Abstract
Epigenetics studies genomic modifications carrying information independent of DNA sequence heritable through cell division. In 1940, Waddington coined the term “epigenetic landscape” as a metaphor for pluripotency and differentiation, but methylation landscapes have not yet been rigorously computed. By using principles of statistical physics and information theory, we derive epigenetic energy landscapes from whole-genome bisulfite sequencing data that allow us to quantify methylation stochasticity genome-wide using Shannon’s entropy and associate entropy with chromatin structure. Moreover, we consider the Jensen-Shannon distance between sample-specific energy landscapes as a measure of epigenetic dissimilarity and demonstrate its effectiveness for discerning epigenetic differences. By viewing methylation maintenance as a communications system, we introduce methylation channels and show that higher-order chromatin organization can be predicted from their informational properties. Our results provide a fundamental understanding of the information-theoretic nature of the epigenome that leads to a powerful approach for studying its role in disease and aging.
INTRODUCTION
In his seminal work, Waddington employed deterministic differential equations to define epigenetics as the emergence of a phenotype that can be perturbed by the environment but whose endpoints are predetermined by genes1. However, growing appreciation for the role of epigenetic stochasticity in development and disease2–5 has led to simple probabilistic models of epigenetic landscapes that account for randomness in DNA methylation by adding a “noise” term to deterministic models6,7. Some authors have also characterized methylation stochasticity using the notion of epipolymorphism4,5, a form of non-additive Tsallis entropy whose measurement is limited to a small portion of the genome and can underestimate heterogeneity in WGBS data (Supplementary Note).4,5
Here we take a foundational approach to understanding the nature of epigenetic information using principles of statistical physics and information theory that organically incorporate stochasticity into the mathematical framework, and apply this approach on diverse whole-genome bisulfite sequencing (WGBS) datasets. In contrast to metaphorical “Waddingtonian” landscapes, we present a rigorous derivation of epigenetic potential energy landscapes that encapsulate the higher-order statistical properties of methylation, fully capturing behavior that is opaque to customary mean-based summaries.
We quantify methylation stochasticity using Shannon’s entropy and provide a powerful information-theoretic methodology for distinguishing epigenomes using the Jensen-Shannon distance between sample-specific energy landscapes associated with stem cells, tissue lineages and cancer. Moreover, we establish a relationship between entropy and topologically associating domains that allows prediction of their boundaries from WGBS samples. We also introduce methylation channels as models of DNA methylation maintenance and show that their informational properties can effectively predict high-order chromatin organization using machine learning. Lastly, we introduce a sensitivity index that quantifies the rate by which environmental perturbations influence methylation stochasticity along the genome.
This merger of epigenetic biology and statistical physics yields many fundamental insights into the relationship between information-theoretic properties of the epigenome and nuclear organization in normal development and disease. Most importantly, it provides novel methods for evaluating informational properties of individual samples and their chromatin structure and for quantifying differences between tissue lineages, aging, and cancer at high resolution genome-wide.
RESULTS
Stochastic epigenetic variation and energy landscapes
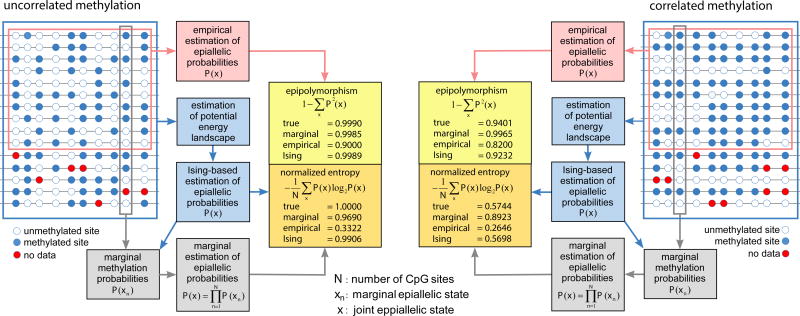
Currently available methods for methylation analysis are predominantly limited to modeling stochastic variation at individual CpG sites while ignoring statistical dependence among neighboring sites8. However, fully characterizing the stochastic and polymorphic nature of epigenetic information requires knowledge of the probability distribution of methylation patterns (epialleles) formed by groups of CpG sites4,5. Presently, this distribution is estimated empirically requiring much higher coverage than what is routinely available in WGBS data (Fig. 1 and Supplementary Note).
Figure 1.
Estimation of epiallelic probabilities, epipolymorphisms, and normalized epiallelic entropies. Multiple WGBS reads within a genomic region are used to form a methylation matrix whose entries represent the methylation status of each CpG site (blue: methylated, white: unmethylated, red: no data). Most methods for methylation analysis estimate marginal probabilities at individual CpG sites using only data within each column of the methylation matrix, which can then be employed to estimate epiallelic probabilities by assuming statistical independence. Empirical estimation of epiallelic probabilities uses only fully observed rows of the methylation matrix, whereas estimation of these probabilities using an Ising potential energy landscape employs all data available in the methylation matrix. At low levels of correlation, a marginal approach to estimating epiallelic probabilities may provide accurate estimation of epipolymorphisms and entropies. However, when high correlation is present, only the Ising-based approach can provide accurate estimates of epipolymorphisms and entropies, while the marginal approach will overestimate these quantities. In this example, empirical estimation of epiallelic probabilities underestimates the true values of epipolymorphisms and entropies regardless of correlation level.
To remedy this problem and better understand the relationship between epigenetic stochasticity and phenotypic variability, we employed an approach based on statistical physics and information theory. We represented methylation within a genomic region containing N CpG sites by a random vector X = [X1, X2,…, XN], where Xn takes value 1 or 0 depending on whether the n-th CpG site is methylated or unmethylated. We then modeled X using the Boltzmann-Gibbs distribution
| (1) |
where U(x) is the energy of the methylation pattern x and
| (2) |
is the partition function.
The function V(x) = U(x) − U(x*), where x* is a methylation pattern with the least energy (ground state), defines a potential energy landscape whose elevation (potential) provides a measure of the improbability of finding the methylation pattern x relative to the most likely pattern x*. This landscape possesses one or more “potential wells” corresponding to local minima of U(x) with each well associated with an attractor representing the most probable methylation pattern to be found within a genomic region among all patterns associated with the well (Fig. 2a). Demethylation and de novo methylation allow methylation patterns to be modified with higher probability for changes that move patterns towards lower potential energy. At steady-state, a genomic region can be associated with a “cloud” of methylation patterns that fluctuate around an attractor, resulting in pattern variability controlled by the width of the potential well. Notably, a potential energy landscape could be associated with two distinct attractors producing “bistable” behavior (Fig. 2a). DNA methylation is subject to this type of behavior, which was found to be associated with gene imprinting (Supplementary Note).
Figure 2.
Potential energy landscapes. (a) Hypothetical monostable and bistable potential energy landscapes illustrating the presence of potential wells that correspond to attractors (green balls), and associated clouds of methylation patterns (brown balls). The magenta balls indicate unlikely methylation patterns drawn towards lower potential energies during maintenance. (b) Potential energy landscapes associated with twelve CpG sites within the CGIs of WNT1 in colon normal and colon cancer and within the CGI of EPHA4 in stem and brain. Each point in the domain of the potential energy landscape marks a methylation pattern, with the point at (0,0) indicating the fully unmethylated state, which is the ground state in both examples. The 212 potential values are distributed over a 64×64 square grid using a 2D version of the Gray code (Online Methods). Violin plots summarize distributions of potential values.
Using the maximum-entropy principle9, we determined an energy function that is consistent with methylation means and “nearest-neighbor” correlations, given by
| (3) |
where α, β, γ are parameters characteristic to the genomic region, ρn is the CpG density, and dn is the distance between CpG sites n and n − 1, leading to the 1D Ising model of statistical physics that takes into account non-cooperative and cooperative factors in methylation (Supplementary Note). This choice encapsulates the notion that methylation depends on two distinct factors: the CpG architecture of the genome, quantified by the CpG densities and distances, and the local biochemical environment provided by the methylation machinery, quantified by parameters α, β, γ. Moreover, it allows computation of potential energy landscapes, joint probabilities of methylation patterns, marginal probabilities at individual CpG sites, and a number of novel measures for methylation analysis (Supplementary Note). Simulated data provided clear evidence that, in contrast to empirically estimating epiallelic probabilities, methylation pattern analysis using the Ising model can consistently produce accurate results using relatively low coverage data (Fig. 1 and Supplementary Note).
