Significance
The CD6 T cell surface glycoprotein regulates T cell activation, and CD6 is a risk gene for autoimmune diseases including multiple sclerosis (MS). Moreover, recent work indicates that CD6 is an attractive target for the development of new therapeutic approaches to autoimmune diseases such as MS. The known ligand of CD6 is CD166 (also termed ALCAM), but CD6–CD166 interactions neither explain CD6-dependent interactions with stromal cell lineages that are critical in organ-targeted autoimmune diseases nor account for effects of CD6-targeted therapeutics in autoimmune diseases. This report definitively establishes CD318 as a second ligand of CD6 and provides evidence for the importance of CD6–CD318 interactions in autoimmune diseases that affect the central nervous system and the synovial lining of joints.
Keywords: CD6, ligand, CD318, autoimmunity, T cell
Abstract
It has been proposed that CD6, an important regulator of T cells, functions by interacting with its currently identified ligand, CD166, but studies performed during the treatment of autoimmune conditions suggest that the CD6–CD166 interaction might not account for important functions of CD6 in autoimmune diseases. The antigen recognized by mAb 3A11 has been proposed as a new CD6 ligand distinct from CD166, yet the identity of it is hitherto unknown. We have identified this CD6 ligand as CD318, a cell surface protein previously found to be present on various epithelial cells and many tumor cells. We found that, like CD6 knockout (KO) mice, CD318 KO mice are also protected in experimental autoimmune encephalomyelitis. In humans, we found that CD318 is highly expressed in synovial tissues and participates in CD6-dependent adhesion of T cells to synovial fibroblasts. In addition, soluble CD318 is chemoattractive to T cells and levels of soluble CD318 are selectively and significantly elevated in the synovial fluid from patients with rheumatoid arthritis and juvenile inflammatory arthritis. These results establish CD318 as a ligand of CD6 and a potential target for the diagnosis and treatment of autoimmune diseases such as multiple sclerosis and inflammatory arthritis.
CD6 is a marker of T cells and an important T-cell regulator (1). Recent genome-wide association studies also identified CD6 as a risk gene for multiple sclerosis (MS) (2–5), an autoimmune disease in which T cells play a vital role in the pathogenesis. CD6 is composed of three extracellular domains (domains 1, 2, and 3), and it functions by interacting with its ligand(s) (6). The domain 3 of CD6 has been shown to be the site that the identified CD6 ligand, CD166, also known as ALCAM (activated leukocyte cell adhesion molecule), binds to (7). However, anti-CD166 antibodies only partially blocked the binding of thymic epithelial cells to CD6-overexpressing COS cells, and mAbs blocking CD6–CD166 interactions do not abolish CD6 function (8, 9). Itolizumab, an anti-CD6 mAb developed in Cuba and approved in India for treating psoriasis, reduces pathogenic T-cell responses in patients with psoriasis, but this mAb binds to domain 1 of CD6 instead of domain 3, and it does not interfere with the CD6–CD166 interaction. Interestingly, UMCD6, a mouse anti-human CD6 mAb that we found highly effective in treating encephalomyelitis (EAE) in CD6 humanized mice, also fails to block the CD6–CD166 interaction. All these studies suggest the existence of an additional CD6 ligand, other than CD166, that binds to domain 1 of CD6, and could be critical for CD6 function in autoimmune conditions. Further studies using a CD6 fusion protein as a bait to pull down CD6-binding proteins from synovial fibroblast surface proteins showed the binding of three polypeptides (10). One of these polypeptides was identified as CD166, and the identities of the other two were unknown (11). A mAb termed 3A11 was developed, and the antigen recognized by this mAb was identified as the new ligand of CD6 that binds to its domain 1 (11, 12). However, attempts to identify the antigen recognized by mAb 3A11 were not previously successful.
CD318 (also known as CDCP1, TRASK, SIMA135, or gp140) is a cell-surface glycoprotein with an apparent molecular mass of ∼140 kDa (13–15). It consists of three extracellular CUB domains, a transmembrane domain, and an intracellular domain. CD318 can be proteolytically cleaved between the two distal CUB domains by certain serine proteases, resulting in different ratios of the ∼140-kDa intact molecule and the ∼80-kDa cleaved product on various cells. Cleaved CD318 is phosphorylated and activated by Src kinase, then the activated CD318 forms a complex with activated β1 integrin and activates FAK/PI3K/Akt motility signaling to promote early tumor dissemination (16). Under normal conditions, CD318 is present on many epithelial cells (17), some hematopoietic cells (18), and mesenchymal stem cells (19). CD318 is also present on many tumor cells (20). Up-regulation of CD318 expression is associated with a poor prognosis for many cancer patients (14, 21–25). Interestingly, a recent study using CD318 KO mice showed that two different oncogene-driven tumors grow much faster in CD318 KO mice than in wild-type (WT) control mice (26). Lack of CD318 in these mice potentially enhances tumor growth by liberating integrin signaling and growth factor receptor cross-talk in unanchored tumor cells (26). So far, all studies on CD318 have been limited to its direct signaling effect in tumor cells, and its possible role in regulating immune responses has never been examined.
In this report, using mass spectroscopy techniques, we identified the antigen recognized by mAb 3A11 as CD318. To validate the proteomics results, we probed proteins immunoprecipitated by mAb 3A11 using established anti-CD318 antibodies and probed recombinant CD318 protein by mAb 3A11 in Western blots. We also compared staining patterns of both mAb 3A11 and established anti-CD318 mAbs by using cells that are known to be positive or negative for CD318, and engineered cells with up-regulated or down-regulated levels of CD318. In addition, we confirmed the binding of CD318 to CD6 by using soluble CD6 protein as a bait in pull-down assays and by staining WT and CD166-deficient cells with the soluble CD6 followed by flow cytometric analyses. We immunized the CD318 KO mice to induce experimental autoimmune EAE and found that CD318 KO mice show ameliorated central nervous system (CNS) injury in association with reduced pathogenic T-cell responses and infiltration into the CNS. Finally, we examined CD318 expression on synovial fibroblasts from rheumatoid arthritis (RA) patients, measured levels of soluble CD318 in synovial fluids from arthritis patients, and studied its potential roles in recruitment and retention of T cells in synovial tissue. Our results showed that CD318 is the CD6 ligand recognized by mAb 3A11 and suggest that CD318 could be a target for the diagnosis and/or treatment of autoimmune diseases such as MS and inflammatory arthritis.
Results
Identification of the Antigen Recognized by mAb 3A11.
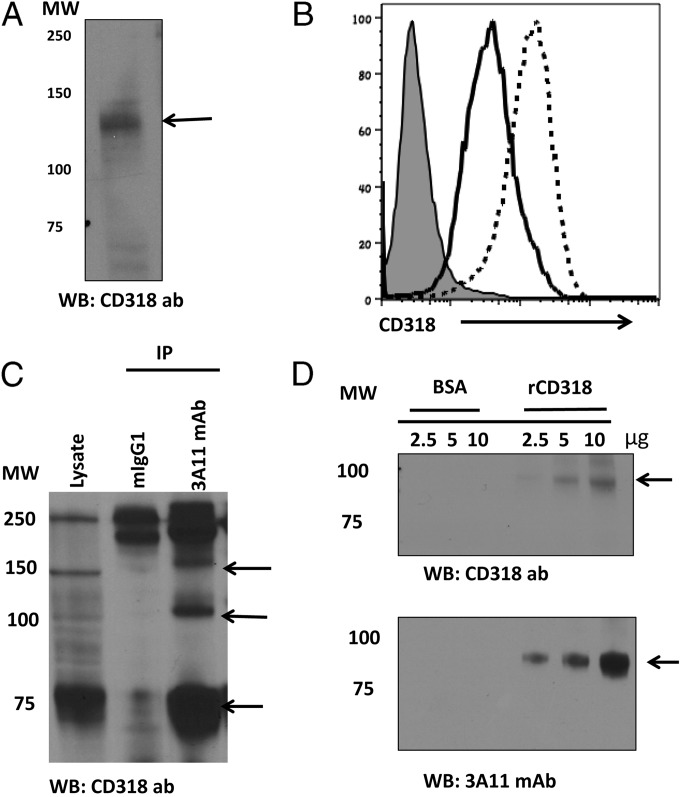
It had been previously established that mAb 3A11 recognizes an uncharacterized CD6 ligand that binds to domain 1 of CD6 (11), the same domain that Itolizumab, the anti-CD6 mAb approved for treating psoriasis in India (27) binds to, suggesting that this new CD6 ligand might have important roles in autoimmune conditions, distinct from the previously identified CD6 ligand, CD166. To determine the identity of the antigen recognized by mAb 3A11, we investigated HBL-100 cell surface proteins pulled down by this mAb by mass spectrum (MS) analysis. We found that CD318-related peptides were abundant in the mAb 3A11 precipitates (Table 1), indicating that the protein recognized by 3A11 could be CD318. We then probed whole HBL-100 cell lysate with an anti-CD318 Ab in Western blot and assessed CD318 expression levels on HBL-100 cells by flow cytometry before and after IFN-γ stimulation. We found that CD318 met the previously established characteristics of the potential mAb 3A11 antigen candidate (11) such as (i) it has a molecular mass of ∼130 kDa (Fig. 1A) and (ii) its expression can be up-regulated by IFN-γ stimulation (Fig. 1B).
Table 1.
