Significance
In light of the recent outbreak of Zika virus (ZIKV), an understanding of the within-host viral dynamics is essential to assess persistence in vivo, transmission risk, and antiviral therapeutics. Using mathematical modeling we find that, in nonhuman primates, the viral dynamics of ZIKV are characterized by a short lifetime of infected cells (<10 h) during which enough viral particles are produced to infect ∼11 other cells. Higher disease burden is associated with changes in natural killer cell subset concentrations and with elevated expression of the cytokine MCP-1, although the mechanisms behind these associations remain unclear. In order for an antiviral treatment to effectively reduce the time to plasma viral clearance therapy should be initiated at the time of infection or given prophylactically.
Keywords: Zika virus, mathematical modeling, viral dynamics, antiviral therapy
Abstract
The recent outbreak of Zika virus (ZIKV) has been associated with fetal abnormalities and neurological complications, prompting global concern. Here we present a mathematical analysis of the within-host dynamics of plasma ZIKV burden in a nonhuman primate model, allowing for characterization of the growth and clearance of ZIKV within individual macaques. We estimate that the eclipse phase for ZIKV, the time between cell infection and viral production, is most likely short (∼4 h), the median within-host basic reproductive number R0 is 10.7, the rate of viral production is rapid (>25,000 virions d−1), and the lifetime of an infected cell while producing virus is ∼5 h. We also estimate that the minimum number of virions produced by an infected cell over its lifetime is ∼5,500. We assess the potential effect of an antiviral treatment that blocks viral replication, showing that the median time to undetectable plasma viral load (VL) can be reduced from ∼5 d to ∼3 d with a drug concentration ∼15 times the drug’s EC50 when treatment is given prophylactically starting at the time of infection. In the case of favipiravir, a polymerase inhibitor with activity against ZIKV, we predict a dose of 150 mg/kg given twice a day initiated at the time of infection can reduce the peak median VL by ∼3 logs and shorten the time to undetectable median VL by ∼2 d, whereas treatment given 2 d postinfection is mostly ineffective in accelerating plasma VL loss in macaques.
Zika virus (ZIKV) is a flavivirus with a ∼10.7-kb positive sense single-stranded RNA genome (1) that is primarily transmitted among humans via a mosquito vector. First identified in Uganda in 1947 (2), it has come to prominence due to an outbreak in 2007 in Micronesia (3) and emergence in Brazil in 2015 (4). Human infection with ZIKV is usually accompanied by a relatively mild, self-limiting fever (3, 5) but can be associated with more severe effects such as Guillain–Barré syndrome (6) and fetal microcephaly (7). Recent reports of non-mosquito-borne transmission (reviewed in ref. 8) including sexual transmission (9, 10) and maternal transmission (11, 12) raise the concern that ZIKV may spread and persist in large parts of the world. Vaccines represent the most effective way to counter the epidemic, with promising results in preclinical studies (13, 14). Despite the fact that human trials have begun, approved vaccines may not become available for years (15). Treatment with antivirals is needed, especially in some populations, such as severely infected patients or individuals for whom viremia remains detectable in the blood or in other compartments for extended periods, as has been observed in pregnancy (16) and in semen (17, 18). So far no antiviral drug has shown a significant effect against ZIKV in vivo. The WHO has highlighted a number of potential drugs that could be relevant against ZIKV (www.who.int/blueprint/priority-diseases/key-action/zika-rd-pipeline.pdf?ua=1) and a high throughput in vitro screen (19) has identified a number of potential small molecules with activity against ZIKV but that will require further testing in animal models.
With that perspective, the establishment of a relevant nonhuman primate (NHP) model (14, 20–22) represents crucial progress toward a better understanding of virus pathogenesis and the development of more efficacious therapeutic strategies. Zika infection in NHPs has recapitulated many of the key clinical findings in human infection, including rapid control of acute viremia, early invasion of the central nervous system, and prolonged viral shedding and fetal pathology in pregnant animals (14, 20, 21, 23, 24). Thus, the study of ZIKV in NHP models can provide insight into viral dynamics as well as providing an essential tool for testing potential antiviral drugs and vaccines (14).
Mathematical modeling of viral dynamics has provided a quantitative understanding of viral dynamics for a number of viruses, including HIV (25, 26), influenza (27), and the flaviviruses hepatitis C virus (28), West Nile virus (WNV) (29), and dengue (30–32). One of the main insights of these models was to provide estimates of parameters that are not easily measurable, such as the half-life (t1/2) of productively infected cells in vivo and the basic reproductive ratio, R0, the number of secondary cells infected by viral production from one infected cell in a population of target cells. In the context of viruses for which antivirals are available, such as HIV and hepatitis C virus, mathematical modeling has been used to optimize and evaluate in silico the impact of antiviral strategies (33–36).
Here we use mathematical modeling to characterize ZIKV dynamics in vivo in NHPs, to explore correlations between viral dynamic parameters and the immunologic response, and to simulate the effects of a potential antiviral agent.
Results
Zika Viral Kinetics.
After s.c. infection of nine rhesus macaques with 106 pfu of a Thai isolate of ZIKV (Materials and Methods), the plasma viral load (VL; data given in SI Appendix, Table S1) was high by day 1 postinfection (>105 RNA copies per mL in all monkeys) and peaked at day 2 (Fig. 1). The peak VL was above 106 copies per mL for all monkeys and above 107 copies per mL for six of the nine monkeys. Following peak viremia, there was a rapid decline in VL, and the VL became undetectable (<200 copies per mL) in eight of the nine monkeys by day 5. The VL in the remaining monkey was undetectable by day 10.
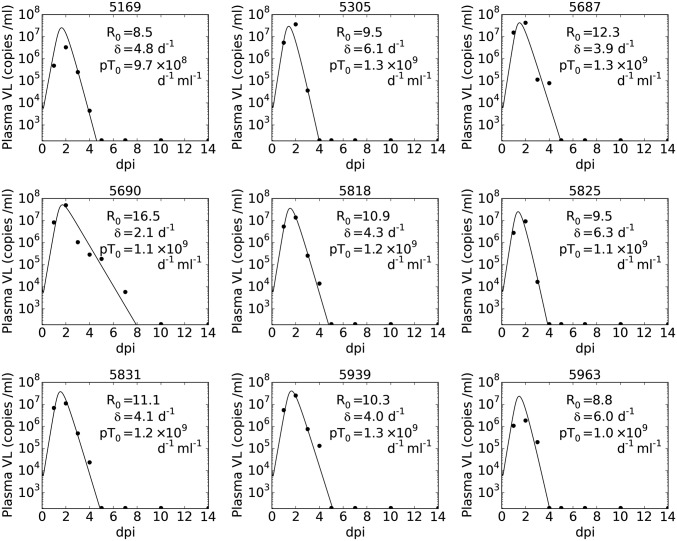
Fig. 1.
