Abstract
Biofilm formation by pathogenic Burkholderia species is a serious complication as it renders the bacteria resistant to antibiotics and host defenses. Using B. thailandensis, we report here a novel redox-sensitive member of the multiple antibiotic resistance regulator (MarR) protein family, BifR, which represses biofilm formation. BifR is encoded as part of the emrB-bif R operon; emrB-bif R is divergent to ecsC, which encodes a putative LasA protease. In Pseudomonas aeruginosa, LasA has been implicated in virulence by contributing to cleavage of elastase. BifR repressed the expression of ecsC and emrB-bif R, and expression was further repressed under oxidizing conditions. BifR bound two sites in the intergenic region between ecsC and emrB-bif R with nanomolar affinity under both reducing and oxidizing conditions; however, oxidized BifR formed a disulfide-linked dimer-of-dimers, a covalent linkage that was absent in BifR-C104A in which the redox-active cysteine was replaced with alanine. BifR also repressed an operon encoding enzymes required for synthesis of phenazine antibiotics, which function as alternate respiratory electron receptors, and inactivation of bif R resulted in enhanced biofilm formation. Taken together, our data suggest that BifR functions to control LasA production and expression of genes involved in biofilm formation, in part by regulating synthesis of alternate electron acceptors that promote survival in the oxygen-limiting environment of a biofilm. The correlation between increased repression of emrB-bif R under oxidative conditions and the formation of a covalently linked BifR dimer-of-dimers suggests that BifR may modulate gene activity in response to cellular redox state.
Graphical abstract
The genus Burkholderia includes species that cause serious plant and animal diseases.1 B. pseudomallei and B. mallei are considered category B priority pathogens as even small doses can cause lethal infection in humans.2 B. cenocepacia is the most serious pathogen in the B. cepacia complex, a group of closely related opportunistic pathogens best known for their ability to cause lung infection in cystic fibrosis (CF) patients. Infection by B. cenocepacia is problematic due to antibiotic resistance that results in chronic infection, deterioration of lung function, and high mortality rates. Numerous virulence factors have been reported to play critical roles in the success of these pathogens, ranging from production of extracellular proteases required for interaction with epithelial cells to quorum sensing signal molecules that regulate expression of virulence gene expression.3,4 B. thailandensis is frequently used as a model system for analysis of such virulence traits, as it is closely related to the pathogenic species, but only rarely causes disease in humans.5
Biofilm formation plays a vital role in the pathogenesis of chronic infections. Biofilm communities, in which sessile bacteria are protected from environmental factors including host defenses and antibiotics, promote persistence and chronic infection. In the CF lung, coinfection by Pseudomonas aeruginosa and B. cenocepacia has particular potential to result in establishment of biofilm communities.6 Among other factors, the initiation of biofilm structures requires surface attachment, and maturation involves production of extracellular polysaccharides.7 The transition from planktonic to biofilm growth is complex and may be triggered by events such as nutrient starvation and changes in temperature and pH, and maintenance depends on production of biofilm matrix components.8,9
Recent reports have also highlighted the role of redox homeostasis, as for example reflected in the NADH/NAD+ ratio, as an important determinant of biofilm morphology.10,11 Within a biofilm, a hypoxic gradient develops as access to oxygen becomes limiting, and alternative respiratory electron receptors such as phenazines may be produced.10,12 The phenazine operon responsible for conversion of chorismic acid to phenazine-1-carboxylic acid (PCA) has likely been distributed by horizontal gene transfer among members of certain bacterial genera, including Pseudomonas and Burkholderia.13
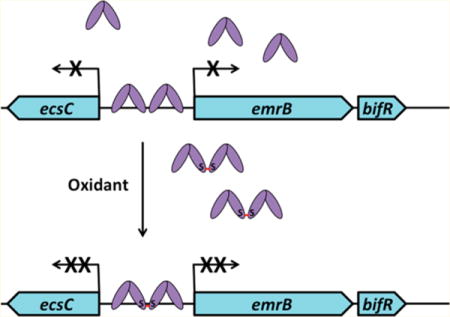
Here we describe a B. thailandensis-encoded redox-sensitive multiple antibiotic resistance regulator (MarR) homologue that controls biofilm formation. On account of this functional role, we propose the name BifR (bio film regulator). While the dimeric BifR represses its own expression, as commonly seen for MarR proteins, repression is more efficient under oxidizing conditions and correlates with formation of a “super-repressor”, a disulfide-bridged dimer-of-dimers. We propose that BifR functions to link cellular redox state to expression of genes with roles in biofilm formation, in part by controlling production of PCA.
MATERIALS AND METHODS
Confirmation of Transposon Mutants
Gene disruption mutants of B. thailandensis E264 in which transposon T23 (ISlacZ-PrhaBo-Tp/FRT) was inserted in emrB-bif R (BTH_I0541–0542; at position 439 of the emrB open reading frame or at position 15 of bif R to generate BTH_I0541–146::ISlacZ-PrhaBo-Tp/FRT and BTH_I0542–129::ISlacZ-PrhaBo-Tp/FRT, respectively) were obtained from the Manoil lab and grown on LB agar plates with 80 μg/mL trimethoprim.14 Single colonies were inoculated in LB medium containing 80 μg/mL trimethoprim and grown overnight. Genomic DNA was isolated from overnight cultures and used as a template for PCR verification of mutants (transposon insertion at the correct location) using primers BTH_clon_Fw, I541_Mut_fw and LacZ_148 (Supplemental Table S1).
Plasmid Construction for Complementation
For the purpose of genetic complementation of transposon mutants, plasmids were constructed that carry emrB or emrB-bif R, each with 106 bp upstream DNA sequence (corresponding to the ecsC-emrB intergenic region as annotated in the NCBI database), cloned into pBBR-MCS5, a gentamycin resistance-encoding derivative of the broad host range cloning vector pBBR1MCS.15 XbaI and KpnI sites were introduced in the upstream and downstream primers, respectively (for EmrB, using primers EmrB_Xba_pBBR_Fw and EmrB_KpnI_pBBR_Rev and for EmrB-BifR, using EmrB_Xba_pBBR_Fw and EmrB_BifR_kpnI_pBBR_Rev; Supplemental Table 1). The PCR product was digested and cloned into pBBR-MCS5. The plasmids were transformed into Escherichia coli Top 10 (Invitrogen) and verified by sequencing.
Conjugative Plasmid Transfer
Plasmids harboring emrB or emrB-bif R were transferred to the B. thailandensis E264 mutant strains by triparental mating. Overnight cultures of the donor (E. coli DH5α with pBBR-MCS5 plasmid containing emrB or emrB-bif R), recipient (emrBΔ-bif RΔ or bif RΔ), and helper (HB101(pRK2013::Tn7)) strains were grown and mixed in a 1:1:2 ratio of donor/recipient/helper strains. The mixed culture was centrifuged to remove LB and residual antibiotics. The pellet was washed four times with 1.0 mL of LB. The entire pellet was resuspended in 200 μL of LB and spotted on a preheated LB agar plate. After overnight incubation, all cells were scraped off and resuspended in 1 mL of LB. Serial dilutions were plated on an LB agar plate containing antibiotics trimethoprim (80 μg/mL), gentamicin (250 μg/mL), and chloramphenicol (8 μg/mL) for selection of trans-conjugants followed by verification by PCR using primers Veri_pBBR_XbaI fw and Rev (Supplemental Table S1).
In Vivo Gene Expression and Operon Confirmation
For RNA isolation, overnight cultures of WT, emrBΔ-bif RΔ, and bif RΔ mutant strains, and the complementation strain bif RΔ e-b were diluted 1:100 in fresh LB medium and grown to OD ≈ 0.5 (exponential phase). Mutant strains emrBΔ-bif RΔ and bif RΔ were grown in LB medium containing trimethoprim (80 μg/mL). Complementation strain bif RΔ e-b was grown in LB medium containing trimethoprim (80 μg/mL), gentamicin (250 μg/mL), and chloramphenicol (8 μg/mL). H2O2 or CuCl2 was added to a final concentration of 1 mM after cells reached OD ≈ 0.5 for analysis of gene expression under oxidizing conditions. The concentrations of H2O2 and CuCl2 were chosen based on a plate sensitivity assay in which these concentrations did not visibly impede growth (data not shown). To determine the effect of redox inactive metal on gene expression, cells were exposed to ZnCl2 and MgCl2 at final concentrations of 1 mM and 30 mM, respectively. Cells were grown for 30 min and harvested by centrifugation. Cells were mixed with ice-cold DEPC-treated water and centrifuged, and the pellet was frozen at −80 °C. Total RNA was isolated using the illustra RNAspin Mini Isolation kit (GE Healthcare). DNA contamination was removed using Turbo DNase (Ambion), and the absence of DNA was verified by PCR. RNA was quantified using NanoDrop (Thermo Scientific).
The cDNA was prepared using 500 ng of total RNA. cDNA was made by mixing either EmrB, Bif R, Nudix, PhzF, or EcsC qPCR primers (EmrB_qpCR_Fw and EmrB_qpCR_Rev, BifR_qPCR_Fw, and BifR_qPCR_Rev, Nudix_qPCR_Fw and Nudix_qPCR_Rev, PhzF_qPCR_Fw and PhZF_qPCR_Rev, or EcsC_qPCR_Fw, and EcsC_qPCR_-Rev; Supplemental Table S1; gene-specific primers were used to increase sensitivity, since only specific transcripts will be reverse-transcribed) in 1× AMV reverse transcriptase buffer with 1 mM MgCl2, 1 mM dNTP, and 10 units of AMV reverse transcriptase (New England Biolabs) in a total reaction volume of 25 μL. The mixture was incubated at 42 °C for 1 h. A ViiA 7 (Applied Biosystems) was used for qPCR using Taq polymerase (New England Biolabs) for amplification and SYBR Green I (Sigma) for detection. For analysis of gene expression in WT, expression of emrB, bif R, nudix, ecsC, and phzF was normalized to the reference gene (glutamate synthase large subunit; BTH_I3014, amplified using primers Glusynlg_qPCR_Fw and Glusynlg_qPCR_Rev) and reported as . The reference gene was selected based on its use as a reference in analysis of the transcriptional response of B. cenocepacia to oxidants, where its expression was found to be stable under different experimental conditions.16 We likewise observed stable expression of BTH_I3014 under our experimental conditions (Supplemental Table S2). For analysis of effect of H2O2, Cu2+, Mg2+, and Zn2+, the data were normalized to the same reference gene, and the abundance of transcript level was calculated using the comparative CT method .17 Data are presented as the mean of three biological replicates (each determined in triplicate) ± SD.
