Significance
Because inaccurate chromosome segregation during meiosis is a leading cause of miscarriage in humans, we seek to understand how homologous chromosomes segregate properly. Meiotic chromosome segregation occurs with fidelity only in the presence of the synaptonemal complex (SC), a protein structure that assembles between homologs and facilitates the occurrence of crossing over. Although some functions of the SC are evolutionarily conserved, the mechanisms underlying its multiple roles during meiosis, as well as organizational variances among different organisms, remain under investigation. Here we combine superresolution and expansion microscopy and find strong evidence that the Drosophila SC comprises two visually distinct layers, perhaps suggesting that each layer connects one sister chromatid from each homologous chromosome.
Keywords: synaptonemal complex, expansion microscopy, meiosis, sister chromatids, structured illumination microscopy
Abstract
The synaptonemal complex (SC), a structure highly conserved from yeast to mammals, assembles between homologous chromosomes and is essential for accurate chromosome segregation at the first meiotic division. In Drosophila melanogaster, many SC components and their general positions within the complex have been dissected through a combination of genetic analyses, superresolution microscopy, and electron microscopy. Although these studies provide a 2D understanding of SC structure in Drosophila, the inability to optically resolve the minute distances between proteins in the complex has precluded its 3D characterization. A recently described technology termed expansion microscopy (ExM) uniformly increases the size of a biological sample, thereby circumventing the limits of optical resolution. By adapting the ExM protocol to render it compatible with structured illumination microscopy, we can examine the 3D organization of several known Drosophila SC components. These data provide evidence that two layers of SC are assembled. We further speculate that each SC layer may connect two nonsister chromatids, and present a 3D model of the Drosophila SC based on these findings.
The synaptonemal complex (SC) is a multiprotein complex assembled between homologous chromosomes during prophase I of meiosis that is critical for the successful execution of meiosis (reviewed in ref. 1). One of the hallmarks of meiotic prophase is the induction of programmed DNA double-strand breaks. The SC facilitates the repair of a subset of these breaks into crossovers. Crossing over creates a physical link between homologous chromosomes, which is required to correctly orient the homologs on the first meiotic spindle, thereby ensuring accurate chromosome segregation.
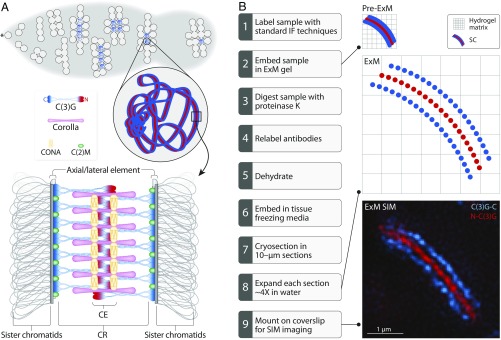
One of the challenges in studying the 3D organization of large, multiprotein structures is the limited resolution of conventional light microscopy. The SC is no exception. Although many SC components are known and have been positioned in two dimensions, a satisfying 3D understanding of the SC remains a major goal of meiotic biology. As shown in Fig. 1A, the SC consists of three parts: lateral elements (LEs), a central region (CR), and a central element (CE). LE proteins, also known as axial elements before the assembly of the CR, run down the length of each homolog in two parallel tracks. Assembly of the LE occurs alongside cohesin and cohesin-like proteins, both of which function in the maintenance of sister-chromatid cohesion and create the chromosome axis (2, 3). The CR includes the proteins that lie within the space between the homologs. Within the CR are the transverse filaments (TFs), which span the region between the two homologs, thereby attaching the LEs together. CE proteins are positioned in the middle of the complex and, together with other CR proteins, are thought to help stabilize the complex. Although the proteins that compose the SC are quite divergent among organisms, the general structure and function are well conserved, and the width between LEs has consistently been measured at ∼90–150 nm in all organisms (4).
Fig. 1.
Drosophila SC biology and expansion protocol. (A) At the anterior tip (asterisk) of the germarium, a cystoblast undergoes four incomplete mitotic divisions to produce a 16-cell interconnected cyst. Euchromatic SC assembly begins in these 16-cell cysts and quickly reaches full length in up to four nuclei within each cyst. At the posterior end of the germarium, only one nucleus in the cyst retains full-length SC. Previous studies demonstrated that C(2)M (green) localizes in the LE; C(3)G homodimers interact to span the CR of the SC, with the C termini of C(3)G (blue) localizing to the LEs and the N termini (red) localizing in the CE; Corolla (pink) lies in the CR; and CONA (yellow) localizes to the CE (reviewed in ref. 1). (B) To adapt ExM to SIM, samples must be prepared and sectioned as shown. The image shows the C (blue) and N (red) termini of C(3)G.
Although the overall 2D structure of the SC is fairly well characterized, the 3D organization—especially the subtle differences among organisms—is not well understood. Some of the most convincing evidence for a multilayered SC configuration has been obtained from the beetle Blaps cribrosa. This organism builds an organized structure of symmetrically arranged CE and TF proteins to form a lattice, several distinct layers of which are joined together via occasional fibrous bridges (5, 6). Mammals, particularly rat and mouse, also are thought to have a multilayered SC, although the structure is not as well defined as the lattice structure assembled in Blaps (5, 7, 8). In yeast, some SC components are added at a faster rate than they are turned over, suggesting that yeast also may have some layering of SC components (9, 10). Finally, in flies, electron microscopy (EM) serial reconstructions have suggested that the SC may have multiple layers (5, 11, 12), although attempts to elucidate its 3D structure have thus far been inconclusive.
The ability to combine genetic screens, immuno-EM analysis, and superresolution microscopy have made Drosophila a particularly useful model for the study of SC components and their 2D organization within the structure (Fig. 1A). The TF protein, C(3)G, forms a homodimer that interacts at its N-terminal end with the N terminus of another C(3)G homodimer. The C terminus of each C(3)G homodimer is positioned within the LE (13, 14), although how the C terminus interacts with the LE and other chromosome axis proteins is unclear. Immuno-EM analysis of the SC protein Corona (CONA) suggests that it runs as two parallel tracks on the outer edges of the CE (15). CONA interacts with a second TF-like protein known as Corolla, as shown by yeast two-hybrid analysis (16). Corolla localizes to the CR of the SC in a tract-like manner, but superresolution microscopy studies have been unable to distinguish among several possible models for a precise localization of Corolla within the CR (16). Immuno-EM analysis of the LE protein C(2)M positions it directly adjacent to the C terminus of C(3)G (14). Based on its interaction with the cohesin component SMC3, C(2)M is thought to be a kleisin subunit of the cohesin complex (17); however, a recent study suggests that C(2)M may function in interhomolog cohesion rather than in sister chromatid cohesion (18, 19).
Although recent work using superresolution technologies such as structured illumination microscopy (SIM) and stochastic optical reconstruction microscopy (STORM) has provided additional information about SC structure (8, 10, 20), we are nearing the limit of what even superresolution microscopy methods can reveal about the detailed structure of this crucial complex. One solution to this impasse is to increase the size of the complex itself. Until recently, this proposal perhaps seemed unattainable; however, the advent of a new method, termed expansion microscopy (ExM), has allowed researchers to do exactly this: to increase the size of a structure of interest, effectively allowing insight beyond superresolution (21–24).
To better understand the organization of the Drosophila SC components, we modified recent ExM protocols to render them compatible with SIM (ExM SIM) (21–24). Using this combination of methods, we enlarged the SC from a preexpansion LE spacing of ∼120 nm to a postexpansion LE spacing of ∼480 nm—an approximate fourfold expansion of the structure—and then used SIM to visualize it. The SC substructure as revealed by ExM SIM appears to form in two layers that are mirror images of one another. We can now position the known components of the Drosophila SC more accurately in 3D within the structure, which allows us to create a new model of the SC. In addition, we propose that each layer of SC might connect one sister chromatid from each homolog.
Results
Combining ExM with SIM.
