Significance
Although C-type inactivation in potassium channels is crucial in many biological signaling processes, its mechanism is debated. We address a hypothesis that K+ affinity reduction and ion loss due to allosterically coupled activation is a major factor in promoting inactivation. We show that in KcsA, activation by proton binding at the intracellular pH sensor can significantly reduce K+ affinity at the extracellular selectivity filter, which would be expected to contribute to current cessation (inactivation). This allosteric coupling is substantially reduced by mutating a residue that connects the inner transmembrane helix hinge (involved in activation) to the base of the selectivity filter, thus providing clear evidence for the allosteric coupling network and explaining why this mutant is inactivationless.
Keywords: allostery, inactivation, potassium channel, solid-state NMR, membrane protein
Abstract
The slow spontaneous inactivation of potassium channels exhibits classic signatures of transmembrane allostery. A variety of data support a model in which the loss of K+ ions from the selectivity filter is a major factor in promoting inactivation, which defeats transmission, and is allosterically coupled to protonation of key channel activation residues, more than 30 Å from the K+ ion binding site. We show that proton binding at the intracellular pH sensor perturbs the potassium affinity at the extracellular selectivity filter by more than three orders of magnitude for the full-length wild-type KcsA, a pH-gated bacterial channel, in membrane bilayers. Studies of F103 in the hinge of the inner helix suggest an important role for its bulky sidechain in the allosteric mechanism; we show that the energetic strength of coupling of the gates is strongly altered when this residue is mutated to alanine. These results provide quantitative site-specific measurements of allostery in a bilayer environment, and highlight the power of describing ion channel gating through the lens of allosteric coupling.
Allostery is a major paradigm describing control of cellular function via molecular conformations and ensembles of conformations (1, 2). In systems that function in from signaling to metabolic regulation and other contexts (3, 4), ligand binding in distal pockets modulates protein function via long-range molecular rearrangements. A canonical expression of allostery is the effect of binding of one ligand on the affinity of another in a distant pocket. Detailed models such as Monod–Wyman–Changeux (MWC) (5), Koshland–Nemethy–Filmer (KNF) (6), and the extended population shift theory (7) provide theoretical frameworks to understand allosteric coupling. These models are most meaningful when detailed thermodynamic, kinetic, and structural data can be obtained under functionally relevant conditions, which is generally a challenging objective. NMR has the potential to provide quantitative probes of the thermodynamic and kinetic details of allosteric behaviors in a site-specific and structurally detailed fashion.
Potassium channels show classic transmembrane allosteric regulation, controlling their mean open time. This ubiquitous family of membrane proteins controls the flow of the potassium ions through membranes, a fundamental step for many physiological functions (8, 9). High selectivity and conduction rate (107 ions per s−1) (10) have been attributed to the selectivity filter, with its strongly conserved signature sequence (TVGYG) (11, 12). Ion flux is controlled by channel inactivation (8, 13), which causes the spontaneous cessation of ion flow after the channels are activated. KcsA is a model system for potassium channels and shares functional similarities with several voltage-gated potassium channels (8, 14).
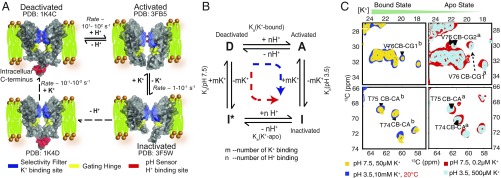
A kinetic hypothesis for channel activation and C-type inactivation is shown in Fig. 1A. The deactivated resting state has permeant ions loaded in the filter so the channel is conducting in the extracellular gate, but has its transmembrane helices in a compact closed state, occluding flow at the intracellular gate. Protons are known to bind to the pH sensor in the deactivated state (15), causing the helical bundle to open (16) and a concomitant surge in current through the channel. This transiently populated conductive form is called the activated state. Thereafter, a spontaneous structural change inactivates the channel, and the current gradually drops to reach a plateau with very low channel open probability. The formation of the inactivated state is reversible and in slow equilibrium with other states (17). Inactivation is distinct from deactivation, which is the reverse of activation. In contrast to activation and deactivation, mutations that perturb inactivation in KcsA (and in Shaker channels) (18, 19) are clustered in region near the selectivity filter, distinct from the activation domain and presumably sensitive to K+ ion binding (20, 21). The kinetics of inactivation are dependent on permeant ions; higher concentrations of K+, or of other permeant ions like Rb+, can significantly slow the rate of inactivation (22, 23), whereas there is little dependence on the activating stimulus (pH for KcsA as it activates at low pH; e.g., pH 3–4) (22, 24). The formation of the inactivated state is assumed to be an allosteric response to activation (25) because of its spontaneous kinetics following activation. Analogous regulatory mechanisms are known to be important in a range of biological contexts, such as regulating the frequency and duration of firing in excitable cells (26) associated with QT syndrome in cardiac tissue, and other pathologies (9, 27).
Fig. 1.
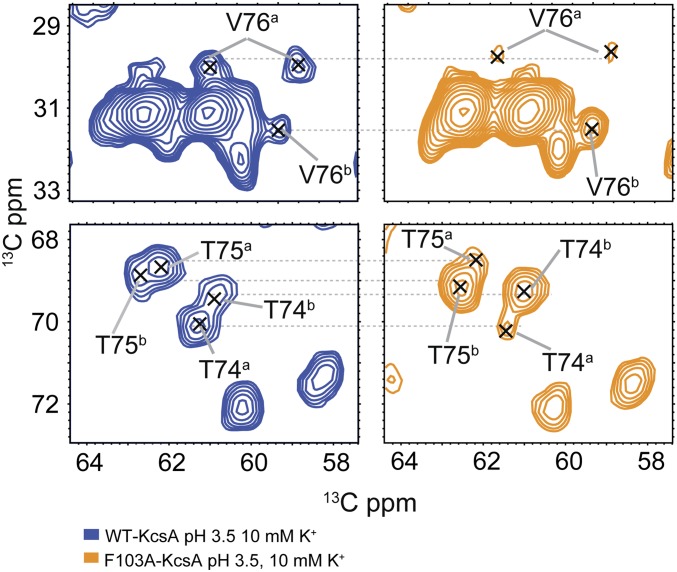
(A) Structural transitions in KcsA during channel function. In each structure, the K+ binding selectivity filter (including conserved residues T74, T75, V76, G77, Y78, and G79) is highlighted in blue; the pH sensor (E118, E120, H25, mutated in 3FB5 and 3F5W) where protons bind during activation is highlighted in red; and the hinge of the inner helix, a site of significant conformational dynamics during activation and allosteric coupling, is highlighted in yellow. The resting state, designated as deactivated (PDB ID code 1K4C) has K+ ions bound in the selectivity filter and is conductive, but has a closed intracellular gate, with a deprotonated pH sensor and the TM2 bundle crossed and occluded. This stable species is observed in numerous X-rays structures of KcsA. Following a drop in pH, the activated state, the only conductive state in functional assays, is populated transiently. In this state both the selectivity filter and the intracellular gate are conductive (PDB ID code 3FB5). The activated state slowly and spontaneously decays to an inactivated state (PDB ID code 3F5W). Our working hypothesis for inactivation is that the inactivated state differs from the activated state by loss of the K+ ions in the selectivity filter and associated conformational and hydration changes. Electrophysiology studies