Using a maximum-likelihood approach (Online Methods), we estimated methylation potential energy landscapes from WGBS data corresponding to 35 diverse samples that allowed for detailed local profiling throughout the methylome (Supplementary Table 1). This analysis, for example, asserts that most methylation patterns associated with the CpG island (CGI) of WNT1 in normal colon exhibit high potential values (Fig. 2b), implying little variability from the pattern with the lowest potential (attractor), which coincides with the fully unmethylated pattern in this case. The accompanying violin plot shows that any deviation from the attractor towards a pattern of higher potential will rapidly be “funneled” back towards the attractor, leading to low methylation stochasticity. However, most methylation patterns in colon cancer manifest much lower potential values than in normal colon (Fig. 2b), implying a significant gain in pattern variability and increased methylation stochasticity in cancer. Similarly, most methylation patterns associated with the CGI of EPHA4, a key developmental gene, had low potential values in stem cells (Fig. 2b), implying significant pattern variability from the attractor, which also coincides with the fully unmethylated state. In contrast, EPHA4 shows higher potential values in the brain (Fig. 2b), yielding lower pattern variability and lower methylation stochasticity than in stem cells.
Epigenetic entropy comprehensively quantifies methylation stochasticity
To facilitate genome-wide analysis of methylation information, we partitioned the genome into non-overlapping genomic units and performed methylation analysis at a resolution of one genomic unit. Consistent with the length of DNA within a nucleosome (~146 bp), we chose genomic units of 150 bp each, which strikes a balance between leveraging as much information as possible within a genomic unit and performing high-resolution methylation analysis. We then quantified methylation within each genomic unit using the methylation level (average methylation), whose probability distribution is calculated from the Ising model, and used its mean and normalized Shannon entropy to measure methylation stochasticity (Online Methods).
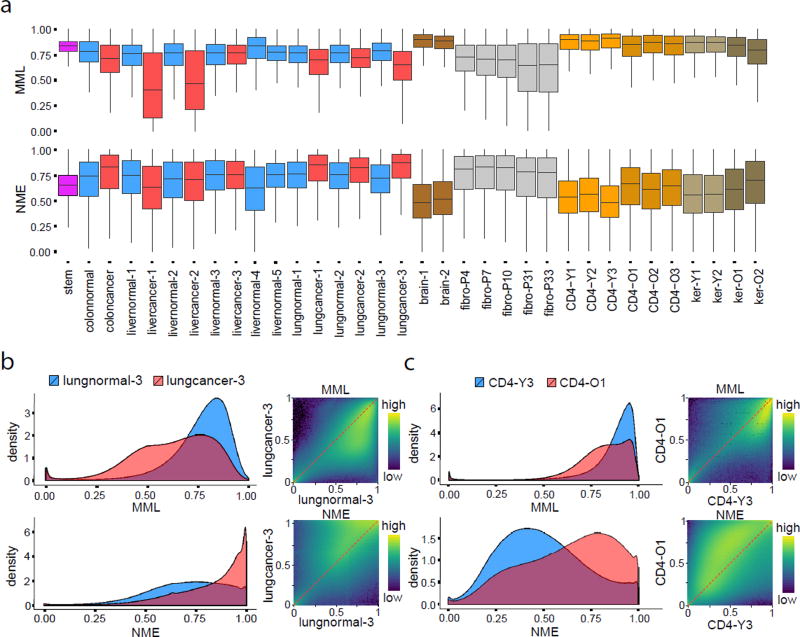
In agreement with the literature10, the mean methylation level was globally higher in stem cells and brain tissues than in normal colon, lung and four livers, and the same was true for CD4+ lymphocytes and skin keratinocytes, while it was reduced in cancer and was progressively lost in cell culture (Fig. 3a,b). We also observed low normalized methylation entropy in stem and brain cells and CD4+ lymphocytes and skin keratinocytes associated with young subjects, and a global increase of entropy in most cancers but not in two liver cancer samples with profound hypomethylation accompanied by a less entropic methylation state (Fig. 3a). While differential entropy changes in cancer were often associated with changes in mean level (Supplementary Fig. 1a), this was not always true (Supplementary Fig. 1b,c), proving that changes in stochasticity are not necessarily related to changes in mean methylation, which demands that both be assessed when interrogating biological samples (Supplementary Fig. 2). Furthermore, genome-wide mean level and entropy distributions over selected genomic features demonstrate lower and more variable values within CGIs and transcription start sites (TSSs) compared to other genomic features, such as shores, exons, introns, etc. (Supplementary Fig. 3a,b).
Figure 3.
Mean methylation level and normalized entropy. (a) Boxplots of genome-wide distributions of mean methylation level (MML) and normalized methylation entropy (NME) values in all samples used in this study. The boxes show the 25% quantile, the median, and the 75% quantile, whereas each whisker has a length of 1.5× the interquartile range. (b) Genome-wide 1D and 2D log10 transformed MML and NME densities associated with lung normal/cancer show global MML loss in cancer accompanied by gain in entropy. (c) CD4+ lymphocytes from older subjects exhibit global loss of MML, accompanied with gain in entropy.
Consistent with a previous analysis comparing newborns to centenarians11, global hypomethylation was found in all three CD4 samples from older people compared to three samples from younger individuals, and the same was true in skin keratinocytes (Fig. 3a,c). This was accompanied by gain in methylation entropy, with changes more pronounced in entropy than mean (Fig. 3a,c). Although passage number in fibroblasts was also associated with global hypomethylation, this was more pronounced than in CD4 samples, whereas entropy loss was globally observed at later passages (Fig. 3a), in stark contrast to global gain in entropy observed in old CD4 samples. We also assessed changes in methylation stochasticity while accounting for biological, statistical, and technical variability, which confirmed that aged CD4 cells often exhibit changes in entropy (Supplementary Note). Using the Jensen-Shannon informational distance (Online Methods), we investigated differences between young and old in the CD4 samples, as well as dissimilarities with passage in cultured fibroblasts. Our results (Supplementary Note) suggest that increasing fibroblast culture passage inaccurately models methylation stochasticity in aging, which is associated with global gain in informational dissimilarity, driven by increased entropy.
Informational distances delineate lineages and identify developmentally critical genes
Previous studies indicated that epigenetic discordance within gene regulatory elements, such as enhancers, might fully account for observed epigenetic dissimilarities between two samples12,13. However, by computing genome-wide distributions of Jensen-Shannon distances within several genomic features, we did not find consistent containment of epigenetic dissimilarity within a particular feature (Supplementary Fig. 4). Therefore, to understand the relationship between epigenetic information and phenotypic variation, we used the Jensen-Shannon informational distance to precisely quantify epigenetic discordance between pairs of samples (Online Methods). We then asked if we could distinguish colon, lung, and liver from each other and from matched cancers, as well as from stem, brain, and CD4+ lymphocytes, for computational feasibility limiting to 17 representative samples and visualizing using multidimensional scaling (Online Methods). We found the samples falling into clear categories based on developmental germ layers, equidistant from stem cells, and with cancers well separated from normal (Fig. 4).
Figure 4.
Informational distances and lineages. Visualization of genomic dissimilarity between seventeen diverse cell and tissue samples using multidimensional scaling (equally scaled axes with a horizontal break), evaluated using the Jensen-Shannon distance, displays grouping of samples into clear categories based on lineage. Endoderm (colon, lung, liver), mesoderm (CD4), and ectoderm (brain) derived tissues are located roughly equidistant from stem cells (dashed circle). Cancerous tissues are well separated from normal tissues and stem cells, with two liver cancers being far removed from their matched normal counterparts.
Given the interesting relationship between the stem cell sample and the three germ layers, we examined genes that had differences in mean methylation level in stem cells compared to differentiated tissues and genes that possessed epigenetic discordance quantified by the Jensen-Shannon distance. We ranked genes based on absolute differences in mean methylation level within their promoters, as well as using the Jensen-Shannon distance (Supplementary Table 2 and Online Methods). Some genes known to be involved in development and differentiation (e.g., FOXD3, SALL1, SOX2, and ZIC1 when comparing stem to lung) had relatively small changes in mean methylation yet large Jensen-Shannon distances, affirming that the probability distributions of methylation levels within their promoters were different, despite little differences in mean. We further explored whether non-mean related methylation differences could identify genes between sample groups that are occult to existing mean-based analyses by employing a relative ranking scheme that assigned higher score to genes with larger Jensen-Shannon distance but smaller absolute change in mean methylation level (Online Methods). In the stem cell to brain comparison, for example, many key genes (IGF2BP1, FOXD3, NKX6-2, SALL1, EPHA4, ASCL2, and OTX1) topped the relative ranking list (Supplementary Table 2a), with GO enrichment analysis14 identifying key process categories associated with stem cell maintenance and brain cell development (Supplementary Table 3a). Moreover, 30 significant GO process categories using the relative scheme showed 10-fold or greater enrichment, compared to 5 GO categories using the mean-based scheme. We obtained similar results when comparing stem cells to lung, with the relative scheme identifying key developmental processes and genes in both mesodermal and stem cell categories (Supplementary Tables 2b, 3b). Comparing stem cells to CD4+ lymphocytes uncovered enrichment for immune-related functions, driven by differential mean level, and many developmental and morphogenesis process categories, predominantly driven by the Jensen-Shannon distance (Supplementary Tables 2c, 3c).
