Abstract
The cell surface marker CD133 has been used to describe a revised model of adult human hematopoiesis with hematopoietic stem cells and multipotent progenitors (HSCs/MPPs: CD133+CD45RA−CD34+) giving rise to a lympho-myeloid primed progenitor (LMPPs: CD133+CD45RA+CD34+) and an erythro-myeloid progenitor (EMPs: CD133lowCD45RA−CD34+). Since adult and fetal hematopoietic stem and progenitor cells (HSPCs) differ in their gene expression profile, differentiation capabilities, and cell surface marker expression, we were interested whether the reported segregation of lineage potentials in adult human hematopoiesis would also apply to the human fetal liver (FL). CD133 expression was easily detected on human FL cells, and the defined hematopoietic subpopulations were similar to the findings with adult HSPCs. Fetal HSPCs were enriched for EMPs and HSCs/MPPs, which were primed towards erythro-myeloid differentiation. However, the frequency of multipotent CD133+CD45RA−CD34+ HSPCs was much lower than previously reported and comparable to umbilical cord blood. We found that engraftment in NSG mice was mostly driven by LMPPs, confirming recent findings that repopulation in mice is not a unique feature of multipotent HSCs/MPPs. Thus, our data challenges the general assumption that the human FL contains a greater percentage of multipotent HSCs/MPPs than any adult HSC source, and the mouse model may have to be re-evaluated with regard to the type of readout it provides.
Keywords: Human hematopoiesis; Fetal liver; CD34, CD133; multipotent HSC/MPP; SCID-repopulating cells (SRC)
Introduction
Assessment of CD133 on hematopoietic stem and progenitor cell (HSPC) from umbilical cord blood (UCB) and bone marrow (BM) has recently been used to describe a revised model of human hematopoiesis (Figure 1A) [1–3]. These studies showed that CD133+CD34+CD45RA− cell fractions are highly enriched for hematopoietic stem cells and multipotent progenitor cells (HSCs/MPPs) giving rise to CD133+CD34+CD45RA+ lympho-myeloid primed progenitors (LMPPs) and CD133lowCD34+CD45RA− erythro-myeloid restricted progenitors (EMP, Figure 1A and B) [4]. This model further proposed that multipotent HSCs/MPPs could easily be identified as CD133-expressing cells with erythroid differentiation potential.
Figure 1. Phenotypical and functional in vitro characterization of FL-, UCB-, and PBSC- derived CD34+ subpopulations.
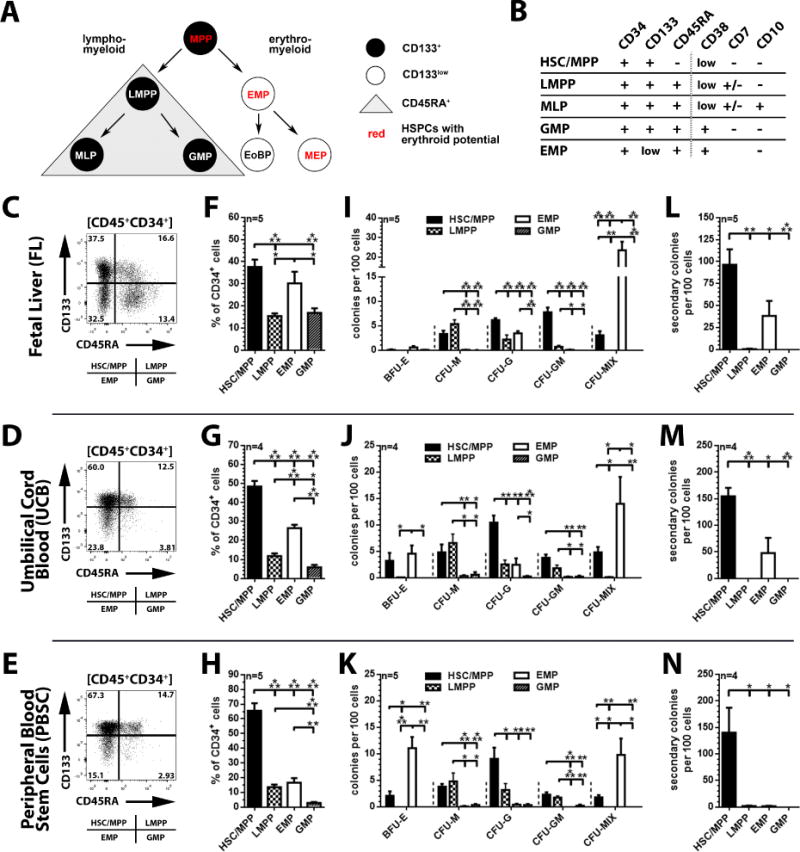
(A) Revised model of human hematopoiesis (BM and UCB) with HSCs/MPPs giving rise to either lympho-myeloid or erythro-myeloid progenitor cells. HSPCs with erythroid differentiation potential are highlighted in red. Abbreviations: MPP: multipotent progenitor; LMPP: lympho-myeloid primed progenitor; EMP: erythro-myeloid progenitor; MLP: multilymphoid progenitor; GMP: granulocyte (neutrophil)-macrophage progenitor; EoBP: eosinophil-basophil progenitor; MEP: megakaryocyte-erythrocyte