show that protein can recover from the inactivated state to the deactivated state at high pH, here represented by the dashed lines through a putative state inactivated* or I* that is “closed at both gates,” meaning both deprotonated at the pH sensor and K+ deplete or possibly by way of the activated state. (B) A thermodynamic cycle for H+ and K+ ion binding in the coupling network of KcsA. The blue dashed and angled arrow represents the activation followed by C-type inactivation as observed in pH jump electrophysiology experiments; the red dashed and angled arrow represents our observations of the result of lowering [K+] at constant neutral or slightly elevated pH (35). The allosteric coupling factor defines the strength of the allosteric coupling and is calculated as: α = Kapparent(3.5)/Kapparent(7.5) or α′ = Ka(K+ bound)/Ka(K+ apo). (C) Similar changes in chemical shift implying similar structural transitions were seen for the selectivity filter of KcsA. Similar chemical shifts are seen in the selectivity filter for K+-bound state, regardless of pH; similar chemical shifts are seen in the selectivity filter for K+-apo state, regardless of pH marker peaks V76 (CB-CG1/CG2), T75 (CB-CA), T74 (CB-CA) obtained from 13C–13C 2D correlation spectra are shown for a variety of [K+] and pH values. (Left) Contrasts KcsA at pH 7.5 (the deactivated state, in yellow) to that at pH 3.5 (activated state, in blue); both experiments are at high ambient [K+] (bound states). (Right) Contrasts the channel at pH 7.5 (inactivated* state, in red) to pH 3.5 (inactivated state, in cyan); both experiments are at low ambient [K+] (apo states). There are significant chemical shift changes between high [K+] (yellow and blue) and low [K+](red and gray) conditions, due to ion release and associated structural changes in the selectivity filter. Excellent agreement in the overlay of this region of the spectra comparing neutral vs. acidic pH illustrates that the structure transition at the selectivity filter at neutral and acidic pH are very similar and the selectivity filter is clearly intact throughout this pH range, i.e., the structure of selectivity filter is K+ dependent, and pH does not directly perturb it. Superscripts refer to the K+ apo and bound states respectively.
Motivated by its significance, efforts have been made to illustrate the mechanism of inactivation, and the structure of the inactivated state (23, 28–30). The inactivated state is believed to have a distinct structure from the deactivated or activated state (22, 24). Kinetic signatures and mutation profiles suggest that inactivation involves ion release in the selectivity filter, due to a large allosteric change in affinity as indicated in the thermodynamic cycle, shown in Fig. 1B; we probe this hypothesis for the nature of inactivation in this work. Cast in this way, the inactivation process is a classic allosteric effect of one binding event on the affinity for a second ligand in a distant pocket. A number of studies have already suggested that the inactivated state is similar to a low-K+ state (28, 31, 32). The putative loss of K+ ions and associated structural change at the selectivity filter would logically lead to cessation of ion flow: in the “knock-on” mechanism (33), steady-state K+ binding and facile flow are essential for conduction. An alternate mechanism involves dilation of the selectivity filter due to changes at the outer mouth of the pore (12, 30). Recently, based on studies of a semisynthetic channel, it has also been emphasized that ion occupancy at a specific site at the selectivity filter leads to inactivation (34). In support of this hypothesis, structures of the selectivity filter at low and at high ambient [K+] show that there is a protein conformational change associated with K+ binding (11). This hypothesis is also supported by a series of crystal structures of constitutively open mutants of KcsA (30), recent simulations (19, 23), properties of mutants near the base of the selectivity filter (19, 23), and by NMR experiments showing that the otherwise tightly bound ion is in fact released when the channel is activated by a pH drop (32, 35, 36). Moreover, we showed that the acid coupled to K+ ion binding is not the proximal E71, but rather the distal E118 and associated residues in the pH sensor (35, 37), consistent with other studies cited above. It is noteworthy that the intracellular pH sensor (activation gate) and the extracellular selectivity filter (inactivation gate) are quite remote (>30 Å), so elucidating the molecular pathway for communication between the coupled domains is of great interest.
To confirm participation of specific residues, we probed the strength of the transmembrane allosteric coupling in KcsA using solid-state NMR (SSNMR), contrasting wild-type and site-specific mutants. These site-specific binding measurements, carried out in an authentic bilayer environment, are an important step toward gaining a comprehensive understanding of the molecular network that mediates this allosteric coupling and will be a vital benchmark for further investigations of coupling. The NMR shifts provide rich structural information for the underlying states. This study is a unique quantitative and site-specific analysis of allosteric coupling in a membrane system by NMR.
Results
SSNMR Shifts Probe KcsA Structures in Its Apo and Ion Bound States.
NMR chemical shift measurements provide highly specific fingerprints for the structures of proteins, probing conformation, and binding. We used chemical shifts to probe KcsA, with particular focus on the selectivity filter. We contrasted spectra at near neutral (pH 7.5) vs. low (3.5) pH, at a broad range of [K+], to achieve the four limiting states in Fig. 1B, in functionally relevant lipid and buffer conditions. Consistent with the fact that these channels function at low pH, the spectra for acidic samples display narrow linewidths, comparable to those observed at neutral pH, and the vast majority of resolved sites show chemical shifts that are indistinguishable from those at neutral pH (Fig. 1C), indicating that KcsA is folded at pH 3.5.
Similar selectivity filter structural transitions are observed at neutral and acidic pH.
Previously, we characterized K+-dependent changes in the selectivity filter at neutral pH (38) that correspond to the high K+ (conductive) and low K+ (collapsed) states described by crystallography (39). Several NMR reporters were identified, including T74 (CA, CB, CG), T75 (CA, CB, CG), and V76 (CB, CG1, CG2). Here, we observe a similar transition between high and low K+ states at low pH, each of which shows highly similar structures to its corresponding counterpart at neutral pH (Fig. 1C); this means that the large difference in pH does not perturb the structure of selectivity filter directly, and that its capability to bind ions is intact. Moreover, the NMR markers for K+ binding are valid also at low pH.
pH sensor residues E118 and E120 are protonated at pH 3.5 across a broad range of K+ concentrations.
