetics) to determine the status of the HR system. A tumour with a score >42 was considered HRD-positive. Primary endpoint was progression-free survival (PFS); secondary endpoints were patient-reported outcome (Functional Assessment of Cancer Therapy – Ovarian Symptom Index questionnaire and the European Quality of Life Scale, the EQ-5D-5L questionnaire), chemotherapy-free interval (time from the last platinum dose until initiation of the next anti-cancer therapy), time to first and second subsequent therapy (time from treatment randomization in the current study to the start date of the first or second subsequent anti-cancer therapies, respectively), PFS2 (time from randomization until progression to subsequent treatment after study treatment or death), time to second subsequent therapy and overall survival (OS).
A total of 553 patients were enrolled, with a median age from 57 to 63 years; 203 in the gBRCAm cohort, 350 in the non-gBRCAm cohort, of whom 201 and 345 received the treatment. Among the non-gBRCAm patients, 162 were HRD-positive and 134 were HRD-negative. Of the 162 HRD-positive patients, 47 exhibited a somatic mutation of BRCA. Treatment with niraparib was associated with significantly longer PFS than placebo in gBRCAm patients: 21 months versus 5.5 months (HR 0.27). PFS was longer also in the HRD-positive subgroup of the non-gBRCA cohort (12.9 versus 3.8 months, HR 0.38) and in the overall non-gBRCAm cohort (9.3 versus 3.9 months, HR 0.45).
Chemotherapy-free interval and time to first subsequent therapy were significantly longer in the niraparib group. Although data were not completely mature, results about PFS2 showed that niraparib was better than the placebo group. Data about time to second subsequent therapy and OS were not mature for analysis. Exploratory analysis was conducted to detect the effect of somatic BRCA mutations in the HRD-positive population. HRD-positive BRCA wild-type patients showed a median PFS of 9.3 months, while PFS for placebo was only 3.7 months (HR 0.38). The HR was similar to that of the overall HRD-positive population. Patients with HRD-positive and somatic BRCA mutation disease achieved similar results to the gBRCAm cohort (20.9 mo versus 11 mo; HR 0.27). Also, for HRD-negative sBRCA wild-type the administration of niraparib led to an increase in median PFS (6.9 months versus 3.8 months; HR 0.58).
All patients in the niraparib group showed almost an adverse event. No death occurred during study treatment. Follow-up analysis showed that three patients (one patient in the niraparib group and two in the placebo group) died from myelodysplastic syndrome of acute myeloid leukaemia. Five patients (1.4%) in the niraparib group developed myelodysplastic syndrome; while two patients in the placebo group developed myelodysplastic syndrome and acute myeloid leukaemia. Incidence of grade 3–4 adverse events was 77.4% in the niraparib group and 22.9% in the placebo group.
Companion diagnostics
All developing biopharmaceutical and biotechnological companies developing PARPis have interests in the development of companion diagnostics to identify PARPis-selective patients. Myriad’s BRACAnalysis CDxTM is the only FDA-approved test to determine olaparib treatment eligibility. The same test is used for veliparib, while niraparib and talazoparib eligibility is determined with myChoice HRD.TM Rucaparib uses Foundation Medicine’s NGS-based CDx.