When comparing differentiated tissues, mean-based GO analysis resulted in highly enriched process categories, mostly related to differentiated function, such as cellular regulation and signaling. However, the relative scheme resulted in highly enriched categories largely related to development and differentiation (Supplementary Tables 2d–f, 3d–f), likely due to relative rankings being low for genes that are only methylated in one cell type. When comparing lung normal to cancer, the relative scheme produced a larger number of highly enriched process categories than mean-based analysis, and these were again related to developmental morphogenesis categories (Supplementary Tables 2g, 3g). There were 40 significant GO process categories with 10-fold or greater enrichment when using the relative scheme, compared to 7 GO categories when using the mean-based scheme. These results show that major changes may occur in the probability distributions of methylation levels associated with developmentally critical genes and that the shape of these distributions, rather than their means per se, may be related to pluripotency and fate lineage determination in development and cancer.
Lastly, by assessing a relationship between transcription factor binding and Jensen-Shannon distances in development, we found a strong association between Jensen-Shannon distances and PRC2 binding within enhancer regions (Supplementary Note). This raises the intriguing possibility that the PRC2 complex not only influences the mean behavior of DNA methylation, as has been established previously15, but most importantly it controls the stochastic behavior of methylation in a developmentally relevant and targeted manner.
Entropy blocks predict TAD boundaries
Topologically associating domains (TADs) are highly conserved structural features of the genome across tissue types and species16–18. Loci within these domains tend to interact frequently with each other, with much less frequent interactions taking place between loci within adjacent domains. Although genome-wide detection of TAD boundaries is experimentally challenging, these boundaries can be reasonably predicted from histone mark ChIP-seq data (CTCF, H3k4me1) using a computational approach19. We therefore examined the possibility of locating TAD boundaries genome-wide using WGBS data.
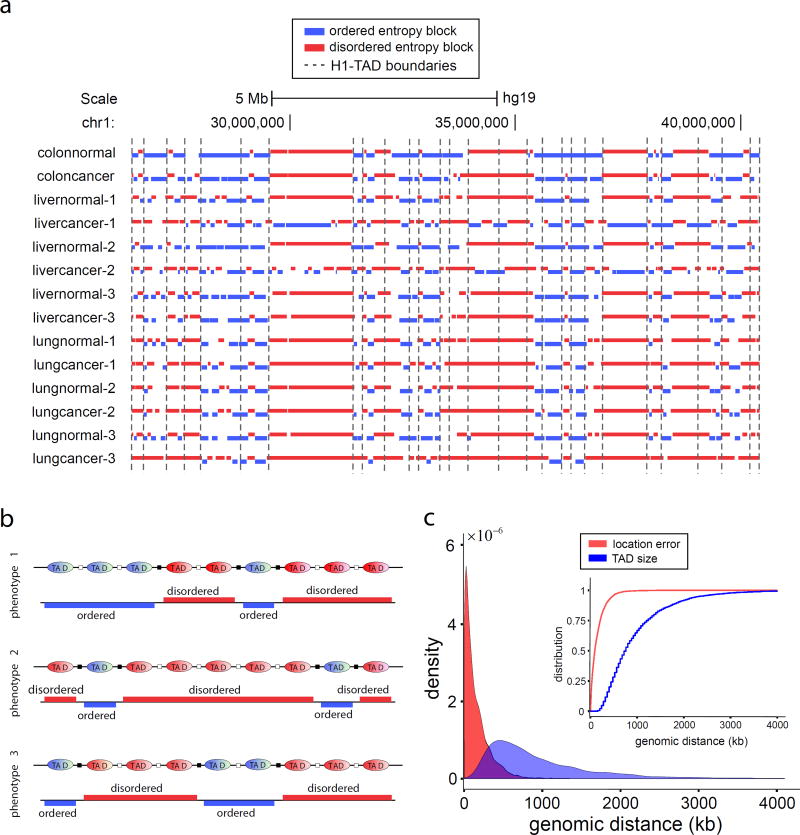
In many of our samples, known TAD boundary annotations were visually proximal to boundaries of entropy blocks, large genomic regions of consistently low or high normalized entropy values (Fig. 5a, Supplementary Fig. 5, and Online Methods). We thus hypothesized that TAD boundaries may be located within genomic regions that separate successive entropy blocks. As a first test, we computed entropy blocks in the stem data and identified 404 regions predictive of TAD boundaries (Online Methods). We then found that 5862 annotated TAD boundaries in H1 stem cells16 were located within these predictive regions or were close in a statistically significant manner, and correctly identified 6% of the annotated TAD boundaries (362 out of 5862) using 90% of the computed predictive regions (Online Methods and Supplementary Note).
Figure 5.
Entropy blocks and TAD boundaries. (a) In the normal/cancer panel, a subset of known TAD boundary annotations in H1 stem cells appear to be correlated with boundaries of entropy blocks (blue: ordered, red: disordered), suggesting that TADs may maintain a consistent level of methylation entropy within themselves. (b) Regions of entropic transitions can be used to identify the location of some TAD boundaries (black squares). Since TADs are cell-type invariant, the location of more TAD boundaries can be identified using additional WGBS data corresponding to distinct phenotypes. (c) Probability densities and cumulative probability distributions (insert) of the TAD boundary location error and TAD sizes. The cumulative probability distributions imply that the probability of the location error to be smaller than K base pairs is greater than the probability that the TAD size is smaller than K, for every K. Therefore, the location error is smaller than the TAD size in a well-defined statistical sense, known as stochastic ordering.
Since TADs are thought to be cell-type invariant16,18, we can predict the location of more TAD boundaries by combining information from entropy blocks derived from additional phenotypes (Fig. 5b). We therefore computed entropy blocks using WGBS data from 17 different cell types, determined predictive regions for each cell type, and combined these regions to form a single list (6687 predictive regions) that encompasses information from all cell types (Online Methods). Moreover, we combined the TAD boundary annotations for H1 stem cells with available annotations for IMR90 lung fibroblasts16 and obtained a total of 10,276 “ground-truth” annotations. We then obtained results similar to the case of stem cells with TAD boundaries falling within identified predictive regions did so significantly more often than expected by chance. This resulted in 62% correct identification of the annotated TAD boundaries (6369 out of 10,276) derived from 95% of computed predictive regions (Online Methods and Supplementary Note), which can be further improved by including additional phenotypes.
We further support these predictions by pointing to the relatively small errors obtained when locating TAD boundaries at the centers of predictive regions in comparison to TAD sizes. This is demonstrated by estimating the probability density and the corresponding cumulative probability distribution of location errors and TAD sizes (Fig. 5c), and the fact that the median location error was an order of magnitude smaller than the median TAD size (92-kb vs. 760-kb). Together, these results provide strong statistical evidence of an underlying relationship between TADs and entropy blocks, according to which a TAD is associated with consistently low or high normalized entropy values, which can be used to computationally identify TAD boundaries accurately from WGBS data genome-wide.
Methylation channels explain epigenetic memory maintenance
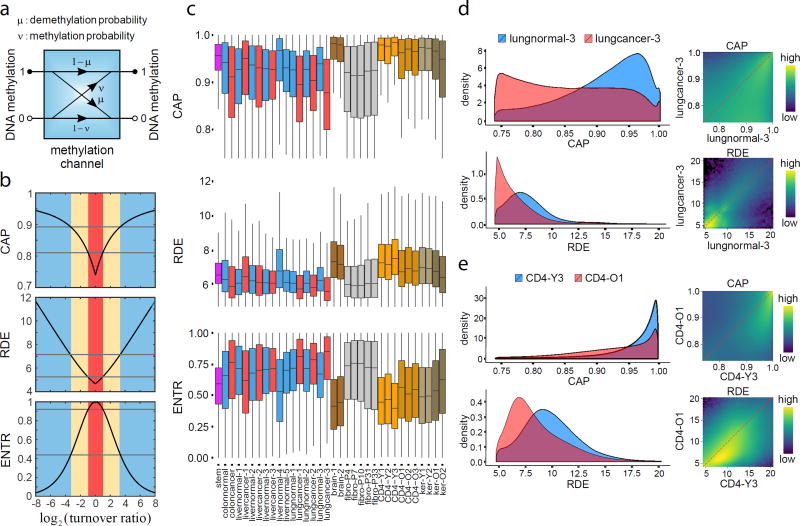
To investigate epigenetic memory maintenance from an information-theoretic perspective, we modeled this process using a methylation channel, which quantifies transitions of the binary methylation state at each CpG site of the genome using the local probabilities of demethylation and de novo methylation (Fig. 6a and Supplementary Note). This results in a model for methylation maintenance known in information theory as the asymmetric-noise binary communication channel20.