progenitor. (B) Phenotype of functionally distinct hematopoietic stem and progenitor cell subpopulations. (C, D and E) Flow cytometric analysis of MACS-purified FL-, UCB-, and PBSC-derived CD34+ cells after exclusion of dead cells/debris (FSC/SSC) and non-hematopoietic/endothelial CD45− cells; subdivision of CD45+CD34+ cells into CD45+CD34+CD133+CD45RA− HSC/MPP−, CD45+CD34+CD133+CD45RA+ LMPP−, CD45+CD34+CD133lowCD45RA− EMP−, and CD45+CD34+CD133lowCD45RA+ GMP-enriched fractions. (F, G and H) Frequency of HSCs/MPPs, LMPPs, EMPs and GMPs within FL-, UCB-, and PBSC-derived CD34+ populations. (I, J and K) Frequency of erythroid (BFU-E: burst forming unit-erythrocyte), myeloid (CFU [colony-forming unit]-M [macrophage], -G [granulocyte] and -GM [granulocyte-macrophage]), and erythro-myeloid (CFU-MIX = granulocyte-erythrocyte-megakaryocyte-macrophage) colonies obtained from sort-purified FL, UCB and PBSC CD34-subpopulations. (L, M and N) Primary CFCs of HSC/MPP, LMPP, EMP, and GMP populations were harvested and replated. Secondary colony-forming potential represents total colony count without discrimination of colony-subtypes.
Previous studies describing this revised model of human hematopoiesis were mostly performed with adult HSPCs derived from steady state bone marrow (BM), GCSF-mobilized peripheral blood stem cells (PBSCs) and umbilical cord blood (UCB). Since adult and fetal HSPCs differ in their gene expression profile, differentiation capabilities, and cell surface marker expression [5–7], we wished to determine whether the revised model of hematopoiesis would also apply to the human fetal liver (FL).
Methods
Human FL (gestational week 17–20, n=5) was acquired from Advanced Bioscience Resources (Alameda, CA), UCB kindly provided by Colleen Delaney (Fred Hutchinson Cancer Research Center), and PBSCs acquired from the Core Center of Excellence in Hematology (Shelly Heimfeld, Fred Hutchinson Cancer Research Center) after informed consent according to the Declaration of Helsinki. FL tissue was dissociated and CD34+ cells from all sources enriched by immunomagnetic separation (IMS). CD34+-enriched populations were flow-cytometrically characterized and sort-purified by means of CD34 (clone 563), CD45RA (clone 5H9) and CD133 (clone AC133) expression on a FACSAria IIu. Sort-purified CD34-subpopulations were cultured in StemSpan (Stemcell Technologies, Vancouver, British Columbia, Canada) supplemented with SCF, TPO and FLT3-L (100 ng/ml each) and 1% pen/strep (Life Technologies, Grand Island, NY).
For Colony-forming cell (CFC) assays 1000–1200 sorted cells were seeded into 3.5 ml MethoCult H4434 (Stemcell Technologies). Hematopoietic colonies were scored after 12–14 days. Arising colonies were identified as colony-forming unit- (CFU-) macrophage (M), granulocyte (G), granulocyte-macrophage (GM) and burst forming unit-erythrocyte (BFU-E). Colonies consisting of erythroid and myeloid cells were scored as CFU-MIX.
