Abstract
By genetic analysis of Caenorhabditis elegans mutants defective in yolk uptake, we have identified new molecules functioning in the endocytosis pathway. Here we describe a novel J-domain–containing protein, RME-8, identified by such genetic analysis. RME-8 is required for receptor-mediated endocytosis and fluid-phase endocytosis in various cell types and is essential for C. elegans development and viability. In the macrophage-like coelomocytes, RME-8 localizes to the limiting membrane of large endosomes. Endocytosis markers taken up by the coelomocytes rapidly accumulate in these large RME-8–positive endosomes, concentrate in internal subendosomal structures, and later appear in RME-8–negative lysosomes. rme-8 mutant coelomocytes fail to accumulate visible quantities of endocytosis markers. These observations show that RME-8 functions in endosomal trafficking before the lysosome. RME-8 homologues are found in multicellular organisms from plants to humans but not in the yeast Saccharomyces cerevisiae. These sequence homologies suggest that RME-8 fulfills a conserved function in multicellular organisms.
INTRODUCTION
Intracellular trafficking of endocytosed molecules requires an elaborate network of organelles (Mellman, 1996; Mukherjee et al., 1997). The compartments within the endocytic network were established in mammalian cells by studying the trafficking kinetics of ligands and receptors (such as transferrin and transferrin receptor, epidermal growth factor [EGF], and EGF-receptor, and low-density lipoprotein [LDL] and LDL receptor) and of fluid-phase markers. Ligands and receptors are often internalized through clathrin-coated pits and vesicles and then are targeted to early or sorting endosomes by vesicle fusion. In the sorting endosome, many ligands can dissociate from their receptors because of a low pH environment. The ligands bound for degradation are then sorted and targeted to the late endosome (Dunn and Maxfield, 1992). Recycling receptors are sorted to an endocytic recycling compartment, from which they recycle back to the plasma membrane (van der Sluijs et al., 1992). In the late endosome, ligands and other lysosome targeted molecules are further sorted, concentrated, and delivered to the lysosome for enzymatic degradation (Futter et al., 1996).
Each of these trafficking steps occurs in a specific vesicular compartment. Despite the continuous flow of endocytosed molecules, these compartments are characterized by unique morphologies, sizes, intravesicular pH levels, membrane proteins, and lipid compositions, although these characteristics can vary with different cell types (Mellman, 1996; Mukherjee et al., 1997; Odorizzi et al., 2000). In mammalian cells, sorting endosomes have a tubular-vesicular morphology with an internal pH ∼6.0 (Mukherjee et al., 1997). Late endosomes are similar in size to sorting endosomes but have characteristic internal vesicles formed by the invagination of late endosomal membranes (Hirsch et al., 1968; Futter et al., 1996). Two alternative models were proposed to describe the transport of contents from sorting endosomes through late endosomes to lysosomes. According to the maturation model, sorting endosomes mature to become late endosomes by losing sorting endosome markers and by acquiring late endosome markers (Dunn and Maxfield, 1992; van Deurs et al., 1993). Late endosomes further mature to become lysosomes. A vesicular transport model predicts that endosomes are stable subcellular compartments and that the endosomal contents move from one compartment to another by means of small transport vesicles (Gruenberg et al., 1989; Gruenberg and Maxfield, 1995). In both models, many macromolecules are expected to function together in the formation and maintenance of these unique membrane structures and in the trafficking of endocytic contents through them. Most of these macromolecules remain to be identified, particularly in multicellular organisms.
A few molecular markers are found to label each of these compartments, and they are frequently used to define specific compartments in mammalian cells (Mellman, 1996; Mukherjee et al., 1997). Studies of a class of small GTPases called Rabs that are associated with different vesicular compartments provide a framework for understanding membrane trafficking mechanisms (Olkkonen and Stenmark, 1997). It has been proposed that Rabs provide the specificity for the targeting of vesicles from one compartment to another by regulating specific fusion of homo- or heterogeneous vesicles. For example, Rab5 primarily associates with early endosomes in mammalian cells and functions in the fusion of endocytic vesicles with early endosomes (Gorvel et al., 1991). Rab5 regulates the specificity of membrane fusion by recruiting another early endosome-associated protein EEA1 (Simonsen et al., 1998; Christoforidis et al., 1999).
Studies of yeast mutants defective in endocytosis and intracellular trafficking have defined trafficking compartments similar to those in mammalian cells (Bryant and Stevens, 1998; Wendland et al., 1998). Taking advantage of the ease of genetic manipulation and biochemical analysis in yeast, many genes have been assigned to functional steps in the trafficking pathway. Some of these yeast genes have sequence homologues in multicellular organisms. For example, the yeast homolog of rab5, VPS21, and the yeast homolog of EEA1, Vac1, function in a similar molecular mechanism to rab5 and EEA1, albeit in a different transport pathway in the Golgi-to-endosome stage of protein transport (Peterson et al., 1999; Odorizzi et al., 2000). Drosophila sequence homologues of the yeast VPS18 and VPS33, Deep orange and Carnation, function in a protein complex in the late endosome in both species (Sevrioukov et al., 1999). However, the full extent of the mechanistic similarities of endocytic trafficking between yeast and multicellular organisms remains to be determined.
To gain more insights into the molecular mechanisms of these trafficking steps in multicellular organisms, we performed genetic screens for mutants defective in endocytosis in Caenorhabditis elegans (Grant and Hirsh, 1999). These screens were based on an assay that monitors the receptor-mediated endocytosis of yolk proteins by growing oocytes. Several mutant strains were isolated that contain mutations in the yolk receptor, a member of the LDL receptor–related protein family (Grant and Hirsh, 1999). Several other mutant strains were isolated that show defective endocytosis in many cell types in addition to the oocyte, indicating that these genes encode endocytosis factors common to many cell types.
Here, we describe the molecular and phenotypic characterization of one of these mutants, rme-8. We found that RME-8 is required for both receptor-mediated and fluid-phase endocytosis and that it is likely to function in an endocytic trafficking event before transport into lysosomes. Because RME-8 is conserved in multicellular organisms, the mechanisms of endosomal trafficking revealed by studying it in C. elegans are likely to be generally applicable to other multicellular organisms. The discovery of RME-8 also indicates that our genetic screens can readily identify new genes required for endocytosis.
MATERIALS AND METHODS
General Methods and Microscopy
Worm cultures, genetic crosses, and other C. elegans methods were performed according to standard protocols (Brenner, 1974; Wood, 1988). Strains carrying rme-8(b1023) were grown at the permissive temperature (15°C) at which the only phenotype observed was the defect in YP170::GFP (green fluorescent protein) uptake. The other phenotypes of rme-8 (defective fluid-phase endocytosis, molting defect, and lethality) were examined when worms were shifted to the restrictive temperature (25°C). Strains without rme-8(b1023) were grown at 20°C. Confocal images were obtained on a Zeiss LSM 410 confocal microscope (Carl Zeiss, Inc, Thornwood, NY) and processed with Adobe PhotoShop 5.0 (Adobe Systems, San Jose, CA).
Endocytosis Assays
Two assays were used to monitor the steady-state endocytosis in C. elegans: YP170::GFP endocytosis and GFP fluid-phase endocytosis. The YP170::GFP endocytosis assay detects the receptor-mediated endocytosis of yolk protein into oocytes (Grant and Hirsh, 1999). The GFP fluid-phase endocytosis assay detects the endocytosis of pseudocoelomic GFP (Fares and Greenwald, 2001). In this assay, a secreted form of GFP is synthesized in the body wall muscle from an integrated transgenic array arIs37[pmyo-3::ssGFP] and secreted into the body cavity, where the GFP is nonspecifically endocytosed primarily by coelomocytes.