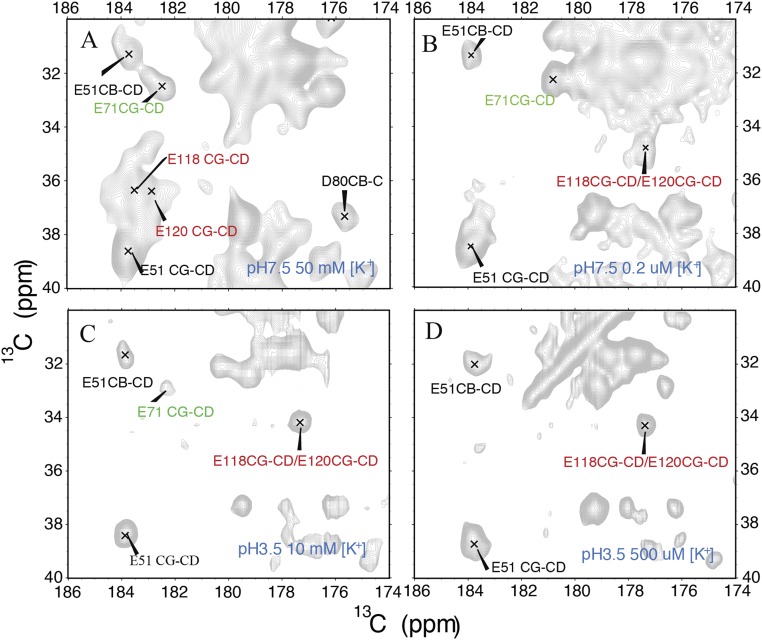
The pH sensor binds protons (15, 36) and causes channel activation. Previous studies show that the effective pKa value for activation is 5 or 5.5 (15). Accordingly, the E118 and E120 CG-CD NMR cross-peaks show diagnostic chemical shift changes associated with significant protonation at pH 3.5 (32, 35); at neutral pH, the peaks are characteristic of a deprotonated anionic state (Fig. S1), unless the ambient [K+] is significantly lowered to cause a coupled pronation process (35).
Fig. S1.
NMR spectra of the glutamic acid residues in the intracellular pH sensor controlling the activation gate of KcsA show that the pH gate residues Glu118 and Glu120 are protonated at pH 3.5 across a broad range of [K+], whereas at pH 7.5, the behavior is more complex, and at high [K+], the pH sensor residues are deprotonated as expected, but at low [K+], due to coupling between K+ ion release and H+ binding, the sensor residues are protonated based on the NMR spectra. (A–D) Spectra for samples at (respectively) pH 7.5 and 50 mM [K+], pH 7.5 and 0.2 μM [K+], pH 3.5 and 10 mM [K+], and pH 3.5 and 500 μM [K+]. The signals associated with Glu118 and Glu120’s CG-CD correlation show comparable chemical shifts in B–D, indicating that they are protonated, whereas in A they show clear indication of deprotonation. We conclude that the local environment of these residues are similar at acidic pH across a range of [K+], and also at neutral pH if the [K+] is very low (35).
Potassium Ion Affinity at Acidic pH: Quantifying the Strength of Allosteric Coupling.
To confirm strong allosteric coupling of the two binding events, and the participation of specific residues in the allosteric process, we developed a SSNMR approach to analysis of binding strength and applied it to this system. Although the low K+ and high K+ structures (and NMR shifts) of the selectivity filter show little sensitivity to pH, we demonstrate here that the energetics of the transition, as characterized by the affinity of the selectivity filter for K+ ions, are dramatically dependent on pH. Such a pH dependence was suggested by previous studies but has not been studied systematically or quantified in a membrane environment (28, 40). Our four-state thermodynamic formulation allows the magnitude of the allosteric coupling to be quantitatively recast as the difference in the equilibrium K+ binding affinity at high vs. low pH. We are able to measure the K+ affinity in a site-specific manner and in a native bilayer environment using solid-state NMR. Because the K+ binding event is coupled with structural changes, the apparent potassium affinity (Kapparent) is measured. This titration study is rich with structural information because it is based on NMR shifts.
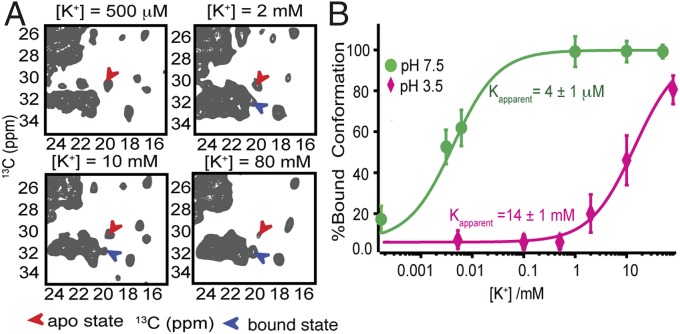
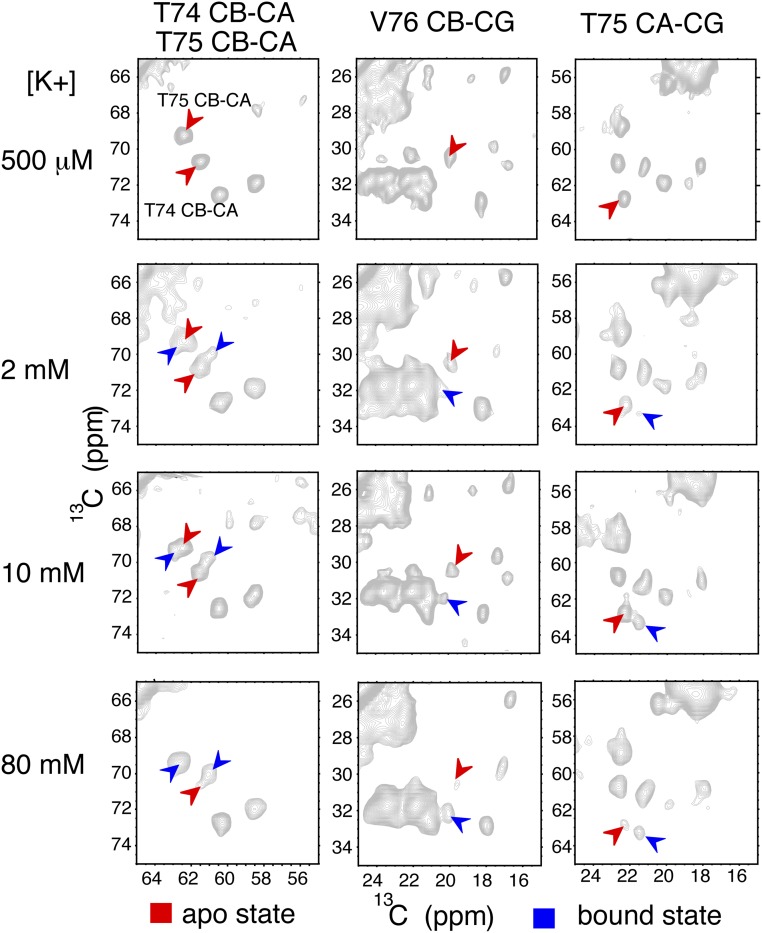
We measured Kapparent, contrasting acidic pH (3.5) vs. neutral pH (7.5) by quantifying the relative populations of K+ bound vs. apo states using the “marker” cross-peaks identified above, including T74 (CB-CA), T75 (CB-CA, CA-CG), and V76 (CA-CG1, CG2) (Fig. 2A and Fig. S2). These residues were selected because of their robust signal and good resolution with respect to other signals in 2D planes. Fitting the titration data (using a fixed Hill coefficient of 1) yielded a Kapparent of 14 ± 1 mM at pH 3.5 at 0 °C; in contrast, the Kapparent at neutral pH is 4 ± 1 μM, under otherwise comparable conditions (Fig. 2B). The temperature control for these measurements is improved relative to prior measurements (38) due to the use of an Efree probe with reduced sample heating (41). We calculated the allosteric coupling factor alpha, defined as the ratio of the potassium affinities contrasting open vs. closed activation gate (i.e., at low vs. neutral pH); there is a remarkable shift of the Kapparent by more than three orders of magnitude (α = 3,500) over the pH range where the channel converts from the deactivated to the inactivated state (7.5–3.5).