Best fit of model (Eq. 1, solid lines) to the observed plasma VL (black circles), with fixed k = 6 d−1, c = 10 d−1, V(0) = 104 mL−1, T(0) = 105 mL−1, and fixed ω = 0.4 for fitted parameters R0, δ, and p (see text for further details). When virus was not detectable in a sample it is displayed at a value equal to the limit of detection, 200 RNA copies per mL. Subplot titles indicate animal ID.
Viral Kinetic Model and Parameter Estimates.
Using nonlinear mixed effects modeling we fit eight different models to the plasma VL data, including ones with immune effects via measured IFN-α, natural killer (NK) cell, or monocyte chemoattractant protein 1 (MCP-1) dynamics. Using model selection theory (37) we were unable to distinguish models incorporating explicit immune effects from a target cell-limited model, because all models had similar Bayesian information criterion (BIC) values (details are given in SI Appendix). We chose to use a simple model, a target cell-limited model with an eclipse phase as given by Eq. 1 in Materials and Methods, because it allowed us to test the effects of antiviral therapy. We considered fits of this model with different fixed values of V(0) (effective initial plasma VL concentration), k (rate of transition to productive infection), and c (rate of viral clearance). The model fit with V(0) = 104/mL and k = 6 d−1 was chosen based on the BIC across all values of c tested (SI Appendix, Figs. S1A and S2A). With these parameter values, the quality of model fit with c = 5 d−1, 10 d−1, or 15 d−1 was broadly indistinguishable (within ∼2 BIC points, SI Appendix, Fig. S1A), and we selected the model with c = 10 d−1 to analyze further. We also considered the effect of the initial target cell density, T(0) or T0, selecting T0 = 105/mL (SI Appendix and SI Appendix, Fig. S1 C and D) and ω, the SD of the random effect term, selecting ω = 0.4 (SI Appendix and SI Appendix, Fig. S1B). With these values, the fitting process provided an estimate of the median basic reproductive ratio R0 of 10.7 (SI Appendix, Fig. S2B) with a relative SE (r.s.e) of 18% and the productively infected cell death rate, δ, was estimated as 4.5 d−1 (SI Appendix, Fig. S2C, r.s.e. 15%). The estimated value of p is dependent on T0, the initial target cell density, and only the product pT0 can be reliably estimated (38), with the fitting process estimating pT0 as 1.2 × 109 d−1⋅mL−1 (SI Appendix, Fig. S2D). The individual parameters derived from this population fit provide estimated VLs for each monkey as shown in Fig. 1, and the predicted target cell dynamics for each individual monkey (SI Appendix, Fig. S3) show that target cells are largely but not completely eliminated by infection. Estimates of the value of R0 for each monkey range between 8.5 and 16.5 (Fig. 1). The goodness of fit and estimated parameters for each combination of V(0) and k are shown in SI Appendix, Fig. S2, and under the model averaging approach (Materials and Methods) the distribution of R0 in the population had a median of 10.5 with range 5.3–19.5 (5th–95th percentiles). We also estimated parameters R0, δ, and pT0 by fitting the model to each individual monkey using a nonlinear least-squares approach (SI Appendix, Fig. S4). This approach gave values of R0 that ranged between 5.3 and 25.9 and that have a very similar median (10.6) to that estimated by population modeling.
Correlation Between Plasma VL and VL in Other Compartments.
There was a statistically significant positive correlation, after Bonferroni correction for multiple testing, between the area under the log10 VL curve (AUC) between days 0 and 14 in plasma calculated by the model and in saliva (SI Appendix, Fig. S5A, Spearman correlation, P = 0.05). There were no other statistically significant correlations at the P = 0.05 level between modeled AUC in plasma and AUC in other compartments (SI Appendix, Fig. S5 B–E) or between peak log10 VL in plasma as predicted by the model and peak log10 VL in other compartments (SI Appendix, Fig. S6).
Immunological Markers During ZIKV Infection.
We tested for correlations between immune cell subset or cytokine concentrations during ZIKV infection and the modeled plasma VL. None of the Spearman correlations were statistically significant after correction for multiple testing, but we considered those cell subsets and cytokines with the strongest correlations with VL to look for suggestions of an immune response to ZIKV infection that we could further examine via model fitting.
Of the cell subsets, NK CD16+ CD69+ cells showed the strongest positive correlation (r = 0.83 between subset concentration at day 0 and peak log10 VL, SI Appendix, Fig. S7 A and B), and NK CD16− CD56+ cells (SI Appendix, Table S4) show the strongest negative correlation (r = −0.83 between subset concentration at day 2 and log10 VL AUC between days 0 and 14, SI Appendix, Fig. S7 C and D). Biologically we might expect NK cells to aid in clearance of infected cells, but model fitting incorporating the NK CD16− CD56+ subset did not provide strong support for or against these cells controlling ZIKV VL (Model G, SI Appendix, Table S5).
Of the cytokines, MCP-1 (SI Appendix, Table S2) showed the strongest positive correlation (r = 0.88 between MCP-1 concentration at day 1 and estimated infected cell half-life, SI Appendix, Fig. S7 E and F). A mechanism by which MCP-1 recruits target cells might explain the observed positive correlation, but model fitting (Model H, SI Appendix, Table S5) does not provide strong evidence for or against this hypothesis. Additionally, we hypothesized that IFN-α (SI Appendix, Fig. S8 and Table S3) might be involved in immune control of VL, despite the absence of a correlation with VL (SI Appendix, Fig. S9). Models incorporating an IFN-α response (Models D–F, SI Appendix, Table S5) were all indistinguishable from our basic model, Eq. 1, using model selection theory.
Evaluating the Effect of an Antiviral Treatment.