The trafficking kinetics of fluid-phase markers in coelomocytes was performed as follows. Texas Red–conjugated BSA (TR-BSA; Sigma, St. Louis, MO) was injected at 1 mg/ml into the pseudocoelomic space in the pharyngeal region of adult worms that were semidehydrated on a dried agarose pad immersed in oil (Mello et al., 1991). Injected worms were then rehydrated in M9 buffer and transferred to a seeded NGM plate at 20°C. At different time points (e.g., 5 min, 10 min, etc.), the injected worms were transferred to an ice-chilled NGM plate. This incubation on ice efficiently stops the intracellular trafficking of endocytosed molecules. Worms were then mounted in 1% paraformaldehyde on agarose pads for confocal microscopy. Separate injected worms were used for different time points. For each time point, similar results were obtained with more than five coelomocytes of different worms.
Genetic Mapping and Molecular Cloning
rme-8(b1023) was isolated in a genetic screen for ts mutants defective in YP170::GFP uptake with the use of N-ethyl-N-nitrosourea (ENU) as the mutagen (Grant and Hirsh, 1999). ENU was known to induce a higher frequency of transversions than that of EMS (De Stasio et al., 1997). rme-8 was first mapped around stP124 on chromosome I by STS mapping (Williams et al., 1992). rme-8 was placed between unc-13 and lin-10 by the following three 3-factor mapping results: dpy-5 (57) rme-8 (22) unc-29; SEM-4 (6) rme-8 (0) unc-13; and unc-13 (29) rme-8 (5) lin-10. At the same time, rme-8 was also localized to the same region by mapping with five deficiencies on chromosome I: nDf24, nDf25, mnDf111, and hDf9 deleted rme-8. qDf16, ozDf5, and sDf6 did not delete rme-8. In these genetic mappings, both YP170::GFP endocytosis and lethality phenotypes were scored, and they were found to cosegregate, indicating a tight genetic linkage.
Eight predicted genes (D20005.1, R11A5.1, T22C1.6, H05L14.2, F18C12.1, F18C12.2, C09H6.1, and K04G2.3; The C. elegans Sequencing Consortium, 1998) located between unc-13 and lin-10 were selected to test their endocytosis functions with the YP170::GFP endocytosis assay. The activity of each of these genes was disrupted by RNA interference (RNAi; Fire et al., 1998) in worms expressing YP170::GFP. Representative EST clones (kindly provided by Y. Kohara, National Institute of Genetics, Mishima, Japan) derived from each of these predicted genes were used to make dsRNA by in vitro transcription. dsRNA was injected at 200–400 μg/ml into the syncytial region of the hermaphrodite gonad. Injected hermaphrodites were assayed for YP170::GFP uptake defects, and their progeny were analyzed for other phenotypes such as developmental arrest or morphological defects.
To identify rme-8 containing DNA fragments, cosmids (C54G4, T22C1, F22E4, F20G4, T04A2, D1085, F18C12, and K04G2; 10–20 μg/ml; clones kindly provided by A. Coulson, Sanger Center, United Kingdom) together with the dominant transgenic marker rol-6(su1006) (plasmid pRF4, 50 μg/ml) were microinjected into rme-8(b1023) worms to establish transgenic strains at 15°C (Mello et al., 1991). Several independent extrachromosomal arrays for each cosmid were tested for rescue of the lethal phenotype at 25°C. Only F18C12 rescued the lethality (2/2 lines rescued). A subcloned 16.3-kb FspI fragment (plasmid F18C12.2FspI), containing only F18C12.2 and its promoter region, rescued the lethal phenotype of rme-8(b1023) (3/3 lines rescued) as well as the coelomocyte GFP endocytosis defect. To identify mutations in rme-8(b1023), the F18C12.2 genomic region was amplified by PCR in five fragments with a Hi-Fidelity PCR kit (Boehringer Mannheim, Indianapolis, IN) and sequenced by automated DNA sequencing. The sequence was compared with the wild-type genomic sequence from the C. elegans Genome Sequencing Project. The A to T transversion in b1023 was confirmed by sequencing a different PCR product with the use of different primers.
The RME-8 cDNA sequence was determined by comparing the genomic sequence from the Genomic Sequencing Project and the sequences obtained by sequencing EST clone yk212c7, reverse transcription (RT)-PCR fragments and 5′-RACE (rapid amplification of 5′ cDNA ends) products. By RT-PCR, the trans-spliced leader SL1 (Krause and Hirsh, 1987) but not SL2 (Spieth et al., 1993) together with an internal primer (R3653: 5′-CGAAGACCAGAATATGGAAC-3′) amplified a 1.2-kb fragment. Sequencing this RT-PCR product revealed that SL1 was spliced to an exon that follows a consensus splice acceptor sequence. Similar result was obtained by performing 5′-RACE (Life Technologies-BRL, Rockville, MD) with the same internal primer. The assembled RME-8 coding region is 6837 bp, with a 420-bp 3′UTR, derived from 23 exons. Based on the sequences from two other ESTs, yk250d6 and yk398h12, an alternatively spliced form of RME-8, may exist that lacks the 24-bp-long exon 19. The J-domain was identified by searching the Pfam protein motif database. The IWN repeats were identified by with the use of the MEME program (http://meme.sdsc.edu/meme/website/meme.html), which was designed to recognize repeats.
The RME-8 homologues were identified by performing BLASTP and TBLASTN searches with RME-8 sequence against the nonredundant sequences in the GenBank. Homology was found in EST KIAA0678, which contains the C-terminal half of putative human RME-8. No genomic sequence for this clone is available presently. The Drosophila Genome Sequencing Project predicted only the first two exons for Drosophila RME-8 (Adams et al., 2000). The full-length Drosophila RME-8 was identified by TBLASTN, with C. elegans RME-8 against the complete Drosophila genomic sequence (Adams et al., 2000). The intron–exon boundaries of Drosophila RME-8 were predicted on the basis of its identity and similarity to C. elegans RME-8. This splicing pattern was further confirmed and refined based on the gene prediction program GeneMark (http://genemark.biology.gatech.edu/GeneMark). This prediction is supported by the presence of several matching Drosophila EST sequences in GenBank (LD15941, LD11777, LD41107, LP02307, LD15569, GM07668, and LD15941). The predicted Drosophila RME-8 coding region is 7221 bp long derived from 21 exons. The plant RME-8 from Arabidopsis thaliana was from the predicted gene AC005168 in GenBank.
The C. elegans RME-8 cDNA was deposited in GenBank (Accession number AF372457).
Antibody Production, Immunochemistry, and RME-8::GFP Reporter Constructs
An RME-8 C-terminal fragment that contains amino acids from positions 2064 to 2270 was amplified by PCR from yk212c7 (forward primer: F212BAM2, 5′-GCATGGATCCAGCCTTTGTTGAACTGGTCC-3′; reverse primer: R212XHO2, 5′-GCATCTCGAGTTGATGCATTGGTGTGTGCTG-3′) and cloned into the 6xHis expression vector pET24b (Novagen, Madison, WI) at BamHI and XhoI sites. The His-tagged recombinant protein was expressed in Escherichia coli strain BL21 and purified with a Nickel agarose column under denaturing conditions according to the manufacturer's instructions (Qiagen, Chatsworth, CA). The protein was further gel-purified by SDS-PAGE. Gel slices containing the denatured protein from the SDS-PAGE were excised and used for antibody production in rabbits at Covance Immunological Services (Denver, PA). Specific antibodies were affinity-purified on a Nickel agarose column (Gu et al., 1994). Total worm proteins were extracted in 2 volumes of SDS loading buffer by boiling for 15 min and used for SDS-PAGE. Western blot analysis was performed with the use of the ECL system (Amersham, Arlington Heights, IL).