Identification of CDCP1/CD318 protein by LC-MS/MS
| No. | Peptide | Mr(expt) | Mr(calc) | ppm | Score |
| 124–130 | TFIWDVK | 907.4825 | 907.4804 | 2 | 26 |
| 148–166 | QIGPGESCPDGVTHSISGR | 1,952.9027 | 1,952.9011 | 1 | 56 |
| 281–300 | VEYYIPGSTTNPEVFKLEDK | 2,328.1589 | 2,328.1525 | 3 | 22 |
| 344–353 | IYVVDLSNER | 1,206.6258 | 1,206.6244 | 1 | 40 |
| 354–362 | AMSLTIEPR | 1,016.5331 | 1,016.5324 | 1 | 27 |
| 397–406 | ISFLCDDLTR | 1,238.5976 | 1,238.5965 | 1 | 43 |
| 457–466 | LSLVLVPAQK | 1,066.6765 | 1,066.6750 | 1 | 57 |
| 519–530 | TFAPSFQQEASR | 1,367.6484 | 1,367.6470 | 1 | 58 |
| 561–566 | TPNWDR | 787.3624 | 787.3613 | 1 | 37 |
| 582–592 | DQVACLTFFK | 1,227.5974 | 1,227.5958 | 1 | 46 |
| 604–612 | AFMIIQEQR | 1,150.5824 | 1,150.5805 | 2 | 67 |
Fig. 1.
CD318 as the potential antigen recognized by mAb 3A11. (A) Probing the HBL-100 cell lysates with a commercial anti-CD318 Ab. Cell lysate were separated by SDS/PAGE and probed with a commercial anti-CD318 Ab in Western blot, showing a ∼135-kDa band (arrow). Data are representative of two independent experiments. (B) CD318 up-regulation in response to IFN-γ stimulation. HBL-100 cells were stimulated with human IFN-γ for 72 h, and CD318 expression levels were analyzed by flow cytometry following staining with a commercial anti-CD318 mAb (Clone CUB1). Thin shaded line, isotype control; thick line, without stimulation; dotted line, with stimulation. Data are representative of three independent experiments. (C) Probing the mAb 3A11 immunoprecipitates with the anti-CD318 Ab. HBL-100 cell lysates were immunoprecipitated with the same concentrations of mAb 3A11 or mIgG1 control, then the immunoprecipitates were separated by SDS/PAGE and probed with a commercial anti-CD318 Ab. The arrows indicate the full length and different isoforms of CD318 that immunoprecipitated. Data are representative of two independent experiments. (D) Three different concentrations (2.5, 5, 10 μg) of rCD318 or BSA control were separated by SDS/PAGE and probed with either mAb 3A11 (Lower) or a commercial anti-CD318 Ab (Upper).
Western Blots Using a Commercial Anti-CD318 Antibody and Recombinant CD318.
To validate the MS results, we performed Western blotting of the above immunoprecipitates by using a commercial anti-CD318 antibody (Pierce) and found that these antibodies detected three bands (Fig. 1C), including an ∼140-kDa band and an ∼80-kDa band, in the mAb 3A11 immunoprecipitates, but not the control IgG1 immunoprecipitates. In addition, we prepared recombinant soluble CD318 (rCD318) by synthesizing an artificial gene coding for the extracellular domains of CD318 with a C-terminal 6XHis-tag and cloning it into the expression vector pcDNA3.1. After transfecting the expression construct into 293 cells, we purified the rCD318 in the culture supernatants by nickel affinity chromatography following published protocols (13) and verified the protein by Western blot using an anti-His tag antibody. We then probed the rCD318 and the same amount of BSA with mAb 3A11 or an established anti-CD318 antibody in Western blots and found that both the mAb 3A11 and the anti-CD318 antibody selectively recognized rCD318 but not the BSA (Fig. 1D).
Flow Cytometric Analysis of Cells Normally Expressing or Not Expressing CD318 Using both mAb 3A11 and a Commercial Anti-CD318 mAb.
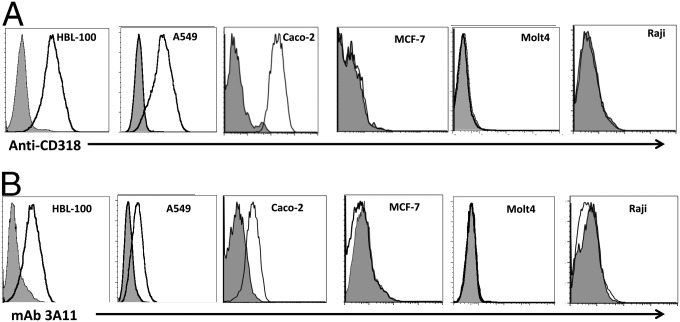
CD318 has been reported to be present on A549 (14), HBL-100 (28), and Caco2 (29) cells, but not on MCF-7 (30), Molt-4 (31), or Raji cells (14). We analyzed all of these cell lines with a commercial anti-CD318 mAb (Fig. 2A) or mAb 3A11 (Fig. 2B) and found highly similar staining patterns, suggesting that mAb 3A11 and the anti-CD318 mAb recognize the same antigen.
Fig. 2.
The anti-CD318 mAb and mAb 3A11 have an identical staining pattern on cells previously known to express or lack CD318 expression on the cell surface. Reported CD318-positive cells (HBL-100, A549, and Caco-2) and negative cells (MCF-7, Molt-4, and Raji) were stained either with the anti-CD318 mAb (A) or mAb 3A11 (B) and analyzed by flow cytometry. Data are representative of three independent experiments. Shaded histogram: isotype controls (mIgG2b for CD318 staining and mIgG1 for mAb 3A11 staining). Open histogram represents CD318 (A) or mAb 3A11 (B) staining.
Flow Cytometric Analysis of Engineered Cells with CD318 Overexpression or Down-Regulation.
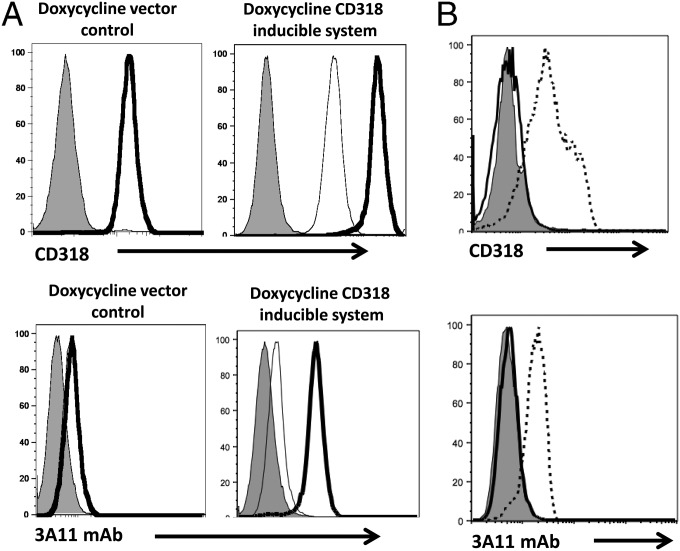
We could not entirely exclude the slight possibility in the above-described flow cytometry experiments that mAb 3A11 and the anti-CD318 mAb might recognize different antigens that happen to have the same expression pattern on the various examined cell lines. To address this issue, we studied the transfected MDA-468 cells that overexpress CD318 after doxycycline induction (13, 32) and the transfected MDA-468 cells knocked down for CD318 expression using shRNA (32) by flow cytometry using the commercial anti-CD318 mAb and mAb 3A11. We found that, consistent with previous reports (32), following doxycycline treatment, the expression of CD318 increases above basal levels in MDA-468 cells expressing the CD318 inducible system and not in cells expressing empty vector (control) (Fig. 3A). These assays also showed that staining of these cells expressing CD318 with mAb 3A11 resulted in exactly the same pattern as seen with the anti-CD318 mAb (Fig. 3A), whereas in CD318 knockdown cells, neither antibody showed detectable staining of the cell surface.
Fig. 3.
The anti-CD318 mAb and mAb 3A11 have an identical staining pattern on cells engineered to up-regulate or down-regulate CD318 expression. (A) The anti-CD318 mAb and mAb 3A11 staining on cells overexpressing CD318. MDA-468 cells transfected with vector alone (control) or a doxycycline-inducible CD318 expressing construct were incubated with doxycycline overnight, then stained either with CD318 Ab (Upper) or mAb 3A11 (Lower) and analyzed by flow cytometry. Shaded histograms, isotype controls; thin open histograms, basal expression level of anti-CD318 mAb/mAb 3A11 staining before doxycycline induction; thick open histograms, level of anti-CD318 mAb/mAb 3A11 staining after doxycycline induction. Data are representative of three independent experiments. (B) The anti-CD318 mAb and mAb 3A11 staining on cells with CD318 knocked down. MDA-468 WT and CD318 knockdown cells were stained with either CD318 mAb (Upper) or mAb 3A11 (Lower) and analyzed by flow cytometry. Thin lines, isotype controls; dashed lines, WT cells stained with the anti-CD318 mAb or mAb 3A11; thick lines, CD318 knocked down cells stained with the anti-CD318 mAb and mAb 3A11. Data are representative of three independent experiments.
CD6 Binding Analysis on Cells Expressing both CD166 and CD318, or CD318 Alone.
