Abstract
Cancer cells elude anti-tumour immunity through multiple mechanisms, including upregulated expression of ligands for inhibitory immune checkpoint receptors1,2. Phagocytosis by macrophages plays a critical role in cancer control3–6. Therapeutic blockade of signal regulatory protein (SIRP)-α, an inhibitory receptor on macrophages, or of its ligand CD47 expressed on tumour cells, improves tumour cell elimination in vitro and in vivo7–10, suggesting that blockade of the SIRPα–CD47 checkpoint could be useful in treating human cancer11–14. However, the prophagocytic receptor(s) responsible for tumour cell phagocytosis is(are) largely unknown. Here we find that macrophages are much more efficient at phagocytosis of haematopoietic tumour cells, compared with non-haematopoietic tumour cells, in response to SIRPα–CD47 blockade. Using a mouse lacking the signalling lymphocytic activation molecule (SLAM) family of homotypic haematopoietic cell-specific receptors, we determined that phagocytosis of haematopoietic tumour cells during SIRPα–CD47 blockade was strictly dependent on SLAM family receptors in vitro and in vivo. In both mouse and human cells, this function required a single SLAM family member, SLAMF7 (also known as CRACC, CS1, CD319), expressed on macrophages and tumour cell targets. In contrast to most SLAM receptor functions15–17, SLAMF7-mediated phagocytosis was independent of signalling lymphocyte activation molecule-associated protein (SAP) adaptors. Instead, it depended on the ability of SLAMF7 to interact with integrin Mac-1 (refs 18–20) and utilize signals involving immunoreceptor tyrosine-based activation motifs21,22. These findings elucidate the mechanism by which macrophages engulf and destroy haematopoietic tumour cells. They also reveal a novel SAP adaptor-independent function for a SLAM receptor. Lastly, they suggest that patients with tumours expressing SLAMF7 are more likely to respond to SIRPα–CD47 blockade therapy.
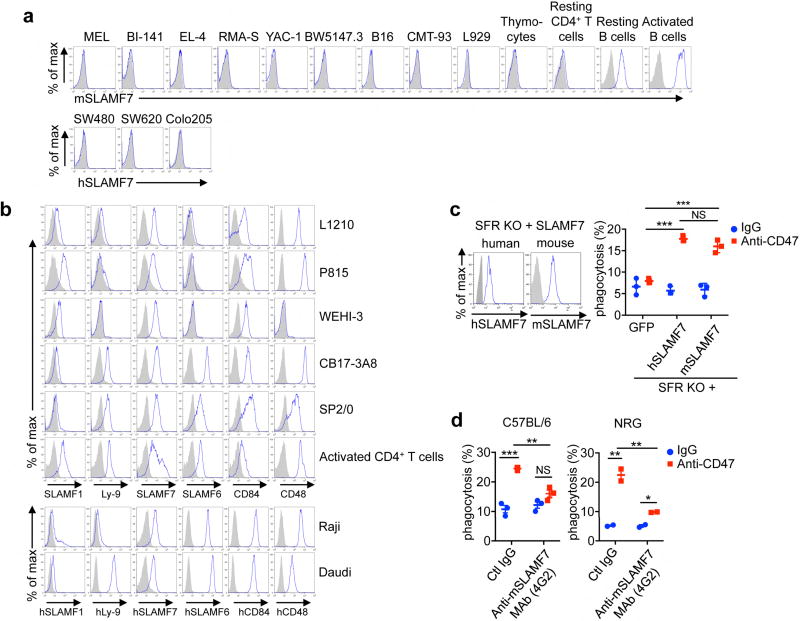
Bone-marrow-derived macrophages (BMDMs) were tested for phagocytosis of various target cells, with blocking anti-CD47 antibodies or control immunoglobulin-G (IgG), using several assays (Fig. 1a – c and Extended Data Fig. 1a, b). Augmented phagocytosis was seen with mouse B-cell lineage and myeloid tumour cell lines L1210, CB17–3A8, SP2/0, P815, and WEHI-3, treated with anti-CD47 antibodies compared with control IgG (Fig. 1d). This was seen with intact antibodies or F(ab′ )2 fragments of antibodies, implying that it was Fc receptor (FcR)-independent (Fig. 1e). Similar results were obtained with peritoneal macrophages (Fig. 1f). Anti-CD47 antibodies had no direct effect on tumour cells (Extended Data Fig. 1c – f). No effect of anti-CD47 on phagocytosis was seen with T-cell tumour lines, the mouse erythroleukemia (MEL) cell line, and non-haematopoietic cell lines, even though they expressed CD47 (Fig. 1g and Extended Data Fig. 2a). Increased phagocytosis by anti-human CD47 antibodies was also seen with human B-cell tumour cell lines Raji and Daudi, but not with several non-haematopoietic human tumour cells (Fig. 1h). F(ab′ )2 fragments of mouse-originated anti-human CD47 antibodies were used to avoid triggering mouse FcRs. Non-transformed activated mouse T cells bearing the CD4 molecule on their surface (CD4+ T cells), but not thymocytes, freshly isolated CD4+ T cells and B cells, and activated B cells, also displayed increased phagocytosis in response to anti-CD47 antibodies (Fig. 1i). Thus, some, but not all, haematopoietic tumour cells and normal cells displayed enhanced phagocytosis during SIRPα–CD47 blockade, in an FcR-independent manner.
Figure 1. Macrophages phagocytose a subset of haematopoietic cells.
a, Phagocytosis assay. Ctrl, control. b, Phagocytosis (arrows) of L1210 (green) was assessed by fluorescence microscopy. Scale bars, 50 µm. c, Same as b, except using confocal microscopy. Macrophages, red; targets, green. Arrow, non-phagocytosed cell. Scale bars, 5 µm. d, Same as b, using various mouse haematopoietic tumour cell lines as targets. e, Same as d, using F(ab′)2 fragments of antibodies. f, Same as d, using peritoneal macrophages. g, Same as d, using other mouse targets. NS, not significant. h, Same as d, using human targets. i, Same as d, using normal mouse haematopoietic target cells. *P < 0.05; **P < 0.01; ***P < 0.001 (two-tailed Student’s t-tests). Results pooled from five (L1210, P815, WEHI-3), three (CB17–3A8), or four (SP2/0) (d), three (e, i), two (f), three (MEL, BI-141, BW5147.3), or four (EL-4, RMA-S, YAC-1, B16, CMT-93, L929) (g), and four (h) mice studied in independent experiments. Each symbol represents one mouse. All data are means ± s.e.m. Photographs are representative of six (b) or two (c) independent experiments. See also Extended Data Figs 1 and 2.
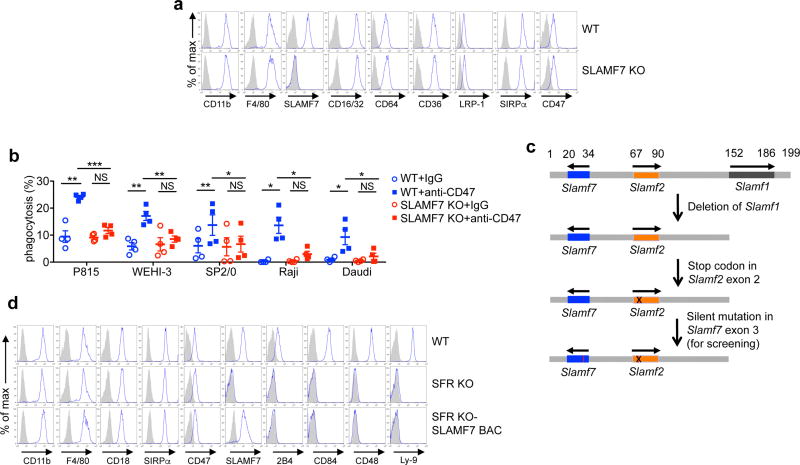
Phagocytosis of tumour cells in response to SIRPα–CD47 blockade was proposed to be mediated by the LRP-1 receptor, which can recognize calreticulin on tumour cells23. However, phagocytosis of L1210 and P815 cells was equivalent in control and LRP-1 knockout (KO) macrophages, implying an alternative mechanism (Extended Data Fig. 2b, c). Since CD47 antibody blockade had the greatest effect on haematopoietic targets, we tested the involvement of SLAM family receptors (SFRs), a group of homotypic receptors expressed solely on haematopoietic cells15–17. Macrophages from a mouse lacking all SFRs (SFR KO mouse) had normal differentiation markers (Extended Data Fig. 3a). However, they failed to display increased phagocytosis with anti-CD47 antibodies, either intact or as F(ab′ )2 fragments (Fig. 2a – c and Extended Data Fig. 3b, c). This defect was seen with mouse and human targets, and with BMDMs and peritoneal macrophages. SFR KO macrophages also displayed a marked defect in phagocytosis of CD47 KO L1210 cells, compared with wild-type (WT) macrophages, in the absence of added anti-CD47 antibodies (Fig. 2d and Extended Data Fig. 3d). SFR KO macrophages exhibited normal phagocytosis of several other types of target, including IgG-opsonized L1210 cells (Extended Data Fig. 3e – g).
Figure 2. SLAM receptors are required for phagocytosis of haematopoietic cells.
a, Same as Fig. 1d, using macrophages from WT or SFR KO mice. b, Same as a, using human targets. c, Same as Fig. 1e, using SFR KO macrophages. d, Phagocytosis of parental or CD47 KO L1210. e, Intraperitoneal tumour clearance assay using L1210 cells. f, Same as e, except WT mice were injected with liposomes containing clodronate or phosphate-buffered saline (PBS). g, Growth of L1210 injected subcutaneously in RAG-1 KO or RAG-1 SFR dKO mice. D, day. *P <0.05; **P <0.01; ***P <0.001; ****P <0.0001 (two-tailed Student’s t-tests). Results pooled from a total of eight (L1210), six (P815), seven (WEHI-3) or five (CB17–3A8, SP2/0) (a), five (b, d), three (c), and two (f) mice studied in independent experiments; six mice from five independent experiments (e); and 11 mice from two of four independent experiments (g). Each symbol represents one mouse. For g, bar graphs represent mean volumes. All data are means ± s.e.m. See also Extended Data Figs 3 and 4 and Source Data for this figure are available in the online version of the paper.