Two protocols were used for immunofluorescence staining. For staining gonads, ovaries were dissected from adult hermaphrodites and processed for staining as in Grant and Hirsh (1999). For whole mount staining, mix-staged worms were collected for fixation and permealization (Bettinger et al., 1996). In both protocols, purified primary antibody against RME-8 was applied to samples at 1:50 dilution for overnight incubation at 20°C. Anti-yolk protein mouse monoclonal antibodies (01C103 and pIIaA3, kindly provided by S. Strome, University of Indiana, Bloomington, IN)) were used as 1:100 dilutions. Secondary antibodies that are conjugated with Cy3 (Jackson ImmunoResearch Laboratories, West Grove, PA) were applied at 1:500 dilutions at 20°C. Stained samples were mounted in Slowfade Light (Molecular Probes, Eugene, OR) on an agarose pad and examined immediately.
RME-8::GFP reporters were constructed by inserting GFP into two existing unique restriction enzyme sites in the RME-8 coding region: AgeI in exon 9 or MluI site in exon 21. PCR-amplified GFP (S65C variant from plasmid pPD117.01, Fire et al., personal communication) engineered with AgeI or MluI sites were inserted into the corresponding unique sites of plasmid F18C12.2FspI. The resulting RME-8 gene fusions (RME-8::GFP/AgeI and RME-8::GFP/MluI) contain the full-length RME-8 protein. Extrachromosomal arrays carrying each of these fusions (20 μg/ml) were established with rol-6(su1006) (50 μg/ml) as the transgenic marker in rme-8(b1023) worms to test their abilities to rescue the lethal phenotype. RME-8::GFP/AgeI failed to rescue (0/5 lines rescued), whereas RME-8::GFP/MluI rescued the lethal phenotype (2/3 lines rescued). Each of these fusions (20 μg/ml) together with rol-6(su1006) (50 μg/ml) were also introduced into wild-type N2 worms to establish both extrachromosomal arrays and integrated transgenic lines. Worms carrying RME-8::GFP/AgeI, either extrachromosomal arrays or integrated lines, showed diffuse GFP expression in the cytoplasm (integrated line: strain DH1334: bIs32[rme-8::GFP/AgeI; rol-6]; our unpublished results). Worms carrying RME-8::GFP/MluI, both extrachromosomal arrays and integrated lines, showed GFP expression localized to punctate structures similar to those observed with antibody staining. This construct is referred to as RME-8::GFP, and an integrated line (strain DH1336: bIs34[rme-8::GFP/MluI; rol-6]) was used in experiments described in the text. When crossed into rme-8(b1023) background, this transgene rescued the coelomocyte GFP endocytosis defect (our unpublished results).
RESULTS
rme-8 is an Essential Gene Required for Multiple Cell Types in C. elegans
As in many animals, C. elegans yolk proteins are synthesized in somatic tissues and are taken up into vesicles of the oocyte by receptor-mediated endocytosis (Schneider, 1996). Taking advantage of this phenomenon, we used a transgene carrying a fusion of the C. elegans major yolk protein YP170 with GFP (YP170::GFP) to monitor endocytosis in live animals (Grant and Hirsh, 1999). Like the endogenous yolk proteins, YP170::GFP is synthesized in the adult intestine, secreted into the pseudocoelom from which it is endocytosed by growing oocytes, and stored in yolk granules. With the use of classical genetic screens for mutants defective in YP170::GFP uptake, both viable and temperature-sensitive (ts) lethal mutants were isolated (Grant and Hirsh, 1999). Here, we have analyzed one ts lethal mutant, b1023, which we assigned the gene name rme-8 (rme for receptor-mediated endocytosis defective).
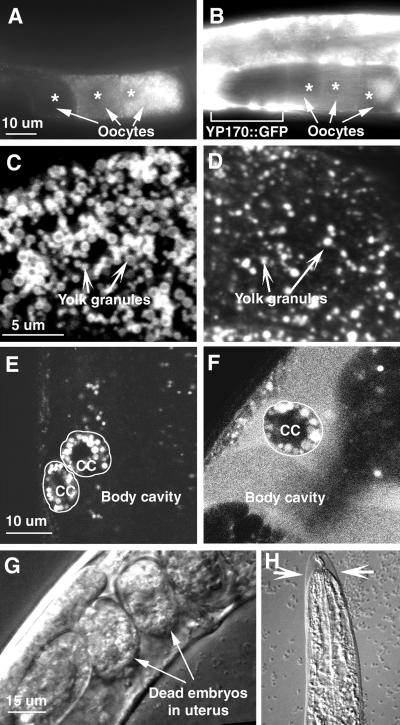
rme-8(b1023) is defective for yolk protein uptake. In wild-type animals, YP170::GFP was efficiently cleared from the pseudocoelom by growing oocytes (Figure 1A). In rme-8(b1023) worms, YP170::GFP accumulated in the pseudocoelomic space, suggesting that the YP170::GFP uptake by oocytes was defective (Figure 1B). This uptake defect was not specific to YP170::GFP; the uptake of endogenous yolk proteins was also reduced, as evidenced by abundant pseudocoelomic yolk was visible as oily fluid in rme-8 mutants (our unpublished results). In contrast to other phenotypes described below, the YP170::GFP and endogenous yolk uptake defect was not ts. The defect in yolk endocytosis led to the formation of fewer and smaller yolk-containing vesicles (Figure 1, C and D). This defective yolk granule biogenesis indicates that RME-8 is required for the formation of yolk granules, which are specialized lysosomal compartments. These small yolk-containing vesicles could be small mature yolk granules equivalent to lysosomes or immature yolk granules equivalent to an earlier endocytic compartment.
Figure 1.
rme-8 is an essential gene required for endocytosis in multiple cells. rme-8(b1023) mutants display defects in receptor-mediated endocytosis (A–D) and fluid-phase endocytosis (E and F) and show developmental arrest (G and H). In all experiments, worms were increased at the permissive temperature and shifted to the restrictive temperature for 24 h. (A and B) Adult hermaphrodites of wild-type (A) and rme-8(b1023) (B) carrying the YP170::GFP transgene. Arrows point to the three most full-grown oocytes (asterisks labeling the positions of nuclei) with the most mature one on the right. In wild-type, YP170::GFP is efficiently endocytosed and stored in oocytes with the highest level in the most full-grown oocyte. In rme-8(b1023), YP170::GFP has reduced storage in oocytes, and an accumulation in the pseudocoelomic space (e.g., the large white patches indicated by the bracket). (C and D) Confocal micrographs of yolk granules in wild-type (C) and rme-8(b1023) (D) oocytes. Wild-type oocytes have abundant yolk in dense large yolk granules. In contrast, rme-8(b1023) oocytes have much reduced yolk in sparse small vesicles. Yolk proteins were visualized by immunofluorescence staining. Confocal micrographs were taken by focusing on the cortical region of the most mature oocytes. (E and F) Endo cytosis of pseudocoelomic GFP in wild-type (E) and rme-8(b1023) worms (F). In wild-type, the body cavity is dark and void of GFP. Two adjacent coelomocytes (cc, cell boundaries marked with circles) have abundant vesicles containing GFP. In rme-8(b1023), GFP accumulated in the body cavity. The single coelomocyte (cc, cell boundary marked with a circle) in this focal plane has some GFP-containing vesicles. (G and H) Developmental defects in rme-8(b1023). Adult hermaphrodites often contain a few dead embryos (G), and larvae usually arrest in the next molt (H, an arrested L3 larva; arrows point to the unshed cuticle).
