Significance
Viruses are adept at exploiting the host cytoskeleton to facilitate various aspects of their replication. Among host cytoskeletal regulators, diaphanous-related formins (DRFs) coordinate actin nucleation and microtubule (MT) stabilization in response to various environmental signals. Here, we uncover a function of DRFs and show that human immunodeficiency virus type 1 (HIV-1) exploits DRFs to coordinate the disassembly of the viral capsid shell, or “uncoating,” with induction of MT stabilization and virus transport. These two functions of DRFs during HIV-1 infection were genetically separable and independent of actin. Our findings suggest that HIV-1 coopts discrete functions of DRFs to coordinately control uncoating and MT-based virus transport, mimicking how DRFs naturally function to coordinate host actin and MT dynamics.
Keywords: diaphanous-related formin, HIV-1, HIV-1 uncoating, viral trafficking, stable microtubules
Abstract
Diaphanous (Dia)-related formins (DRFs) coordinate cytoskeletal remodeling by controlling actin nucleation and microtubule (MT) stabilization to facilitate processes such as cell polarization and migration; yet the full extent of their activities remains unknown. Here, we uncover two discrete roles and functions of DRFs during early human immunodeficiency virus type 1 (HIV-1) infection. Independent of their actin regulatory activities, Dia1 and Dia2 facilitated HIV-1–induced MT stabilization and the intracellular motility of virus particles. However, DRFs also bound in vitro assembled capsid–nucleocapsid complexes and promoted the disassembly of HIV-1 capsid (CA) shell. This process, also known as “uncoating,” is among the most poorly understood stages in the viral lifecycle. Domain analysis and structure modeling revealed that regions of Dia2 that bound viral CA and mediated uncoating as well as early infection contained coiled-coil domains, and that these activities were genetically separable from effects on MT stabilization. Our findings reveal that HIV-1 exploits discrete functions of DRFs to coordinate critical steps in early infection and identifies Dia family members as regulators of the poorly understood process of HIV-1 uncoating.
The intracellular transport and subcellular localization of macromolecular cargos is mediated by actin and microtubule (MT) networks and their associated motors. As large cargos themselves, viruses also depend upon these host trafficking networks throughout their lifecycle and have evolved an array of strategies to manipulate cytoskeletal functions (1, 2). Like many other viruses, human immunodeficiency virus type 1 (HIV-1), the retrovirus that causes AIDS, exploits the actin cytoskeleton for short-range movement at the cell periphery followed by long-range transport on MT networks toward the nucleus (3). Although the events immediately after HIV-1 entry into the cell are among the least understood aspects of the viral life cycle, it is known that after the viral core is released into the cytosol, the viral RNA genome is reverse transcribed into double-stranded DNA, which then enters the nucleus through nuclear pores and integrates into the host chromosome (4). The HIV-1 core is a conical structure composed of ∼1,500 molecules of capsid (CA) protein oligomerized into hexameric structures (5). Given that the intact HIV-1 core is too large to pass through nuclear pores, disassembly of the CA shell, a poorly understood process called “uncoating,” is an important step in the early viral life cycle (6). However, whether uncoating occurs in the cytoplasm (7, 8) and/or at the nuclear pore (9–11) remains contentious. This process is intricately linked to reverse transcription, which can trigger uncoating (12–14). Similar to uncertainty surrounding uncoating, it has also been proposed that reverse transcription can occur during transport on MTs (15) or upon reaching the nucleus (16). Indeed, although the core is proposed to shield the viral genome from cytosolic sensors (17, 18), this function could conceivably be performed by the reverse transcription machinery and formation of the preintegration complex. Indeed, live cell imaging has suggested that HIV-1 uncoating may occur in the cytoplasm, but that some CA is retained (11). In addition, CA can be detected in the nucleus (19–21) and influence integration (22, 23), establishing that at least some CA remains somehow associated with the viral genome during postnuclear entry events. Despite uncertainty over where uncoating and reverse transcription occur, it is known that HIV-1 particles exhibit MT-based bidirectional motility during transport to the nucleus (15, 16, 24). Notably, targeted gene silencing of either inward or outward MT motors (dynein and kinesin-1, respectively), pharmacological blockade of motor function, or disruption of MT networks all impair trafficking of HIV-1 and delay uncoating (15, 24–26). These results indicate that MTs and motors are involved in long-range transport of viral cores, and that trafficking is somehow linked to capsid stability. However, although MTs are clearly involved in HIV-1 infection through effects on virus transport and capsid stability, potential links between these processes and how they are coordinated remain unclear.
We, and others have recently shown that HIV-1 stimulates the formation of stable MT subsets to promote early infection (27, 28). Indeed, whereas it is well established that long-range transport is mediated by a highly dynamic network of MTs, the formation of a small subset of stable, posttranslationally modified MTs can act as specialized tracks for vesicle trafficking (29, 30). Although these posttranslational modifications, which include acetylation and detyrosination, do not impart stability per se, they accumulate on stable MTs and are thought to enable recognition by specific motor proteins. MTs are polarized filaments whose minus ends nucleate from a MT-organizing center (MTOC), whereas their plus ends radiate toward the plasma membrane. MT dynamics and stabilization are regulated by an array of specialized proteins that associate with the plus ends of growing MTs, called plus end tracking proteins (+TIPs) (31). Although many +TIPs can directly bind MTs, their recruitment to growing plus ends requires the end-binding protein 1 (EB1) (31, 32). EB1’s recruitment of other +TIPs creates functional modules at the ends of growing MTs, facilitating interactions with the cell cortex, organelles, and cargos, as well as inducing localized signaling and MT stabilization at specific subcellular sites (31, 33). Among these pathways, diaphanous-related formins (DRFs) are core effectors of RhoA-mediated effects on MT stability (34, 35). The DRF family contains three isoforms, Dia1, Dia2, and Dia3. The most widely studied isoform is the mouse ortholog of human Dia1, mDia1, which shares 90.3% sequence identity to its human counterpart. Notably, DRFs regulate both actin nucleation and MT stability, functioning as downstream effectors of the Rho family of small GTPases and enabling coordinated remodeling of actin and MT networks. Prompted by our earlier discovery that EB1 acts as a critical mediator of HIV-1–induced MT stabilization and early infection (27), here we show that the Dia family members, Dia1 and Dia2, coordinate discrete processes during early HIV-1 infection through distinct effects on MT stability and CA disassembly. Whereas DRFs facilitated MT stabilization and HIV-1 transport independently of their effects on actin, we also identify previously unknown and functionally distinct properties of DRFs as host cofactors that bind HIV-1 cores and regulate uncoating. Mimicking the function of DRFs in coordinating actin and MT dynamics, our findings suggest that DRFs are exploited by HIV-1 to coordinate its trafficking and uncoating.
Results
Dia1 and Dia2 Promote HIV-1 Infection.
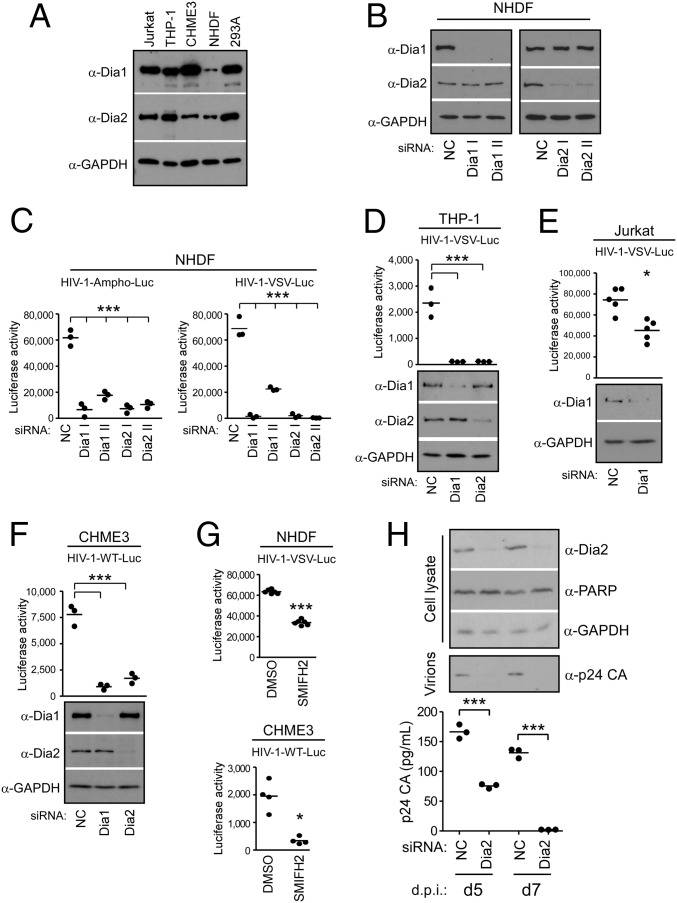
Whereas EB1 is critical for HIV-1–induced MT stabilization and the translocation of the incoming viral cores to the nucleus (27), the underlying mechanisms and contribution of stable MTs remain unclear. Given the established role of the Rho–Dia–EB1 pathway in stable MT formation (34, 35), we tested whether Dia1 and Dia2 were also involved in infection by HIV-1. To do this test, we first verified that Dia1 and Dia2 were expressed in biologically relevant cell types, including T cells (Jurkat), monocytes (THP-1), and microglial cells (CHME3), as well as in primary normal human dermal fibroblasts (NHDFs) and human embryonic kidney cells (293A) (Fig. 1A). To test whether endogenous Dia1 and/or Dia2 could influence cellular susceptibility to infection, primary NHDFs were treated with control, nontargeting siRNAs or siRNAs targeting Dia1 or Dia2. siRNA-treated cells were then infected with HIV-1 carrying a luciferase or ZsGreen reporter gene and pseudotyped with either a MuLV amphotropic envelope, which mediates entry by fusion at the cell surface, or vesicular stomatitis virus G envelope glycoprotein (VSV-G), which mediates entry by endocytosis. Infecting cells with both pseudotyped viruses allowed us to assess potential effects on the entry of virus through either of the two pathways used by HIV-1 (36, 37). Depletion of Dia1 or Dia2 using either of two independent siRNAs suppressed infection by HIV-1 pseudotyped with either envelope compared with control siRNA-treated cultures (Fig. 1 B and C and Fig. S1 A and B). Whereas Dia1 or Dia2 siRNAs did not induce apoptosis (Fig. S1 C and D), further ruling out potential off-target effects, siRNA-resistant Dia1 rescued infection in Dia1 siRNA-treated cells (Fig. S1E). This result indicated these DRFs promote early HIV-1 infection regardless of the route of viral entry. To verify these results in natural target cells for HIV-1 infection, Dia1 or Dia2 were depleted in THP-1 cells that were differentiated into macrophages, as well as Jurkat or CHME3 cells, followed by infection with HIV-1 containing a luciferase reporter and pseudotyped with either VSV-G (HIV-1–VSV–Luc) or wild-type HIV-1 envelope (HIV-1–WT–Luc). In all cases, DRF depletion again suppressed HIV-1 infection (Fig. 1 D–F). To independently confirm these siRNA-based observations, we tested the effects of SMIFH2, a small molecule competitive inhibitor that targets the formin homology 2 (FH2) domain and blocks the ability of formins to interact with downstream targets (38). Treatment of NHDF or CHME3 cells with SMIFH2 was found to inhibit early HIV-1 infection compared with DMSO-treated control cells (Fig. 1G). Finally, validating findings using pseudotyped HIV-1, CHME3 4 × 4 cells, expressing higher levels of CD4 and CXCR4 (39), were depleted of Dia2 followed by infection with WT transmitted/founder isolate (JR-CSF derived) HIV-1. Similar to findings above, Dia depletion had no adverse effects on cell viability, as determined by PARP-1 levels, but resulted in a reduction of the release of p24 CA into culture supernatants from cells infected with WT HIV-1, as determined by either Western blot (WB) analysis or ELISA over time (Fig. 1H). These findings established that DRFs were required for early HIV-1 infection in a variety of cell types, including natural target cells, independently of the route of viral entry.
Fig. 1.
Dia1 and Dia2 promote early HIV-1 infection in human cells. (A) Western blot (WB) showing the expression of Dia1 and Dia2 with GAPDH loading control in Jurkat, THP-1, CHME3, NHDF, and 293A cells. (B and C) NHDFs were transfected with negative control (NC) siRNA or two independent siRNAs targeting Dia1 (Dia1 I and Dia1 II) or Dia2 (Dia2 I and Dia2 II). Cells were either lysed and analyzed by WB (B) using antibodies against Dia1, Dia2, and GAPDH (loading control) or infected with HIV-1–VSV–Luc or HIV-1–Ampho–Luc (C). Levels of infection were determined by measurements of luciferase activity. Data are shown as scatterplot with mean, n = 3. (D–F) THP-1 cells differentiated into macrophages (D), Jurkat cells (E), or CHME3 cells (F) were transfected with NC, Dia1, or Dia2 siRNAs. Cells were either lysed and analyzed by WB using antibodies against Dia1, Dia2, and GAPDH (loading control) or infected with HIV-1–VSV–Luc (D and E) or HIV-1–WT–Luc (F) followed by measurements of luciferase activity. Data are shown as scatterplot with mean, n = 3, 5, and 3, respectively. (G) NHDF and CHME3 cells were treated with equal amounts of DMSO solvent control or SMIFH2 (10 μM), followed by infection with HIV-1–VSV–Luc or HIV-1–WT–Luc. Levels of infection were determined by measurements of luciferase activity. Data are shown as scatterplot with mean, n = 6 and 4, respectively. (H) CHME3 4 × 4 cells treated with NC or Dia2 siRNAs were infected with JR-CSF–derived HIV-1 followed by measurements of Dia2, PARP-1, or GAPDH in cell lysates as well as the release of p24 CA-containing HIV-1 particles into culture supernatants by either WB analysis or p24 ELISA at indicated days postinfection (d.p.i.). Similar results were obtained in at least three independent experiments. Statistical analysis of all data was performed using one-way ANOVA and posttest or t test, *P ≤ 0.05 and ***P ≤ 0.001, respectively.