Although the SC can be visualized using various superresolution microscopy techniques, these techniques have so far been unable to completely resolve the 3D localization of individual SC components (7, 16, 20). Using ExM methods, we can expand the SC in 3D, allowing us to more accurately position SC proteins. For example, ExM SIM imaging with a biological (unexpanded) xy resolution of 100 nm, applied to a fourfold-expanded sample, would effectively give a biological resolution of 25 nm, and, owing to the limits of optical physics, a biological resolution in the z dimension of 50–60 nm (25). However, a fourfold expansion presents a major challenge in performing conventional superresolution imaging, such as SIM (26). The distance between the expanded sample and the objective lens increases dramatically following expansion, making superresolution imaging nearly impossible. One way to address this issue is to section the expanded sample; however, when a sample contains a large amount of water, any histological sectioning process becomes extremely difficult. We exploited the ability of the ExM gel matrix to be contracted and expanded by dehydrating the digested samples in the gel, thereby making it possible to histologically section the sample (Fig. 1B) (22–24). Following this, the sections were expanded in water and mounted on coverslips, with the assumption that some of the expanded sections have SC sufficiently close to the coverslip surface to permit superresolution imaging. This modification made the expanded sample compatible with SIM imaging (Fig. 1 B and C).
Two recent studies have shown that the SC progressively becomes more stable throughout the pachytene stage of prophase I, likely through the posttranslational modification of SC proteins (27, 28). This gradual switch to a more stable SC could potentially alter the structure as it progresses through meiosis. So, for this study, we analyzed only nuclei that had full-length SC and were within the germarium (regions 2a–2b; Fig. 1A), which would correspond to the early pachytene to early–mid pachytene stages of prophase I. To prevent the analysis of the late prophase stages, we removed the later oocyte stages following the formaldehyde fixing of dissected ovaries (Materials and Methods).
By using ExM SIM on SC sections, we clearly separated the components of the SC within the structure (Fig. 2). The C(3)G C-terminal antibody, C(3)G-C, which effectively labels the LE–CR boundary, has two tracks ∼480 nm apart, indicating that we have expanded the SC by approximately four times its normal width of ∼120 nm (16). To visualize the LE, we used a transgene overexpressing a C-terminally HA-tagged version of C(2)M. Consistent with previous immuno-EM analysis, ExM SIM localizes C(2)M to the outer edge of the SC (14). As has been previously shown using other superresolution microscopy techniques, Corolla, CONA, and the N terminus of C(3)G [N-C(3)G] all lie within the CR of the SC (14–16). Their positions within the CR did not change following the fourfold expansion, demonstrating that this degree of expansion does not disrupt the relative positions of proteins within the SC.
Fig. 2.
ExM SIM images of approximately fourfold-expanded SC showing partial z projections of the SC-containing nuclei labeled for C(3)G-C (blue) and one of C(2)M (green), Corolla (pink), CONA (yellow), or N-C(3)G (red). Dashed boxes designate the region of the image shown in the zoomed-in view, arrows indicate regions in which splitting of the protein is observed in x, and arrowheads indicate regions in which splitting of the protein is observed in z. (Scale bars represent expanded distances.)
Visualization of a Dual-Layered SC.
The fourfold-expanded samples presented the opportunity to search for new structural insights into the SC. Throughout the expanded nuclei, we observed segments of SC that appeared flat (referred to as type 1 SC), in which the two tracks of C(3)G-C were easily observed (Fig. 2). In other regions, the SC appeared to be turning on its side (referred to as type 2 SC), which was observed in regions where the SC was bending to follow the chromosomes within the nucleus (Fig. 2). Turning and twisting morphologies of the SC have been observed before in many different organisms, and it has been hypothesized that the twisting may play a role in recombination through an as-yet unclear mechanism (3, 7, 20, 29). Although the turning morphologies that we observed do not appear to be regular or helical, it is possible that some of them contain levels of torsion that may be necessary for recombination.
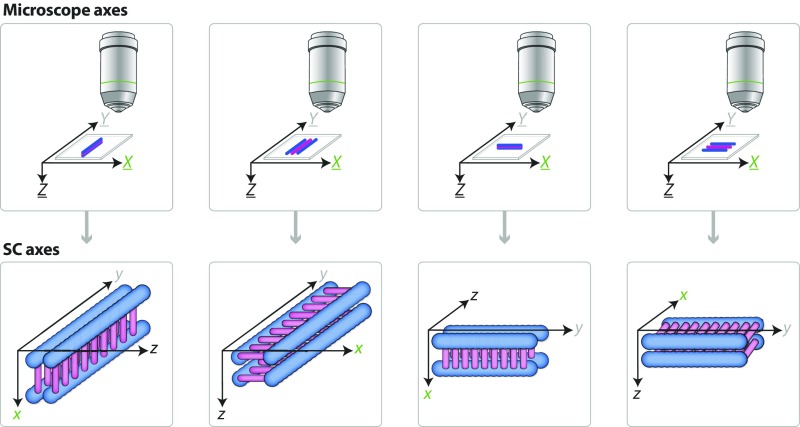
Throughout this paper, references to the SC dimensions are referred to as x, y, and z (in lowercase), with the x-axis representing the region from one LE to the other, the y-axis representing the SC length running parallel to the chromatin, and the z-axis representing the depth of the SC (Fig. S1). The microscope dimensions are referred to as X, Y, and Z (in uppercase), where the X- and Y-axes correspond to the width and length of the microscope stage, respectively, and the Z-axis represents the depth or the vertical movement the stage travels when acquiring a 3D stack image (Fig. S1). When the SC turns on its side, as in type 2 SC images, it effectively puts the z view of the SC into the XY plane of the microscope, which provides better resolution than can be obtained from the microscope’s Z view.
Fig. S1.
Diagram of the microscope and the SC axes. The X, Y, and Z axes of the microscope correspond to the width (X) and length (Y) of the stage and the vertical movement of the stage during 3D image stack acquisition (Z). The x, y, and z axes of the SC correspond to the width (x), length (y), and depth (z) of the SC. Diagrammed are the various rotations that the SC can undergo with the appropriate axes labeled on top of the SC. Note that the microscope dimensions never change position, only the SC dimensions change as the SC rotates. Blue represents the LE of the SC, and pink represents the CR of the SC.
Surprisingly, Corolla, CONA, and N-C(3)G all displayed two tracks when the z plane was captured, suggesting that the SC may have two layers (Fig. 2). To determine whether these two layers were simply an artifact of the expansion protocol, we examined a subset of the proteins using a different superresolution technique, stimulated emission depletion (STED), on unexpanded SC samples (Fig. S2). We observed two SC tracks via this method as well, verifying that the layers in the ExM SIM samples were real and not merely artifacts of the expansion process.
Fig. S2.
STED image of unexpanded SC labeling the C(3)G C termini (blue) and Corolla (pink). Regions a and b are two areas in which the SC has turned on its side, showing that Corolla is splitting in z. Dashed boxes designate the region of the image shown in the zoomed-in view, and arrowheads indicate regions where Corolla splitting is observed.
ExM SIM Reveals the 3D SC Organization.
To further characterize both the x and z profiles of the SC, we used ImageJ to trace segments of type 1 and type 2 SC (Fig. 3A). Using a custom ImageJ plugin, we straightened both types of segments along the y-axis to create a straightened 3D image of the traced SC segment. On this straightened 3D image, we projected along the y-axis over a manually selected uniform region to create an average xz profile of that region. For the type 2 SC fragments, we also rotated the average profile to position the x-axis at the bottom for simplified viewing (Materials and Methods).
Fig. 3.