Fig. 2.
Differences in K+ binding at neutral vs. acidic pH values for WT-KcsA. (A) 13C–13C correlation spectra at pH 3.5 show changes in the populations of apo vs. bound state as the [K+] increases; the cross-peaks (V76 CB-CG1) were integrated to calculate the relative populations (using these and additional peaks shown in Fig. S2; Materials and Methods). (B) A significant change in potassium ion affinity (α = 3,500) is observed comparing channels with an open activation gate measured at pH 3.5 (magenta) vs. a closed gate measured at pH 7.5 (green). The Kd values calculated by fitting the data to a noncooperative binding equation (Hill coefficient n = 1) are 4 ± 1 μM for pH 7.5 and 14 ± 1 mM at pH 3.5.
Fig. S2.
Marker peaks in NMR spectra showing the structural transition at the selectivity filter at acidic pH 3.5 as the environmental [K+] changes from 500 μM to 80 mM. The volumes of cross-peaks indicated by arrows are used to calculate the bound state population ratio and Kapparent calculations in Fig. 2B.
The thermodynamic cycle (Fig. 1B) also dictates that if the ambient pH affects the equilibrium K+ affinity, then the ambient K+ level must also alter the apparent pKa of the coupled pH sensor. Prior work showed that pH gate residues E118 and E120 bind protons as the [K+] is lowered to about 0.2 μM at pH 7.5, which indicates a large pKa shift in the pH sensor as a function of K+ (35), and confirms the identity of the pH sensor as the coupled general base (whereas in contrast, the proximal E71 remains protonated in all [K+]) (28). The K+-dependent pKa shift in E118 and E120 has not been fully quantified, but it is clear that the pKa is altered by at least 3 pH units comparing low and high potassium conditions, because at low [K+] the pKa must be 8.0 or higher, and at high [K+] the pKa value is known to be ∼4–5 and is consistent with our experimental results (22, 36).
The free energy of allosteric coupling between K+ and H+ binding was estimated from the pH dependence of the K+ Kapparent values: ∆∆G = −RTln [Kapparent(pH 7.5)/Kapparent(pH 3.5)] = −19 ± 2 kJ⋅mol−1. A tentative value estimated from the pKa shift with [K+] would similarly be ∆∆G = −2.303RT∆pKa = −16 kJ⋅mol−1. We regard these energies as estimates; they were calculated using a simplified binding model that treats both K+ and H+ binding as single-ligand, noncooperative events. The stoichiometry and cooperativity of the binding events is certainly more complex and the details remain debated (42–44).
Due to the relatively large pH range used, the phenomenological shift in binding affinity can include a number of effects. We argue that a large portion of it is allosteric coupling. Guoy–Chapman analysis suggests that the differences with respect to previous micelle measurements are too large to be explained by K+ ions concentrating at a negatively charged lipid interface (an ∼10-fold effect) (37). Nonspecific effects of pH on the lipid bilayer and protein structure and thermodynamics could certainly also contribute to the altered affinity, although our data show that KcsA remains folded throughout this range. Finally, a large and specific effect of site-specific mutation involving the nonionizable F103 (discussed below) provides strong support that the altered affinity is dominated by contacts in the protein that result in nonadditive energies of binding (19, 23).
Kinetics of Ion Release Are Slow Across All Conditions Studied.
Inactivation has a clear kinetic signature, and is a remarkably slow process (on the seconds timescale) (22). The origin of the kinetic barrier is debated; molecular dynamics simulations suggest that changes in solvation associated with the conformational change in the filter present a kinetic bottleneck (17). We observe clear evidence for slow conformational exchange between the K+ bound and depleted states at both acidic and neutral pH. At both conditions, intermediate K+ concentrations result in sharply resolved NMR lines corresponding to two states of the filter and indicating slow exchange (100 ms or slower, which is well below estimates of koff based on the affinities measured here) (38). The relative populations are significantly affected by temperature changes in situ (Table S1), supporting the assumption that the system is in slow dynamic equilibrium during the NMR experiments. The slow kinetics observed here are in contrast to those observed by solution NMR in detergent micelles at higher temperatures and at pH 6.0 (45), but are consistent with the slow inactivation kinetics observed in electrophysiology (22, 24).
Table S1.
Summary of K+ affinities at various pH conditions and temperatures for WT-KcsA and F103A mutant
| Construct | pH | Temperature, K | K+ Kapparent, mM | RTln(Kapparent), KJ/mol |
| WT | 3.5 | 273 | 14 ± 1 | −9.7 ± 0.2 |
| WT | 3.5 | 293 | 1.0 ± 0.2 | −16.8 ± 0.5 |
| WT | 7.5 | 258 | 0.003 ± 0.001 | −27.3 ± 0.7 |
| WT | 7.5 | 270 | 0.004 ± 0.001 | −27.9 ± 0.6 |
| WT | 7.5 | 283 | 0.005 ± 0.002 | −28.7 ± 0.9 |
| F103A | 7.5 | 273 | 0.002 ± 0.001 | −29.8 ± 1.0 |
| F103A | 7.5 | 290 | 0.0006 ± 0.0005 | −34.5 ± 2.0 |
| F103A | 3.5 | 273 | 0.035 ± 0.015 | −18.1 ± 2.0 |
The affinities were computed by simulating data using a Hill equation (Materials and Methods).
Confirming Candidates in the Allosteric Coupling Pathway: Studies of the Inactivationless F103A.
F103 has been previously suggested as a possibly important allosteric residue. Prior evidence from crystallography and molecular simulations suggested that steric interactions between T74 at the base of the selectivity filter, F103 in the TM2 “hinge” (second transmembrane helix hinge), and I100 from a neighboring monomer are involved in the allosteric network (19, 23, 30). Experimental support for this role consisted in changes in inactivation. In contrast, here we provide a direct experimental test of its contribution to allosteric coupling. The quantitative contribution of F103 to the allosteric coupling was probed by using a mutant, F103A. We speculated that if the F103A mutant “loosens” this network, the energetic coupling between the pH gate and the selectivity filter will become much weaker, rendering K+ affinity at the selectivity filter relatively insensitive to environmental pH and, in particular, we predicted that it would not release K+ at low pH.