While BRACAnalysis CDxTM, as the name explains, evaluates only BRCA, myChoiceHRDTM, developed by the same company, evaluates loss of heterozygosity (LOH) beyond BRCA and can be considered an enhancement of BRACAnalysis CDxTM. It is an NGS-based assay that assesses BRCA1/2 sequences, and genomic scarring (HRD score), composed by LOH, telomeric allelic balance (TAI) and large-scale transitions (LST). HRD is correlated with alterations in BRCA1/2, PTEN, FANCM and RAD51C.31 High levels of TAI are correlated with DNA repair defects. All BRCA1/2 mutated cancer have high levels of LST. It seems that LST scores indicate HRD better than BRCA status. In addition, LST signature is inexpensive. Recently, retrospective data from an ovarian cancer cohort showed that dichotomized individual components (LST, TAI, LOH) to the combined biomarkers have a significant correlation for the combined HRD score with PFS and OS.30 HR threshold of myChoice HRDTM can identify 95% of patients with mutations in BRCA1/2 and other HR genes, and who have a higher likelihood of responding to treatment with DNA-damaging agents. In a study by Telli and colleagues,34 a positive myChoice HRDTM result with a threshold of 42 was showed to be related with response to neoadjuvant chemotherapy in patients with triple-negative breast cancer (TNBC) or BRCA1/2 mutated breast cancer. Using the same validated endpoint of the trial by Telli and colleagues, the myChoice HRDTM test was used to predict response to chemotherapy in a Geparsixto population: patients with positive myChoice HRDTM test had a higher rate of tumour response.35 The same threshold was used in the niraparib phase III trial: all the BRCAm patients were identified except one, supporting the use of this companion diagnostic to select sensitive patients.36
Discussion
Niraparib and olaparib are the main PARPis studied in ovarian cancer. In trials both drugs have showed to improve PFS in women with recurrent platinum-sensitive disease, with manageable side effects. However, beneficial effects in terms of OS have not been adequately demonstrated by the addition of olaparib, while data about OS with niraparib are still immature. More data are required to decide on the clinical application and strategies for the use of these drugs.
The European Medicines Agency (EMA) approved olaparib for monotherapy for maintenance treatment of adult patients with platinum-sensitive relapsed BRCA-mutated (germline and/or somatic) HGSOC, fallopian tube or primary peritoneal cancer who are in response (complete response (CS) or PR) to platinum-based chemotherapy in 2014. Meanwhile, niraparib has received label by the FDA in all platinum-sensitive patients (PR and CR) regardless of histology, BRCA and HRD status.
In the absence of more refined understanding of PARPis action, BRCA1/2 mutation status has been the most extensively studied predictor of PARPis sensitivity to date.
However, trials of maintenance treatment of olaparib and niraparib are quite different. In the olaparib phase II trial by Lederman and colleagues,37 patients were enrolled only on the basis of their platinum sensitivity; indeed there was no initial selection for BRCA mutations, and mutation status was initially unknown in the majority of cases (64%). However, retrospective analysis indicated that 136 patients (51%) were positive for BRCA1 or 2; this group achieved a PFS of 11.2 months, while the placebo-only group achieved 4.3 months (HR 0.18). In wild-type BRCA patients, olaparib showed more modest results, as expected: mPFS 7.4 months versus 5.5 months (HR 0.54). No significant differences were seen in OS analysis.38
The other phase II trial by Oza and colleagues39 was different, because olaparib was introduced in the treatment with chemotherapy and continued in monotherapy after the completion of chemotherapy: so, one arm had a standard chemotherapy treatment with carboplatin and paclitaxel and another had carboplatin, paclitaxel and olaparib. In this case, BRCA status was known for few patients at the beginning of the trial but, as expected, the addition of olaparib had very good effect on BRCAm patients. Overall, olaparib plus chemotherapy resulted in an mPFS of 12.2 versus 9.6 months for chemotherapy alone. For BRCAm patients the HR was 0.21. No differences were seen in OS.
In the NOVA trial, the population was initially selected for BRCA1/2 mutations and also the evaluation of HRD was introduced to investigate the effect of the drug in all potentially sensitive patients. Inclusion criteria required that patients have received at least four cycles of platinum-based chemotherapy, with the possible addition of bevacizumab,41,42 with CR or PR before starting niraparib. Among the patients enrolled in the trial, only about 25% in each cohort have had a previous treatment with bevacizumab. Data about efficacy of niraparib are biased by these strategies; moreover, it is not correct to talk of niraparib maintenance therapy, but rather of switch maintenance. What we have seen is that niraparib showed very interesting results in gBRCAm patients, but also showed to be effective in wtBRCA HRD-positive patients and in wtBRCA HRD-negative patients, with an HR similar to that of the olaparib trial (HR 0.58). Results in the HRD-positive population with sBRCA mutation need to be further investigated, because this population was small in the trial.