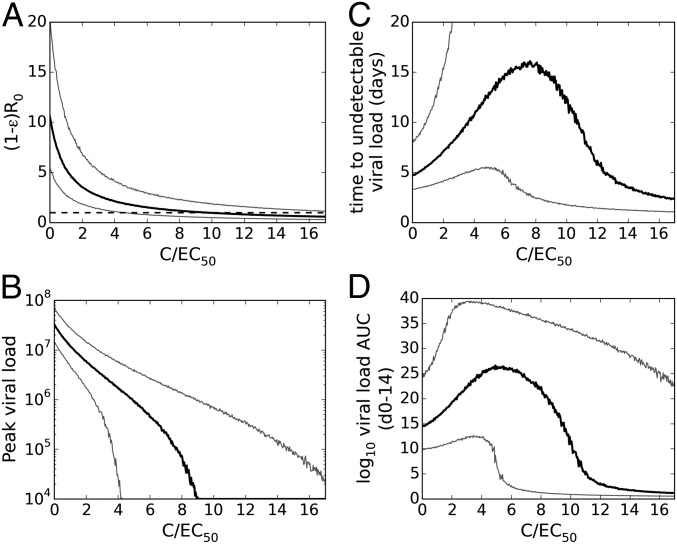
For an antiviral that reduces the viral production rate, the effect on the VL dynamics can be assessed using a target cell-limited model if the EC50 against ZIKV and the free drug concentration are known. Because we do not know how lowered VL due to antiviral therapy will influence immune responses, models incorporating the measured immune markers in these data are not suitable for studying the effect of an antiviral. The model, Eq. 1, predicts that when the drug concentration C is low relative to its EC50, R0 declines almost linearly with increasing drug concentration (Fig. 2A). However, once the drug concentration is well above the EC50 further increases in C have little effect. To reduce the median value of R0 to below 1, meaning no establishment of infection, a constant free drug concentration 9.7 times the EC50 (i.e., 9.7 EC50) is required. Similarly, the peak VL decreases monotonically with increasing C (Fig. 2B).
Fig. 2.
The predicted effect of a constant concentration, C, of an antiviral drug initiated at the time of infection that reduces viral production rate p from infected cells by a factor 1 − ε [where ε = C/(EC50 + C) is the drug effectiveness] on the (A) basic reproductive ratio, (B) peak VL, (C) time until the VL becomes undetectable (below 200 RNA copies per mL), and (D) log10 VL AUC. At each value on the x axis, 5,000 viral dynamic parameter sets were sampled from the estimated parameter distributions as determined by mixed effects model fitting. Black lines represent the median value across simulations and gray lines represent the 5th and 95th percentiles. In A, the horizontal black dashed line represents R0 = 1.
Despite the monotonic reduction in R0 with increasing C, the time until VL becomes undetectable initially increases with C before declining at higher ratios of C to EC50 (Fig. 2C). This initial increase at low drug concentrations is due to the effect of the drug in reducing the number of viral particles produced and therefore the rate at which target cells become productively infected. Consequently, the time until the target cell population is substantially depleted is increased in the model, leading to a prolonged viral infection, albeit at lower levels than without treatment. The median time until the VL becomes undetectable is maximal, at 16.1 d, when C is 7.6 EC50 and becomes lower than the time with no drug (4.7 d) when C is 12.8 EC50, reaching 3 d when C is ∼15 EC50. The VL AUC exhibits similar behavior as constant C is increased (Fig. 2D). Whether infection will be prolonged at low drug concentrations in reality is unclear because an adaptive immune response most likely will arise and limit the infection (23).
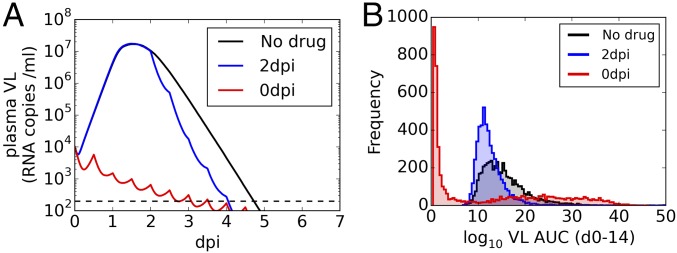
To model the effects of an antiviral drug with activity against ZIKV under more realistic conditions of changing drug concentration we examined favipiravir, an antiviral drug, approved for human use, with activity against many RNA viruses (39) and with known pharmacokinetics (PK) in monkeys (40). We used a previously developed PK model that shows differences between male and female monkeys (SI Appendix), assumed a 150 mg/kg twice daily dosing regime initiated at the time of infection, and chose PK (for male monkeys) and viral kinetic parameters randomly from their estimated distributions to simulate VL dynamics for 5,000 monkeys. The peak median VL was substantially reduced by ∼3 logs for male monkeys and the time to first undetectable median VL was shortened by ∼2 d (Fig. 3A, red). Initiating treatment 2 d after infection had essentially no effect on peak log10 VL and only shortened the time to undetectable VL by ∼0.5 d (Fig. 3A, blue). Assessing the reduction in VL burden by the AUC calculated between 0 and 14 d postinfection (dpi) (Fig. 3B) shows a reduction in median log10 VL AUC from 14.5 d copies per mL with no drug (Fig. 3B, black) to 4.7 d copies per mL (with large variation between simulations) when treatment is initiated at the time of infection (Fig. 3B, red). When treatment is initiated 2 d after infection, the effect on median log10 VL AUC in less substantial (Fig. 3B, blue). The equivalent analysis for female monkeys shows a less substantial effect on VL (SI Appendix, Fig. S10).
Fig. 3.
The predicted effect in male macaques of a 150 mg/kg twice daily regime of favipiravir, initiated at time of infection (red) or 2 d after infection (blue) on (A) median VL and (B) distribution of log10 VL AUCs. In A, the horizontal black dashed line represents the limit of detection of the experimental assay, 200 RNA copies per mL. In B, median AUC values are 14.5 (no drug), 11.8 (2 dpi), and 4.7 (0 dpi). Data shown are from 5,000 repeated simulations, each including independent simulations of the PK profile and VL dynamics determined by parameters R0, δ, and p selected at random from the population distribution predicted by mixed effects model fitting.
Discussion
This study presents an analysis of the within host dynamics of ZIKV in NHPs. After s.c. infection with 106 pfu of a Thai isolate, ZIKV is found at high concentrations in the plasma at day 1 and peaks at day 2 in these animals, followed by a rapid decline. These rapid viral kinetics resemble those observed in influenza A infections in humans, where VL tends to peak 2–3 dpi and declines to undetectable levels by 6–7 dpi in many individuals (27). This similarity motivated the use of a target cell-limited viral infection model developed by Baccam et al. (27) for influenza to fit the ZIKV plasma VL data, and the use of similar methods of analysis as in ref. 41.