To verify the operon, Bif R primer (BifR_qPCR_Rev; Supplemental Table S1) was mixed with the RNA, and cDNA was prepared as described above. cDNA was amplified with the Emr B primers (EmrB_qpCR_Fw, EmrB_qpCR_Rev; Supplemental Table S1), and the PCR product was electrophoresed in a 1% (w/v) agarose gel and visualized by ethidium bromide staining.
Growth and Colony Morphology
To determine if mutations altered growth, overnight cultures of WT, emrBΔ-bif RΔ, and bif RΔ mutant strains and the corresponding complementation strains were grown to an OD600 of ∼2.0. Overnight cultures were diluted 1:100 in LB broth, and absorbance of all the strains was recorded at 1 h intervals. Data are representative of three replicates. Absorbance data were plotted on log10 scale.
For inspection of colony morphology, overnight cultures of WT, emrBΔ-bif RΔ, and bif RΔ mutant strains and the corresponding complementation strains were grown to an OD600 of ∼2.0. Ten microliters of cells were spotted on an LB agar plate. Colony morphology was analyzed after incubation for 48–72 h at 37 °C.
Elastase Activity
To assess the elastin cleavage activity, overnight cultures of WT and emrBΔ-bif RΔ mutant strains were grown to an OD600 of ∼2.0. Ten microliters of cells were spotted on preheated LB agar plates containing 0.1% (w/v) insoluble elastin (from bovine neck ligament; Sigma). Elastin cleavage activity (clearing zone on a cloudy background caused by insoluble elastin) was analyzed after incubation for ∼5 days.
Pellicle Formation and Biofilm Assay
To assess if biofilms (pellicles) formed at the air–liquid interface, overnight cultures were diluted 1:500 in 3 mL of LB medium. Conjugative plasmids were maintained using selective antibiotics. Culture tubes were kept stationary for 72–96 h at room temperature. Pellicles were assayed by visual inspection of the air–liquid interface of culture tubes. Quantification of biofilm (adherent to the surface of culture tubes) was performed as described.18 Briefly, crystal violet dye was used to stain the biofilm, and DMSO was used to dissolve the crystal violet-stained biofilm. To quantify the biofilm, absorbance was recorded at 560 nm. The data represent the mean (±SD) of three separate cultures. To compare growth of the stationary cultures, the absorbance of cells in liquid culture was measured, following which the pellicle was resuspended in the culture; the absorbance was again measured, and cells were plated for determination of CFU.
Preparation of BifR and Mutant Protein
The gene encoding BifR was amplified from B. thailandensis E264 genomic DNA using primers BifR_clon_Fw and BifR_clon_-Rev (Supplemental Table S1). The PCR product was digested with NdeI and EcoRI and cloned into pET28b, and the plasmid was verified by sequencing. To create the C104A substitution, an overhanging primer technique was used to amplify the whole plasmid using primers C104A Fw and Rev (Supplemental Table 1).19 The parental plasmid was digested using DpnI and mutant plasmid was gel purified and transformed into E. coli TOP 10 (Invitrogen) and verified by sequencing.
The resulting plasmids (WT and C104A) were transformed into E. coli BL21(DE3)pLysS. Cells were grown at 37 °C in LB media with 50 μg/mL kanamycin. An overnight culture was diluted 1:100 and grown to OD600 ≈ 0.6, and protein expression was induced by the addition of 1 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) for 1 h. Cells were pelleted at 4 °C and stored at −80 °C. The cell pellets were thawed for 1 h on ice and resuspended in chilled buffer containing 50 mM Tris-Cl (pH 7.0), 300 mM NaCl, 2 mM DTT. To 5 mL cell suspension, lysozyme (1.5 mg/mL), 50 μL of 10× DNase I buffer, and 2 μL of DNase I were added. The reaction was incubated for 2 h, and lysate was prepared by centrifugation at 10000g for 80 min at 4 °C. The supernatant was collected and mixed with HIS-Select Nickel Affinity beads (Sigma) previously equilibrated with wash buffer containing 10 mM imidazole for 1 h at 4 °C. After 1.5 h of incubation, the mixture was transferred to a gravity flow column, and protein was eluted with a gradient of imidazole from 10 mM to 250 mM. Peak fractions, which contained pure protein were pooled and buffer exchanged to wash buffer containing 10% glycerol. Protein was concentrated using Amicon centrifugal filter device (Millipore). The purity of protein was verified using Coomassie blue-stained SDS-PAGE gel. Concentration was calculated using the BCA protein assay kit (Pierce).
To assess oxidation by t-butyl hydroperoxide (tBHP), cumene hydroperoxide (CHP), hydrogen peroxide, and CuCl2, the protein was incubated with increasing concentration of oxidants in a total volume of 10 μL. Reactions were incubated for 15 min and terminated by adding Laemmli sample buffer without β-mercaptoethanol, and protein was subjected to SDS-PAGE.
To determine the effect of DNA binding on BifR oxidation, protein was titrated with increasing concentration of 57 bp DNA (10–60 μM DNA corresponding to stoichiometric conditions; [DNA] ≫ Kd). Reactions were incubated for 30 min and then supplemented with 500 μM CuCl2. After 15 min, reactions were terminated by addition of Laemmli sample buffer without β-mercaptoethanol, and protein was subjected to SDS-PAGE.
For oligomeric state determination, the protein was cross-linked with 0.5% glutaraldehyde in a total volume of 10 μL. The protein was incubated for 30 min, and an equal volume of Laemmli sample buffer was added to terminate the reaction. The cross-linked proteins were subjected to SDS-PAGE.
Size Exclusion Chromatography
To determine oligomeric state of BifR, a Superdex 75 5/150 GL column with mobile phase buffer consisting of 50 mM Tris (pH 7.0) and 150 mM NaCl was used, and the column was calibrated with markers carbonic anhydrase (29 kDa), bovine serum albumin (66 kDa), alcohol dehydrogenase (150 kDa), beta amylase (200 kDa), and blue dextran (2000 kDa) (Sigma). A standard curve was obtained as a plot of Ve/Vo as a function of the log10 of molecular weight (where Ve and Vo represent the retention volume of the protein and void volume of the column, respectively).20 Reduced protein (with DTT) or protein oxidized with CuCl2 was run on the column.
Western Blot
To determine if oxidation of BifR occurs in vivo, Western Blots were performed. E. coli BL21(DE3)pLysS cells harboring plasmids encoding BifR or BifR-C104A were grown at 37 °C to OD600 of 0.6. Protein expression was induced by addition of 1 mM IPTG. After 30 min, 1 mM H2O2 was added and cells were incubated an additional 30 min; control cultures received no H2O2. Cells were harvested and stored at −80 °C. The cell pellets were thawed and resuspended in 50 mM sodium phosphate buffer (pH 7.0) with 300 mM NaCl. To 5 mL of cell suspension, lysozyme (1.5 mg/mL), 50 μL 10× DNase I buffer, and 2 μL of DNase I were added. Reactions were incubated for 2 h for lysis. Supernatants were collected after centrifugation at 10000g for 1 h. Fifteen microliters of supernatant was resolved on 18% SDS-PAGE gels, and the resolved proteins were transferred to polyvinylidene fluoride membrane. Anti-his-tag mAb (A00088; GenScript) was added at a dilution of 1:3000, followed by incubation with secondary antibody (goat antimouse IgG (H+L)–HRP conjugate, 172–1011, BioRad) used at a dilution of 1:5000. The blots were developed using Opti-4CN substrate kit (BioRad).
DNA Binding Assays
The apparent dissociation constant Kd was determined using electrophoretic mobility shift assays (EMSA). Synthetic oligonucleotides representing 57 bp emrB-bif R promoter region with the two identified palindromes at the center were purchased and purified by denaturing polyacrylamide gel electrophoresis. The top strand of the operator region was radiolabeled using γ-32P-ATP and T4 polynucleotide kinase. To form duplex DNA, the top strand was annealed with the complementary strand by heating at 90 °C, followed by slow cooling. The 355 bp operator DNA was amplified using primers 355 Fw and Rev (Supplemental Table 1) and radiolabeled using γ-32P-ATP and T4 polynucleotide kinase. DNA (0.8 nM) and protein were mixed in binding buffer (0.5 M Tris-HCl pH 8.0, 250 mM NaCl, 0.1 M EDTA, 0.1 mM dithiothreitol, 0.05% Brij58, 10 μg/mL BSA, and 5% glycerol) and incubated at room temperature for 30 min. Before loading the reaction mixture, 10% native polyacrylamide gels (39:1 acrylamide/bis-acrylamide) were prerun for 30 min in 0.5× Tris borate EDTA (45 mM Tris borate (pH 8.3) and 1 mM Na2EDTA) at room temperature. After electrophoresis, gels were dried, and free DNA and protein–DNA complexes were visualized using a Storm 840 phosphorimager (GE Healthcare). Densitometric data were obtained with Image-Quant 5.1 and analyzed using KaleidaGraph 4.0 (Synergy Software). The data were fitted using the equation (where nH is the Hill coefficient, Kd is the apparent equilibrium dissociation constant approximating half-maximal saturation of the DNA (not the affinity for a single site), and [X] is the protein concentration). Data are reported as mean ± SD of three experiments.