rme-8(b1023) is defective in fluid-phase endocytosis in a ts manner as determined in an in vivo assay. In this assay, GFP protein is secreted from the body wall muscle into the pseudocoelom (Fares and Greenwald, 2001). This pseudocoelomic GFP is efficiently endocytosed by coelomocytes of wild-type worms and degraded in lysosomes, and therefore GFP does not accumulate in the pseudocoelom (Figure 1E). The presence of intracellular GFP-containing vesicles in coelomocytes reflects the balance between continuing endocytosis and lysosomal degradation. In rme-8(b1023) mutants, GFP uptake was normal at the permissive temperature. After rme-8(b1023) worms were shifted to the restrictive temperature, GFP accumulated in the pseudocoelom, indicating a reduction in GFP uptake (Figure 1F).
rme-8(b1023) worms also show a ts lethal phenotype. We analyzed the developmental functions of rme-8 by performing temperature shift experiments with rme-8(b1023) mutants. At the permissive temperature, rme-8(b1023) worms displayed normal development, morphology, and a normal brood size (our unpublished results). When shifted to the restrictive temperature, rme-8(b1023) embryos and larvae soon arrested their development, suggesting a continuous requirement for rme-8 activity. Adult hermaphrodites shifted to the restrictive temperature produced only dead embryos (Figure 1G). Larvae shifted to the restrictive temperature usually successfully molted once and then arrested during the next molt (Figure 1H). After 2 d at the restrictive temperature, adult worms became sluggish, and many of their cells began to show morphological abnormalities as judged by Nomarski optics. Most adults died 3 to 4 d after an upshift to the restrictive temperature. Thus, RME-8 is essential for cellular functions and development.
RME-8 is a Conserved J–Domain-containing Protein
We identified the rme-8 gene by positional cloning. We mapped rme-8 between unc-13 and lin-10 on chromosome I (see MATERIALS AND METHODS). The expression of predicted genes residing in the unc-13 to lin-10 interval were disrupted by RNAi (Fire et al., 1998) in hermaphrodites carrying a YP170::GFP transgene. Only disruption of the expression of F18C12.2 reduced the uptake of YP170::GFP by oocytes and increased the accumulation of YP170::GFP in the pseudocoelom, defects also seen in rme-8(b1023) mutants. Most progeny of hermaphrodites subjected to F18C12.2 RNAi died as embryos. Some of the F18C12.2 RNAi embryos hatched before the RNAi effect reached its full potency but arrested in molting. The similarity of these RNAi phenotypes to those observed with rme-8(b1023) mutants suggested that F18C12.2 corresponds to the rme-8 gene. Three additional lines of evidence have confirmed this identification. First, transgenes carrying either the entire cosmid F18C12 or a subclone containing only F18C12.2 rescued the coelomocyte endocytosis defect, as well as the lethality and molting phenotypes associated with the mutant strain rme-8(b1023) (see MATERIALS AND METHODS). Second, we identified an A-to-T transversion in F18C12.2 from rme-8(b1023) worms. This mutation resulted in a Ser substituting Arg 916, which is a conserved residue among RME-8 homologues. Third, we found reduced levels of RME-8 protein in b1023 mutants at the restrictive temperature (see below).
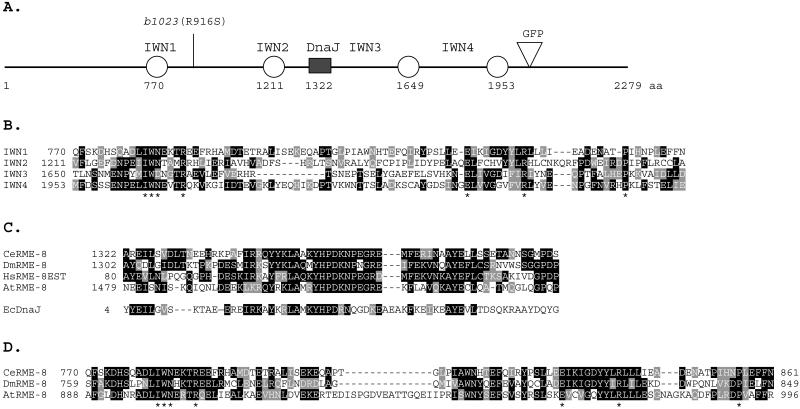
We determined the sequence of the rme-8 coding region by analyzing different cDNAs (see MATERIALS AND METHODS). The rme-8 message is trans-spliced with trans-spliced leader sequence SL1 and encodes a predicted protein of 2279 amino acids (Figure 2A). RME-8 has a J-domain in its central region (Figure 2, A and C). The J-domain is ∼60 amino acids containing an invariant core tripeptide (HPD) flanked by additional conserved amino acids (Silver and Way, 1993; Kelley, 1998). The J-domain was first identified in the E. coli DnaJ protein and since then in many other proteins with diverse functions. RME-8 falls into a group of J-domain–containing proteins that lack a glycine-rich spacer and CxxCxGxG repeats found in canonical DnaJ proteins. The RME-8 group of J-domain proteins includes yeast Sec63, which is a transmembrane protein required in protein secretion, and auxilin, which is a cytoplasmic protein required for uncoating clathrin-coated vesicles (Kelley, 1998; Ungewickell et al., 1995). There is no sequence similarity between RME-8 and Sec63 or RME-8 and auxilin outside the J-domain. C. elegans does have a true auxilin homolog with function similar to mammalian auxilin (Greener et al., 2001). We also identified a repeated sequence in RME-8 that is present in four copies. Each repeat is ∼90 amino acids long with seven invariant residues (Figure 2, A and B) that are also conserved among RME-8 homologues (Figure 2D). We named these repeats IWN repeats with the use of symbols of the first three invariant residues.
Figure 2.
RME-8 structure and sequence conservation. Conserved residues for sequence alignments (B–D) are shaded black and similar amino acids are shaded gray. (A) Domain structure of RME-8, showing a central DnaJ-domain (filled box) and four novel IWN repeats (circles). rme-8(b1023) has an amino acid substitution (R916S), as shown on top. In the RME-8::GFP fusion reporter, GFP (inverted triangle) is inserted after the fourth IWN repeat. (B) Alignment of the four IWN repeats in C. elegans RME-8. Invariant amino acids present in all IWN repeats are marked with asterisk at the bottom. These residues are also conserved in IWN repeats of other RME-8 homologues (see D for IWN1). (C) Sequence conservation among the J-domains of RME-8 homologues from C. elegans (CeRME-8), humans (HsRME-8EST), Drosophila (DmRME-8), and Arabidopsis (AtRME-8). The J-domain from E. coli DnaJ protein (EcDnaJ) is shown for comparison at the bottom. (D) Sequence conservation of the IWN1 region of RME-8 homologues.
We found RME-8 homologues in the genomes of Arabidopsis, Drosophila, and humans, but not in genomes of S. cerevisiae or S. pombe. RME-8 homologues contain a J-domain and IWN repeats located at positions similar to those in C. elegans RME-8. The amino acid sequence identity among these homologues extends throughout the entire protein but with a higher degree of identity around the J-domain, the IWN repeats, and several other distinct regions (Figure 2, C and D; our unpublished results). The C-terminal region of RME-8 has 42% identity and 61% similarity to a human EST, KIAA0678, encoding 1047 amino acids. This EST includes the J-domain (Figure 2C), IWN3, and IWN4, and thus it likely represents the C-terminal part of human RME-8. The full-length RME-8 has 37% identity and 56% similarity to Drosophila RME-8, whereas it has 26% identity and 42% similarity with Arabidopsis RME-8. None of these RME-8 homologues has been studied functionally. These RME-8 homologues form a new family of J-domain proteins that are conserved among multicellular organisms.