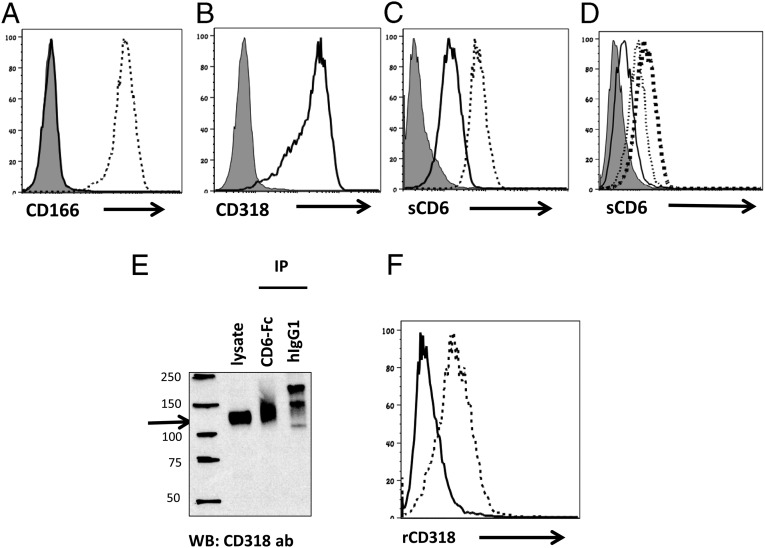
After confirming that CD318 is the protein recognized by mAb 3A11 in the above experiments, we tested whether CD318 binds to CD6, as suggested by previous studies. We have already shown that soluble CD6 protein can be used to stain cells expressing CD6 ligands in flow cytometric assays. The human fibrosarcoma cell line HT-1080 expresses both CD318 and CD166 (33, 34), and we generated an HT-1080 CD166 KO cell line by CRISPR/Cas9 technology to exclude the previously known CD6–CD166 interaction (Fig. 4A). We first confirmed that our HT-1080 CD166 KO cell line expresses CD318 but not CD166 (Fig. 4B), then stained the WT and CD166 KO cells with the soluble CD6 protein. We found that, in the absence of CD166, the binding of CD6 to the surface of these cells was significantly reduced but still evident (Fig. 4C), further evidence that CD6 has ligand(s) other than CD166. We then carried out a competitive binding assay by using our prepared soluble rCD318 protein and found that binding of CD6 to the CD166 KO cells was reduced by rCD318 in a dose-dependent manner (Fig. 4D). In addition, we incubated soluble CD6-Fc protein or the same amount of purified human IgG1 with the CD166 KO cell lysates and probed the CD6-precipitated proteins with a commercial anti-CD318 antibody in Western blot. We found that CD6-Fc protein, but not the control human IgG1 protein, pulled down a protein that was recognized by the anti-CD318 antibody (Fig. 4E). Finally, we stained transfected CHO cells expressing human CD6 on the surface and control CHO cells with the soluble rCD318 and found that rCD318 binds to human CD6-expressing CHO cells but not the control CHO cells (Fig. 4F). These results demonstrate that CD6 binds to CD318.
Fig. 4.
CD6 interacts with CD318. (A) HT1080 CD166 KO cells are deficient of CD166. The CD166 KO cells were analyzed for CD166 expression by flow cytometry. Thin line, isotype control; thick line, CD166 KO cells stained with an anti-CD166 mAb; thin dash line, WT HT1080 cells stained with the anti-CD166 mAb. Data are representative of five independent experiments. (B) HT1080 CD166 KO cells express CD318. The CD166 KO cells were analyzed for CD318 expression by flow cytometry using the anti-CD318 mAb. Thin line, isotype control; thick line, CD166 KO cells stained with the anti-CD318 mAb. Data are representative of five independent experiments. (C) CD6 binds to WT and CD166 KO cells. WT and CD166 KO cells were incubated with the same concentrations of human IgG1 (control) or recombinant human CD6-Fc fusion protein (1 µM), followed by detecting the cell surface-bound CD6 using an Alexa488-anti-human IgG Ab. Thin line, isotype control; thick line, CD166 KO cells stained with CD6; thin dashed line, WT cells stained with CD6. Data are representative of six experiments. (D) Binding of CD6 onto CD166 KO cells is competitively inhibited by soluble CD318. CD166 KO cells were incubated with recombinant human CD6-Fc fusion protein (1 µM) in the presence of different concentrations of recombinant soluble CD318 (0, 1, and 3 µM), then level of cell surface-bound CD6 was quantitated by flow cytometric analysis after staining the cells with an Alexa488-anti-human IgG Ab. Thin shaded line: isotype control; thick dash line, no rCD318; thin dash line, 1 µM CD318; thin line, 3 µM rCD318. The numbers in parentheses represent the molar concentrations of either rHCD6 or rCD318. Data are representative of six independent experiments. (E) CD6 immunoprecipitates CD318 from the CD166 KO cell lysates. CD166 KO cell lysates were immunoprecipitated with the same concentrations of either soluble CD6 or human IgG1, then the immunoprecipitates were separated by SDS/PAGE and probed with the anti-CD318 Ab, showing that CD6 selectively pulled down CD318 (arrow). Data are representative of eight independent experiments. (F) Soluble CD318 binds to CHO cells that express CD6 on the surface. Control CHO cells (solid line) and CHO cells expressing human CD6 (dotted line) were incubated with recombinant CD318. After washing, the binding of CD318 on the cell surface was assessed by flow cytometry after staining the cells with the anti-CD318 mAb.
CD318 KO Mice Are Protected in EAE.
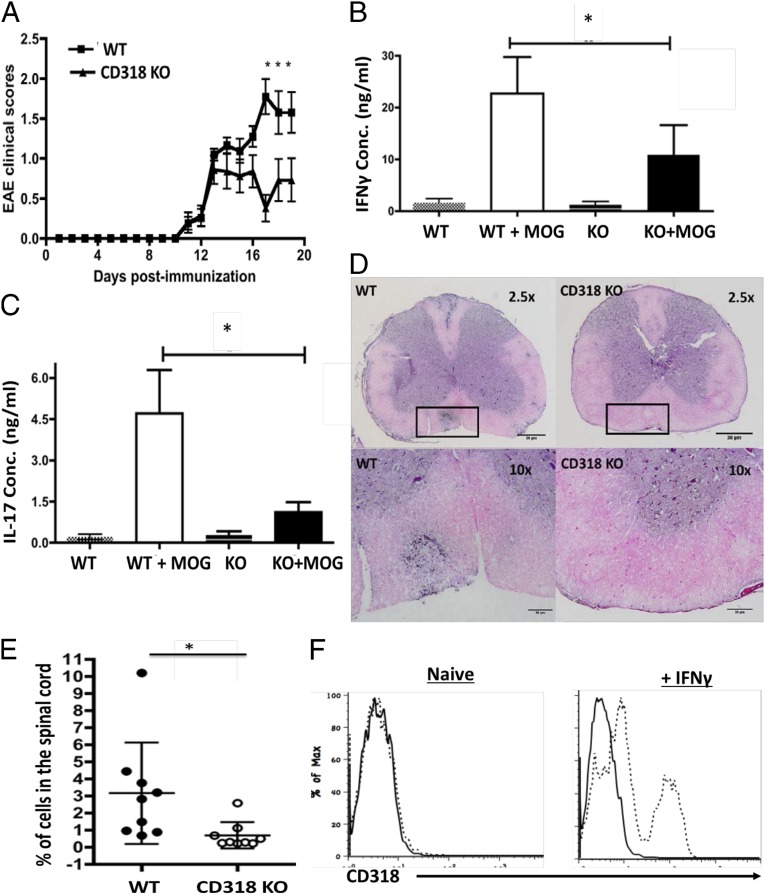
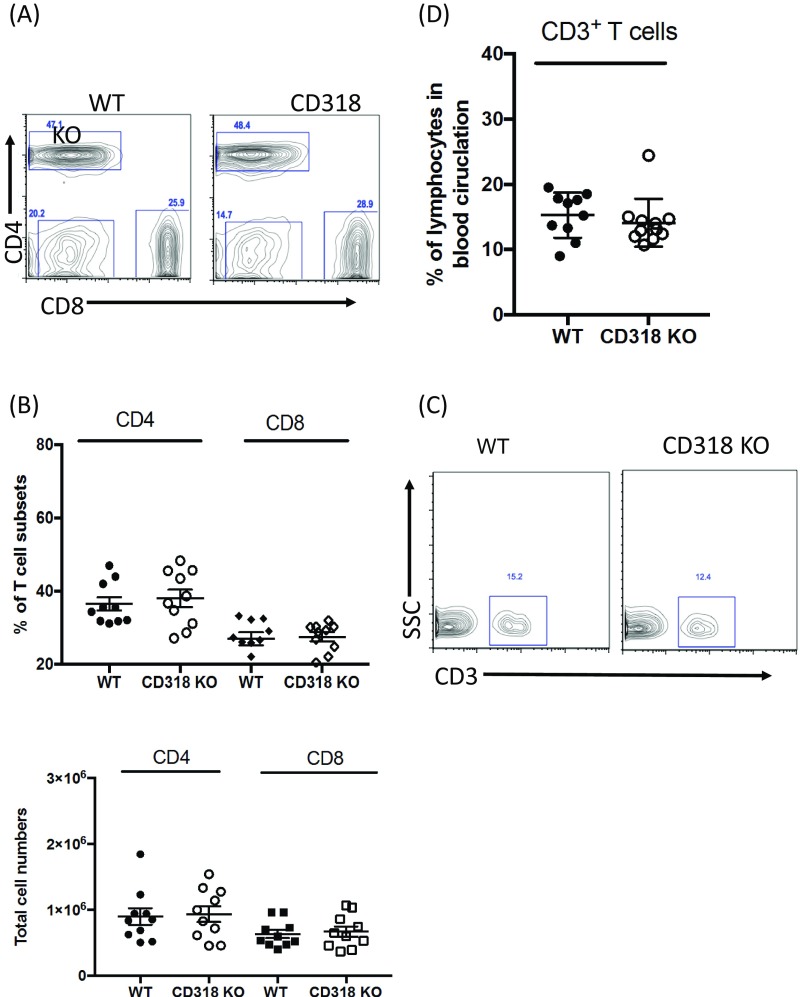
Although the potential role of CD318 in immune regulation has never been studied, our data showing that it is a ligand for CD6, a molecule that is important in the pathogenesis of EAE and MS (35), suggests that it might have a previously unknown immunoregulatory role and might regulate the development of EAE/MS. To test this hypothesis, we induced EAE in matched WT and CD318 KO mice by immunizing them with MOG35–55 peptide in Complete Freund’s Adjuvant (CFA) plus pertussis toxin and found that, like CD6 KO mice (35), CD318 KO mice had attenuated disease severity in EAE (Fig. 5A). These CD318 KO mice in EAE showed reduced MOG-specific Th1 (Fig. 5B) and Th17 (Fig. 5C) responses and had significantly decreased inflammation (Fig. 5D) and CD4+ T-cell infiltration (Fig. 5E) in the spinal cord. Experiments using isolated brain microvascular endothelial cells from WT mice showed that although CD318 was not detectable on these cells under constitutive conditions, after IFN-γ stimulation, CD318 expression on them was highly induced (Fig. 5F). These results reveal a previously unknown role for CD318 in immune regulation, providing further evidence that it is important for CD6 function in EAE. The CD318 KO mice had normal size and cell subset composition of their lymphoid organs (Fig. S1).
