Abstract
The mesenteric lymph nodes (MLN) and the liver are exposed to microbes and microbial products from the gastrointestinal (GI) tract, making them immunologically unique. The GI tract and associated MLN are sites of early viral replication in human immunodeficiency virus (HIV) infection and the MLN are likely important reservoir sites that harbor latently-infected cells even after prolonged antiretroviral therapy (ART). The liver has been shown to play a significant role in immune responses to lentiviruses and appears to play a significant role in clearance of virus from circulation. Nonhuman primate (NHP) models for HIV and Acquired Immunodeficiency Syndrome (AIDS) closely mimic these aspects of HIV infection and serial longitudinal sampling of primary sites of viral replication and the associated immune responses in this model will help to elucidate critical events in infection, pathogenesis, and the impact of various intervention strategies on these events. Current published techniques to sample liver and MLN together involve major surgery and/or necropsy, which limits the ability to investigate these important sites in a serial fashion in the same animal. We have previously described a laparoscopic technique for collection of MLN. Here, we describe a minimally invasive laparoscopic technique for serial longitudinal sampling of liver and MLN through the same two port locations required for the collection of MLN. The use of the same two ports minimizes the impact to the animals as no additional incisions are required. This technique can be used with increased sampling frequency compared to major abdominal surgery and reduces the potential for surgical complications and associated local and systemic inflammatory responses that could complicate interpretation of results. This procedure has potential to facilitate studies involving NHP models while improving animal welfare.
Keywords: Medicine, Issue 123, Mesenteric, Lymph node, Liver, Hepatic, Macaque, Laparoscopic, Laparoscopy, Refinement, Minimally invasive, SIV, HIV
Introduction
The gastrointestinal (GI) tract is the body's largest mucosal surface and is exposed to a myriad of antigens derived from food, pathogens, and endosymbiotic bacterial communities commonly referred to as the microbiome1,2. Mesenteric lymph nodes (MLN) line the GI tract and are a principal site of immune function for promoting inflammatory or tolerogenic responses towards these diverse antigens. The physical organization of the MLN creates a compartmentalized system that can respond to antigens locally without eliciting systemic responses3. Similarly, blood from the GI tract drains to the liver by way of the portal vein before returning to circulation, and is thus exposed to microbes and microbial products that have translocated from the GI tract into the lamina propria and entered the bloodstream4. The liver also functions as a secondary lymphoid organ and has a large number of immune cells, including specialized macrophages, to remove translocated microbes5,6. Thus, the MLN and the liver are the primary immune organs exposed to commensal and pathogenic bacteria from the GI tract, as well as the milieu of antigens from other sources, making them unique and important from an immunological perspective.
The MLN are critical sites for evaluation of the virologic and immunologic effects of human immunodeficiency virus (HIV) or pathogenic simian immunodeficiency virus (SIV) infection, and are likely involved in early dissemination of SIV following intrarectal challenge. Gut-associated lymphoid tissue is known to be a major site of persistent HIV replication, despite effective control of viremia by modern antiretroviral therapies (ART)7. In SIV infection models, MLN have been shown to be important reservoirs of latent virus and may be the primary reservoir8,9. The liver is also important in lentiviral infection as evidenced by the accumulation of SIV specific CD8+ T cells in the livers of macaques during acute SIV infection10. Further, it is the major organ responsible for clearing the virus from circulation11.
HIV and SIV infection are associated with altered GI microbiota, disrupted GI epithelial integrity, and increased translocation of microbes and microbial products from the colon into the periphery and circulation12,13. These processes are associated with local and systemic immune activation, and increased morbidity and mortality in HIV infected individuals14. In SIV infection, translocated bacteria have been observed in the MLN15, whereas accumulation of microbial products in the liver has been shown to result in inflammation and damage to the organ16. Thus, in the context of HIV and SIV infections, the MLN and liver can be highly informative for understanding the inflammatory processes driven by GI-resident bacteria.
Here, we present a minimally invasive laparoscopic technique for serial sampling of the MLN and liver in non-human primates (NHPs). We demonstrate successful performance of this technique on two healthy female rhesus macaques (RMs), sampling each animal two times, with 160 days in between each surgery. We go on to use these samples to evaluate and compare key leukocyte and lymphocyte populations within each organ using flow cytometry, and demonstrate highly consistent data between the two time points.
Protocol
Animals were housed and cared for in Association for the Assessment and Accreditation of Laboratory Animal Care international (AAALACi) accredited facilities, and all animal procedures were performed according to protocols approved by the Institutional Animal Care and Use Committee (IACUC) of University of Washington and Washington National Primate Research Center.