The early peak VL (2 dpi) makes it is unlikely that an adaptive immune response has been mounted and is responsible for the postpeak viral decline. However, innate immune responses may play a role, as has been suggested for dengue (31), and cytokines such as type I IFN could place some uninfected cells into an antiviral state, protecting them from infection. Fitting models that include IFN-α, using its measured dynamics, did not improve the fit to the data sufficiently to justify including these innate immune responses in the model, consistent with the observation that IFN-α is not correlated with VL (SI Appendix, Fig. S9) and that ZIKV disrupts IFN signaling via STAT2 degradation (42–44). Similarly, the modeling did not strongly support or reject including an NK cell-mediated innate immune response. It may be that these data do not contain enough information to allow inference about the presence of an innate immune response, and additional VL measurements at earlier time points or after infection with lower doses may allow more detailed assessment of the contribution of the innate immune system to control of ZIKV in these animals. It may be that inclusion of immune data could reduce the unexplained individual variability of the parameters, but these data do not contain enough information to allow us to estimate this. Given the positive relationship between the expression of MCP-1, a cytokine implicated in recruitment of cells to the site of infection (45), and the predicted infected cell half-life (SI Appendix, Fig. S7F), we also considered a viral dynamics model in which the target cell population was increased in an MCP-1–dependent manner (model H, SI Appendix, Table S5). In fitting this model to the VL data from each monkey, the model BIC suggested that the available data cannot support or reject the hypothesis that MCP-1 affects the VL via this mechanism.
We highlight the point that these data do not contain enough information to allow for either acceptance or rejection of the hypothesis that an immune response is responsible for control of plasma ZIKV in these animals. Our modeling approach is to select the simplest model from those that provide equivalently good fits to the data, and as such we have performed analysis without explicit inclusion of an immune effect.
The initial viral infection is characterized by a basic reproductive ratio (R0) of 10.7, suggesting that early after infection each infected cell on average infects ∼11 other cells. To attain rapid spread, we estimate the mean eclipse phase for ZIKV (i.e., the time between infection and viral production) is ∼4 h. This estimated eclipse phase length is shorter than that for WNV in mice (29) and that estimated for influenza A in humans (27). In vitro ZIKV viral production from infected human skin cells was detected at 6 h, the earliest time examined (46). We also estimate that once cells begin producing virus they live an average of 5.3 h to yield a total lifespan, which includes the eclipse phase, of slightly less than 10 h.
Using the model, Eq. 1, we can only estimate the product pT0 (38), where T0 is the initial target cell density and p is the rate of viral production from a productively infected cell. Because the target cells for ZIKV are not currently fully characterized, we have no a priori estimate of T0. However, we can constrain p because the amount of infectious virus made over an infected cell’s productive lifetime of 5.3 h needs to be greater than R0. For a French Polynesian isolate of ZIKV it has been estimated that 500–1,000 virions correspond to 1 pfu (20). Thus, with R0 ∼11, an infected cell needs to produce at a minimum 500 × 11= 5,500 virions over 5.3 h. This implies a daily production rate p ≥ 25,000 virions d−1. From our model fitting, we estimated that pT0 = 1.2 × 109 d−1⋅mL−1, so that with p ≥ 25,000 virions d−1 we estimate T0 ≤ 4.8 × 104/mL. Our finding of a low initial target cell density combined with the broad range of cell types that ZIKV has been shown to be able to productively infect in vitro (44, 46–50) might suggest a role of an early innate response, not captured in these data, placing potential target cells into an antiviral state, thus reducing the effective target cell population.
The early peak viremia and rapid viral decay in this NHP model, along with the limited longitudinal sampling, means that the chosen value of V(0) and the estimate of R0 should be taken cautiously. Plasma samples taken during the first few hours postinfection would allow for a more accurate assessment of the likely value of V(0) and would give insight into how rapidly a s.c. infection is trafficked to the blood. Also, studies with different inoculum sizes would help in determining a relationship between dose and V(0). The number of data points available also limits our ability to estimate the distribution of parameters found in the population, because these data are unable to support fitting the variance of the estimated parameter distributions. Additional animals as well as more frequent sampling would improve the estimates of these distributions. The lack of VL AUC correlation between the plasma and semen compartments (SI Appendix, Fig. S5D) might be due to the small number of data points available in this study, or it might indicate that different mechanisms affect the VL in the semen, which would restrict our ability to assess sexual transmission risk from plasma samples. Overall, the persistence of virus in saliva and semen (17) and the prolonged presence in the plasma of pregnant macaques and humans (16, 20) as well as in a child born to ZIKV-infected parents (51) remains to be explained.
In VL dynamics from a study with different ZIKV strains and with lower inoculum doses (23) it was seen that peak VL tended to be later than in these data, and a model incorporating immune control from IFN-α (reducing viral production) and antibodies (reducing viral infectivity) was able to improve the fit of the model. The estimated value of δ was found to be very similar to this study, whereas the estimated value of R0 was much lower (∼3). This discrepancy between the estimated values of R0 can be attributed to a later peak with lower inoculum doses as well as a higher estimate of V(0) with equivalent inoculum doses, highlighting the need for early samples to assess the true initial plasma viral concentration.
Because antiviral drugs against ZIKV are being tested in rhesus macaques (52) we investigated the effect of an antiviral drug at concentration C that reduces the production of virus by a factor of 1 – C/(EC50 + C). Increasing the drug concentration reduces R0, but we estimate that a drug concentration of at least 12.8 EC50 is required to reduce the time to undetectable VL (Fig. 2), where these results are based on our target cell limitation model assumptions. This approach can allow for rapid screening of potential repurposed drug candidates: if a drug is unable to reach this concentration with a safe dose, it is unlikely to have a substantial positive impact of the VL dynamics, although both differences between EC50 in vitro and in vivo and differences between the VL dynamics in the NHP model and in humans need to be accounted for. We identified favipiravir, a small-molecule antiviral, as capable of reducing the median viral burden in NHP infections in simulations and found that substantial reduction can be achieved for the majority, but not all, of simulations when treatment is started at the time of infection. The PK of favipiravir is highly variable in NHPs (40), resulting in the broad distribution of VL AUCs seen in our simulations (Fig. 3B), although the sustained infections seen in our simulations when the PK is unfavorable, giving low drug concentrations, would likely be mitigated by later immune responses. However, if an antiviral strategy is to have any substantial effect on peak VL, time to undetectable viremia or VL AUC it needs to be initiated before peak viremia. Because peak plasma VL occurs very early in this NHP model, this effectively means that treatment needs to be given prophylactically. In humans where lower viremia levels are often observed (e.g., refs. 18, 53, and 54), as well as when lower inoculum doses are used in NHPs (14) the viral dynamics will be altered and the peak of plasma viremia will likely be delayed, affecting the assessment of antiviral impact and potentially allowing for later antiviral treatment initiation.