NSG mice (NOD.Cg-PrkdcscidIl2rgtmlWjl/SzJ) within 48 hours of birth received a radiation dose of 150 cGy followed 4 hours later by a 30μL intrahepatic injection of 1×106 FL CD34+ cells. Beginning at 8 weeks post injection, blood samples were collected biweekly and analysed by flow cytometry for expression of human lineage markers CD45 (clone 2D1), CD3 (clone UCHT1), CD4 (clone RPA-T4), CD8 (clone SK1), CD14 (clone M5E2), and CD20 (clone 2H7). After 18–24 weeks, animals were sacrificed, lymphoid tissues harvested and dissociated through 70μm filters for analysis.
All animal studies were carried out at Fred Hutchinson Cancer Research Center in compliance with the approved IACUC protocol #1864.
All data are given as mean ± standard error of the mean (SEM). Significance analyses was performed with the paired Student t test (*: p < 0.05; **: p < 0.01; ***: p <0.001).
Results/Discussion
Assessment of CD133 within FL-derived HSPCs allowed for objective discrimination of HSCs/MPPs (CD133+CD45RA−CD34+) which give rise to lympho-myeloid primed progenitors (LMPPs: CD133+CD45RA+CD34+), erythro-myeloid progenitors (EMPs: CD133lowCD45RA−CD34+) and granulocyte-macrophage progenitors (GMPs: CD133lowCD45RA+CD34+) (Figure 1C), similar to adult/postnatal HSC-sources (Figure 1D–E and [3]). Consistent with its biological function [3, 8–10] and in contrast to PBSCs and BM, which exhibit a strong bias towards more primitive CD133+CD45RA− (PBSCs) or erythro-myeloid CD133lowCD45RA− (BM) progenitors (Figure 1G–H and [3]), FL-derived CD34+ cells were enriched for phenotypical EMPs (30.2 ± 5.2%) as well as their more primitive ancestors (HSC/MPP: 37.6 ± 3.4%, Figure 1F).
Within UCB, PBSCs, and BM, HSPCs with colony-forming unit (CFU) granulocyte-macrophage (CFU-GM) potential were exclusively found within HSC/MPP and LMPP populations, whereas erythro-myeloid restricted CD133lowCD45RA− EMPs were enriched for erythrocyte-granulocyte-macrophage (CFU-MIX) progenitors (Figure 1J–K and [2]). CFU-GM potential in the FL was only detected in CD133+CD45RA− HSCs/MPPs (7.7 ± 0.9%) as well as CD133+CD45RA+ LMPP fractions (0.7 ± 0.3%), whereas CD133lowCD45RA− EMPs were highly enriched for CFU-MIX progenitors with a frequency 1.7 to 2.4-fold higher than in UCB, PBSCs and BM (23.4 ± 4.6%, Figure 1I). In contrast to previous publications reporting up to 200-fold higher frequency of HSCs in the FL than in UCB and BM [5, 11, 12], the frequency of ‘true’ multipotent HSCs/MPPs (3.1 ± 0.8%, Figure 1I), defined as CD133+ cells with CFU-MIX potential [2], was lower than in UCB (4.8 ± 1.0%, Figure J) and only 2-fold higher than in PBSCs and BM (1.8 ± 0.5%, Figure K).
Enrichment of more primitive HSPCs in the CD133+CD45RA− cell fraction was further confirmed by secondary colony-forming potential (96.4 ± 17.6%, Figure 1L–N), whereas LMPPs and GMPs entirely lacked the ability to form secondary CFCs (Figure 1L–N). Surprisingly, primary CFC assays from FL and UCB-derived EMPs contained a substantial fraction of progenitor cells with secondary colony-forming potential that has not been observed for PBSCs (38.1 ± 17.1% and 47.5 ± 28.5%, Figure 1L–N).