Some considerations have to be made: somatic mutation testing is more laborious and less reproducible than germline testing; while in germline testing genes are evaluated in healthy cells, in somatic testing the evaluation is carried out in cancer cells, which are very heterogeneous. Obtaining high-quality DNA from tumour cells is quite difficult and depends on well-preserved and well-sized specimens. Finally, as we have said before, cancer cells are heterogeneous by definition and intra-tumour heterogeneity could lead to a change in biomarkers, like sBRCA, over time. Further evaluations are needed.
In the NOVA trial, initial division was used with germline evaluation, while HRD was evaluated in tissue samples using myChoice; all the evaluations made about differences between germline and somatic testing have to be kept in mind.
Surprisingly, even if the population was selected in a different way – to have a greater population who could benefit from PARPis treatment, not only gBRCAm but also a HRD-positive subgroup – patients receiving niraparib had a significantly longer PFS than those receiving placebo regardless of gBRCA mutations or HRD status. The effect of niraparib in the BRCAwt population is clearly visible; indeed 20% of this population showed more than 18 months of benefit from niraparib administration.
However, authors did not report how many cycles (average/ranging) were administered; moreover, in the study a dose reduction was allowed (300 → 200 → 100 mg), but, unlike the olaparib trial, efficacy was not reported at different doses: data about dose reductions are awaited.
These results could be an important success for the drug, making niraparib the first choice in the treatment of EOC, not only in gBRCAm EOC, but also in BRCAwt. However, some patients with BRCAm EOC do not respond or develop resistance. A possible explanation could be the following: secondary genetic and epigenetic events (such as secondary BRCA1/2 mutations) that restore functional HR in HR-deficient tumours; suppression of NHEJ via loss of 53BP1 or other mechanisms which leads to PARPis resistance but not platinum resistance; increased expression of p-glycoprotein efflux transporter mediating multi-drug resistance. On the other hand, we have a BRCAwt HR-non-deficient population that responds to niraparib: how can this happen?
In consideration of the setting in which niraparib is administered, which is maintenance treatment, BRCA status and HRD, even if they may provide essential information about treatment efficacy, do not meet the clinical definition of platinum sensitivity. Maybe the myChoice test does not investigate some other gene involved in the HR system – that, we still do not know. Other studies are awaited to confirm this result.
In a future scenario, PARPis will be administered to all EOC patients, and BRCA and HRD testing will not be the indicators of eligibility, but biomarkers of treatment efficacy. Our opinion is that initial evaluation should be somatic testing on tumour samples. If a mutation is found, patients will be referred for genetic counselling to investigate whether this mutation is somatic or germline. We want to underline that a germline mutation also has consequences for the lives of the patient’s relatives.
In consideration of the manageable side effects of long-term administration, niraparib could be considered a well-tolerated drug to use in combination treatment: a possible strategy to improve outcomes in EOC could be the combination with another maintenance treatment that has already showed some results, like bevacizumab; the creation of neo-antigens could potentiate the action of immunotherapy (Table 2). It is confirmed that checkpoint inhibitors, and in particular anti-PD1 antibodies, are well tolerated and effective in EOC. Immunotherapy may play a significant role in future clinical management, improving the prognosis of EOC. First results about efficacy in platinum-resistant EOC with nivolumab have already been published.40
Table 2.
Ongoing trials with niraparib.