1. Surgical Preparation, Anesthesia, and Analgesia
- Sedation and Induction
- Sedate the macaque with an intramuscular injection of 10 mg/kg ketamine and 0.015 mg/kg dexmedetomidine (combined in one syringe). Ensure absence of withdrawal reflexes prior to removal from the cage.
- Anesthesia Support and Monitoring
- Insert a peripheral venous catheter and provide intravenous isotonic fluids (such as Lactated Ringers Solution) at a rate of 5 mL/kg/h throughout anesthesia and surgery. Apply sterile eye lubricant to prevent corneal dryness.
- Provide analgesia, such as a sustained release formulation of buprenorphine (0.2 mg/kg, subcutaneously). NOTE: An NSAID (such as meloxicam or ketoprofen) may also be provided during anesthetic recovery, provided that the experimental protocol permits NSAID administration and that animal is normotensive and well-hydrated during anesthesia.
- Monitor anesthetic depth (e.g. jaw tone), heart rate, respiration rate, EKG, blood pressure, pulse oxygenation, end tidal CO2, and body temperature throughout anesthesia and surgery. NOTE: When the abdomen is insufflated, the animal may hypoventilate. Provide mechanical ventilation if respiration rate decreases, or the end tidal CO2 levels is above 45 mmHg.
- Following surgery, administer atipamezole (0.15 mg/kg, intramuscularly) for dexmedetomidine reversal.
- Surgical Preparation
- Perform multiple warm water enemas to reduce the volume of feces in the colon to aid visualization and intestinal manipulation during the procedure20.
- Use clippers with a number 40 blade to remove hair from the surgical field. Shave from the costal ridge to the pubis and from the left to right flank.
- Aseptically prepare the surgical field using appropriate antiseptics, such as alternating chlorhexidine-alcohol or povidone-iodine-alcohol scrubs.
- Position the animal on the operating table on its dorsum, at an angle halfway between dorsal and right lateral recumbency. Tie the arms and legs with rope in an extended position. Provide a final chlorhexidine and alcohol prep once the animal moves to the operating room and is positioned for surgery.
2. Surgery
Note: For further information of laparoscopy in small animals, see reference21.
- Laparoscopic Instrument Preparation
- Drape the animal with a sterile fenestrated drape.
- With the help of a non-sterile assistant, wrap the digital camera with the sterile camera drape and attach the cable to the tower. Attach the rigid scope to the camera head.
- Attach the sterile light cable to the rigid scope and connect the cable to the light source. White balance the camera according to the manufacturer's instructions.
- Attach the sterile insufflator tubing to the insufflator.
- Laparoscopic Entry
- Using a #15 scalpel, make a ~ 5 mm stab incision through the skin to the depth of the abdominal musculature, approximately 1 - 2 cm to the left of the umbilicus.
- Using the non-dominant hand, lift up the abdominal wall cranial to the site of the stab incision. With the dominant hand, place the tip of the Veress needle through the incision and angle the tip cranially. NOTE: The penetration angle of the Veress needle will vary based on the body condition of the animal. For instance, in a fat animal, the Veress needle will need to be inserted nearly perpendicular to the abdominal wall. In a thin animal, the Veress needle should be angled cranially to reduce risk of contacting the viscera.
- Holding the shaft (not the hub) of the Veress needle, firmly push the needle to penetrate the abdominal wall into the abdominal cavity. A distinct "click" and decrease in resistance will be felt as the sharp tip penetrates the peritoneum and the blunt tip of the Veress needle springs forward into the abdominal cavity. NOTE: If the needle passes appropriately into the abdominal cavity, there will be little resistance when it is advanced and withdrawn 1 - 2 cm.
- Connect the CO2 insufflator tubing to the needle and open the stopcock. Pressurize the abdomen to no more than 10-12 mmHg to create a pneumoperitoneum. NOTE: The abdomen has achieved ideal insufflation when it has expanded symmetrically and there is a loss of the normal sharp contour of the costal margins. The pressure should provide adequate space for insertion of the cannula/trocar without risk of contacting the viscera.
- Cannula Placement
- Once the abdomen is fully insufflated, make a ~ 5 - 7 mm incision with a #15 scalpel blade through the skin of the left cranial abdomen to the abdominal musculature ~ 2 - 4 cm caudal to the last rib.
- Place a 5 mm trocar-cannula assembly through the skin incision and angle it ~ 45 - 60 degrees caudomedially. Penetrate the abdominal cavity to a depth of 1-2 cm. Screw the threaded cannula into the abdomen by rotating the cannula (~ 0.5 - 1 cm further) to a depth of 1.5 - 3 cm, and remove the trocar.