Treatment of WT mice with anti-CD47 antibodies, but not control IgG, also resulted in elimination of L1210 cells introduced in the peritoneal cavity (Fig. 2e). This effect was not seen in SFR KO mice (Fig. 2e and Extended Data Fig. 4a, b). L1210 clearance was abrogated by clodronate, implying that it was macrophage-mediated (Fig. 2f and Extended Data Fig. 4c–e). Additionally, when L1210 cells were inoculated subcutaneously in RAG-1 (recombination-activating gene-1) KO mice, anti-CD47 antibodies significantly reduced tumour growth, compared with control IgG (Fig. 2g and Extended Data Fig. 4f, g). The anti-CD47 antibody-dependent effects were absent in RAG-1 SFR double (d)KO mice (Fig. 2g and Extended Data Fig. 4f, g). Hence, SFRs were necessary for macrophages to eliminate tumour cells in response to SIRPα–CD47 blockade in vitro and in vivo. Anti-CD47 antibodies delayed, but did not abolish, tumour growth in mice (Fig. 2g), suggesting that, as single agent, these antibodies cannot completely control tumour growth in vivo.
Five of the six SFRs, namely SLAMF1, 2B4, SLAMF7, Ly-9, and CD84, are expressed on macrophages (Extended Data Fig. 3a). SLAMF7 single KO macrophages, but not SLAMF1, 2B4, Ly-9, or CD84 single KO macrophages, had a defect in phagocytosis, nearly comparable to that of SFR KO macrophages (Fig. 3a and Extended Data Fig. 5a, b). Moreover, the defect in phagocytosis of SFR KO macrophages was rescued by re-expression of mouse Slamf7 using transgenesis (Fig. 3b, c and Extended Data Fig. 5c, d). As SLAMF7 is a homotypic receptor (that is, self-ligand)15–17,24, we also asked whether target cells susceptible to enhanced phagocytosis in response to anti-CD47 antibodies expressed SLAMF7. All susceptible target cells expressed SLAMF7 (Fig. 3d). In contrast, none of the non-susceptible target cells, except normal B cells, did (Extended Data Fig. 6a). Susceptible targets also expressed other SFR ligands, but macrophages lacking these SFRs showed no defect in phagocytosis (Fig. 3a and Extended Data Fig. 6b). We also found that, unlike WT activated CD4+ T cells, SLAMF7 KO activated CD4+ T cells did not display increased phagocytosis by WT macrophages with anti-CD47 antibodies (Fig. 3e). Likewise, when injected intravenously in WT mice, SLAMF7 KO CD4+ T cells were less efficiently cleared from the blood in response to anti-CD47 antibodies, compared with WT CD4+ T cells (Fig. 3f). As human target cells were susceptible to phagocytosis by mouse macrophages (Fig. 1h), we also expressed human SLAMF7 in SFR KO macrophages. Expression of human SLAMF7 conferred an enhanced phagocytosis response during CD47 antibody blockade (Extended Data Fig. 6c). Lastly, anti-mouse SLAMF7 antibodies 4G2, but not control antibodies, interfered with anti-CD47 antibody-enhanced engulfment of L1210 cells (Extended Data Fig. 6d). Similarly, anti-human SLAMF7 antibody 162 blocked the augmented capacity of human blood-derived macrophages to engulf Raji cells in response to anti-CD47 antibodies (Fig. 3g). Therefore, SLAMF7 expression on macrophages and tumour cells was required to endow mouse and human macrophages with the capacity to phagocytose haematopoietic cells in the presence of anti-CD47 antibodies.
Figure 3. SLAMF7 is necessary and sufficient for phagocytosis of haematopoietic cells.
a, Same as Fig. 1d, using macrophages lacking individual SFRs and L1210. b, c, Same as Fig. 1d (b) and Fig. 2e (c), using macrophages from SFR KO mice reconstituted with Slamf7 bacterial artificial chromosome (BAC) transgene and L1210. d, Expression of SLAMF7 (blue lines); prefixes m, mouse; h, human. Filled curves: isotype controls. e, Phagocytosis of activated WT or SLAMF7 KO CD4+ T cells by WT macrophages. f, Residual WT and SLAMF7 KO CD4+ T cells in blood of WT mice. Left, representative dot plot; right, quantification. g, Phagocytosis of Raji cells by human monocytes/macrophages, in the presence of anti-hSLAMF7 162 or control IgG. *P < 0.05; **P < 0.01; ***P < 0.001 (two-tailed Student’s t-tests). Results pooled from a total of eight (SLAMF7 KO) or three (all other KO mice) (a) and three (b, c, e) mice studied in independent experiments; six mice in three experiments (f); and three healthy human donors studied in one experiment (g). Each symbol represents one mouse or healthy donor. All data are means ± s.e.m. Flow cytometry profiles are representative of six independent experiments (d). See also Extended Data Figs 5 and 6 and Source Data for this figure are available in the online version of the paper.
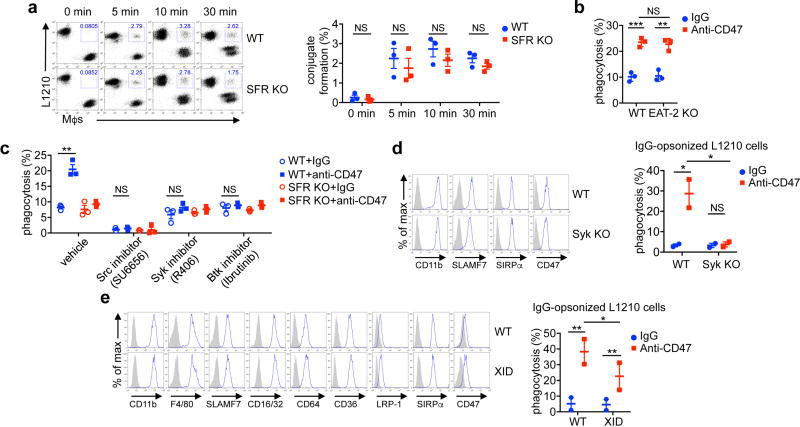
SLAMF7 might augment adhesion between macrophages and target cells. However, there was no defect in conjugate formation between SFR KO macrophages and L1210 cells in the presence of anti-CD47 antibodies, although a phagocytosis defect was seen at a later time point (Fig. 4a and Extended Data Fig. 7a). SLAMF7 might also promote signals triggering phagocytosis. In confocal microscopy studies of conjugates in the presence of anti-CD47 antibodies, actin polarization towards target cells, a key step during phagocytosis, was markedly reduced in SFR KO macrophages (Fig. 4b). Hence, SLAMF7 did not detectably enhance adhesion to targets, but did stimulate cytoskeletal reorganization required for phagocytosis.
Figure 4. SLAMF7 controls actin polarization and promotes phagocytosis independently of SAP adaptors.
a, Conjugate formation (left) and phagocytosis (right) of L1210 by macrophages. b, Actin polarization (arrows) in macrophages incubated with L1210 detected by immunofluorescence. Left, representative examples of fully polarized and non-polarized conjugates. Right, quantitation from a total of 130 conjugates in three experiments. Scale bars, 5 µm. c, Same as Fig. 1d, using SFR KO macrophages expressing green fluorescent protein (GFP) alone or in combination with WT or Y→ F mSLAMF7, and L1210. Left, expression of SLAMF7 was determined by flow cytometry; right, quantification. Blue lines, anti-SLAMF7; filled curves, control antibodies. d, e, Same as Fig. 1d, using macrophages from Syk KO (d) or X-linked immunodeficient (e) mice, and L1210. Representative anti-Syk immunoblot is shown in d (left). **P < 0.01; ***P < 0.001 (two-tailed Student’s t-tests). Results pooled from a total of four (a, left), three (a, right), three (b), five (c), or four (d, e) independent mice studied in independent experiments. Each symbol represents one mouse. In b, bars represent mean numbers of conjugates with fully polarized actin. All data are means ± s.e.m. Uncropped blots can be seen in Supplementary Fig. 1. See also Extended Data Fig. 7.
To elucidate how SLAMF7 mediated these effects, we first expressed SLAMF7 in which three intra-cytoplasmic tyrosines (Y) were mutated to phenylalanines (F) (Y→ F mutations) in SFR KO macrophages (Fig. 4c). These tyrosines couple SLAMF7 to effectors, including the SAP adaptor EAT-2 (refs 15–17, 25). The Y→ F mutations had no impact on phagocytosis (Fig. 4c). In agreement, phagocytosis was unaffected in EAT-2 KO macrophages (Extended Data Fig. 7b). Therefore, SLAMF7 promoted phagocytosis by a novel SAP adaptor-independent mechanism. We analysed the contributions of various signalling effectors in SLAMF7-dependent phagocytosis. Pharmacological inhibitors of three classes of kinases6,21, namely the Src family, Syk, and Btk kinases, abrogated the enhanced phagocytosis in response to anti-CD47 antibodies (Extended Data Fig. 7c). The enhanced phagocytosis of L1210 cells with anti-CD47 antibodies was also abrogated in Syk KO macrophages (Fig. 4d), and reduced in macrophages from mice with X-linked immunodeficiency26, which carry a loss-of-function mutation in Btk (Fig. 4e). Loss of Syk or the X-linked immunodeficiency mutation had no impact on macrophage markers, although they caused defects in phagocytosis of IgG-opsonized targets, as expected22 (Extended Data Fig. 7d, e). Hence, SLAMF7-dependent phagocytosis required Src, Syk, and Btk kinases, and this effect was macrophage-intrinsic for at least Syk and Btk.