To determine the specificity of protein–DNA complex, protein mixed with labeled DNA was challenged with increasing concentrations of unlabeled 355 bp operator DNA. Protein with labeled DNA was also titrated with increasing concentrations of nonspecific pET28b DNA. Reactions were incubated at room temperature for 30 min and processed as described above.
To assess the effect of oxidant on protein–DNA binding, protein was mixed with CuCl2 in a ratio of 1:50 and incubated on ice for 15 min. EMSA was performed with the oxidized protein, and the apparent dissociation constant was determined as described above.
To determine the effect of potential inducer on DNA binding, DNA (0.8 nM) and 1 nM BifR were mixed in binding buffer (50 mM Tris-HCl pH 8.0, 50 mM NaCl, 0.1 M EDTA, 0.1 mM dithiothreitol, 0.05% Brij58, 10 μg/mL BSA, and 5% glycerol) to which increasing concentration of salicylate was added. Reactions were incubated for 30 min and electrophoresed as described above.
RESULTS
BifR Regulates Expression of Divergently Oriented Genes ecsC and emrB-bif R
The B. thailandensis E264 genomic locus BTH_I0541–BTH_I0542 (new locus tag BTH_RS14955-RS14960) is annotated as an operon encoding a predicted EmrB efflux pump and a member of the MarR family of transcription factors. It is divergently oriented to BTH_I0540, (new locus tag BTH_RS14950) annotated as encoding EcsC protein (Figure 1A). Many EcsC homologues are annotated as putative LasA protease (Ensembl genomes family: 215938 protease domain). In Pseudomonas, disruption of quorum sensing has been correlated with reduced expression of virulence genes, including LasA protease.21 LasA is a secreted zinc-binding metalloprotease with restricted specificity that possesses elastolytic and staphylolytic activity.22,23 The short 106 bp intergenic region between start codons of ecsC and emrB suggests that promoters for the divergent genes may be overlapping. The entire locus is conserved in other Burkholderia species, including the pathogenic species, and Ortholuge predicts that the encoded proteins are orthologs.24
Figure 1.
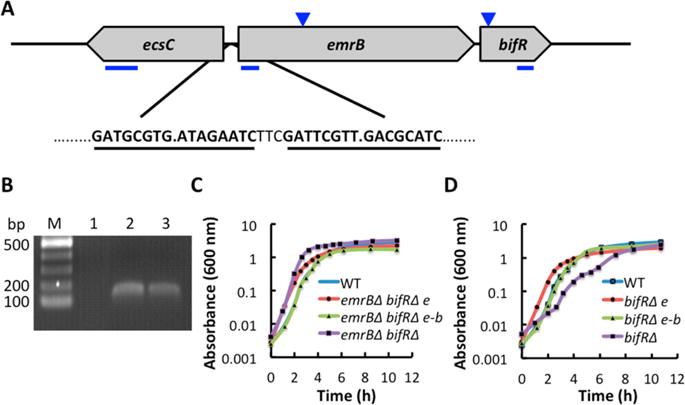
B. thailandensis-encoded emrB-bif R operon. (A) The emrB-bif R operon (BTH_I0541–0542/BTH_RS14955–14960) is oriented divergently to a gene annotated as ecsC (BTH_I0540/BTH_RS14950). Two imperfect palindromes in the intergenic region are shown bold and underlined. Blue lines beneath the arrows depicting open reading frames represent the positions of PCR amplicons used for qRT-PCR. Inverted triangles indicate positions of transposon insertion. (B) Verification of emrB-bif R operon by PCR amplification of emrB fragment. Lane 1, negative control using RNA as template; lane 2, positive control using genomic DNA as template; lane 3, cDNA synthesized using bif R-specific primer as template. (C) Growth curve of wild-type, emrBΔ-bif RΔ and the corresponding mutant strains complemented with emrB (e) or emrB-bif R (e-b). (D) Growth curve of wild-type, bif RΔ, and the corresponding complemented strains; growth curves are representative of triplicate cultures. The doubling time for WT was ∼25 min, compared to ∼20 min for emrBΔ-bif RΔ and ∼55 min for bif RΔ.
On the basis of the role of the BTH_I0542-encoded MarR homologue in regulation of biofilm formation detailed below, we propose the name BifR. Inspection of the intergenic region between ecsC and emrB-bif R revealed two conserved, imperfect palindromes consisting of 8 bp half-sites, with the two palindromes separated by 3 bp (Figure 1A). These palindromes are putative binding sites for BifR.25,26 Predicted promoter elements overlap with the identified palindromes, suggesting that BifR will repress the divergently oriented genes (Supplemental Figure S1). To confirm the predicted emrB-bif R operon, cDNA was prepared using a primer specific to the bif R open reading frame, followed by PCR amplification using emrB-specific primers. As shown in Figure 1B, a product of the expected size was obtained, confirming the annotated operon.
We obtained and verified disruptant strains in which a transposon was inserted at position 15 of bif R or at position 439 of the emrB open reading frames, respectively.14 Since bif R and emrB are encoded as part of an operon, transposon insertion in emrB is polar and is expected to interfere with bif R expression as well; the corresponding disruptant strain is therefore referred to as emrBΔ-bif RΔ. In contrast, disruption of bif R (bif RΔ) is not expected to prevent basal emrB expression.
The emrBΔ-bif RΔ strain was seen to exhibit a modestly increased growth rate (Figure 1C). Complementation with emrB only was not sufficient to restore the growth rate to wild-type levels, whereas complementation with emrB-bif R resulted in a growth rate equivalent to that of wild-type (Figure 1C). By contrast, inactivation of bif R led to a reduced growth rate; complementation with emrB resulted in a growth rate higher than that observed for wild-type cells (consistent with the elevated growth rate when emrB was expressed in emrBΔ-bif RΔ), while complementation with emrB-bif R restored the growth rate to that of wild-type cells (Figure 1D).
The transcript levels of ecsC, emrB, and bif R were determined in the wild type strain (Figure 2A). Transcript levels of ecsC, emrB, and bif R were 0.2 ± 0.01, 0.3 ± 0.02, 0.2 ± 0.01, respectively, with relative levels of emrB and bif R reflecting the expected reduction in transcript yield for the second gene in the operon.27 To assess the predicted regulation by BifR, expression of ecsC and emrB was analyzed in the bif RΔ strain. The relative transcript level in the bif RΔ strain was higher for both ecsC (10.5 ± 0.5) and emrB (16.1 ± 3.6; Figure 2A), reflecting an ∼50-fold increased expression relative to wild-type. This shows that BifR functioned as a repressor of the divergent genes. As expected from the polar transposon insertion in emrB, little expression of bif R was detected in emrBΔ-bif RΔ compared to wild-type (bif R transcript level 0.1 ± 0.02). Complementation of bif RΔ with plasmid-encoded emrB-bif R resulted in marked repression of ecsC and emrB (Figure 2A), verifying the role of BifR as a repressor.
Figure 2.
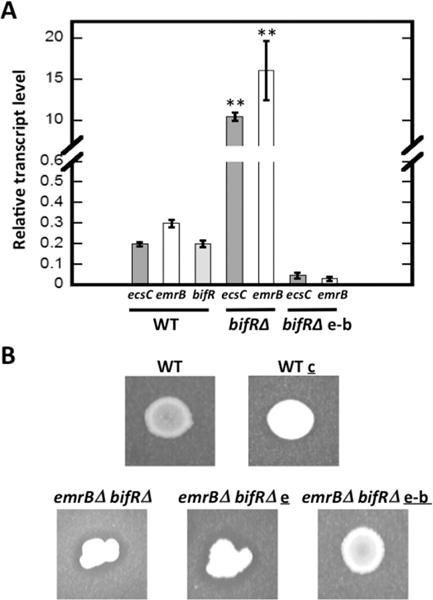
Regulation of gene expression. (A) Relative transcript level of ecsC, emrB, and bif R in wild type (WT) strain and ecsC and emrB in bif RΔ and bif RΔ e-b strain (complemented with emrB-bif R) calculated using relative to reference gene encoding glutamate synthase large subunit (BTH_I3014/BTH_RS27550). Error bars represent standard deviation of three separate experiments (each with three technical replicates). Asterisks represent statistical significance from WT based on Student’s t test (p < 0.001). (B) Elastin cleavage activity in WT and emrBΔ-bif RΔ strains and emrBΔ-bif RΔ complemented with emrB (emrBΔ-bif RΔ e) or with emrb-bif R (emrBΔ-bif RΔ e-b); WT c carries vector without genes inserted. Plates contain insoluble elastin and clearing zone reflects elastin cleavage.
To assess if upregulation of ecsC led to increased production of a functional LasA protease, we inoculated plates containing insoluble elastin with wild-type and emrBΔ-bif RΔ cultures. A comparison showed a distinct clearing zone only around emrBΔ-bif RΔ (Figure 2B). Restoration of the wild-type phenotype required complementation with bif R (strain emrBΔ-bif RΔ e-b), consistent with repression of ecsC by BifR. This indicates that the ecsC gene encodes a functional LasA protease and that its upregulation in the emrBΔ-bif RΔ strain leads to significantly increased extracellular elastase activity.
Oxidized BifR Forms a Dimer of Dimers
Having verified the role of BifR in repressing expression of ecsC and emrB-bif R, we examined its predicted properties in vitro. BifR (Mw ≈ 18 kDa) shares 28% sequence identity with P. aeruginosa-encoded MarR family regulator pa3341 (2fbh), and BifR was modeled using SWISS-MODEL and 2fbh as template. The model reflects the obligate MarR dimer with DNA recognition helices (Figure 3A; green) positioned for interaction in consecutive DNA major grooves; all MarR homologues share this general fold, suggesting that the model is a reasonable approximation of the BifR structure despite the relatively low sequence conservation.25,26 Each BifR monomer has a single Cys residue located in the middle of helix 4 that connects the helix-turn-helix DNA-binding domain to the dimerization region, placing the two cysteines far apart in the dimer and on opposite faces of the dimer when viewed with the DNA-recognition helices at the bottom (arrows; Figure 3A). B. thailandensis bif R was cloned, expressed in E. coli, and purified to apparent homogeneity (Figure 3B). BifR was quite stable, unfolding in a one-step melting transition with Tm = 66.1 °C (Supplemental Figure S2). Cross-linking of BifR with glutaraldehyde (which cross-links lysine residues) resulted in formation of a cross-linked species with Mw corresponding to a dimer, suggesting that reduced BifR exists as a dimer (Figure 3B, lane 2).