Fig. 5.
CD318 KO mice are protected in EAE. (A) WT and CD318 KO mice were immunized with MOG35–55 to induce EAE, and the development of EAE was monitored daily by clinical scoring. Combined results were from five individual experiments. WT, n = 21; CD318 KO, n = 25; data are mean ± SEM, *P < 0.05, (B and C). At the end of the experiments, splenocytes were collected and incubated with or without 10 μg/mL MOG35–55 peptide for 72 h. Levels of IFN-γ (B) and IL-17 (C) in the culture supernatants were measured by respective ELISA. Combined results were from three individual experiments, WT, n = 17; CD318 KO, n = 16; data are mean ± SEM, *P < 0.05. (D) Representative histology images of spinal cord sections from WT (Left) and CD318 KO (Right) mice in EAE, showing significantly reduced inflammation in the CD318 KO mouse spinal cords. Spinal cords were collected at the end of the experiment and processed for H&E staining. Squares in Upper show the areas that were amplified in Lower. (E) Flow cytometric analysis of infiltrated CD4+ T cells in spinal cord from WT and CD318 KO mice in EAE, showing significantly reduced CD4+ T-cell infiltration in the spinal cords of the CD318 KO mice in EAE. Spinal cords were collected at the end of the experiment, minced, and digested with collagenase. Single-cell suspension was prepared after Percoll centrifugation, stained with an anti-mouse CD4 mAb, and analyzed by a flow cytometer. (F) Mouse BMECs do not constitutively express CD318 but do after IFN-γ stimulation. Primary BMECs were isolated from WT mice, characterized, incubated without (Left) or with (Right) IFN-γ for 48 h, and stained with sheep anti-mouse CD318 IgG (dotted line) or control IgG (solid line), then analyzed on a flow cytometer.
Fig. S1.
Intact T-cell compartment in the lymph node and peripheral blood of CD318 KO mice. (A) A representative frequency of CD4+ and CD8+ T cells in WT and CD318 KO inguinal lymph nodes. Data are representative of 10 mice. (B) The summary of the frequency and total cell numbers of CD4+ and CD8+ T cells in WT and CD318 KO inguinal lymph node. n = 10 in each group. (C) A representative frequency of CD3+ T cells in WT and CD318 KO peripheral blood. (D) The summary of frequency of CD3+ T cells in the peripheral blood of WT and CD318 KO mice. n = 10 in each group.
Measurement of CD318 Levels in Synovial Tissues from RA and Osteoarthritis Patients by ELISA.
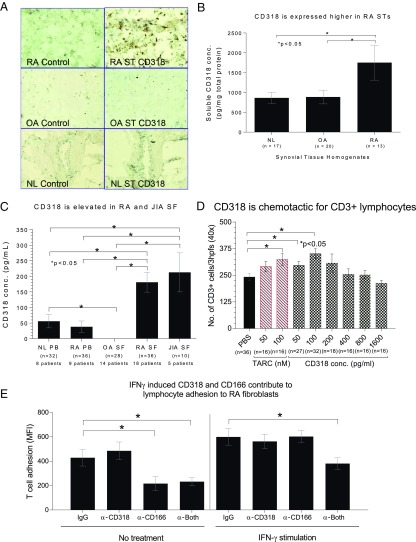
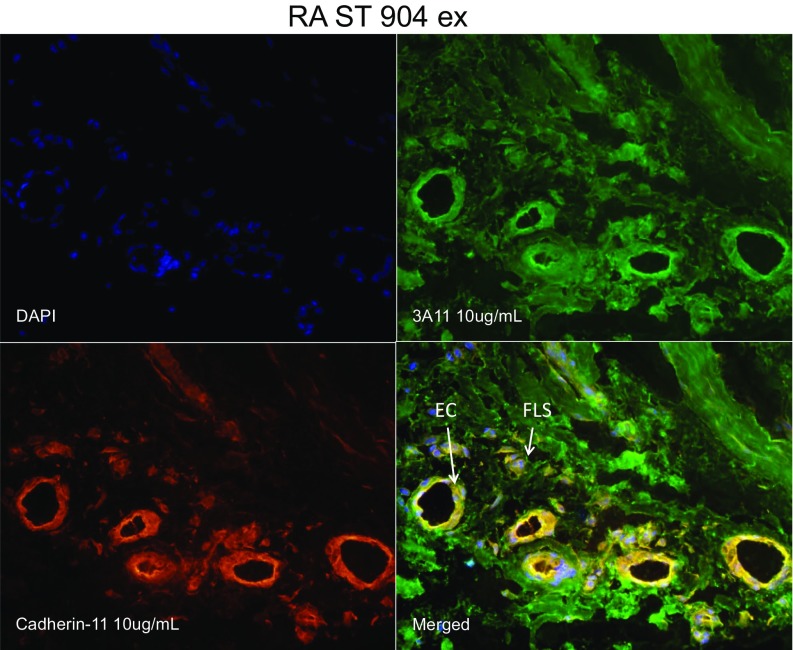
We have established previously that the antigen recognized by the mAb 3A11 (now shown to be CD318) is highly expressed in synovial fibroblasts from RA patients after IFN-γ stimulation. To explore a potential role for CD318 in the pathogenesis of arthritis, we first carried out immunohistochemistry (IHC) staining for CD318 in synovial tissue sections of RA, osteoarthritis (OA), and nonrelevant controls. We found that CD318 is more strongly expressed in RA synovial tissues (Fig. 6A). Two-color immunofluorescence microscopy indicated that CD318 is expressed on both fibroblasts and endothelial cells in synovial tissue (Fig. S2). We then homogenized synovial tissues from patients without or with RA or OA and measured levels of total CD318 in them by ELISA. We found that CD318 was detectable in lysates from all of the synovial tissues examined. Whereas levels of CD318 were comparable between the control groups without arthritis and with OA, levels of CD318 in synovial tissues from RA patients were significantly higher (Fig. 6B).
Fig. 6.
CD318 is a potential biomarker for inflammatory arthritis and chemotactic for T cells. (A) CD318 is highly expressed in synovial tissues from RA patients. Synovial tissue sections from RA, OA, and nonrelevant controls (NL) were stained with either mAb 3A11 (mouse anti-human CD318) or the same amount of control IgGs, then slides were examined under a microscope. (B) Levels of total CD318 are elevated in synovial tissues from RA patients. Synovial tissue from patients with RA (n = 13), OA (n = 20), and normal synovial tissues (Ctrl, n = 17) were homogenized, and levels of total CD318 were analyzed by ELISA. (C) Levels of soluble CD318 are significantly higher in synovial fluids from patients with RA (n = 36) or JIA (n = 10) than in those from patients with OA (n = 28). Sr, serum; SF, synovial fluid. (D) Soluble CD318 is chemotactic to T cells. T-cell migration toward various concentrations of CD318 (50, 100, 200, 400, 800, and 1,600 pg/mL) was assessed in Boyden chemotaxis chambers. PBS was the negative control, and TARC was the positive control. Readings represent the number of cells migrating through the membrane (the sum of three high power 40x fields per well, averaged for each quadruplicate well). (E) T-cell adhesion to IFN-γ–stimulated synovial fibroblasts in the presence of mouse IgG (control), anti-CD318, anti-CD166, or both was measured by a Synergy plate reader.
Fig. S2.
Immunofluorescence microscopy of synovial tissue. To further define the cellular CD318 expression in RA and OA synovium, synovial tissue (ST) slides were fixed with cold acetone for 20 min and blocked with 20% FBS and 5% goat serum for 1 h at 37 °C. Rabbit anti-human cadherin-11 antibody (GeneTex) and mouse anti-human CD318 antibody (3A11 antibody made in house) were used at 10 µg/mL for 1-h incubation at 37 °C. Nonspecific rabbit (Invitrogen) and mouse IgG (BD Biosciences) were used as respective antibody controls. STs were then washed and incubated for an additional hour with 10 µg/mL fluorescently conjugated goat anti-mouse/rabbit secondary antibodies [Alexa Fluor 555 (PE) for cadherin-11 binding; and 488 (FITC) for CD318 binding (Invitrogen)]. For nuclear staining, DAPI was used at 1:5,000 diluted in PBS. Images were taken at 400× magnification. Anti-CD318 positive synoviocytes are shown by fluorescent green staining, and cadherin-11 staining is shown by fluorescent red. Yellow cells are a result of a merger of the green and red fields showing dual fluorescence of cadherin-11 and CD318 expression on the same cells within the ST.
Measurement of Soluble CD318 Levels in Sera and Synovial Fluids from RA and OA Patients by ELISA.
Soluble CD318 has been found in cancer cell culture supernatants and in urine samples from men with high-risk prostate cancer (13). We first measured levels of soluble CD318 in sera from RA patients and healthy donors and found that levels of soluble CD318 in the sera were low, barely above the sensitivity limit of the ELISA that we used (Fig. 6C). We also examined synovial fluids from patients with RA, juvenile inflammatory arthritis (JIA), and OA by the same ELISA, and found that levels of soluble CD318 were significantly higher in synovial fluids than in serum and that levels of soluble CD318 were significantly higher in synovial fluids from both RA and JIA patients than in those from the OA patients (CD318 was not detectable in synovial fluids from OA patients) (Fig. 6C). These data suggest that soluble CD318 is produced within the joints, especially in RA and JIA.
T-Cell Chemotaxis in Response to Soluble CD318.
The gradient between serum and synovial fluid levels of soluble CD318, and the elevated levels of CD318 in synovial fluids from patients with RA and JIA but not in OA, led us to assess the possibility that soluble CD318 might be chemotactic for T lymphocytes. Using a modified Boyden chamber assay, we found that peripheral blood T cells migrated in response to soluble CD318 as a single stimulus, with a peak response at a concentration that approximated the difference between the mean RA serum and RA synovial fluid soluble CD318 levels (Fig. 6D).