The mathematical modeling approach in this study describes the characteristics of the viral dynamics of ZIKV as observed in the plasma of an NHP model and provides a tool by which to assess, in silico, the effect of candidate antivirals.
Materials and Methods
ZIKV-Infected Rhesus Macaques.
Ten Indian-origin rhesus macaques (five male and five female) were infected s.c. with 106 pfu of a Thai isolate of ZIKV as described in ref. 21. Samples were collected from blood (plasma), urine, saliva, cerebrospinal fluid, semen, and vaginal secretions for 4 wk postinfection. Samples were taken regularly, and viral RNA levels in samples were assessed using real-time PCR, with a limit of detection of 200 copies per mL. Additionally, peripheral blood mononuclear cells (PBMC) were analyzed from the same blood samples. PBMC samples were stained with fluorescent antibody mixes and flow cytometry used to quantify the concentration of T-cell, B-cell, and NK subsets. The set of immune cells measured includes total lymphocytes, eight T-cell subsets, three B-cell subsets, and four NK cell subsets, and both total concentration and activated (as indicated by expression of CD69) concentration were measured (21). Cytokine expression in plasma samples was also measured by multiplex bead immunoassays at days 0, 1, 3 and 7. All animals were housed at Bioqual Inc. or the Wisconsin National Primate Research Center (WNPRC). The described studies were approved by the Institutional Animal Care and Use Committee of Bioqual Inc. or WNPRC, respectively.
Viral Dynamic Model.
Plasma viral dynamics in nine of the 10 animals were characterized using a standard target cell-limited model with an eclipse phase originally developed to analyze influenza infection kinetics (27). One animal in which there were only two plasma VLs above the detection limit of the assay was excluded from this analysis. The model with an eclipse phase was used in preference to a model without an eclipse phase (SI Appendix). The model is given by the following system of ordinary differential equations:
| [1] |
In this model, target cells (T) are infected by free virus (V) at a rate proportional to both virus and target cell concentrations, as given by the law of mass action term βVT. After infection, the cell enters an eclipse phase, denoted I1, where it is infected but does not produce virus. It then transitions to a productively infected state (I2) at rate k, that is, 1/k is the average duration of the eclipse phase. Productively infected cells are cleared at per capita rate δ, incorporating the observed cytopathic effects of ZIKV (55, 56), and produce virus at rate p per cell. Free virus is cleared at rate c per virion. Because of the short-term nature of the infection as measured in blood with a peak VL at day 2 in all nine animals, target cell replenishment was ignored in the model.
The model predicts an initial decline of virus until the end of the eclipse phase of the first infected cells, followed by an exponential increase until target cells become limiting. At the end of this expansion phase, the target cells are largely but not completely depleted (SI Appendix, Fig. S3), the number of new infections is low, and the loss of infected cells largely outnumbers the number of new cell infections. Consequently, the number of infected cells, and hence the total viral production, declines. Mathematical analysis shows that this decline is exponential with a rate proportional to the infected cell death rate, δ, when it is much smaller than c and k (57). In this model all parameters were assumed to be constant, but in principle they could vary over time. Thus, for instance, an adaptive immune response, if present, could be associated with an increasing value of δ over time. One can derive from this model the basic reproductive number, R0, representing the number of secondary infected cells that would be created by a single infected cell introduced into a population of T0 target cells, given by R0 = βpT0 /δc. The model was reparametrized as a function of R0, which we constrained to be larger than 1 to ensure productive infection. More details on viral dynamics models during acute infection can be found in refs. 57–59. We additionally tested a number of other potential models, including a model without an eclipse phase and models incorporating innate immune responses (SI Appendix). Model selection was based on BIC, with more complex models only being accepted if an improvement in BIC of >2 points was observed (60).
Fixed Parameters.
Because not all model parameters can be estimated using only VL data (38) a number of parameters had to be fixed (details are given in SI Appendix). First the number of nonproductively and productively infected cells at time of infection is assumed to be 0 [I1(0) = I2(0) = 0], and the number of target cells at the time of infection, T(0), is assumed to be 105 per mL. As shown in ref. 38, for a standard viral dynamic model the viral production rate p and the initial target cell density T(0) are dependent, and with measurements of VL only one is able to be estimated. Here the assumed value of T(0) was chosen to give biologically reasonable estimates of p (SI Appendix). Note that although the value of T(0) was chosen to be plausible it does not affect our main findings regarding the basic reproductive number R0, the half-life of infected cells, and the effect of antiviral treatment. Additionally, we do not assume that all target cells and infection events are in blood, but we model the effective impact of target cell infection on the plasma VL as has been done, for example, in models of hepatitis viruses (28), whose target cells are mainly or exclusively in the liver. After initial model fitting to determine the effect of the rate of clearance of free virus, c, on the model fit (details are given in SI Appendix) we set c to a fixed value of 10 d−1.
Estimated Parameters.
The macaques were inoculated s.c. with 106 pfu ZIKV. Assuming that 1 pfu represents between 500 and 1,000 copies of viral RNA (20) and that a typical macaque plasma volume is about 300 mL, the inoculum would at most generate between 1.7 × 106 and 3.3 × 106 viral RNA copies per mL of plasma. Because it is likely that much of the early infection occurs in the skin and that only a small proportion of the inoculum is transported to the blood immediately after infection, we used values of V(0) between 103 and 105 viral RNA copies per mL in our model fitting. Here V(0) should be interpreted as an effective initial viral concentration.
The eclipse phase duration for ZIKV is unknown. We let k be 4, 6, or 8 d−1 (representing a mean eclipse phase of 6, 4, or 3 h). We restricted our range of possible eclipse phase lengths to short times because the Zika plasma VL can be as high as 107 RNA copies per mL by 1 d postinfection, suggesting rapid viral replication and hence a short eclipse phase. Further, in vitro experiments have observed ZIKV viral production 6 h postinfection (46).