Figure 3.
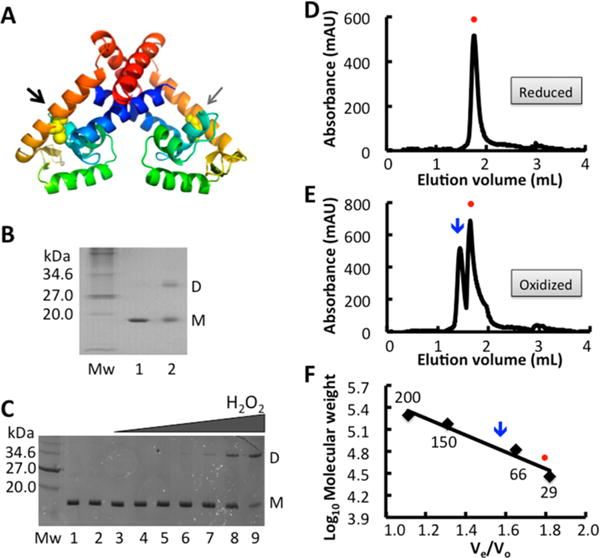
Dimer formation by BifR. (A) BifR model based on the structure of MarR family regulator pa3341 (2fbh). Model was created using SwissModel in automated mode; 2fbh was selected as the template with the greatest sequence similarity to BifR, and the generated model had a Global Model Quality Estimation (GMQE) score of 0.63. Both monomers are colored blue to red (amino-terminus to carboxy-terminus). The cysteines in each monomer are in yellow sphere representation, with the cysteine in the left monomer toward the front of the protein (black arrow) and the cysteine in the right monomer facing the back (gray arrow). (B) Purified BifR electrophoresed on an 18% SDS-PAGE gel. Mw, molecular weight marker (kDa); lane 1 BifR; lane 2, BifR cross-linked with 0.5% (v/v) glutaraldehyde. Monomer (M) and dimer (D) identified at the right. (C) BifR titrated with increasing concentration of hydrogen peroxide. Lane Mw is molecular weight marker (kDa); lane 1, air oxidized protein, lane 2, reduced protein; lanes 3–9, BifR with hydrogen peroxide (10 μM to 2 mM). Monomer (M) and dimer (D) identified at the right. (D) Elution of reduced BifR from size exclusion column. (E) Size-exclusion chromatogram of BifR previously incubated with CuCl2, resulting in a mixture of reduced (dimer; circle) and oxidized (dimer-of-dimers; arrow) species. (F) Elution of reduced and oxidized BifR from size exclusion column indicated by circle and arrow, respectively. Mw of standards are indicated (in kDa). The standard curve was generated using Ve/Vo as the function of log10 of molecular weight of these standards.
To determine if protein oxidation alters physicochemical properties, reduced BifR was incubated with increasing concentrations of H2O2. BifR was oxidized as evidenced by appearance of a band near the Mw of a dimer in the SDS-PAGE gel (Figure 3C). BifR was also oxidized with organic oxidants tert-butyl hydroperoxide (tBHP) and cumene hydroperoxide (CHP; data not shown). That the dimeric species observed by protein oxidation migrated slightly slower than dimer obtained by glutaraldehyde cross-linking suggests a more extended conformation. On the basis of the location of cysteines in dimeric BifR, we infer that oxidation resulted in formation of a trans-dimer between two monomers in separate BifR cis-dimers.
Size exclusion chromatography was used to determine the oligomeric state of reduced and oxidized BifR. Reduced protein eluted as a single species with Mw 35.2 kDa (theoretical molecular weight of dimeric BifR is 36.4 kDa; Figure 3D). Oxidized protein eluted as two separate species, one with Mw ≈ 69.5 kDa (theoretical molecular weight of BifR dimer-of-dimers is 72.8 kDa) and another with Mw ≈ 35.2 kDa (Figure 3E,F). A significant presence of higher-order oligomers was not detected. This is consistent with SDS-PAGE results and suggests that BifR exists as a dimer in the reduced form and as a dimer-of-dimers when oxidized.
Cu2+-catalyzed oxidation of BifR also resulted in dimer formation (Figure 4A), and it was found to be more efficient compared to other oxidants (tBHP, CHP, and H2O2). To assess the inference that dimer formation was due to disulfide bond formation, a mutant was created in which Cys was replaced with Ala (BifR-C104A), and the protein was purified to apparent homogeneity. Addition of Cu2+ did not result in any dimer formation (Figure 4B). To determine if BifR forms a dimer-of-dimers in vivo, expression of the His6-tagged protein was induced in E. coli (under aerobic conditions) and BifR was detected by Western blotting using antibody to the tag. As shown in Figure 4D, BifR dimers were readily detected, whereas no dimerization of BifR-C104A was observed.
Figure 4.
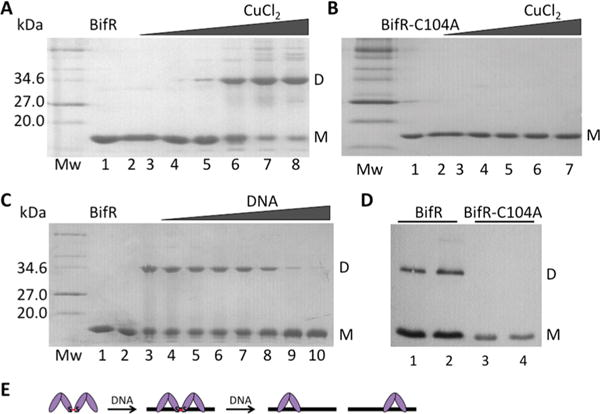
BifR oxidation by copper. In panels (A–C), left lane is protein marker (Mw; in kDa), lane 1 is air-oxidized protein, and lane 2 is reduced protein. (A) BifR with increasing concentration of CuCl2 (lanes 3–8; 5 μM to 2.5 mM). (B) BifR-C104A with increasing concentration of CuCl2 (lanes 3–7; 5 μM to 1 mM). (C) Lane 3, BifR oxidized with 500 μM CuCl2; lanes 4–10, BifR with increasing concentration of operator DNA (10–60 μM) followed by addition of 500 μM CuCl2. (D) Western blot of lysate from E. coli BL21(DE3)-pLysS using antibody to His6-tagged BifR (lanes 1 and 2) and BifR-C104A (lanes 3 and 4); lanes 2 and 4 correspond to cultures grown in the presence of 1 mM H2O2 for 30 min. Monomer (M) migrates with a Mw ≈ 18 kDa and dimer (D) migrates with a Mw ≈ 36 kDa; monomer and dimer is indicated to the right of each image. (E) Interpretation of data from panel (C) in which absence of cross-linked species requires a stoichiometric excess of DNA.
As noted above (Figure 1A), the ecsC-emrB intergenic region contains two side-by-side palindromes that might serve as binding sites for BifR. To determine if DNA-binding affects association of BifR dimers, BifR was titrated with 57 bp DNA containing both palindromes followed by oxidation with Cu2+. At a DNA concentration equivalent to that of BifR (10 μM DNA; this corresponds to stoichiometric conditions under which all BifR will bind cognate DNA based on the nM affinity of BifR, as discussed below), no change in dimer formation was observed (compare lanes 3 and 4). In contrast, increasing the DNA concentrations above that of BifR resulted in a decrease in dimer formation when DNA was in stoichiometric excess over BifR (Figure 4C, lanes 8–10). These data suggest that two BifR dimers can associate in the absence of DNA (as also seen by size exclusion chromatography, Figure 3E) and that they can bind side-by-side to the identified palindromes, an arrangement in which oxidation still leads to disulfide bond formation between trans-dimers. Addition of excess DNA likely results in each BifR dimer binding to separate DNA molecules, reducing formation of BifR dimer-of-dimers and precluding disulfide bond formation.
The Cu2+-catalyzed protein oxidation suggested possible metal binding by BifR. Since metal-binding has the potential to affect protein stability, the thermal stability of BifR was determined using differential scanning fluorometry.20 Compared to reduced BifR, copper(II)-oxidized BifR had a lower melting temperature (Tm = 54.4 °C, Supplemental Figure S2B and Table 1). Other oxidants (tBHP, CHP, H2O2) had little effect on protein stability, likely reflecting the more efficient oxidation by copper(II) compared to the other oxidants. Zinc(II)-binding modestly reduced protein stability (Tm = 61.1 °C). Addition of magnesium(II) to BifR had no effect on protein stability (Tm = 66.2 °C). Metal-binding to BifR was further confirmed using the metal chelator 4-(2-pyridylazo) resorcinol (PAR) that forms complex with various divalent metals, including Zn2+ (Supplemental Figure S2E).20 These data verified binding of divalent metal ions (Zn2+) to BifR. While BifR-C104A was slightly more stable than wild-type BifR, protein stability was not altered by Cu2+, consistent with Cys-oxidation by Cu2+.
Table 1.
Thermal Stability of BifR and BifR-C104Aa
| BifR Tm (°C) | BifR-C104A Tm (°C) | |
|---|---|---|
| BifR | 66.1 ± 0.2 | 68.9 ± 0.1 |
| tBHP (1:100) | 64.0 ± 0.3 | nd |
| CHP (1:100) | 65.6 ± 1.2 | nd |
| H2O2 (1:100) | 64.1 ± 0.1 | nd |
| ZnCl2 | 61.1 ± 0.9 | 71.4 ± 0.1 |
| CuCl2 | 54.4 ± 0.1 | 67.9 ± 0.1 |
| MgCl2 | 66.2 ± 0.2 | nd |
nd, not determined. Mean ± SD of three replicates.