Contributions of CD6 Ligands to Adhesion Between T Cells and Synovial Fibroblasts.
Previous work has suggested that a CD6 ligand other than CD166 could contribute to adhesion between human T lymphocytes and various IFN-γ–treated nonhematopoietic cell types (10, 11, 36). To evaluate the roles of CD166 and CD318 in interactions between T cells and synovial fibroblasts, we performed adhesion assays by using fluorescently tagged T cells and synovial fibroblasts that were or were not precultured with IFN-γ. In these assays, we found that CD6 ligands were significantly involved in the adhesion of T cells to synovial fibroblasts (Fig. 6E). Without IFN-γ pretreatment of the synovial fibroblasts, only CD166 was functionally important in these adhesion assays, consistent with the minimal expression of CD318 on these cells. Interestingly, when IFN-γ–treated synovial fibroblasts were used, both ligands were functional, and adhesion was substantially interrupted only when both were simultaneously masked with monoclonal antibodies. These results are consistent with important functional roles for both CD6 ligands in synovial tissue in vivo.
Discussion
In this report, using MS analysis, we identified abundant CD318-derived peptides in mAb 3A11-immunoprecipiated proteins. CD318 also met the previously published criteria of a 3A11 antigen, suggesting that CD318 is the antigen recognized by mAb 3A11. We confirmed the MS results by probing the mAb 3A11-immunoprecipiated proteins with a commercial anti-CD318 antibody, and by producing and probing recombinant CD318 with mAb 3A11 in Western blots. In addition, we found that mAb 3A11 and the established CD318 mAb have the same staining patterns on cells known to naturally express or lack CD318, and on cells engineered to overexpress or knockdown CD318 expression. Using soluble CD6, soluble CD318, cells that express both CD166 and CD318 or CD318 alone, and transfected CHO cells expressing CD6 in flow cytometric analyses and pull-down experiments, we confirmed that CD318 binds to CD6. Finally, we found that CD318 is abundant in synovial tissues and whereas levels of total CD318 are similar in synovial tissues from controls and OA patients, they are significantly elevated in RA patients. Moreover, a soluble form of CD318 can be readily detected in synovial fluids from patients with inflammatory (RA, JIA) but not noninflammatory (OA) arthritis, and these levels are appreciably higher than those in the serum. Of particular interest, both the membrane-bound and soluble forms of CD318 are functionally active in vitro, in adhesion and chemotaxis assays, respectively, that are arguably relevant to components of the pathogenesis on joint inflammation in vivo.
The distinct patterns of expression of CD318 compared with CD166 are striking—whereas CD166 is expressed widely on a broad range of hematopoietic and nonhematopoietic cells, including activated T lymphocytes, CD318 expression appears to be confined to nonhematopoietic lineages, such as fibroblasts, keratinocytes, epithelial cells, and a variety of neoplastic cells (10, 11, 34, 36). The one possible exception to the lack of CD318 expression on hematopoietic cells is a subset of cord blood hematopoietic progenitors (37). Thus, engagement of CD6 by CD318 is an unusual example of a ligand–receptor interaction between a lymphocyte-specific cell surface glycoprotein that can participate in T-cell activation (CD6) and a molecule (CD318) that is found only on cells that are traditionally considered not to be components of the immune system. This interaction points to an ability of T cells to specifically receive and recognize distinct signals from “nonimmune system” tissue cells that may be important in organ-targeted autoimmune diseases, such as synovial fibroblasts and keratinocytes.
The functional roles of CD318 versus CD166 must be in part distinct given the marked differences in the distribution of these molecules. With the identification of CD318 as a second ligand of CD6, exploration of the consequences of its binding to CD6 is still at an early stage. Membrane-anchored CD318 appears to be a mediator of T-cell adhesion to tissue cells, such as synovial cells and keratinocytes, and our data suggest that under some conditions, both CD6 ligands, CD166 and CD318, can cooperatively participate in adhesion to T cells. These two CD6 ligands appear to recognize distinct epitopes of CD6, raising the possibility that CD6 can be engaged simultaneously by both of its ligands to form a trimolecular complex. Structural demonstration of such a complex during interactions between intact cells should be an important goal for future experiments. Early studies of CD6 identified distinct immunologic epitopes that appeared to mediate different functional effects on T cells (38), and more recent work has localized the binding domains on CD6 and CD166 that are involved in the CD6–CD166 interaction (39).
CD6 was identified as a risk gene for MS (2–5), and we recently showed that CD6 KO mice are protected from EAE and that treating mice with an anti-CD6 mAb ameliorates CNS injury in EAE (35), suggesting that CD6 could be a new target for treating MS. Interestingly, although treating WT mice with a mAb against CD166, the first identified ligand of CD6, ameliorates EAE (40), CD166 KO mice show exacerbated EAE potentially due to reduced expression of blood–brain barrier (BBB) junctional proteins, leading to increased permeability of CNS blood vessels in the CD166 KO mice (41). Different from the CD166 KO mice, we found that CD318 KO mice were protected in EAE, in association with reduced MOG-specific T-cell responses and decreased CNS T-cell infiltration, suggesting that CD318 is an important ligand for CD6. We also isolated primary brain microvascular endothelial cells (BMECs) from WT mice and examined CD318 expression on this critical component of the BBB by flow cytometry. We found that in contrast to CD166, which is highly expressed on the mouse BMECs (40, 41), CD318 is not constitutively expressed on these cells at all (Fig. 5F). Whether anti-CD318 antibodies could still allow full binding of CD166 to CD6, (and vice versa), and whether binding of either ligand is affected by disease-related CD6 polymorphisms or splice variants, are among the interesting issues for further investigation.
The signal transduction pathways that are activated by CD318 binding to CD6, with or without concurrent binding of CD166 to CD6, remain to be elucidated, both downstream of CD6 in the T lymphocyte and downstream of CD318 and CD166 in non-T cells that express these ligands. Signal transduction events in synovial fibroblasts generated by cell-cell adhesion events that involve CD6 and CD318 are likely to be distinct from pathways activated by antiadhesive effects of the intracellular domain of CD318 that have been described in cancer cells (42). In cancer cells, CD318 is phosphorylated and can enhance downstream phosphorylation events, in part linked to signaling through the epidermal growth factor receptor (43, 44). Moreover, cleavage of full-length 135-kDa CD318 by serine proteases creates a smaller membrane-retained 70-kDa form that either associates with membrane integrins or homodimerizes, and generates downstream signaling events that enhance cancer cell invasiveness and metastasis (16, 44). Cleavage of CD318 can be blocked by dexamethasone (45). Like cancer cells, RA synovial fibroblasts are locally invasive and are stimulated by a variety of growth factors. Moreover, intraarticular injection of corticosteroid ameliorates joint inflammation. It will be important to elucidate the potential roles of CD318 in migration and invasion of synovial fibroblasts and to assess the effect of engagement of CD318 by CD6 on these processes.
On synovial fibroblasts and keratinocytes, CD318 is up-regulated by IFN-γ. However, on cancer cells, its expression is increased following engagement of the epidermal growth factor (EGF) receptor by EGF (46). In cancer, CD318 is also up-regulated by hypoxia-inducible factor 2α, which is also potentially relevant to CD318 expression on synovial fibroblasts because inflamed synovium is a hypoxic environment. Methylation of sites near CD318 has been proposed as a critical element of epigenetic control of its expression. In bone marrow stromal cells, reciprocal CD146+CD318− and CD146−CD318+ subsets of marrow fibroblasts have been identified that have distinct patterns of gene expression (47); whether this finding is also true in synovium or other tissues is as yet unknown.
The elevated levels of soluble CD318 in inflamed synovial tissue and fluid (RA and JIA) raise questions regarding its function in joint inflammation. Our data indicate that soluble CD318 is chemotactic for T cells, which are not present in normal synovial tissue, but which accumulate in large numbers in RA and JIA synovium through mechanisms that are as yet not fully defined. Importantly, the concentration at which soluble CD318 is chemotactic corresponds to the in vivo concentration gradient between RA serum and RA synovial fluid, indicating that this in vitro assay is likely to be physiologically relevant. Whether soluble CD318 is derived by protease-mediated shedding from the synovial fibroblast surface or by secretion of soluble CD318 from the synovial fibroblasts is as yet unknown. The chemotactic effects of soluble CD318 resemble in some respects chemotactic properties of CD13, another membrane protein on synovial fibroblasts that also is present at high concentrations as a soluble molecule in inflammatory joint fluid (48). Neither CD13 nor CD318 show structural resemblance to conventional chemokines, but there is evidence that CD13, like classical chemokines, signals through a G protein-coupled receptor (48).
Although biologic therapeutics have led to important improvements in the treatment of RA and JIA, these agents impair host defenses to various pathogens and do not selectively target molecular interactions that are more important in pathogenic autoimmunity compared with normal immune responses. Identification of CD318 as a ligand of CD6 creates a potential therapeutic target at the level of the T-cell/synovial fibroblast interaction that is not relevant to T-cell interactions with professional antigen-presenting cells in lymphoid organs.
CD318 has been proposed as a novel molecular target for treatment of malignant neoplasms (30, 49, 50); the realization that it is engaged by CD6 will create a perspective from which to assess such possibilities. An anti-CD6 monoclonal antibody has been administered to 12 patients with multiple sclerosis, with insufficient clinical data from this series to assess efficacy (51). Our recent (35) and current data could prompt further evaluation of this approach to treating multiple sclerosis. Moreover, our data could also prompt consideration of CD318 as a therapeutic target in autoimmune diseases.
Materials and Methods
Animals.
Wild-type (WT) and CD318 KO mice (C57BL/6 background) were ordered from Jackson Laboratory and maintained under pathogen-free conditions in the animal facility of Lerner Research Institute, Cleveland Clinic.