For each combination of V(0) and k, the distribution of other parameters across the population of monkeys was estimated using a nonlinear mixed effects approach (61). Under this approach, each parameter is assumed to be log-normally distributed in the population and can be expressed as , where θ is the median value in the population and ηi is the individual random effect, which is normally distributed as N(0, ω2), accounting for variability between individuals in the population. Because the variance of this random-effect term could not be estimated with the small number of animals in this study, we set it to a fixed value for fitted parameters R0, δ, and p (SD ω = 0.4 for each parameter), corresponding to a coefficient of variation just above 40% for all fitted parameters. We also tested the effect of varying the SD of the random effect (details are given in SI Appendix) and found that the models are equally supported (within 2 BIC points) for ω = 0.2, 0.3, and 0.4 (SI Appendix, Fig. S1B) and chose ω = 0.4 to allow the most parameter variation. Estimates for R0, δ, and p (given a fixed value of T0) were then estimated using the stochastic approximation of the expectation maximization algorithm implemented in Monolix (monolix.lixoft.com/), with data below the limit of detection accounted for as described in ref. 62. The combination of V(0) and k that provides the lowest BIC was carried forward for further analyses, and the individual parameters for each monkey were obtained using empirical Bayes estimates.
Using these individual parameters, the individual VL profiles were reconstructed, allowing us to calculate the area under the VL curve (AUC) for each animal. In this study, the plasma AUC is calculated as the area between the modeled log10 VL curve and the log10 detection limit of the experimental assay, calculated using the trapz function of NumPy in Python (63). The AUCs for other compartments were calculated in a similar fashion, using observed rather than modeled VLs.
To account for the fact that a different value of R0 could have been obtained with different pairs of values for V(0) and k we also assessed the distribution of R0 in the population by using a “model averaging” approach (64). We assume that R0 is distributed as a weighted sum of Gaussian distributions, each corresponding to one combination of V(0) and k considered, with weight given by e-BIC/2. The mean of the Gaussian is given by the estimated population R0 with that V(0) and k, and the SD is the SE of that distribution. We then sampled 106 values of R0 from this weighted sum of Gaussians and reported the median and 5th–95th percentiles.
Additionally, a simple individual fitting approach was used to confirm that conclusions similar to those in the mixed effects model with fixed variances were reached. For each monkey, the free model parameters were fitted to the observed VL via a nonlinear least squares approach using a differential evolution algorithm implemented in the DEoptim package in R (65). The parameters estimated by this approach across all nine monkeys were found to be in the same range as the distributions estimated from the mixed effects approach.
ZIKV VL in Anatomic Compartments.
The correlations between ZIKV dynamics in the blood and the other tissue compartments were explored by comparing, for each NHP, the modeled AUC in the blood with the observed AUC in each other compartment, and the modeled peak VL in the blood with the observed peak VL in each other compartment. The correlations were assessed via a Spearman correlation coefficient (adjusted for multiple testing using the Bonferroni correction) and significance was assessed at the P = 0.05 level.
Immune Markers.
Concentrations of a number of lymphocyte subsets and cytokines were measured in each of the monkeys during infection, and we refer to these as immune markers (see ref. 21 for details). For each monkey, we calculated Spearman correlations between characteristics of the immune markers and characteristics of the VL dynamics, as described in SI Appendix. We adjusted for multiple testing using the Bonferroni correction. To identify immune markers to be incorporated into our viral dynamic models, we considered those immune markers that demonstrated the strongest correlations with VL and seemed biologically relevant. We used NK CD16− CD56+ cells and the cytokine MCP-1 in models G and H (SI Appendix). Additionally, we considered correlations between the concentration of IFN-α and the VL in each monkey, because IFN-α may contribute to immune control of ZIKV. IFN-α dynamics were incorporated in models D–F (SI Appendix).
Effect of an Antiviral Treatment on Plasma VL.
Using repeated simulations with parameters selected at random from the fitted estimates of the parameter distributions, we explored the effects of an antiviral drug on the viral dynamics for different ratios of an assumed constant drug concentration (C) to the drug EC50, where EC50 is the concentration providing a 50% viral growth inhibition in vivo. We assumed the drug reduced the viral production rate, p, by a factor (1− ε) where ε = C/(EC50 + C) is the drug antiviral effectiveness. Then we calculated the impact on the basic reproductive number R0, peak VL, time to undetectable VL, and VL AUC for different (constant) values of C/EC50.
Effect of Favipiravir on ZIKV Plasma VL.
Our group identified favipiravir (T-705), a broad-spectrum RNA polymerase inhibitor (66), as a potential ZIKV inhibitor. The EC50 values of favipiravir against ZIKV strains was assessed as detailed in SI Appendix and provided values between 2.7 µg/mL and 6.6 µg/mL. Because the PK of favipiravir in cynomolgus macaques have been described in detail (40), we aimed to evaluate the effects of the drug on the VL dynamics by assuming the PK in rhesus macaques was similar to that described in Mauritian cynomolgus macaques (SI Appendix) and performing simulations as follows.
We simulated a dosing regimen including two loading doses of 250 mg/kg at hours 0 and 12, followed by 12 hourly maintenance doses of 150 mg/kg for 14 d. This dosing regimen was chosen because it has been shown to give no serious abnormalities in NHP, while maintaining a trough concentration at day 7 greater than the EC50 against ZIKV (40). Different treatment initiation times were tested, with dosing starting either at the time of infection or 2 dpi (median time of viral peak). For each scenario, 5,000 PK and viral kinetic parameters were drawn from their estimated distributions, giving simulated trajectories of response. Because the PK in male and female animals differed, this process was done for males and females separately. To summarize the simulations, the median of the VLs at each time point and the distribution of the AUC of log10 VL between 0 and 14 dpi over the 5,000 trajectories were reported for males and females separately.
Supplementary Material
Acknowledgments
We thank France Mentré for advice on the model averaging approach. Portions of this work were done under the auspices of the US Department of Energy under Contract DE-AC52-06NA25396. This work was also supported by NIH Grants R01-AI078881, R01-OD0110955, and R01-AI028433 (to A.S.P.) and the European Union’s Horizon 2020 research and innovation program under Grants 666092 (to J.G.) and 734548 (to X.d.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704011114/-/DCSupplemental.