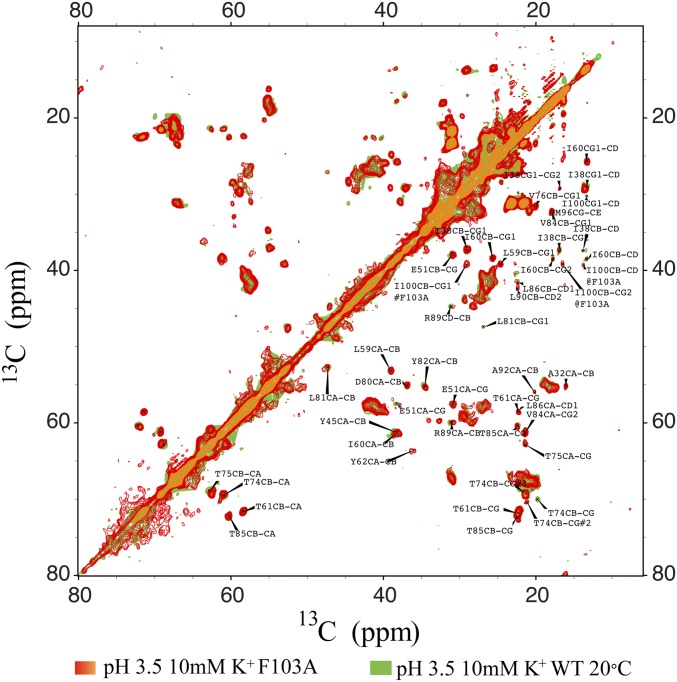
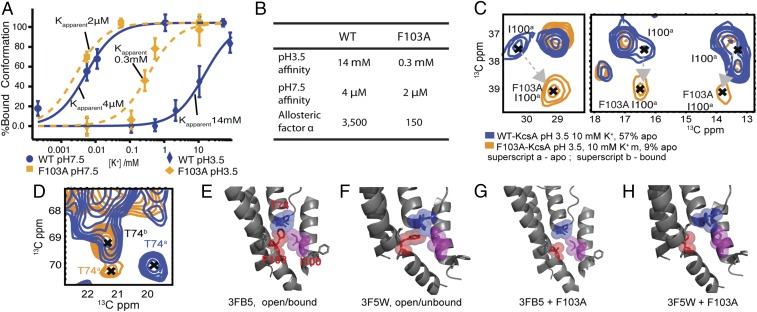
F103A-KcsA was expressed in good yield and forms a stable tetramer in the bilayer; it exhibits NMR chemical shifts that indicate an overall homologous structure to the WT channel (Figs. S3 and S4), but shows perturbations for specific residues, such as I100, which is proximal to F103. As we noted above, I100 is also suspected to be important in the coupling network (19, 46). We speculate that the dramatic chemical shift perturbation is caused by the change in environment of I100 in that there is a strong interaction between I100 and F103 in wild-type when the channel gets activated that is absent in F103A (Fig. 3 C–H). The side chain CG of T74 is also sensitive to the mutation F103A: in WT-KcsA, CG shows a large chemical shift change in the transition from bound to apo state; in contrast, in F103A, the CG chemical shift does not change even though the backbone shows the same structural transition (Fig. 3 C–H). These observations are consistent with the hypothesis that steric contacts occur between the aromatic ring of F103 and the T74 sidechain for WT-KcsA during inactivation, as suggested by the recently published mechanism for allosteric coupling in KcsA developed from crystallography and molecular dynamics simulations (30).
Fig. S3.
Spectra of WT-KcsA (blue) are compared with those of the F103A mutant (orange), exhibiting marker peaks in the selectivity filter. Superscripts refer to the K+ apo and bound states respectively. The chemical shift values for WT and F103A are nearly identical in both the bound and apo states, which simplifies our analysis (no reassignment needed) and indicates that F103A minimally perturbs the structure (see Fig. S4 for the full overlay). In contrast, the relative populations of the states change significantly between F103A (pH 3.5 10 mM K+, 9% apo) and wild type (pH 3.5 10 mM K+, 57% apo).
Fig. S4.
Comparison of 2D homonuclear 13C spectra of wild-type KcsA vs. F103A shows that the chemical shift changes displayed in F103A are specific to the hinge residues. As illustrated by this overlay, the great majority of peaks in the wild-type channel are identical to the peaks in F103A. Specific exceptional changes have been seen in I100 and T74, however, as are discussed in the text and Fig. 3.
Fig. 3.
F103 is a critical residue in the allosteric pathway. (A) The potassium ion titration curves of mutant F103A are compared at neutral and acidic pH with those for the wild-type channel to contrast the affinity and coupling changes. (B) The allosteric coupling strength is significantly weaker for mutant F103A compared with WT-KcsA. (C) Specific chemical shift perturbations are observed for the F103A mutant, compared with the WT-KcsA, underscoring the important interactions between F103 and I100 and with T74. Peaks in spectrum are I100 (CB-CG1, CB-CG1 CB-CD), respectively. In WT-KcsA, I100 shows a perturbation in the peaks associated with the bound, putatively activated state: the peaks are missing or broadened due to dynamics or heterogeneity. Similarly, the bound state peaks for I100 in F103A mutant, where allostery has been reduced, are also missing; but the apo state peaks shows significant chemical shift change, especially at CB. (D) Similarly T74 sidechain correlations show changes in the apo state (putative inactivated state) between the wild-type and the F103A mutant. (E–H) Crystal structure analysis reveals that the steric contact among T74, I100, and A103 is reduced in the F103A mutant compared with WT-KcsA. Steric contact between F103 (red) and I100 (magenta) occurs between monomers in the tetrameric structure. Contact between F103 (red) and T74/T75 (blue), particularly in the apo state, can be seen for the WT sample in both the bound state (E) (PDB ID code 3FB5) and in the apo state (F) (PDB ID code 3F5W). Our data are consistent with a model in which the clash is largest for the activated state. This steric clash is relieved when the bulky F103 sidechain is mutated to alanine (G and H) (made with PyMol).
More importantly, consistent with our predictions, the mutant channel F103A indeed exhibits much weaker allosteric coupling. Fig. S3 contrasts 13C–13C correlation spectra of key residues at the selectivity filter for WT vs. F103A samples (under inactivating, i.e., low pH and moderately high [K+]). Compared with the WT-KcsA, which significantly populates the K+-depleted state under these conditions, the F103A mutant remains bound to K+. We conducted a similar titration study on this F103A mutant and, as the results show in Fig. 3A, the K+ affinity of the F103A (Kapparent = 2 ± 1 μM) is comparable to that of WT-KcsA at a neutral pH, whereas at acidic pH, the affinity is much tighter (Kapparent = 0.32 ± 0.10 mM), which indicates a significantly reduced allosteric coupling strength (Fig. 3B) (α = 160; contrasted with 3,500 for WT-KcsA under the same conditions). These results are interesting in connection with electrophysiology studies on mutant E71H-F103A, where the mutant channel shows significant conductance despite the presence of the strongly inactivating mutation E71H (30). We note that these data provide strong additional support for the hypothesis that ion loss is a key step of the inactivation process: F103A is expected to be virtually inactivationless from prior studies, whereas our own NMR results on F103A indicate it does not release K+ ions upon activation.
Discussion
We offer insights into the slow allosteric transition underlying C-type inactivation, a vital gating mechanism in potassium channels that partly controls mean open time. This titration study, carried out in an authentic lipid environment, allowed us to prepare and stabilize functionally critical states, presumably closely related to the intermediates of electrophysiology experiments. Detailed information from SSNMR, such as chemical shifts, therefore helps to form a more direct connection between structure and function. Our results involving the role of F103 directly disprove a long-standing hypothesis that C-type inactivation is associated with protonation effects proximal to the selectivity filter, and provide strong support for the allosteric hypothesis wherein large-scale conformational events during intracellular activation are responsible for the affinity change at the extracellular selectivity filter.