- Remove the insufflator tubing from the Veress needle, turn off the stopcock, and remove the needle. Connect the insufflator tubing to the cannula and open the stopcock to maintain a pneumoperitoneum at no more than 10 - 12 mmHg.
- Insert the rigid scope through the cannula and into the abdominal cavity.
- Under camera visualization, continue to make a small incision with a #15 scalpel blade through the abdominal musculature and peritoneum at the previous skin incision used for Veress needle insertion. The camera is used to visualize the abdominal side of the incision to avoid vessels and minimize bleeding. NOTE: Ensure the incision through the muscle and peritoneum is large enough to facilitate entry of the probe and exteriorization of the mesentery (~ 0.5 cm). A larger incision may be required to exteriorize mesentery in animals with excessive mesenteric fat.
- Place the animal in the Trendelenburg position21 with the feet elevated at approximately 15 - 30 degrees above the head. This is optional but may allow easier access to mesenteric lymph nodes of the colon, as the small intestinal tract will move to a more cranial position.
- Mesenteric Lymph Node Biopsy
- Under camera visualization, insert a solid probe through the abdominal incision made in step 2.3.5.
- Place the probe between the omentum and bowel and use a sweeping motion to gently sweep the omentum off the bowel to allow visualization of the underlying mesentery. Continuing to use the sweeping motion from the caudal to cranial abdomen. The omentum can be placed in the right cranial abdomen so it is out of the operative field.
- Use the probe to move the descending colon towards the left or right abdominal wall. This will allow better visualization of the mesentery to aid in the search for mesenteric lymph nodes.
- Use the probe to sweep through the mesentery to locate the mesenteric lymph nodes along the colon and mesenteric vessels. NOTE: Colonic nodes run along the mesenteric border of the colon, left colic nodes can be found near the branch points of the vessels, and the inferior mesenteric nodes can be found along the inferior mesenteric vessels. Mesenteric lymph nodes are easily visible in lean animals. When buried in mesenteric fat they will often lend a slightly raised, glistening appearance to the overlying mesentery and may be more a more grayish tan color then surrounding mesenteric fat. Additionally, mesenteric lymph nodes tend to lie along lymphatics and mesenteric vasculature.
- Once a lymph node is identified, remove the probe and insert the Maryland ratcheted forceps into the abdominal cavity. Use the forceps to grasp the mesentery adjacent the target node. Pull the mesentery towards the incision, and remove the scope from the cannula.
- Turn off the insufflator and leave the cannula stopcock open to depressurize the abdomen to facilitate exteriorization of the mesentery. Pull the mesentery outside of the body through the incision. Once exteriorized, grasp the mesentery with curved mosquito hemostats or thumb forceps to ensure access through the incision.
- Identify the lymph node(s) within the exteriorized mesentery by palpating for the firmer lymph node and/or direct visualization of the grayer color or nodular appearance of the lymph node.
- Once identified, make a small opening (~ 3 - 5 mm) in the mesentery near the node using curved hemostats. Free attachments to the mesentery using blunt dissection with the curved hemostats. Ligate the vasculature with 4-0 non-absorbable monofilament suture as necessary. Use sharp dissection with a scalpel to remove the lymph node. Place the nodes in Roswell Park Memorial Institute (RPMI) 1640 medium on ice.
- Examine the exteriorized mesentery for bleeding. If bleeding is noted, provide appropriate hemostasis by occluding the vessels with hemostats or ligatures (4-0 non-absorbable monofilament suture), or by use of cautery. Release the mesentery and insufflate the abdomen back to 10 - 12 mmHg to allow the mesentery to return to the abdominal cavity.
- Repeat steps 2.4.1 through 2.4.8 to obtain additional lymph node biopsies.
- Liver Biopsy
- Return the table to a horizontal position.
- Place a threaded cannula into the abdomen through the incision previously used for lymph node biopsy collection (incision made in step 2.3.5). Ensure the incision is large enough (~ 5 - 7 mm) to allow for cannula placement without trocar insertion. Insert the rigid scope through this cannula toward the cranial abdominal cavity and visualize the cranial abdomen and liver.
- Under camera visualization, insert the biopsy forceps through the cannula previously placed in the left cranial abdomen (incision made in step 2.3.1).
- To obtain liver biopsies, place the biopsy forceps beneath the selected liver lobe margin with the movable part of the jaw facing the liver, open the forceps, and allow the margin to fall into the open jaws. Ensure that no omentum is between the liver and biopsy forceps. Adjust the position of the forceps and close the instrument over the desired sample region.