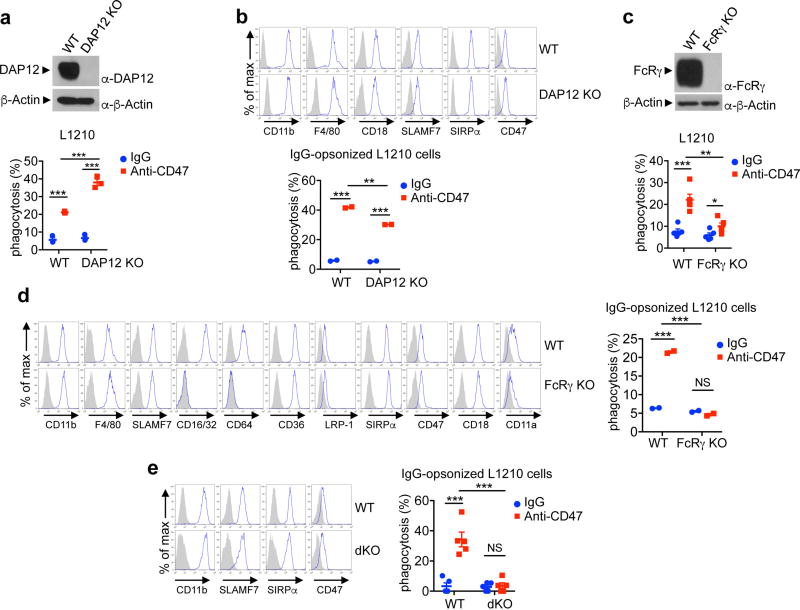
These findings suggested that SLAMF7 might promote phagocytosis by using immunoreceptor tyrosine-based activation motif (ITAM)-containing proteins, which mediate immune cell activation via Src, Syk, and Btk kinases21,22. Macrophages express two ITAM-containing proteins, DAP12 (DNAX-activation protein of 12 kDa) and FcR-associated-γ (FcRγ)22. Whereas no defect in anti-CD47-dependent phagocytosis of L1210 was seen in DAP12 KO macrophages (Extended Data Fig. 8a, b), a partial defect was seen in FcRγ KO macrophages (Extended Data Fig. 8c, d). Macrophages lacking both FcRγ and DAP12 displayed a complete defect (Fig. 5a). Absence of DAP12 and FcRγ had no impact on macrophage markers, with the exception of FcRs CD64 and CD16, which were absent in FcRγ KO macrophages, as described27 (Extended Data Fig. 8b, d, e). Defects in FcR-mediated phagocytosis were also seen in macrophages lacking FcRγ22 (Extended Data Fig. 8d, e). Thus, SLAMF7-dependent phagocytosis was dependent on contributions from FcRγ and DAP12.
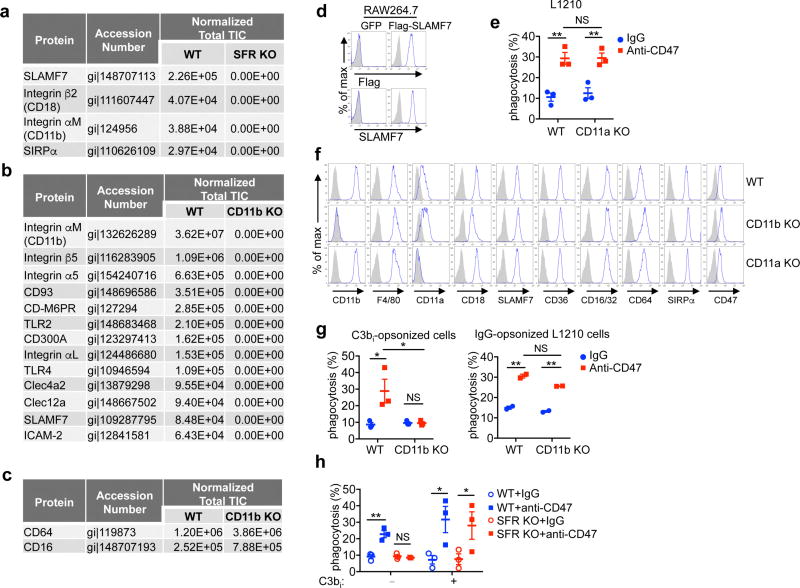
Figure 5. SLAMF7-dependent phagocytosis requires ITAMs and Mac-1.
a, Same as Fig. 1d, using macrophages from FcRγ-DAP12 dKO mice and L1210. Left, representative anti-FcRγ and anti-DAP12 immunoblots; right, quantification. b, Co-immunoprecipitation of SLAMF7 and CD11b (Mac-1) in RAW264.7 expressing GFP alone or with Flag-tagged SLAMF7 (Flag–SLAMF7). IP, immunoprecipitation. c, Co-localization of SLAMF7 and CD11b in RAW264.7 cells expressing GFP alone or with Flag– SLAMF7 assessed by immunofluorescence. Two examples of conjugates for each cell type are shown at top and bottom. Scale bars, 5 µm. d, Same as Fig. 1d, using WT macrophages incubated with antibodies against integrins or control IgG. e, Same as Fig. 1d, using CD11b KO macrophages and L1210. **P < 0.01; ***P < 0.001 (two-tailed Student’s t-tests). Results pooled from a total of three (a, d) or five (e) mice studied in independent experiments. Each symbol represents one mouse. All data are means ± s.e.m. Immunoblots (b) and photographs (c) are representative of five and three independent experiments, respectively. Uncropped blots can be seen in Supplementary Fig. 1. See also Extended Data Figs 8 and 9.
ITAM-containing proteins are typically associated with transmembrane receptors, which recognize extracellular ligands21,22. SLAMF7 lacks a charged transmembrane domain residue that is needed to bind ITAM-containing subunits. To assess if SLAMF7 might interact with FcRγ and DAP12 through other receptors, SLAMF7 was immunoprecipitated from WT macrophages, and associated proteins were identified by mass spectrometry. SLAMF7 immunoprecipitates contained two integrin proteins, α-subunit CD11b (αM) and β-subunit CD18 (β2), which constitute Mac-1 (refs 18–20) (Extended Data Fig. 9a). SIRPα was also identified as a SLAMF7-associated protein. Mac-1 and SIRPα were absent from anti-SLAMF7 immunoprecipitates from SFR KO macrophages. Conversely, SLAMF7 was identified in anti-CD11b immunoprecipitates from WT, but not CD11b KO, macrophages (Extended Data Fig. 9b). Anti-CD11b immunoprecipitates from WT macrophages also contained other receptors, but no other SFRs. As the other receptors found in CD11b immunoprecipitates were not seen in SLAMF7 immunoprecipitates, the complexes of CD11b with SLAMF7 or these other receptors were presumably independent. CD64 and CD16 were equally present in anti-CD11b immunoprecipitates from WT and CD11b KO macrophages, implying that these co-immunoprecipitations were non-specific (Extended Data Fig. 9c).
Mac-1 is known to interact with FcRγ and DAP12 (refs 21, 22). It has multiple broadly expressed ligands such as ICAM-1 and promotes phagocytosis of various types of target, including pathogens. It is also known as complement receptor 3 (CR3), owing to its ability to bind targets opsonized by inactive C3b complement (C3bi)18–20,28. The association between SLAMF7 and CD11b and their co-localization on the cell surface were confirmed by immunoblot (Fig. 5b) and confocal microscopy (Fig. 5c), respectively, using the macrophage cell line RAW264.7 expressing Flag-tagged SLAMF7 (Extended Data Fig. 9d). Co-localization of SLAMF7 with CD11b was especially dense at areas of cell–cell contact and associated with foci of SLAMF7 accumulation (Fig. 5c).
Antibody blockade of CD11b or CD18, but not of integrins CD11a (αL; LFA-1), CD11c (αX), CD29 (β1), or CD61 (β3), significantly attenuated the enhancement of L1210 phagocytosis by anti-CD47 antibodies (Fig. 5d). Anti-CD47 antibody-induction of L1210 cell phagocytosis was also severely compromised in CD11b KO macrophages, but not in CD11a KO macrophages (Fig. 5e and Extended Data Fig. 9e, f). In keeping with the ability of Mac-1 to bind C3bi28, CD11b KO macrophages displayed reduced anti-CD47 antibody-induced phagocytosis of C3bi-opsonized, but not of IgG-opsonized, L1210 cells (Extended Data Fig. 9g). SFR KO macrophages did not display a defect in phagocytosis of C3bi-opsonized L1210 cells in the presence of anti-CD47 (Extended Data Fig. 9h). Therefore, Mac-1 expression on macrophages was required for SLAMF7-dependent phagocytosis of tumour cells, but Mac-1 did not display a reciprocal requirement of SLAMF7 for phagocytosis of C3bi-opsonized targets.
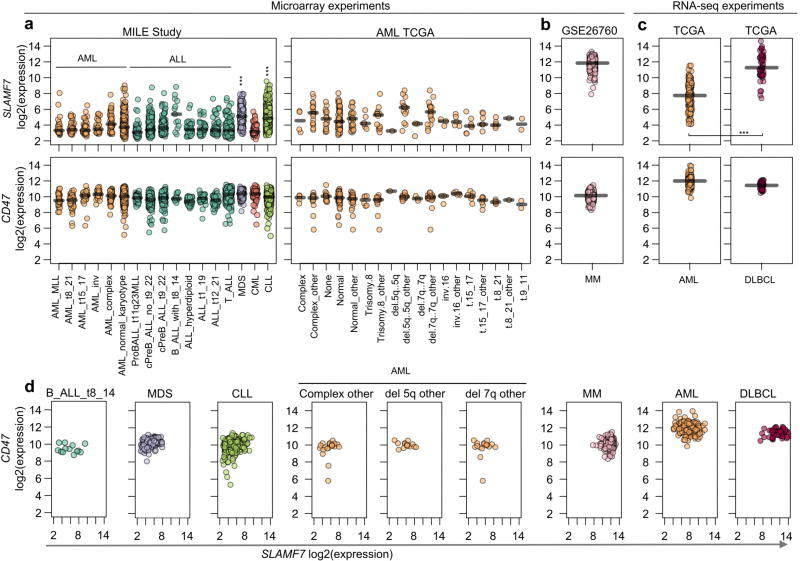
To ascertain which human haematological tumours may be susceptible to SLAMF7-dependent phagocytosis during SIRPα–CD47 blockade therapy, expression datasets of primary human haematological malignancies were extracted for SLAMF7 and CD47. Acute myelogenous leukaemia, acute lymphocytic leukaemia, and chronic myelogenous leukaemia expressed lower median levels of SLAMF7, although some individual samples had higher levels (Extended Data Fig. 10a, b). In contrast, chronic lymphocytic leukaemia, myelodysplastic syndrome, multiple myeloma, and diffuse large B-cell lymphoma had higher levels of SLAMF7 (Extended Data Fig. 10a – c). CD47 was uniformly expressed at high levels in all tumour types, including samples displaying higher levels of SLAMF7 (Extended Data Fig. 10a – d). Thus, some haematological cancers, in particular chronic lymphocytic leukaemia, myelodysplastic syndrome, multiple myeloma, and diffuse large B-cell lymphoma, frequently co-expressed high levels of SLAMF7 and CD47.