Fig. S1.
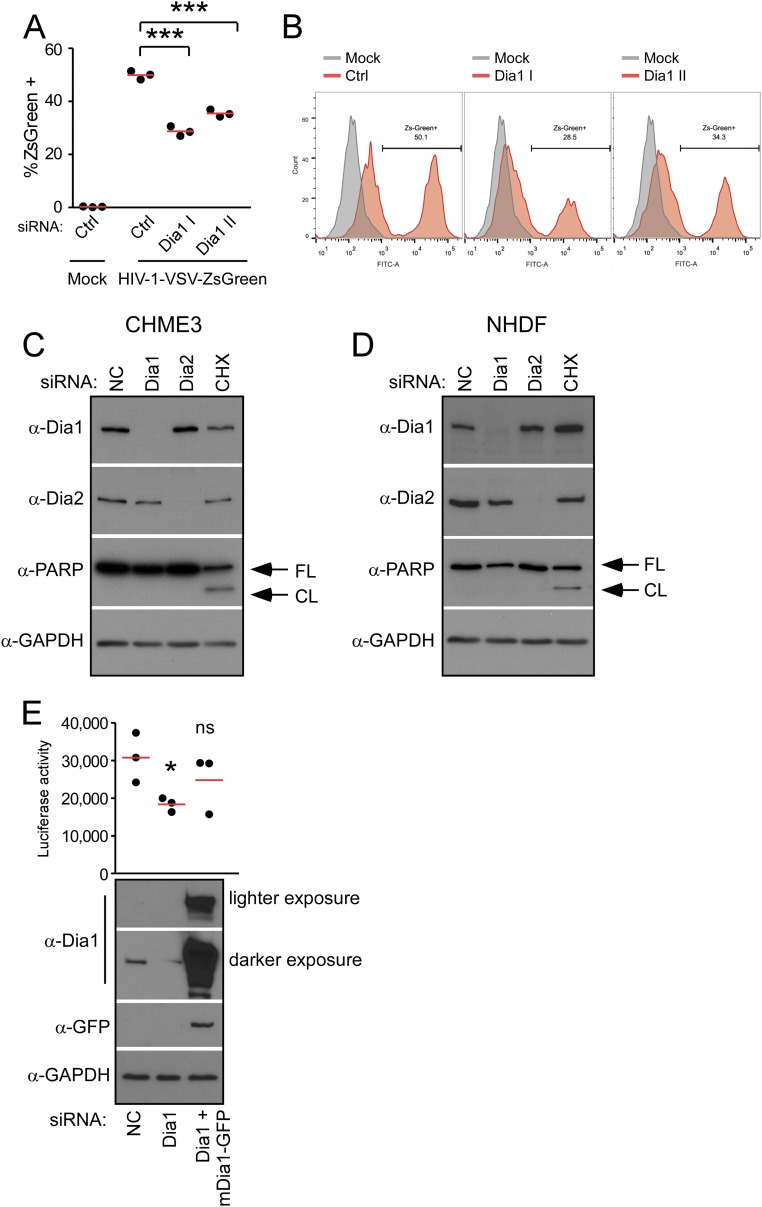
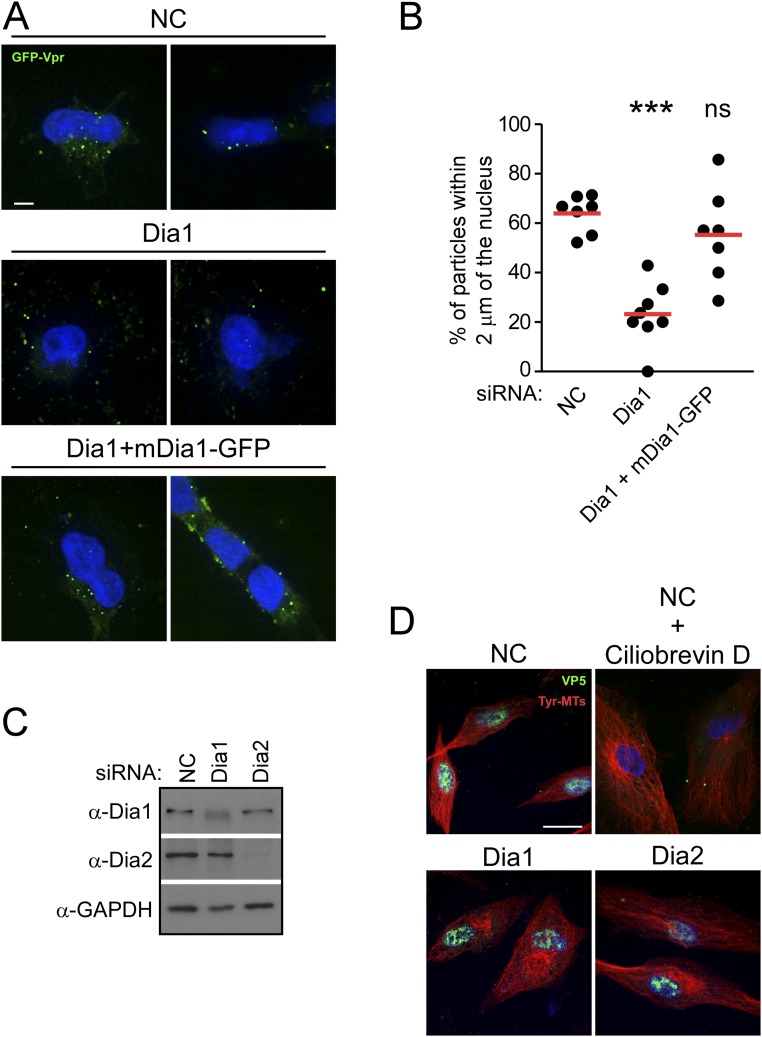
(A and B) Depletion of Dia1 or Dia2 reduces HIV-1 infection carrying ZsGreen marker. NHDF cells were transfected with two independent siRNAs targeting Dia1 (Dia1 I and Dia1 II) or negative control (Ctrl) and infected with HIV-1–VSV–ZsGreen virus, where ZsGreen is a fluorescent reporter. The cells were then detached and fixed for FACS analysis of ZsGreen expression. (A) The mean of the percent of ZsGreen+ cells. Data are shown as scatterplot with mean, n = 3. Statistical significance was determined using one-way ANOVA and posttest; ***P < 0.001. (B) Fluorescence histograms overlaying the ZsGreen fluorescence versus mock control in Ctrl, Dia1 I, and Dia1 II transfected NHDF cells. (C and D) Depletion of Dia1 or Dia2 does not induce apoptosis. CHME3 or NHDF cells were transfected with control siRNA (NC) or siRNA targeting Dia1 and Dia2. At 72 h after transfection, cell lysates were analyzed for expression levels of Dia1, Dia2, and the apoptotic marker Poly (ADP ribose) polymerase (PARP) by WB using specific antibodies. Full-length (FL) and cleaved (CL) forms of PARP are indicated at Right. NC transfected cells were treated with 200 μg/mL cycloheximide (CHX) for 13 h (CHME3) (C) or overnight (NHDF) (D) to induce the onset of apoptosis and included as a positive control to demonstrate detection of PARP cleavage in cells entering apoptosis. GAPDH was used as a loading control using anti-GAPDH antibody. Similar results were obtained in at least three independent experiments. (E) HIV-1 infectivity is restored in Dia1 siRNA-treated cells by siRNA-resistant Dia1. The 293T cells were treated with control siRNA (NC) or Dia1 siRNA followed by transfection with a siRNA-resistant GFP-tagged Dia1 (mDia1-GFP) construct 48 h later. The next day, cells were either infected with HIV-1–VSV–Luc followed by measurements of luciferease activity or lysed and subjected to WB analysis using antibodies specific to Dia1, GFP, or GAPDH (as loading control) (E). Data are shown as scatterplot with mean, n = 3. *P ≤ 0.05, ***P ≤ 0.001. ns, not significant.
Dia1 and Dia2 Promote an Early Postentry Stage of HIV-1 Infection Through Effects on Stable MTs.
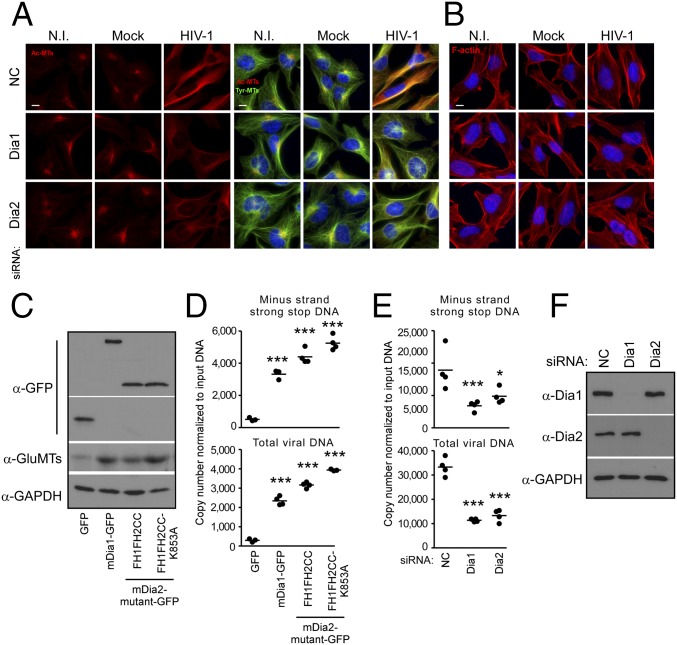
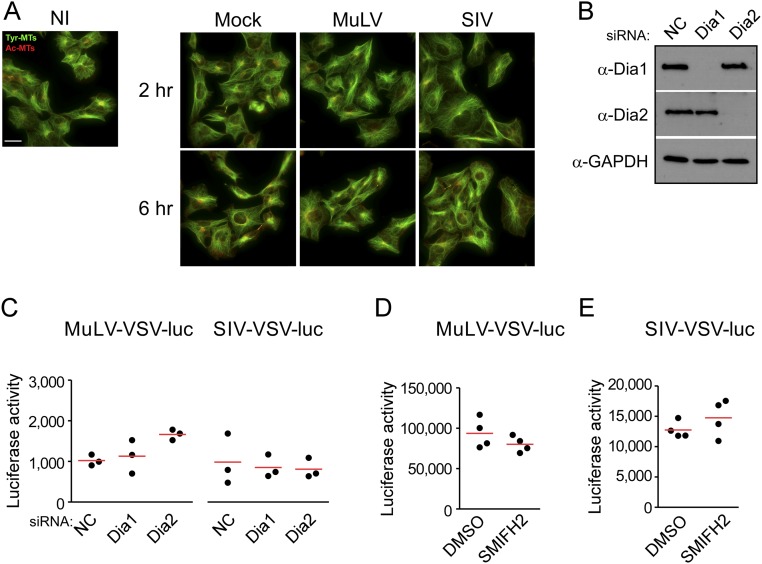
To determine whether DRFs were required for HIV-1–induced MT stabilization, siRNAs were used to deplete Dia1 or Dia2 in CHME3 cells before infection. Immunofluorescence (IF) analysis showed that in control siRNA-treated cultures, HIV-1 induced the stabilization of MTs, detected as an increase in the level of acetylated MTs (Ac-MTs), and this induction was reduced by depletion of Dia1 or Dia2 (Fig. 2A). These findings indicate that DRFs facilitate HIV-1–induced MT stabilization. Furthermore, IF analysis also confirmed that Dia1 or Dia2 depletion did not adversely affect actin fibers in CHME3 cells before or during HIV-1 infection (Fig. 2B), suggesting that DRFs promote HIV-1 infection independently of actin. To determine whether MT stabilization and a requirement for DRFs was specific to HIV-1, we infected CHME3 cells with two distinct VSV-G pseudotyped luciferase reporter retroviruses, MuLV or simian immunodeficiency virus (SIV). IF analysis also showed that in contrast to HIV-1, infection by these nonhuman retroviruses did not induce MT stabilization (Fig. S2A). Moreover, depletion of Dia1 or Dia2 or treatment with SMIFH2 (Fig. S2 B–E, respectively) had no effect on infection by either pseudotyped MuLV or SIV, indicating that the induction of MT stabilization and requirement for DRFs for early infection were specific to HIV-1 and likely related to Dia’s effects on MT stability.
Fig. 2.