Figure 6.
Information-theoretic properties of methylation channels. (a) A methylation channel maintains the methylation state at a CpG site (1: methylated; 0: unmethylated) using four conditional probabilities (μ: demethylation probability; ν: de novo methylation probability). (b) Theoretical curves of the capacity (CAP), relative dissipated energy (RDE), and input/output entropy (ENTR) of a methylation channel in terms of the log2 ratio of the probability of de novo methylation to the probability of demethylation (turnover ratio). Methylation maintenance by a high capacity methylation channel (CAP ≥ 0.89) dissipates significant energy (RDE ≥ 7.125), achieving high reliability (probability of error ≤ 0.0073) and an ordered methylation state (ENTR ≤ 0.44). Conversely, methylation maintenance by a low capacity methylation channel (CAP ≤ 0.81) dissipates less energy (RDE ≤ 5.25), achieving lower reliability (probability of error ≥ 0.026) and a disordered methylation state (ENTR ≥ 0.92). These thresholds correspond to entropy levels used to identify ordered and disordered genomic units that build entropy blocks (Online Methods). (c) Boxplots of genome-wide distributions of capacities, relative dissipated energies, and entropies at individual CpG sites show global differences among cell types. The boxes show the 25% quantile, the median, and the 75% quantile, whereas each whisker has a length of 1.5× the interquartile range. (d) Genome-wide 1D and 2D log10 transformed capacities and relative dissipated energies associated with lung normal/cancer show global channel capacity loss in cancer accompanied by a reduction in dissipated energy. (e) Aging CD4+ lymphocytes exhibit global loss of capacity and dissipated energy as well.
A methylation channel is limited to transmitting a maximum amount of information, quantified by its information capacity20. Moreover, appreciable consumption of free energy, which must be dissipated to the surroundings in the form of heat, is required to achieve high transmission reliability essential for normal cellular function. We assessed methylation reliability using the notion of relative dissipated energy, identified approximate relationships between channel capacity, relative dissipated energy, and methylation entropy (Supplementary Note), and predicted that highly reliable methylation maintenance is achieved through high capacity methylation channels that produce less entropic methylation at the expense of higher energy consumption (Fig. 6b). This establishes the influence of methylation channels on epigenetic memory by providing a fundamental link between their information-theoretic properties and the nature of epigenetic memory maintenance.
We computed capacities, relative dissipated energies, and entropies genome-wide in individual samples and comparative studies (Fig. 6c and Supplementary Fig. 3c,d). We observed global loss of capacity and relative dissipated energy in colon and lung cancer, accompanied by global gain in CpG entropy (Fig. 6c,d), although this was not necessarily true in liver cancer (Fig. 6c). Moreover, brain cells, CD4+ lymphocytes, and skin keratinocytes exhibited high capacities and relative dissipated energies, with loss in older individuals, whereas stem cells had a narrow range of relatively high capacities and relative dissipated energies (Fig. 6c,e). By ordering genes in terms of normalized methylation entropy within their promoters in stem cells (Supplementary Table 5), we discovered many genes to be characterized by high information capacity and relative dissipated energy, such as FOXO3, TGIF1, SATB2, IGF2BP1, SMAD7, ZIC2, SALL1, and SOX2, which are involved in stem cell regulation, pluripotency, and differentiation21–28. We also found that the methylation state within CGIs and TSSs is maintained by methylation channels whose capacities are overall higher than within shores, shelves, open seas, exons, introns and intergenic regions, and this is accomplished by higher energy consumption (Supplementary Fig. 3c,d). These results highlight an information-theoretic view of epigenetic organization that explains methylation stochasticity in a way that is consistent with the need of cells to manage limited energy resources in a strategic manner. According to this model, reliable transmission of methylation information within critical regions of the genome is facilitated by high capacity methylation channels that result in low methylation stochasticity at the cost of high energy consumption. However, methylation transmission within other regions of the genome is transmitted by low capacity methylation channels that consume less energy but produce higher levels of methylation stochasticity.
Information-theoretic prediction of chromatin changes in development and cancer
Recent work in chromatin organization has found the existence of cell-type specific compartments A and B, known to be associated with gene-rich transcriptionally active open chromatin and gene-poor transcriptionally inactive closed chromatin, respectively16,18,29. Although identifying compartments A/B is experimentally challenging29, this can sometimes be achieved computationally30. We therefore sought to identify compartments A/B in individual WGBS samples from local informational properties of the methylome.
By comparing Hi-C data from EBV cells to our methylation channels, we observed enrichment of low information capacity, low relative dissipated energy, and high normalized methylation entropy within compartment B, and the opposite was globally true for compartment A (Fig. 7a,b). This suggested the possibility of predicting compartments A/B from informational properties of methylation maintenance. We therefore employed a random forest regression model to learn the informational structure of compartments A/B from available “ground-truth” data (Online Methods). We achieved reliable prediction, with cross-validated average correlation of 0.82 and average agreement of 91% between predicted and true A/B signals using a calling margin of 0.2 (Supplementary Fig. 6a and Online Methods), suggesting that a small number of local information-theoretic properties of methylation maintenance can be highly predictive of large-scale chromatin organization.
Figure 7.
Information-theoretic prediction of large scale chromatin organization. (a) Analysis of Hi-C and WGBS data shows that maintenance of the methylation state within compartment B (blue) in EBV cells is mainly performed by low information capacity methylation channels that dissipate low amounts of energy and result in a relatively disordered and less methylated state than in compartment A (brown). (b) Violin plots of genome-wide distributions of information capacity (CAP), relative dissipated energy (RDE), normalized methylation entropy (NME), and mean methylation level (MML) demonstrate the attractiveness of these quantities as features for predicting compartments A/B using WGBS data from single samples. The boxes show the 25% quantile, the median, and the 75% quantile, whereas each whisker has a length of 1.5× the interquartile range. (c) Hierarchical clustering of samples using the net percentage of A/B compartment switching as a dissimilarity measure. At a given height, a cluster is characterized by lower overall compartment switching than an alternative grouping of samples. (d) UCSC genome browser images of two chromosomal regions show significant overlap of compartment B in normal lung (blue) with hypomethylated blocks, LADs, and LOCKs. Gain in Jensen-Shannon distance (JSD) is observed within compartment B (blue) in lung samples during carcinogenesis.
Consistent with the fact that compartments A/B are cell-type specific, and in agreement with the finding of extensive A/B compartment reorganization during early stages of development31, we observed many differences in predicted compartments between tissues and in carcinogenesis (Supplementary Fig. 6b–f). We also found from methylation data that our predicted compartment transitions corresponded often to TAD boundaries identified from Hi-C data.31 (Supplementary Fig. 6b). We also quantified observed differences in compartments A/B by computing percentages of switching in all sample pairs (Supplementary Table 5 and Online Methods) and clustered the samples by using the net percentage of A/B switching as a dissimilarity measure (Fig. 7c and Online Methods). The clusters had 31/34 samples grouped in a biologically meaningful manner, providing evidence that A/B switching, as determined by methylation information, can accurately quantify phenotypic differences in the samples. Notably, stem cell differentiation is associated with high levels of chromatin reorganization (Fig. 7c), whereas differentiated lineages and cancer are clustered together but are distinguished from each other. Moreover, fibroblasts form one cluster, whereas young CD4 samples form their own, and the same is true for skin.
Normal samples exhibit strikingly different chromatin organization from matched cancer samples (Fig. 7c). Previous studies found large hypomethylated blocks in cancer that are remarkably consistent across tumor types32. These blocks correspond closely to large-scale regions of chromatin organization, such as lamin-associated domains (LADs) and large organized chromatin K9-modifications (LOCKs)3,33. Consistent with our observations on the information-theoretic properties of compartment B and of carcinogenesis (Fig. 6c and Fig. 7a,b), we asked whether hypomethylated blocks are associated mainly with compartment B (Online Methods). We found (Fig. 7d) significant overlap with compartment B in normal lung, and the same was true for LADs and LOCKs (Supplementary Table 6). Compartment B in normal tissue exhibited regions of large Jensen-Shannon distances (Fig. 7d), suggesting that considerable epigenetic changes may occur within this compartment during carcinogenesis, which is further supported by the genome-wide distributions of the Jensen-Shannon distance values between normal/cancer within compartments A/B in normal (Supplementary Fig. 7). The observed association of compartment B in normal tissue with hypomethylated blocks and large Jensen-Shannon distances indicates that compartment B demarcates genomic regions with methylation information that is more likely to be degraded in cancer.