The mechanisms by which therapeutic agents opposing immune tolerance mediate their anti-tumour effect remain poorly understood1,2,11,12,14. Here we show that macrophages selectively phagocytosed haematopoietic tumour cells in response to SIRPα–CD47 checkpoint blockade, in an FcR-independent manner. Phagocytosis was mediated by the homotypic, haematopoietic cell-specific receptor SLAMF7 (refs 15–17, 24, 25). It also required expression of integrin Mac-1, and of ITAM-containing subunits FcRγ and DAP12 (refs 21, 22). Our finding that Mac-1-blocking antibodies prevented phagocytosis of haematopoietic tumour cells implies that, like SLAMF7, Mac-1 plays a direct role in target cell recognition, although the ligands of Mac-1 in this setting are unidentified. Thus, we propose that SLAMF7 synergizes with Mac-1 both to recognize ligands on target cells and to generate signals leading to phagocytosis. Normal B cells, which highly express SLAMF7 and CD47, are not susceptible to enhanced phagocytosis during SIRPα–CD47 blockade. This is possibly due to lack of relevant ligands for Mac-1, or expression of ligands for inhibitory receptors other than SIRPα.
Given the broad tissue distribution and wide range of Mac-1 ligands, the more narrowly expressed SLAMF7 probably determines which tumour cells are engulfed by phagocytes in response to SIRPα–CD47 blockade. Hence, patients with SLAMF7-positive malignancies may benefit more from SIRPα–CD47 blockade therapy. Our analyses of public datasets and other published data (reviewed in ref. 24) show that SLAMF7 is more expressed in haematological cancers such as chronic lymphocytic leukaemia, myelodysplastic syndrome, diffuse large B-cell lymphoma, and multiple myeloma. Clinical trials testing the impact of SIRPα–CD47 blockade in some of these malignancies are underway (https://clinicaltrials.gov).
METHODS
Mice
Mice lacking all SFRs (SFR KO), SLAMF7 (Slamf7−/−), or 2B4 (Slamf4−/−) were described elsewhere29. In essence, SFR KO mice were created by deletion of the entire 400 kilobase (kb) Slam locus in Bruce 4 C57BL/6 embryonic stem cells. Mice were subsequently backcrossed to the C57BL/6 background for six to ten generations. To produce mice lacking SLAMF1 (Slamf1−/−), DNA fragments encoding SLAMF1 were amplified by PCR from a BAC clone derived from 129S1/Sv mice. The 5′ and 3′ genomic fragments were then cloned on either side of neo in the vector pJA1617. DNA was then transfected into the 129 mouse embryonic stem-cell line R1, and cells were selected with G418. After removal of the neo cassette by transient transfection of the Flpe recombinase, positive clones were injected into blastocysts. The resulting chimaeric mice were crossed with 129S1/Sv mice. Slamf7 BAC transgenic mice (SLAMF7 BAC Tg) were generated using the BAC clone RP23–145F9. The BAC clone was modified using DNA recombineering, to eliminate the Slamf1 gene and introduce a stop codon in Slamf2, the gene coding for CD48 (Extended Data Fig. 5c). It was then injected into B6-C3H F1 fertilized oocytes to generate SLAMF7 BAC Tg mice. Mice were then bred with SFR KO mice to create SFR KO-SLAMF7 BAC Tg mice. Mice lacking Ly-9 (Slamf3−/−) in the 129S1/Sv background, and mice lacking EAT-2 (Sh2d1b1−/−) in the C57BL/6 back-ground, were described previously30,31. The following mice were obtained from The Jackson Laboratory (Bar Harbor, Maine, USA): CD84 KO (Slamf5−/−); CD11b KO (Itgam−/−); CD11a KO (Itgal−/−), CD47 KO (Cd47−/−); LRP-1 conditional KO (Lrp1fl/fl); Lyz2-Cre; RAG-1 KO (Rag1−/−); NRG (NOD;Rag1−/−IL2Rγc−/−), which are NOD congenic mice lacking T cells, B cells, and natural killer (NK) cells; X-linked immunodeficient mice (in the CBA/CaHN background), which carry a loss-of-function point mutation in Btk26; and T-cell-deficient mice (B6.129P2-Tcrbtm1Mom Tcrdtm1Mom/J). Mice lacking Syk in bone marrow cells were generated by transplantation of fetal liver from Syk−/− mice into irradiated RAG-deficient mice32. Mice lacking FcRγ (Fcer1g−/−) were obtained from Taconics (Hudson, New York, USA). Mice lacking DAP12 (Tyrobp−/−) were provided by T. Takai33. Mice devoid of FcRγ and DAP12 (Fcer1g−/−;Tyrobp−/−) were reported elsewhere34. All mice were maintained in the C57BL/6 background, unless specified. They were also kept in a specific pathogen-free environment. Either males or females were used, typically 8–12 weeks of age. Littermates were used as controls in all experiments, except for the studies involving Syk KO, FcRγ KO, FcRγ–DAP12 dKO, and X-linked immunodeficient mice, where WT syngeneic mice were used. Animal experimentation was approved by the Animal Care Committee of the Institut de recherches cliniques de Montréal and performed as defined by the Canadian Council of Animal Care (A.V.), or by the Institutional Animal Care and Use Committee of the University of California at San Francisco, in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (C.A.L.). For experiments with mice, sample size was chosen on the basis of previous studies in this field and to achieve statistical significance. No randomization or blinding was performed. No animals were excluded from the analyses.
Cells and retroviral infection
Mouse BMDMs were produced as described elsewhere35. In brief, femora and tibiae were flushed with tissue culture medium and propagated in bacterial Petri dishes for ~7 days, in medium supplemented with 30% (v/v) L929 cell-conditioned medium as a source of colony-stimulating factor 1. Thioglycollate-elicited peritoneal macrophages were generated as outlined elsewhere35. In our initial studies, we used L1210 (B-cell lymphocytic leukaemia), P815 (mastocytoma), SP2/0 (multiple myeloma), the v-Abl-induced B-cell line CB17–3A8 (v-Abl-transformed B-cell leukaemia), WEHI-3 (myelomonocytic leukaemia), the BW5147-derived T-cell hybridoma BI-141, EL-4 (T-cell lymphoma), RMA-S (T-cell lymphoma), YAC-1 (thymoma), B16 (melanoma), CMT-93 (rectal carcinoma), RAW264.7 (v-Abl-transformed monocyte/ macrophage), and L929 (immortalized fibroblast) from stocks described previously30,35–38. Moreover, Raji (B-cell lymphoma) and Daudi (B-cell lymphoma) were provided by J. Di Noia, whereas Colo205 (colon carcinoma), SW480 (colon carcinoma), and SW620 (colon carcinoma) were provided by N. Rivard. Cells were sporadically tested for mycoplasma and found to be negative. Haematopoietic cells were authenticated by flow cytometry. Subsequently, L1210 (CCL-219), P815 (TIB-64), WEHI-3 (TIB-68), SP2/0 (CRL-1581), BW5147.3 (TIB-47), EL-4 (TIB-39), YAC-1 (TIB-160), B16 (CRL-6475), CMT-93 (CCL-223), L929 (CCL-1), Raji (CCL-86), Daudi (CCL-213), Colo205 (CCL-222), SW480 (CCL-228), and SW620 (CCL-227) were freshly obtained from American Type Culture Collection (Manassas, Virginia, USA), while MEL (96121718) and RAW264.7 (91062702) were freshly obtained from the European Collection of Authenticated Cell Cultures (Sigma-Aldrich, St. Louis, Missouri, USA). The American Type Culture Collection and the European Collection of Authenticated Cell Cultures authenticated these cell lines and showed that they were negative for mycoplasma. RMA-S was obtained from the institute that initially generated this cell line (provided through B. Chambers). They were mycoplasma-negative. No cell lines used in this paper are listed in the database of commonly misidentified cell lines maintained by the International Cell Line Authentication Committee. CD47 KO L1210 cells were generated using CRISPR–Cas-mediated genome editing, using the guide RNA sequences CACCGAGCAACAGCGCCGCCGCCAA and CACCGTTGGCGGCGGCGCTGTTGCT. Thymocytes and freshly isolated CD4+ T cells were isolated as detailed previously36. Activated CD4+ T cells were obtained by stimulating purified splenic CD4+ T cells with concanavalin A (4 µg ml−1; Sigma-Aldrich) for 2 days, followed by IL-2 (50 U ml−1) for 3 days. Unstimulated B cells were obtained by isolating total splenocytes from T-cell-deficient mice, whereas activated B cells were obtained by stimulating purified B cells with lipopolysaccharide (5µ g ml−1) for 5 days. Purity of T cells and B cells was greater than 90%. BMDMs or RAW264.7 cells were infected with retroviruses (using the vector pFB–GFP) encoding various SLAMF7 proteins, in combination with GFP. Retroviruses encoding GFP alone were used as control. For BMDMs, GFP-positive cells were purified by cell sorting 48 h after infection and propagated for an additional 48 h in growth medium, before experimentation. For RAW264.7, GFP-positive cells were isolated by cell sorting, when sufficient numbers of cells were available. The constructs encoding mouse SLAMF7 Y→ F mutant (Y261F;Y266F;Y281F) and Flag-tagged mouse SLAMF7 were created by PCR. L1210 derivatives expressing Tac (CD25) were generated by transfection, using a plasmid (pSRα-puro) encoding a cytoplasmic domain-deleted version of CD25 fused to the transmembrane domain of 2B4. Transfected cells were selected in medium containing puromycin and purified by cell sorting. Human blood samples were collected from healthy donors following informed consent for the McGill University Health Centre Institutional Review Board-approved research protocol GEN10–256. Peripheral blood mononuclear cells were then isolated using Ficoll-Paque PLUS (GE Healthcare, Burlington, Ontario, Canada), according to the manufacturer’s protocol. Peripheral blood mononuclear cells were seeded for 1–3 h at 37 °C in tissue culture dishes containing serum-free RPMI medium. After gentle washes, adherent cells (which mostly represented monocytes) were cultured in RPMI medium supplemented with 10% human serum (Valley Biomedical, Winchester, Virginia, USA). Medium was changed on days 3 and 6. Cells were used for experimentation on day 7.