Cell Culture.
The HBL-100, Raji, A549, Molt4, and MCF, wild type (WT) HT-1080, and CD166 knockout (KO) cell lines were cultured in RPMI 1640 supplemented with 10% FBS, l-glutamine, penicillin/streptomycin, and Na-pyruvate. WT MDA-468 and CD318 knockdown cell lines and transfected CHO cells expressing human CD6 on their surface were cultured in DMEM supplemented with 10% FBS, l-glutamine, penicillin/streptomycin, Na-pyruvate, and 300 μg/mL G418. MDA-468 expressing empty vector or doxycycline-inducible CD318 was also cultured in the same media described above with Zeocin in place of G418. Caco-2 cells were also cultured in the same media described in the absence of selection pressure. MDA-468 expressing vector control and doxycycline-inducible CDCP1 were stimulated with 100 ng/mL doxycycline overnight (32).
CD166 Knockout Cell Line Development.
CD166 was knocked out in the HT-1080 cells by using CRISPR/Cas 9 technology. In brief, RNA (AGACGGTGGCGGAGATCAAG, Horizon Discovery) was transfected into cells by lipofection. Transfection efficiency more than 20% is considered successful. Efficiency was monitored by the surrogate marker, GFP. Cells lacking cell-surface CD166 were selected by fluorescence-activated cell sorting using an antibody against the extracellular domain of CD166 (R&D Systems; MAB656). Cell population was further purified by single-cell colony formation in soft agar. After identification of pure, CD166-negative clonal populations, five clones were selected at random and mixed to create the CD166-KO cell line.
Antibodies.
3A11 mAb was developed and characterized (10, 11). mAbs against human CD166, human CD318 (clone: CUB1), and mouse IgG2b isotype control were all obtained from Biolegend. The polyclonal anti-CD318 antibody was obtained from Thermo Scientific. Recombinant human CD6 was obtained from R&D. Recombinant mouse CD6 was described (10). Purified human IgG1 was obtained from Sigma-Aldrich. Alexa 488-conjugated donkey anti-mouse IgG and Alexa 488-conjugated donkey anti-human IgG were both obtained from ImmunoResearch. Alexa 488-conjugated polyclonal anti-human CD6 was obtained from R&D. FITC-conjugated mouse IgG isotype control was obtained from Biolegend.
Immunoprecipitations.
For 3A11 mAb IP, HBL-100 breast carcinoma cells were biotinylated by using E-Z link sulfo-NHS-LC biotin and subsequently lysed in Nonidet P-40 lysis buffer (Invitrogen) containing 0.1% SDS (Fisher Scientific), 0.1% deoxycholic acid (Sigma-Aldrich), and one cOmplete tablet of protease inhibitor (Roche) on ice for 30 min. Immunoprecipitation was performed overnight at 4 °C by using either mouse IgG1 or 3A11 mAb. Antigen–antibody complexes were pulled down by using protein (A+G). Following antigen-recombinant protein complex pull down, the samples were boiled for 5 min in 2× Laemmli sample buffer (Bio-Rad). For Western Blot, samples were loaded onto an SDS/PAGE and following electrophoresis, proteins were transferred to a polyvinylidene difluoride (PVDF) membrane for Western blotting. The membrane was incubated for 1 h at room temperature with blocking buffer containing 5% BSA and 0.05% Tween 20. Following blocking, the protein was detected by using streptavidin-HRP conjugate and visualized by using the chemiluminescent substrate ECL (Amersham Biosciences). In some experiments, HBL-100 carcinoma cells were not biotinylated before the preparation of cell lysates for immunoprecipitation with either mouse IgG1 or 3A11 mAb as described above. Following blocking of the PVDF membrane, the membrane was incubated with CDCP1 (Thermo Scientific) primary antibody and followed by HRP-conjugated goat anti-rabbit (Southern Biotech). The protein was visualized by using ECL Western Blot detection reagent (Amersham Biosciences).
Recombinant Soluble CD6 IP.
HT1080 CD166-KO cells were lysed in 0.5% Nonidet P-40 (Roche) lysis buffer on ice for 30 min. Lysates were immunoprecipitated with either purified human IgG1 or recombinant mouse CD6 overnight at overnight at 4 °C. Protein A/G-agarose beads was then added to the samples for 3 h. Following antigen-recombinant protein complex pull down, the samples were boiled for 5 min in 2× Lamini sample buffer (Bio-Rad). For Western blot, samples were loaded onto an SDS/PAGE and following electrophoresis, proteins were transferred to a PVDF membrane for Western blotting. Following blocking of the PVDF membrane, the membrane was incubated with an anti-CDCP1 (Thermo Scientific) primary antibody and followed by HRP-conjugated goat anti-rabbit IgGs (Southern Biotech).
In-Gel Digestions and Mass Spectrometry.
Proteins immunoprecipitated by mAb 3A11 were separated by SDS/PAGE. Bands at the appropriate size were excised and destained with 50% acetonitrile in 100 mM ammonium bicarbonate followed by 100% acetonitrile. Cysteine residues were first reduced by incubating the sample with 20 mM DTT at room temperature for 60 min, and then alkylated with 50 mM iodoacetamide for 30 min in the dark. The gel pieces were washed with 100 mM ammonium bicarbonate, dehydrated in acetonitrile, dried in a SpeedVac centrifuge, and then rehydrated in 50 mM ammonium bicarbonate containing sequencing grade modified trypsin for overnight digestion at 37 °C. The resulting proteolytic peptides were extracted from the gel with 50% acetonitrile in 5% formic acid, dried, and reconstituted in 0.1% formic acid for liquid chromatography (LC)-MS/MS analysis.
The digests were analyzed by LC-MS/MS using Orbitrap Elite Hybrid Mass Spectrometer (Thermo Electron), equipped with a Waters nanoAcquity UPLC system. The spectra were acquired in the positive ionization mode by data-dependent methods consisting of a full MS scan at 120,000 resolution and MS/MS scans of the 20 most abundant precursor ions in ion traps by collision-induced dissociation at normalized collision energy of 35%. A dynamic exclusion function was applied with a repeat count of 2, repeat duration of 30 s, exclusion duration of 45 s, and exclusion size list of 500. The obtained data were submitted for a database search by using Mascot Daemon (Matrix Science). Carbamidomethylation of Cys was set as a fixed modification, whereas oxidation of Met was selected as variable modifications. The mass tolerance was set as 10 ppm for precursor ions and 0.8 Da for productions. SwissProt (July 2014) database (546,000 sequences; 194,259,968 residues) was used for searching against the taxonomy of human (20,210 sequences). The significance threshold P value was set to <0.05. Proteins hits with at least two unique peptides at Mascot score >20 were considered to be identified.
Flow Cytometric Staining.
For CD318, CD166, and 3A11 mAb cell surface staining, cells were stained with anti-human CD318, anti-human CD166, and 3A11 mAb, respectively, on ice for 30 min. Following 3A11 mAb cell surface staining cells, these cells were washed and subsequently stained with the secondary antibody Alexa 488-conjugated donkey anti-mouse IgG and analyzed by flow cytometry. For soluble CD6 cell surface staining, HT1080 CD166-KO cells were incubated with 1 μM recombinant human CD6-Ig or human IgG1 at 4 °C for 45 min. After 45 min, the cells were washed and subsequently stained with Alexa 488-conjugated donkey anti-human IgG at 4 °C for 30 min, washed, and analyzed by flow cytometry. In some experiments, HBL-100 cells or synovial fibroblasts were stimulated with 1,000 U/mL human IFN-γ for 72 h before analyzing the expression of CD318 by flow cytometry.
For rCD318 binding to control or human CD6-expressing CHO cells, these cells were incubated with rCD318 at 4 °C for 45 min. After 45 min, the cells were washed and subsequently stained with PE-conjugated mAb against human CD318 at 4 °C for 30 min, washed, and analyzed by flow cytometry.
Production of Recombinant CD318 Extracellular Domains and Western Blots.
Gene sequence encoding for the extracellular domains of CD318 with a C-terminal 6XHis-tag was synthesized (Genscript) and cloned into the expression vector pcDNA3.1. After transfection of the expression construct into 293 cells for transient expression, recombinant CD318 in the culture supernatant was purified by nickel affinity chromatography following published protocols. For Western blots, the same amount of either recombinant CD318 or BSA was separated by SDS/PAGE and transferred to a PVDF membrane, then probed with either the 3A11 mAb or a rabbit anti-human CD318 antibody, followed by either rat anti-mouse HRP conjugate or goat anti-rabbit HRP conjugate, respectively. Protein bands were visualized by using the chemiluminescent substrate ECL.
Induction of EAE.
EAE was induced by active immunization, and disease severity was assessed by assigning clinical scores following published protocols (52, 53). In brief, 8- to 10-wk-old female mice were immunized at the base of the tail and in both thighs with 200 μg of mouse MOG35–55 peptide (custom synthesized by GenScript USA) emulsified in CFA (Difco Laboratories) that had been supplemented with Mycobacterium tuberculosis strain H37Ra to 4 mg/mL. Pertussis toxin (0.2 µg; List Biologic Laboratories) was injected i.p. immediately after immunization and the following day. Clinical severity was assessed daily with a 0–5 scoring system (0, no signs; 1, flaccid tail; 2, impaired righting reflex and/or gait; 3, partial hind limb paralysis; 4, total hind limb paralysis; 5, moribund or dead).
Histological Analysis of Spinal Cords.
Following killing of the mice, spinal cords were removed and fixed in 10% formalin in PBS buffer for 24 h, then embedded in paraffin. Sections were cut at 5 μm on a microtome and stained with hematoxylin and eosin (H&E) to assess CNS inflammatory infiltrates by following established protocols.
Analysis of CD4+ T-Cell Infiltration in the Spinal Cord of EAE Mice.