- Maintain pressure for approximately 20 - 30 s to ensure hemostasis. With the forceps closed, remove the sample by gently pulling the forceps up through the cannula. This will gently tear the biopsy from the liver. Place liver biopsies in RPMI 1640 medium on ice. NOTE: The liver biopsy size is approximately 5 mm x 2 mm x 2 mm using 5 mm biopsy forceps.
- Examine the biopsy site for hemorrhage using camera visualization. If hemorrhage is observed, place a saline moistened sterile cotton swab through the cannula and apply pressure to the biopsy site. Alternatively, pack the biopsy sites with sterile gel foam to aid hemostasis.
- Repeat steps 2.5.3 through 2.5.5 to obtain additional liver biopsies. NOTE: Here, 3 biopsies per animal per procedure were collected, which is sufficient for all downstream analyses.
- Surgical Closure
- Examine the abdominal cavity for bleeding. NOTE: This has not occurred to date. However, if excessive bleeding is noted, determine the site of hemorrhage and provide appropriate hemostasis via pressure with sterile cotton swabs or the use of gel foam.
- Turn off the insufflator and open the cannula stopcocks. Apply gentle manual pressure to the abdomen to fully depressurize the peritoneal cavity. Remove the cannulas.
- Close each of the abdominal wall incisions with 1 to 2 simple interrupted sutures per site. 4-0 absorbable suture is recommended.
- Perform a splash block by placing 0.1-0.3 cc bupivacaine in each incision for local analgesia prior to skin closure.
- Close the skin with 1 - 2 buried interrupted sutures (4-0 absorbable suture is recommended) per site. Tissue glue may be applied.
3. Liver Biopsy Processing and Lymphocyte Analysis
- Store several pieces of liver tissue (approximately 0.5 mm x 0.5 mm x 0.5 mm in size) in cryopreservation tubes, in a preferred RNA storage solution, and 10% formalin for future molecular and histological analyses.
- Place the remaining liver biopsies in 50 mL of RPMI 1640 medium supplemented with 1x penicillin/streptomycin, 40 µg/mL of a commercially available collagenase solution, and 4 µg/mL DNAse in a sterile 250 mL plastic cup with a lid. Stir vigorously using a magnetic stir bar and a stir plate for 1 h at 37 °C.
Evenly distribute by pouring the digested tissue. Grind the digested tissue (from step 3.1.1) over two 70 µm filters fit into two 50 mL conical tubes into a single cell suspension using the end of a sterile 5 mL syringe. Bring the volume of both tubes to 50 mL in RPMI 1640 medium supplemented with 10% fetal bovine serum and 1x penicillin/streptomycin (R10).
- Centrifuge the cells at 840 x g for 6 min at 4 °C. Decant the supernatant. Concentrate the cells into a single 50 mL conical tube by suspending cells from both tubes in 5 mL R10 and combining into one 50 mL conical tube. Bring volume to 50 mL using R10. Count cells using a hemocytometer to determine cellular yield.
- Centrifuge the cells at 840 x g for 6 min at 4 °C. Resuspend the cells in 1 mL R10, distribute evenly into two 5 mL round-bottom polystyrene tubes and bring the volume of each tube to 4 mL with R10, targeting for >500,000 cells in each tube.
Centrifuge the cells at 840 x g for 6 min at 4 °C. Decant the supernatant and resuspend the cell pellet in 4 mL phosphate buffered saline (PBS). Centrifuge at 840 x g for 6 min at 4 °C.
Decant the supernatant and resuspend the cells in 100 µL of PBS by vortexing gently. Add 1.5 µL of a fixable dead cell stain. Incubate at room temperature for 5 min.
Add the following undiluted surface staining antibodies in titers recommended by the manufacturer: CD45-PerCP (clone D058-1283), CD3-PE-CF594 (clone SP34-2), CD20-PE-Cyanin5 (clone 2H7), CD4-BV605 (clone OKT4), CD8-BV570 (clone RPA-T8), and CD14-BV785 (clone M5E2). Incubate at 4 °C for 20 min.
Add 4 mL PBS and centrifuge at 840 x g for 6 min at 4 °C. Decant the supernatant and resuspend the cells in 250 µL of 1% paraformaldehyde in PBS. Caution: Paraformaldehyde is toxic. Wear appropriate personal protective equipment.
Analyze the cell populations using a flow cytometer as described in reference22.