Analysis of average x and z profiles of SC proteins. (A) Image analysis workflow. Segments were traced of either flat SC with two clearly observable C(3)G-C tracks (type 1 SC) or turned SC where two C(3)G-C tracks were no longer distinguishable (type 2 SC). In type 1 SC, the microscope axes (XYZ) match the SC axes (xyz); however, in type 2 SC, the SC is turned on its side, placing the SC x-axis along the Z-axis of the microscope and the SC z-axis into the XY microscope plane. Using ImageJ, segments were straightened in 3D along the y-axis of the SC and each slice was then projected along the y-axis to create the average z profile. For type 2 SC, images were rotated to position the x-axis of the SC on the bottom for ease of viewing. Line profiles were drawn on the averaged xz images along either the x-axis (type 1 SC) or the z-axis (type 2 SC), as shown. Then line profiles were averaged together to plot the average distribution of fluorescence intensity. (Scale bars: expanded distances, 250 nm.) (B and C) Representative averaged xz images for type 1 SC (B) and type 2 SC (C) labeled for C(3)G-C (blue), Corolla (pink), CONA (yellow), N-C(3)G (red), and C(2)M (green). The variation observed in these images reflects the distortion from the microscope Z-axis; for a perfect image, the SC must lie completely flat with its side to the microscope, but type 2 SC frequently turns and twists and thus shows more variability than type 1 images. (Scale bars: expanded distances, 250 nm.) (D and E) Multiple line profiles along the x-axis [D: N-C(3)G, n = 21 SC fragments from 8 nuclei; C(3)G-C, n = 21 SC fragments from 8 nuclei; Corolla, n = 7 SC fragments from 6 nuclei; CONA, n = 9 SC fragments from 7 nuclei; C(2)M, n = 9 SC fragments from 4 nuclei] or z-axis [E: N-C(3)G, n = 15 SC fragments from 8 nuclei; C(3)G-C, n = 10 SC fragments from 8 nuclei; Corolla, n = 12 SC fragments from 6 nuclei; CONA, n = 12 SC fragments from 7 nuclei; C(2)M, n = 15 SC fragments from 4 nuclei] were averaged together and then mirrored to generate the distribution of the SC components along the axes. Error bars indicate SE. For both distributions, an expansion factor correction was applied (Materials and Methods) to determine the approximate unexpanded distances in nm. (F) Modeled positions of C(3)G-C (blue), Corolla (pink), CONA (yellow), N-C(3)G (red), and C(2)M (green) based on the line profiles in D and E. (Scale bar: biological distance, 50 nm.)
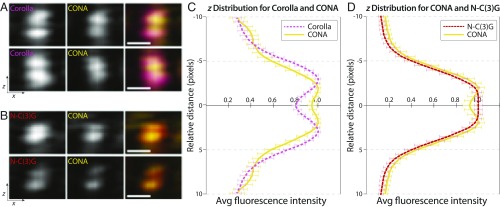
It is clear from the averaged xz view of type 1 SC that Corolla, CONA, and N-C(3)G are positioned in the middle of the SC between the two C(3)G-C spots (Fig. 3B), as described previously (14–16). The C(2)M spots are shifted slightly outside of the two C(3)G-C spots, further supporting previous immuno-EM analysis indicating that C(2)M is positioned slightly adjacent to C(3)G (14). For type 2 SC, the averaged xz images showed that Corolla, CONA, and N-C(3)G could each be resolved as two spots separated in z (Fig. 3C), supporting our hypothesis that the SC comprises two layers. Although it is possible that averaging may falsely display the appearance of two spots if the SC twists back and forth along the dimension being averaged, we were able to observe two tracks of Corolla, CONA, and N-C(3)G along the y-axis of the straightened type 2 image before averaging. For this reason, we are confident that averaging along the y-axis is not erroneously giving the appearance of two spots.
For C(3)G-C and C(2)M, type 2 xz images displayed a range of spots from two to four, with three spots most commonly observed (Fig. 3C). One way to explain this variability is that type 2 SC segments must be perfectly flat in the XY plane of the microscope when the z plane of the SC is displayed to allow for the resolution of all four spots of each protein. Even at fourfold expansion, we are nearly at the resolution limit of the microscope; thus, if the SC is off-axis by even a small amount, the fluorescence from C(3)G-C or C(2)M will begin to overlap, resulting in the observation of two or three spots in the averaged xz images.
To more accurately evaluate the positions of SC components relative to one another, we drew line profiles on each averaged xz image of both type 1 SC (to obtain average x profiles) and type 2 SC (to obtain average z profiles) (Fig. 3A). Each line profile consisted of a straight line through the middle of the spots along the x-axis (type 1) or z-axis (type 2). Because all four proteins exhibited mild variability in xz images in type 1 and type 2 SC, we averaged the line profiles of similar groups of images together to obtain a more accurate distribution of each protein.
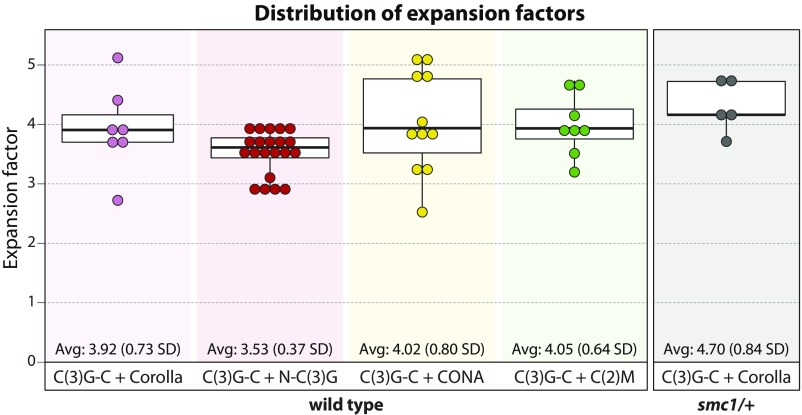
Any type 2 xz image that did not display two spots in z was excluded from this analysis. Corolla, CONA, and N-C(3)G all displayed two spots in z, and thus no images were excluded. For C(2)M and C(3)G-C, one or two lines were drawn depending on whether two or four spots were apparent. To report the distances for the line profiles on a biological scale, we calculated an expansion factor for each image based on the previously reported SIM SC width of 120 nm between the two C(3)G-C tracks in x (16) (Fig. S3 and Materials and Methods).
Fig. S3.
Boxplots displaying the expansion factor values for each dataset analyzed in Fig. 3 and Fig. S4. Average expansion factors and SDs are shown at the bottom of each plot.
Upon quantification, we found that in the average x profiles of type 1 SC, Corolla occupied a much wider area of the CR than either CONA or N-C(3)G (Fig. 3D and Table S1). The average z profile analysis of type 2 SC indicated that Corolla also lay outside of CONA and N-C(3)G in z (Fig. 3E and Table S1) (P = 0.034 and 8.7 × 10−5, respectively). In addition, CONA and N-C(3)G in both type 1 and type 2 SC appeared to be in nearly the same position in both x and z (Fig. 3 D and E and Table S1). In x, CONA and N-C(3)G lay directly in the center of the SC. Although CONA showed a slightly larger x distribution than N-C(3)G, the difference between these distributions was not statistically significant (P = 0.10) (Fig. 3C and Table S1). In z, both CONA and N-C(3)G displayed two peaks, matching Corolla’s distribution in z (Fig. 3E). Thus, in the xz orientation, the two SC layers appeared as mirror images, with Corolla outside of the other CE components on each SC, in both x and z (Fig. 3E).
Table S1.
Average biological distances between two tracks of an SC protein in x and/or z and the FWHM in x and z for the distributions in Fig. 3 D and E
| Protein | Average distance between tracks in x, nm | FWHM in x, nm | Average distance between tracks in z, nm | FWHM in z, nm |
| C(2)M | 141.5 ± 0.7 | 65.6 ± 1.0 | 65.2 ± 1.5 | 56.5 ± 1.4 |
| C(3)G-C | 120 | 46.7 ± 0.5 | 55.7 ± 3.4 | 60.2 ± 2.5 |
| Corolla | NA | 76.7 ± 2.0 | 63.1 ± 0.7 | 50.3 ± 0.8 |
| CONA | NA | 46.7 ± 1.2 | 56.2 ± 3.7 | 56.8 ± 3.0 |
| N-C(3)G | NA | 38.3 ± 0.3 | 49.7 ± 3.4 | 52.7 ± 2.3 |
| Corolla (smc1/+) | NA | ND | 70.4 ± 0.4 | 47.3 ± 0.5 |
Data are mean ± SE. NA, not applicable; ND, not determined.
The C(3)G-C x distribution was consistent with previous descriptions of C(3)G-C localization in xy (14) (Fig. 3D). Specifically, the average x profile positioned C(3)G-C on opposite ends of the SC, adjacent to the LE protein C(2)M, where it has been shown to localize by EM (14). C(2)M was shifted slightly outside of C(3)G-C on either side of the SC in x by ∼21.5 nm (P < 0.001) (Fig. 3D and Table S1), consistent with previous localization data (14). Given the variation between individual C(3)G-C and C(2)M profiles in type 2 xz images, we centered and averaged a large number of the line profiles together, as we did with CONA, Corolla, and N-C(3)G (Materials and Methods). Again, the out-of-focus (presumably third and fourth) peaks observed in the images should average out, leaving the true z profile. This profile indeed shows two distinct peaks separated in z for both C(3)G-C and C(2)M (Fig. 3E). Surprisingly, similar to what was observed in the x distribution, C(2)M also was positioned outside of C(3)G-C in z by ∼9.6 nm (P = 0.006) (Table S1). These results suggest that, like the CR, the LE also forms two layers.