Dia1 and Dia2 are required for HIV-1–mediated MT stabilization at an early postentry stage of infection. (A and B) CHME3 cells were transfected with negative control (NC), Dia1, or Dia2 siRNA and then noninfected (N.I.), mock infected, or infected with HIV-1–VSV at multiplicity of infection (MOI) 3. Samples were fixed after 6 h and stained for acetylated tubulin (Ac-MTs) and tyrosinated tubulin (Tyr-MTs) (A) or F-actin (B), together with Hoescht to stain the nucleus. Images are shown for representative fields. (Scale bar, 10 μm.) Similar results were obtained in at least three independent experiments. (C and D) Effects of Dia overexpression on HIV-1 DNA synthesis. CHME3 cells were transfected with control GFP, mDia1-GFP, or GFP-tagged truncation mutants of mDia2 (FH1FH2CC or FH1FH2CCK853A). At 48 h postinfection (h.p.i.), cells were either lysed and analyzed by WB using antibodies against GFP, detyrosinated tubulin (Glu-MTs), and GAPDH (as loading control) (C) or infected with HIV-1–VSV–puro (D). Low-molecular Hirt DNA was isolated at 24 h.p.i., and levels of viral MSS DNA and total viral DNA in samples were measured by qPCR using primers specific to MSS and puromycin, respectively (D). Copy numbers were calculated and normalized to input DNA in each sample. (E and F) Effects of Dia knockdown on HIV-1 DNA synthesis. (E) Measurements of early reverse transcription products in infected CHME3 cells transfected with NC, Dia1, or Dia2 siRNAs as described in D. (F) WB analysis showing the endogenous levels of Dia1, Dia2, or GAPDH (as loading control) in samples from E. Data are shown in D and E as scatterplot with mean, n = 4. Statistical analysis of all data was performed using one-way ANOVA and posttest, *P ≤ 0.05 and ***P ≤ 0.001, respectively.
Fig. S2.
Dia1 and Dia2 do not regulate MT stabilization and infection by nonhuman retroviruses. (A) CHME3 cells were left noninfected (NI) or infected with mock virus, MuLV–VSV–Luc or SIV–VSV–Luc. Then samples were fixed at the indicated hours post infection (h.p.i.) and stained for tyrosinated tubulin (Tyr-MTs) and acetylated tubulin (Ac-MTs) MTs. (Scale bar, 40 μm.) Similar results were obtained in at least three independent experiments. (B and C) CHME3 cells were transfected with siRNA targeting Dia1, Dia2, or negative control (NC). Cells were either lysed for WB to verify knockdown using antibodies against Dia1, Dia2, and GAPDH (as loading control) (B) or infected with MuLV or SIV virus carrying a luciferase reporter pseudotyped with VSVG envelope (C). Levels of infection were determined by measurements of luciferase activity. Data are shown as scatterplot with mean, n = 3. (D and E) CHME3 cells were treated with DMSO (0.1%) or 10 μM SMIFH2, and infected with (D) MuLV or (E) SIV virus carrying a luciferase reporter pseudotyped with VSVG envelope. Levels of infection were determined by luciferase assay. Similar results were obtained in four independent experiments. Data are shown as scatterplot with mean, n = 4.
To independently test whether DRFs influenced infection through their effects on MTs versus actin, and to determine the point in HIV-1 infection that was affected, CHME3 cells were transfected with plasmids expressing GFP control or GFP-tagged forms of mDia. These tagged Dia forms included full-length mDia1, a constitutively active truncation mutant of mDia2 (FH1FH2CC), as well as a point mutant of this constitutively active form of mDia2 (FH1FH2CCK853A), which harbors a K853A point mutation that prevents interaction with and regulation of actin, limiting its activity to MTs (40). WB analysis confirmed expression of the GFP-tagged mDia1 and mDia2 forms, which, in line with previous findings in mouse 3T3 cells (40), also enhanced stable MT levels in human CHME3 cells, as detected by an increase in detyrosinated MTs (also known as Glu-MTs, as detyrosination exposes a glutamic acid at the C terminus of tubulin) (Fig. 2C). To determine the point in infection that was affected by each Dia form, transfected cultures were infected with HIV-1–VSV and the levels of minus-strand strong stop (MSS) DNA, the first detectable retroviral DNA, as well as total viral DNA were determined (41). Quantitative real-time PCR (qPCR) analysis of viral MSS DNA and total viral DNA showed that overexpression of each mDia form enhanced the generation of early reverse transcription products, indicating that DRFs function before or at the initiation of reverse transcription (Fig. 2D). In line with this finding, knockdown of human Dia1 or Dia2 in CHME3 cells resulted in a reduction in early reverse transcription products (Fig. 2 E and F). Furthermore, the fact that reverse transcription was increased by the mDia2 mutant, FH1FH2CCK853A, demonstrated that DRFs promote very early stages of HIV-1 infection of natural target cells through effects on stable MTs and independently of its actin regulatory activity.
Dia1 and Dia2 Regulate the Trafficking of Incoming HIV-1 Particles.
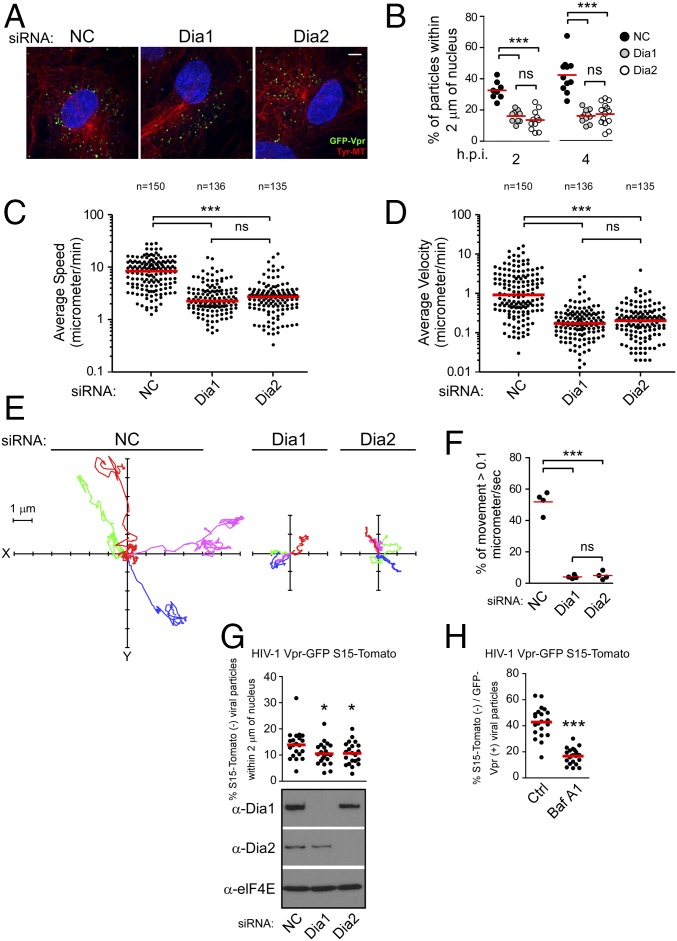
Given HIV-1’s requirement for DRFs to induce the formation of stable MT networks, we next tested whether Dia depletion impacted the transport of incoming HIV-1 particles toward the nucleus. This test was initially done using fixed imaging approaches. NHDFs treated with either control, Dia1, or Dia2 siRNAs were infected with HIV-1–VSV carrying GFP-labeled Vpr (GFP-Vpr) (15) and fixed at 2 or 4 h postinfection (h.p.i.). Samples were then stained for GFP and tyrosinated MTs, whereas the nucleus was stained using Hoechst, and the number of viral particles within 2 µm of the nucleus was measured (24) (Fig. 3). Whereas the number of viral particles within 2 μm of the nucleus increased over time in control siRNA-treated cells, the majority of viral particles remained scattered and distant from the nucleus in Dia1- or Dia2-depleted cells (Fig. 3 A and B). Ruling out potential off-target effects, although viral particles failed to reach the nucleus in Dia1 siRNA-treated cells compared with controls, translocation to the nucleus was rescued in Dia1 siRNA-treated cultures expressing siRNA-resistant Dia1 (Fig. S3 A and B). In addition, depletion of Dia1 or Dia2 did not affect the transport of HSV-1 to the nucleus, another viral cargo that uses dynein-mediated transport but on dynamic rather than stable MTs (42) (Fig. S3 C and D), and whose replication and spread we have shown is unaffected by Dia depletion (43). These findings suggested that DRFs mediate the movement of incoming HIV-1 cores toward the nucleus. To verify that virus motility was indeed suppressed, the effects of DRF depletion in CHME3 cells on the motility of HIV-1 pseudotyped with WT envelope whose core was labeled using GFP-Vpr was examined using live cell microscopy (24) (Fig. 3 C–F). Whereas rapid, long-range movement of HIV-1 particles was readily observed in control siRNA-treated cells (Movie S1), depletion of either Dia1 or Dia2 (Movies S2 and S3, respectively) potently suppressed HIV-1 motility. Analysis of individual virus particle trajectories revealed that in Dia1 or Dia2 knockdown cells, there was a significant reduction in average speed and average velocity relative to control siRNA-treated cells (Fig. 3 C and D and Movies S4–S6). Particle tracking also allowed us to determine whether HIV-1 particles exhibited MT-based movement, characterized as greater than 0.1 μm/s (15, 16, 24). Only ∼5% of viral particles in Dia1- or Dia2-depleted cells moved at greater than 0.1 μm/s, whereas most exhibited slower, short-range movements characteristic of either actin-based motility or free diffusion (Fig. 3 E and F). This finding was in stark contrast to the ∼50% of viral particles exhibiting MT-based movement in control siRNA-treated cells. Finally, to distinguish viral particles that had fused into the cytoplasm from those that might have been nonproductively endocytosed, cells treated with control or Dia siRNAs were infected with VSV-G pseudotyped HIV-1 containing GFP-Vpr (labeling cores) and S15-Tomato (labeling viral membranes) (24, 44). Importantly, these infections were performed using virus preparations that were >90% double labeled. Depletion of Dia1 or Dia2 decreased the number of fused (shed the S15-Tomato and appear green) viral particles within 2 μm of the nucleus compared with control (Fig. 3G). Treatment of cells with the fusion inhibitor, bafilomycin, resulted in a significant reduction in the percentage of green viral particles in the cell (Fig. 3H), further confirming that these represented fused virus, similar to previous reports (24, 44, 45). Altogether, fixed and live cell imaging approaches established that DRFs promote MT-based retrograde trafficking of HIV-1 cores to the nucleus. Importantly, the motility of cellular cargos such as mitochondria was not affected in Dia1- or Dia2-depleted CHME3 cells (Movies S7–S9), demonstrating that DRF depletion did not result in gross defects in host cargo transport and specifically promoted the trafficking of HIV-1, but not other viruses tested above. This finding established DRFs as key mediators of HIV-1–induced MT stabilization and long-range movement of viral cores to the nucleus.
Fig. 3.
Dia1 and Dia2 promote trafficking of HIV-1 particles to the nucleus. (A and B) Dia1 and Dia2 depletion inhibits the translocation of viral particles to the nucleus. NHDFs treated with NC, Dia1, or Dia2 siRNAs were infected with HIV-1–VSV–GFP-Vpr at MOI 1. Samples were fixed and stained (A) for Tyr-MTs, GFP, and nuclei (Hoescht). Representative images are shown. (Scale bar, 10 μm.) (B) Quantification of the percentage virions within 2 μm of the nucleus in infected cells at indicated times postinfection, represented as scatterplot with mean with ≥600 virus particles quantified in ≥20 cells per sample. (C–F) Depletion of Dia1 and Dia2 impairs the trafficking of HIV-1. CHME3 cells transfected with negative control (NC), Dia1, or Dia2 siRNAs were infected with HIV-1–WT containing GFP-Vpr followed by live cell imaging over a period of 5–10 min at one frame per second using a spinning-disk confocal microscope. Particle tracking was used to track and quantify virus movement in trajectories. (C) The average speed (cumulative distance traveled/time tracked) of virus particles. (D) The average velocity (displacement/time tracked) of virus particles. (C and D) A total of ≥135 trajectories were analyzed (n value above treatment) from ≥10 movies per treatment and are shown as scatterplot with median. (E) Four representative viral trajectories where the virus particle started moving at x = 0, y = 0. (F) The percent of movement (micrometers per second) of virus particles in E that is associated with microtubules (>0.1 μm/s). Similar results were obtained in four independent experiments. Data are shown as scatterplot with mean. (G) CHME3 cells were treated with control, Dia1, or Dia2 siRNAs followed by infection with HIV-1–VSV containing GFP-Vpr and S15-Tomato. At 1 h.p.i. cells were fixed and GFP and Tomato signals were acquired using a spinning-disk confocal microscope. The percent fused (S15-Tomato−, GFP+) viral particles within 2 μm of the nucleus was quantified in n ≥ 21 cells and an average of 95 viral particles per cell and is shown as scatterplot with mean. (Lower) Knockdown levels analyzed by WB using antibodies against Dia1, Dia2, or eIF4E (as loading control) before infection. (H) Control siRNA-transfected cells (Ctrl) were treated with Bafilomycin A1 (Baf A1) for 2 h during spinoculation followed by infection as described and processed in G. Data are shown as scatterplot with mean, n = 21. Statistical analysis was determined using one-way ANOVA with posttest or t test, *P ≤ 0.05 and ***P ≤ 0.001, respectively; ns, not significant.
Fig. S3.