Antibodies
For flow cytometry or blocking, the following monoclonal antibodies were used. Anti-CD11b (M1/70), anti-F4/80 (BM8), anti-CD18 (M18/2), anti-CD11a (M17/4), anti-CD29 (HMb1–1), anti-CD11c (N418), anti-CD61 (2C9.G3), anti-CD16/32 (93), anti-CD36 (72–1), anti-mouse CD47 (Miap301), and anti-human CD47 (B6H12) were from eBioscience (San Diego, California, USA). Anti-mouse CD47 (Miap301) was also obtained from Biolegend (San Diego, California, USA). Anti-CD64 (monoclonal antibody X54–5/7.1), anti-human CD47 (monoclonal antibody CC2C6), anti-human SLAMF7 (monoclonal antibody 162.1), anti-human SLAMF1 (A12 (7D4)), anti-human Ly-9 (HLy-9.1.25), anti-human NTB-A (NT-7), anti-human CD84 (CD84.1.21), and anti-human CD48 (BJ40) were from Biolegend. Anti-LRP-1 (monoclonal antibody 5A6) was from Novus Biologicals (Littleton, Colorado, USA). Anti-CD11b (monoclonal antibody 5C6) was from AbD Serotec (Kidlington, UK). Anti-SIRPα (monoclonal antibody P84) was from BD Biosciences (Mississauga, Ontario, Canada). Antibodies directed against mouse SFRs and CD48 were described previously36. Anti-human SLAMF7 monoclonal antibody 162 was reported elsewhere39. F(ab′)2 fragments of anti-mouse CD47 monoclonal antibody (Miap301), anti-human CD47 monoclonal antibody (B6H12), anti-human SLAMF7 monoclonal antibody (162), and control IgG were generated using pepsin (Sigma-Aldrich), according to standard protocols. Purity and integrity of F(ab′)2 fragments were confirmed by protein gel electrophoresis. For immunoprecipitations and immunoblots, the following antibodies were used: anti-Syk and anti-SLAMF7 rabbit antisera (generated in our laboratory40,41), anti-β-actin (C4; Santa Cruz Biotechnology, Santa Cruz, California, USA), anti-DAP12 (D7G1X; New England Biolabs, Ipswich, Massachusetts, USA), anti-FcRγ (PM068; MBL International, Woburn, Massachusetts, USA), anti-CD11b (EPR1344; Abcam, Toronto, Ontario, Canada), and anti-LRP-1 (monoclonal antibody 5A6; Abcam). For immunofluorescence, the following antibodies were used: anti-Flag (monoclonal antibody M2; Sigma-Aldrich), anti-CD11b (monoclonal antibody EPR1344; Abcam), and anti-β-actin (monoclonal antibody AC-74, Sigma-Aldrich).
In vitro phagocytosis assays
For the microscopy-based assay, 5 × 104 macrophages were seeded overnight in a 24-well tissue culture plate. The next day, target cells were washed and labelled with 2.5 µM of carboxyfluorescein succinimidyl ester (CFSE), using a CFSE Cell Proliferation Kit (C34554; Life Technologies, Burlington, Ontario, Canada). After incubating macrophages in serum-free medium for 2 h, 2 × 105 CFSE-labelled target cells were added to the macrophages, in the presence of anti-CD47 antibodies or control IgG (10 µg ml−1). After incubation for 2 h at 37°C, macrophages were extensively washed and imaged with an inverted microscope (Carl Zeiss Axiovert S100 TV). The phagocytosis efficiency was calculated as the number of macrophages containing CFSE+ target cells per 100 macrophages. For the flow cytometry-based assay, macrophages were prepared and then incubated with Tac-expressing L1210 cells as targets, as detailed for the microscopy assay, except that targets were labelled with 0.2 µM of CFSE. Once the phagocytosis period was completed, all cells in the well were collected in the presence of Accutase. Cells were then stained on ice for 30 min with APC-conjugated anti-human Tac (CD25) and phycoerythrin-conjugated monoclonal antibody F4/80, and analysed by flow cytometry. After gating on F4/80+Tac− cells (which included macrophages but excluded non-phagocytosed Tac-positive L1210 cells), phagocytosis efficiency was determined as the percentage of F4/80+Tac− cells containing CFSE-derived green fluorescence (detected in FL1 channel). For the pHrodo-based assay, macrophages were prepared and incubated with targets as detailed for the microscopy assay, except that targets were labelled with 100 ng ml−1 of pHrodo Green AM Intracellular pH Indicator (Thermo Fisher Scientific, Waltham, Massachusetts, USA), according to the manufacturer’s protocol. pHrodo dyes are non-fluorescent at neutral pH and become fluorescent in acidic environments such as phagolysosome. Once the phagocytosis period was completed, all cells in the well were collected in the presence of Accutase. They were then stained with APC-conjugated anti-F4/80 and analysed by flow cytometry. Phagocytosis efficiency was determined as the percentage of F4/80+ cells containing pHrodo-derived green fluorescence (detected in FL1 channel). For phagocytosis of IgG-opsonized tumour cells, L1210 cells expressing Tac (CD25) were opsonized with anti-Tac monoclonal antibody 7G7 (a mouse IgG2a) for 1 h at 37°C, before the microscopy-based phagocytosis assay. For phagocytosis of C3bi-opsonized tumour cells, L1210 cells were incubated with C5-deficient human serum (Sigma-Aldrich) for 1 h at 37 °C, before the microscopy-based phagocytosis assay. For phagocytosis of apoptotic thymocytes, thymocytes (2 × 107 cells per millilitre) from 4- to 8-week-old C57BL/6 mice were treated at 37 °C for 10 h with 1 µM of dexamethasone (Sigma-Aldrich), which caused about 70% of cells to become apoptotic (annexin V-positive). Cells were then labelled with 2.5 µM of CFSE and incubated with macrophages at a ratio of 20:1 for 30 min at 37 °C. Phagocytosis was monitored by microscopy. For endocytosis of immune complexes, biotinylated mouse IgG (eBioscience) was inactivated at 65 °C for 30 min and mixed with phycoerythrin-coupled streptavidin (eBioscience) on ice for 30 min, to form immune complexes. Then, IgG immune complexes were incubated with macrophages at 37 °C for 30 min. A solution of PBS (pH 2.5) was added to the cell mixture for 1 min at 4 °C, to remove non-endocytosed immune complexes. Cells were subsequently washed, fixed with 4% paraformaldehyde, and analysed by flow cytometry. For phagocytosis of bacteria, GFP-expressing Escherichia coli (DH5α) were cultured overnight at 37 °C. They were then mixed with macrophages at a ratio of 100:1 and incubated at 37 °C for 15 min or 30 min. After washing, the mixture was digested with lysozyme at 37 °C for 30 min, to remove non-phagocytosed bacteria. Then, cells were washed, fixed with 4% paraformaldehyde, and analysed by flow cytometry. For phagocytosis of opsonized sheep red blood cells (sRBCs), sRBCs (MP Biomedicals, Santa Ana, California, USA) were washed with cold PBS and opsonized with rabbit anti-sRBC IgG (MP Biomedicals) for 1.5 h at 37 °C. sRBCs were then labelled with PKH26 (Sigma-Aldrich), according to the manufacturer’s protocol. Macrophages and sRBCs were incubated at a ratio of 1:10 at 37°C for 30 min. After lysing non-phagocytosed sRBCs with RBC lysis buffer (Sigma-Aldrich), macrophages were washed, fixed with 4% paraformaldehyde, and analysed by flow cytometry. For phagocytosis of mouse RBCs, freshly isolated mouse RBCs from WT or CD47 KO mice were labelled with PKH26. After incubating macrophages in serum-free medium for 2 h, 1 × 106 mouse RBCs were added to the macrophages, as detailed for the fluorescence microscopy assay. After incubation for 30 min at 37 °C, non-phagocytosed RBCs were lysed with RBC lysis buffer and macrophages were imaged by fluorescence microscopy. For experiments with pharmacological inhibitors, macrophages were incubated with the following pharmacological inhibitors: Btk family kinase inhibitor, ibrutinib (10 nM; Selleckchem, Burlington, Ontario, Canada); Syk kinase inhibitor, R406 (750 nM; Calbiochem, Burlington, Ontario, Canada); or Src kinase inhibitor, SU6656 (100 nM; 572635; Calbiochem). Inhibitors were added to macrophages 1 h before and during the phagocytosis assay. They had no deleterious impact on cell viability, as verified by staining cells with propidium iodide and annexin V (data not shown).
Intraperitoneal tumour clearance assay
Mice (6–8 weeks old) were injected intraperitoneally with 1.5 ml of 4% (w/v) thioglycollate medium (BD Biosciences). After 4 days, 5 × 106 CFSE-labelled tumour cells (L1210; in 200 µl of PBS) were injected intraperitoneally, in the presence of anti-CD47 or control IgG. After 24 h, cells in the peritoneal cavity were collected using cold PBS washing buffer containing 2% fetal bovine serum and 1 mM EDTA. Numbers of remaining CFSE-positive target cells were quantified by flow cytometry. Numbers of macrophages were also determined, by staining with anti-F4/80. A fixed number of fluorescent beads (1 × 104; 7.58 µm in diameter; Flow Cytometry Absolute Count Standard, Full Spectrum; Bangs Laboratories, Fishers, Indiana, USA) was added to one-twentieth of the cell suspension before flow cytometry. Equivalent numbers of fluorescent beads (2 × 102) were acquired for standardization of cell numbers. In some experiments, mice were injected intraperitoneally on days − 1 and 3 with 200 µl of liposomes containing clodronate or PBS (http://www.clodronateliposomes.com/; Amsterdam, The Netherlands), to deplete macrophages.
Subcutaneous tumour transplantation assay
One million L1210 cells (in some cases, expressing GFP) were injected subcutaneously into the right flank of 6- to 10-week-old RAG-1 KO or RAG-1 SFR dKO mice. RAG-1 KO mice were used to avoid T-cell-mediated rejection of L1210 cells, which are derived from a mouse strain (DBA) different from that of SFR KO mice (C57BL/6). Starting on day 4, mice were injected daily intraperitoneally with 200 µg of control rat IgG2a or rat anti-mouse CD47 (Miap301). Tumour volume was measured every day using a calliper and the formula (length × width2)/2. Antibody treatment was stopped at day 11 and mice were immediately killed. Tumour size limit allowed was 1.5 cm in diameter. Experiments were terminated when or before this size was reached. Tumours were then dissected and weighed. Volumes were also assessed. Then, tumours were sliced into small pieces and pressed through a strainer using the plunger end of a syringe. Cells were washed twice with cold PBS with 2% fetal bovine serum. Total cell numbers were determined, while live cells were enumerated by staining with trypan blue to exclude dead cells. Tumour cells were detected by flow cytometry, using the marker GFP, while immune cells were detected by staining with the relevant antibodies and flow cytometry.
T-cell adoptive transfer
Concanavalin A-activated WT and SLAMF7 KO CD4+ T cells were labelled with 5 µM of CFSE or CellTrace Violet (CTV; Life Technologies), respectively. Then, a 1:1 mixture of WT and SLAMF7 KO T cells was injected intravenously into WT mice, along with control IgG or anti-CD47 antibodies. After 24 h, peripheral blood mononuclear cells were isolated and the presence of CFSE-positive or CTV-positive cells was detected by flow cytometry.