Based on these findings, we hypothesized that the four C(3)G-C and C(2)M spots (two for each LE) might well represent the presence of two stacked SCs, each connecting two nonsister chromatids, or one sister from each homolog. If this were the case, then moving sister chromatids farther away from each other in z would also increase the z distance between the two SC layers. We postulated that decreasing the amount of the cohesin component SMC1 by one-half might result in a “looser” axis, causing the sister chromatids to be positioned slightly farther away from each other. Because homozygotes for this deficiency are lethal, we used SMC1 deficiency heterozygotes (smc1/+) for these experiments. By SIM, smc1/+ assembled full-length SC that appeared indistinguishable from wild type (WT) in xy (Fig. S4A). However, further examination by ExM SIM revealed that in z, the distance between Corolla foci was increased significantly, by ∼10.9 nm, compared with WT (P < 0.001) (Fig. S4B and Table S1).
Fig. S4.
ExM SIM in an SMC1 deficiency heterozygote (smc1/+). (A) SIM images of WT SC (Top) and smc1/+ SC (Bottom) labeling the C(3)G C termini (blue) and Corolla (pink). (B) The average distribution along the z-axis from ExM SIM images of WT and smc1/+. The analysis was performed as described in Fig. 3. WT: n = 12 SC fragments from six nuclei; smc1/+: n = 12 SC fragments from four nuclei.
Considering all of the foregoing results together, we modeled the relative positions of the SC components in x and z (Fig. 3F). This representation shows two layers of SC, separated by a gap of 55.7 nm (SE 3.4). We further propose, that each SC layer connects two nonsister chromatids, or one sister chromatid from each homolog (Discussion). These SC-connected chromatids are referred to as homologous sister pairs hereinafter.
Ultrastructural Examination of Individual CR SC Components.
When calculating the expansion factor, any uncertainty in this calibration adds uncertainty to the comparison of distances between these proteins; slight differences in the expansion factor, and our ability to determine it, can affect the distance reported. Consequently, we verified the orientation of proteins in the z dimension of the SC by directly comparing Corolla and CONA or CONA and N-C(3)G in two-color samples (Fig. 4 A and B and Materials and Methods). Although this allowed direct comparison, these data were considered without a calibration for expansion factor; thus, we present it with dimensionless units on the length axis. Importantly, these comparisons further illustrate that the Corolla distribution is 32% wider than CONA and again position Corolla on the outer edge of CONA in z (P = 3.7 × 10−7) (Fig. 4C). Corolla (554 aa) is a larger protein than CONA (207 aa); therefore, it is not surprising that Corolla displays a wider distribution than CONA in x. CONA was spaced 7.5% wider than N-C(3)G in z, consistent in orientation with Fig. 3, but once again, this difference was not significant (P = 0.085) (Fig. 4D).
Fig. 4.
Relative positions of CONA, Corolla, and N-C(3)G. (A) Representative averaged xz images of Corolla (pink) and CONA (yellow). (B) Representative averaged xz images of N-C(3)G (red) and CONA (yellow). (C) Multiple line profiles along the z-axis were averaged together and then mirrored to generate the z distribution of Corolla (n = 10 SC fragments from 3 nuclei) and CONA (n = 9 SC fragments from 3 nuclei). (D) Multiple line profiles along the z-axis were averaged together and then mirrored to generate the z distribution of CONA (n = 11 SC fragments from 5 nuclei) and N-C(3)G (n = 10 SC fragments from 5 nuclei). An expansion factor correction could not be applied to these samples (Materials and Methods); therefore, the relative distance is in pixel units. Error bars indicate SE. (Scale bars: expanded distances, 250 nm.)
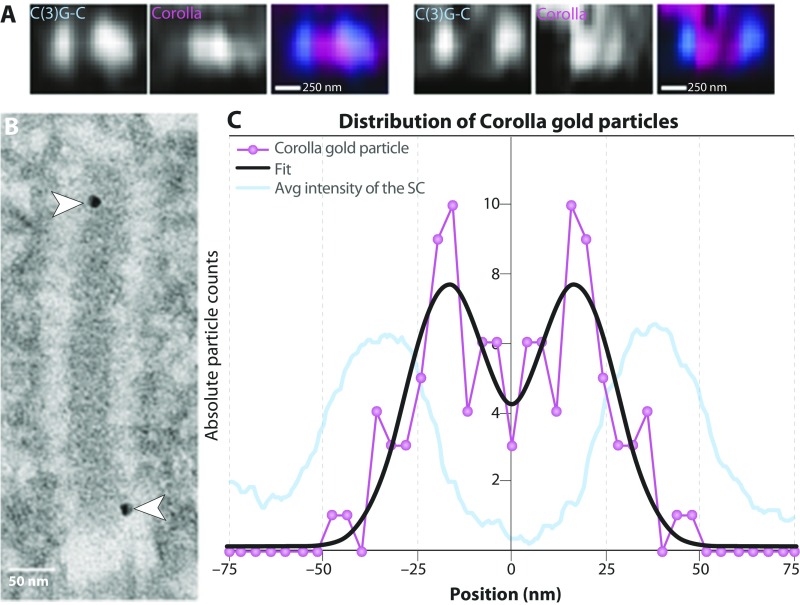
In addition, a previous SIM study suggested that, based on its width, the CR protein Corolla may exist as two tracks in the xy plane (16). In fact, although it was not evident in the average x profile of Corolla in the present study (Fig. 3D), several individual datasets did display two Corolla tracks within the CR in x (Fig. S5A). To further investigate this, we used immuno-EM to localize Corolla more precisely within the SC (Fig. S5B). The immuno-EM distribution of gold particles indicated that Corolla is indeed two tracks centered 34.3 nm (SE 1.4) apart, with each broad Corolla track 25.0 nm (SE 1.8) wide (Fig. S5C). Furthermore, some of the Corolla gold particles localized within the electron-dense CE of the SC (Fig. S5B). This localization into the CE may indicate the region of Corolla that is interacting with CONA; however, more experiments are needed to map this interaction.
Fig. S5.
Analysis of the distribution of Corolla in xy. (A) Averaged xz images of C(3)G C termini (blue) and Corolla (pink) from type 1 SC. (Scale bars represent expanded distances.) (B) Immuno-EM image of Corolla labeled with gold particles. Arrowheads mark the positions of the gold particles. (C) Histogram of the distribution of Corolla gold particles (n = 51) based on immuno-EM. Overlaid on the histogram is the average intensity of the SC to illustrate the locations of the LEs in relation to the position of the Corolla gold particles. This average intensity is determined by drawing a line profile across the SC in the EM images and then measuring the intensity of the image across that line. These intensity measurements are then averaged together to give the average intensity of the SC.
For CONA, a previous immuno-EM localization study used an anti-GFP antibody to an overexpressed C-terminal CONA-Venus construct to localize CONA as two parallel tracks running adjacent to the CE (15). Attempts to use ExM SIM to visualize an anti-GFP antibody to the CONA-Venus tag also revealed two tracks (Fig. S6). Although this construct is fully functional (30), the Venus tag is restricted to the C-terminal portion of the protein. Thus, this localization may represent only the approximate position of the C terminus of CONA and not necessarily the position of other domains of CONA within the SC.
Fig. S6.
ExM SIM of CONA-Venus. (A and B) Representative partial z projection images of type 1 SC segments from flies expressing CONA-Venus under the Nanos Gal4 driver with C(3)G C termini (blue) and CONA-Venus (yellow). Arrowheads indicate regions were CONA-Venus is splitting in xy. (A′) Averaged xz images of C(3)G C termini (blue) and CONA-Venus (yellow) from the type 1 SC segment in A. (Scale bar: expanded distance, 250 nm.) (B′) Averaged xz images of C(3)G C termini (blue) and CONA-Venus (yellow) from the type 1 SC segment in B. (Scale bar: expanded distance, 250 nm.)
For this reason, in the present study, we used the native CONA protein at endogenous levels with a polyclonal antibody made to the whole CONA protein to determine the localization of CONA. In contrast with the previous EM study, our present experiments showed a single track of CONA in x (Fig. 3 B and D). The contrast between the CONA-Venus data and the polyclonal antibody data suggests that the polyclonal antibody may recognize more epitopes along CONA than the C-terminal region. Thus, CONA may be positioned as two tracks in the SC, with the C-terminal end at the edge of the CE and the rest of the protein extending into the CE. In this arrangement, CONA would be visualized as one track if the two CONA proteins either are too close to resolve the distance between them or are touching in the center of the CE. We are currently unable to distinguish between these two possibilities.