(A and B) Translocation of HIV-1 cores is restored in Dia1-expressing cells in the presence of Dia1 siRNA. The 293T cells treated with control siRNA (NC) or Dia1 siRNA were transfected with a siRNA-resistant GFP-Dia1 construct (mDia1-GFP) after 48 h, followed by infection with HIV-1–GFP–Vpr at MOI 1. Samples were fixed and stained for GFP and nuclei (Hoescht) (A). Representative images are shown. (Scale bar, 10 μm.) (B) Quantification of the percentage of virions within 2 μm of the nucleus in infected cells at 4 h.p.i., represented as scatterplot with mean; ≥155 virus particles were quantified in ≥11 cells per sample. Statistical significance was determined using one-way ANOVA and posttest, ***P < 0.001; ns, not significant. (C and D) Depletion of Dia1 or Dia2 does not affect translocation of HSV-1 to the nucleus. NHDFs were treated with NC, Dia1, or Dia2 siRNAs. After 72 h, the cells were either (C) lysed and subjected to WB analysis using antibodies specific to Dia1, Dia2, or GAPDH (as loading control) or (D) infected with HSV-1 strain F at MOI 30 for 5 h followed by fixing and staining for the major capsid protein VP5 (green) and tyrosinated tubulin (Tyr-MTs). Nuclei were stained with Hoechst. Representative wide-field images are shown. As a control for the role of MT-based movement in HSV-1 infection, NC siRNA treated NHDF were treated with the dynein inhibitor, ciliobrevin D, which blocks HSV-1 transport to the nucleus (42). (Scale bar, 10 μm.)
Dia1 and Dia2 Promote Uncoating of Incoming HIV-1 Cores.
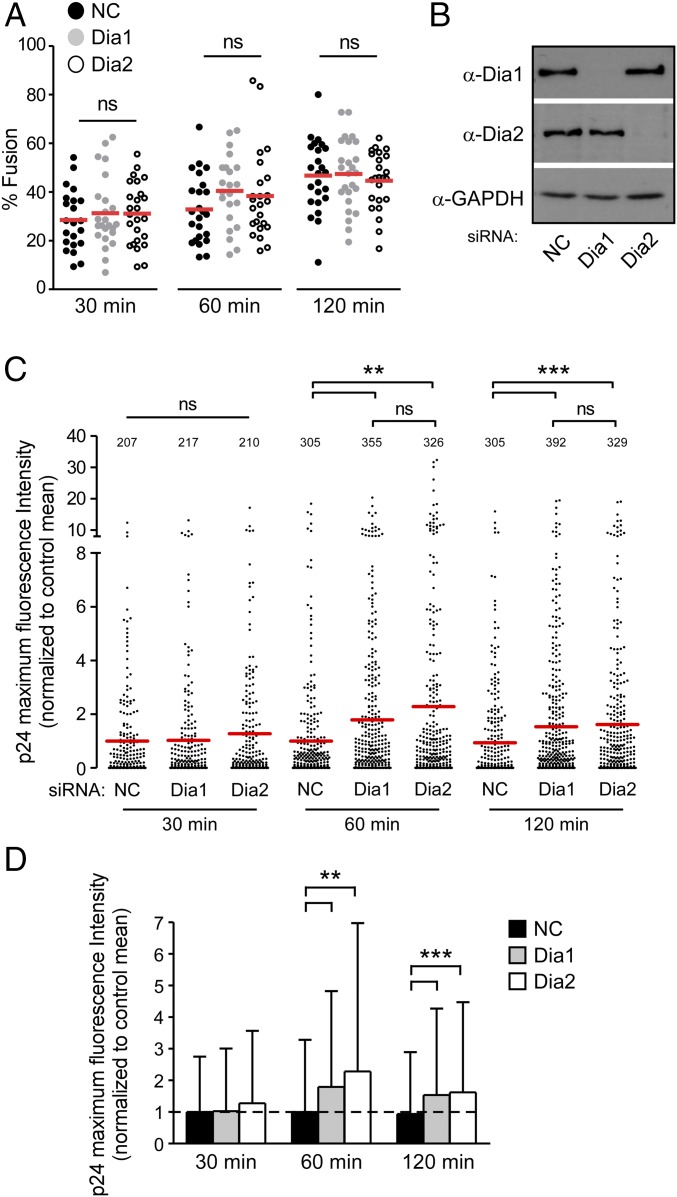
Given that DRFs enhanced the accumulation of early reverse transcription products and promoted the trafficking of HIV-1 particles, we next determined whether Dia1 or Dia2 influenced HIV-1 uncoating. We used two independent assays widely used to measure uncoating. First, we used an in situ fluorescence microscopy uncoating assay that determines the levels of p24 CA protein that remains associated with virus particles that have fused into the cytosol (46). CHME3 cells were treated with control, Dia1, or Dia2 siRNAs followed by infection with the double-labeled HIV-1 (S15-Tomato/GFP-Vpr) described above. Fixed cells were then stained for p24. Imaging the relative loss of S15-Tomato–labeled envelope showed that virus fusion was not significantly affected by depletion of either Dia1 or Dia2 compared with controls (Fig. 4 A and B). This finding was in line with earlier findings that Dia1 or Dia2 depletion affected infection independently of the viral envelope used in multiple cell types (Fig. 1 C–F and H), demonstrating a postentry block to infection. Quantification of p24 staining intensities associated with GFP-Vpr+/S15-Tomato− viral particles (24) revealed that depletion of Dia1 or Dia2 led to a significant increase in the amount of p24 associated with fused virions compared with that of control siRNA-treated cultures at either 1 or 2 h.p.i. (Fig. 4 C and D). This finding suggested that CA loss, or uncoating, was delayed in DRF-depleted CHME3.
Fig. 4.
Dia1 and Dia2 regulate HIV-1 uncoating as measured by in situ uncoating assay. (A–D) CMHE3 cells transfected with negative control (NC), Dia1, or Dia2 siRNA were infected with GFP-Vpr and S15-Tomato–labeled VSV-G pseudotyped HIV-1 virus. Virions were analyzed for the presence of p24, GFP, and S15 as described in Materials and Methods. (A) Quantification of the percentage fused (GFP+, S15-Tomato−) virus particles at the indicated times postinfection. Data are shown as scatterplot with mean; ≥200 virus particles were analyzed in ≥22 fields of view (≥30 cells) per siRNA treatment at each time point. (B) Cells were also lysed and analyzed by WB using antibodies against Dia1, Dia2, and GAPDH (loading control). (C) The maximum p24 intensity of each virus particle is shown in a scatterplot where the data were normalized to the mean of the control at each time point and the mean of each group is shown in red. (D) Bar graph shows the means of the maximum fluorescence intensities of p24 with SD. Dotted line across bar graph indicates control mean for reference. Statistical analysis for all data was determined using one-way ANOVA with posttest, **P ≤ 0.01 and ***P ≤ 0.001, respectively; ns, not significant.
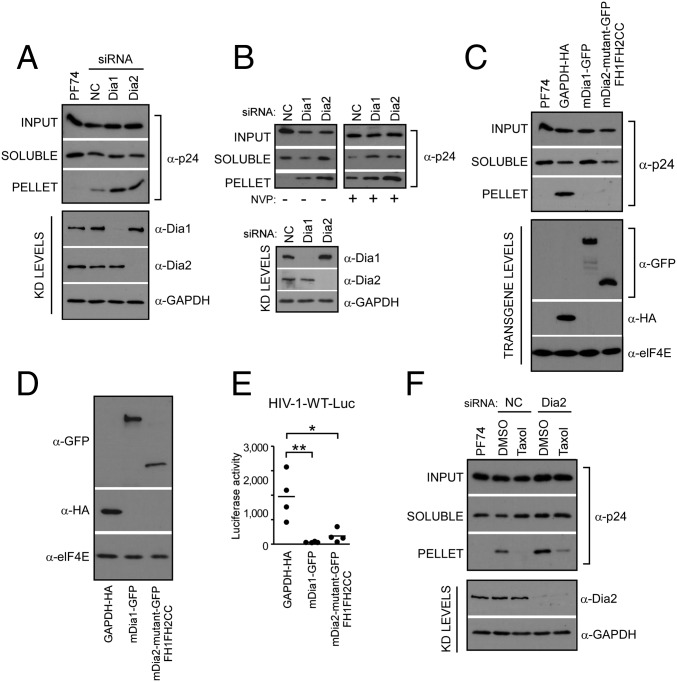
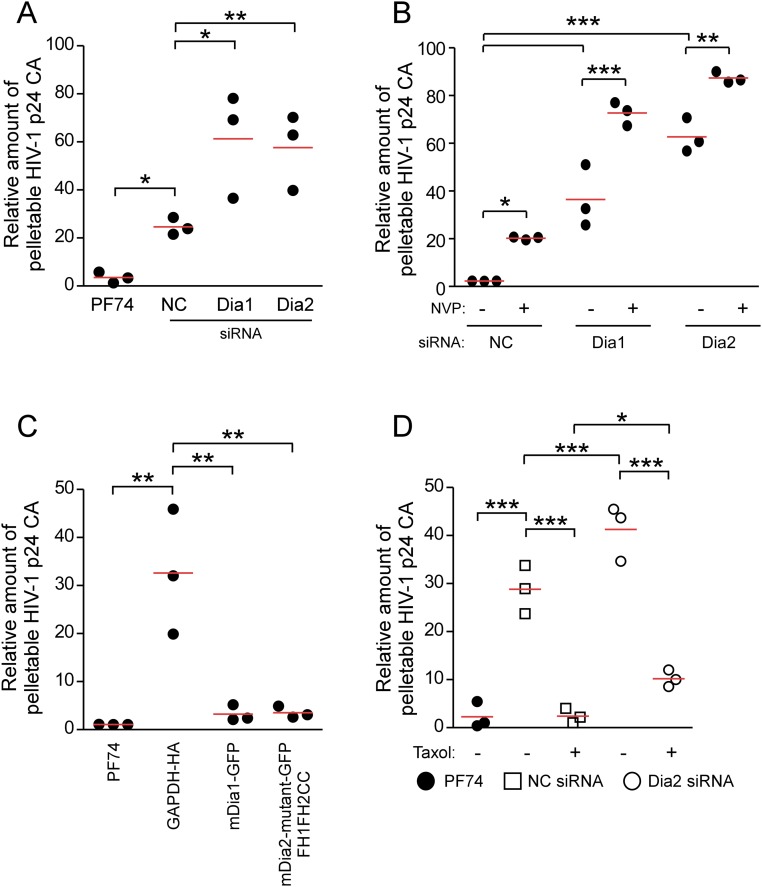
To independently confirm these findings using in situ uncoating assays, a second, fate-of-capsid assay in which intact HIV-1 cores are sedimented from infected cell lysates through a sucrose gradient was used (47). This assay was performed at 3 h.p.i. in cells either depleted of Dia1 or Dia2, or in cells expressing GFP-tagged Dia1 or the GFP-tagged constitutively active mutant of Dia2 (FH1FH2CC), which promotes MT stabilization. As a control for detection of effects on uncoating, cells were also treated with PF74, a small molecule that destabilizes capsids at high concentrations (48). In line with results from the in situ fluorescence assays, knockdown of Dia1 or Dia2 increased the recovery of intact pelletable HIV-1 cores compared with control siRNA-treated cultures, whereas PF74 destabilized HIV-1 cores (Fig. 5A and Fig. S4A). Given that reverse transcription and uncoating are coupled processes (12–14), we then tested whether the impaired uncoating detected in Dia-depleted cells was a direct effect or a secondary consequence of an effect on reverse transcription by repeating these knockdown experiments in the presence or absence of the reverse transcription inhibitor, Nevirapine. Similar to previous reports, inhibition of reverse transcription resulted in impaired uncoating in control siRNA-treated cultures. However, Dia1 or Dia2 depletion exerted additive effects on uncoating in Nevirapine-treated cells compared with effects in control siRNA-treated samples (Fig. 5B and Fig. S4B). This finding demonstrated that Dia did not affect uncoating indirectly through effects on reverse transcription, in line with its ability to bind HIV-1 cores in assays below. Confirming these siRNA-based findings, overexpression of mDia1 or constitutively active mDia2 resulted in the opposite effect to depletion, decreasing the levels of pelletable cores compared with control cells (Fig. 5C and Fig. S4C). This decrease in pelletable cores resulting from Dia overexpression suggested that Dia accelerated the rate of uncoating. Either accelerating or slowing the natural rate of uncoating has been shown to have negative consequences for HIV-1 infectivity (18, 49, 50). In line with this finding, a decrease in infectivity with HIV-1–WT–Luc was observed in CHME3 cells expressing mDia1 or the constitutively active form of mDia2 (FH1FH2CC) compared with control lines (Fig. 5 D and E).
Fig. 5.
Dia1 and Dia2 regulate HIV-1 uncoating as measured by fate of capsid assay. (A and B) The 293A (A) or CHME3 (B) cells transfected with negative control (NC), Dia1, or Dia2 siRNA were infected with HIV-1–VSV-Luc in the absence (A and B) or presence (B) of Nevirapine (NVP) (10 μM) and the amounts of soluble and particulate capsid were determined at 3 h.p.i. (Lower) Knockdown (KD) levels analyzed by WB using antibodies against Dia1, Dia2, and GAPDH (loading control) before infection. (C) The 293A cells were transfected with HA-tagged GAPDH and GFP-tagged mDia1 or mDia2 FH1FH2CC. At 48 h posttransfection, cells were either infected with HIV-1 followed by measurements of the amount of soluble and particulate capsid or lysed for WB analysis as described in A. (D and E) Effects of mDia overexpression on HIV-1 infection. CHME3 cells were transfected with control GAPDH-HA, mDia1-GFP, or the GFP-tagged mDia2 mutant (FH1FH2CC). Cells were either lysed and analyzed by WB using antibodies against HA, GFP, and elF4E (as loading control) (D) or infected with HIV-1–WT–Luc (E) and levels of infection were determined by measurements of luciferase activity. Similar results were obtained in four independent experiments. Data are shown as scatterplot with mean, n = 4. (F) The 293A cells transfected with negative control (NC) or Dia2 siRNA were treated with equal volumes of DMSO solvent control or 100 nM taxol. Cultures were then infected and assayed for soluble and particulate capsid as described in A.
Fig. S4.