BifR Binds Specifically to the Intergenic Region between ecsC and emrB-bif R
To determine BifR binding to the intergenic region between ecsC and emrB-bif R, a 355 bp DNA was used in electrophoretic mobility shift assay (EMSA). BifR bound this DNA, forming two complexes C1 and C2 (Figure 5A) with an apparent dissociation constant (Kd) of 1.4 ± 0.1 nM and Hill coefficient (nH) 1.1 ± 0.02 under reducing conditions (Table 2). Complex C1 was faint but detectable at lower protein concentrations, and the predominant complex was the slower migrating C2. Considering the ability of BifR to form dimer-of-dimers and the presence of side-by-side palindromes, we speculate that C2 corresponds to binding of two BifR dimers. At higher protein concentrations, complexes migrated slightly slower, perhaps reflecting nonspecific binding. Oxidation of BifR prior to incubation with DNA did not change the DNA binding as reflected in Kd = 0.9 ± 0.03 nM and nH = 1.1 ± 0.1. BifR-C104A had modestly reduced DNA binding with Kd = 6.8 ± 1.6 nM and nH = 0.9 ± 0.1 (Figure 5B,D). Since C104A is located at a position that is not predicted to contact DNA directly, it is more likely that the mutation led to a structural rearrangement in the protein that is communicated to the DNA binding lobes, a reasonable inference given the position of C104 in helix 4 that connects the DNA-binding and dimerization regions of the protein.
Figure 5.
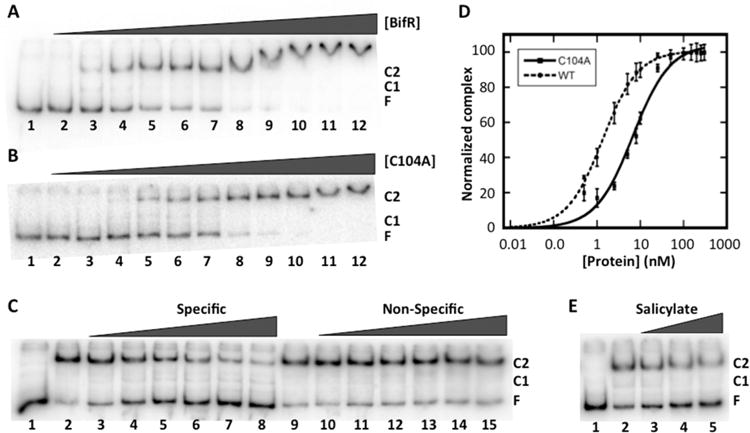
BifR binds specifically to the emrB-bif R promoter DNA. (A) EMSA showing 355 bp operator DNA (0.8 nM) titrated with increasing concentration of BifR (lane 2, 0.5 nM; lane 3, 1 nM; lane 4, 2.5 nM; lane 5, 5 nM; lane 6, 8 nM; lane 7, 10 nM; lane 8, 25 nM; lane 9, 50 nM; lane 10, 100 nM; lane 11, 150 nM; lane 12, 250 nM); reaction in lane 1 contains DNA only. Free DNA is identified as F and complexes are identified as C1 and C2 at the right. (B) EMSA showing 355 bp operator DNA (0.8 nM) titrated with increasing concentration of BifR-C104A (lane 2, 0.5 nM; lane 3, 1 nM; lane 4, 2.5 nM; lane 5, 5 nM; lane 6, 8 nM; lane 7, 10 nM; lane 8, 25 nM; lane 9, 50 nM; lane 10, 100 nM; lane 11, 150 nM; lane 12, 250 nM). (C) BifR (1.3 nM) bound to labeled operator DNA challenged with unlabeled 355 bp operator DNA (0.8–45 nM, lanes 3–8) or equivalent concentration of nonspecific DNA pET28b (lanes 10–15). Reaction in lane 1 contains free DNA; reactions in lanes 2 and 9 contain no competitor DNA. (D) Fractional complex plotted as a function of BifR (●, dashed line) and BifR-C104A (■, solid line) concentration. Error bars represent the standard deviation of three independent repeats. (E) BifR-DNA complexes titrated with increasing concentration of salicylate (1–3 mM; lanes 3–5). Reaction in lane 2 contains no inducer. Reaction in lane 1 contains DNA only.
Table 2.
Apparent Dissociation Constant of BifRa
| conditions | Kd (nM) | nH | |
|---|---|---|---|
| 355 bp | reduced | 1.4 ± 0.1 | 1.1 ± 0.01 |
| oxidized | 0.9 ± 0.03 | 1.1 ± 0.1 | |
| 57 bp | reduced | 10.8 ± 1.8 | 0.7 ± 0.1 |
| oxidized | 4.2 ± 0.4 | 0.5 ± 0.1 | |
| zinc-bound | 10.1 ± 2.5 | 0.8 ± 0.1 |
Data represent mean ± SD from three experiments.
Using a shorter 57 bp DNA containing the two identified palindromes, the apparent dissociation constant for BifR binding (∼10 nM; Table 2) was higher than that determined for binding to 355 bp DNA, perhaps because the macroscopic binding constant for binding to the latter DNA construct included a contribution from the additional complex C3 observed at higher protein concentrations. A 2-fold increase in the binding affinity was observed on binding of oxidized compared to reduced BifR to 57 bp DNA, whereas zinc binding had no effect on DNA binding affinity (Table 2). Mg2+-binding to BifR also did not change the DNA binding (data not shown).
BifR binding to the intergenic DNA was specific; addition of unlabeled intergenic DNA effectively competed for BifR binding (Figure 5C, lanes 3–8, whereas addition of excess nonspecific DNA (Figure 5C, lanes 10–15) did not reduce complex formation.
DNA binding by MarR family proteins is often attenuated by ligand binding, an event that is associated with induction of gene expression.26,28 For clues to the BifR ligand, we used RaptorX-Binding to predict ligand-binding sites of BifR based on the predicted three-dimensional model created by RaptorX.29 Modeling predicts that BifR can bind to salicylate, a ligand previously shown to bind several MarR homologues.30–32 To determine the ability of salicylate to bind BifR and cause attenuated DNA binding, increasing concentration of salicylate was added to reactions containing 0.8 nM DNA and 1.0 nM BifR. While salicylate did reduce DNA binding, the effect was only observed at millimolar concentration, suggesting that a structurally related compound could be a natural inducer (Figure 5E).
Expression of emrB and bif R Is Further Repressed under Oxidizing Conditions
Considering the formation of a covalently linked dimer-of-dimers on oxidation of BifR, we determined the mRNA level of emrB and bif R in cells grown in the presence of hydrogen peroxide or Cu2+. Addition of 1 mM H2O2 resulted in an ∼2.5-fold repression of emrB (relative transcript level 0.4 ± 0.01) and ∼10-fold repression of bif R (relative transcript level 0.1 ± 0.03) compared to unsupplemented cultures (Figure 6A). Growth in the presence of 1 mM Cu2+ likewise resulted in repression of both emrB (0.6 ± 0.02) and bif R (0.1 ± 0.04). This would be consistent with oxidized BifR being a more efficient repressor of emrB and bif R in vivo. Addition of 1 mM ZnCl2 and 30 mM MgCl2 had no effect on expression of emrB (1.0 ± 0.2 and 0.8 ± 0.1, respectively) and bif R (0.9 ± 0.2 and 1.4 ± 0.4, respectively) (Figure 6A). This is consistent with the observation that Zn2+ and Mg2+ had no effect on DNA binding by BifR, and it suggests that Cu2+ is the physiologically relevant BifR-binding metal ion.
Figure 6.
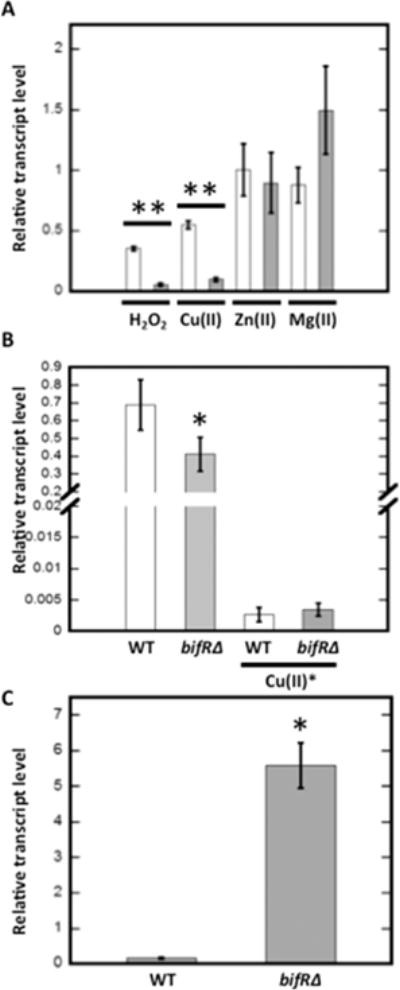
Regulation of gene expression by metals and oxidants. (A) Relative abundance of transcript level of emrB (open bars) and bif R (gray bars) after addition of 1 mM H2O2, 1 mM CuCl2, 1 mM ZnCl2, or 30 mM MgCl2. The transcript level was calculated using relative to the reference gene glutamate synthase large subunit, and the comparative CT method was used to calculate relative abundance with reference to the transcript level of unsupplemented control samples. (B) Relative transcript level of Nudix pyrophosphatase (BTH_I2657/BTH_RS25735) in WT and bif RΔ strains calculated using relative to the reference gene glutamate synthase large subunit. The relative abundance of transcript level of the gene encoding Nudix pyrophosphatase in 1 mM CuCl2 treated WT and bif RΔ strains was calculated using relative to the reference gene glutamate synthase large subunit and the comparative CT method was used to calculate relative abundance with reference to the transcript level of unsupplemented control samples (in samples marked by asterisk). (C) Relative transcript level of PCA biosynthetic gene phzF in WT and bif RΔ strain calculated using relative to reference gene encoding glutamate synthase large subunit. Error bars represent standard deviation of three experiments. Asterisks represent statistical significance from unsupplemented WT based on Student’s t test (*, p < 0.05 and **, p < 0.001 for both emrB and bif R expression in the presence of oxidants).