RME-8 is Ubiquitously Expressed and Localized to Vesicular Structures
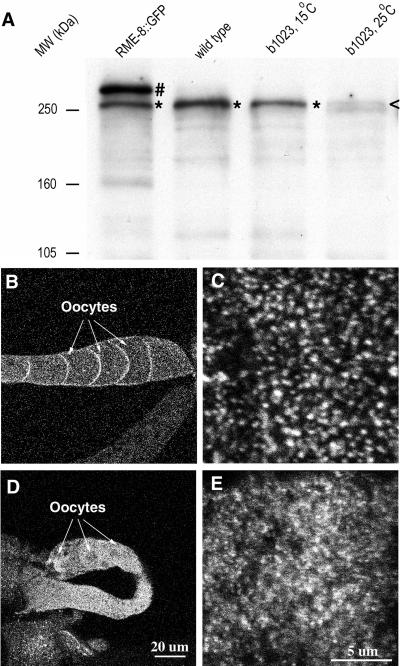
We studied the expression pattern and subcellular localization of RME-8 by immunofluorescence staining with specific anti–RME-8 antibodies and by with the use of an RME-8::GFP reporter expressed under the control of the rme-8 promoter (Figure 2A). By Western blot analysis, the anti–RME-8 antibodies specifically recognized a protein band in total worm lysates similar in size to the predicted RME-8 protein (Figure 3A). This band was greatly reduced in the b1023 mutant worms shifted to the restrictive temperature. An additional larger band corresponding to the size of the predicted RME-8::GFP protein was observed in the RME-8::GFP strain. Transgenes carrying the RME-8::GFP fusion protein rescued multiple rme-8(b1023) phenotypes (see MATERIALS AND METHODS). Similar expression patterns and subcellular localizations were observed by both antibody staining and RME-8::GFP reporter expression, except that RME-8::GFP was not expressed in the germline, most likely because of a general silencing of transgenes in the C. elegans germline (Kelly and Fire, 1998). These observations suggest that this RME-8::GFP fusion is an accurate reporter of endogenous RME-8 expression and subcellular localization.
Figure 3.
Detection of RME-8 by Western blot analysis (A) and RME-8 localization in ovary by indirect immunofluorescence staining (B–E). A protein band (*) corresponding to the size of the predicted RME-8 (260 kDa) was detected in wild type (A). This band was present in rme-8(b1023) worms reared at the permissive temperature, but it was greatly reduced (<) in rme-8(b1023) worms that were shifted to the restrictive temperature for 48 h. In addition to the wild-type band, a larger band (No.) corresponding the size of the predicted RME-8::GFP protein (290 kDa) was observed in the RME-8::GFP strain. Equal amounts of total worm proteins were loaded on each lane. (B–E) Confocal images of wild-type (B and C) and rme-8(b1023) (D and E) ovaries stained with anti–RME-8 antibodies. Images were obtained with the same exposure parameters focusing on the middle focal plane at low magnification (B and D) or on the cortical focal plane at high magnification (C and E). Wild-type RME-8 is localized to the oocyte cortex (B) on punctate structures (C). Mutant RME-8 carrying the b1023 mutation shows a diffuse cytoplasmic staining (D) and less localization to the punctate structures (E). Worms are shifted to the restrictive temperature for 24 h.
We examined RME-8 expression in dissected adult ovaries by immunofluorescence. Consistent with the requirement of RME-8 in yolk endocytosis, RME-8 was highly expressed in oocytes (Figure 3). RME-8 staining was concentrated in the cortex of oocytes beneath the plasma membrane (Figure 3B). At high magnification, RME-8 staining was localized to punctate structures that were 0.2–0.4 μm in diameter (Figure 3C). RME-8 staining in the rme-8(b1023) missense mutants was diminished and mostly diffuse in the oocyte cytoplasm with little localization to cortical punctate structures (Figure 3, D and E). Similar mislocalization was also observed in other cell types in the mutant worms (our unpublished results).
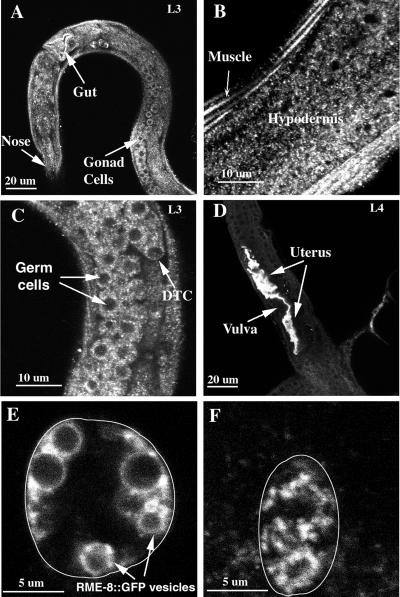
We examined RME-8 expression in other cell types by whole mount immunostaining. Consistent with our genetic analysis, which showed that rme-8 activity is required in many cells, RME-8 is expressed in almost all cells (Figure 4A). In all the cells examined, RME-8 staining was localized to cytoplasmic punctate structures similar to those seen in the oocytes. RME-8 was highly expressed in the lateral hypodermis, muscle (Figure 4B), and the developing germ cells in larvae (Figure 4C). RME-8 staining was extremely abundant in the developing uterus of L4 worms (Figure 4D), suggesting that the developing uterus may require an especially high level of RME-8 activity.
Figure 4.
RME-8 is expressed in many cell types throughout development. RME-8 expression was detected by immunofluorescence antibody staining with affinity-purified antibodies (A–D) or by GFP signal from an RME-8::GFP reporter (E and F). RME-8 staining is present in many cells in an L3 larva (A). Particularly, RME-8 staining is abundant in the hypodermis and muscle (B), the developing hermaphrodite gonad (C), and the developing uterus (D). The exposure for the image in D was ∼10-fold lower than in other images to show the RME-8 staining in the uterus; consequently, staining in other cells was underexposed. RME-8::GFP is localized to heterogeneously sized large vesicles in a coelomocyte in a live animal (E). These RME-8::GFP-labeled large vesicles collapse to small puncta after a standard antibody staining process (F). The cell boundaries of coelomocytes are outlined in E and F. DTC, distal tip cell of the developing gonad.
Similar expression patterns of RME-8 in somatic cells were observed with the RME-8::GFP reporter. In most cells, RME-8::GFP-labeled small puncta with sizes similar to the puncta stained with RME-8 antibodies (our unpublished results). In live coelomocytes, RME-8::GFP labeled the limiting membranes of larger vesicles, with diameters ranging from 0.5 to 4.0 μm (Figure 4E). By serial thin-section confocal microscopy, these vesicles appeared to be round regardless of their sizes, but after fixation (Bettinger et al., 1996), these RME-8::GFP-labeled vesicles appeared smaller, as if they had collapsed (Figure 4F).
RME-8 Functions in Endosomal Trafficking
The localization of RME-8 to the limiting membrane of vesicles in coelomocytes suggests that RME-8 may function in endosomes. We tested this hypothesis by studying the intracellular trafficking of endocytosis markers in the coelomocytes of live worms. Coelomocytes are macrophage-like cells that presumably function as scavengers in the pseudocoelom (Wood, 1988). Coelomocytes actively take up endocytosis markers, such as free GFP and fluorescently labeled proteins or dextrans, when they are delivered into the pseudocoelom by targeted gene expression or microinjection (for an example of free GFP, see Figure 1C). Once taken up by coelomocytes, these markers travel through the endocytic compartments and eventually reach lysosomes, where the digestible peptides are degraded and the indigestible fluorescent molecules persist. This process is analogous to the endocytosis and trafficking of similar fluid-phase markers in mammalian and Drosophila cells (van Deurs et al., 1993; Sevrioukov et al., 1999).