Finally, Anderson et al. (14) suggested that the N-terminal ends of C(3)G may interact and overlap either head-to-head or side-by-side in the middle of the SC. This overlap was calculated to be ∼25 nm. However, following the fourfold expansion, we were still unable to resolve two peaks for N-C(3)G along the x-axis (Fig. 3D). Therefore, we were unable to differentiate between the possible ways in which the N termini of C(3)G interact.
Discussion
Since it was first described in the 1950s, the structure of the SC has been studied in many different organisms (4, 31). Decades of research have revealed extensive amino acid sequence diversity among the structural proteins that compose the SC and substantial differences in the number of proteins required for assembling the structure. However, there appears to be strong conservation of the overall structure of the SC, as well as its essential function in chromosome segregation during meiosis. The diversity found within the structural components of the SC presents a challenge for determining its organization. Not only are the proteins involved difficult to identify using an amino acid conservation approach, but there appear to be many ways to assemble the SC (reviewed in ref. 1). Some organisms, such as mammals, yeast, and flies, use pairs of TF homodimers [SYCP1, Zip1, and C(3)G, respectively] to span the CR of the SC, whereas worms use multiple SC components (SYP1–4) staggered across the CR (13, 32–34). In addition, multiple SC components are thought to make up the CE in both mice (SYCE1–3, TEX12) and yeast (Ecm11, Gmc2), whereas only one CE protein (CONA) has been identified in flies (30, 35–39).
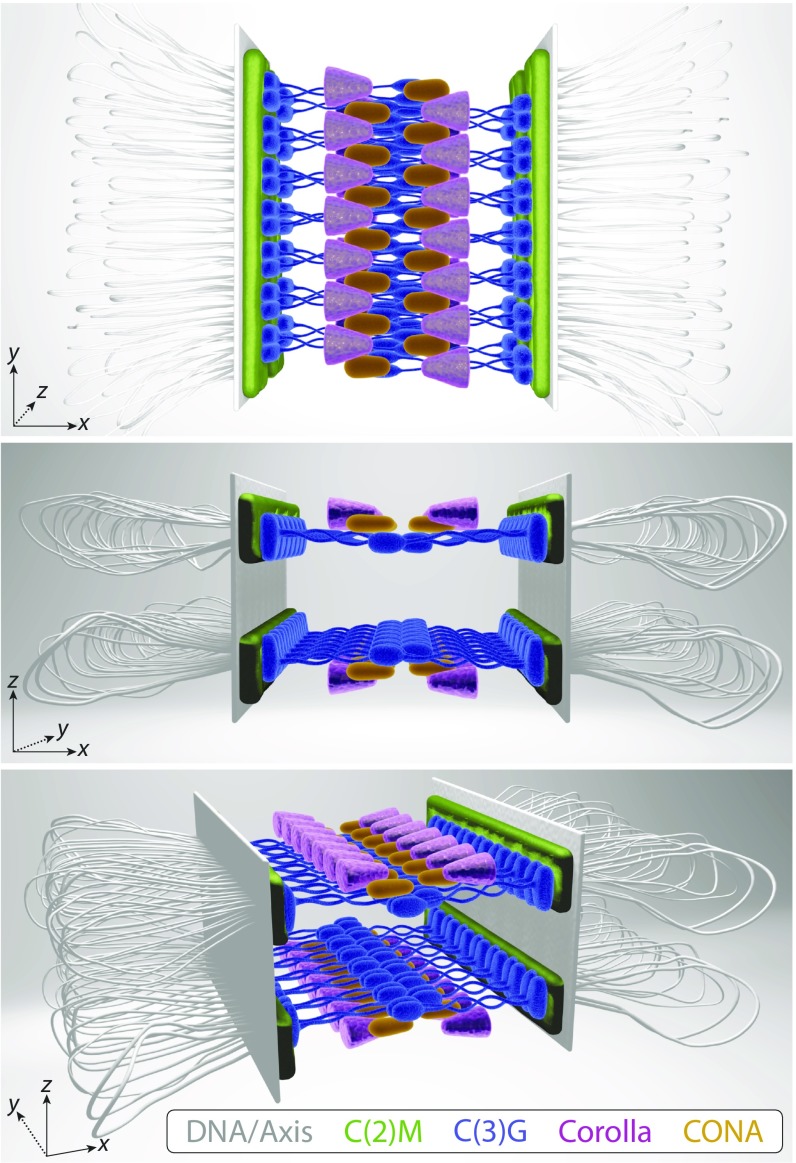
Although the 2D structure of the SC is well established in flies, only recent advancements in superresolution microscopy have allowed for the 3D study of SC organization. Using ExM SIM allowed us to achieve an effective resolution of ∼25 nm in XY and ∼60 nm in Z. By merging the new data obtained by ExM SIM from this study with previous data, we are better able to position the Drosophila SC components relative to one another and present an updated SC model (Fig. 5 and Movie S1) (13–16, 30). These combined data suggest that there are two layers of SC, each assembled between homologous sister pairs. The two layers are mirror images of each other, with Corolla located above and below the CR. Furthermore, the LE protein C(2)M is adjacent to the C termini of C(3)G and is positioned slightly above and below the C-terminal ends of C(3)G in the mirrored SC layers. As represented in our model, Corolla forms two parallel tracks in xy that extend in toward the edges of the CE. CONA also forms two parallel tracks, which likely extend farther into the CE than previously thought, perhaps even touching (15). Because Corolla and CONA are known to interact (16), it is possible that CONA might localize between Corolla and the C(3)G N termini; however, future experiments are needed to confirm this localization. We have drawn the Corolla and CONA interaction as a zipper-tooth pattern within the CR, but it is also possible that the proteins face each other in a mirror image pattern; there is no evidence to distinguish between these options. Both CONA and the C(3)G N termini appear to occupy the same region of the CE, and thus we suspect that they may directly or indirectly interact, but again, further studies are needed to investigate this.
Fig. 5.
A 3D model of the Drosophila SC showing two mirrored SC layers in z, each connecting one sister chromatid of each homologous chromosome. C(2)M (green), Corolla (pink), and CONA (yellow) assemble in two tracks in xy, and C(3)G (blue) spans the distance between homologs. Although Corolla and CONA are known to interact and C(2)M is suspected to interact with the C termini of C(3)G, these interactions have yet to be mapped. Chromosome axis proteins (gray) were not directly examined in this study. [Illustration by Ryan Kramer (artist).]
The observation of SC between homologous sister pairs has been documented previously in many ultrastructural analyses of both sectioned and spread SCs (reviewed in ref. 3). A study in barley demonstrated that the TF protein ZYP1 can be visualized in a vertically stacked, two-layer configuration when the SC is captured in a lateral or cross-sectional view using SIM (40). An EM study of pigeon spermatocytes observed in a few cases splitting of the two sister chromatid cores encased in a substance of the lateral component (LE) (41). In addition, more compelling evidence for splitting of sister chromatid axes was observed in the EM spreads of hamster spermatocytes where the sister chromatids assemble dual subaxes that run parallel to the SC axis (42). This splitting has been explained as being the result of a predetermined transition stage during prophase I, a meiotic mutant phenotype, or an artifact of fixation or preparation of the chromosomes (reviewed in ref. 3). However, our observations suggest that this splitting may be indicative of two discrete SCs connecting homologous sister pairs. Indeed, a recent superresolution study of the axis and LE proteins in Caenorhabditis elegans proposed a model wherein the axis components are layered and mirrored with respect to one another, such that one complex of proteins is bound to each sister chromatid (43). Although we did not directly analyze axis proteins in the present study, our proposed model of the Drosophila SC corresponds well with the model of C. elegans axis structure. Thus, although the proteins involved in assembling the SC of both Drosophila and worms contain high amino acid sequence diversity, the overall structure of the SC might be extremely similar.