BifR repressed emrB-bif R, and this repression was enhanced under oxidizing conditions when BifR forms a covalently linked dimer-of-dimers, indicating that repression by BifR is modulated by cellular redox state. Querying the B. thailandensis genome sequence with the BifR consensus sequence (GATg/tCGTNNa/tc/aGc/aATC) using Pattern Locator33 yielded a number of potential BifR sites. However, only one instance of two adjacent sites was observed in addition to the site in the emrB-bif R promoter. Those potential adjacent BifR sites were in the promoter of BTH_I2657, which encodes a Nudix pyrophosphatase, an enzyme that is predicted to regulate NAD+/NADH ratios. Measurement of relative mRNA levels in wild-type and bif RΔ strains showed modestly reduced expression (1.8-fold) of the gene encoding Nudix in the bif RΔ strain, suggesting that BifR may function as an activator (Figure 6B). The location of the predicted BifR sites ∼200 bp upstream of the annotated start codon is consistent with this inference. While nudix expression was barely detectable in cells grown in the presence of Cu2+, this repression was independent of BifR.
Focusing on genes encoding proteins with a predicted link to cellular redox state, we noted a single palindrome modestly resembling the BifR consensus sequence in the promoter driving expression of the phz operon BTH_I0953–I0949. This operon is predicted to encode proteins responsible for synthesis of phenazine-1-carboxylic acid (PCA). The transcript level of phzF (BTH_I0949) was analyzed in wild-type and bif RΔ cells. Relative transcript levels of phzF in wild-type and bif RΔ strains were 0.2 ± 0.02 and 5.6 ± 0.6, respectively, reflecting an ∼28-fold upregulation in bif RΔ (Figure 6C).
BifR Represses Biofilm Formation
The BifR-mediated repression of the phz operon suggested its potential role in control of biofilm growth. As shown in Figure 7, both emrBΔ-bif RΔ and bif RΔ strains exhibited increased biofilm formation. Wild-type and mutant strains were kept stationary for 48–96 h, at which time a pellicle had formed at the air–liquid interface in both mutant strains (Figure 7A). Quantitation of biofilm formation by crystal violet staining showed that both mutant strains emrBΔ-bif RΔ (1.8 ± 0.2) and bif RΔ (1.6 ± 0.1) produced significantly more biofilm compared to wild-type (0.8 ± 0.1) (Figure 7B; each bar corresponds to the strain used for the pellicle picture directly above it). The increased biofilm phenotype was restored to wild-type levels in both mutants when complemented with the entire emrB-bif R locus (emrBΔ-bif RΔ/e-b (0.7 ± 0.1) and bif RΔ/e-b (0.6 ± 0.2)). Since emrB is disrupted in emrBΔ-bif RΔ but highly expressed in bif RΔ, we infer that regulation by BifR of genes other than emrB is responsible for the increased biofilm phenotype. This interpretation was further confirmed by the observation that complementation with emrB only was insufficient to restore the biofilm formation to WT levels in emrBΔ-bif RΔ and bif RΔ strains; crystal violet quantitation of biofilm formation in emrBΔ-bif RΔ/e and bif RΔ/e showed absorbances of 1.4 ± 0.2 and 1.1 ± 0.1, respectively. A measurement of growth in the stationary cultures by determination of absorbance or CFU confirmed the modestly increased growth rate of emrBΔ-bif RΔ compared to wild-type (Figure 1C and Supplemental Figures S5 and S6), suggesting that the increased cell count may contribute to increased biofilm formation in this strain.
Figure 7.
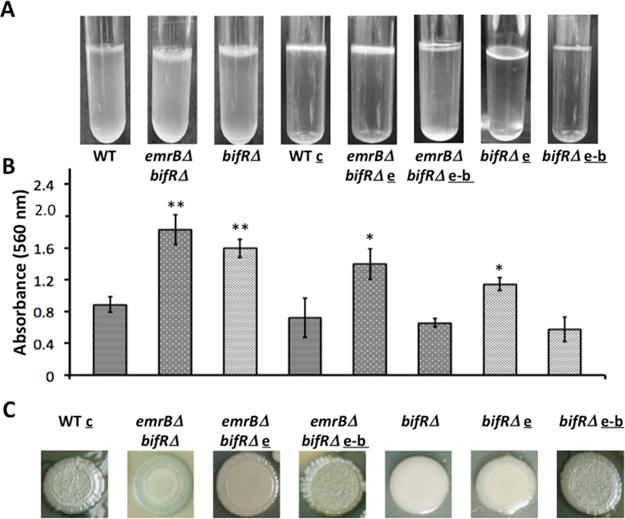
Deletion of bif R leads to increased biofilm formation. (A) Pellicle formation in static cultures. (B) Quantitation of biofilm using crystal violet staining; corresponding pictures of pellicle formation and strain identification are directly above each bar. Error bars represent standard deviation from three separate cultures. (C) Colony morphology of WT, bif RΔ, emrBΔ-bif RΔ, and corresponding complemented strains. WT and mutant strains were complemented with empty pBBR-MCS5 (denoted c, for WT); e, pBBR-MCS5 encoding emrB; e-b, pBBR-MCS5 encoding emrB-bif R. Asterisks represent statistical significance from WT based on Student’s t test (**, p < 0.001 and *, p < 0.05).
Colony morphology of the mutant strains was also examined. Wild-type cells formed rugose colonies; as noted above, such wrinkling has for example been attributed to the need for extended surface area to enhance access to molecular oxygen and to maintain cellular redox homeostasis.11 By contrast, bif RΔ cells formed completely smooth colonies (Figure 7C). This colony morphology was not changed on complementation with emrB, only complementation with emrB-bif R restored the rugose morphology (bif RΔ/e-b). The observed change in colony morphology (particularly for the bif RΔ mutant for which the growth rate is slower than WT (Figure 1D)) points to altered production of matrix components. Phenazines are synthesized from chorismic acid via anthranilate, which in B. pseudomallei is linked to regulation of biofilm formation; anthranilate is structurally similar to salicylate, which modestly attenuated DNA binding by BifR (Figure 5C).34 However, DNA binding by BifR was not attenuated by anthranilate (data not shown).
DISCUSSION
DNA Binding and Oligomerization by BifR
Two adjacent palindromes are present in the emrB-bif R promoter, predicting side-by-side binding of two BifR dimers (Figure 8). This inference is supported by preferred formation of the slower migrating Complex 2 in EMSA (Figure 5) and with the ability of excess DNA to prevent formation of disulfide bonds between two BifR dimers (Figure 4C). The distance between the centers of these palindromes is 19 bp, corresponding to ∼1.8 turns or 63 Å assuming B-form DNA. This distance corresponds well to the width of MarR proteins,31 suggesting that two BifR dimers bound at adjacent palindromes would indeed make contact, as evidenced by disulfide bond formation between adjacent DNA-bound BifR dimers. The 1.8 helical turns between centers of palindromes in B-form DNA also predict that two BifR dimers are slightly offset relative to each other to center on cognate sites that do not exactly line up on the same face of the helix; alternatively a DNA distortion may be induced on protein binding that alters the relative positions of cognate sites. Considering that the two cysteine residues in a BifR dimer face the “front” and “back” of the dimer, respectively (Figure 3A), the two dimers would be expected to be offset relative to each other to accommodate a disulfide bond between trans-dimers (Supplemental Figure S4). This organization of the BifR dimer-of-dimers would be consistent with the position of cognate DNA sites.
Figure 8.
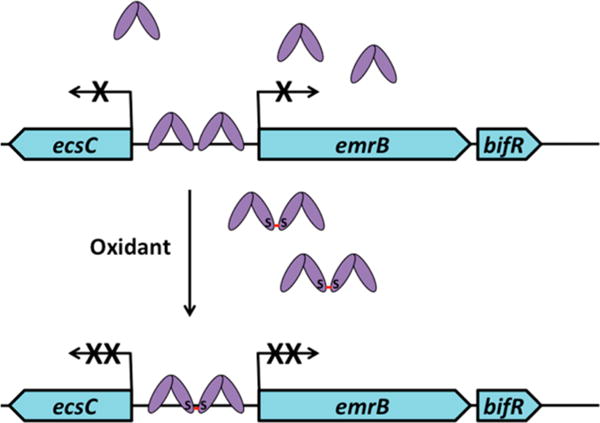
Model of BifR-mediated gene regulation. BifR (purple) exists as a dimer under reducing conditions. Two BifR dimers bind adjacent palindromes. Upon oxidation, a disulfide bond links two BifR dimers; in this condition, repression of gene activity is more efficient, perhaps because RNA polymerase fails to displace promoter-bound BifR.
BifR forms a homodimer, yet oxidation did not result in formation of oligomeric assemblies greater than a dimer-of-dimers (Figure 3E). In other MarR homologues, negative cooperativity of ligand-binding has been reported in which occupancy of one ligand-binding pocket is communicated via the dimer interface to reduce the affinity of ligand for the second site.28,35–37 Similarly, we speculate that disulfide-bond formation involving the cysteine in one BifR monomer results in conformational changes that are communicated to the second monomer, disfavoring its interaction with another BifR dimer, thereby limiting the oligomeric assembly. This may also explain why complete conversion of BifR to dimeric species was not observed, even with a large excess of oxidant (Figures 3C and 4A).