At the end of EAE experiments, mice were perfused through the left ventricle by using 100 mL of HBSS, then spinal cords were dissected, washed by HBSS, and digested with 10 mg/mL collagenase d (Roche) at 37 °C for 45 min with shaking every 15 min. Cells released from the digestion were passed through a 70-µm cell strainer and separated by using 38% Percoll. After this process, single-cell suspension was prepared and stained with an anti-CD4 mAb (Biolegend) followed by flow cytometric analysis.
Th1 and Th17 Recall Assays.
Splenocytes were collected immediately following killing of the mice. After lysing the red blood cells, 0.4 × 106 splenocytes were incubated with or without 10 µg/mL of the MOG35–55 peptide in 100 µL of RPMI medium with 10% FBS in each well of a 96-well plate. After 72 h, IFN-γ and IL-17a levels in the culture supernatants were measured by respective ELISA (Biolegend) following manufacturer-provided protocols.
Analysis of CD318 Expression on BMECs.
Mouse BMEC were isolated by following a published protocol (54). In brief, mouse brains were isolated, and the brainstems, cerebella, thalami, and meninges were removed under a dissecting microscope. The remaining tissue was minced and digested by 5 mg/mL collagenase CLS2 (Worthington Biochemical) in DMEM for 1 h at 37 °C, then washed with 20% BSA-DMEM and centrifuged at 1,000 × g for 20 min at 4 °C. The pellet was resuspended in 1 mg/mL collagenase/dispase (Worthington Biochemical) and incubated for another 1 h at 37 °C. After the final washing, the resultant cells were cultured in endothelial cell medium (PeproTech). The isolated BMEC purity was determined by flow cytometric analysis after staining the cells with an anti-CD34 mAb (Biolegend). For CD318 expression detection, the cells were stimulated with or without 1,000 U/mL of IFN-γ, then incubated with 10 μg/mL sheep anti-mouse IgG (R&D Systems) or sheep IgGs, then analyzed by a flow cytometer.
Synovial Tissue and Synovial Fluid Specimens.
Synovial tissue specimens were obtained from patients with RA (n = 13) and OA (n = 20) at the time of arthroplasty. Normal synovial tissues were obtained from cadavers (n = 17). A portion of each tissue was homogenized for ELISAs. Synovial fluids were obtained at therapeutic arthrocentesis from patients with RA (n = 36), OA (n = 28), and JIA(n = 10). In all cases, synovial tissue and fluid specimens were excess materials obtained at procedures performed for clinical indications.
IHC Staining.
Human ST sections were fixed in cold acetone and then treated with 3% peroxidase in 0.1 M Tris. Tissues were blocked with 3% horse serum. The sections were incubated with the 3A11 mAb (mouse anti-human CD318 IgG) or purified mouse IgG (MyBioSource) for an additional hour. Then, a 1:100 dilution of horse anti-mouse biotinylated secondary antibody (Vector) was added to the tissue sections and incubated at room temperature. Then VECTASTAIN Elite ABC HRP Kit (Vector) was added at a 1:10,000 dilution. Finally, diaminobenzidine tetrahydrochloride substrate (Vector) was added to the sections. The sections were then counterstained with Harris’ hematoxylin and dipped in saturated lithium carbonate solution for bluing and then mounted with coverslips.
ELISA of Soluble CD318.
Levels of soluble CD318 in sera and synovial fluids were measured by an ELISA kit (R&D Systems) per the manufacturer’s protocol.
T-Cell Chemotaxis Assay.
CD3+ T-cells were isolated by using a RosetteSep lymphocyte isolation kit (Stem Cell Technologies) and instructions were followed per the manufacturer’s protocol. Cytospin examination was performed to ensure that the isolated cells were lymphocytes.
Chemotaxis membranes were coated with type IV collagen (Sigma-Aldrich) used at 1 μg/mL overnight and dried. Forty microliters of varying concentrations of CD318 (50, 100, 200, 400, 800, and 1,600 pg/mL) were added to wells in the bottom of the chamber. T cells (10 × 106 cells per mL in PBS, 0.04 mL containing 400,000 T cells per sample) were placed in the top wells of a 48-well Boyden chemotaxis chamber. PBS served as the negative control in this experiment and TARC, a known T-cell chemoattractant (55), was the positive control. After 18 h, the membranes were then removed and stained with Diff-Quik (Thermo Fisher Scientific). Readings represent the number of cells migrating through the membrane (the sum of three high power 40× fields per well, averaged for each quadruplicate well).
Adhesion Assay.
CD3+ T-cells were isolated by using a RosetteSep lymphocyte isolation kit (Stem Cell Technologies) and were stained with carboxyfluorescein succinimidyl ester (Thermo Fisher Scientific) at 5 μM. RA synovial fibroblasts at culture passage 4–8 were stimulated with 1,000 U/mL IFN-γ (Cell Signaling Technologies) in 10% FBS in culture media or in 10% FBS for 3 d. Fibroblasts were then incubated with mouse IgG, anti-CD318, anti-CD166, or both anti-CD318 and anti-CD166. T cells were added at 50,000 cells per well onto fibroblasts and incubated for 1 h at room temperature. Intensity was measured with a Synergy plate reader.
Statistics.
EAE clinical scores were evaluated with two-way ANOVA and Bonferroni’s post hoc testing. One-tailed P values <0.05 were considered significant. MOG recall T-cell assays were evaluated with nonparametric t test. One-tailed P values <0.05 were considered significant.
Study Approval and Animal Use.
All procedures involving mice were approved by the Institutional Animal Care and Use Committee of Cleveland Clinic, and all were done in accordance with the US Department of Health and Human Services Guide for the Care and Use of Laboratory Animals, and institutional guidelines. Human tissues samples were collected as part of study no. 2002–0875, which was approved by the University of Michigan Institutional Review Board. All subjects provided informed consent before their participation in the study.
Acknowledgments
This work is supported in part by NIH Grants R01 EY025373 and R01 AR061564 (to F.L.), T32 CA009592 and F31 CA189764 (to K.E.H.), and R01 CA143081 (to A.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704008114/-/DCSupplemental.
References
- 1.Pinto M, Carmo AM. CD6 as a therapeutic target in autoimmune diseases: Successes and challenges. BioDrugs. 2013;27:191–202. doi: 10.1007/s40259-013-0027-4. [DOI] [PubMed] [Google Scholar]
- 2.International Multiple Sclerosis Genetics Consortium The genetic association of variants in CD6, TNFRSF1A and IRF8 to multiple sclerosis: A multicenter case-control study. PLoS One. 2011;6:e18813. doi: 10.1371/journal.pone.0018813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swaminathan B, et al. Validation of the CD6 and TNFRSF1A loci as risk factors for multiple sclerosis in Spain. J Neuroimmunol. 2010;223:100–103. doi: 10.1016/j.jneuroim.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 4.De Jager PL, et al. International MS Genetics Consortium Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41:776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heap GA, et al. Genome-wide analysis of allelic expression imbalance in human primary cells by high-throughput transcriptome resequencing. Hum Mol Genet. 2010;19:122–134. doi: 10.1093/hmg/ddp473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aruffo A, Melnick MB, Linsley PS, Seed B. The lymphocyte glycoprotein CD6 contains a repeated domain structure characteristic of a new family of cell surface and secreted proteins. J Exp Med. 1991;174:949–952. doi: 10.1084/jem.174.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowen MA, et al. Cloning, mapping, and characterization of activated leukocyte-cell adhesion molecule (ALCAM), a CD6 ligand. J Exp Med. 1995;181:2213–2220. doi: 10.1084/jem.181.6.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zimmerman AW, et al. Long-term engagement of CD6 and ALCAM is essential for T-cell proliferation induced by dendritic cells. Blood. 2006;107:3212–3220. doi: 10.1182/blood-2005-09-3881. [DOI] [PubMed] [Google Scholar]
- 9.Nair P, Melarkode R, Rajkumar D, Montero E. CD6 synergistic co-stimulation promoting proinflammatory response is modulated without interfering with the activated leucocyte cell adhesion molecule interaction. Clin Exp Immunol. 2010;162:116–130. doi: 10.1111/j.1365-2249.2010.04235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joo YS, et al. Evidence for the expression of a second CD6 ligand by synovial fibroblasts. Arthritis Rheum. 2000;43:329–335. doi: 10.1002/1529-0131(200002)43:2<329::AID-ANR12>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 11.Saifullah MK, et al. Expression and characterization of a novel CD6 ligand in cells derived from joint and epithelial tissues. J Immunol. 2004;173:6125–6133. doi: 10.4049/jimmunol.173.10.6125. [DOI] [PubMed] [Google Scholar]
- 12.Alonso-Ramirez R, et al. Rationale for targeting CD6 as a treatment for autoimmune diseases. Arthritis (Egypt) 2010;2010:130646. doi: 10.1155/2010/130646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhatt AS, Erdjument-Bromage H, Tempst P, Craik CS, Moasser MM. Adhesion signaling by a novel mitotic substrate of src kinases. Oncogene. 2005;24:5333–5343. doi: 10.1038/sj.onc.1208582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scherl-Mostageer M, et al. Identification of a novel gene, CDCP1, overexpressed in human colorectal cancer. Oncogene. 2001;20:4402–4408. doi: 10.1038/sj.onc.1204566. [DOI] [PubMed] [Google Scholar]
- 15.Hooper JD, et al. Subtractive immunization using highly metastatic human tumor cells identifies SIMA135/CDCP1, a 135 kDa cell surface phosphorylated glycoprotein antigen. Oncogene. 2003;22:1783–1794. doi: 10.1038/sj.onc.1206220. [DOI] [PubMed] [Google Scholar]