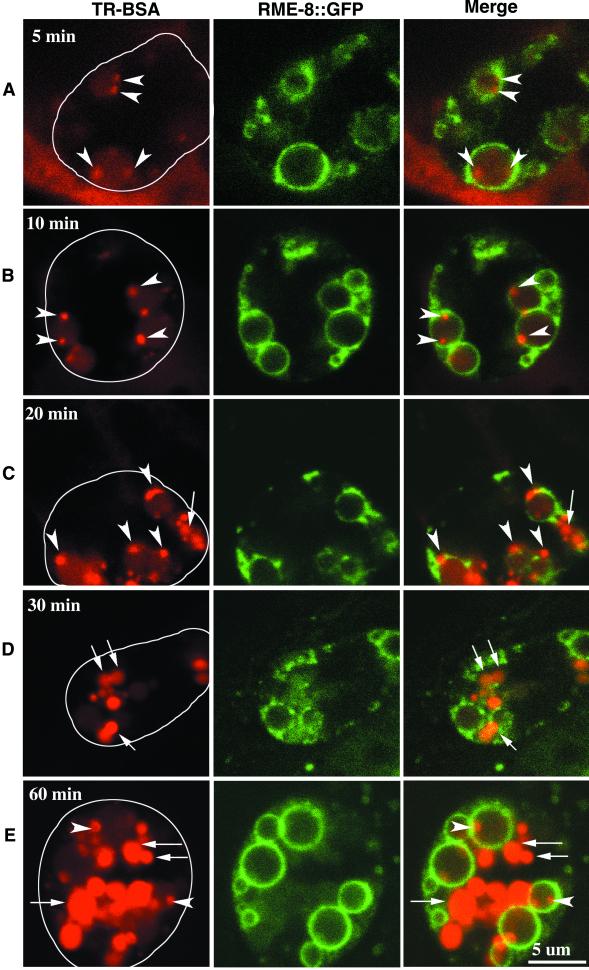
We microinjected the fluid-phase endocytosis marker TR-BSA into the pseudocoelom of adult worms and monitored its trafficking in coelomocytes at sequential time points. Similar trafficking kinetics were observed in wild-type worms either expressing (Figure 5) or not expressing RME-8::GFP (our unpublished results; Figure 6A). At early times, TR-BSA was rapidly endocytosed and accumulated in the RME-8::GFP-labeled vesicles of different sizes (Figure 5, A and B, 5 and 10 min after injection). During these early time points, TR-BSA accumulated exclusively in RME-8::GFP-labeled vesicles, indicating that all endocytosed TR-BSA passed through RME-8::GFP-labeled vesicles before reaching its final destination. Concurrent with the endocytosis of TR-BSA, a few small areas (∼0.5 μm in diameter) of concentrated TR-BSA gradually appeared inside the RME-8::GFP-labeled vesicles (arrowheads in Figure 5, A and B). We presume that these areas of concentrated TR-BSA are associated with membrane structures, and we refer to them as concentrating vesicles. Twenty minutes after injection, these concentrating vesicles became brighter and larger, indicating that more TR-BSA accumulated in them, whereas the surrounding area enclosed by RME-8::GFP still had only a low level of TR-BSA (Figure 5C). These observations suggested that the endocytosed TR-BSA is continuously concentrated into the concentrating vesicles. Also ∼20 min after injection, some of the concentrating vesicles appeared outside of the RME-8::GFP-labeled vesicles (arrow in Figure 5C). Thirty minutes after injection, more of these concentrating vesicles appeared outside of the RME-8::GFP vesicles (arrows in Figure 5D). These concentrating vesicles outside of the RME-8::GFP vesicles were larger than those inside the RME-8::GFP vesicles, and they were usually smaller than the RME-8::GFP vesicles. These RME-8::GFP-negative, late-stage, TR-BSA–containing vesicles had uniformly concentrated TR-BSA. Their morphologies and the endocytosed contents remained constant over time, indicating that they were lysosomes. By 60 min after injection, TR-BSA filled lysosomes were predominant. Meanwhile, RME-8::GFP-labeled vesicles contained a low level of TR-BSA and a few concentrating vesicles (Figure 5E), indicating a continued endocytosis and delivery of TR-BSA into RME-8::GFP-labeled vesicles. After 24 h, TR-BSA was depleted from the pseudocoelom and was found only in the lysosomes (our unpublished results). The transient accumulation of fluid-phase markers inside the RME-8::GFP-labeled vesicles shows that RME-8 labels an intermediate endosomal compartment between the plasma membrane and the lysosome.
Figure 5.
Endocytic trafficking in wild-type coelomocytes. Endocytosis marker TR-BSA was microinjected into the body cavity (where coelomocytes are located), and TR-BSA taken up by coelomocytes was examined by confocal microscopy at 5, 10, 20, 30, and 60 min after injection (A–E). Cross sections of a single coelomocyte (cell boundary outlined on the left image) at a given time point are shown in each row, with TR-BSA (red) on the left, RME-8::GFP (green) in the middle, and the merged image on the right. Endocytosed TR-BSA appears rapidly inside RME-8::GFP-labeled vesicles (A, 5 min; best viewed in red only). Some TR-BSA fills RME-8::GFP-labeled vesicles at a low level, whereas some TR-BSA is concentrated into small areas (arrowheads; A, 5 min; B, 10 min). These areas containing concentrated TR-BSA increase in size over time and begin to appear in RME-8::GFP-negative vesicles (arrow) by ∼20 min after injection (C). Over a longer time, more TR-BSA appears in RME-8::GFP-negative vesicles (arrows; D, 30 min; E, 60 min). At these time points, newly endocytosed TR-BSA appears in RME-8::GFP-labeled vesicles that have characteristic concentrating vesicles (arrowheads).
Figure 6.
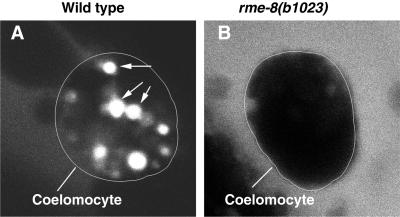
Endocytic trafficking is blocked in rme-8(b1023) mutant coelomocytes. Similar trafficking experiments were done as in Figure 5, but in worms that did not carry the RME-8::GFP reporter. Worms were shifted to the restrictive temperature for 48 h before being used for the trafficking experiments. At 30 min after injection, there was little or no uptake of TR-BSA in rme-8(b1023) mutant coelomocytes (B), whereas abundant TR-BSA–containing vesicles (arrows) were present in wild-type coelomocytes (A). Coelomocyte boundary is outlined.
To determine how RME-8 functions in the endosomes, we analyzed the trafficking of TR-BSA in rme-8(b1023) worms. b1023 is a ts loss-of-function allele of rme-8. Longer exposure of b1023 worms to restrictive temperature leads to lower levels of RME-8, as can be seen by the disappearance of RME-8 in a Western blot (Figure 3A). rme-8 worms shifted to the restrictive temperature for 48 h failed to accumulate TR-BSA in their coelomocytes 30 min after injection (Figure 6B). By contrast, at the same time after injection, abundant TR-BSA–containing vesicles accumulated in coelomocytes of wild-type worms (Figure 6A). Even several hours after injection, TR-BSA failed to accumulate in coelomocytes of mutants (our unpublished results). These experiments show that the block of endocytosis in rme-8(b1023) occurs early, before or during the formation of RME-8–labeled endosomes.
rme-8(b1023) mutants expressing secreted GFP started to accumulate GFP in their pseudocoeloms 24 h after a shift to the restrictive temperature, consistent with a reduction in endocytosis. Coelomocytes in these animals still contained GFP-filled vesicles similar to those seen in the wild type (Figure 1F). Similarly, analysis of TR-BSA uptake in the coelomocytes of rme-8 mutants shifted to the restrictive temperature for 24 h as opposed to 48 h failed to identify a kinetic difference from wild type (our unpublished results). A more sensitive endocytosis assay will be necessary to detect subtle reductions in those trafficking steps that result from partial reduction of RME-8 activity.