Within adult stem cell sources, HSC/MPP have been shown to divide asymmetrically giving rise to a LMPP and EMP daughter cell [4]. Thus we wished to determine whether FL-derived HSCs/MPPs would also serve as precursors of LMPPs and EMPs. We sort-purified all four CD34 subpopulations, cultured them for three days and then analyzed the progeny by flow cytometry (Figure 2A–C). Similar to UCB and PBSCs (Figure 2B–C and [4]), only HSCs/MPPs from FL gave rise to LMPPs, EMPs and GMPs (Figure 2A). In contrast to UCB and PBSC-derived HSC/MPP fractions which predominantly give rise to LMPPs within similar growth conditions (Figure 2B–C and [4]), the majority of FL-derived HSCs/MPPs differentiated into EMPs (Figure 2A). To confirm that FL-HSCs/MPPs can give rise to functional LMPPs and EMPs, arising daughter cells were plated into CFC assays. As expected, HSC/MPP-derived progeny from all three tissue sources showed lineage potentials similar to freshly isolated subpopulations (Figure 1D–F).
Figure 2. Phenotypical and functional characterization of culture-derived FL, UCB, and PBSC CD34+ subpopulations.
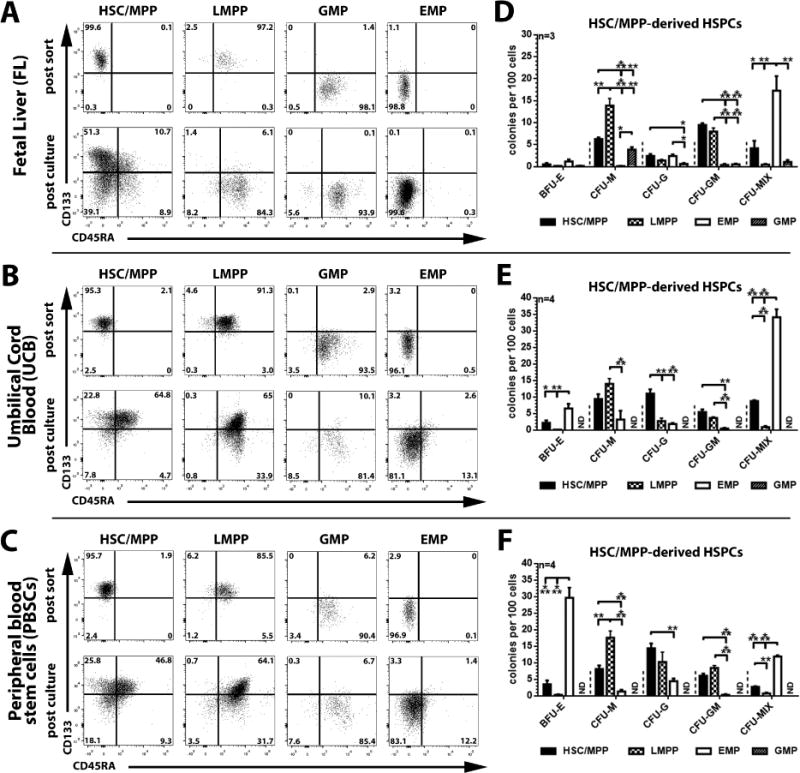
(A–C) Sort-purified FL-, UCB-, and PBSC-derived HSC/MPP, LMPP, EMP, and GMP subpopulations (post sort, upper row) were cultured in StemSpan supplemented with SCF, TPO and Flt3-L (100ng/ml each). Phenotype, composition, and proliferation of arising progeny was analyzed on day 3 (post culture, lower row). (D–F) On day 3 of culture, HSC/MPP-derived CD133/CD45RA-subpopulations were sort-purified, introduced into colony-forming cell assay and ability to form different colony-subtypes quantified (all statistics mean ± SEM; *: p<0.05, **: p<0.01, ***: p<0.001).
The percentage of phenotypical lympho-myeloid progenitors (LMPPs) and primitive HSCs/MPPs was low in FL-derived HSPCs and there was a strong bias toward erythro-myeloid differentiation among the HSC/MPP population. Thus, we further analyzed the lymphoid differentiation potential of three independent FL CD34+ HSPC samples in neonatal NSG-mice. We transplanted unfractionated FL CD34+ populations to investigate simultaneously the influence and the support of the murine BM environment on the selection and maintenance of distinct HSPC subpopulations. Stable engraftment of human CD45+ cells in the peripheral blood (PB) ranging from 28 up to 89% was detected after 8–18 weeks (Figure 3A) containing human B cells (CD20+), T cells (CD3+/CD4+/CD8+), as well as monocytes (CD14+) (Figure 3B and D). Multi-lineage engraftment was further confirmed in the thymus, spleen, and BM (Figure 3C), which were enriched for human T cells (47.5 ± 9.1%), B cells (43.7 ± 9.0%) or CD34+ progenitor cells (7.4 ± 2.2%), respectively (Figure 3E–G).