Representative Results
Detailed methods for processing of laparoscopically collected MLN, as well as cellular yields, and leukocyte frequencies in these organs have been previously reported13. Figure 1 shows cellular yield and viability data comparing the MLN and liver biopsies collected at two time points 160 days apart from two healthy female RMs using this laparoscopic technique. We found that cellular yield from the MLN was significantly higher than from the liver (P = 0.0021), with an average of 33.5 x 106 and 6.74 x 106 cells obtained, respectively. Dead cell staining and analysis by flow cytometry indicated that >90% of the cells isolated from both organs were viable.
Using flow cytometry, we evaluated and compared the abundance of leukocytes and lymphocytes in the MLN and liver. A representative gating scheme for the liver is shown in Figure 2, where we first gated on single cells to exclude aggregates, followed by selection of only viable cells, then selection of CD45+ cells, and finally CD3+ cells. From the CD3+ population, we evaluated abundances of CD4+ and CD8+ T cells, while we evaluated abundances of CD14+ and CD20+ cells from the CD3- populations. Representative flow cytometry plots showing CD45+ and CD3+ populations from the MLN and liver of both animals at Day 0 and Day 160 are shown in Figure 3, while Figure 4 shows leukocyte and lymphocyte abundances for the two samplings of the two animals. The MLN showed significantly higher abundances of CD45+ cells than did the liver (average 76.9% and 7.66% of viable cells, respectively, P= 0.0002). Not surprisingly, the MLN showed significantly higher proportions of CD3+ T cells (P < 0.0001) and CD4+ T cells (P = 0.0004). Conversely, the liver was significantly enriched for CD14+ leukocytes compared to the MLN (P = 0.0036). The MLN showed a higher average of CD8+ T cells, but this was not significant. Surprisingly, for these two animals both organs demonstrated similar abundances of CD20+ B cells. The cellular yields and abundances of different leukocyte and lymphocyte subsets from the MLN in this study confirmed previously reported values23.
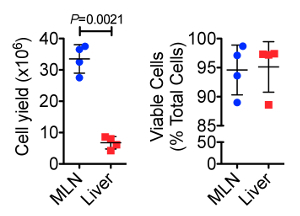 Figure 1. Total Cellular Yield (top) and Cell Viability (bottom) from Serially Collected MLN and Liver Biopsies. The MLN showed significantly higher cell yields than did the liver. However, both sample types provided ample cells for downstream analyses. Cell viability, measured by flow cytometrically using a fixable dead cell amine stain, showed that both sample types typically yielded > 90% cell viability after tissue dissociation. Error bars represent standard deviation between samples. Please click here to view a larger version of this figure.
Figure 1. Total Cellular Yield (top) and Cell Viability (bottom) from Serially Collected MLN and Liver Biopsies. The MLN showed significantly higher cell yields than did the liver. However, both sample types provided ample cells for downstream analyses. Cell viability, measured by flow cytometrically using a fixable dead cell amine stain, showed that both sample types typically yielded > 90% cell viability after tissue dissociation. Error bars represent standard deviation between samples. Please click here to view a larger version of this figure.
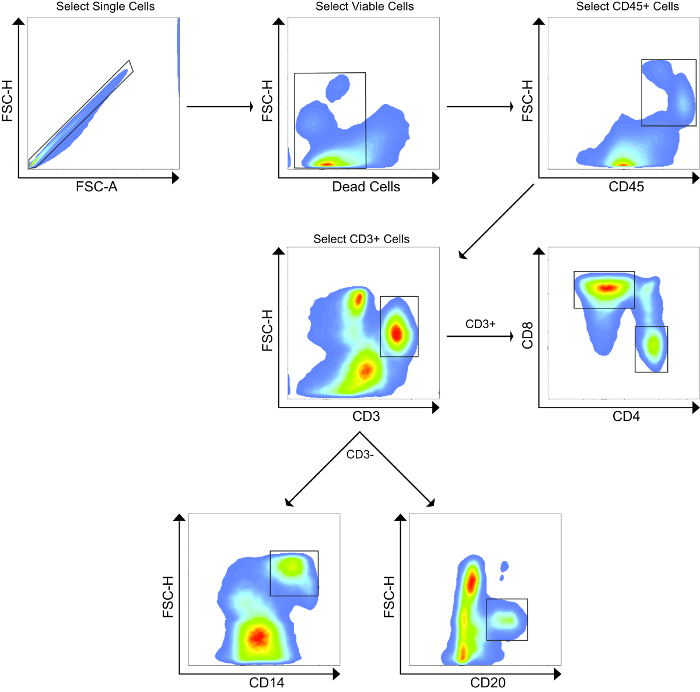 Figure 2. Representative Flow Cytometry Gating Strategy. First, aggregated cells were excluded, followed by selection of only viable cells after staining using a fixable dead cell amine stain. Next, CD45+ leukocytes were selected, followed by selection of CD3+ T cells. From the CD3+ population, CD4+ and CD8+ T cells were analyzed. From the CD3- population, CD14+ and CD20+ cells were analyzed. This gating strategy was also used for the MLN. Please click here to view a larger version of this figure.