Quantification of the levels of pelletable p24 from fate of capsid uncoating assay presented in Fig. 5. (A) Densitometric quantification of the relative amount of pelletable p24 CA from Fig. 5A. (B) Densitometric quantification of the relative amount of pelletable p24 CA from Fig. 5B. (C) Densitometric quantification of the relative amount of pelletable p24 CA from Fig. 5C. (D) Densitometric quantification of the relative amount of pelletable p24 CA from Fig. 5F. Data are shown as scatterplot with mean, n = 3. Statistical analysis of all data was performed using one-way ANOVA and posttest, with statistical significance; *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001, respectively.
The fact that uncoating and infectivity were affected by DRFs suggested that they might do so through their effects on stable MTs. To determine whether Dia regulated early infection in a stable MT-dependent manner, control or Dia2 siRNA-treated cells were treated with taxol to induce MT stabilization. The fate-of-capsid assay was then used to monitor capsid stability during infection. In control siRNA-treated cultures, taxol accelerated uncoating compared with DMSO controls (Fig. 5F and Fig. S4D). This acceleration demonstrated that MT stabilization could promote uncoating, and was consistent with reports that MT depolymerization using nocodazole reduces uncoating (26). However, in Dia2-depleted cells significant amounts of pelletable capsid remained in taxol-treated samples (Fig. 5F and Fig. S4D). This finding suggested that DRFs promoted uncoating in part through MT stabilization, but might also regulate uncoating in a second, MT-independent manner. This observation prompted us to determine whether DRFs might physically associate with HIV-1 cores to influence uncoating.
Dia1 and Dia2 Associate with in Vitro Assembled Capsid–Nucleocapsid Complexes.
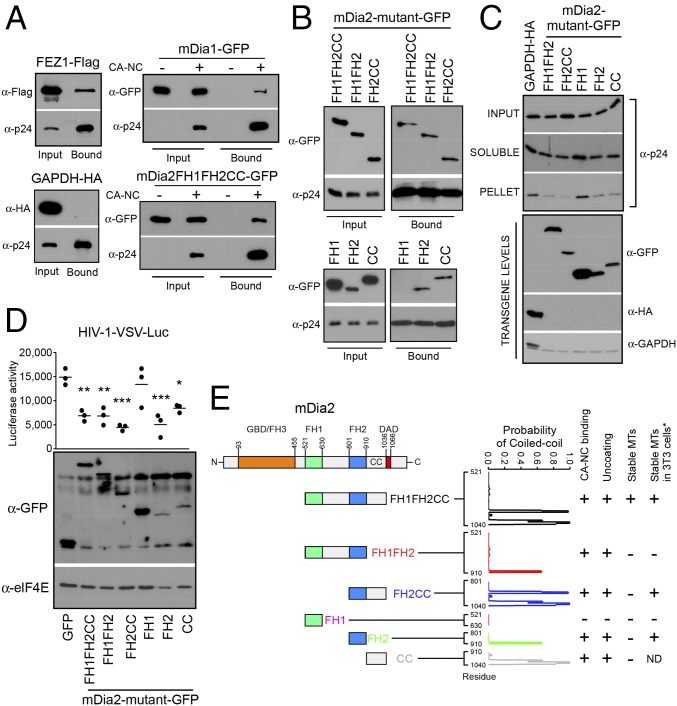
To test the possibility that Dia1 or Dia2 had the capacity to bind HIV-1 cores, we measured their association with in vitro assembled HIV-1 capsid–nucleocapsid (CA–NC) complexes (51, 52). Two control proteins were included in these experiments: FEZ1, which we recently showed to bind HIV-1 cores served as a positive control (24), and GAPDH served as a negative control. Extracts from cells expressing Flag-tagged FEZ1, HA-tagged GAPDH, GFP-tagged mDia1, or the GFP-tagged constitutively active mutant of mDia2 (FH1FH2CC) were mixed with in vitro assembled CA–NC that were then sedimented by centrifugation over a 70% sucrose cushion (Fig. S5A). To control that proteins did not simply pellet independently, extracts were also pelleted in the absence of in vitro assembled CA–NC cores. As expected, Flag-tagged FEZ1 bound assembled CA–NC (24), whereas HA-tagged GAPDH control did not (Fig. 6A). Notably, both mDia1 and mDia2 were detected in association with pelleted HIV-1 CA–NC complexes and were not pelleted in the absence of CA–NC complexes (Fig. 6A). Overall, these findings showed that Dia proteins can associate with CA–NC complexes, and that this may underlie a second mechanism by which these formins influence the rate of HIV-1 uncoating.
Fig. S5.
(A) Quality control of the in vitro assembled CA–NC complexes. In vitro assembled CA–NC complexes were spinoculated onto coverslips for 1 h at 1,200 × g and fixed, followed by staining using anti-p24 CA antibodies. A representative image of the assembled CA–NC tubes are shown, which were as described previously (51). (Magnification: 100×.) (B) Quantification of the levels of pelletable p24 from fate of capsid uncoating assay. Densitometric quantification of the relative amount of pelletable p24 CA from fate of capsid assay in corresponding panel from Fig. 6C. Data are shown as scatterplot with mean, n = 3. One-way ANOVA and posttest were used to determine statistical significance; *P < 0.05. ns, not significant.
Fig. 6.
mDia interacts with in vitro assembled HIV-1 CA–NC complexes and regulates uncoating. (A and B) Binding of mDia to in vitro assembled HIV-1 CA–NC complexes. The 293T cells were transfected with plasmids expressing Flag-tagged FEZ1 (FEZ1-Flag) as positive control, HA-tagged GAPDH as negative control, and GFP-tagged mDia1 or mDia2 FH1FH2CC (A) or GFP-tagged truncation mutants of mDia2 containing FH1FH2CC, FH1FH2, FH2CC, FH1, FH2, and CC (B). Cells were lysed 36 h after transfection and lysates incubated at room temperature for 1 h with in vitro assembled HIV-1 CA–NC complexes. Samples were taken either before (input) or after sedimentation through a 70% sucrose cushion (bound) and analyzed by WB using indicated antibodies. (C) The 293A cells were transfected with control HA-tagged GAPDH or GFP-tagged forms of mDia2 consisting of FH1FH2, FH2CC, FH1, FH2, or CC domains. Cells were infected with HIV-1–VSV-Luc and the amounts of soluble and particulate capsid determined using the fate-of-capsid assay (Upper). Before infection, cells were lysed and analyzed by WB to determine transgene levels using anti-GFP, HA, and GAPDH antibodies (Lower). (D) CHME3 cells were transfected with plasmids expressing GFP control or GFP-tagged truncation mutants of mDia2: FH1FH2CC, FH1FH2, FH2CC, FH1, FH2, or CC. At 48 h posttransfection, cells were either infected with HIV-1–VSV–Luc followed by measurement of luciferase activity or lysed and subjected to WB analysis using anti-GFP and anti-elF4E (as loading control) antibodies. Data are shown as scatterplot with mean, n = 3. Statistical analysis was determined using one-way ANOVA with posttest, *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001, respectively. (E) Schematic of the domains of mDia2 and domains studied (Left). Summary of whether each Dia2 construct binds CA–NC, affects uncoating, or induces stable MTs. * indicates results from previously published data (34) for comparison with our findings. The probability that each mutant will form a coiled coil is shown and was calculated using the COILS program, with the residue marked on the x axis.
Distinct Domains of Dia Are Important for Binding CA–NC Complexes and Regulating Uncoating.
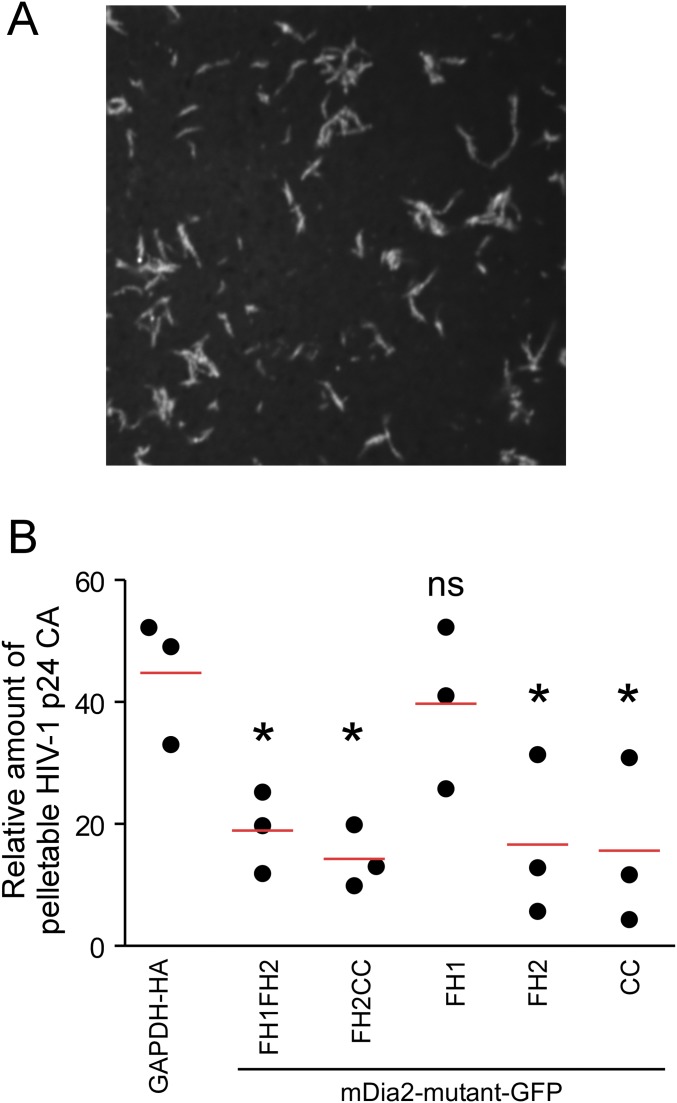
To understand how Dia association with HIV-1 cores might regulate uncoating, and to explore potential links with Dia’s effects on stable MTs, we examined a variety of Dia mutants. We first determined the domains in DRFs that regulate interactions with in vitro assembled CA–NC complexes. The ability of the active mutant of mDia2 containing FH1FH2CC to bind assembled capsids suggested that the FH1, FH2, and/or coiled-coil (CC) domains mediate this interaction (Fig. 6A). To further elucidate which domains were important for binding, extracts from cells expressing five different GFP-tagged portions of mDia2 containing either FH1FH2, FH2CC, FH1, FH2, or CC (34) were incubated with assembled CA–NC and sedimented by centrifugation over a 70% sucrose cushion (51, 52). Notably, FH1FH2, FH2CC, FH2, and the CC domain-containing forms of Dia2 all pelleted with assembled CA–NC complexes, whereas the FH1 domain did not (Fig. 6B). This result indicates that the FH2 and CC domains alone are sufficient to bind assembled capsids, whereas the FH1 was both dispensable and unable to bind. To test whether the interaction between DRFs and CA–NC had a biological impact on capsid stability during infection, the same truncated mutants of mDia2 were expressed in cells and the fate of viral cores was monitored using the fate-of-capsid assay (47). Notably, whereas comparable levels of pelletable intact capsid were recovered from cells expressing control GAPDH or the FH1 domain, which does not bind CA–NC, all forms of mDia2 that bound assembled capsid (FH1FH2, FH2CC, FH2, and CC) also decreased the amount of pelletable cores (Fig. 6C and Fig. S5B). Importantly, the effect of each Dia2 mutant on HIV-1 infectivity (Fig. 6D) correlated with their effects on both uncoating and their ability to bind in vitro assembled CA–NC complexes.
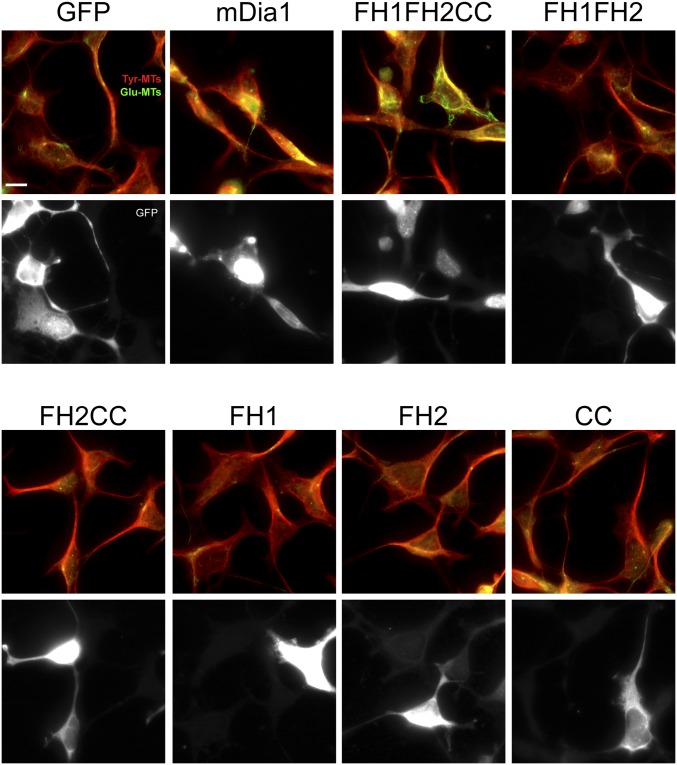
To determine whether the effects of these Dia2 mutants on HIV-1 uncoating correlated with their effects on stable MT formation, CHME3 cells were transfected with GFP control, GFP-mDia1, or GFP-tagged mDia2 domains: FH1FH2, FH2CC, FH1, FH2, or CC. Transfected cells were then fixed and stained for tyrosinated tubulin (Tyr-MTs) and detryosinated tubulin (Glu-MTs), which distinguish dynamic and stable MT networks, respectively (Fig. S6). When effects on uncoating, early infection, and ability to induce stable MT formation were tabulated (Fig. 6 C and D and Fig. S6), the FH1 domain that failed to promote uncoating and early infection also failed to promote stable MT formation. However, only the FH1FH2CC domain that regulated uncoating and early infection was also able to induce MT stabilization, whereas FH1FH2, FH2CC, FH2, and CC domains that enhanced uncoating and early infection were unable to induce stable MTs in CHME3 cells. These data were in agreement with previously published effects of these domains on MT stabilization, with the exception of FH2CC and FH2, which induced MT stabilization in mouse cells (34). Regardless of these slight differences for these particular domains, both studies found that the FH1FH2 domain was unable to stabilize MTs; yet this domain retained effects on uncoating (Fig. 6E). Moreover, the CC domain displayed a similar divergence in effects on stable MTs and uncoating, demonstrating that DRFs had the capacity to bind HIV-1 capsids and regulate uncoating independently of their effects on stable MTs. To evaluate potential structural similarities that might exist between the Dia2 domains that bind in vitro assembled CA–NC complexes, we analyzed whether each mutant had the potential to form CC domains, which are present in the limited number of known capsid-binding proteins (41, 53, 54). Using the COILS program of Lupas et al. (55) and Lupas (56) to calculate the probability that each sequence will adopt a CC conformation, we determined that the FH1 domain, which does not bind to CA–NC and is not able to regulate capsid stability, had zero probability of containing a CC domain, whereas each of the mutants that bound CA–NC and regulated uncoating and early infection had a high probability of containing one or more CC domains (Fig. 6E). These findings suggested that DRFs have the capacity to interact with viral cores through their multiple CC domains, enabling them to regulate capsid stability through dual modes, both independently of as well as through their effects on MT stability.