- 16.Casar B, et al. In vivo cleaved CDCP1 promotes early tumor dissemination via complexing with activated β1 integrin and induction of FAK/PI3K/Akt motility signaling. Oncogene. 2014;33:255–268. doi: 10.1038/onc.2012.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spassov DS, Baehner FL, Wong CH, McDonough S, Moasser MM. The transmembrane src substrate Trask is an epithelial protein that signals during anchorage deprivation. Am J Pathol. 2009;174:1756–1765. doi: 10.2353/ajpath.2009.080890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conze T, et al. CDCP1 is a novel marker for hematopoietic stem cells. Ann N Y Acad Sci. 2003;996:222–226. doi: 10.1111/j.1749-6632.2003.tb03249.x. [DOI] [PubMed] [Google Scholar]
- 19.Bühring HJ, et al. CDCP1 identifies a broad spectrum of normal and malignant stem/progenitor cell subsets of hematopoietic and nonhematopoietic origin. Stem Cells. 2004;22:334–343. doi: 10.1634/stemcells.22-3-334. [DOI] [PubMed] [Google Scholar]
- 20.Uekita T, Sakai R. Roles of CUB domain-containing protein 1 signaling in cancer invasion and metastasis. Cancer Sci. 2011;102:1943–1948. doi: 10.1111/j.1349-7006.2011.02052.x. [DOI] [PubMed] [Google Scholar]
- 21.Perry SE, et al. Expression of the CUB domain containing protein 1 (CDCP1) gene in colorectal tumour cells. FEBS Lett. 2007;581:1137–1142. doi: 10.1016/j.febslet.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 22.Uekita T, et al. CUB-domain-containing protein 1 regulates peritoneal dissemination of gastric scirrhous carcinoma. Am J Pathol. 2008;172:1729–1739. doi: 10.2353/ajpath.2008.070981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Awakura Y, et al. Microarray-based identification of CUB-domain containing protein 1 as a potential prognostic marker in conventional renal cell carcinoma. J Cancer Res Clin Oncol. 2008;134:1363–1369. doi: 10.1007/s00432-008-0412-4. [DOI] [PubMed] [Google Scholar]
- 24.Razorenova OV, et al. VHL loss in renal cell carcinoma leads to up-regulation of CUB domain-containing protein 1 to stimulate PKCdelta-driven migration. Proc Natl Acad Sci USA. 2011;108:1931–1936. doi: 10.1073/pnas.1011777108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong CH, et al. Phosphorylation of the SRC epithelial substrate Trask is tightly regulated in normal epithelia but widespread in many human epithelial cancers. Clin Cancer Res. 2009;15:2311–2322. doi: 10.1158/1078-0432.CCR-08-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spassov DS, Wong CH, Wong SY, Reiter JF, Moasser MM. Trask loss enhances tumorigenic growth by liberating integrin signaling and growth factor receptor cross-talk in unanchored cells. Cancer Res. 2013;73:1168–1179. doi: 10.1158/0008-5472.CAN-12-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jayaraman K. Biocon’s first-in-class anti-CD6 mAb reaches the market. Nat Biotechnol. 2013;31:1062–1063. doi: 10.1038/nbt1213-1062b. [DOI] [PubMed] [Google Scholar]
- 28.Seidel J, Bruemann C, Kunc K, Possinger K, Lueftner D. Evaluation of CUB-domain-containing protein (CDCP1)-expression as predictive marker of adhesion-independent cell survival in breast cancer cell lines. Cancer Res. 2009;69:6167. [Google Scholar]
- 29.Wortmann A, et al. Cellular settings mediating Src Substrate switching between focal adhesion kinase tyrosine 861 and CUB-domain-containing protein 1 (CDCP1) tyrosine 734. J Biol Chem. 2011;286:42303–42315. doi: 10.1074/jbc.M111.227462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kollmorgen G, et al. Antibody mediated CDCP1 degradation as mode of action for cancer targeted therapy. Mol Oncol. 2013;7:1142–1151. doi: 10.1016/j.molonc.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ikeda JI, et al. Epigenetic regulation of the expression of the novel stem cell marker CDCP1 in cancer cells. J Pathol. 2006;210:75–84. doi: 10.1002/path.2026. [DOI] [PubMed] [Google Scholar]
- 32.Spassov DS, et al. Phosphorylation of Trask by Src kinases inhibits integrin clustering and functions in exclusion with focal adhesion signaling. Mol Cell Biol. 2011;31:766–782. doi: 10.1128/MCB.00841-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lunter PC, et al. Activated leukocyte cell adhesion molecule (ALCAM/CD166/MEMD), a novel actor in invasive growth, controls matrix metalloproteinase activity. Cancer Res. 2005;65:8801–8808. doi: 10.1158/0008-5472.CAN-05-0378. [DOI] [PubMed] [Google Scholar]
- 34.Miyazawa Y, et al. CUB domain-containing protein 1, a prognostic factor for human pancreatic cancers, promotes cell migration and extracellular matrix degradation. Cancer Res. 2010;70:5136–5146. doi: 10.1158/0008-5472.CAN-10-0220. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, et al. CD6 as a potential target for treating multiple sclerosis. Proc Natl Acad Sci USA. 2017;114:2687–2692. doi: 10.1073/pnas.1615253114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singer NG, et al. CD6 dependent interactions of T cells and keratinocytes: Functional evidence for a second CD6 ligand on gamma-interferon activated keratinocytes. Immunol Lett. 1997;58:9–14. doi: 10.1016/s0165-2478(97)02707-7. [DOI] [PubMed] [Google Scholar]
- 37.Takeda H, Fujimori Y, Kai S, Ogawa H, Nakano T. CD318/CUB-domain-containing protein 1 expression on cord blood hematopoietic progenitors. Exp Ther Med. 2010;1:497–501. doi: 10.3892/etm_00000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bott CM, Doshi JB, Morimoto C, Romain PL, Fox DA. Activation of human T cells through CD6: Functional effects of a novel anti-CD6 monoclonal antibody and definition of four epitopes of the CD6 glycoprotein. Int Immunol. 1993;5:783–792. doi: 10.1093/intimm/5.7.783. [DOI] [PubMed] [Google Scholar]
- 39.Chappell PE, et al. Structures of CD6 and its ligand CD166 give insight into their interaction. Structure. 2015;23:1426–1436. doi: 10.1016/j.str.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cayrol R, et al. Activated leukocyte cell adhesion molecule promotes leukocyte trafficking into the central nervous system. Nat Immunol. 2008;9:137–145. doi: 10.1038/ni1551. [DOI] [PubMed] [Google Scholar]
- 41.Lécuyer MA, et al. Dual role of ALCAM in neuroinflammation and blood-brain barrier homeostasis. Proc Natl Acad Sci USA. 2017;114:E524–E533. doi: 10.1073/pnas.1614336114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spassov DS, Ahuja D, Wong CH, Moasser MM. The structural features of Trask that mediate its anti-adhesive functions. PLoS One. 2011;6:e19154. doi: 10.1371/journal.pone.0019154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He Y, Harrington BS, Hooper JD. New crossroads for potential therapeutic intervention in cancer - intersections between CDCP1, EGFR family members and downstream signaling pathways. Oncoscience. 2016;3:5–8. doi: 10.18632/oncoscience.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wright HJ, et al. CDCP1 cleavage is necessary for homodimerization-induced migration of triple-negative breast cancer. Oncogene. 2016;35:4762–4772. doi: 10.1038/onc.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Law ME, et al. Glucocorticoids and histone deacetylase inhibitors cooperate to block the invasiveness of basal-like breast cancer cells through novel mechanisms. Oncogene. 2013;32:1316–1329. doi: 10.1038/onc.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong Y, et al. The cell surface glycoprotein CUB domain-containing protein 1 (CDCP1) contributes to epidermal growth factor receptor-mediated cell migration. J Biol Chem. 2012;287:9792–9803. doi: 10.1074/jbc.M111.335448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iwata M, Torok-Storb B, Wayner EA, Carter WG. CDCP1 identifies a CD146 negative subset of marrow fibroblasts involved with cytokine production. PLoS One. 2014;9:e109304. doi: 10.1371/journal.pone.0109304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morgan R, et al. Expression and function of aminopeptidase N/CD13 produced by fibroblast-like synoviocytes in rheumatoid arthritis: Role of CD13 in chemotaxis of cytokine-activated T cells independent of enzymatic activity. Arthritis Rheumatol. 2015;67:74–85. doi: 10.1002/art.38878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He Y, et al. Elevated CDCP1 predicts poor patient outcome and mediates ovarian clear cell carcinoma by promoting tumor spheroid formation, cell migration and chemoresistance. Oncogene. 2016;35:468–478. doi: 10.1038/onc.2015.101. [DOI] [PubMed] [Google Scholar]
- 50.Wortmann A, He Y, Deryugina EI, Quigley JP, Hooper JD. The cell surface glycoprotein CDCP1 in cancer–insights, opportunities, and challenges. IUBMB Life. 2009;61:723–730. doi: 10.1002/iub.198. [DOI] [PubMed] [Google Scholar]
- 51.Hafler DA, et al. Immunologic responses of progressive multiple sclerosis patients treated with an anti-T-cell monoclonal antibody, anti-T12. Neurology. 1986;36:777–784. doi: 10.1212/wnl.36.6.777. [DOI] [PubMed] [Google Scholar]
- 52.Li Q, Huang D, Nacion K, Bu H, Lin F. Augmenting DAF levels in vivo ameliorates experimental autoimmune encephalomyelitis. Mol Immunol. 2009;46:2885–2891. doi: 10.1016/j.molimm.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abdul-Majid KB, et al. Screening of several H-2 congenic mouse strains identified H-2(q) mice as highly susceptible to MOG-induced EAE with minimal adjuvant requirement. J Neuroimmunol. 2000;111:23–33. doi: 10.1016/s0165-5728(00)00360-x. [DOI] [PubMed] [Google Scholar]
- 54.Ruck T, Bittner S, Epping L, Herrmann AM, Meuth SG. Isolation of primary murine brain microvascular endothelial cells. J Vis Exp. 2014;(93):e52204. doi: 10.3791/52204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Imai T, et al. The T cell-directed CC chemokine TARC is a highly specific biological ligand for CC chemokine receptor 4. J Biol Chem. 1997;272:15036–15042. doi: 10.1074/jbc.272.23.15036. [DOI] [PubMed] [Google Scholar]