The observation of two SC layers raises the possibility of “linker” proteins that might span the two SC layers. An EM study in Drosophila showed that the CE exhibits a three-layered structure when viewed laterally (12), suggesting that two individual chromatid subaxes may be separated by an interlayer region (3). Although we did not observe three layers, it is likely that as-yet unidentified proteins are involved in assembling the SC. It is highly possible that these unknown proteins make up this previously observed third layer of the CE, and perhaps this layer provides a connection between the two SC layers that we observed. The search continues to uncover the full complement of SC and SC-associated proteins. Only then can we fully understand the architecture and mechanics of this essential protein structure.
Materials and Methods
Drosophila Stocks and Reagents.
All Drosophila stocks were maintained at 25 °C on standard food. The WT stock used for all assays was y w; pol. Other stocks used were Pnos-Gal4::VP16 [on X and 3] (44), conaA12/TM3 (30), UASp-CONAVenus; conaf04903/TM3 (30), UASp-C(2)M-HAT40 (14), and w1118; Df(3R)Exel6197/TM6B (Bloomington stock no. 7676).
Primary antibodies were as follows: mouse monoclonal anti-C(3)G C-terminal [1A8–1G2, 5G4–1F1, and 1G5–2F7, generated from C(3)G peptide containing residues 565–743; a mixture of all three was used with each at 1:500 dilution] (14), rabbit polyclonal anti-C(3)G C-terminal [from Mary Lilly, generated from C(3)G peptide containing residues 442–743, used at 1:3,000) (45), rabbit polyclonal anti-C(3)G N-terminal [HL4287, from Mary Lilly, generated from C(3)G peptide containing residues 1–135, used at 1:3,000] (14), affinity-purified rabbit polyclonal anti-Corolla (used at 1:2,000) (16), guinea pig polyclonal anti-CONA (used at 1:500) (30), rabbit anti-GFP (Abcam; AB6556, used at 1:1,000) and rat anti-HA clone 3F10 (Sigma-Aldrich; 11867423001, used at 1:100). The following conjugated secondary antibodies were purchased from Thermo Fisher Scientific and used at 1:500: goat anti-mouse Alexa Fluor 488 (A-11001), goat anti-mouse Alexa Fluor 555 (A-21424), goat anti-rabbit Alexa Fluor 488 (A-11008), goat anti-rabbit Alexa Fluor 555 (A-21429), goat anti-rabbit Alexa Fluor 594 (A-11037), and goat anti-guinea pig Alexa Fluor 488 (A-11073). Anti-mouse Atto 647N secondary antibody (50185-1ML-F) was purchased from Sigma-Aldrich and used at 1:500. Background buster buffer (NB306) was purchased from Innovex Biosciences, and antibody diluent reagent solution (003118) was purchased from Thermo Fisher Scientific.
Paraformaldehyde (16%) was obtained from Electron Microscopy Sciences. Acryloyl-X, SE (AcX, A-20770), tetramethylethylenediamine (TEMED, 17919), and ammonium persulfate (APS, 17874) were purchased from Thermo Fisher Scientific. DAPI (9542), guanidine hydrochloride (G3272), methacrylic acid N-hydroxysuccinimide ester (MA-NHS, 730300), N,N′-methylenebisacrylamide (146072), 4-hydroxy-2,2,6,6-tetramethylpiperidine 1-oxyl (4-hydroxy-TEMPO, 97%, 176141), sodium acrylate (97%, 408220), anhydrous DMSO, sodium bicarbonate, EDTA, magnesium chloride, Triton X-100, and sodium borohydride were obtained from Sigma-Aldrich. The 40% acrylamide (1610140) and 2% bis-acrylamide (1610142) solutions were purchased from Bio-Rad. Proteinase K (P8107S) was purchased from New England BioLabs.
Immunohistochemistry.
Whole-mount Drosophila ovaries were prepared as described previously (46), with minor modifications. Following washings after a 2-h incubation with secondary antibody, the germarium tip tissues were incubated for at least 6 h in freshly made 0.1 mg/mL AcX in PBS at room temperature (RT), and then washed three times for 10 min each in PBS before proceeding with the ExM protocol (see below). For STED imaging, the sample was not expanded, and the ovaries were prepared as described previously (46), except that DAPI not was added before mounting in ProLong Gold (P36930; Thermo Fisher Scientific). In addition, to improve the quality of the STED images, only Alexa Fluor 594 and Atto 647N secondary antibodies were used in these samples. For immuno-EM, samples were prepared and imaged as described previously (46), with minor modifications. The secondary antibodies used were goat anti-mouse Alexa Fluor 488 and the F(ab′) 2 fragment of goat anti-rabbit IgG UltraSmall Gold (Electron Microscopy Sciences; 25360, used at 1:50).
ExM and Postdigestion Immunofluorescence.
The processes for tissue gelation, digestion, and expansion were similar to those reported previously (21–23, 47) (“proExM Protocol for Tissues” at expansionmicroscopy.org). In brief, before gelation, germarium tips were incubated in monomer with inhibitor reagent [1× PBS, 2 M NaCl, 2.5% (wt/wt) acrylamide, 0.15% (wt/wt) N,N′-methylenebisacrylamide, 8.625% (wt/wt) sodium acrylate, and 4-hydroxy-TEMPO 0.01% (wt/wt)] for 30 min at 4 °C. The tissue was transferred into the cap of a 500-µL Eppendorf tube, and as much of the excess monomer solution was removed as possible, with care taken to not let the tissue dry out. New monomer solution with APS and TEMED (referred to as gelling solution) at concentrations of 0.2% (wt/wt) was added for gelation, and the tissue was incubated at 4 °C for 10 min. Then fresh gelling solution was added, and the tissue was incubated at 4 °C for another 20 min, then at 37 °C for 2 h. Once gelation was complete, freshly made digestion buffer (1× TAE buffer, 0.5% Triton X-100, and 0.8 M guanidine HCl) containing 8 U/mL proteinase K was added. The gel block was twisted off the tube wall and digested for 4 h at 37 °C.
After the digestion, we performed a postdigestion immunofluorescence step. We found that in samples in which we did not perform the second antibody labeling following the digestion, the fluorescence signal was either too weak to allow for SIM or was completely absent (Fig. S7A). When we added only primary antibodies before digestion and added secondary antibodies postdigestion, we observed only very weak fluorescent staining (Fig. S7B), and when we added both primary and secondary antibodies, only postdigestion showed a complete absence of detectable fluorescent signal (Fig. S7C). We speculate that the protease digestion step may destroy some of the predigestion antibody complexes. Presumably, predigestion antibody labeling protects the interacting residues on the target protein from the protease digestion, and the postdigestion antibody labeling allows for those residues to be rebound by the antibody complex, thereby increasing the fluorescence signal (Fig. S7D). Thus, a combination of predigestion and postdigestion antibody labeling was used for all samples.
Fig. S7.
Spinning-disk confocal images of expanded SC in which antibodies were added predigestion and/or postdigestion. All of the antibody conditions were imaged on the same day with the same exposure time and laser power, illustrating that the predigestion and postdigestion labeling has the strongest fluorescent signal. The DNA stain DAPI was used to determine the location of the nuclei within each image. (Scale bars: 5 µm.) (A) Only predigestion labeling of C(3)G-C and Corolla with primary and secondary antibodies. (B) Predigestion labeling of C(3)G-C and Corolla with primary antibodies and postdigestion labeling of secondary antibodies. (C) Only postdigestion labeling of C(3)G-C and Corolla with primary and secondary antibodies. (D) Predigestion and postdigestion labeling of C(3)G-C and Corolla with primary and secondary antibodies.
For postdigestion immunofluorescence, the gels were rinsed well in PBS with 0.1% Tween (PBST) after removal from digestion buffer. Then the primary antibody, diluted in PBST, was incubated with the gels overnight at 4 °C. After three washes of 20 min each in PBST, secondary antibodies, diluted in PBST, were added for 1 h at RT, followed by six 20-min washes in PBST.
The postdigestion, labeled gels were dehydrated by washing with 30% sucrose three times for 15 min each. The gel block was then placed in 30% sucrose solution and allowed to sink to the bottom of the tube. (This step can be done overnight at RT.) Once sunk to the bottom, the gel block loses enough water to the surrounding solution to allow it to be embedded in tissue-freezing media (TFM). The dehydrated gel block was transferred to a Petri dish with TFM to remove the excess sucrose solution from the surface of the gel block. Immediately following the wash, the block was transferred to a mold with fresh TFM and embedded at −70 °C.