Fig. S6.
The effects of mDia1 and truncated mutants of mDia2 on stable MT formation in human cells. The 293A cells were transfected with GFP, mDia1-GFP, or GFP-tagged truncation mutants of mDia2 containing FH1FH2CC, FH1FH2, FH2CC, FH1, FH2, and CC. At 48 h after transfection, cells were fixed and stained for tyrosinated tubulin (Tyr-MTs) and detyrosinated tubulin (Glu-MTs). (Scale bar, 20 μm.) Similar results were obtained in at least three independent experiments.
Discussion
The formins Dia1 and Dia2 regulate actin and MT dynamics, thereby coordinating rearrangements in these two key cytoskeletal networks during a range of cellular processes (57, 58). Here, we identify a function for DRFs in binding and regulating HIV-1 core uncoating. In addition, we illustrate how the virus exploits distinct Dia domains, mimicking Dia’s ability to coordinate actin and MT dynamics, to couple HIV-1 core uncoating with MT stabilization and trafficking to the nucleus. These findings not only contribute to our limited understanding of how +TIPs regulate virus infection, but also identify activities for the CC domains of DRFs.
Although it is well established that MT networks are important for infection by many viruses (1, 2), our understanding of how MTs are regulated and the potential role of stable MT subsets during infection remains limited. Whereas several viruses can promote infection by targeting signal pathways that alter MT dynamics (59–65), the role of downstream effector +TIPs has remained largely unexplored. We, and others, have recently shown that HIV-1 rapidly stimulates the formation of stable MT subsets to facilitate early infection (27, 28). HIV-1 particles were also found to localize to stable MTs (27). Moreover, both early infection and HIV-induced MT stabilization required EB1 and its binding partner, Kif4, and their depletion blocked the accumulation of HIV-1 particles at the nucleus in fixed imaging analysis (27). Stable MTs are thought to be selectively recognized by kinesin motors and serve as specialized tracks for cargo trafficking (29, 30). Notably, we have recently shown that HIV-1 exploits kinesin-1 for retrograde transport to the nucleus (24). Cumulatively, these lines of evidence suggest that HIV-1 exploits stable MT subsets during early infection. Kif4 functions as part of the Dia MT stabilization pathway, prompting us to test DRFs contributions to early HIV-1 infection. DRFs were found to be required for MT stabilization and MT-based transport of HIV-1 cores in living cells. Whereas several studies have shown that actin remodeling and Dia regulate late stages of HIV-1 replication and cell-to-cell transmission (66–69), and that RhoA is required for early HIV-1 infection in T cells (70, 71), Dia’s ability to regulate MT networks to influence HIV-1 infection remained unknown. The availability of Dia mutants that specifically induce MT stabilization without regulating actin allowed us to genetically separate these activities and further show the specific importance of MT stabilization to early HIV-1 infection. Moreover, infection by retroviruses that did not induce MT stabilization was unaffected by Dia depletion. We recently showed that HSV-1 does not induce MT stabilization until late in infection, and does so through distinct pathways that do not require Dia1 (43). Without inducing MT stabilization and unaffected by Dia1 depletion, incoming HSV-1 particles exploit dynamic MTs and a distinct set of EB1-associated +TIPs (42). By contrast, another herpesvirus, Kaposi’s sarcoma-associated herpesvirus (KSHV) induces the formation of stable MTs in a Dia-dependent manner (61), suggesting that Dia’s ability to regulate stable MTs makes it an important host factor for viruses that use these specific networks. However, it remains unclear whether Dia contributes to actin- or MT-based processes that facilitate KSHV infection. Overall, these findings illustrate how different viruses exploit distinct MT subsets to mediate their transport to the nucleus, and our findings using Dia mutants demonstrate that MT stabilization does indeed contribute to early HIV-1 infection.
The DRFs were found to be important not just for virus-induced MT stabilization and HIV-1 trafficking, but also for HIV-1 uncoating. Whereas our results show that the induction of stable MTs regulates HIV-1 uncoating, in line with suggestions that core disassembly is coupled to reverse transcription and likely influenced by the movement of viral cores on MTs by motor proteins, Dia’s effects on uncoating could also be genetically separated from its effects on MT stabilization. As such, Dia has two clearly distinct functions during early HIV-1 infection. Dia also associated with HIV-1 cores, and could promote both reverse transcription and the disassembly of incoming HIV-1 capsids, independently of MT stabilization. These findings in combination with the fact that Dia depletion can exert additive effects on uncoating when reverse transcription is chemically blocked, suggest that Dia most likely functions by directly binding incoming HIV-1 cores and affecting uncoating, which in turn affects reverse transcription. Whereas the precise timing and location of HIV-1 uncoating remains contentious, it is clear that the process is carefully controlled on both a spatial and temporal level (6, 72). This allows reverse transcription of the viral genome, while at the same time protecting it from intracellular viral sensors (17, 18). Disturbing the kinetics of uncoating, either by increasing the stability of viral cores or by chemical induction of premature uncoating is detrimental to infection (18, 49, 50, 73, 74). Partial loss of CA from viral cores in the cytoplasm, but detection of some CA in the nucleus, suggests that uncoating is gradual and begins in the cytoplasm (11, 12, 19, 20, 75). Adding to this complexity, the uncoating process is intricately linked to reverse transcription of the viral genome, and events following the first strand transfer during reverse transcription trigger uncoating (12–14). Given their interaction with in vitro assembled CA–NC complexes and ability to enhance both reverse transcription and uncoating, our findings identify DRFs as important host determinants or cofactors that facilitate the uncoating and reverse transcription processes. Whereas few host factors that enhance reverse transcription have been identified, and overexpression of proteins can have unanticipated effects, the opposing effects of DRF overexpression versus depletion were in agreement with Dia proteins regulating uncoating and reverse transcription. There is precedent for this observation, as overexpression of another coiled-coil containing protein, PDZD8, that binds HIV-1 capsids and regulates uncoating also promotes reverse transcription (41, 54). Moreover, through distinct domains, the DRFs also facilitate HIV-induced MT stabilization and trafficking to the nucleus. Indeed, given the fact that HIV-1 transport, reverse transcription, and uncoating are so intricately intertwined, it is difficult to discern the individual contributions to infection made by these discrete functions of DRFs. However, by regulating MT dynamics, virus transport, and uncoating, DRFs position themselves at the center of this intricate early stage of infection, serving as key factors that likely allow the virus to couple the timing of uncoating with the onset of MT stabilization and virus transport.
Whereas Dia isoforms have functional redundancy in regulating the actin cytoskeleton, they have nonredundant functions in MT capture. mDia1 has been shown to function in cell polarization and migration, axonal outgrowth, and exocrine vesicle secretion in the apical membrane, whereas mDia2 is involved in filopodia formation, cytokinesis, asymmetric cell division, and endosome trafficking in fibroblasts (57). These unique and shared properties may explain why both Dia1 and Dia2 are required for infection. Alternatively, redundancy will only be evident for proteins whose combined abundance is in excess of requirement. If the combined abundance of Dia1 and Dia2 is not in excess, this would also explain the lack of apparent redundancy in our system. These multidomain proteins share core structural features, including a central proline rich formin homology 1 (FH1) and FH2 domain, as well as dimerization domains (DDs) and CC domains (57). In addition, a C-terminal diaphanous autoregulatory domain (DAD) interacts with its own N-terminal FH3 recognition domain, resulting in autoinhibition. Binding of Rho GTPases to the N-terminal GTPase-binding domain (GBD) releases this autoinhibition, resulting in Dia activation. Once activated, the DRFs regulate both the actin and MT cytoskeletons through their FH2 domain. However, several reports have demonstrated that these functions are genetically separable and that mDia can stabilize MTs independently of its actin nucleation activity (40, 76, 77), allowing for the coordinated regulation of both cytoskeletal networks. Likewise, we show that the ability of DRFs to stabilize MTs and interact with incoming HIV-1 cores is also genetically separable, and that the latter function correlates with the presence of CC domains in portions of DRFs that can regulate uncoating. Nearly all formins have a predicted CC domain that overlaps with the C-terminal portion of the FH2 domain and extends toward the C terminus (78). The function of the C-terminal CC is not well understood, but in a related formin, FHOD1 was shown to be important for self-oligomerization (79). Here, we identify a function for the CC domains in DRFs, as all fragments of Dia that bound HIV-1 capsid and regulated uncoating exhibited a high probability of containing CC domains. CCs are formed from heptad repeats and play a key role in determining a proteins 3D structure and ability to form multimeric structures (80, 81). Interestingly, the small number of proteins known to interact with in vitro assembled CA–NC complexes or the Gag polyprotein, which contains capsid, all contain CC domains (24, 41, 53, 54). Moreover, the CC domain of PDZD8 was shown to be required for binding to HIV-1 Gag (41). In the case of the DRFs, HIV-1 appears to exploit their CC domains to control core stability, whereas independently exploiting their MT stabilizing activity to coordinate the processes of HIV-1 uncoating with virus transport. This reveals previously unknown functions for CC domains in the DRFs, and identifies these formins as critical host cofactors exploited by HIV-1 during early infection.
Materials and Methods
Cells and Viruses.
NHDFs were purchased from Lonza. The 293T, 293A, CHME3, and CHME3 4 × 4 cells were described previously (39, 82). Jurkat and THP-1 cell lines were kindly provided by Thomas Hope, Northwestern University, Chicago. THP-1 cells were differentiated into macrophages by the addition of PMA (30 ng/mL) to cultures overnight. To generate HIV-1 carrying a luciferase reporter or ZsGreen marker, pNL4-3.Luc.R−.E− plasmid (AIDS Reagent Repository no. 3418) or pNL4-3.ZsGreen.R−.E−, were transfected into 293T cells together with constructs expressing VSV-G (pVSV-G) or MuLV amphotropic (pHit456) envelope glycoprotein, or HIV-1 CCR5 tropic (pCI-env) envelope glycoprotein using polyethylenimine (PEI: Polysciences) as described (27). WT HIV-1 was generated by transfection of 293T cells with infectious clone pYK-JRCSF (AIDS Reagent Repository no. 2708). HIV-1 virus containing GFP-Vpr pseudotyped with WT HIV-1 envelope was prepared by PEI transfection with 6 μg pNL4-3.Luc.R−.E−, 3 μg pCI-env, and 1 μg GFP-Vpr (HIV-1–GFP-Vpr–WT) as described (24). HIV-1 virus containing GFP-Vpr and S15-Tomato pseudotyped with VSV-G was generated using 5.7 μg pNL4-3.Luc.R−.E−, 2.9 μg VSV-G, 1.4 μg GFP-Vpr, and 0.44 μg S15-Tomato (24). Mock virus preparations were generated by transfecting cells without DNA.
siRNA and Plasmid Transfections.
The expression construct for C′ terminally Flag-tagged FEZ1 and C′ terminally human influenza HA-tagged GAPDH was described previously (82). Expression constructs for full-length mDia1, and truncation mutants of mDia2 including FH1FH2CC, FH1FH2CCK853A, FH1FH2, FH2CC, FH1, FH2, and CC were described before (34, 40, 83). Plasmid transfections were performed using 2 μg DNA and 4 μL Lipofectamine 2000 (Life Technologies) per manufacturer’s instructions or 22.5 μg of PEI and 10 μg pDNA in a 10-cm dish. For transient knockdown, CHME3, NHDF, or 293A cells were transfected with 100 pmol of siRNA duplexes per well in a 12-well plate using oligofectamine RNAiMAX transfection reagent (Invitrogen) (82). Differentiated THP-1 cells were washed twice and maintained in complete media without antibiotics for an additional 24 h before siRNA transfection as described above. siRNAs used were: ID nos. 146615 and 15997 (Dia1 I and Dia1 II, respectively), ID nos. 146689 and 146691 (Dia2 I and Dia2 II, respectively), or ID nos. AM46355 and AM46357 (NC, control nontargeting siRNA) from Ambion. For all experiments except in Fig. 1 B and C, siRNAs Dia1 I and Dia2 I were used. After knockdown, cells were seeded, infected, and processed as described in Results.