Figure 3. Engraftment potential of FL-derived CD34+ cells.
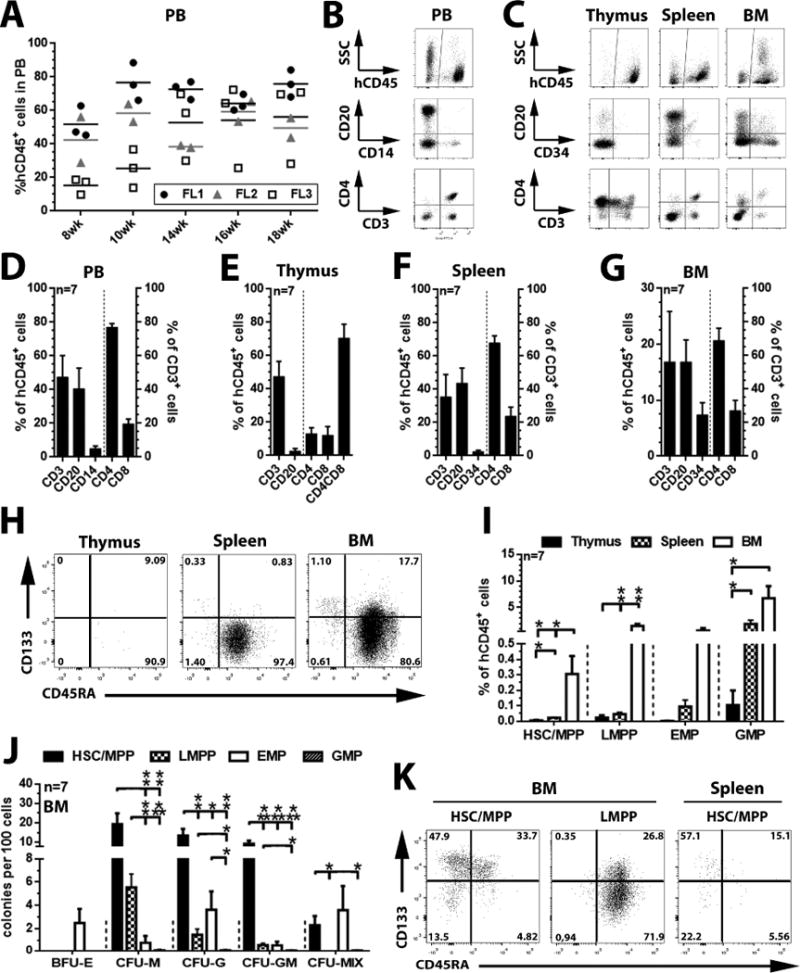
(A) Flow-cytometric quantification of human CD45+ (hCD45+) cells in the peripheral blood (PB) of transplanted mice 8–18 weeks post transplant. (B and C) Representative flow-cytometric analysis of human B cells (CD20+), monocytes (CD14+), T cells (CD3+/CD4+/CD8+/CD4+CD8+) and CD34+ progenitor cells in PB, thymus, spleen, and bone marrow (BM) of reconstituted mice (week 18). (D–G) Average frequency of human B cells, T cells, monocytes, and CD34+ cells within hCD45+ fractions (left axis) as well as T cell-subtypes within human CD3+ cell populations (right axis) (H) Representative flow-cytometric analysis and (I) frequency of the HSC/MPP-, LMPP-, EMP-, and GMP subpopulations in thymus-, spleen-, and BM-resident CD34+ fractions (week 18). (J) Colony-forming potential of sort-purified human HSC/MPP-, LMPP-, EMP-, and GMP-enriched subpopulations from murine BM. (K) Sort-purified human HSC/MPP- and LMPP-enriched subpopulations from murine BM samples (post sort) were cultured in StemSpan supplemented with SCF, TPO and Flt3-L (100ng/ml each) and composition/proliferation of arising progeny analyzed on day 3 post culture (all statistics mean ± SEM; *: p<0.05, **: p<0.01, ***: p<0.001).