| Trial (ClinicalTrials.gov identifier) | Title | Recruitment |
|---|---|---|
| NCT01847274 | A maintenance study with niraparib versus placebo in patients with platinum sensitive ovarian cancer | Completed |
| NCT02354131 | Niraparib and/or niraparib-bevacizumab combination against bevacizumab alone in HRD platinum sensitive ovarian cancer | Recruiting |
| NCT01227941 | MK-4827 in combination with pegylated liposomal doxorubicin in participants with advanced solid tumors and ovarian cancer (MK-4827-011) | Terminated |
| NCT02655016 | A study of niraparib maintenance treatment in patients with HRD-positive advanced ovarian cancer following response on front-line platinum-based chemotherapy | Recruiting |
| NCT02354586 | A study of niraparib in patients with ovarian cancer who have received three or four previous chemotherapy regimens | Recruiting |
| NCT02657889 | Study of niraparib in combination with pembrolizumab (MK-3475) in patients with triple-negative breast cancer or ovarian cancer | Recruiting |
| NCT00749502 | A study of MK4827 in participants with advanced solid tumors or hematologic malignancies (MK-4827-001 AM8) | Completed |
Efficacy of anti-PD1 is well established in other tumours, like lung cancer, kidney cancer, urothelial cancer and melanoma; moreover, high mutational loads are associated with improved survival in melanoma patients but are not predictive of response to anti-PD-1 therapy, suggesting that other genomic and non-genomic features also contribute to response patterns on PD-1 checkpoint blockade therapy. Hugo and colleagues analysed the somatic mutanomes and transcriptomes of pre-treatment melanoma biopsies to identify factors that may influence innate sensitivity or resistance to anti-PD-1 therapy, finding that overall high mutational load is associated with improved survival, and tumours from responding patients are enriched for mutations in the DNA repair gene BRCA2. Thus, BRCA2 loss-of-function mutations, which are expected to produce defects in HR and DNA DSB repair, may produce specific mutational signatures or unknown effects (e.g. induction of cell death) that contribute to anti-PD-1 responsiveness.41,42
Moreover, considering PARPis mechanism of action, their use could further increase the mutational loads in BRCAm EOC patients; therefore, it would be very interesting to evaluate the effectiveness of the combination of PARPis and anti-PD1 in BRCAm EOC patients, as is ongoing in some trials.
Our knowledge about all mechanisms involved in the control of DNA damage is still incomplete. PARP are ubiquitous enzymes in the nucleus of the cells and we can certainly state that it is the first line of defence when DNA is damaged. Inhibiting the PARP family could cause more DNA damage than we want to, having some carcinogenic consequences, as we saw with the incidence of myelodysplastic syndrome in patients under olaparib or niraparib. In our opinion, it could be interesting to understand backup pathways that could cause less DNA damage, causing cell death. Particularly, it could be fascinating to target backup pathways that are less active in normal cells; in this area of investigation one possible backup pathway could be driven by the Rad52 protein.43–45
Acknowledgments
This work was realized on behalf of Consorzio Interuniversitario Nazionale per la Bio-Oncologia (CINBO) – Via dei Vestini, 31, 66100 Chieti, Italy.
Footnotes
Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Conflict of interest statement: The authors declare that there is no conflict of interest.
Contributor Information
Davide Caruso, Department of Medico-Surgical Sciences and Biotechnologies, University of Rome ‘Sapienza’, Latina, Italy.
Anselmo Papa, Department of Medico-Surgical Sciences and Biotechnologies, University of Rome ‘Sapienza’, Corso della Repubblica 79, 04100, Latina, Italy.
Silverio Tomao, Department of Medico-Surgical Sciences and Biotechnologies, University of Rome ‘Sapienza’, Latina, Italy.
Patrizia Vici, Division of Medical Oncology 2, ‘Regina Elena’ National Cancer Institute, Rome, Italy.
Pierluigi Benedetti Panici, Department of Gynaecology and Obstetrics, University of Rome ‘Sapienza’, Rome, Italy.
Federica Tomao, Department of Gynaecology and Obstetrics, University of Rome ‘Sapienza’, Rome, Italy; Department of Gynecology, University of Heraklion, Heraklion, Greece.