Adhesion assays
For the microscopy-based assay, BMDMs (2 × 105) were labelled with CTV and plated overnight onto cover slips. The next day, target cells (L1210) were labelled with CFSE. Macrophages and target cells were then mixed at a 1:4 ratio in serum-free culture medium and incubated for 30 min at 37 °C to allow conjugate formation. Cells were subsequently washed extensively to remove unconjugated cells. Slides were then analysed by LSM 710 laser-scanning confocal microscope (Carl Zeiss Canada, Toronto, Ontario, Canada). Conjugates between BMDMs and target cells were counted. For the flow cytometry-based assay, macrophages were labelled on ice with APC-conjugated anti-F4/80 monoclonal antibody BM8, while target cells were labelled in a similar way using 0.5 µ M CFSE. After washing, cells were resuspended at a concentration of 2 × 106 cells per millilitre. Conjugate formation was analysed by mixing macrophages with targets, and by incubating the mixture for various periods of time at 37 °C, as described31. Conjugates were detected by flow cytometry.
Stimulation of target cells with anti-CD47 antibodies
To detect apoptosis, 2 × 105 L1210 or P815 cells were incubated in 24-well plates in the presence of 10 µg ml−1 of control rat IgG or rat anti-CD47 antibodies. Cells were collected the next day and cell death was examined by staining with annexin V and propidium iodide, using an Annexin V Apoptosis Detection Kit APC (eBioscience). To measure proliferation, 2 × 105 L1210 or P815 cells were labelled with 5 µM CFSE and incubated overnight in cell culture medium containing 10 µg ml−1 of control rat IgG or rat anti-CD47 antibodies. Cells were collected the next day and CFSE intensity was analysed by flow cytometry. To probe calcium flux, L1210 or P815 cells (2 × 106 cells per sample) were loaded with the calcium indicator dye Indo-1, and then stimulated or not with rat anti-CD47 antibodies and rabbit anti-rat antibodies. Calcium flux was assessed by flow cytometry, using the FL4/FL5 fluorescence ratio. Ionomycin served as positive control. To study protein tyrosine phosphorylation, 2 × 106 L1210 or P815 cells were stimulated or not with rat anti-CD47 antibodies plus rabbit anti-rat antibodies. Protein tyrosine phosphorylation was detected by immunoblotting of total cell lysates with anti-phosphotyrosine antibodies (monoclonal antibody 4G10).
Immunoprecipitations, mass spectrometry, immunoblots
Immunoprecipitations were performed as described42, using Brij99 as detergent in lysis buffer. Mass spectrometry was performed by the Institut de recherches cliniques de Montréal Proteomics Core Facility, as outlined elsewhere43. In brief, immunoprecipitated proteins were digested with trypsin (Promega, Madison, Wisconsin, USA) and analysed by liquid chromatography–tandem mass spectrometry on an LTQ Orbitrap Velos (ThermoFisher Scientific, Bremen, Germany) equipped with a Proxeon nanoelectrospray ion source. A gradient of 100 min was used for liquid chromatography separation, and standard proteomics parameters were used for the mass spectrometers. Protein database searching was performed with Mascot 2.5 (Matrix Science) and data analysis was conducted using Scaffold (version 3.6). The following criteria were used to select potentially relevant SLAMF7 interactors: (1) present in SLAMF7 immunoprecipitates from WT, but not from SFR KO, macrophages; (2) observed in a minimum of four of the five independent SLAMF7 immunoprecipitates from WT macrophages; and (3) a receptor known to regulate macrophage activation. The following criteria were used to select potentially relevant CD11b interactors: (1) present in CD11b immunoprecipitates from WT, but not from CD11b KO, macrophages; (2) observed in a minimum of five of the six independent CD11b immunoprecipitates from WT macrophages; and (3) a receptor known to regulate macrophage function. Immunoblots were performed as reported elsewhere44.
Immunofluorescence
RAW264.7 cells expressing Flag–SLAMF7 or GFP alone (3 × 105) were seeded onto glass cover slips in six-well plates. The next day, cells were fixed in PBS containing 4% formaldehyde. Subsequently, they were washed twice with PBS and permeabilized for 15 min at 4 °C in PBS containing 0.1% Triton X-100. After additional washes, non-specific staining was prevented by blocking for 30 min in PBS supplemented with 5% bovine serum albumin. Cells were washed again and incubated for 1 h with anti-CD11b rabbit monoclonal antibody EPR1344, and anti-Flag mouse monoclonal antibody M2. After further washing, cells were incubated for 1 h with Alexa Fluor 594-coupled goat anti-rabbit IgG and Alexa Fluor 488-coupled goat anti-mouse IgG (Thermo Fisher Scientific). After three additional washes, cover slips were mounted over glass slides using fluorescent mounting medium (Dako, Markham, Ontario, Canada). Data were acquired using a laser-scanning confocal microscope LSM-710. To study actin polarization, BMDMs (2 × 105) were stained with CTV and seeded onto cover slips. The next day, target cells (L1210) were stained with CFSE. Macrophages and targets were then mixed at a 1:4 ratio in serum-free culture medium and incubated for 30 min at 37 °C to allow conjugate formation. Cells were subsequently fixed, washed, and permeabilized, and non-specific staining was blocked, as detailed above for RAW264.7 cells. Then, cells were washed and incubated for 1 h with anti-actin mouse monoclonal antibody AC-74 (Sigma-Aldrich). After further washing, they were incubated for 1 h with Alexa Fluor 594-coupled goat anti-mouse IgG (Thermo Fisher Scientific). Slides were then processed and analysed by confocal microscopy, as detailed above for RAW264.7 cells. Conjugates with full polarization of actin at the area of contact between the macrophage and the target cell were quantitated.
Gene expression dataset analyses
Expression of SLAMF7 and CD47 RNA was analysed using public datasets of human haematological tumours. Normalized data for samples of leukaemia were extracted from the MILE Study45, whereas normalized data for acute myelogenous leukaemia were obtained from The Cancer Genome Atlas (TCGA). In both cases, data were downloaded through Bloodspot46 (http://www.bloodspot.eu). Data for various samples of multiple myeloma were extracted from GSE26760 (ref. 47), using the ‘GEOquery’ R package. Lastly, data for various samples of acute myelogenous leukaemia and diffuse large B-cell lymphoma were directly extracted from TCGA, using the ‘TCGAretriever’ R package. Data were plotted and statistical analyses (unpaired Student’s t-tests (two-tailed)) were performed using the R software package.
Statistical analyses
Except for the gene expression dataset analyses, unpaired Student’s t-tests (two-tailed) were performed using Prism 6 (GraphPad Software). Sample sizes in experiments were chosen on the basis of sizes reported in this field and to achieve statistical significance.
Data availability
Source Data are provided with the online version of the paper. All other data are available from the corresponding authors upon reasonable request.
Extended Data
Extended Data Figure 1. Phagocytosis assays and direct impact of anti-CD47 on target cells.
a, b, Same as Fig. 1b, except phagocytosis assessed by flow cytometry using Tac-expressing L1210 (a) or pHrodo dye (b). Mϕs, macrophages. c, Cell death (in the absence of added macrophages) was examined by staining with annexin V and propidium iodide (PI), and flow cytometry. d, Cell proliferation was studied by CFSE dilution and flow cytometry. MFI, mean fluorescence intensity. e, Ca2+ fluxes were analysed using the Ca2+ indicator dye Indo-1, and flow cytometry. Ionomycin served as positive control. Time of addition of stimuli is shown by arrow. f, Protein tyrosine phosphorylation was detected by anti-phosphotyrosine (pTyr) immunoblotting. Representative of four (a), three (b–d), and two (e, f) independent experiments. Uncropped blots can be seen in Supplementary Fig. 1.
Extended Data Figure 2. Cell-surface receptors on target cells.
a, Expression of CD47 (blue lines); prefixes m, mouse; h, human. Filled curves, isotype controls. b, c, Expression of LRP-1 in BMDMs from LRP-1 KO mice (Lrp1fl/fl;Lys2-Cre) and mice expressing Lys2-Cre alone (as control) was verified by immunoblot (b), while phagocytosis was determined as detailed for Fig. 1d (c). *P < 0.05; **P < 0.01 (two-tailed Student’s t-tests). Results pooled from a total of three (L1210) and two (P815) independent mice (c). Each symbol represents one mouse. All data are means ± s.e.m. Flow cytometry profiles are representative of five (L1210, P815, CB17-3A8, WEHI-3, SP2/0, activated CD4+ T cells, Raji, Daudi), three (MEL, BI-141, EL-4, RMA-S, YAC-1, BW5147.3, B16, CMT-93, L929, thymocytes, resting CD4+ T cells, resting B cells, activated B cells), and two (SW480, SW620, Colo205) experiments (a). Immunoblots are representative of one experiment (deletion of the LRP-1-encoding gene was shown by genotyping in three experiments (data notshown)) (b). Uncropped blots can be seen in Supplementary Fig. 1.
Extended Data Figure 3. SLAM receptors are required for phagocytosis of haematopoietic cells, but not of other targets.
a, Expression of various cell surface markers, including SLAM receptors (blue lines). Filled curves, isotype controls. b, Phagocytosis analysed as detailed for Extended Data Fig. 1a, b, using a flow cytometry-based assay (top) or the pHrodo-based assay (bottom). Left, representative experiments; right, quantification. c, Same as Fig. 1d, using peritoneal macrophages. d, Expression of CD47 (blue lines) on parental and CD47 KO L1210 cells. Filled curves, isotype controls. e, Phagocytosis of IgG-containing immune complexes (I.C.), GFP-expressing E. coli, or IgG-opsonized sRBCs was examined by flow cytometry (blue lines). Filled curves, BMDMs in the absence of phagocytosis. Phagocytosis of apoptotic thymocytes was analysed using microscopy-based assay. f, Phagocytosis of RBCs from WT or CD47 KO mice (mRBCs) was analysed by microscopy. g, Phagocytosis of IgG-opsonized L1210 was analysed. *P < 0.05; **P < 0.01 (two-tailed Student’s t-tests). Results pooled from a total of four (top) and three (bottom) (b), three (left) and two (right) (c), three (e, g), and two (f) mice in independent experiments. Each symbol represents one mouse. All data are means ± s.e.m. Flow cytometry profiles are representative of four (a) and three (d; e, left) independent experiments.