We quantitatively probed the allosteric coupling strength for wild-type channel in membranes using these pH-dependent ion titrations. We observe a difference of three orders of magnitude in K+ affinity over the typically used pH range, which indicates substantial transmembrane coupling. Quantification of the potassium affinities of KcsA under various conditions has been the subject for several previous studies, using different methods. Functional methods suggest that the affinity of the selectivity filter for K+ at pH 7.5 (deactivated channel) is ∼29 μM (47), and inactivation at pH 3.5 is associated with ion loss with an affinity of ∼0.9 mM (32). A different picture emerged in isothermal titration calorimetry experiments in detergents, which showed the potassium affinity to be 0.41 mM at pH 7.5 (43). Shimada and colleagues (36) reported elegant solution NMR experiments with detergent-stabilized KcsA showing that the potassium affinity changes from 6 to 50 mM between pH 3.2 and 6.7 at 45 °C; the pH dependence of potassium affinity in these studies is much weaker than estimates based on functional studies. Our experiments show a pH dependence that is much larger than these previously reported, and are more compatible with the available functional data and with the hypothesis that ion loss is the essential step of inactivation. This allosteric effect of pH on protein function is analogous to other systems, for example the case of Yeast pyruvate kinase, in which pH modulates protonation state of various ionizable residues to ultimately allosterically affect the reactivation rates (48). We presume the difference between the solution NMR or thermocalorimetric affinity measurements for KcsA and these SSNMR studies is due to our use of membrane bilayers, and conclude that dramatically stronger allosteric coupling occurs in the bilayer. The importance of the membrane environment for membrane protein integrity and function has been reported in numerous studies (49–52).
We used analogous measurement of allosteric coupling strength to test the involvement of an important residue in the allosteric network. These results show that replacement of the bulky aromatic sidechain of F103 with the smaller sidechain of alanine greatly reduces the allosteric coupling presumably by diminishing the energy penalty caused the clash between F103 sidechain to I100 T74, which would destabilize the activated state. In contrast to functional studies, the thermodynamic characterization of the coupling energy leaves us with a clear picture for why F103A does not inactivate: loss of K+ ion is not expected for F103A after channel activation.
Through this study we also stabilized the activated state of a wild-type functional KcsA at low pH and high [K+], with both the activation gate open (protonated pH sensor) and inactivation gate open (K+ bound in the selectivity filter). The activated state is normally an evanescent or metastable intermediate in functional studies of KcsA, and has been difficult to stabilize for structural characterization, requiring use of multiple mutations and other alterations in the system. Stabilizing this species enables structural and dynamics studies by NMR in native bilayers.
NMR has been particularly powerful for studying allosteric coupling, because of the rich structural and dynamic information it can provide (53–57). Chemical shift analysis has been used to identify residues that participate in long-range response to ligand binding (54). The ability of NMR to probe the slightly populated or “invisible” states in coupled systems (55) also holds great potential for detecting minor population involved in allostery and monitoring its population shift to facilitate biological functions (56). NMR studies of allostery have highlighted the possibility of dynamically driven allostery, as in the case of the catabolite activator protein to dissect the entropy and enthalpy contributions to an allosteric DNA binding in a WT and mutation (57). These prior studies feature proteins in solution, but it is still a formidable task to study transmembrane allostery, which is ubiquitous in critical biological systems, including G protein-coupled receptor, kinases, EGFR, ion channels, and many other signaling systems, and has been resistant to quantitative mechanistic investigations (53–58). The present study helps to pave the way for similarly exciting studies of allosteric membrane proteins by solid-state NMR. We envision that approaches demonstrated in this study will be helpful for understanding other systems, of relevance to human health.
Materials and Methods
NMR Sample Preparation.
13C–15N uniformly enriched WT-KcsA and the mutant F103A-KcsA were prepared by overexpression in Escherichia coli JM83 cells in M9 minimum media using a tetracycline-regulated expression vector pask90, as described previously (31, 59). Both constructs have an N-terminal hexahistidine tag. The KcsA-F103A mutant was prepared using a QuikChange Lighting Site-Directed Mutagenesis kit (Agilent Technologies) and verified by gene sequencing (Genewiz). F103A-KcsA is less stable as a tetramer compared with WT-KcsA, as indicated by SDS gels, but stays as a tetramer revealed by size-exclusion chromatography. KcsA was reconstituted into liposomes by reducing the detergent concentration through dialysis; the liposomes contain 9:1 ratio of 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,2-dioleoyl-sn-glycero-3-phospho-l-serine by mass, and 1:1 ratio of protein:lipid by mass. To prepare samples at acidic pH values, proteoliposomes were first dialyzed at room temperature against a 50-mM Tris buffer at pH 7.5 with the desired potassium concentrations and then further dialyzed at 4 °C against 10 mM sodium citrate buffer at pH 3.5 with the appropriate potassium concentration. NaCl was added to achieve a consistent total concentration ([K+] + [Na+]) of 50 mM to keep the ionic strength constant in the titration series (except the sample with 80 mM [K+]). After dialysis, liposomes were centrifuged using a Sorvall benchtop centrifuge at 4,125 rpm (3,697 × g, 4 °C) for 40 min. The pH values of the supernatants after centrifugation were verified to be within 0.1 unit of the desired pH value. The proteoliposomes were then subject to three freeze–thaw cycles to remove excess water. Finally, samples were packed into 3.2-mm Bruker rotors for NMR measurement.
NMR Data Collection and Processing.
The details of data collection can be found in SI Materials and Methods. All spectra were measured at 0 ± 2 °C if the temperature is not otherwise specifically indicated. All spectra were processed in NMRpipe (60) with −30 Hz Lorentzian and 90 Hz Gaussian apodization; Sparky (61) was used for spectral visualization and integration of cross-peaks. The populations of the bound states for each residue were quantified by integrating the bound and apo cross-peaks of T74 (CB-CA), T75 (CB-CA, CA-CG), and V76 (CB-CG, CG2) on a 13C–13C 2D correlation (Fig. S2). The normalized population ratios of protein were calculated by averaging the population ratios of reporter residues T74, T75, and V76. The normalized bound-state percentage was defined as integral(bound)/[integral(bound) + integral(apo)] × 100% for each residue. Normalized populations were fit to the binding expression below with Hill coefficient, n, equal to 1 (noncooperative), where θ is the normalized population: . The reported errors are dominated by the fitting error (Kd), which is roughly on the same scale of errors from peak integration and averaging.
SI Materials and Methods
NMR Data Collection and Processing.
NMR spectra were collected on a Bruker Avance 750 MHz (17.6 T) spectrometer, or a Bruker Avance III 600 MHz (14.1 T) spectrometer with a 3.2-mm Efree probe at magic-angle spinning (MAS) frequencies of 12 or 14 kHz. The dipolar assistant recoupling resonance pulse sequence (62) was used for 13C–13C correlation experiments with a 15-ms mixing period. Sample temperature was calibrated based on an external reference using methanol chemical shift (63) to correct for heating during MAS spinning and RF pulsing. All spectra were measured at 0 ± 2 °C if the temperature is not otherwise specifically indicated. Carbon chemical shifts were referenced externally to the downfield adamantane line at 40.48 ppm.