A distinct subpopulation of CD34+ cells could also be found within the spleen (2.05 ± 0.75%), with the vast majority of progenitor cells being phenotypically committed GMPs (1.89 ± 0.69%) and only trace amounts of HSCs/MPPs (0.021 ± 0.005%, Figure 3H and I). Compared to the spleen, the murine BM contained phenotypical HSCs/MPPs (0.30 ± 0.12%), LMPPs (1.52 ± 0.39%), EMPs (0.64 ± 0.47%), and GMPs (6.74 ± 2.25%, Figure 3H and I). Similar to cultured cells, sort-purified human CD34+ subpopulations from the murine BM exhibited colony-forming potential with LMPPs restricted to myeloid lineages (G/M/GM), EMP biased to erythroid lineages (BFU-E), and HSC/MPP showing full erythro-myeloid differentiation capabilities (Figure 3J). However, CD34+ subpopulations from the spleen exhibited no differentiation potential in CFC assays (data not shown).
Interestingly, the frequency of BM-resident multipotent CD133+CD45RA− with CFU-MIX potential [4] was very low (Figure 3J) with a calculated frequency of 0.007 ± 0.003% (2.29% [CFU-Mix frequency] of 0.30% CD133+CD45RA− HSPCs [Figure 3I]) in the bone marrow-resident human CD45+ cell fraction. Considering the high frequency of human CD45+ cell in the peripheral blood of engrafted mice, this suggests the observed engraftment was predominantly driven by committed progenitors, e.g. LMPPs (1.52 ± 0.39%) and GMPs (6.74 ± 2.25%). Even though the calculated frequency of engrafted multipotent HSPC was very low, sort-purified human CD133+CD45RA−CD34+ HSPCs from the murine BM were able to give rise to phenotypical and functional LMPPs and EMPs following ex vivo culture (Figure 3K).
In summary, we demonstrate that the assessment of CD34 in combination with CD133 and CD45RA allows for the characterization of functionally distinct hematopoietic subpopulations in the human FL, similar to findings with adult stem cell sources. The FL is enriched for massively expanding EMPs and erythro-myeloid primed HSCs/MPPs, which is expected based on the tissue’s primary function [8–10]. However, we observed a lower frequency of multipotent HSPCs than previously reported, challenging the general assumption that human FL contains greater numbers of multipotent HSCs/MPPs than adult stem cell sources [11–13]. Even though we transplanted an unfractionated FL-CD34+ population, engraftment of FL-HSPCs in mice and production of mature blood cells in the periphery were likely driven by committed progenitor cells (LMPPs), confirming our recent finding that repopulation in mice is not a unique feature of multipotent HSCs/MPPs only [1, 2]. Therefore, quantification of SCID-repopulating cells (SRCs), the gold standard for HSC research and surrogate for multipotent HSCs/MPPs, may have to be re-evaluated with regard to the type of readout it provides [14].
Highlights.
No increase of multipotent HSCs in the human fetal liver compared to adult tissues
Segregation of lympho-myeloid and erythro-myeloid lineages in fetal hematopoiesis
Fetal liver HSCs/MPPs are primed towards erythro-myeloid lineage specification
NOD/SCID mouse engraftment is mostly driven by lympho-myeloid restricted LMPPs
Acknowledgments
Shirley Chan, Kina McAllister and Morgan Giese assisted performing the experiments. Umbilical cord blood was kindly provided by Colleen Delaney. Peripheral blood stem cells were kindly provided by Shelly Heimfeld.
Funding:
This work was supported in part by grants to HPK from the National Institutes of Health, Bethesda, MD: HL115128 and HL098489. HPK is a Markey Molecular Medicine Investigator and received support as the inaugural recipient of the José Carreras/E. Donnall Thomas Endowed Chair for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authorship Contributions
HPK is the principal investigator and oversaw the design, data analysis and manuscript writing. S. R. conceived the study, designed and performed the experiments and wrote the manuscript. K. H. performed the experiments and wrote the manuscript. All authors discussed the data and contributed to the manuscript.
