Abstract
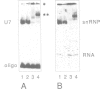
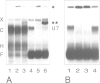
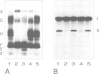
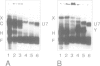
Histone RNA 3' end formation occurs through a specific cleavage reaction that requires, among other things, base-pairing interactions between a conserved spacer element in the pre-mRNA and the minor U7 snRNA present as U7 snRNP. An oligonucleotide complementary to the first 16 nucleotides of U7 RNA can be used to characterize U7 snRNPs from nuclear extracts by native gel electrophoresis. Using similar native gel techniques, we present direct biochemical evidence for a stable association between histone pre-mRNA and U7 snRNPs. Other complexes formed in the nuclear extract are dependent on the 5' cap structure and on the conserved hairpin element of histone pre-mRNA, respectively. However, in contrast to the U7-specific complex, their formation is not required for processing. Comparison of several authentic and mutant histone pre-mRNAs with different spacer sequences demonstrates that the formation and stability of the U7-specific complex closely follows the predicted stability of the potential RNA-RNA hybrid. However, this does not exclude a stabilization of the complex by U7 snRNP structural proteins.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Bond U. M., Yario T. A., Steitz J. A. Multiple processing-defective mutations in a mammalian histone pre-mRNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes Dev. 1991 Sep;5(9):1709–1722. doi: 10.1101/gad.5.9.1709. [DOI] [PubMed] [Google Scholar]
- Cotten M., Gick O., Vasserot A., Schaffner G., Birnstiel M. L. Specific contacts between mammalian U7 snRNA and histone precursor RNA are indispensable for the in vitro 3' RNA processing reaction. EMBO J. 1988 Mar;7(3):801–808. doi: 10.1002/j.1460-2075.1988.tb02878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Stunnenberg H. G., Birnstiel M. L. Biochemical complementation with RNA in the Xenopus oocyte: a small RNA is required for the generation of 3' histone mRNA termini. Cell. 1983 Oct;34(3):823–828. doi: 10.1016/0092-8674(83)90539-1. [DOI] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gick O., Krämer A., Keller W., Birnstiel M. L. Generation of histone mRNA 3' ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 1986 Jun;5(6):1319–1326. doi: 10.1002/j.1460-2075.1986.tb04362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gick O., Krämer A., Vasserot A., Birnstiel M. L. Heat-labile regulatory factor is required for 3' processing of histone precursor mRNAs. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8937–8940. doi: 10.1073/pnas.84.24.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann I., Birnstiel M. L. Cell cycle-dependent regulation of histone precursor mRNA processing by modulation of U7 snRNA accessibility. Nature. 1990 Aug 16;346(6285):665–668. doi: 10.1038/346665a0. [DOI] [PubMed] [Google Scholar]
- Konarska M. M. Analysis of splicing complexes and small nuclear ribonucleoprotein particles by native gel electrophoresis. Methods Enzymol. 1989;180:442–453. doi: 10.1016/0076-6879(89)80116-8. [DOI] [PubMed] [Google Scholar]
- Leibold E. A., Munro H. N. Cytoplasmic protein binds in vitro to a highly conserved sequence in the 5' untranslated region of ferritin heavy- and light-subunit mRNAs. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2171–2175. doi: 10.1073/pnas.85.7.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier V. S., Böhni R., Schümperli D. Nucleotide sequence of two mouse histone H4 genes. Nucleic Acids Res. 1989 Jan 25;17(2):795–795. doi: 10.1093/nar/17.2.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry K. L., Oh R., Steitz J. A. Each of the conserved sequence elements flanking the cleavage site of mammalian histone pre-mRNAs has a distinct role in the 3'-end processing reaction. Mol Cell Biol. 1989 Jul;9(7):3105–3108. doi: 10.1128/mcb.9.7.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry K. L., Steitz J. A. Both conserved signals on mammalian histone pre-mRNAs associate with small nuclear ribonucleoproteins during 3' end formation in vitro. Mol Cell Biol. 1987 May;7(5):1663–1672. doi: 10.1128/mcb.7.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry K. L., Steitz J. A. Identification of the human U7 snRNP as one of several factors involved in the 3' end maturation of histone premessenger RNA's. Science. 1987 Dec 18;238(4834):1682–1687. doi: 10.1126/science.2825355. [DOI] [PubMed] [Google Scholar]
- Müllner E. W., Neupert B., Kühn L. C. A specific mRNA binding factor regulates the iron-dependent stability of cytoplasmic transferrin receptor mRNA. Cell. 1989 Jul 28;58(2):373–382. doi: 10.1016/0092-8674(89)90851-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaufele F., Gilmartin G. M., Bannwarth W., Birnstiel M. L. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3' end of H3 messenger RNA. 1986 Oct 30-Nov 5Nature. 323(6091):777–781. doi: 10.1038/323777a0. [DOI] [PubMed] [Google Scholar]
- Soldati D., Schümperli D. Structural and functional characterization of mouse U7 small nuclear RNA active in 3' processing of histone pre-mRNA. Mol Cell Biol. 1988 Apr;8(4):1518–1524. doi: 10.1128/mcb.8.4.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber C., Soldati D., Lüscher B., Schümperli D. Histone-specific RNA 3' processing in nuclear extracts from mammalian cells. Methods Enzymol. 1990;181:74–89. doi: 10.1016/0076-6879(90)81113-9. [DOI] [PubMed] [Google Scholar]
- Strub K., Birnstiel M. L. Genetic complementation in the Xenopus oocyte: co-expression of sea urchin histone and U7 RNAs restores 3' processing of H3 pre-mRNA in the oocyte. EMBO J. 1986 Jul;5(7):1675–1682. doi: 10.1002/j.1460-2075.1986.tb04411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub K., Galli G., Busslinger M., Birnstiel M. L. The cDNA sequences of the sea urchin U7 small nuclear RNA suggest specific contacts between histone mRNA precursor and U7 RNA during RNA processing. EMBO J. 1984 Dec 1;3(12):2801–2807. doi: 10.1002/j.1460-2075.1984.tb02212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasserot A. P., Schaufele F. J., Birnstiel M. L. Conserved terminal hairpin sequences of histone mRNA precursors are not involved in duplex formation with the U7 RNA but act as a target site for a distinct processing factor. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4345–4349. doi: 10.1073/pnas.86.12.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E. A., Gilmartin G. M., Nevins J. R. Poly(A) site efficiency reflects the stability of complex formation involving the downstream element. EMBO J. 1991 Jan;10(1):215–219. doi: 10.1002/j.1460-2075.1991.tb07938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]