The tissue blocks were sectioned at 10-μm thickness using a CryoStar NX70 Cryostat (Thermo Fisher Scientific; objective temperature −41 to −44 °C and blade temperature −35 °C) and transferred to pure water for at least 5 min to expand the sample fourfold. The fully expanded cryosections were picked up with a #1.5 coverslip and mounted on glass slides in water for SIM imaging. The Alexa Fluor 488, Alexa Fluor 555, and Atto 647N secondary antibodies worked well with the protocol, but the Alexa Fluor 647 and Alex Fluor 633 secondary antibodies did not. We stored the Atto 647N-labeled samples for <1 d before imaging; however, the Alexa Fluor 488- and Alexa Fluor 555-labeled samples can be stored in a humidifier chamber with pure water at 4 °C for several weeks without much loss of fluorescent signal.
Microscopy and Image Analysis.
The testing of predigestion and postdigestion antibody staining was done using a PerkinElmer Spinning Disk Confocal System with a C-Apochromat 40×/1.2 W Corr M27 objective. Alexa Fluor 488, Alexa Fluor 555 and DAPI were excited by 488-nm, 561-nm, and 405-nm lasers, respectively. These images were obtained at the same laser power (488 nm at 40.5%, 561 nm at 40.5%, and 405 nm at 80%) and exposure time (488 nm for 200 ms, 561 nm for 200 ms, and 405 nm for 200 ms) to be able to observe the changes in antibody labeling with each condition.
STED images were acquired with a Leica SP8 confocal microscope system equipped with a STED lens (HC PL APO 93×/1.30 GLY objective). Excitation of Alexa Fluor 594 was performed with a pulsed tunable laser set at 594 nm, and excitation of Atto 647N was performed at 647 nm. The depletion laser for both colors was a STED 775-nm pulsed laser. All images were deconvolved with Huygens professional software.
SIM acquisition was performed similarly as described previously (16). All SIM images were acquired with an Applied Precision OMX Blaze (GE Healthcare). A 60× 1.42 numerical aperture Plan Apo oil objective was used, and emission photons were collected by PCO Edge sCMOS cameras, each dedicated to one specific channel. Color alignment of xy direction was performed using the color alignment slide provided by GE Healthcare, and z direction was calibrated by 100-nm TetraSpeck beads (Life Technologies). SIM images were reconstructed using softWoRx software (GE Healthcare) with a Wiener filter of 0.001. To minimize spherical aberration due to a large focal plane depth (48), different immersion oils with a refractive index (RI) of 1.524–1.530 were chosen to optimize SIM image quality. The RI value was calculated and determined using the lens information tool in softWoRx 6.5.2 (GE Healthcare).
A full tutorial on image alignment and averaging of superresolution data, both for the SC and the yeast spindle pole body (49), is available online (research.stowers.org/imagejplugins/spasim.html). In brief, following acquisition of SIM data, images were aligned manually based on the previously known 2D positions of the SC proteins (14, 16, 30), regions of SC were traced in 3D in ImageJ (Fig. 3), and spots along the region were saved in the ROI manager. The plugin “roi 2 traj3D jru v1” was used to generate a line profile. This profile was straightened along the y-axis using “thick 3D polyline profile jru v1” and then averaged over z for a manually selected well-straightened region (Fig. 3).
For alignment and averaging, line profiles were generated along x or z using a line average over 4 pixels. Because both C(3)G-C and C(2)M displayed a range of two to four spots in each type 2 xz image, line profiles were drawn only on the sides of the SC that displayed two spots. If either side of C(3)G-C or C(2)M showed only one spot, then that side was excluded from our analysis; thus, we may be overestimating the distance between the two foci. For Corolla, CONA, C(2)M, C(3)G-C, and N-C(3)G profiles generated along z and for the C(3)G-C and C(2)M x profiles, a fit to two Gaussians was carried out, and profiles were averaged after aligning to the center of the two peaks (Fig. 3 D and E). For Corolla, CONA, and N-C(3)G generated along x, the line profiles were fit to a single Gaussian distribution and aligned to this center (Fig. 3D). To generate the average profiles shown in Fig. 3, the average line profiles were fit to determine the separation of the width of the peaks. The line profiles in Fig. 4 were generated in a similar manner as in Fig. 3.
To show the line profiles in Fig. 3 in real units, it was necessary to correct for the expansion factor (Fig. S3). For each average profile of a region of C(3)G-C, a line profile was generated across the x dimension. This line profile was fit to two Gaussians, and the spacing between the two Gaussians (generally 400–500 nm) was normalized to 120 nm, a distance previously reported for SIM data of unexpanded C(3)G-C (16). The expansion factor was calculated for each dataset independently and was consistent across datasets, with an average of 3.82 ± 0.62 (Fig. S3).
ImmunoGold images were imported into ImageJ, and polyline profiles were drawn down the center of the synaptonemal complexes where visible. These images were then straightened and centered by assuming that each line segment was a rectangle and concatenated into a long carpet. Then points were placed in the center of each gold spot, and a histogram of x positions was created. Finally, the two sides of the histogram were averaged together and mirrored to obtain the final histogram. Histogram fitting was performed in the same way as the single particle averaging-SIM profiles.
Averaged superresolution profiles and ImmunoGold distribution histograms were fit to 1D multi-Gaussian functions by nonlinear least squares as described by Burns et al. (49), using the open source tools described above. Error analysis of the fit parameters was performed using a Monte Carlo approach with 100 random simulations, also as described by Burns et al. (49). SE propagation methods were used to calculate errors of derivative parameters (distance and width). Distance differences and their errors were also calculated by error propagation, and P values for the distance of these differences from 0 were determined using a two-tailed t test.
3D Model.
The 3D model in Fig. 5 was generated in Cinema 4D R18 Studio, and Movie S1 was edited in Final Cut Pro X.
Data Availability.
Primary data files for images presented in this paper are publicly accessible at www.stowers.org/research/publications/odr. Custom ImageJ plugins used for data analysis are available at research.stowers.org/imagejplugins/zipped_plugins.html.
Supplementary Material
Acknowledgments
We thank S. McKinney (Stowers Institute for Medical Research) for assistance with the STED imaging, and A. Miller for editorial assistance and figure preparation. We also thank Cathleen Lake and the members of the R.S.H. laboratory for helpful discussions. R.S.H. is supported by the Stowers Institute for Medical Research and is an American Cancer Society Research Professor.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705623114/-/DCSupplemental.