DISCUSSION
RME-8–mediated Endocytic Functions in C. elegans
RME-8 is a newly identified protein that is required for endocytic functions in many cells in C. elegans. It is required for receptor-mediated yolk endocytosis in the oocyte and for fluid-phase endocytosis in the coelomocyte. RME-8 is essential for viability and is expressed in many cell types during development. These observations indicate that RME-8 is important for endocytosis in many different processes.
RME-8 activity may be more critical in certain cell types than in others, as reflected in the differential temperature sensitivity of mutant phenotypes and the different expression levels of RME-8 in various cell types. At the permissive temperature, the ts mutant strain rme-8(b1023) is healthy and displays normal fluid-phase endocytosis by coelomocytes, but it has defective yolk uptake by oocytes. RME-8 carrying the b1023 mutation probably functions at the permissive temperature at a reduced level that is sufficient for fluid-phase endocytosis in coelomocytes but insufficient for yolk endocytosis in oocytes. Yolk endocytosis is one of the more dramatic examples of endocytosis (Schneider, 1996). Oocytes probably require high RME-8 activity for the rapid uptake of large amounts of yolk. At the restrictive temperature, rme-8(b1023) is defective in endocytosis in the coelomocyte and shows both embryonic and larval lethality.
A molting defect is also associated with the developmental arrest of rme-8 mutants. In C. elegans, the advance of one larval stage to the next requires the growth of a new cuticle and shedding of the old one. Arrested rme-8 mutants usually fail to shed their cuticle. Thus, RME-8 may be required for the detachment or degradation of old cuticle from the hypodermis. LRP-1, an LDL receptor–related protein similar to mammalian gp330/megalin, is known to be required for molting. Like RME-8, LRP-1 is expressed in the hypodermal cells (Yochem et al., 1999). Mutants lacking either RME-8 or LRP-1 show similar molting defects. Thus, RME-8 and LRP-1 may both perform endocytic functions required for molting.
RME-8 in Endosomal Trafficking
Endocytosis is a complex cellular function that involves a network of intracellular vesicular structures. RME-8 is localized to the periphery of large vesicles in coelomocytes. These vesicles appear to be endosomes, based on the transient accumulation of fluid-phase markers in them (see a model in Figure 7). Fluid-phase markers begin to accumulate in these RME-8 vesicles in ∼5 min and by 20 min become highly concentrated into smaller compartments inside the RME-8–labeled vesicles. By 30 min, most of the endocytosed fluid-phase marker appears in lysosomes that are not labeled by RME-8. These trafficking kinetics resemble those seen in the uptake of LDL into late endosomes by CHO cells or Hep2 cells (Dunn and Maxfield, 1992; Ghosh et al., 1994).
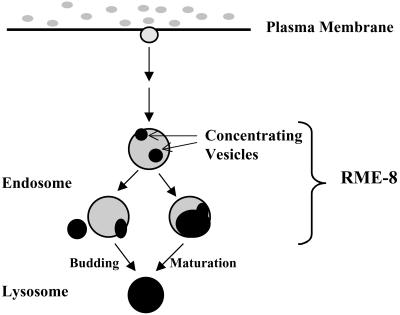
Figure 7.
A model illustrating RME-8 function in endosomal trafficking in the C. elegans coelomocyte. Lysosome-bound endocytosis markers such as TR-BSA are internalized and delivered to RME-8–labeled endosomes. TR-BSA is depicted as gray to dark areas with a degree of darkness, indicating relative concentration of TR-BSA. It is unclear how many steps of intracellular trafficking are involved before TR-BSA enters RME-8–labeled endosomes (see DISCUSSION). In the RME-8–labeled endosome, TR-BSA is gradually concentrated into small concentrating vesicles (dark area). These concentrating vesicles are then delivered to the RME-8 negative lysosomes, after either a vesicular transport model (left branch of the pathway) in which the concentrating vesicles bud off and fuse with existing lysosomes or a maturation model (right branch of the pathway) in which the entire RME-8–positive endosomes become lysosomes.
It remains unresolved whether RME-8 functions in the early or late endosomes or both. In the absence of RME-8 activity in rme-8(b1023) mutants, endocytosis markers fail to accumulate in endosomes. This suggests that RME-8 is required either for a very early uptake function or for the formation or function of endosomes. Because RME-8 is present on the surface of endosomes that contain low levels of endocytosed markers as well as endosomes that contain concentrating vesicles, RME-8 may also function in a later step, such as the formation of the concentrating vesicles or the exit of endocytic contents to the lysosome.
The formation of concentrating vesicles in RME-8–labeled endosomes resembles an endosomal concentration process observed in multivesicular bodies (MVBs) in mammalian cells (Dunn and Maxfield, 1992; van Deurs et al., 1993; Futter et al., 1996). MVBs are late endosomes that contain internal vesicles with concentrated endocytosed molecules. These internal vesicles together with aggregated endocytosed molecules are most abundant in late stage endosomes that are about to be delivered to lysosomes. The similarity of these two concentration processes suggests that the concentrating vesicles inside RME-8–labeled endosomes may be equivalent to the aggregates of internal vesicles observed in late MVBs. Electron microscopic studies in the C. elegans coelomocyte will be necessary to substantiate this proposal.
The exit of concentrating vesicles from RME-8–labeled endosomes remains mechanistically unresolved. Two alternative mechanisms can be envisioned (Figure 7). Concentrating vesicles could bud off from the RME-8–labeled endosomes and be targeted to the lysosomes, as described in the vesicular transport model (Gruenberg et al., 1989; Gruenberg and Maxfield, 1995). In this model, RME-8–labeled endosomes would be expected to be stable compartments, or the entire RME-8–labeled endosome including its concentrating vesicles could mature into a lysosome, which is consistent with the maturation model (Dunn and Maxfield, 1992; van Deurs et al., 1993). In this case, RME-8 would be transiently associated with the endosome.
Molecular Function of RME-8 and Its Evolutionary Conservation in Multicellular Organisms
The central J-domain is the most distinctive feature of the RME-8 sequence. J-domains are found in many proteins from bacteria to humans. They perform a variety of subcellular functions that involve protein unfolding or refolding (Silver and Way, 1993; Kelley, 1998). J-domains interact with proteins belonging to the heat shock 70 (Hsc70) family. For example, the J-domain protein auxilin functions with an Hsc70 to disassemble the clathrin coat of coated vesicles (Ungewickell et al., 1995). By analogy, perhaps the J-domain of RME-8 functions in protein folding or unfolding and catalyzes the shedding of endosomal protein coats. Because RME-8 is primarily localized to the limiting membrane of endosomes, RME-8 itself could be a protein coat, and its J-domain could function to stimulate the removal of RME-8 from the endosome.
We found highly conserved RME-8 homologues in multicellular organisms from animals to plants. These homologues share conserved domains, for example, the J-domain and the IWN repeats, a high percentage of sequence identity, and a relatively constant length and positioning of the various domains. These conserved features suggest that RME-8 functions in a process that is common to various multicellular organisms. The absence of an RME-8 homolog in yeast is striking and suggests that fundamental differences in endocytic trafficking may exist between yeast and multicellular organisms, perhaps to accommodate the special requirements for intercellular communication in multicellular organisms during development and homeostasis.
ACKNOWLEDGMENTS
We thank H. Fares, I. Greenwald for sharing unpublished information and C. elegans strains; L. Pedraza and W. Pryzlecki for excellent technical assistance; Y. Kohara and A. Coulson for cDNA and cosmid clones, respectively; H. Fares, I. Greenwald, S. Emmons, and O. Hobert for comments on this manuscript. Many of the strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Center for Research Resources of the National Institutes of Health. This work was supported by grants from the March of Dimes Foundation (FY00-284) and the National Science Foundation (MCB-0078129).