References
- 1.Lei J, et al. Crystal structure of Zika virus NS2B-NS3 protease in complex with a boronate inhibitor. Science. 2016;353:503–505. doi: 10.1126/science.aag2419. [DOI] [PubMed] [Google Scholar]
- 2.Dick GW, Kitchen SF, Haddow AJ. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952;46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 3.Duffy MR, et al. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med. 2009;360:2536–2543. doi: 10.1056/NEJMoa0805715. [DOI] [PubMed] [Google Scholar]
- 4.Campos GS, Bandeira AC, Sardi SI. Zika virus outbreak, Bahia, Brazil. Emerg Infect Dis. 2015;21:1885–1886. doi: 10.3201/eid2110.150847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brasil P, et al. Zika virus outbreak in Rio de Janeiro, Brazil: Clinical characterization, epidemiological and virological aspects. PLoS Negl Trop Dis. 2016;10:e0004636. doi: 10.1371/journal.pntd.0004636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao-Lormeau V-M, et al. Guillain-Barré Syndrome outbreak associated with Zika virus infection in French Polynesia: A case-control study. Lancet. 2016;387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ventura CV, Maia M, Bravo-Filho V, Góis AL, Belfort R., Jr Zika virus in Brazil and macular atrophy in a child with microcephaly. Lancet. 2016;387:228. doi: 10.1016/S0140-6736(16)00006-4. [DOI] [PubMed] [Google Scholar]
- 8.Grischott F, Puhan M, Hatz C, Schlagenhauf P. Non-vector-borne transmission of Zika virus: A systematic review. Travel Med Infect Dis. 2016;14:313–330. doi: 10.1016/j.tmaid.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Hills SL, et al. Transmission of Zika virus through sexual contact with travelers to areas of ongoing transmission — continental United States, 2016. MMWR Morb Mortal Wkly Rep. 2016;65:215–216. doi: 10.15585/mmwr.mm6508e2. [DOI] [PubMed] [Google Scholar]
- 10.Davidson A, Slavinski S, Komoto K, Rakeman J, Weiss D. Suspected female-to-male sexual transmission of Zika virus — New York City, 2016. MMWR Morb Mortal Wkly Rep. 2016;65:716–717. doi: 10.15585/mmwr.mm6528e2. [DOI] [PubMed] [Google Scholar]
- 11.Meaney-Delman D, et al. Zika virus infection among U.S. pregnant travelers — August 2015–February 2016. MMWR Morb Mortal Wkly Rep. 2016;65:211–214. doi: 10.15585/mmwr.mm6508e1. [DOI] [PubMed] [Google Scholar]
- 12.Dupont-Rouzeyrol M, Biron A, O’Connor O, Huguon E, Descloux E. Infectious Zika viral particles in breastmilk. Lancet. 2016;387:1051. doi: 10.1016/S0140-6736(16)00624-3. [DOI] [PubMed] [Google Scholar]
- 13.Larocca RA, et al. Vaccine protection against Zika virus from Brazil. Nature. 2016;536:474–478. doi: 10.1038/nature18952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbink P, et al. Protective efficacy of multiple vaccine platforms against Zika virus challenge in rhesus monkeys. Science. 2016;353:1129–1132. doi: 10.1126/science.aah6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferguson NM, et al. Countering the Zika epidemic in Latin America. Science. 2016;353:353–354. doi: 10.1126/science.aag0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suy A, et al. Prolonged Zika virus viremia during pregnancy. N Engl J Med. 2016;375:2611–2613. doi: 10.1056/NEJMc1607580. [DOI] [PubMed] [Google Scholar]
- 17.Mansuy JM, et al. Zika virus in semen and spermatozoa. Lancet Infect Dis. 2016;16:1106–1107. doi: 10.1016/S1473-3099(16)30336-X. [DOI] [PubMed] [Google Scholar]
- 18.Barzon L, et al. Infection dynamics in a traveller with persistent shedding of Zika virus RNA in semen for six months after returning from Haiti to Italy, January 2016. Euro Surveill. 2016;21:30316. doi: 10.2807/1560-7917.ES.2016.21.32.30316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu M, et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med. 2016;22:1101–1107. doi: 10.1038/nm.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dudley DM, et al. A rhesus macaque model of Asian-lineage Zika virus infection. Nat Commun. 2016;7:12204. doi: 10.1038/ncomms12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osuna CE, et al. Zika viral dynamics and shedding in rhesus and cynomolgus macaques. Nat Med. 2016;22:1448–1455. doi: 10.1038/nm.4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aliota MT, et al. Heterologous protection against Asian Zika virus challenge in rhesus macaques. PLoS Negl Trop Dis. 2016;10:e0005168. doi: 10.1371/journal.pntd.0005168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aid M, et al. Zika virus persistence in the central nervous system and lymph nodes of rhesus monkeys. Cell. 2017;169:610–620.e14. doi: 10.1016/j.cell.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams Waldorf KM, et al. Fetal brain lesions after subcutaneous inoculation of Zika virus in a pregnant nonhuman primate. Nat Med. 2016;22:1256–1259. doi: 10.1038/nm.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ribeiro RM, et al. Estimation of the initial viral growth rate and basic reproductive number during acute HIV-1 infection. J Virol. 2010;84:6096–6102. doi: 10.1128/JVI.00127-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perelson AS. Modelling viral and immune system dynamics. Nat Rev Immunol. 2002;2:28–36. doi: 10.1038/nri700. [DOI] [PubMed] [Google Scholar]
- 27.Baccam P, Beauchemin C, Macken CA, Hayden FG, Perelson AS. Kinetics of influenza A virus infection in humans. J Virol. 2006;80:7590–7599. doi: 10.1128/JVI.01623-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neumann AU, et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282:103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 29.Banerjee S, Guedj J, Ribeiro RM, Moses M, Perelson AS. Estimating biologically relevant parameters under uncertainty for experimental within-host murine West Nile virus infection. J R Soc Interface. 2016;13:20160130. doi: 10.1098/rsif.2016.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clapham HE, Tricou V, Van Vinh Chau N, Simmons CP, Ferguson NM. Within-host viral dynamics of dengue serotype 1 infection. J R Soc Interface. 2014;11:6. doi: 10.1098/rsif.2014.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ben-Shachar R, Koelle K. Minimal within-host dengue models highlight the specific roles of the immune response in primary and secondary dengue infections. J R Soc Interface. 2015;12:20140886. doi: 10.1098/rsif.2014.0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clapham HE, et al. Modelling virus and antibody dynamics during dengue virus infection suggests a role for antibody in virus clearance. PLOS Comput Biol. 2016;12:e1004951. doi: 10.1371/journal.pcbi.1004951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenbloom DIS, Hill AL, Rabi SA, Siliciano RF, Nowak MA. Antiretroviral dynamics determines HIV evolution and predicts therapy outcome. Nat Med. 2012;18:1378–1385. doi: 10.1038/nm.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Louie M, et al. Determining the relative efficacy of highly active antiretroviral therapy. J Infect Dis. 2003;187:896–900. doi: 10.1086/368164. [DOI] [PubMed] [Google Scholar]