Allosteric Coupling and Related Thermodynamic Parameters.
We calculated the free energy for conformational transitions at neutral and acidic pH based on Eq. S1, which relates the measured affinity (Kd), to the free energy (∆G) of the transition.
| [S1] |
The strength of the allosteric coupling is related to the difference in the relative free energy of the transition at neutral vs. acidic pH, as described in Eq. S2.
| [S2] |
Based on our measurements of K+ affinities of 4 ± 1 μM for pH 7.5 and 14 ± 1 mM at pH 3.5, and making the simplifying assumption that the Hill coefficient is 1 for both K+ and H+, the ∆∆G representing the allosteric coupling is −19 ± 2 kJ⋅mol−1, at a temperature T = 273K (R is the universal gas constant). This value represents a lower bound on the allosteric coupling strength, because we assume that the binding involves a single K+ and a single H+. The stoichiometry of the binding reaction is debated and difficult to measure (11, 33, 42). An equation of allosteric coupling energy for n K+ binding will be
| [S3] |
Acknowledgments
We thank members of A.E.M.’s group: Dr. Wylie and Rivkah Rogawski for helpful discussions, and Dr. Ivan Sergeyev and Dr. Wenbo Li for NMR techniques. We also thank the laboratory of Crina Nimigean for the PASK90 plasmid; Dr. Dorothy Kim and Dr. David Pason for the help of electrophysiology measurement; and Dr. Boris Itin at the New York Structural Biology Center (NYSBC) for support with NMR instrumentation. A.E.M. is a member of NYSBC, a Strategically Targeted Academic Research center supported by the New York State Office of Science, Technology, and Academic Research. This work was supported by NIH Grant NIH R01 GM088724 (to A.E.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701330114/-/DCSupplemental.
References
- 1.Nussinov R, Tsai C-J. Allostery in disease and in drug discovery. Cell. 2013;153:293–305. doi: 10.1016/j.cell.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 2.Cui Q, Karplus M. Allostery and cooperativity revisited. Protein Sci. 2008;17:1295–1307. doi: 10.1110/ps.03259908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helmstaedt K, Krappmann S, Braus GH. Allosteric regulation of catalytic activity: Escherichia coli aspartate transcarbamoylase versus yeast chorismate mutase. Microbiol Mol Biol Rev. 2001;65:404–421. doi: 10.1128/MMBR.65.3.404-421.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.May LT, Leach K, Sexton PM, Christopoulos A. Allosteric modulation of G protein-coupled receptors. Annu Rev Pharmacol Toxicol. 2007;47:1–51. doi: 10.1146/annurev.pharmtox.47.120505.105159. [DOI] [PubMed] [Google Scholar]
- 5.Monod J, Wyman J, Changeux J-P. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 6.Koshland DE, Jr, Némethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 7.Wrabl JO, et al. The role of protein conformational fluctuations in allostery, function, and evolution. Biophys Chem. 2011;159:129–141. doi: 10.1016/j.bpc.2011.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yellen G. The voltage-gated potassium channels and their relatives. Nature. 2002;419:35–42. doi: 10.1038/nature00978. [DOI] [PubMed] [Google Scholar]
- 9.Jentsch TJ. Neuronal KCNQ potassium channels: Physiology and role in disease. Nat Rev Neurosci. 2000;1:21–30. doi: 10.1038/35036198. [DOI] [PubMed] [Google Scholar]
- 10.Baukrowitz T, Yellen G. Use-dependent blockers and exit rate of the last ion from the multi-ion pore of a K+ channel. Science. 1996;271:653–656. doi: 10.1126/science.271.5249.653. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 12.Doyle DA. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 13.Salkoff L, Butler A, Ferreira G, Santi C, Wei A. High-conductance potassium channels of the SLO family. Nat Rev Neurosci. 2006;7:921–931. doi: 10.1038/nrn1992. [DOI] [PubMed] [Google Scholar]
- 14.Choe S. Potassium channel structures. Nat Rev Neurosci. 2002;3:115–121. doi: 10.1038/nrn727. [DOI] [PubMed] [Google Scholar]
- 15.Thompson AN, Posson DJ, Parsa PV, Nimigean CM. Molecular mechanism of pH sensing in KcsA potassium channels. Proc Natl Acad Sci USA. 2008;105:6900–6905. doi: 10.1073/pnas.0800873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu YS, Sompornpisut P, Perozo E. Structure of the KcsA channel intracellular gate in the open state. Nat Struct Biol. 2001;8:883–887. doi: 10.1038/nsb1001-883. [DOI] [PubMed] [Google Scholar]
- 17.Ostmeyer J, Chakrapani S, Pan AC, Perozo E, Roux B. Recovery from slow inactivation in K+ channels is controlled by water molecules. Nature. 2013;501:121–124. doi: 10.1038/nature12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cordero-Morales JF, et al. Molecular driving forces determining potassium channel slow inactivation. Nat Struct Mol Biol. 2007;14:1062–1069. doi: 10.1038/nsmb1309. [DOI] [PubMed] [Google Scholar]
- 19.Pan AC, Cuello LG, Perozo E, Roux B. Thermodynamic coupling between activation and inactivation gating in potassium channels revealed by free energy molecular dynamics simulations. J Gen Physiol. 2011;138:571–580. doi: 10.1085/jgp.201110670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng WWL, McCoy JG, Thompson AN, Nichols CG, Nimigean CM. Mechanism for selectivity-inactivation coupling in KcsA potassium channels. Proc Natl Acad Sci USA. 2011;108:5272–5277. doi: 10.1073/pnas.1014186108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cordero-Morales JF, et al. Molecular determinants of gating at the potassium-channel selectivity filter. Nat Struct Mol Biol. 2006;13:311–318. doi: 10.1038/nsmb1069. [DOI] [PubMed] [Google Scholar]
- 22.Chakrapani S, Cordero-Morales JF, Perozo E. A quantitative description of KcsA gating I: Macroscopic currents. J Gen Physiol. 2007;130:465–478. doi: 10.1085/jgp.200709843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cuello LG, et al. Structural basis for the coupling between activation and inactivation gates in K(+) channels. Nature. 2010;466:272–275. doi: 10.1038/nature09136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakrapani S, Cordero-Morales JF, Perozo E. A quantitative description of KcsA gating II: Single-channel currents. J Gen Physiol. 2007;130:479–496. doi: 10.1085/jgp.200709844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao L, Mi X, Paajanen V, Wang K, Fan Z. Activation-coupled inactivation in the bacterial potassium channel KcsA. Proc Natl Acad Sci USA. 2005;102:17630–17635. doi: 10.1073/pnas.0505158102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasmusson RL, et al. Inactivation of voltage-gated cardiac K+ channels. Circ Res. 1998;82:739–750. doi: 10.1161/01.res.82.7.739. [DOI] [PubMed] [Google Scholar]
- 27.Sanguinetti MC, Tristani-Firouzi M. hERG potassium channels and cardiac arrhythmia. Nature. 2006;440:463–469. doi: 10.1038/nature04710. [DOI] [PubMed] [Google Scholar]