Entropic sensitivity quantifies epigenetic responsiveness to environmental variability
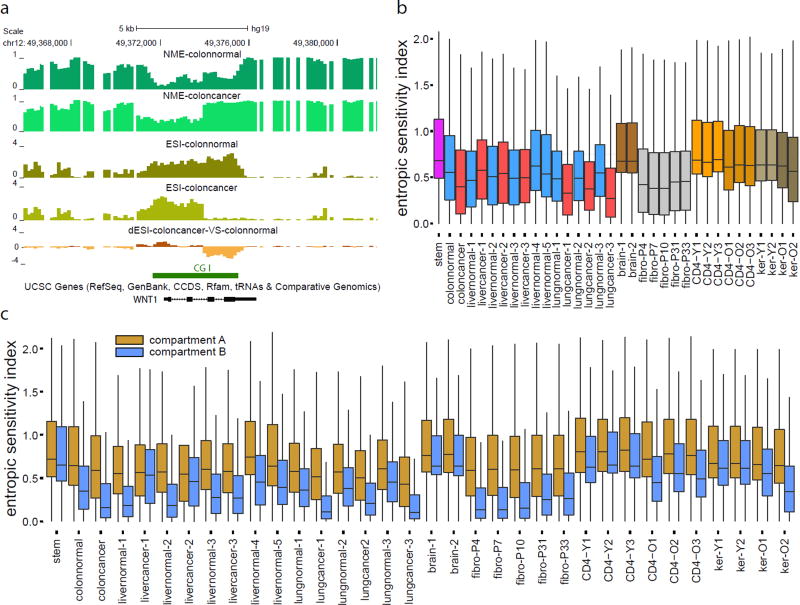
Epigenetic changes integrate environmental signals with genetic variation to modulate phenotype. We therefore sought to investigate the influence of environmental exposure on methylation stochasticity. We viewed environmental variability as a process that directly influences the parameters of the methylation potential energy landscape and employed a probabilistic approach that allowed us to compute from WGBS data a sensitivity index that quantifies the rate by which environmental perturbations influence methylation entropy along the genome (Fig. 8, Supplementary Figs. 3e and 8, and Supplementary Note). For example, we observed entropic sensitivity within a CGI associated with WNT1 in normal colon, with a portion gaining entropy and losing sensitivity in the matched cancer sample (Fig. 8a).
Figure 8.
Entropic sensitivity distributions in single samples and comparative studies. (a) Gain in entropy and loss of entropic sensitivity is observed within a portion of the CGI associated with WNT1. NME: normalized methylation entropy, ESI: entropic sensitivity index, dESI: differential entropic sensitivity index. (b) Boxplots of genome-wide distributions of entropic sensitivity display global differences across cell types. (c) Boxplots of genome-wide distributions of entropic sensitivity within compartment A (brown) and compartment B (blue) show appreciably higher entropic sensitivity within compartment A than within compartment B. The boxes in (b) & (c) show the 25% quantile, the median, and the 75% quantile, whereas each whisker has a length of 1.5× the interquartile range.
Globally, we observed differences in entropic sensitivity among tissues (Fig. 8b, Supplementary Fig. 8), with stem and brain having higher levels of entropic sensitivity than the rest of the samples. Since the brain cells are significantly methylated (Fig. 3a), high levels of entropic sensitivity would predict that brain can exhibit high rates of demethylation in response to environmental stimuli, consistent with recent data showing that the DNA demethylase Tet3 acts as a synaptic activity sensor that epigenetically regulates neural plasticity through active demethylation34. Colon and lung cancer exhibited global loss of entropic sensitivity (Fig. 8b, Supplementary Fig. 8a), whereas gain was noted in liver cancer. Moreover, CD4+ lymphocytes and skin keratinocytes exhibited global loss of entropic sensitivity in older individuals (Fig. 8b, Supplementary Fig. 8b), while cultured fibroblasts had lower sensitivity. Notably, we observed higher and more variable entropic sensitivity values within CGIs and at TSSs compared to other genomic features, such as shores, exons, and introns (Supplementary Fig. 3e). However, some unmethylated CGIs exhibited low entropic sensitivity, whereas changes in entropic sensitivity within CGIs were observed between normal and cancer, as well as in older individuals (Supplementary Fig. 8c–h). Notably, differences in entropic sensitivity were not simply due to entropy itself, as many regions of low entropy had small sensitivity values, while other such regions displayed high values (Supplementary Fig. 8c–e,g). Lastly, we ordered genes in terms of entropic sensitivity within their promoters (Online Methods) and found many key developmental regulators or environmental sensors to be associated with high entropic sensitivity in stem and colon (Supplementary Table 7).
Entropic sensitivity within compartment A was higher than in compartment B in all samples (Fig. 8c), consistent with the notion that the transcriptionally active compartment A should be more responsive to stimuli. Moreover, observed differences between normal and cancer were largely confined to compartment B (Fig. 8c). Substantial loss of entropic sensitivity was observed in compartment B in older CD4+ lymphocytes and skin keratinocytes, but not in compartment A. In contrast, cell culture gained sensitivity within compartment B (Fig. 8c).
To further investigate entropic sensitivity changes between tissues, we ranked genes according to differential entropic sensitivity within their promoters between colon normal and cancer (Supplementary Table 8 and Online Methods). Several highly ranked genes were found to code for LIM-domain proteins, including LIMD2, and are implicated in colon and other types of cancer, such as QKI, HOXA9, a canonical rearranged homeobox gene35 that is dysregulated in cancer, and FOXQ1, which is overexpressed and enhances tumorigenicity of colorectal cancer36. Together, these results indicate that environmental exposure may influence epigenetic stochasticity in cells with sensitivity that varies along the genome and between compartments in a cell-type specific manner. This presents the intriguing possibility that disease, environmental exposure, and aging are associated with substantial changes in entropic sensitivity, thus compromising integration of environmental cues regulating cell growth and function.
DISCUSSION
Our information-theoretic approach to epigenomics utilizing the Ising model of statistical physics has shown that a formal approach to methylation analysis can precisely extract and quantify the information content of experimental data to yield fundamental insights into epigenetic behavior. We provided a formal definition of potential energy landscapes, characterized intrinsic epigenetic stochasticity, rigorously derived epigenetic entropy and methylation channels, associated chromatin organization with informational properties of methylation, and estimated entropic sensitivity to environmental conditions. We also developed high-resolution computational tools for analyzing stochasticity in WGBS methylation data with low (10–20×) coverage, for quantifying epigenetic distances using normal/disease pairs that could be crucial in personalized medicine, and for predicting 3D chromatin structure from individual methylation samples in health and disease.
Shannon entropy varied markedly among tissues, across the genome and among features of the genome. Entropy was increased with aging in skin and blood, but not in cell culture, suggesting a link between increased entropy and epigenetic aging. Jensen-Shannon distances precisely quantified epigenetic discordances between individual samples, demonstrating that cancer is informationally distant from both stem cells and normal tissues, thus providing a potential clinical advantage of identifying specific differences between two samples. Importantly, epigenetic discordance was found to be associated with changes in entropy or large Jensen-Shannon distances and not necessarily with differences in mean methylation, and should be routinely used in epigenetic analysis.
We discovered that TAD boundaries are potential transition points between high and low entropy blocks and that information-theoretic properties of methylation channels could effectively predict chromatin organization in terms of compartments A/B. Computed compartments B demonstrated lower capacity, lower relative dissipated energy, higher Shannon entropy, lower entropic sensitivity, and larger JSD values in carcinogenesis, as well as significant overlap with hypomethylated blocks, LOCKs and LADs. Moreover, A/B switching accurately quantified differences in phenotype, with marked switching in development and carcinogenesis. Finally, some cancers and aging were associated with global loss of entropic sensitivity that could be related to the autonomous nature of tumor cells and the well-known reduced physiological plasticity of aging.
This study demonstrates a relationship between chromatin structure, methylation channels, and entropic sensitivity that may maximize an organism’s efficiency in storing epigenetic information and help explain developmental plasticity. In this model, pluripotent stem cells require relatively high energy to maintain high capacity methylation channels within a portion of the genome, achieving reduced methylation stochasticity. Other regions characterized by increased entropic sensitivity are associated with highly deformable potential energy landscapes, which may correspond to differentiation branch points, as metaphorically suggested by Waddington. After differentiation, some large genomic domains, such as regions associated with pluripotency, need not maintain high channel capacities and energy consumption, with their sequestration providing increased energy efficiency with the cost of high epigenetic stochasticity and reduced responsiveness.
Furthering this model, our observation that compartment B exhibits reduced energy expenditure and channel capacity, thus failing to accurately maintain methylation information, explains the observed significant overlap of compartment B in normal tissue with hypomethylated blocks in cancer, implying that compartment B is more dysregulated than compartment A in carcinogenesis, in agreement with the observed higher JSD values. We therefore hypothesize that cancer cells gain a micro-evolutionary advantage upon reorganization of dysregulated B domains, thereby amplifying epigenetic stochasticity to increase plasticity and adaptability beyond that of the primary tissue.