REFERENCES
- Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Bettinger JC, Lee K, Rougvie AE. Stage-specific accumulation of the terminal differentiation factor LIN-29 during Caenorhabditis elegans development. Development. 1996;122:2517–2527. doi: 10.1242/dev.122.8.2517. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant NJ, Stevens TH. Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol Mol Biol Rev. 1998;62:230–247. doi: 10.1128/mmbr.62.1.230-247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforidis S, McBride HM, Burgoyne RD, Zerial M. The Rab5 effector EEA1 is a core component of endosome docking. Nature. 1999;397:621–625. doi: 10.1038/17618. [DOI] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- De Stasio E, Lephoto C, Azuma L, Holst C, Stanislaus D, Uttam J. Characterization of revertants of unc-93(e1500) in Caenorhabditis elegans induced by N-ethyl-N-nitrosourea. Genetics. 1997;147:597–608. doi: 10.1093/genetics/147.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn KW, Maxfield FR. Delivery of ligands from sorting endosomes to late endosomes occurs by maturation of sorting endosomes. J Cell Biol. 1992;117:301–310. doi: 10.1083/jcb.117.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares H, Greenwald I. Regulation of endocytosis by cup-5, the Caenorhabditis elegans mucolipin-1 homologue. Nat Genet. 2001;28:64–68. doi: 10.1038/ng0501-64. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Futter CE, Pearse A, Hewlett LJ, Hopkins CR. Multivesicular endosomes containing internalized EGF-EGF receptor complexes mature and then fuse directly with lysosomes. J Cell Biol. 1996;132:1011–1023. doi: 10.1083/jcb.132.6.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh RN, Gelman DL, Maxfield FR. Quantification of low density lipoprotein and transferrin endocytic sorting HEp2 cells using confocal microscopy. J Cell Sci. 1994;107:2177–2189. doi: 10.1242/jcs.107.8.2177. [DOI] [PubMed] [Google Scholar]
- Gorvel JP, Chavrier P, Zerial M, Gruenberg J. rab5 controls early endosome fusion in vitro. Cell. 1991;64:915–925. doi: 10.1016/0092-8674(91)90316-q. [DOI] [PubMed] [Google Scholar]
- Grant B, Hirsh D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol Biol Cell. 1999;10:4311–4326. doi: 10.1091/mbc.10.12.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greener T, Grant B, Zhang Y, Wu X, Greene LE, Hirsh D, Eisenberg E. Caenorhabditis elegans auxilin: a J-domain protein essential for clathrin-mediated endocytosis in vivo. Nat Cell Biol. 2001;3:215–219. doi: 10.1038/35055137. [DOI] [PubMed] [Google Scholar]
- Gruenberg J, Griffiths G, Howell KE. Characterization of the early endosome and putative endocytic carrier vesicles in vivo and with an assay of vesicle fusion in vitro. J Cell Biol. 1989;108:1301–1316. doi: 10.1083/jcb.108.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J, Maxfield FR. Membrane transport in the endocytic pathway. Curr Opin Cell Biol. 1995;7:552–563. doi: 10.1016/0955-0674(95)80013-1. [DOI] [PubMed] [Google Scholar]
- Gu J, Stephenson CG, Iadarola MJ. Recombinant proteins attached to a nickel-NTA column: use in affinity purification of antibodies. Biotechniques. 1994;2:257–262. [PubMed] [Google Scholar]
- Hirsch JG, Fedorko ME, Cohn ZA. Vesicle fusion and formation at the surface of pinocytic vacuoles in macrophages. J Cell Biol. 1968;38:629–632. doi: 10.1083/jcb.38.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley WL. The J-domain family and the recruitment of chaperone power. Trends Biochem Sci. 1998;23:222–227. doi: 10.1016/s0968-0004(98)01215-8. [DOI] [PubMed] [Google Scholar]
- Kelly WG, Fire A. Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans. Development. 1998;125:2451–2456. doi: 10.1242/dev.125.13.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Ghosh RN, Maxfield FR. Endocytosis. Physiol Rev. 1997;77:759–803. doi: 10.1152/physrev.1997.77.3.759. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD. Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem Sci. 2000;25:229–235. doi: 10.1016/s0968-0004(00)01543-7. [DOI] [PubMed] [Google Scholar]
- Olkkonen VM, Stenmark H. Role of Rab GTPases in membrane traffic. Int Rev Cytol. 1997;176:1–85. doi: 10.1016/s0074-7696(08)61608-3. [DOI] [PubMed] [Google Scholar]
- Peterson MR, Burd CG, Emr SD. Vac1p coordinates Rab and phosphatidylinositol 3-kinase signaling in Vps45p-dependent vesicle docking/fusion at the endosome. Curr Biol. 1999;9:159–162. doi: 10.1016/s0960-9822(99)80071-2. [DOI] [PubMed] [Google Scholar]
- Schneider WJ. Vitellogenin receptors: oocyte-specific members of the low-density lipoprotein receptor supergene family. Int Rev Cytol. 1996;166:103–137. doi: 10.1016/s0074-7696(08)62507-3. [DOI] [PubMed] [Google Scholar]
- Sevrioukov EA, He JP, Moghrabi N, Sunio A, Kramer H. A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol Cell. 1999;4:479–486. doi: 10.1016/s1097-2765(00)80199-9. [DOI] [PubMed] [Google Scholar]
- Silver PA, Way JC. Eukaryotic DNAJ homologs and the specificity of Hsp70 activity. Cell. 1993;74:5–6. doi: 10.1016/0092-8674(93)90287-z. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Lippe R, Christoforidis S, Gaullier JM, Brech A, Callaghan J, Toh BH, Murphy C, Zerial M, Stenmark H. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394:494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- Spieth J, Brooke G, Kuersten S, Lea K, Blumenthal T. Operons in C. elegans: polycistronic mRNA precursors are processed by trans-splicing of SL2 to downstream coding regions. Cell. 1993;73:521–532. doi: 10.1016/0092-8674(93)90139-h. [DOI] [PubMed] [Google Scholar]
- Ungewickell E, Ungewickell H, Holstein SE, Lindner R, Prasad K, Barouch W, Martin B, Greene LE, Eisenberg E. Role of auxilin in uncoating clathrin-coated vesicles. Nature. 1995;378:632–635. doi: 10.1038/378632a0. [DOI] [PubMed] [Google Scholar]
- van der Sluijs P, Hull M, Webster P, Male P, Goud B, Mellman I. The small GTP-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell. 1992;70:729–740. doi: 10.1016/0092-8674(92)90307-x. [DOI] [PubMed] [Google Scholar]
- van Deurs B, Holm PK, Kayser L, Sandvig K, Hansen SH. Multivesicular bodies in HEp-2 cells are maturing endosomes. Eur J Cell Biol. 1993;61:208–224. [PubMed] [Google Scholar]
- Wendland B, Emr SD, Riezman H. Protein traffic in the yeast endocytic and vacuolar protein sorting pathways. Curr Opin Cell Biol. 1998;10:513–522. doi: 10.1016/s0955-0674(98)80067-7. [DOI] [PubMed] [Google Scholar]
- Williams BD, Schrank B, Huynh C, Shownkeen R, Waterston RH. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics. 1992;131:609–624. doi: 10.1093/genetics/131.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W. The Nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Yochem J, Tuck S, Greenwald I, Han M. A gp330/megalin-related protein is required in the major epidermis of Caenorhabditis elegans for completion of molting. Development. 1999;126:597–606. doi: 10.1242/dev.126.3.597. [DOI] [PubMed] [Google Scholar]