According to EMSA, the binding mode and affinity for the adjacent sites in the emrB-bif R promoter are comparable for reduced and oxidized protein. That the mutant BifR-C104A binds with reduced affinity speaks to the ability of changes in helix 4 to result in altered DNA binding. Reduced and oxidized BifR may likewise exhibit differences in DNA binding that are not distinguishable by EMSA, but are significant in terms of control of gene activity. Such changes in DNA binding mode have been previously reported; for example, the Bacillus subtilis-encoded redox-sensitive MarR homologue HypR binds to DNA with the same affinity in oxidized and reduced conditions, yet only oxidized HypR functions to activate gene expression.38 Similarly, the MarR homologue PecS from the plant pathogen Pectobacterium atrosepticum binds DNA with equivalent affinity when increasing pH from 7.4 to 8.3, but it represses gene expression only at the higher pH, a difference attributed to PecS modifying promoter DNA topology under repressive conditions.39
The most effective BifR oxidant is Cu2+ (Figure 4). Addition of either ZnCl2 or MgCl2 to BifR did not result in altered DNA-binding affinity, nor did addition of ZnCl2 or MgCl2 induce any change in gene expression in vivo (Figure 6). Zn2+-bound BifR had reduced thermal stability, an effect not seen on Mg2+-binding, indicating that BifR does bind specific metal ions. Assuming preferred binding to the native folded protein (of lowest energy), binding of metal or other ligand would be expected to be stabilizing. Another possibility is that metal-binding necessitates a conformational change to accommodate the metal (induced fit), in which case a reduced stability may be observed if the metal-induced conformation is a result of disrupting stabilizing interactions (unfolding). In addition, the protein will sample an ensemble of conformational states, resulting in an ensemble-averaged Tm. Therefore, it is also conceivable that a conformational selection takes place in which metal preferentially binds to (and stabilizes) an accessible subpopulation of states for which the Tm is lower than the ensemble average, thereby shifting the population toward the conformation with the highest affinity for the metal.40 Taken together, the observation that metal-bound, reduced BifR has lower thermal stability than apo-BifR suggests that metal-bound protein adopts a different less-stable conformation, perhaps through a combination of conformational selection and induced fit. As far as oxidation by Cu2+ is concerned, this also results in a destabilization. It is a reasonable inference that this also is due to a conformational change in the protein to accommodate the disulfide that renders it less stable. Since only Cu2+ induces changes in gene expression in vivo, we surmise that Cu2+ is the physiologically relevant metal ion.
BifR-Mediated Gene Regulation Is Linked to Redox State
The increased expression of ecsC and emrB in bif RΔ indicates that BifR is a repressor (Figure 1), as expected based on the position of palindromic sequences. On addition of H2O2 or Cu2+, a further repression of emrB and bif R gene expression is observed (Figure 6). Since oxidized BifR forms a disulfide-linked dimer-of-dimers, we propose that oxidized BifR functions as a “super-repressor” that competes more efficiently with RNA polymerase for DNA binding (Figure 8). Repression of emrB under oxidizing conditions was previously reported in biofilm-grown B. cenocepacia (BCAL0861; ∼2.3-fold down-regulation with both hydrogen peroxide and sodium hypochlorite, consistent with the ∼2.5-fold repression of emrB observed in B. thailandensis), suggesting a conserved regulatory mechanism for repression.16
As noted above, the genomic locus that includes the divergently oriented ecsC gene that encodes LasA protease is conserved among Burkholderia spp., including B. cenocepacia. In P. aeruginosa, LasA is an important virulence factor that is involved in cleavage of elastin, which is a major component of connective tissue; by contributing to the degradation of this physical barrier to infection, LasA enhances the invasiveness of the bacteria.23,41 The discovery that BifR controls expression of a functional LasA with elastolytic activity (Figure 2B) therefore has particular implications for virulence of Burkholderia species that colonize the CF lung.
The link between BifR-mediated gene regulation and cellular redox state is further suggested by the observation that BifR modestly activates expression of a gene encoding a Nudix phosphohydrolase (Figure 6). Nudix enzymes are ubiquitous and hydrolyze a variety of nucleotide derivatives. According to CombFunc, the Nudix hydrolase under BifR control is predicted to be an NAD+ hydrolase.42 The ratio of NAD+ to NADH is a signal for metabolic redox state and linked to gene activity. In P. syringae and M. tuberculosis, for example, deletion of a gene encoding an NADH-hydrolyzing Nudix enzyme is associated with reduced swarming and reduced biofilm.43,44 Activation of Nudix hydrolase expression by BifR may therefore contribute to maintaining NAD+/NADH ratios.
In P. aeruginosa, colony morphology has been shown to depend on redox-active phenazines, with failure to synthesize phenazines associated with a more rugose phenotype that facilitates oxygen diffusion; phenazines function as alternate respiratory electron acceptors within a biofilm when oxygen becomes limiting.10 Accordingly, upregulation of the phz operon would be expected under hypoxic conditions, whereas it should be repressed under aerobic conditions (where BifR forms a dimer-of-dimers). Since the phz operon was significantly upregulated in the bif RΔ strain, we infer that increased production of PCA in the bif RΔ strain may contribute to the smooth colony morphology.
Several MarR family transcription factors have been shown to undergo a cysteine oxidation that alters gene regulation. As noted above, HypR is induced by oxidation to activate gene expression.38 By contrast, other redox-sensitive MarR homologues such as OhrR and SarZ are released from DNA upon oxidation, leading to increased gene expression.45–47 The formation of a “super-repressor” by interdimer cross-linking of BifR protomers reveals a novel mode of redox-mediated gene regulation by MarR family proteins.
BifR Controls Biofilm Formation
Biofilm formation is increased in bif RΔ strains, regardless of emrB expression. The simplest interpretation of this observation is that BifR represses expression of genes involved in biofilm formation. When emrB is highly expressed, as in bif RΔ and emrBΔ-bif RΔ complemented with emrB, slightly lower biofilm formation is observed compared to emrBΔ-bif RΔ (Figure 7). One possibility for this phenotype is that EmrB exports an agent that slows biofilm formation or promotes its dispersal (according to TrSSP (http://bioinfo.noble.org/TrSSP), substrates for EmrB are predicted to be sugars/anions). While high levels of emrB expression appear to reduce biofilm formation marginally, a more apparent phenotype is the enhanced growth exhibited by the emrBΔ-bif RΔ mutant. The slightly increased biofilm formation may therefore be a consequence of the increased growth rate.
The “colony biofilm” on agar plates allows inspection of biofilm development over time. The hypoxic gradient that develops as oxygen diffusion becomes limited may be alleviated by colony wrinkling, which increases surface area. If the terminal electron acceptor O2 becomes limiting, growth is adversely affected and the intracellular redox state becomes reduced, as reflected in a higher ratio of NADH to NAD+ (and corresponding to a condition under which BifR would be reduced and exist as a dimer). The generation of alternate electron acceptors such as phenazines have been shown to attenuate colony wrinkling in P. aeruginosa.10 On the basis of the significant upregulation of the phenazine biosynthetic operon in bif RΔ combined with the smooth colony phenotype characteristic of the mutant (Figures 7 and 8), we propose that BifR plays a significant role in linking cellular redox state to biofilm formation, in large part by controlling PCA synthesis.
CONCLUSIONS
Our data suggest that BifR represses the expression of ecsC and emrB-bif R and that oxidation of BifR transforms it into a disulfide-bridged “super-repressor”. The BifR-mediated repression of the PCA biosynthetic phz operon rationalizes the link between BifR and cellular redox state; when O2 is limiting and the intracellular environment becomes reducing, the BifR-mediated repression of the phz operon may be lessened. The observation that ecsC encodes a functional extracellular protease with elastolytic activity has implications for pathogenic strains such as B. cenocepacia in which the ecsC-emrB-bif R locus is conserved.
Supplementary Material
Acknowledgments
We thank Huangen Ding for the use of his FPLC and Marcia Newcomer for the use of her spectrophotometer. We also thank Jong H. Ham and Inderjit Kaur for helper strain and pBBR-MCS5 vector. This work was supported by National Institutes of Health Grant 1R15GM107825 (to A. Grove). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.7b00103.
Predicted promoter elements for divergent genes; metal-binding by BifR; DNA binding by CuCl2-oxidized BifR; cartoon of BifR dimer-of-dimers; absorbance of WT, emrBΔ-bif RΔ, and emrBΔ-bif RΔ e-b; log 10(CFU/mL) of WT, emrBΔ-bif RΔ, and emrBΔ-bif RΔ e-b after mixing the biofilm; table showing CT values for qRT-PCR reference gene; sequences of primers used (PDF)
ORCID
Ashish Gupta: 0000-0002-1800-2755
Anne Grove: 0000-0002-4390-0354
Notes
The authors declare no competing financial interest.