The stochastic nature and properties of DNA methylation and their close relationship with chromatin structure raise the intriguing possibility that epigenetic information is carried by a population of cells as a whole, and that this information not only helps to achieve and maintain a differentiated state but also to mediate developmental plasticity throughout the life of an organism.
ONLINE METHODS
Samples for whole-genome bisulfite sequencing
We used previously published WGBS data corresponding to 10 samples, which included H1 human embryonic stem cells37, normal and matched cancer cells from colon and liver12, keratinocytes from skin biopsies of sun protected sites from younger and older individuals38, and EBV-immortalized lymphoblasts39. We also generated WGBS data corresponding to 25 samples that included normal and matched cancer cells from liver and lung, pre-frontal cortex, cultured HNF fibroblasts at 5 passage numbers, and sorted CD4+ T-cells from younger and older individuals, all with IRB approval. We obtained pre-frontal cortex samples from the University of Maryland Brain and Tissue Bank, which is a Brain and Tissue Repository of the NIH NeuroBioBank. Peripheral blood mononuclear cells (PBMCs) were isolated from peripheral blood collected from healthy subjects and separated by using a Ficoll density gradient separation method (Sigma-Aldrich). CD4+ T-cells were subsequently isolated from PBMCs by positive selection with MACS magnetic bead technology (Miltenyi). Post-separation flow cytometry assessed the purity of CD4+ T-cells to be at 97%. Primary neonatal dermal fibroblasts (Mycoplasma-free) were acquired from Lonza and cultured in Gibco's DMEM supplemented with 15% FBS (Gemini BioProducts).
DNA isolation
We extracted genomic DNA from samples using the Masterpure DNA Purification Kit (Epicentre). High molecular weight of the extracted DNA was verified by running a 1% agarose gel and by assessing the 260/280 and 260/230 ratios of samples on Nanodrop. Concentration was quantified using Qubit 2.0 Fluorometer (Invitrogen).
Generation of WGBS libraries
For every sample, 1% unmethylated Lambda DNA (Promega, cat # D1521) was spiked-in to monitor bisulfite conversion efficiency. Genomic DNA was fragmented to an average size of 350 bp using a Covaris S2 sonicator (Woburn, MA). Bisulfite sequencing libraries were constructed using the Illumina TruSeq DNA Library Preparation kit protocol (primers included) or NEBNext Ultra (NEBNext Multiplex Oligos for Illumina module, New England BioLabs, cat # E7535L) according to the manufacturer’s instructions. Both protocols use a Kapa HiFi Uracil+ PCR system (Kapa Biosystems, cat # KK2801).
For Illumina TruSeq DNA libraries, gel-based size selection was performed to enrich for fragments in the 300–400 bp range. For NEBNext libraries, size selection was performed using modified AMPure XP bead ratios of 0.4× and 0.2×, aiming also for an insert size of 300–400 bp. After size-selection, the samples were bisulfite converted and purified using the EZ DNA Methylation Gold Kit (Zymo Research, cat # D5005). PCR-enriched products were cleaned up using 0.9× AMPure XP beads (Beckman Coulter, cat # A63881).
Final libraries were run on the 2100 Bioanalyzer (Agilent, Santa Clare, CA, USA) using the High-Sensitivity DNA assay for quality control purposes. Libraries were then quantified by qPCR using the Library Quantification Kit for Illumina sequencing platforms (cat # KK4824, KAPA Biosystems, Boston, USA), using 7900HT Real Time PCR System (Applied Biosystems) and sequenced on the Illumina HiSeq2000 (2×100 bp read length, v3 chemistry according to the manufacturer’s protocol with 10× PhiX spike-in) and HiSeq2500 (2×125 bp read length, v4 chemistry according to the manufacturer’s protocol with 10× PhiX spike-in).
Quality control and alignment
FASTQ files were processed using Trim Galore! v0.3.6 (Babraham Institute) to perform single-pass adapter- and quality-trimming of reads, as well as running FastQC v0.11.2 for general quality check of sequencing data. Reads were then aligned to the hg19/GRCh37 genome using Bismark v0.12.3 and Bowtie2 v2.1.0. Separate mbias plots for read 1 and read 2 were generated by running the Bismark methylation extractor using the “mbias_only” flag. These plots were used to determine how many bases to remove from the 5' end of reads. The number was generally higher for read 2, which is known to have poorer quality. The amount of 5' trimming ranged from 4 bp to 25 bp, with most common values being around 10 bp. BAM files were subsequently processed with Samtools v0.1.19 for sorting, merging, duplicate removal and indexing.
FASTQ files associated with the EBV sample were processed using the same pipeline described for the in-house samples. BAM files associated with the normal colon and liver samples, obtained from Ziller et al12, could not be assessed using the Bismark methylation extractor due to incompatibility of the original alignment tool (MAQ) used on these samples. We therefore followed the advice of the authors and trimmed 4 bp from all reads for those files.
Genomic features and annotations
Files and tracks bear genomic coordinates for hg19. CGIs were obtained from Wu et al40. CGI shores were defined as sequences flanking 2-kb on either side of islands, shelves as sequences flanking 2-kb beyond the shores, and open seas as everything else. The R Bioconductor package “TxDb.Hsapiens.UCSC.hg19.knownGene” was used for defining 3UTRs, 5UTRs, exons, introns and transcription start sites (TSSs). Promoter regions were defined as sequences flanking 2-kb on either side of TSSs. A curated list of enhancers was obtained from the VISTA enhancer browser41 by downloading all human (hg19) positive enhancers that had reproducible expression in at least three independent transgenic embryos. Hypomethylated blocks (colon and lung cancer) were obtained from Timp et al32, whereas H1 stem cell LOCKs and Human Pulmonary Fibroblast (HPF) LOCKs were obtained from Wen et al42. LAD tracks associated with Tig3 cells derived from embryonic lung fibroblasts were obtained from Guelen et al43. Gene bodies were obtained from the UCSC genome browser, H1 and IMR90 TAD boundaries were obtained from Bing Ren’s Lab at UCSD, and BED files for Hi-C data processed into compartments A and B were provided by Fortin and Hansen.
Estimation and display of potential energy landscapes
We partitioned the genome into consecutive non-overlapping regions of equal size and estimated the parameters θ of the potential energy landscape within a region by maximizing the average log-likelihood , with x1, x2, …, xM being M independent observations of the methylation state (i.e., WGBS sequencing reads) within the region. To take into account partially observed methylation states, we replaced P(xm | θ) by the joint probability distribution over only those sites at which methylation information is available, which we calculated by marginalizing P(xm | θ) over these sites. After extensive experimentation, we considered 3-kb estimation regions by striking a balance between estimation and computational performance. To avoid statistical overfitting, we did not model regions with less than 10 CpG sites. We also ignored regions with not enough data for which less than 2/3 of the CpG sites were observed or the average depth of coverage was less than 2.5 observations per CpG site. We finally performed optimization using the multilevel coordinate search (MCS) algorithm44, which was chosen due to its superior performance among the derivative-free global optimization algorithms we tested (such as simulated annealing).
To visualize a potential energy landscape as a 3D plot, we used the 2D version of the Gray code45. According to this method, we placed all possible 2N binary-valued methylation states within a genomic region with N CpG sites on a 2D plane in a manner so that states located adjacent to each other in the east/west and north/south directions differ in only one bit. We then obtained a 3D plot by assigning to each state its potential value.
Computation of probability distribution of methylation level
We calculated the probability distribution P(l) of the methylation level within a genomic unit with N CpG sites from the Ising probability distribution P(x) of the methylation patterns within the genomic unit using P(l) = Σx∈Q(Nl) P(x) where Q(Nl) is the set of all methylation patterns with exactly Nl methylated CpG sites. In the rarer case when N was too large to make direct summation tractable, we used the method of maximum entropy to approximate P(l)46 by estimating the first four non-central moments of the methylation level L using Monte Carlo.
Normalized methylation entropy
We quantified methylation stochasticity within a genomic unit with N CpG sites using the normalized methylation entropy h = H/log2(N + 1), where H = − Σl P(l)log2P(l) is the informational (Shannon) entropy20 of the methylation level. The normalized methylation entropy ranges between 0 and 1, taking its maximum value when all methylation levels within a genomic unit are equally likely (fully disordered sate) regardless of the number of CpG sites, and achieving its minimum value only when a single methylation level is observed (perfectly ordered state).