References
- 1. Quinn M, Babb P, Brock A. Cancer trends in England and Wales 1950–1999. London: The Stationery Office, 2001. [Google Scholar]
- 2. Shepherd JH. Revised FIGO staging for gynaecological cancer. Br J Obstet Gynaecol 1989; 96: 889–892. [DOI] [PubMed] [Google Scholar]
- 3. De Angelis R, Sant M, Coleman MP, et al. Cancer survival in Europe 1999–2007 by country and age: results of EUROCARE-5: a population-based study. Lancet Oncol 2014; 15: 23–34. [DOI] [PubMed] [Google Scholar]
- 4. Chao S-Y, Chiang J-H, Huang A-M, et al. An integrative approach to identifying cancer chemoresistance-associated pathways. BMC Med Genomics 2011; 4: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mavaddat N, Barrowdale D, Andrulis IL, et al. Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA). Cancer Epidemiol Biomarkers Prev 2012; 21: 134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Risch HA, McLaughlin JR, Cole DE, et al. Prevalence and penetrance of germline BRCA1 and BRCA2 mutations in a population series of 649 women with ovarian cancer. Am J Hum Genet 2001; 68: 700–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Risch HA, McLaughlin JR, Cole DEC, et al. Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: a kin-cohort study in Ontario, Canada. J Natl Cancer Inst 2006; 98: 1694–1706. [DOI] [PubMed] [Google Scholar]
- 8. Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005; 434: 913–917. [DOI] [PubMed] [Google Scholar]
- 9. Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005; 434: 917–921. [DOI] [PubMed] [Google Scholar]
- 10. Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature 2012; 481: 287–294. [DOI] [PubMed] [Google Scholar]
- 11. Davar D, Beumer JH, Hamieh L, et al. Role of PARP inhibitors in cancer biology and therapy. Curr Med Chem 2012; 19: 3907–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O’Brien PJ. Catalytic promiscuity and the divergent evolution of DNA repair enzymes. Chem Rev 2006; 106: 720–752. [DOI] [PubMed] [Google Scholar]
- 13. Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009; 461: 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene 2006; 25: 5864–5874. [DOI] [PubMed] [Google Scholar]
- 15. Chambon P, Weill JD, Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem Biophys Res Commun 1963; 11: 39–43. [DOI] [PubMed] [Google Scholar]
- 16. Dantzer F, de La Rubia G, Ménissier-De Murcia J, et al. Base excision repair is impaired in mammalian cells lacking poly(ADP-ribose) polymerase-1. Biochemistry (Mosc) 2000; 39: 7559–7569. [DOI] [PubMed] [Google Scholar]
- 17. Amé JC, Rolli V, Schreiber V, et al. PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem 1999; 274: 17860–17868. [DOI] [PubMed] [Google Scholar]
- 18. Dantzer F, Schreiber V, Niedergang C, et al. Involvement of poly(ADP-ribose) polymerase in base excision repair. Biochimie 1999; 81: 69–75. [DOI] [PubMed] [Google Scholar]
- 19. Fortini P, Pascucci B, Parlanti E, et al. The base excision repair: mechanisms and its relevance for cancer susceptibility. Biochimie 2003; 85: 1053–1071. [DOI] [PubMed] [Google Scholar]
- 20. Andrabi SA, Dawson TM, Dawson VL. Mitochondrial and nuclear cross talk in cell death: parthanatos. Ann N Y Acad Sci 2008; 1147: 233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ledermann JA, Drew Y, Kristeleit RS. Homologous recombination deficiency and ovarian cancer. Eur J Cancer Oxf Engl 1990 2016; 60: 49–58. [DOI] [PubMed] [Google Scholar]
- 22. Saleh-Gohari N, Bryant HE, Schultz N, et al. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol Cell Biol 2005; 25: 7158–7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ashworth A. A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J Clin Oncol 2008; 26: 3785–3790. [DOI] [PubMed] [Google Scholar]
- 24. Audeh MW. Novel treatment strategies in triple-negative breast cancer: specific role of poly(adenosine diphosphate-ribose) polymerase inhibition. Pharmacogenomics Pers Med 2014; 7: 307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer 2004; 4: 814–819. [DOI] [PubMed] [Google Scholar]