Luciferase Assay.
NHDF, CHME3, Jurkat, or differentiated THP-1 cells seeded in 12- or 24-well plates were infected with different dilutions of HIV-1–VSV–Luc, HIV-1–Ampho–Luc, or HIV-1–WT–Luc followed by lysis of the cells 48–72 h.p.i. and measurements of luciferase activity using the Luciferase Assay System (Promega) (27). For formin inhibitor experiments, NHDF or CHME3 cells were treated with 0.1% DMSO or 10 μM SMIFH2 (Sigma-Aldrich, S4826) for the duration of infection.
Virion Yield Assay.
For measurements of replication-competent HIV-1 virion yields, CHME3 4 × 4 cells were mock infected or infected with high titer JR-CSF-derived HIV-1. Supernatants were collected and filtered and cells were lysed at 5 and 7 d.p.i. Physical particle yields were determined by the measurements of p24 CA into culture supernatants by either WB analysis using anti–HIV-1–Pr55/p24/p17 antibodies or p24 ELISA. Cell lysates were subjected to WB analysis for detection of the indicated cellular proteins.
qPCR Measurement of Viral DNA.
To quantify viral DNA, cells were infected at high multiplicity of infection (MOI) with HIV-1–VSV–puro. Hirt DNA was isolated 24 h postinfection and the amount of HIV-1 MSS DNA and total viral DNA were measured using primers specific to MSS and puromycin, respectively (41). The numbers of target copies were calculated using standard curves.
Western Blotting and Immunofluorescence.
Antibodies used for WB were Dia1 (610848) and eIF4E (610269) from BD Biosciences; Dia2 (5474) from Cell Signaling; Flag (F7425), HA (H6098), and acetylated tubulin (T7451) from Sigma; HIV-1 p55/24/17 (ab63917), HIV-1-p24 (ab9071), tyrosinated tubulin [YL1/2] (ab6160) and GFP (ab13970) from Abcam; GAPDH (sc-25778) from Santa Cruz; detyrosinated tubulin (AB3201) from Millipore; HIV-1 p24 (AG3.0) from NIH AIDS Reagent Program (4121), and Phalloidin-647 from Biotium. Cells were lysed in Laemmli sample buffer. For WB analysis, all primary antibodies were used at a 1:1,000 dilution in 3% BSA TBS-T overnight at 4 °C and detected using the appropriate HRP-conjugated secondary. For IF analysis, cells were grown on glass coverslips and either fixed with ice-cold methanol (when performing IF of MTs) or 4% paraformaldehyde (PFA) (USB, Affymetrix), blocked and permeabilized as described (27). Samples were stained with primary antibody overnight at 4 °C, washed, and incubated with the appropriate Alexa Fluor-conjugated secondary for 1 h at room temperature. Nuclei were stained with Hoechst 33342. Coverslips were mounted with FluorSave reagent (Calbiochem). Images were acquired using a Leica DMI6000 B motorized spinning-disk confocal microscope with a Yokogawa CSU-X1 A1 confocal head. Confocal microscopy was used to image the translocation of viral particles and in situ uncoating using a Hamamatsu ImagEM X2 EM-CCD camera run by MetaMorph imaging software (Molecular Devices). Wide-field microscopy was used to image the cytoskeleton using a Hamamatsu C11440, Hamamatsu C4742-95 or a Photometrics Prime 95B camera run by MetaMorph imaging software (Molecular Devices). Imaging and quantification of HIV-1–GFP-Vpr–VSV particles within 2 μm of the nucleus in NHDF cells was performed as described (27).
In Vitro Capsid Binding Assay.
The HIV-1 CA–NC expression, purification, and assembly were performed as previously described (51). HIV-1 CA–NC particles were assembled in vitro by diluting the CA–NC protein to a concentration of 10 mg/mL in 50 mM Tris⋅HCl (pH 8.0), 0.5 M NaCl, and 2 mg/mL DNA oligo-(TG)50 as described previously (24, 84). Human 293T cells were transfected with the mammalian expression vectors encoding HA-tagged GAPDH, Flag-tagged FEZ1, GFP-tagged mDia1, and GFP-tagged mDia2 truncated mutants, as described above. Cell lysates were incubated with in vitro assembled CA–NC (input) followed by ultracentrifugation through a 70% sucrose cushion. The pellet was resuspended in SDS/PAGE loading buffer (bound). WB was used to detect GFP, HA, or Flag. Pelleting of capsids in samples was confirmed by WB analysis of HIV-1 CA–NC protein using anti-p24 antibody (Abcam, 63917) (24).
Fate-of-Capsid Assay.
The fate-of-capsid assay was performed as described previously (47, 85). Cells treated with siRNAs or transfected with HA-tagged GAPDH, GFP-tagged mDia1, or GFP-tagged mutants of mDia2 were infected with HIV-1–VSV–Luc in CO2-independent medium (Gibco) containing 5% FBS (Gibco), 2 mM glutamine (Corning), and 1 mM sodium pyruvate (Gibco). Infection was synchronized by spinoculating at 16 °C for 1 h at 1,200 × g. For drug-treated experiments, cells were treated with either DMSO (0.1%), 10 μM PF74 (Sigma-Aldrich), 100 nM taxol (Sigma-Aldrich), or 10 μM Nevirapine (AIDS Reagent Repository no. 4666), the media were replaced with fresh media supplemented with the respective drug after spinoculation, and the cells were incubated at 37 °C for 3 h. The cells were detached by pronase (Sigma-Aldrich), pelleted, and lysed in hypotonic lysis buffer using 30 strokes in a 7-mL Dounce homogenizer with pestle B (loose) as described. The cleared lysate was layered onto a 50% (wt/vol) sucrose cushion and centrifuged at 100,000 × g for 2 h at 4 °C. Following centrifugation, 30 μL of the topmost portion of the supernatant (soluble fraction) was collected and diluted in 30 μL 2× Laemmli buffer. The pellet (insoluble capsid) was resuspended in 30 μL 1× Laemmli. All samples were then subjected to WB analysis to detect HIV-1 p24 using anti–HIV-1 p55/24/17 (Abcam, 63917). To detect transgene and knockdown levels, some cells were lysed before infection and subjected to SDS/PAGE and WB.
In Situ Uncoating Assay.
The in situ uncoating assay, generation, and harvest of double-labeled virus, and determination of background levels and labeling efficiency were performed essentially as previously described (24, 44, 46). CHME3 cells grown on glass coverslips were infected with S15-Tomato/GFP-Vpr–labeled HIV-1 (>90% virus particles were double labeled) via spinoculation at 16 °C for 1 h at 1,200 × g. Cells were then incubated at 37 °C until fixed at various times points with 4% PFA. Coverslips were stained with AG3.0 (monoclonal anti-p24) and Hoechst 33342. Cells were imaged using a Leica DMI6000 B motorized spinning-disk confocal microscope with a Yokogawa CSU-X1 A1 confocal head and Hamamatsu ImagEM X2 EM-CCD camera run by MetaMorph imaging software (Molecular Devices). Individual virus particles were then analyzed to measure the maximum p24 fluorescence intensity associated with each virus particle that had fused and entered the cytoplasm. Virus particles that did not productively fuse and become endocytosed were S15-Tomato+ and GFP-Vpr+, whereas virus particles that had productively fused into the cytoplasm were S15-Tomato− and GFP-Vpr+. The percentage of fusion was determined by dividing the number of fused virus particles by the number of virus particles in each of the fields. Changes in p24 intensity during infection were determined as described (44). The mean of the maximum p24 fluorescence intensity of each virus particle in control siRNA-treated cells was calculated for each time point. Data were then normalized within each time point by dividing the maximum p24 fluorescence intensity of each virus particle by the mean of the control siRNA-treated cells for the 30-, 60-, and 120-min time points.
Live Imaging and Analysis.
Live cell video microscopy was performed using a motorized spinning-disk confocal (described above) and environmental chamber (3i Intelligent Imaging Innovations or InVivo Scientific). CHME3 cells plated on collagen-coated glass 35-mm culture dishes (MatTek, P35G-1.5–14C) were infected with HIV-1–GFP-Vpr–WT in CO2-independent media via spinoculated as above. Cells were washed once and supplemented with fresh media containing the oxygen scavenger Oxyrase (Oxyrase Company), and sodium DL-lactate (Sigma-Aldrich) before imaging. Images were acquired using a Leica DMI6000 B motorized spinning-disk confocal microscope with Yokogawa CSU-X1 A1 confocal head and a Hamamatsu ImagEM X2 EM-CCD camera run by MetaMorph imaging software (Molecular Devices) at one frame per second (fps) from 5 to 10 min. FIJI (86) and the MosaicSuite plugin was used for multiple particle detection and tracking from digital videos (87). Tracked viral trajectories were then exported to Excel for conversion from distances traveled in pixels to micrometers and calculation of speed and displacement for each tracked virus particle. The average speed and velocity of virus particles were calculated as the mean of the total distance traveled (μm)/time tracked (min) and the displacement (μm)/time tracked (min), respectively. All particles with a lifetime less than 20 s were excluded from the calculations. Over 130 particles were tracked per sample, with 5–15 particles being tracked per cell. Mitochondrial trafficking was imaged using MitoTracker (Thermo Fisher), as previously described (42).
SI Materials and Methods
For generation of VSV-G pseudotyped SIV-Luc (SIVmac-based viral vector carrying a luciferase reporter), 293T cells were cotransfected with pSIVmac239-Luc-E− (88) (a kind gift from Nathaniel Landau) and the VSV-G expressing construct pVSV-G (41). The same protocol was used for preparation of VSV-G pseudotyped MuLV viral vectors containing a luciferease marker (MuLV–VSV–Luc) using the MoMuLV Gag-Pol expressing vector pCMVintron (27) and the luciferase expressing vector pFB–Luc (Stratagene). HSV-1 strain F was grown and titrated in Vero cells (43).
Supplementary Material
Acknowledgments
We thank Gregg Gunderson, Francesca Bartolini, Thomas Hope, Olivier Schwartz, Nathaniel Landau, and Joseph Sodroski for providing reagents; Barbie Ganser-Pornillos and Kayla Schipper for technical help; and Derek Walsh for help in editing the manuscript. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH: Nevirapine; pNL4-3.Luc.R−.E− from Nathaniel Landau, New York University School of Medicine, New York; HIV-1 p24 (AG3.0) from Jonathan Allan, Southwest Foundation for Biomedical Research, San Antonio, TX; and pYK-JR-CSF from Irvin Chen and Yoshio Koyanagi, Institute for Frontier Life and Medical Sciences, Kyoto University, Kyoto. This study was supported by NIH Grants P01GM105536, R01GM101975, and R01NS099064 (to M.H.N.) and NIH Grant T32AI007476-19 (to M.K.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700247114/-/DCSupplemental.