Quantifying differential behavior in methylation level
To quantify differences in the probability distributions of the methylation level within a genomic unit between two samples, we employed the Jensen-Shannon distance47
where P1 and P2 are the probability distributions of the methylation level within the genomic unit in the first and second samples, Q = (P1 + P2)/2 is the average of the two probability distributions, and
is the relative entropy or Kullback-Leibler divergence20. The Jensen-Shannon distance simultaneously encapsulates any difference in the distribution (including mean methylation and entropy) by measuring dissimilarities between probability distributions of methylation level within a genomic unit across two samples. It is a normalized distance metric, taking values between 0 and 1, which equals 0 only when the two probability distributions P1 and P2 are identical and reaches its maximum value of 1 when the supports of the two distributions do not intersect each other.
Epigenetic distances, multidimensional scaling, and gene ranking
We quantified the epigenetic discordance between two samples by calculating a dissimilarity value defined as the average of all Jensen-Shannon distance values computed genome-wide. To visualize epigenetic similarities or dissimilarities between samples, we computed the epigenetic distances between all pairs of samples, formed the corresponding dissimilarity matrix, and employed a 2D representation, using multidimensional scaling based on Kruskal’s non-metric method, to find a 2D configuration of points whose inter-point distances approximately correspond to the epigenetic dissimilarities among the samples.
To rank genes based on the absolute difference in mean methylation level within their promoters, we centered a 4-kb window at the transcription start site of each gene in the genome, computed the absolute difference in mean methylation level within each genomic unit that overlaps this window, and scored the gene by averaging these values. We used the same method to rank genes based on the Jensen-Shannon distance. We also ranked genes using a relative scheme that assigned a higher score to genes with larger Jensen-Shannon distances but smaller absolute differences in mean methylation level. We did so by scoring a gene using the ratio of its ranking in the mean-based list to its ranking in the list obtained by the Jensen-Shannon distance.
Computation of entropy blocks
Computation of entropy blocks requires detection of ordered and disordered blocks; i.e., large genomic regions of consistently low or high normalized methylation entropy values. To effectively summarize methylation entropy in a single sample, we computed the normalized methylation entropy h within each genomic unit and classified it into one of three classes: ordered (0 ≤ h ≤ 0.44), weakly ordered/disordered (0.44 ≤ h ≤ 0.92), and disordered (0.92 ≤ h ≤ 1). We determined the threshold values by investigating the relationship between the normalized methylation entropy within a genomic unit that contains one CpG site and the ratio of the probability p of methylation to the probability 1 − p of unmethylation at that site. To this end, we focused on the odds ratio r = p/(1 − p) and considered the methylation level to be “ordered” if r ≥ 10 or r ≤ 1/10 (i.e., if the probability of methylation is at least 10× larger than the probability of unmethylation, and likewise for the probability of unmethylation), in which case, p ≥ 0.9091 or p ≤ 0.0909, which correspond to a maximum normalized methylation entropy threshold of 0.44. Moreover, we considered the methylation level to be “disordered” if 1/2 ≤ r ≤ 2 (i.e., if the probability of methylation is no more than 2× the probability of unmethylation, and likewise for the probability of unmethylation), in which case, 0.3333 ≤ p ≤ 0.6667, which corresponds to a minimum normalized methylation threshold of 0.92.
To compute entropy blocks, we slid a window of 500 genomic units (75-kb) along the genome and labeled the window as being ordered or disordered if at least 75% of its genomic units were effectively classified as being ordered or disordered, respectively. We then determined ordered or disordered blocks by taking the union of all ordered or disordered windows and by removing discordant overlappings.
Prediction of TAD boundaries
Using entropy blocks computed for a given sample, we identified predictive regions of the genome that might contain TAD boundaries by detecting the space between successive entropy blocks with distinct labels (ordered or disordered). For example, if an ordered block located at chr1: 1–1000 were followed by a disordered block at chr1: 1501–2500, then chr1: 1001–1500 was deemed to be a predictive region. To reduce false identification of predictive regions, we did not consider successive entropy blocks of the same type, since the genomic space between two such entropy blocks may be due to missing data or other unpredictable factors. To control the resolution of locating a TAD boundary, we only considered gaps smaller than 50-kb. This resulted in a resolution of an order of magnitude smaller than the mean TAD size (~900-kb). To combine predictive regions obtained from methylation analysis of several distinct epigenotypes, we computed the “predictive coverage” of each base pair by counting the number of predictive regions that contained the base pair. We then combined predictive regions by grouping consecutive base pairs whose predictive coverage was at least 4. We subsequently applied this method on WGBS data corresponding to 17 distinct cell and tissue types (stem, colonnormal, coloncancer, livernormal-1, livercancer-1, livernormal-2, livercancer-2, livernormal-3, livercancer-3, lungnormal-1, lungcancer-1, lungnormal-2, lungcancer-2, lungnormal-3, lungcancer-3, brain-1, brain-2), and analyzed our results using “GenometriCorr”48, a statistical package for evaluating the correlation of genome-wide data with given genomic features. Finally, we considered a boundary prediction to be “correct” when the distance of a “true” TAD boundary from the center of a predictive region was less than the first quartile of the “true” TAD width distribution (Fig. 5c insert – green).
A/B compartment prediction and analysis
Genome-wide prediction of A/B compartments was performed by a random forest regression model. We trained this model using a small number of available Hi-C data associated with EBV and IMR90 samples49, as well as A/B tracks produced by the method of Fortin and Hansen using long-range correlations computed from pooled 450k array data associated with colon cancer, liver cancer, and lung cancer samples30. Due to the paucity of currently available Hi-C data, we included the Fortin-Hansen data in order to increase the number of training samples and improve the accuracy of performance evaluation. We first paired the Hi-C and Fortin-Hansen data with WGBS EBV, fibro-P10, and colon cancer samples, as well as with samples obtained by pooling WGBS liver cancer (livercancer-1, livercancer-2, livercancer-3) and lung cancer (lungcancer-1, luncancer-2, lungcancer-3) data. We subsequently partitioned the entire genome into 100-kb bins (to match the available Hi-C and Fortin-Hansen data), and computed eight information-theoretic features of methylation maintenance within each bin (median values and interquartile ranges of information capacity, relative dissipated energy, normalized methylation entropy, and mean methylation level). By using all feature/output pairs, we trained a random forest model using the R package “randomForest” with its default settings, except that we increased the number of trees to 1000. We then applied the trained random forest model on each WGBS sample and produced A/B tracks that approximately identified A/B compartments associated with the samples. Since regression takes into account only information within a 100-kb bin, we averaged the predicted A/B values using a three-bin smoothing window and removed from the overall A/B signal its genome-wide median value, as suggested by Fortin and Hansen30.
To test the accuracy of the resulting predictions, we employed 5-fold leave-one-out cross validation, which involved training using four sample pairs and testing on the remaining pair for all five combinations. We evaluated performance by computing the average correlation as well as the average percentage agreement between the predicted and each of the “ground-truth” A/B signals within 100-kb bins at which the absolute values of the predicted and “ground-truth” signals were both greater than a calling margin, where we used a non-zero calling margin to remove unreliable predictions. We finally calculated agreement by testing whether the predicted and the “ground-truth” A/B values within a 100-kb bin had the same sign.
For each pair of WGBS samples, we computed the percentage of A to B compartment switching by dividing the number of 100-kb bin pairs for which an A prediction is made in the first sample and a B prediction is made in the second sample by the total number of bins for which A/B predictions were available in both samples, and similarly for the case of B to A switching. We summed these percentages and formed a matrix of dissimilarity measures, which we then used as an input to a Ward error sum of squares hierarchical clustering scheme50, which we implemented using the R package “hclust” by setting the method variable to “ward.D2”.
To test the significance of overlapping of hypomethylated blocks, LADs, and LOCKs with compartment B, we used available hypomethylated blocks, LOCKs, and LADs, and predicted compartment B data for the lungnormal-1, lungnormal-2, and lungnormal-3 samples, which best match the previous tracks. To evaluate enrichment of hypomethylated blocks (and similarly for LADs and LOCKs) within compartment B, we defined two binary (0–1) random variables R and B for each genomic unit of the genome, such that R = 1 if the genomic unit overlaps a block, and B = 1 if the genomic unit overlaps compartment B. We then tested against the null hypothesis that R and B are statistically independent by applying the χ2-test on the 2 × 2 contingency table for R and B and calculated the odds ratio (OR) as a measure of enrichment.
Entropy sensitivity and gene ranking
We ranked genes based on entropic sensitivity and its differences between a test and a reference sample within their promoters. We did so by centering a 4-kb window at the transcription start site of each gene in the genome, computed the value or the absolute difference in the value of the entropic sensitivity index within each genomic unit that “touches” this window, and scored the gene by averaging these values.