- 26. De Summa S, Pinto R, Sambiasi D, et al. BRCAness: a deeper insight into basal-like breast tumors. Ann Oncol 2013; 24(Suppl. 8): viii13–viii21. [DOI] [PubMed] [Google Scholar]
- 27. AlHilli MM, Becker MA, Weroha SJ, et al. In vivo anti-tumor activity of the PARP inhibitor niraparib in homologous recombination deficient and proficient ovarian carcinoma. Gynecol Oncol 2016; 143: 379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zaremba T, Curtin NJ. PARP inhibitor development for systemic cancer targeting. Anticancer Agents Med Chem 2007; 7: 515–523. [DOI] [PubMed] [Google Scholar]
- 29. Wang L, Mason KA, Ang KK, et al. MK-4827, a PARP-1/-2 inhibitor, strongly enhances response of human lung and breast cancer xenografts to radiation. Invest New Drugs 2012; 30: 2113–2120. [DOI] [PubMed] [Google Scholar]
- 30. Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011; 474: 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pennington KP, Walsh T, Harrell MI, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res 2014; 20: 764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sandhu SK, Schelman WR, Wilding G, et al. The poly(ADP-ribose) polymerase inhibitor niraparib (MK4827) in BRCA mutation carriers and patients with sporadic cancer: a phase 1 dose-escalation trial. Lancet Oncol 2013; 14: 882–892. [DOI] [PubMed] [Google Scholar]
- 33. Mirza MR, Monk BJ, Herrstedt J, et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N Engl J Med 2016; 375: 2154–2164. [DOI] [PubMed] [Google Scholar]
- 34. Telli ML, Timms KM, Reid J, et al. Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin Cancer Res 2016; 22: 3764–3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Minckwitz GV, Timms K, Untch M, et al. Prediction of pathological complete response (pCR) by homologous recombination deficiency (HRD) after carboplatin-containing neoadjuvant chemotherapy in patients with TNBC: results from GeparSixto. J Clin Oncol 2015; 33, http://meetinglibrary.asco.org/content/147752-156 (accessed 6 February 2017). [Google Scholar]
- 36. Wilcoxen KM, Becker M, Neff C, et al. Use of homologous recombination deficiency (HRD) score to enrich for niraparib sensitive high grade ovarian tumors. J Clin Oncol 2015; 33, http://meetinglibrary.asco.org/content/147040-156 (accessed 6 February 2017). [Google Scholar]
- 37. Ledermann J, Harter P, Gourley C, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med 2012; 366: 1382–1392. [DOI] [PubMed] [Google Scholar]
- 38. Ledermann J, Harter P, Gourley C, et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol 2014; 15: 852–861. [DOI] [PubMed] [Google Scholar]
- 39. Oza AM, Cibula D, Benzaquen AO, et al. Olaparib combined with chemotherapy for recurrent platinum-sensitive ovarian cancer: a randomised phase 2 trial. Lancet Oncol 2015; 16: 87–97. [DOI] [PubMed] [Google Scholar]
- 40. Hamanishi J, Mandai M, Ikeda T, et al. Safety and antitumor activity of anti-PD-1 antibody, nivolumab, in patients with platinum-resistant ovarian cancer. J Clin Oncol 2015; 33: 4015–4022. [DOI] [PubMed] [Google Scholar]
- 41. Hugo W, Zaretsky JM, Sun L, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 2016; 165: 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Holloman WK. Unraveling the mechanism of BRCA2 in homologous recombination. Nat Struct Mol Biol 2011; 18: 748–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Delahaye-Sourdeix M, Oliver J, Timofeeva MN, et al. The 12p13.33/RAD52 locus and genetic susceptibility to squamous cell cancers of upper aerodigestive tract. PLoS One 2015; 10: e0117639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shi T-Y, Yang G, Tu X-Y, et al. RAD52 variants predict platinum resistance and prognosis of cervical cancer. PLoS One 2012; 7: e50461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Choi BH, Chen Y, Dai W. Chromatin PTEN is involved in DNA damage response partly through regulating Rad52 sumoylation. Cell Cycle Georget Tex 2013; 12: 3442–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]