References
- 1.Compant S, Nowak J, Coenye T, Clement C, Ait Barka E. Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol Rev. 2008;32:607–626. doi: 10.1111/j.1574-6976.2008.00113.x. [DOI] [PubMed] [Google Scholar]
- 2.Galyov EE, Brett PJ, DeShazer D. Molecular insights into Burkholderia pseudomallei and Burkholderia mallei pathogenesis. Annu Rev Microbiol. 2010;64:495–517. doi: 10.1146/annurev.micro.112408.134030. [DOI] [PubMed] [Google Scholar]
- 3.Mahenthiralingam E, Urban TA, Goldberg JB. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol. 2005;3:144–156. doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- 4.Drevinek P, Mahenthiralingam E. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin Microbiol Infect. 2010;16:821–830. doi: 10.1111/j.1469-0691.2010.03237.x. [DOI] [PubMed] [Google Scholar]
- 5.Deshazer D. Virulence of clinical and environmental isolates of Burkholderia oklahomensis and Burkholderia thailandensis in hamsters and mice. FEMS Microbiol Lett. 2007;277:64–69. doi: 10.1111/j.1574-6968.2007.00946.x. [DOI] [PubMed] [Google Scholar]
- 6.Bragonzi A, Farulla I, Paroni M, Twomey KB, Pirone L, Lore NI, Bianconi I, Dalmastri C, Ryan RP, Bevivino A. Modelling co-infection of the cystic fibrosis lung by Pseudomonas aeruginosa and Burkholderia cenocepacia reveals influences on biofilm formation and host response. PLoS One. 2012;7:e52330. doi: 10.1371/journal.pone.0052330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanley NR, Lazazzera BA. Environmental signals and regulatory pathways that influence biofilm formation. Mol Microbiol. 2004;52:917–924. doi: 10.1111/j.1365-2958.2004.04036.x. [DOI] [PubMed] [Google Scholar]
- 8.Fazli M, Almblad H, Rybtke ML, Givskov M, Eberl L, Tolker-Nielsen T. Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ Microbiol. 2014;16:1961–1981. doi: 10.1111/1462-2920.12448. [DOI] [PubMed] [Google Scholar]
- 9.Laverty G, Gorman SP, Gilmore BF. Biomolecular Mechanisms of Pseudomonas aeruginosa and Escherichia coli Biofilm Formation. Pathogens. 2014;3:596–632. doi: 10.3390/pathogens3030596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dietrich LE, Okegbe C, Price-Whelan A, Sakhtah H, Hunter RC, Newman DK. Bacterial community morphogenesis is intimately linked to the intracellular redox state. J Bacteriol. 2013;195:1371–1380. doi: 10.1128/JB.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okegbe C, Price-Whelan A, Dietrich LE. Redox-driven regulation of microbial community morphogenesis. Curr Opin Microbiol. 2014;18:39–45. doi: 10.1016/j.mib.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Kern SE, Newman DK. Endogenous phenazine antibiotics promote anaerobic survival of Pseudomonas aeruginosa via extracellular electron transfer. J Bacteriol. 2010;192:365–369. doi: 10.1128/JB.01188-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitzpatrick DA. Lines of evidence for horizontal gene transfer of a phenazine producing operon into multiple bacterial species. J Mol Evol. 2009;68:171–185. doi: 10.1007/s00239-009-9198-5. [DOI] [PubMed] [Google Scholar]
- 14.Gallagher LA, Ramage E, Patrapuvich R, Weiss E, Brittnacher M, Manoil C. Sequence-defined transposon mutant library of Burkholderia thailandensis. mBio. 2013;4:e00604–13. doi: 10.1128/mBio.00604-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, 2nd, Peterson KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 16.Peeters E, Sass A, Mahenthiralingam E, Nelis H, Coenye T. Transcriptional response of Burkholderia cenocepacia J2315 sessile cells to treatments with high doses of hydrogen peroxide and sodium hypochlorite. BMC Genomics. 2010;11:90. doi: 10.1186/1471-2164-11-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 18.Merritt JH, Kadouri DE, O’Toole GA. Growing and analyzing static biofilms. Curr Protoc Microbiol. 2005;1 doi: 10.1002/9780471729259.mc01b01s00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grove A, Kushwaha AK, Nguyen KH. Determining the role of metal binding in protein cage assembly. Methods Mol Biol. 2015;1252:91–100. doi: 10.1007/978-1-4939-2131-7_9. [DOI] [PubMed] [Google Scholar]
- 21.Choi SC, Zhang C, Moon S, Oh YS. Inhibitory effects of 4-hydroxy-2,5-dimethyl-3(2H)-furanone (HDMF) on acyl-homoserine lactone-mediated virulence factor production and biofilm formation in Pseudomonas aeruginosa PAO1. J Microbiol. 2014;52:734–742. doi: 10.1007/s12275-014-4060-x. [DOI] [PubMed] [Google Scholar]
- 22.Gustin JK, Kessler E, Ohman DE. A substitution at His-120 in the LasA protease of Pseudomonas aeruginosa blocks enzymatic activity without affecting propeptide processing or extracellular secretion. J Bacteriol. 1996;178:6608–6617. doi: 10.1128/jb.178.22.6608-6617.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cowell BA, Twining SS, Hobden JA, Kwong MS, Fleiszig SM. Mutation of lasA and lasB reduces Pseudomonas aeruginosa invasion of epithelial cells. Microbiology. 2003;149:2291–2299. doi: 10.1099/mic.0.26280-0. [DOI] [PubMed] [Google Scholar]
- 24.Winsor GL, Khaira B, Van Rossum T, Lo R, Whiteside MD, Brinkman FS. The Burkholderia Genome Database: facilitating flexible queries and comparative analyses. Bioinformatics. 2008;24:2803–2804. doi: 10.1093/bioinformatics/btn524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perera IC, Grove A. Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J Mol Cell Biol. 2010;2:243–254. doi: 10.1093/jmcb/mjq021. [DOI] [PubMed] [Google Scholar]
- 26.Grove A. MarR family transcription factors. Curr Biol. 2013;23:R142–143. doi: 10.1016/j.cub.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 27.Lim HN, Lee Y, Hussein R. Fundamental relationship between operon organization and gene expression. Proc Natl Acad Sci U S A. 2011;108:10626–10631. doi: 10.1073/pnas.1105692108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta A, Grove A. Ligand-binding pocket bridges DNA-binding and dimerization domains of the urate-responsive MarR homologue MftR from Burkholderia thailandensis. Biochemistry. 2014;53:4368–4380. doi: 10.1021/bi500219t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kallberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, Xu J. Template-based protein structure modeling using the RaptorX web server. Nat Protoc. 2012;7:1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilkinson SP, Grove A. HucR, a novel uric acid-responsive member of the MarR family of transcriptional regulators from Deinococcus radiodurans. J Biol Chem. 2004;279:51442–51450. doi: 10.1074/jbc.M405586200. [DOI] [PubMed] [Google Scholar]
- 31.Saridakis V, Shahinas D, Xu X, Christendat D. Structural insight on the mechanism of regulation of the MarR family of proteins: high-resolution crystal structure of a transcriptional repressor from Methanobacterium thermoautotrophicum. J Mol Biol. 2008;377:655–667. doi: 10.1016/j.jmb.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 32.Alekshun MN, Levy SB. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J Bacteriol. 1999;181:4669–4672. doi: 10.1128/jb.181.15.4669-4672.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mrazek J, Xie S. Pattern locator: a new tool for finding local sequence patterns in genomic DNA sequences. Bioinformatics. 2006;22:3099–3100. doi: 10.1093/bioinformatics/btl551. [DOI] [PubMed] [Google Scholar]
- 34.Butt A, Halliday N, Williams P, Atkins HS, Bancroft GJ, Titball RW. Burkholderia pseudomallei kynB plays a role in AQ production, biofilm formation, bacterial swarming and persistence. Res Microbiol. 2016;167:159–167. doi: 10.1016/j.resmic.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 35.Wilkinson SP, Grove A. Negative cooperativity of uric acid binding to the transcriptional regulator HucR from Deinococcus radiodurans. J Mol Biol. 2005;350:617–630. doi: 10.1016/j.jmb.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 36.Deochand DK, Perera IC, Crochet RB, Gilbert NC, Newcomer ME, Grove A. Histidine switch controlling pH-dependent protein folding and DNA binding in a transcription factor at the core of synthetic network devices. Mol BioSyst. 2016;12:2417–2426. doi: 10.1039/c6mb00304d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liguori A, Malito E, Lo Surdo P, Fagnocchi L, Cantini F, Haag AF, Brier S, Pizza M, Delany I, Bottomley MJ. Molecular Basis of Ligand-Dependent Regulation of NadR, the Transcriptional Repressor of Meningococcal Virulence Factor NadA. PLoS Pathog. 2016;12:e1005557. doi: 10.1371/journal.ppat.1005557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palm GJ, Khanh Chi B, Waack P, Gronau K, Becher D, Albrecht D, Hinrichs W, Read RJ, Antelmann H. Structural insights into the redox-switch mechanism of the MarR/DUF24-type regulator HypR. Nucleic Acids Res. 2012;40:4178–4192. doi: 10.1093/nar/gkr1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deochand DK, Meariman JK, Grove A. pH-Dependent DNA Distortion and Repression of Gene Expression by Pectobacterium atrosepticum PecS. ACS Chem Biol. 2016;11:2049–2056. doi: 10.1021/acschembio.6b00168. [DOI] [PubMed] [Google Scholar]
- 40.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gambello MJ, Iglewski BH. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wass MN, Barton G, Sternberg MJ. CombFunc: predicting protein function using heterogeneous data sources. Nucleic Acids Res. 2012;40:W466–470. doi: 10.1093/nar/gks489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Modzelan M, Kujawa M, Glabski K, Jagura-Burdzy G, Kraszewska E. NudC Nudix hydrolase from Pseudomonas syringae, but not its counterpart from Pseudomonas aeruginosa, is a novel regulator of intracellular redox balance required for growth, motility and biofilm formation. Mol Microbiol. 2014;93:867–882. doi: 10.1111/mmi.12702. [DOI] [PubMed] [Google Scholar]
- 44.Wolff KA, de la Pena AH, Nguyen HT, Pham TH, Amzel LM, Gabelli SB, Nguyen L. A redox regulatory system critical for mycobacterial survival in macrophages and biofilm development. PLoS Pathog. 2015;11:e1004839. doi: 10.1371/journal.ppat.1004839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee JW, Soonsanga S, Helmann JD. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Natl Acad Sci U S A. 2007;104:8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newberry KJ, Fuangthong M, Panmanee W, Mongkolsuk S, Brennan RG. Structural mechanism of organic hydroperoxide induction of the transcription regulator OhrR. Mol Cell. 2007;28:652–664. doi: 10.1016/j.molcel.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 47.Poor CB, Chen PR, Duguid E, Rice PA, He C. Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus. J Biol Chem. 2009;284:23517–23524. doi: 10.1074/jbc.M109.015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