Supplementary Material
Acknowledgments
We thank Xin Li, Amy Vandiver, and Jeremy Walston for cells and/or FASTQ files; Rakel Trygvadottir, Birna Berndsen, Adrian Idrizi and Colin Callahan for sequencing; Jean-Phillippe Fortin and Kasper Hansen for providing A/B compartment data; Andrew Sullivan and Alex Gimelbrant for access to imprinted gene and MAE datasets; Alexander Meissner and Michael Ziller for access to bisulfite sequencing datasets; and Winston Timp and Kasper Hansen for critical reading of the manuscript. This work was supported by NIH Grants R01CA054348 and DP1ES022579 to A.P.F., NSF Grants CCF-1217213 and CCF-1656201 to J.G., and NIH Grant AG021334 to Jeremy Walston. E.P. was supported by the Medical Scientist Training Program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
URLs. Gene bodies, http://genome.ucsc.edu/; curated list of enhancers, http://enhancer.lbl.gov/; H1 and IMR90 TAD boundaries, http://chromosome.sdsc.edu/mouse/hi-c/download.html; BED files for Hi-C data processed into compartments A/B, https://github.com/Jfortin1/HiC_AB_Compartments.
Accession codes. Whole-genome bisulfite sequencing data and bigWig files of relevant features: GSE86340. URL at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse86340
Software availability. Source code is available at https://github.com/GarrettJenkinson/informME/.
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS
A.P.F., E.P., G.J., and J.G. designed the study. G.J. and J.G. developed the mathematical and computational methods. G.J. wrote the computer code and implemented the methods. A.P.F. and E.P. designed and led the experiments. E.P. procured outside data, performed quality control, preprocessing and bisulfite alignment. G.J., E.P., A.P.F., and J.G. analyzed the data. A.P.F., G.J., and J.G. wrote the manuscript with the assistance of E.P.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Waddington CH. The strategy of the genes. Allen and Unwin; London: 1957. [Google Scholar]
- 2.Feinberg AP, Irizarry RA. Evolution in health and medicine Sackler colloquium: Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci U S A. 2010;107(Suppl 1):1757–64. doi: 10.1073/pnas.0906183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansen KD, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–75. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Landan G, et al. Epigenetic polymorphism and the stochastic formation of differentially methylated regions in normal and cancerous tissues. Nat Genet. 2012;44:1207–14. doi: 10.1038/ng.2442. [DOI] [PubMed] [Google Scholar]
- 5.Shipony Z, et al. Dynamic and static maintenance of epigenetic memory in pluripotent and somatic cells. Nature. 2014;513:115–9. doi: 10.1038/nature13458. [DOI] [PubMed] [Google Scholar]
- 6.Pujadas E, Feinberg AP. Regulated noise in the epigenetic landscape of development and disease. Cell. 2012;148:1123–31. doi: 10.1016/j.cell.2012.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer. 2013;13:497–510. doi: 10.1038/nrc3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bock C. Analysing and interpreting DNA methylation data. Nat Rev Genet. 2012;13:705–19. doi: 10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- 9.Presse S, Ghosh K, Lee J, Dill KA. Principles of maximum entropy and maximum caliber in statistical physics. Rev Mod Phys. 2013;85:1115–41. [Google Scholar]
- 10.Cedar H, Bergman Y. Programming of DNA methylation patterns. Annu Rev Biochem. 2012;81:97–117. doi: 10.1146/annurev-biochem-052610-091920. [DOI] [PubMed] [Google Scholar]
- 11.Heyn H, et al. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci U S A. 2012;109:10522–7. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziller MJ, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–81. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell RE, et al. Enhancer methylation dynamics contribute to cancer plasticity and patient mortality. Genome Res. 2016;26:601–11. doi: 10.1101/gr.197194.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohn F, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–66. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–5. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49:773–82. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J, Marco E, Pinello L, Yuan GC. Predicting chromatin organization using histone marks. Genome Biol. 2015;16:162. doi: 10.1186/s13059-015-0740-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cover TM, Thomas JA. Elements of Information Theory. John Wiley & Sons; New York: 1991. [Google Scholar]
- 21.Savarese F, et al. Satb1 and Satb2 regulate embryonic stem cell differentiation and Nanog expression. Genes Dev. 2009;23:2625–38. doi: 10.1101/gad.1815709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karantzali E, et al. Sall1 regulates embryonic stem cell differentiation in association with nanog. J Biol Chem. 2011;286:1037–45. doi: 10.1074/jbc.M110.170050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu K, et al. The multiple roles for Sox2 in stem cell maintenance and tumorigenesis. Cell Signal. 2013;25:1264–71. doi: 10.1016/j.cellsig.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozair MZ, Noggle S, Warmflash A, Krzyspiak JE, Brivanlou AH. SMAD7 directly converts human embryonic stem cells to telencephalic fate by a default mechanism. Stem Cells. 2013;31:35–47. doi: 10.1002/stem.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gopinath SD, Webb AE, Brunet A, Rando TA. FOXO3 promotes quiescence in adult muscle stem cells during the process of self-renewal. Stem Cell Reports. 2014;2:414–26. doi: 10.1016/j.stemcr.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahaira LG, et al. IGF2BP1 expression in human mesenchymal stem cells significantly affects their proliferation and is under the epigenetic control of TET1/2 demethylases. Stem Cells Dev. 2014;23:2501–12. doi: 10.1089/scd.2013.0604. [DOI] [PubMed] [Google Scholar]
- 27.Lee BK, et al. Tgif1 Counterbalances the Activity of Core Pluripotency Factors in Mouse Embryonic Stem Cells. Cell Rep. 2015;13:52–60. doi: 10.1016/j.celrep.2015.08.067. [DOI] [PubMed] [Google Scholar]
- 28.Luo Z, et al. Zic2 is an enhancer-binding factor required for embryonic stem cell specification. Mol Cell. 2015;57:685–94. doi: 10.1016/j.molcel.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fortin JP, Hansen KD. Reconstructing A/B compartments as revealed by Hi-C using long-range correlations in epigenetic data. Genome Biol. 2015;16:180. doi: 10.1186/s13059-015-0741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dixon JR, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–6. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Timp W, et al. Large hypomethylated blocks as a universal defining epigenetic alteration in human solid tumors. Genome Med. 2014;6:61. doi: 10.1186/s13073-014-0061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berman BP, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2012;44:40–6. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu H, et al. Tet3 regulates synaptic transmission and homeostatic plasticity via DNA oxidation and repair. Nat Neurosci. 2015;18:836–43. doi: 10.1038/nn.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamura T, et al. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat Genet. 1996;12:154–8. doi: 10.1038/ng0296-154. [DOI] [PubMed] [Google Scholar]
- 36.Kaneda H, et al. FOXQ1 is overexpressed in colorectal cancer and enhances tumorigenicity and tumor growth. Cancer Res. 2010;70:2053–63. doi: 10.1158/0008-5472.CAN-09-2161. [DOI] [PubMed] [Google Scholar]
- 37.Schlaeger TM, et al. A comparison of non-integrating reprogramming methods. Nat Biotechnol. 2015;33:58–63. doi: 10.1038/nbt.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vandiver AR, et al. Age and sun exposure-related widespread genomic blocks of hypomethylation in nonmalignant skin. Genome Biol. 2015;16:80. doi: 10.1186/s13059-015-0644-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen KD, et al. Large-scale hypomethylated blocks associated with Epstein-Barr virus-induced B-cell immortalization. Genome Res. 2014;24:177–84. doi: 10.1101/gr.157743.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu H, Caffo B, Jaffee HA, Irizarry RA, Feinberg AP. Redefining CpG islands using hidden Markov models. Biostatistics. 2010;11:499–514. doi: 10.1093/biostatistics/kxq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wen B, et al. Euchromatin islands in large heterochromatin domains are enriched for CTCF binding and differentially DNA-methylated regions. BMC Genomics. 2012;13:566. doi: 10.1186/1471-2164-13-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–51. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 44.Huyer W, Neumaier A. Global optimization by multilevel coordinate search. J. Global Optim. 1999;14:331–355. [Google Scholar]
- 45.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Recipes. The Art of Scientific Computing. Cambridge University Press; Cambridge: 2007. [Google Scholar]
- 46.Mohammad-Djafari A. A Matlab program to calculate the maximum entropy distributions. In: Smith CR, Erickson GJ, Neudorfer PO, editors. Maximum Entropy and Bayesian Methods. Kluwer Academic Publishers; 1991. pp. 221–234. [Google Scholar]
- 47.Lin J. Divergence measures based on the Shannon entropy. IEEE Trans. Inform. Theory. 1991;37:145–151. [Google Scholar]
- 48.Favorov A, et al. Exploring massive, genome scale datasets with the GenometriCorr package. PLoS Comput Biol. 2012;8:e1002529. doi: 10.1371/journal.pcbi.1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–80. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murtagh F, Legendre P. Ward's hierarchical agglomerative clustering method: Which algorithms implement Ward's criterion? J Classif. 2014;31:274–295. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.