References
- 1.Radtke K, Döhner K, Sodeik B. Viral interactions with the cytoskeleton: A hitchhiker’s guide to the cell. Cell Microbiol. 2006;8:387–400. doi: 10.1111/j.1462-5822.2005.00679.x. [DOI] [PubMed] [Google Scholar]
- 2.Dodding MP, Way M. Coupling viruses to dynein and kinesin-1. EMBO J. 2011;30:3527–3539. doi: 10.1038/emboj.2011.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naghavi MH, Goff SP. Retroviral proteins that interact with the host cell cytoskeleton. Curr Opin Immunol. 2007;19:402–407. doi: 10.1016/j.coi.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goff SP. In: Retroviridae: The Retroviruses and Their Replication. Knipe DM, Howley PM, editors. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1871–1939. [Google Scholar]
- 5.Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Curr Opin Struct Biol. 2008;18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell EM, Hope TJ. HIV-1 capsid: The multifaceted key player in HIV-1 infection. Nat Rev Microbiol. 2015;13:471–483. doi: 10.1038/nrmicro3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller MD, Farnet CM, Bushman FD. Human immunodeficiency virus type 1 preintegration complexes: Studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fassati A, Goff SP. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J Virol. 2001;75:3626–3635. doi: 10.1128/JVI.75.8.3626-3635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arhel NJ, et al. HIV-1 DNA Flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO J. 2007;26:3025–3037. doi: 10.1038/sj.emboj.7601740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou L, et al. Transportin 3 promotes a nuclear maturation step required for efficient HIV-1 integration. PLoS Pathog. 2011;7:e1002194. doi: 10.1371/journal.ppat.1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francis AC, Marin M, Shi J, Aiken C, Melikyan GB. Time-resolved imaging of single HIV-1 uncoating in vitro and in living cells. PLoS Pathog. 2016;12:e1005709. doi: 10.1371/journal.ppat.1005709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hulme AE, Perez O, Hope TJ. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc Natl Acad Sci USA. 2011;108:9975–9980. doi: 10.1073/pnas.1014522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y, Fricke T, Diaz-Griffero F. Inhibition of reverse transcriptase activity increases stability of the HIV-1 core. J Virol. 2013;87:683–687. doi: 10.1128/JVI.01228-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cosnefroy O, Murray PJ, Bishop KN. HIV-1 capsid uncoating initiates after the first strand transfer of reverse transcription. Retrovirology. 2016;13:58. doi: 10.1186/s12977-016-0292-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonald D, et al. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol. 2002;159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arhel N, et al. Quantitative four-dimensional tracking of cytoplasmic and nuclear HIV-1 complexes. Nat Methods. 2006;3:817–824. doi: 10.1038/nmeth928. [DOI] [PubMed] [Google Scholar]
- 17.Rasaiyaah J, et al. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature. 2013;503:402–405. doi: 10.1038/nature12769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Sage V, Mouland AJ, Valiente-Echeverría F. Roles of HIV-1 capsid in viral replication and immune evasion. Virus Res. 2014;193:116–129. doi: 10.1016/j.virusres.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 19.Peng K, et al. Quantitative microscopy of functional HIV post-entry complexes reveals association of replication with the viral capsid. Elife. 2014;3:e04114. doi: 10.7554/eLife.04114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hulme AE, Kelley Z, Okocha EA, Hope TJ. Identification of capsid mutations that alter the rate of HIV-1 uncoating in infected cells. J Virol. 2015;89:643–651. doi: 10.1128/JVI.03043-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen NY, et al. HIV-1 capsid is involved in post-nuclear entry steps. Retrovirology. 2016;13:28. doi: 10.1186/s12977-016-0262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaller T, et al. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog. 2011;7:e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koh Y, et al. Differential effects of human immunodeficiency virus type 1 capsid and cellular factors nucleoporin 153 and LEDGF/p75 on the efficiency and specificity of viral DNA integration. J Virol. 2013;87:648–658. doi: 10.1128/JVI.01148-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malikov V, et al. HIV-1 capsids bind and exploit the kinesin-1 adaptor FEZ1 for inward movement to the nucleus. Nat Commun. 2015;6:6660. doi: 10.1038/ncomms7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pawlica P, Berthoux L. Cytoplasmic dynein promotes HIV-1 uncoating. Viruses. 2014;6:4195–4211. doi: 10.3390/v6114195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lukic Z, Dharan A, Fricke T, Diaz-Griffero F, Campbell EM. HIV-1 uncoating is facilitated by dynein and kinesin 1. J Virol. 2014;88:13613–13625. doi: 10.1128/JVI.02219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabo Y, et al. HIV-1 induces the formation of stable microtubules to enhance early infection. Cell Host Microbe. 2013;14:535–546. doi: 10.1016/j.chom.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez J, et al. Microtubule-associated proteins 1 (MAP1) promote human immunodeficiency virus type I (HIV-1) intracytoplasmic routing to the nucleus. J Biol Chem. 2015;290:4631–4646. doi: 10.1074/jbc.M114.613133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li R, Gundersen GG. Beyond polymer polarity: How the cytoskeleton builds a polarized cell. Nat Rev Mol Cell Biol. 2008;9:860–873. doi: 10.1038/nrm2522. [DOI] [PubMed] [Google Scholar]
- 30.Janke C, Bulinski JC. Post-translational regulation of the microtubule cytoskeleton: Mechanisms and functions. Nat Rev Mol Cell Biol. 2011;12:773–786. doi: 10.1038/nrm3227. [DOI] [PubMed] [Google Scholar]
- 31.Jiang K, Akhmanova A. Microtubule tip-interacting proteins: A view from both ends. Curr Opin Cell Biol. 2011;23:94–101. doi: 10.1016/j.ceb.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 32.Honnappa S, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- 33.Gundersen GG. Evolutionary conservation of microtubule-capture mechanisms. Nat Rev Mol Cell Biol. 2002;3:296–304. doi: 10.1038/nrm777. [DOI] [PubMed] [Google Scholar]
- 34.Wen Y, et al. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol. 2004;6:820–830. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- 35.Morris EJ, Nader GP, Ramalingam N, Bartolini F, Gundersen GG. Kif4 interacts with EB1 and stabilizes microtubules downstream of Rho-mDia in migrating fibroblasts. PLoS One. 2014;9:e91568. doi: 10.1371/journal.pone.0091568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell. 2009;137:433–444. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herold N, et al. HIV-1 entry in SupT1-R5, CEM-ss, and primary CD4+ T cells occurs at the plasma membrane and does not require endocytosis. J Virol. 2014;88:13956–13970. doi: 10.1128/JVI.01543-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rizvi SA, et al. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem Biol. 2009;16:1158–1168. doi: 10.1016/j.chembiol.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donahue DA, et al. SUN2 overexpression deforms nuclear shape and inhibits HIV. J Virol. 2016;90:4199–4214. doi: 10.1128/JVI.03202-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartolini F, et al. The formin mDia2 stabilizes microtubules independently of its actin nucleation activity. J Cell Biol. 2008;181:523–536. doi: 10.1083/jcb.200709029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henning MS, Morham SG, Goff SP, Naghavi MH. PDZD8 is a novel Gag-interacting factor that promotes retroviral infection. J Virol. 2010;84:8990–8995. doi: 10.1128/JVI.00843-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jovasevic V, Naghavi MH, Walsh D. Microtubule plus end-associated CLIP-170 initiates HSV-1 retrograde transport in primary human cells. J Cell Biol. 2015;211:323–337. doi: 10.1083/jcb.201505123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naghavi MH, Gundersen GG, Walsh D. Plus-end tracking proteins, CLASPs, and a viral Akt mimic regulate herpesvirus-induced stable microtubule formation and virus spread. Proc Natl Acad Sci USA. 2013;110:18268–18273. doi: 10.1073/pnas.1310760110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Campbell EM, Perez O, Melar M, Hope TJ. Labeling HIV-1 virions with two fluorescent proteins allows identification of virions that have productively entered the target cell. Virology. 2007;360:286–293. doi: 10.1016/j.virol.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bowman EJ, Siebers A, Altendorf K. Bafilomycins: A class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc Natl Acad Sci USA. 1988;85:7972–7976. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamashita M, Perez O, Hope TJ, Emerman M. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog. 2007;3:1502–1510. doi: 10.1371/journal.ppat.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Y, Luban J, Diaz-Griffero F. The fate of HIV-1 capsid: A biochemical assay for HIV-1 uncoating. Methods Mol Biol. 2014;1087:29–36. doi: 10.1007/978-1-62703-670-2_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi J, Zhou J, Shah VB, Aiken C, Whitby K. Small-molecule inhibition of human immunodeficiency virus type 1 infection by virus capsid destabilization. J Virol. 2011;85:542–549. doi: 10.1128/JVI.01406-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Forshey BM, von Schwedler U, Sundquist WI, Aiken C. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol. 2002;76:5667–5677. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J Virol. 2003;77:5439–5450. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 52.Fricke T, et al. The ability of TNPO3-depleted cells to inhibit HIV-1 infection requires CPSF6. Retrovirology. 2013;10:46. doi: 10.1186/1742-4690-10-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diaz-Griffero F, et al. Requirements for capsid-binding and an effector function in TRIMCyp-mediated restriction of HIV-1. Virology. 2006;351:404–419. doi: 10.1016/j.virol.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 54.Guth CA, Sodroski J. Contribution of PDZD8 to stabilization of the human immunodeficiency virus type 1 capsid. J Virol. 2014;88:4612–4623. doi: 10.1128/JVI.02945-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 56.Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 57.Kühn S, Geyer M. Formins as effector proteins of Rho GTPases. Small GTPases. 2014;5:e29513. doi: 10.4161/sgtp.29513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ridley AJ. Rho GTPase signalling in cell migration. Curr Opin Cell Biol. 2015;36:103–112. doi: 10.1016/j.ceb.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suomalainen M, Nakano MY, Boucke K, Keller S, Greber UF. Adenovirus-activated PKA and p38/MAPK pathways boost microtubule-mediated nuclear targeting of virus. EMBO J. 2001;20:1310–1319. doi: 10.1093/emboj/20.6.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mabit H, et al. Intact microtubules support adenovirus and herpes simplex virus infections. J Virol. 2002;76:9962–9971. doi: 10.1128/JVI.76.19.9962-9971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Naranatt PP, Krishnan HH, Smith MS, Chandran B. Kaposi’s sarcoma-associated herpesvirus modulates microtubule dynamics via RhoA-GTP-diaphanous 2 signaling and utilizes the dynein motors to deliver its DNA to the nucleus. J Virol. 2005;79:1191–1206. doi: 10.1128/JVI.79.2.1191-1206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Veettil MV, et al. RhoA-GTPase facilitates entry of Kaposi’s sarcoma-associated herpesvirus into adherent target cells in a Src-dependent manner. J Virol. 2006;80:11432–11446. doi: 10.1128/JVI.01342-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arakawa Y, Cordeiro JV, Way M. F11L-mediated inhibition of RhoA-mDia signaling stimulates microtubule dynamics during vaccinia virus infection. Cell Host Microbe. 2007;1:213–226. doi: 10.1016/j.chom.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 64.Raghu H, et al. Lipid rafts of primary endothelial cells are essential for Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8-induced phosphatidylinositol 3-kinase and RhoA-GTPases critical for microtubule dynamics and nuclear delivery of viral DNA but dispensable for binding and entry. J Virol. 2007;81:7941–7959. doi: 10.1128/JVI.02848-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roohvand F, et al. Initiation of hepatitis C virus infection requires the dynamic microtubule network: Role of the viral nucleocapsid protein. J Biol Chem. 2009;284:13778–13791. doi: 10.1074/jbc.M807873200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu TC, et al. HIV-1 Nef disrupts the podocyte actin cytoskeleton by interacting with diaphanous interacting protein. J Biol Chem. 2008;283:8173–8182. doi: 10.1074/jbc.M708920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nikolic DS, et al. HIV-1 activates Cdc42 and induces membrane extensions in immature dendritic cells to facilitate cell-to-cell virus propagation. Blood. 2011;118:4841–4852. doi: 10.1182/blood-2010-09-305417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aggarwal A, et al. Mobilization of HIV spread by diaphanous 2 dependent filopodia in infected dendritic cells. PLoS Pathog. 2012;8:e1002762. doi: 10.1371/journal.ppat.1002762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shrivastava A, Prasad A, Kuzontkoski PM, Yu J, Groopman JE. Slit2N inhibits transmission of HIV-1 from dendritic cells to T-cells by modulating novel cytoskeletal elements. Sci Rep. 2015;5:16833. doi: 10.1038/srep16833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Watanabe T, et al. The hematopoietic cell-specific Rho GTPase inhibitor ARHGDIB/D4GDI limits HIV type 1 replication. AIDS Res Hum Retroviruses. 2012;28:913–922. doi: 10.1089/AID.2011.0180. [DOI] [PubMed] [Google Scholar]
- 71.del Real G, et al. Statins inhibit HIV-1 infection by down-regulating Rho activity. J Exp Med. 2004;200:541–547. doi: 10.1084/jem.20040061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Da Silva Santos C, Tartour K, Cimarelli A. A novel entry/uncoating assay reveals the presence of at least two species of viral capsids during synchronized HIV-1 infection. PLoS Pathog. 2016;12:e1005897. doi: 10.1371/journal.ppat.1005897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.De Iaco A, Luban J. Cyclophilin A promotes HIV-1 reverse transcription but its effect on transduction correlates best with its effect on nuclear entry of viral cDNA. Retrovirology. 2014;11:11. doi: 10.1186/1742-4690-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saito A, et al. Roles of capsid-interacting host factors in multimodal inhibition of HIV-1 by PF74. J Virol. 2016;90:5808–5823. doi: 10.1128/JVI.03116-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu H, et al. Evidence for biphasic uncoating during HIV-1 infection from a novel imaging assay. Retrovirology. 2013;10:70. doi: 10.1186/1742-4690-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Palazzo AF, et al. Cdc42, dynein, and dynactin regulate MTOC reorientation independent of Rho-regulated microtubule stabilization. Curr Biol. 2001;11:1536–1541. doi: 10.1016/s0960-9822(01)00475-4. [DOI] [PubMed] [Google Scholar]
- 77.Ishizaki T, et al. Coordination of microtubules and the actin cytoskeleton by the Rho effector mDia1. Nat Cell Biol. 2001;3:8–14. doi: 10.1038/35050598. [DOI] [PubMed] [Google Scholar]
- 78.Wallar BJ, Alberts AS. The formins: Active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 2003;13:435–446. doi: 10.1016/s0962-8924(03)00153-3. [DOI] [PubMed] [Google Scholar]
- 79.Madrid R, et al. Oligomerization of the diaphanous-related formin FHOD1 requires a coiled-coil motif critical for its cytoskeletal and transcriptional activities. FEBS Lett. 2005;579:441–448. doi: 10.1016/j.febslet.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 80.Lupas A. Coiled coils: New structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 81.Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: A highly versatile protein folding motif. Trends Cell Biol. 2001;11:82–88. doi: 10.1016/s0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- 82.Haedicke J, Brown C, Naghavi MH. The brain-specific factor FEZ1 is a determinant of neuronal susceptibility to HIV-1 infection. Proc Natl Acad Sci USA. 2009;106:14040–14045. doi: 10.1073/pnas.0900502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat Cell Biol. 1999;1:136–143. doi: 10.1038/11056. [DOI] [PubMed] [Google Scholar]
- 84.Fricke T, Brandariz-Nuñez A, Wang X, Smith AB, 3rd, Diaz-Griffero F. Human cytosolic extracts stabilize the HIV-1 core. J Virol. 2013;87:10587–10597. doi: 10.1128/JVI.01705-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stremlau M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sbalzarini IF, Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. J Struct Biol. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 88.Yu Q, et al. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J Biol Chem. 2004;279:53379–53386. doi: 10.1074/jbc.M408802200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.