Extended Data Figure 4. Tumour transplantation assays.
a, b, From the experiment depicted in Fig. 2e. Cells were analysed by flow cytometry, in the presence of a fixed number of fluorescent beads to allow quantitation of total cell numbers. Beads are boxed in R1, while L1210 are boxed in R2 (a). Numbers of peritoneal macrophages were determined (b). c, Schematic representation of the experiment presented in Fig. 2f. TG, thioglycollate. d, e, From experiment depicted in Fig. 2f. Cells were analysed as specified for a and b. f, g, Tumours from experiment depicted in Fig. 2g were dissected, weighed, measured, and analysed by flow cytometry. Two RAG-1 KO mice treated with anti-CD47 (mice 9 and 10) showed no clinically detectable tumour when alive. However, upon dissection, small nodules with no detectable mass on the scale were present. These nodules were processed and analysed as for the other tumours. Masses are denoted as ‘0’ in the Source Data. L1210 were GFP+; macrophages were Ly6G−CD11b+NK1.1−; neutrophils were Ly6G+CD11b+NK1.1−; and NK cells were Ly6G−CD11b+NK1.1+. *P < 0.05; **P < 0.01; ***P < 0.001 (two-tailed Student’s t-tests). Results pooled from a total of six mice analysed in five independent experiments (b), two mice (e), or 11 mice in two of four independent experiments (f, g). Each symbol represents one mouse. All data are means ± s.e.m. Dot plots are representative of six (a) or two (d) independent mice. See Source Data.
Extended Data Figure 5. Impact of SLAMF7 on phagocytosis.
a, Expression of various cell surface markers, including SLAMF7 (blue lines). Filled curves, isotype controls. b, Phagocytosis was tested as detailed in Fig. 1d. For human targets (Raji, Daudi), F(ab′)2 fragments of antibodies were used. c, BAC transgenic mice expressing SLAMF7 were generated as detailed in Methods. In brief, the C57BL/6 BAC clone was first truncated at the 3′ end to eliminate the Slamf1 gene. Then, a stop codon (denoted by ‘X’) was introduced in exon 2 of Slamf2, the gene coding for CD48, and a silent mutation (HindIII site; denoted by short vertical red line) was created in Slamf7 to allow screening of transgenic mice. The transcriptional orientation of the Slam genes is depicted by arrows, while the relative positions of the genes in the clone are indicated by their distances from the 5′ end (in kilobases). d, Expression of various cell surface markers, including SLAMF7 (blue lines). Filled curves, isotype controls. *P < 0.05; **P < 0.01; ***P < 0.001 (two-tailed Student’s t-tests). Results pooled from a total of four mice studied in independent experiments (b). Each symbol represents one mouse. All data are means ± s.e.m. Flow cytometry profiles are representative of four (a) and three (d) independent experiments.
Extended Data Figure 6. Expression of SLAMF7 on target cells.
a, b, Expression of various cell surface markers, including SFRs and their ligands (blue lines); prefixes m, mouse; h, human. Filled curves, isotype controls. c, Phagocytosis of L1210 by SFR KO macrophages expressing GFP alone or with SLAMF7 (human or mouse). Left, representative flow cytometry analysis; right, quantification. d, Phagocytosis of L1210 by macrophages from WT C57BL/6 or NRG mice, in the presence of anti-mSLAMF7 monoclonal antibody 4G2 or control rat IgG. Results pooled from a total of three (c), and three (C57BL/6) or two (NRG) (d) independent mice. Each symbol represents one mouse. All data are means ± s.e.m. Flow cytometry profiles are representative of three (MEL, BI-141, EL-4, RMA-S, YAC-1, BW5147.3, B16, CMT-93, L929, thymocytes, resting CD4+ T cells, resting B cells, activated B cells) or two (SW480, SW620, Colo205) (a), and four (b) independent experiments.
Extended Data Figure 7. Mechanism of action of SLAMF7.
a, Formation of conjugates (boxed) between macrophages and L1210 was detected by flow cytometry. Left, representative experiment. The percentages of conjugate formation are indicated above the boxes. Right, quantification. b, Same as Fig. 1d, using EAT-2 KO macrophages and L1210. c, Same as Fig. 1d, using WT or SFR KO macrophages and L1210, in the presence of kinase inhibitors. d, e, Expression of various cell surface markers (blue lines); filled curves, control antibodies. Phagocytosis of IgG-opsonized L1210 was analysed as detailed for Extended Data Fig. 3g. *P < 0.05; **P < 0.01; ***P < 0.001 (two-tailed Student’s t-tests). Results pooled from a total of three (a–c) or two (d, e) mice studied in independent experiments. Each symbol represents one mouse. All data are means ± s.e.m. Flow cytometry profiles representative of two independent experiments (d, left; e, left).
Extended Data Figure 8. Impact of FcRγ and DAP12 on phagocytosis.
a–e, Same as Fig. 1d and Extended Data Fig. 3g, using macrophages from DAP12 KO (a, b), FcRγ KO (c, d), or dKO (e) mice and L1210. a, c, Top, representative anti-DAP12 and anti-FcRγ immunoblots. **P < 0.01; ***P < 0.001 (two-tailed Student’s t-tests). Results pooled from a total of three (a, bottom), two (b, bottom; d, right; e, right), and five (c, bottom) mice studied in independent experiments. Each symbol represents one mouse. All data are means ± s.e.m. Flow cytometry profiles are representative of two independent experiments (b, top; d, left; e, left). Uncropped blots can be seen in Supplementary Fig. 1.
Extended Data Figure 9. Influence of integrins on phagocytosis.
a, Mass spectrometry analysis of anti-SLAMF7 immunoprecipitates. The GenInfo Identifier (gi) accession number and means of the normalized total ion current (TIC) for the potential interactors are shown. b, Same as a, except that CD11b was immunoprecipitated. c, Same as b, except that the data for FcRs CD64 and CD16 are shown. d, Expression of SLAMF7 and Flag (blue lines); filled curves, isotype control antibodies. e, Phagocytosis of L1210 was analysed as detailed for Fig. 1d. f, Expression of various cell surface markers (blue lines); filled curves, isotype control antibodies. g, h, Phagocytosis of L1210 opsonized with C3bi or IgG was analysed as detailed for Extended Data Fig. 3g. *P < 0.05; **P < 0.01 (two-tailed Student’s t-tests). Results pooled from two experiments with a total of five (a) or six (b, c) immunoprecipitates from independent mice, or three (e, g, h) independent experiments. Each symbol represents one mouse. All data are means ± s.e.m. Flow cytometry profiles are representative of four (d) or three (f) independent experiments.
Extended Data Figure 10. Gene expression analyses of SLAMF7 and CD47.
Expression of SLAMF7 and CD47 RNA in human haematological tumours. a, RNA levels of SLAMF7 (top) and CD47 (bottom) in several types and subtypes of leukaemia were analysed, using data obtained from microarray experiments. Data for only one oligonucleotide probe are shown. However, similar findings were made with other SLAMF7 and CD47 probes (data not shown). Each symbol represents a different patient sample. Median expression for a given type or subtype of malignancy is depicted by a horizontal line. For statistical analysis, Student’s t-tests were performed comparing SLAMF7 expression in the combination of all acute myelogenous leukaemia (AML) and acute lymphocytic leukaemia (ALL), versus either myelodysplastic syndrome (MDS) or chronic lymphocytic leukaemia (CLL). CML, chronic myelogenous leukaemia. b, Same as a, except that samples of multiple myeloma (MM) were analysed. c, Same as a, except that samples of acute myelogenous leukaemia and diffuse large B-cell lymphoma (DLBCL) were studied. Moreover, RNA expression was quantitated by RNA sequencing. d, Levels of SLAMF7 and CD47 RNAs for individual samples from selected tumour types, which displayed higher levels of SLAMF7 RNA, were analysed in parallel using dot plots. ***P < 0.001. The values of n, from left to right, are (a) MILE Study: 38, 41, 37, 28, 48, 352, 70, 237, 122, 13, 40, 36, 58, 174, 206, 76, 448; AML TCGA: 4, 20, 16, 91, 27, 6, 14, 1, 14, 3, 17, 3, 5, 7, 7, 6, 1, 2; (b) multiple myeloma: 304; (c) TCGA AML: 173; TCGA diffuse large B-cell lymphoma: 48; (d) 13, 206, 448, 20, 14, 17, 304, 173, 48.
Supplementary Material
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (CIHR; MT-14429, MOP-82906, FDN-143338) and the Canadian Cancer Society Research Institute (CCSRI; grant 018114) to A.V., RO1AI65495, RO1AI68150 and RO1AI113272 from the National Institutes of Health to C.A.L., and the Natural Sciences and Engineering Research Council (NSERC 1050319) to J.S.D. J.C. is recipient of a Fellowship from the Cole Foundation, while S.M. is a fellowship recipient of the RDV Foundation and N.W. was recipient of a Fellowship from Fonds de la recherche du Québec – Santé (FRQ-S). D.C.V. is a Chercheur-clinicien boursier of FRQ-S. J.S.D. holds the Anne and Max Tanenbaum Chair in Molecular Medicine, University of Toronto. A.V. holds the Canada Research Chair in Signaling in the Immune System.
Footnotes
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Information is available in the online version of the paper.
Author Contributions J.C. planned and performed most experiments, interpreted data, and wrote the manuscript. M.-C.Z. planned and performed mouse tumour formation experiments (with J.C.), and interpreted data. M.-C.Z., H.G., and D.D. together confirmed key results of phagocytosis assays, and interpreted data. H.G. performed the assays of anti-CD47 effects on calcium fluxes and protein tyrosine phosphorylation. D.D. performed the microscopy-based conjugate formation assay (with J.C.). H.G., Y.L., and L.-A.P.-Q. generated and characterized reagents, and interpreted data. S.M., I.R., S.Z., M.-E.C.-M., N.W., D.C.V., and M.S. characterized reagents and interpreted data. V.C. performed the bioinformatics analyses of SLAMF7 and CD47 expression C.A.L. and J.S.D. planned experiments, interpreted data, revised the manuscript, and provided funding. A.V. planned experiments, generated reagents, interpreted data, wrote the manuscript, and obtained funding. All authors commented on the manuscript.