- 28.Bhate MP, McDermott AE. Protonation state of E71 in KcsA and its role for channel collapse and inactivation. Proc Natl Acad Sci USA. 2012;109:15265–15270. doi: 10.1073/pnas.1211900109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoshi T, Armstrong CM. C-type inactivation of voltage-gated K+ channels: Pore constriction or dilation? J Gen Physiol. 2013;141:151–160. doi: 10.1085/jgp.201210888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cuello LG, Jogini V, Cortes DM, Perozo E. Structural mechanism of C-type inactivation in K(+) channels. Nature. 2010;466:203–208. doi: 10.1038/nature09153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhate MP, et al. Preparation of uniformly isotope labeled KcsA for solid state NMR: Expression, purification, reconstitution into liposomes and functional assay. Protein Expr Purif. 2013;91:119–124. doi: 10.1016/j.pep.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ader C, et al. Coupling of activation and inactivation gate in a K+-channel: Potassium and ligand sensitivity. EMBO J. 2009;28:2825–2834. doi: 10.1038/emboj.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Köpfer DA, et al. Ion permeation in K+ channels occurs by direct Coulomb knock-on. Science. 2014;346:352–355. doi: 10.1126/science.1254840. [DOI] [PubMed] [Google Scholar]
- 34.Devaraneni PK, et al. Semisynthetic K+ channels show that the constricted conformation of the selectivity filter is not the C-type inactivated state. Proc Natl Acad Sci USA. 2013;110:15698–15703. doi: 10.1073/pnas.1308699110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wylie BJ, Bhate MP, McDermott AE. Transmembrane allosteric coupling of the gates in a potassium channel. Proc Natl Acad Sci USA. 2014;111:185–190. doi: 10.1073/pnas.1319577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imai S, Osawa M, Takeuchi K, Shimada I. Structural basis underlying the dual gate properties of KcsA. Proc Natl Acad Sci USA. 2010;107:6216–6221. doi: 10.1073/pnas.0911270107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhate MP. 2012. Mechanistic studies of ion binding and inactivation in the potassium channel KcsA by solid state NMR. PhD dissertation (Columbia University, New York)
- 38.Bhate MP, Wylie BJ, Tian L, McDermott AE. Conformational dynamics in the selectivity filter of KcsA in response to potassium ion concentration. J Mol Biol. 2010;401:155–166. doi: 10.1016/j.jmb.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang Y, et al. The open pore conformation of potassium channels. Nature. 2002;417:523–526. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- 40.Ader C, et al. A structural link between inactivation and block of a K+ channel. Nat Struct Mol Biol. 2008;15:605–612. doi: 10.1038/nsmb.1430. [DOI] [PubMed] [Google Scholar]
- 41.Gor’kov PL, et al. Using low-E resonators to reduce RF heating in biological samples for static solid-state NMR up to 900 MHz. J Magn Reson. 2007;185:77–93. doi: 10.1016/j.jmr.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 42.Posson DJ, Thompson AN, McCoy JG, Nimigean CM. Molecular interactions involved in proton-dependent gating in KcsA potassium channels. J Gen Physiol. 2013;142:613–624. doi: 10.1085/jgp.201311057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lockless SW, Zhou M, MacKinnon R. Structural and thermodynamic properties of selective ion binding in a K+ channel. PLoS Biol. 2007;5:e121. doi: 10.1371/journal.pbio.0050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewandowski JR, Halse ME, Blackledge M, Emsley L. Direct observation of hierarchical protein dynamics. Science. 2015;348:578–581. doi: 10.1126/science.aaa6111. [DOI] [PubMed] [Google Scholar]
- 45.Chill JH, Louis JM, Baber JL, Bax A. Measurement of 15N relaxation in the detergent-solubilized tetrameric KcsA potassium channel. J Biomol NMR. 2006;36:123–136. doi: 10.1007/s10858-006-9071-4. [DOI] [PubMed] [Google Scholar]
- 46.Peters CJ, Fedida D, Accili EA. Allosteric coupling of the inner activation gate to the outer pore of a potassium channel. Sci Rep. 2013;3:3025. doi: 10.1038/srep03025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piasta KN, Theobald DL, Miller C. Potassium-selective block of barium permeation through single KcsA channels. J Gen Physiol. 2011;138:421–436. doi: 10.1085/jgp.201110684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Susan-Resiga D, Nowak T. The proton transfer step catalyzed by yeast pyruvate kinase. J Biol Chem. 2003;278:12660–12671. doi: 10.1074/jbc.M300257200. [DOI] [PubMed] [Google Scholar]
- 49.Valiyaveetil FI, Zhou Y, MacKinnon R. Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry. 2002;41:10771–10777. doi: 10.1021/bi026215y. [DOI] [PubMed] [Google Scholar]
- 50.Martens C, et al. Lipids modulate the conformational dynamics of a secondary multidrug transporter. Nat Struct Mol Biol. 2016;23:744–751. doi: 10.1038/nsmb.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song Y, Mittendorf KF, Lu Z, Sanders CR. Impact of bilayer lipid composition on the structure and topology of the transmembrane amyloid precursor C99 protein. J Am Chem Soc. 2014;136:4093–4096. doi: 10.1021/ja4114374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poget SF, Cahill SM, Girvin ME. Isotropic bicelles stabilize the functional form of a small multidrug-resistance pump for NMR structural studies. J Am Chem Soc. 2007;129:2432–2433. doi: 10.1021/ja0679836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rivalta I, et al. Allosteric pathways in imidazole glycerol phosphate synthase. Proc Natl Acad Sci USA. 2012;109:E1428–E1436. doi: 10.1073/pnas.1120536109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Selvaratnam R, Chowdhury S, VanSchouwen B, Melacini G. Mapping allostery through the covariance analysis of NMR chemical shifts. Proc Natl Acad Sci USA. 2011;108:6133–6138. doi: 10.1073/pnas.1017311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baldwin AJ, Kay LE. NMR spectroscopy brings invisible protein states into focus. Nat Chem Biol. 2009;5:808–814. doi: 10.1038/nchembio.238. [DOI] [PubMed] [Google Scholar]
- 56.Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 57.Tzeng S-R, Kalodimos CG. Dynamic activation of an allosteric regulatory protein. Nature. 2009;462:368–372. doi: 10.1038/nature08560. [DOI] [PubMed] [Google Scholar]
- 58.LeVine MV, Cuendet MA, Khelashvili G, Weinstein H. Allosteric mechanisms of molecular machines at the membrane: Transport by sodium-coupled symporters. Chem Rev. 2016;116:6552–6587. doi: 10.1021/acs.chemrev.5b00627. [DOI] [PubMed] [Google Scholar]
- 59.Heginbotham L, Kolmakova-Partensky L, Miller C. Functional reconstitution of a prokaryotic K+ channel. J Gen Physiol. 1998;111:741–749. doi: 10.1085/jgp.111.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 61. Goddard TD, Kneller DG, SPARKY 3 (University of California, San Francisco)
- 62.Takegoshi K, Nakamura S, Terao T. 13C–1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344:631–637. [Google Scholar]
- 63.Raiford DS, Fisk CL, Becker ED. Calibration of methanol and ethylene glycol nuclear magnetic resonance thermometers. Anal Chem. 1979;51:2050–2051. [Google Scholar]