Figure 2. Representative Flow Cytometry Gating Strategy. First, aggregated cells were excluded, followed by selection of only viable cells after staining using a fixable dead cell amine stain. Next, CD45+ leukocytes were selected, followed by selection of CD3+ T cells. From the CD3+ population, CD4+ and CD8+ T cells were analyzed. From the CD3- population, CD14+ and CD20+ cells were analyzed. This gating strategy was also used for the MLN. Please click here to view a larger version of this figure.
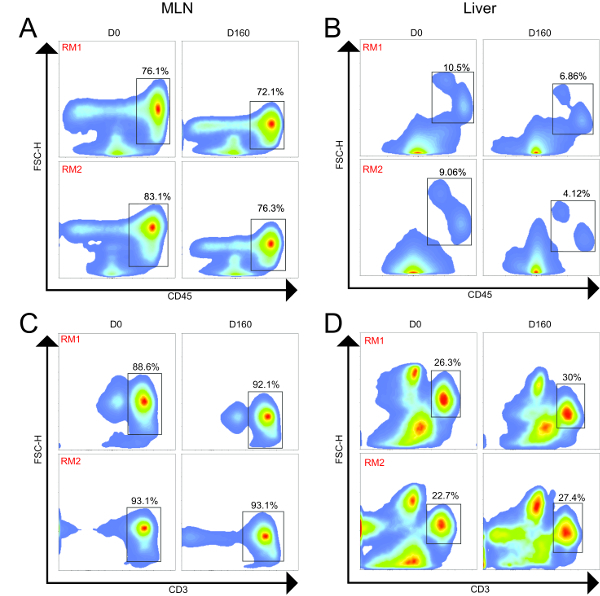 Figure 3. Flow Cytometry Plots from MLN and Liver of Two Animals Longitudinally. MLN and liver samples were collected 160 days apart for both animals. (A). MLN CD45+ cells (leukocytes); (B) Liver CD45+ cells (leukocytes); (C) MLN+ CD3+ cells (T cells); (D) Liver CD3+ cells (T cells). For both animals, the MLN showed higher proportions of CD45+ and CD3+ cells compared to the liver. Across time, the frequencies of these populations were similar for both animals. Please click here to view a larger version of this figure.
Figure 3. Flow Cytometry Plots from MLN and Liver of Two Animals Longitudinally. MLN and liver samples were collected 160 days apart for both animals. (A). MLN CD45+ cells (leukocytes); (B) Liver CD45+ cells (leukocytes); (C) MLN+ CD3+ cells (T cells); (D) Liver CD3+ cells (T cells). For both animals, the MLN showed higher proportions of CD45+ and CD3+ cells compared to the liver. Across time, the frequencies of these populations were similar for both animals. Please click here to view a larger version of this figure.
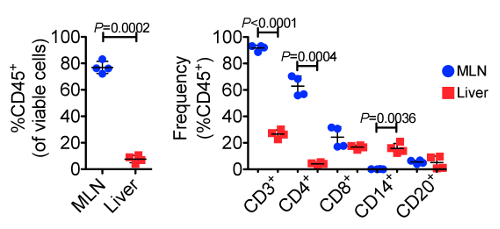 Figure 4. Frequencies of CD45+ Cells (left) and Various Leukocyte and Lymphocyte Subpopulations (right) Across 23 Individual Samplings. The MLNs displayed significantly greater frequencies of CD45+, CD3+, and CD4+ cells, while the liver showed significantly greater frequencies of CD14+ leukocytes, demonstrating the unique functions of each organ. Error bars represent standard deviation between samples. Please click here to view a larger version of this figure.
Figure 4. Frequencies of CD45+ Cells (left) and Various Leukocyte and Lymphocyte Subpopulations (right) Across 23 Individual Samplings. The MLNs displayed significantly greater frequencies of CD45+, CD3+, and CD4+ cells, while the liver showed significantly greater frequencies of CD14+ leukocytes, demonstrating the unique functions of each organ. Error bars represent standard deviation between samples. Please click here to view a larger version of this figure.