The authors declare competing financial interests: details are available in the online version of the paper. Readers are welcome to comment on the online version of the paper. Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Korman AJ, Peggs KS, Allison JP. Checkpoint blockade in cancer immunotherapy. Adv. Immunol. 2006;90:297–339. doi: 10.1016/S0065-2776(06)90008-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161:205–214. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arandjelovic S, Ravichandran KS. Phagocytosis of apoptotic cells in homeostasis. Nat. Immunol. 2015;16:907–917. doi: 10.1038/ni.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon S, Plüddemann A, Martinez Estrada F. Macrophage heterogeneity in tissues: phenotypic diversity and functions. Immunol. Rev. 2014;262:36–55. doi: 10.1111/imr.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schultze JL, Schmidt SV. Molecular features of macrophage activation. Semin. Immunol. 2015;27:416–423. doi: 10.1016/j.smim.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Freeman SA, Grinstein S. Phagocytosis: receptors, signal integration, and the cytoskeleton. Immunol. Rev. 2014;262:193–215. doi: 10.1111/imr.12212. [DOI] [PubMed] [Google Scholar]
- 7.Chao MP, et al. Anti-CD47 antibody synergizes with rituximab to promote phagocytosis and eradicate non-Hodgkin lymphoma. Cell. 2010;142:699–713. doi: 10.1016/j.cell.2010.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Majeti R, et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138:286–299. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaiswal S, et al. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. 2009;138:271–285. doi: 10.1016/j.cell.2009.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Theocharides AP, et al. Disruption of SIRP α signaling in macrophages eliminates human acute myeloid leukemia stem cells in xenografts. J. Exp. Med. 2012;209:1883–1899. doi: 10.1084/jem.20120502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murata Y, Kotani T, Ohnishi H, Matozaki T. The CD47-SIRP α signalling system: its physiological roles and therapeutic application. J. Biochem. 2014;155:335–344. doi: 10.1093/jb/mvu017. [DOI] [PubMed] [Google Scholar]
- 12.Barclay AN, Van den Berg TK. The interaction between signal regulatory protein alpha (SIRPα) and CD47: structure, function, and therapeutic target. Annu. Rev. Immunol. 2014;32:25–50. doi: 10.1146/annurev-immunol-032713-120142. [DOI] [PubMed] [Google Scholar]
- 13.Chao MP, Weissman IL, Majeti R. The CD47-SIRPα pathway in cancer immune evasion and potential therapeutic implications. Curr. Opin. Immunol. 2012;24:225–232. doi: 10.1016/j.coi.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho JM, Danska JS, Wang JC. Targeting SIRP α in cancer. OncoImmunology. 2013;2:e23081. doi: 10.4161/onci.23081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu N, Veillette A. SLAM family receptors in normal immunity and immune pathologies. Curr. Opin. Immunol. 2016;38:45–51. doi: 10.1016/j.coi.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Cannons JL, Tangye SG, Schwartzberg PL. SLAM family receptors and SAP adaptors in immunity. Annu. Rev. Immunol. 2011;29:665–705. doi: 10.1146/annurev-immunol-030409-101302. [DOI] [PubMed] [Google Scholar]
- 17.Calpe S, et al. The SLAM and SAP gene families control innate and adaptive immune responses. Adv. Immunol. 2008;97:177–250. doi: 10.1016/S0065-2776(08)00004-7. [DOI] [PubMed] [Google Scholar]
- 18.ffrench-Constant C, Colognato H. Integrins: versatile integrators of extracellular signals. Trends Cell Biol. 2004;14:678–686. doi: 10.1016/j.tcb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Miranti CK, Brugge JS. Sensing the environment: a historical perspective on integrin signal transduction. Nat Cell Biol. 2002;4:E83–E90. doi: 10.1038/ncb0402-e83. [DOI] [PubMed] [Google Scholar]
- 20.Arnaout MA, Mahalingam B, Xiong JP. Integrin structure, allostery, and bidirectional signaling. Annu. Rev. Cell Dev. Biol. 2005;21:381–410. doi: 10.1146/annurev.cellbio.21.090704.151217. [DOI] [PubMed] [Google Scholar]
- 21.Jakus Z, Fodor S, Abram CL, Lowell CA, Mócsai A. Immunoreceptor-like signaling by β2 and β3 integrins. Trends Cell Biol. 2007;17:493–501. doi: 10.1016/j.tcb.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Hamerman JA, Ni M, Killebrew JR, Chu CL, Lowell CA. The expanding roles of ITAM adapters FcRγ and DAP12 in myeloid cells. Immunol. Rev. 2009;232:42–58. doi: 10.1111/j.1600-065X.2009.00841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chao MP, et al. Calreticulin is the dominant pro-phagocytic signal on multiple human cancers and is counterbalanced by CD47. Sci. Transl. Med. 2010;2:63ra94. doi: 10.1126/scitranslmed.3001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veillette A, Guo H. CS1, a SLAM family receptor involved in immune regulation, is a therapeutic target in multiple myeloma. Crit. Rev. Oncol. Hematol. 2013;88:168–177. doi: 10.1016/j.critrevonc.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Guo H, Cruz-Munoz ME, Wu N, Robbins M, Veillette A. Immune cell inhibition by SLAMF7 is mediated by a mechanism requiring Src kinases, CD45, and SHIP-1 that is defective in multiple myeloma cells. Mol. Cell. Biol. 2015;35:41–51. doi: 10.1128/MCB.01107-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rawlings DJ, et al. Mutation of unique region of Bruton’s tyrosine kinase in immunodeficient XID mice. Science. 1993;261:358–361. doi: 10.1126/science.8332901. [DOI] [PubMed] [Google Scholar]
- 27.Takai T, Li M, Sylvestre D, Clynes R, Ravetch JV. FcRγ chain deletion results in pleiotrophic effector cell defects. Cell. 1994;76:519–529. doi: 10.1016/0092-8674(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 28.Todd RF., III The continuing saga of complement receptor type 3 (CR3) J. Clin. Invest. 1996;98:1–2. doi: 10.1172/JCI118752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo H, et al. Deletion of Slam locus in mice reveals inhibitory role of SLAM family in NK cell responses regulated by cytokines and LFA-1. J. Exp. Med. 2016;213:2187–2207. doi: 10.1084/jem.20160552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu N, et al. A hematopoietic cell-driven mechanism involving SLAMF6 receptor, SAP adaptors and SHP-1 phosphatase regulates NK cell education. Nat. Immunol. 2016;17:387–396. doi: 10.1038/ni.3369. [DOI] [PubMed] [Google Scholar]
- 31.Dong Z, et al. The adaptor SAP controls NK cell activation by regulating the enzymes Vav-1 and SHIP-1 and by enhancing conjugates with target cells. Immunity. 2012;36:974–985. doi: 10.1016/j.immuni.2012.03.023. [DOI] [PubMed] [Google Scholar]
- 32.Elliott ER, et al. Deletion of Syk in neutrophils prevents immune complex arthritis. J. Immunol. 2011;187:4319–4330. doi: 10.4049/jimmunol.1100341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaifu T, et al. Osteopetrosis and thalamic hypomyelinosis with synaptic degeneration in DAP12-deficient mice. J. Clin. Invest. 2003;111:323–332. doi: 10.1172/JCI16923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mócsai A, et al. Integrin signaling in neutrophils and macrophages uses adaptors containing immunoreceptor tyrosine-based activation motifs. Nat. Immunol. 2006;7:1326–1333. doi: 10.1038/ni1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhee I, Davidson D, Souza CM, Vacher J, Veillette A. Macrophage fusion is controlled by the cytoplasmic protein tyrosine phosphatase PTP-PEST/ PTPN12. Mol. Cell. Biol. 2013;33:2458–2469. doi: 10.1128/MCB.00197-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong Z, et al. Essential function for SAP family adaptors in the surveillance of hematopoietic cells by natural killer cells. Nat. Immunol. 2009;10:973–980. doi: 10.1038/ni.1763. [DOI] [PubMed] [Google Scholar]
- 37.Chow LM, et al. Two distinct protein isoforms are encoded by ntk, a csk-related tyrosine protein kinase gene. Oncogene. 1994;9:3437–3448. [PubMed] [Google Scholar]
- 38.Abraham N, Miceli MC, Parnes JR, Veillette A. Enhancement of T-cell responsiveness by the lymphocyte-specific tyrosine protein kinase p56lck. Nature. 1991;350:62–66. doi: 10.1038/350062a0. [DOI] [PubMed] [Google Scholar]
- 39.Bouchon A, Cella M, Grierson HL, Cohen JI, Colonna M. Activation of NK cell-mediated cytotoxicity by a SAP-independent receptor of the CD2 family. J. Immunol. 2001;167:5517–5521. doi: 10.4049/jimmunol.167.10.5517. [DOI] [PubMed] [Google Scholar]
- 40.Latour S, Chow LM, Veillette A. Differential intrinsic enzymatic activity of Syk and Zap-70 protein-tyrosine kinases. J. Biol. Chem. 1996;271:22782–22790. doi: 10.1074/jbc.271.37.22782. [DOI] [PubMed] [Google Scholar]
- 41.Cruz-Munoz ME, Dong Z, Shi X, Zhang S, Veillette A. Influence of CRACC, a SLAM family receptor coupled to the adaptor EAT-2, on natural killer cell function. Nat. Immunol. 2009;10:297–305. doi: 10.1038/ni.1693. [DOI] [PubMed] [Google Scholar]
- 42.Heit B, et al. Multimolecular signaling complexes enable Syk-mediated signaling of CD36 internalization. Dev. Cell. 2013;24:372–383. doi: 10.1016/j.devcel.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cloutier P, Lavallée-Adam M, Faubert D, Blanchette M, Coulombe B. Methylation of the DNA/RNA-binding protein Kin17 by METTL22 affects its association with chromatin. J. Proteomics. 2014;100:115–124. doi: 10.1016/j.jprot.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 44.Veillette A, Bookman MA, Horak EM, Bolen JB. The CD4 and CD8 T cell surface antigens are associated with the internal membrane tyrosine-protein kinase p56lck. Cell. 1988;55:301–308. doi: 10.1016/0092-8674(88)90053-0. [DOI] [PubMed] [Google Scholar]
- 45.Kohlmann A, et al. An international standardization programme towards the application of gene expression profiling in routine leukaemia diagnostics: the Microarray Innovations in LEukemia study prephase. Br. J. Haematol. 2008;142:802–807. doi: 10.1111/j.1365-2141.2008.07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bagger FO, et al. BloodSpot: a database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res. 2016;44(D1):D917–D924. doi: 10.1093/nar/gkv1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source Data are provided with the online version of the paper. All other data are available from the corresponding authors upon reasonable request.