Disclosure of Conflicts of Interest:
The authors declare that there are no conflicts of interest
Author information:
Ph.D. Stefan Radtke, sradtke@fredhutch.org
Ph.D. Kevin G Haworth, khaworth@fredhutch.org
M.D. Hans-Peter Kiem, hkiem@fredhutch.org
References
- 1.Görgens A, Radtke S, Horn PA, Giebel B. New relationships of human hematopoietic lineages facilitate detection of multipotent hematopoietic stem and progenitor cells. Cell Cycle. 2013;12 doi: 10.4161/cc.26900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Görgens A, Radtke S, Möllmann M, et al. Revision of the human hematopoietic tree: granulocyte subtypes derive from distinct hematopoietic lineages. Cell reports. 2013;3:1539–1552. doi: 10.1016/j.celrep.2013.04.025. [DOI] [PubMed] [Google Scholar]
- 3.Radtke S, Görgens A, Kordelas L, et al. CD133 allows elaborated discrimination and quantification of haematopoietic progenitor subsets in human haematopoietic stem cell transplants. British journal of haematology. 2015;169:868–878. doi: 10.1111/bjh.13362. [DOI] [PubMed] [Google Scholar]
- 4.Görgens A, Ludwig AK, Möllmann M, et al. Multipotent hematopoietic progenitors divide asymmetrically to create progenitors of the lymphomyeloid and erythromyeloid lineages. Stem cell reports. 2014;3:1058–1072. doi: 10.1016/j.stemcr.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rollini P, Faes-Van’t Hull E, Kaiser S, Kapp U, Leyvraz S. Phenotypic and functional analysis of human fetal liver hematopoietic stem cells in culture. Stem Cells Dev. 2007;16:281–296. doi: 10.1089/scd.2006.0096. [DOI] [PubMed] [Google Scholar]
- 6.Dorrell C, Gan OI, Pereira DS, Hawley RG, Dick JE. Expansion of human cord blood CD34(+)CD38(-) cells in ex vivo culture during retroviral transduction without a corresponding increase in SCID repopulating cell (SRC) frequency: dissociation of SRC phenotype and function. Blood. 2000;95:102–110. [PubMed] [Google Scholar]
- 7.Ivanovs A, Medvinsky A. In search of human hematopoietic stem cell identity. Cell Stem Cell. 2015;16:5–6. doi: 10.1016/j.stem.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 8.Gekas C, Rhodes KE, Van Handel B, Chhabra A, Ueno M, Mikkola HK. Hematopoietic stem cell development in the placenta. The International journal of developmental biology. 2010;54:1089–1098. doi: 10.1387/ijdb.103070cg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alvarez-Silva M, Belo-Diabangouaya P, Salaun J, Dieterlen-Lievre F. Mouse placenta is a major hematopoietic organ. Development. 2003;130:5437–5444. doi: 10.1242/dev.00755. [DOI] [PubMed] [Google Scholar]
- 10.Ciriza J, Thompson H, Petrosian R, Manilay JO, Garcia-Ojeda ME. The migration of hematopoietic progenitors from the fetal liver to the fetal bone marrow: lessons learned and possible clinical applications. Exp Hematol. 2013;41:411–423. doi: 10.1016/j.exphem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 11.Mikkola HK, Gekas C, Orkin SH, Dieterlen-Lievre F. Placenta as a site for hematopoietic stem cell development. Exp Hematol. 2005;33:1048–1054. doi: 10.1016/j.exphem.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 12.Holyoake TL, Nicolini FE, Eaves CJ. Functional differences between transplantable human hematopoietic stem cells from fetal liver, cord blood, and adult marrow. Experimental hematology. 1999;27:1418–1427. doi: 10.1016/s0301-472x(99)00078-8. [DOI] [PubMed] [Google Scholar]
- 13.Rebel VI, Miller CL, Eaves CJ, Lansdorp PM. The repopulation potential of fetal liver hematopoietic stem cells in mice exceeds that of their liver adult bone marrow counterparts. Blood. 1996;87:3500–3507. [PubMed] [Google Scholar]
- 14.Horn PA, Kiem HP. Expansion of SCID repopulating cells does not prove expansion of hematopoietic stem cells. Blood. 2006;108:771. doi: 10.1182/blood-2006-02-002618. author reply 771–772. [DOI] [PubMed] [Google Scholar]