Discussion
Here, we demonstrate a minimally invasive technique for serial collection of MLN and liver biopsies that had a 100% success rate in these and other animals not described in this study. Furthermore, use of this technique on the same animals across time has not been associated with any adverse events. Indeed, there have been no complications resulting from use of this technique in other cohorts of animals that were infected with SIV, simian/human immunodeficiency virus (SHIV), or Zika virus (unpublished data). Similarly, this surgery has proven successful even in animals that had previously undergone major abdominal surgery, and in thrombocytopenic animals (unpublished data). We demonstrated that these samples can be collected through the same two ports by reversing the camera location when switching from MLN to liver avoiding the need for additional incisions. Factors affecting the collection of MLN have been previously reported13.
Up to four collections of 3 MLN and 3 liver biopsies each have been performed across time on individual animals without complications, changes in the success rates, or alterations to the data collected. Additionally, this laparoscopic MLN/liver collection technique has been successfully combined with other procedures such as collection of peripheral LNs, endoscopic upper and lower GI biopsies, bone marrow aspiration, cerebrospinal fluid collection, and venipuncture at the same anesthetic event allowing for extensive evaluation of virologic and immunologic responses in a serial fashion (unpublished data).
The entire procedure typically takes 30 - 45 min to complete and is accomplished through two ~ 0.5 cm incisions. In obese animals, it may be necessary to make a larger incision (~1 - 1.5 cm) to retrieve MLN through and may require additional time to complete. Animals with extensive mesenteric fat often require significant experience to differentiate MLN from the surrounding fat. MLN are often found along the mesenteric vasculature and seem to be more prevalent near branch points in the vessels. A critical aspect of this technique is that it requires the selected MLN to have sufficient mobility in the mesentery to be exteriorized through the abdominal incision. As a result, this technique cannot be used to collect MLN close to the root of the mesentery. Due to magnification, it is at times easier to identify MLN with the camera, and grasping in close proximity to the MLN can help with locating and identifying the node once it is exteriorized.
Use of the Trendelenburg position may not always be required for MLN collection, and if MLN can be easily retrieved without the use of Trendelenburg position it can make liver biopsies easier as it will prevent the organs from shifting cranially. At times, after placement in Trendelenburg position, the liver is shifted up against the diaphragm and must be gently manipulated back into position prior to biopsy collection. As the port placement for the MLN collection is to the left, liver biopsies are typically taken from the left lateral and left medial liver lobes as they are closer to the cannula.
Collected MLN and liver biopsies provided ample cell yields for a variety of downstream analyses and showed high viability of the collected cells. Here, the cells were stained for flow cytometric analysis of leukocyte and lymphocyte populations, and key differences between these two important immune organs were elucidated. For example, the MLN were highly enriched for CD4+ T cells compared to the liver, owing to the importance of the MLN as central points for gathering and disseminating adaptive immune responses, as well as to the importance of CD4+ T for managing inflammatory and tolerogenic responses to the milieu antigens derived from dietary intake and GI-resident microorganisms24. However, the liver was significantly enriched for CD14+ leukocytes compared to the MLN. CD14 is expressed predominantly on monocytes and macrophages and is a key component of the receptor complex for bacterial lipopolysaccharide (LPS)25. Indeed, increased plasma concentrations of soluble CD14 are associated with microbial translocation and immune activation in HIV and pathogenic SIV infections26. Thus, higher frequencies of CD14+ leukocytes in the liver likely result from a greater bacterial burden in this organ when compared to the MLN.
Here, we demonstrate a rapid and minimally invasive surgical technique for serial collection of MLN and liver biopsies using only two small incisions for laparoscopic entry that has not led to any adverse outcomes. These samples provide a useful route to evaluate key immunological, virological, and microbiological processes that take place in the MLN and liver, which are separate and unique from systemic immunity. Similarly, liver biopsies are critical for the long-term evaluation of the effects of HIV/SIV, drug therapies, and translocated microbes, which often combine to induce significant liver damage16. Further, because the MLN and liver are immune organs exposed to GI microbiota, the ability to evaluate immunity in these organs across time provides an excellent platform for understanding the role that the microbiome plays in maintaining host immune homeostasis. Taken together, evaluation of the liver and MLN is of major importance in the context of vaccine and therapeutic efficacies, SIV viral reservoir clearance, and general mucosal immunity. The ability to collect these samples through a minimally invasive approach means that there is less risk of inflammation related to the procedure than exists with currently published techniques and less impact on the animals' physiology. In the future, we will evaluate the potential to combine the collection of MLN and liver with the collection of spleen through the same two port locations to allow for sampling of another important and distinct part of the immune system that has been shown to play a significant role in a number of disease models.
Disclosures
The authors declare no competing financial interests.
Acknowledgments
This work was supported by the NIH grant number P51OD010425.