- 35.Perelson AS, Guedj J. Modelling hepatitis C therapy–predicting effects of treatment. Nat Rev Gastroenterol Hepatol. 2015;12:437–445. doi: 10.1038/nrgastro.2015.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guedj J, et al. Modeling viral kinetics and treatment outcome during alisporivir interferon-free treatment in hepatitis C virus genotype 2 and 3 patients. Hepatology. 2014;59:1706–1714. doi: 10.1002/hep.26989. [DOI] [PubMed] [Google Scholar]
- 37.Burnham KP, Anderson DR. Model Selection and Multimodel Inference. Springer; New York: 2002. [Google Scholar]
- 38.Stafford MA, et al. Modeling plasma virus concentration during primary HIV infection. J Theor Biol. 2000;203:285–301. doi: 10.1006/jtbi.2000.1076. [DOI] [PubMed] [Google Scholar]
- 39.Furuta Y, et al. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antiviral Res. 2013;100:446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madelain V, et al. Favipiravir pharmacokinetics in nonhuman primates and insights for future efficacy studies of hemorrhagic fever viruses. Antimicrob Agents Chemother. 2016;61:e01305–e01316. doi: 10.1128/AAC.01305-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hadjichrysanthou C, et al. Understanding the within-host dynamics of influenza A virus: From theory to clinical implications. J R Soc Interface. 2016;13:20160289. doi: 10.1098/rsif.2016.0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar A, et al. Zika virus inhibits type-I interferon production and downstream signaling. EMBO Rep. 2016;17:1766–1775. doi: 10.15252/embr.201642627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grant A, et al. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe. 2016;19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bowen JR, et al. Zika virus antagonizes type I interferon responses during infection of human dendritic cells. PLoS Pathog. 2017;13:e1006164. doi: 10.1371/journal.ppat.1006164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ajuebor MN, et al. Endogenous monocyte chemoattractant protein-1 recruits monocytes in the zymosan peritonitis model. J Leukoc Biol. 1998;63:108–116. doi: 10.1002/jlb.63.1.108. [DOI] [PubMed] [Google Scholar]
- 46.Hamel R, et al. Biology of Zika virus infection in human skin cells. J Virol. 2015;89:8880–8896. doi: 10.1128/JVI.00354-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Retallack H, et al. Zika virus cell tropism in the developing human brain and inhibition by azithromycin. Proc Natl Acad Sci USA. 2016;113:14408–14413. doi: 10.1073/pnas.1618029113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quicke KM, et al. Zika virus infects human placental macrophages. Cell Host Microbe. 2016;20:83–90. doi: 10.1016/j.chom.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.El Costa H, et al. ZIKA virus reveals broad tissue and cell tropism during the first trimester of pregnancy. Sci Rep. 2016;6:35296. doi: 10.1038/srep35296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang H, et al. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell. 2016;18:587–590. doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oliveira DBL, et al. Prolonged shedding of Zika virus associated with congenital infection. N Engl J Med. 2016;375:1202–1204. doi: 10.1056/NEJMc1607583. [DOI] [PubMed] [Google Scholar]
- 52.Lim S-Y, et al. 2016 BCX4430, a broad-spectrum adenosine analog direct-acting antiviral drug, abrogates viremia in rhesus macaques challenged with Zika virus. Available at https://idsa.confex.com/idsa/2016/webprogram/Paper60907.html.
- 53.Waggoner JJ, et al. Viremia and clinical presentation in Nicaraguan patients infected with Zika virus, chikungunya virus, and dengue virus. Clin Infect Dis. 2016;63:1584–1590. doi: 10.1093/cid/ciw589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lanciotti RS, et al. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis. 2008;14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Souza BSF, et al. Zika virus infection induces mitosis abnormalities and apoptotic cell death of human neural progenitor cells. Sci Rep. 2016;6:39775. doi: 10.1038/srep39775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barr KL, Anderson BD, Prakoso D, Long MT. Working with Zika and Usutu viruses in vitro. PLoS Negl Trop Dis. 2016;10:e0004931. doi: 10.1371/journal.pntd.0004931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith AM, Adler FR, Perelson AS. An accurate two-phase approximate solution to an acute viral infection model. J Math Biol. 2010;60:711–726. doi: 10.1007/s00285-009-0281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Canini L, Perelson AS. Viral kinetic modeling: State of the art. J Pharmacokinet Pharmacodyn. 2014;41:431–443. doi: 10.1007/s10928-014-9363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith AM, Perelson AS. Influenza A virus infection kinetics: Quantitative data and models. Wiley Interdiscip Rev Syst Biol Med. 2011;3:429–445. doi: 10.1002/wsbm.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raftery AE. Bayesian model selection in social research. Sociol Methodol. 1995;25:111–163. [Google Scholar]
- 61.Pinheiro J, Bates DM. Mixed-Effects Models in S and S-PLUS. Springer; New York: 2000. [Google Scholar]
- 62.Samson A, Lavielle M, Mentré F. Extension of the SAEM algorithm to left-censored data in nonlinear mixed-effects model: Application to HIV dynamics model. Comput Stat Data Anal. 2006;51:1562–1574. [Google Scholar]
- 63.Ascher D, Dubois PF, Hinsen K, Hugunin J, Oliphant T. 1999 Numerical Python. Available at www.lassp.cornell.edu/sethna/DM/Documentation/numpy.pdf.
- 64.Buckland ST, Burnham KP, Augustin NH. Model selection: An integral part of inference. Biometrics. 1997;53:603–618. [Google Scholar]
- 65.Mullen K, Ardia D, Gil D, Windover D, Cline J. DEoptim : An R Package for global optimization by differential evolution. J Stat Softw. 2011;40:1–17. [Google Scholar]
- 66.Sleeman K, et al. In vitro antiviral activity of favipiravir (T-705) against drug-resistant influenza and 2009 A(H1N1) viruses. Antimicrob Agents Chemother. 2010;54:2517–2524. doi: 10.1128/AAC.01739-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.