References
- 1.Cahoon CK, Hawley RS. Regulating the construction and demolition of the synaptonemal complex. Nat Struct Mol Biol. 2016;23:369–377. doi: 10.1038/nsmb.3208. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein P. Multiple synaptonemal complexes (polycomplexes): Origin, structure and function. Cell Biol Int Rep. 1987;11:759–796. doi: 10.1016/0309-1651(87)90157-3. [DOI] [PubMed] [Google Scholar]
- 3.Zickler D, Kleckner N. Meiotic chromosomes: Integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 4.Moses MJ, Counce SJ, Paulson DF. Synaptonemal complex complement of man in spreads of spermatocytes, with details of the sex chromosome pair. Science. 1975;187:363–365. [PubMed] [Google Scholar]
- 5.Schmekel K, Daneholt B. The central region of the synaptonemal complex revealed in three dimensions. Trends Cell Biol. 1995;5:239–242. doi: 10.1016/s0962-8924(00)89017-0. [DOI] [PubMed] [Google Scholar]
- 6.Schmekel K, Wahrman J, Skoglund U, Daneholt B. The central region of the synaptonemal complex in Blaps cribrosa studied by electron microscope tomography. Chromosoma. 1993;102:669–681. doi: 10.1007/BF00650893. [DOI] [PubMed] [Google Scholar]
- 7.Hernández-Hernández A, et al. The central element of the synaptonemal complex in mice is organized as a bilayered junction structure. J Cell Sci. 2016;129:2239–2249. doi: 10.1242/jcs.182477. [DOI] [PubMed] [Google Scholar]
- 8.Qiao H, et al. Interplay between synaptonemal complex, homologous recombination, and centromeres during mammalian meiosis. PLoS Genet. 2012;8:e1002790. doi: 10.1371/journal.pgen.1002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voelkel-Meiman K, Moustafa SS, Lefrançois P, Villeneuve AM, MacQueen AJ. Full-length synaptonemal complex grows continuously during meiotic prophase in budding yeast. PLoS Genet. 2012;8:e1002993. doi: 10.1371/journal.pgen.1002993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voelkel-Meiman K, et al. SUMO localizes to the central element of synaptonemal complex and is required for the full synapsis of meiotic chromosomes in budding yeast. PLoS Genet. 2013;9:e1003837. doi: 10.1371/journal.pgen.1003837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carpenter AT. Electron microscopy of meiosis in Drosophila melanogaster females, I: Structure, arrangement, and temporal change of the synaptonemal complex in wild-type. Chromosoma. 1975;51:157–182. doi: 10.1007/BF00319833. [DOI] [PubMed] [Google Scholar]
- 12.Schmekel K, Skoglund U, Daneholt B. The three-dimensional structure of the central region in a synaptonemal complex: A comparison between rat and two insect species, Drosophila melanogaster and Blaps cribrosa. Chromosoma. 1993;102:682–692. doi: 10.1007/BF00650894. [DOI] [PubMed] [Google Scholar]
- 13.Page SL, Hawley RS. c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 2001;15:3130–3143. doi: 10.1101/gad.935001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson LK, et al. Juxtaposition of C(2)M and the transverse filament protein C(3)G within the central region of Drosophila synaptonemal complex. Proc Natl Acad Sci USA. 2005;102:4482–4487. doi: 10.1073/pnas.0500172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lake CM, Hawley RS. The molecular control of meiotic chromosomal behavior: Events in early meiotic prophase in Drosophila oocytes. Annu Rev Physiol. 2012;74:425–451. doi: 10.1146/annurev-physiol-020911-153342. [DOI] [PubMed] [Google Scholar]
- 16.Collins KA, et al. Corolla is a novel protein that contributes to the architecture of the synaptonemal complex of Drosophila. Genetics. 2014;198:219–228. doi: 10.1534/genetics.114.165290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heidmann D, et al. The Drosophila meiotic kleisin C(2)M functions before the meiotic divisions. Chromosoma. 2004;113:177–187. doi: 10.1007/s00412-004-0305-5. [DOI] [PubMed] [Google Scholar]
- 18.Gyuricza MR, et al. Dynamic and stable cohesins regulate synaptonemal complex assembly and chromosome segregation. Curr Biol. 2016;26:1688–1698. doi: 10.1016/j.cub.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cahoon CK, Hawley RS. Meiosis: Cohesins are not just for sisters any more. Curr Biol. 2016;26:R523–R525. doi: 10.1016/j.cub.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 20.Schücker K, Holm T, Franke C, Sauer M, Benavente R. Elucidation of synaptonemal complex organization by super-resolution imaging with isotropic resolution. Proc Natl Acad Sci USA. 2015;112:2029–2033. doi: 10.1073/pnas.1414814112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen F, Tillberg PW, Boyden ES. Optical imaging: Expansion microscopy. Science. 2015;347:543–548. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chozinski TJ, et al. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat Methods. 2016;13:485–488. doi: 10.1038/nmeth.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tillberg PW, et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat Biotechnol. 2016;34:987–992. doi: 10.1038/nbt.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ku T, et al. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat Biotechnol. 2016;34:973–981. doi: 10.1038/nbt.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gustafsson MG, et al. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 2008;94:4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert TJ, Waters JC. Navigating challenges in the application of superresolution microscopy. J Cell Biol. 2017;216:53–63. doi: 10.1083/jcb.201610011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pattabiraman D, Roelens B, Woglar A, Villeneuve AM. Meiotic recombination modulates the structure and dynamics of the synaptonemal complex during C. elegans meiosis. PLoS Genet. 2017;13:e1006670. doi: 10.1371/journal.pgen.1006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nadarajan S, et al. Polo-like kinase-dependent phosphorylation of the synaptonemal complex protein SYP-4 regulates double-strand break formation through a negative feedback loop. eLife. 2017;6:6. doi: 10.7554/eLife.23437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Libuda DE, Uzawa S, Meyer BJ, Villeneuve AM. Meiotic chromosome structures constrain and respond to designation of crossover sites. Nature. 2013;502:703–706. doi: 10.1038/nature12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Page SL, et al. Corona is required for higher-order assembly of transverse filaments into full-length synaptonemal complex in Drosophila oocytes. PLoS Genet. 2008;4:e1000194. doi: 10.1371/journal.pgen.1000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moses MJ. Chromosomal structures in crayfish spermatocytes. J Biophys Biochem Cytol. 1956;2:215–218. doi: 10.1083/jcb.2.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong H, Roeder GS. Organization of the yeast Zip1 protein within the central region of the synaptonemal complex. J Cell Biol. 2000;148:417–426. doi: 10.1083/jcb.148.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Vries FA, et al. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 2005;19:1376–1389. doi: 10.1101/gad.329705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schild-Prüfert K, et al. Organization of the synaptonemal complex during meiosis in Caenorhabditis elegans. Genetics. 2011;189:411–421. doi: 10.1534/genetics.111.132431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphryes N, et al. The Ecm11-Gmc2 complex promotes synaptonemal complex formation through assembly of transverse filaments in budding yeast. PLoS Genet. 2013;9:e1003194. doi: 10.1371/journal.pgen.1003194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamer G, et al. Progression of meiotic recombination requires structural maturation of the central element of the synaptonemal complex. J Cell Sci. 2008;121:2445–2451. doi: 10.1242/jcs.033233. [DOI] [PubMed] [Google Scholar]
- 37.Schramm S, et al. A novel mouse synaptonemal complex protein is essential for loading of central element proteins, recombination, and fertility. PLoS Genet. 2011;7:e1002088. doi: 10.1371/journal.pgen.1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Costa Y, et al. Two novel proteins recruited by synaptonemal complex protein 1 (SYCP1) are at the centre of meiosis. J Cell Sci. 2005;118:2755–2762. doi: 10.1242/jcs.02402. [DOI] [PubMed] [Google Scholar]
- 39.Bolcun-Filas E, et al. Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLoS Genet. 2009;5:e1000393. doi: 10.1371/journal.pgen.1000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips D, Nibau C, Wnetrzak J, Jenkins G. High-resolution analysis of meiotic chromosome structure and behaviour in barley (Hordeum vulgare L.) PLoS One. 2012;7:e39539. doi: 10.1371/journal.pone.0039539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nebel BR, Coulon EM. The fine structure of chromosomes in pigeon spermatocytes. Chromosoma. 1962;13:272–291. doi: 10.1007/BF00577043. [DOI] [PubMed] [Google Scholar]
- 42.Dresser ME, Moses MJ. Synaptonemal complex karyotyping in spermatocytes of the Chinese hamster (Cricetulus griseus), IV: Light and electron microscopy of synapsis and nucleolar development by silver staining. Chromosoma. 1980;76:1–22. doi: 10.1007/BF00292222. [DOI] [PubMed] [Google Scholar]
- 43.Köhler S, Wojcik M, Xu K, Dernburg AF. Superresolution microscopy reveals the three-dimensional organization of meiotic chromosome axes in intact Caenorhabditis elegans tissue. Proc Natl Acad Sci USA. 2017;114:E4734–E4743. doi: 10.1073/pnas.1702312114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8:243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- 45.Hong A, Lee-Kong S, Iida T, Sugimura I, Lilly MA. The p27cip/kip ortholog dacapo maintains the Drosophila oocyte in prophase of meiosis I. Development. 2003;130:1235–1242. doi: 10.1242/dev.00352. [DOI] [PubMed] [Google Scholar]
- 46.Lake CM, et al. Vilya, a component of the recombination nodule, is required for meiotic double-strand break formation in Drosophila. eLife. 2015;4:e08287. doi: 10.7554/eLife.08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen F, et al. Nanoscale imaging of RNA with expansion microscopy. Nat Methods. 2016;13:679–684. doi: 10.1038/nmeth.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hiraoka Y, Sedat JW, Agard DA. Determination of three-dimensional imaging properties of a light microscope system: Partial confocal behavior in epifluorescence microscopy. Biophys J. 1990;57:325–333. doi: 10.1016/S0006-3495(90)82534-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burns S, et al. Structured illumination with particle averaging reveals novel roles for yeast centrosome components during duplication. eLife. 2015;4:4. doi: 10.7554/eLife.08586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Primary data files for images presented in this paper are publicly accessible at www.stowers.org/research/publications/odr. Custom ImageJ plugins used for data analysis are available at research.stowers.org/imagejplugins/zipped_plugins.html.