References
- Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14(3):141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- Jacobs JP, Braun J. Immune and genetic gardening of the intestinal microbiome. FEBS Lett. 2014;588(22):4102–4111. doi: 10.1016/j.febslet.2014.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson AJ, Smith K. Mesenteric lymph nodes at the center of immune anatomy. J Exp Med. 2006;203(3):497–500. doi: 10.1084/jem.20060227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaikunthanathan T, Safinia N, Lombardi G, Lechler RI. Microbiota, immunity and the liver. Immunol Lett. 2016;171:36–49. doi: 10.1016/j.imlet.2016.01.008. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Heikenwalder M, Ganal-Vonarburg SC. The Liver at the Nexus of Host-Microbial Interactions. Cell Host Microbe. 2016;20(5):561–571. doi: 10.1016/j.chom.2016.10.016. [DOI] [PubMed] [Google Scholar]
- Shuai Z, et al. Adaptive immunity in the liver. Cell Mol Immunol. 2016;13(3):354–368. doi: 10.1038/cmi.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, et al. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J Infect Dis. 2008;197(5):714–720. doi: 10.1086/527324. [DOI] [PubMed] [Google Scholar]
- Horiike M, et al. Lymph nodes harbor viral reservoirs that cause rebound of plasma viremia in SIV-infected macaques upon cessation of combined antiretroviral therapy. Virology. 2012;423(2):107–118. doi: 10.1016/j.virol.2011.11.024. [DOI] [PubMed] [Google Scholar]
- North TW, et al. Viral sanctuaries during highly active antiretroviral therapy in a nonhuman primate model for AIDS. J Virol. 2010;84(6):2913–2922. doi: 10.1128/JVI.02356-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Wang X, Lackner AA, Veazey RS. CD8 down-regulation and functional impairment of SIV-specific cytotoxic T lymphocytes in lymphoid and mucosal tissues during SIV infection. J Leukoc Biol. 2013;93(6):943–950. doi: 10.1189/jlb.1112580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Dailey PJ, Gettie A, Blanchard J, Ho DD. The Liver Is a Major Organ for Clearing Simian Immunodeficiency Virus in Rhesus Monkeys. Journal of Virology. 2002;76(10):5271–5273. doi: 10.1128/JVI.76.10.5271-5273.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon SM, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2014;7(4):983–994. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, et al. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated HIV infection. J Infect Dis. 2009;199(8):1177–1185. doi: 10.1086/597476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler NG, et al. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis. 2011;203(6):780–790. doi: 10.1093/infdis/jiq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klase Z, et al. Dysbiotic bacteria translocate in progressive SIV infection. Mucosal Immunol. 2015;8(5):1009–1020. doi: 10.1038/mi.2014.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans TI, et al. SIV-induced Translocation of Bacterial Products in the Liver Mobilizes Myeloid Dendritic and Natural Killer Cells Associated With Liver Damage. J Infect Dis. 2016;213(3):361–369. doi: 10.1093/infdis/jiv404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish R, Danneman PJ, Brown M, Karas A. Anesthesia and Analgesia in Laboratory Animals. 2 edn. Elsevier; 2011. [Google Scholar]
- Flecknell PA. Laboratory Animal Anaesthesia. 3 edn. Academic Press; 2009. [Google Scholar]
- Tranquilli WJ, Thurnom JC, Grimm KA. Lumb and Jones' Veterinary Anesthesia and Analgesia. Blackwell Publishing; 2007. [Google Scholar]
- Ford RB, Mazzaferro EM. Kirk and Bistner's Handbook of Veterinary Procedures and Emergency Treatment. 9 edn. Elsevier; 2012. [Google Scholar]
- Fransson BA, Mayhew PD. Small Animal Laparoscopy and Thoracoscopy. Wiley Blackwell; 2015. [Google Scholar]
- Hensley-McBain T, et al. Effects of Fecal Microbial Transplantation on Microbiome and Immunity in Simian Immunodeficiency Virus-Infected Macaques. J Virol. 2016;90(10):4981–4989. doi: 10.1128/JVI.00099-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley J, et al. Laparoscopic Technique for Serial Collection of Para-Colonic, Left Colic, and Inferior Mesenteric Lymph Nodes in Macaques. PLoS One. 2016;11(6):0157535. doi: 10.1371/journal.pone.0157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 2015;8(1):80–93. doi: 10.1038/mi.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler-Heitbrock HW, Ulevitch RJ. CD14: cell surface receptor and differentiation marker. Immunol Today. 1993;14(3):121–125. doi: 10.1016/0167-5699(93)90212-4. [DOI] [PubMed] [Google Scholar]
- Brenchley JM, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12(12):1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]


