Significance
Pre-mRNA processing not only enhances the diversity encoded in the genome without the need to increase the number of genes but also provides a means to adjust cellular transcript abundance. Environmental light has a profound effect on transcript accumulation, but how this is partitioned between transcriptional and posttranscriptional processes is largely unknown. Here we describe the identification and characterization of the splicing factor for phytochrome signaling (SFPS), which directly interacts with the photoreceptor phytochrome B. sfps seedlings are hyposensitive to light and display pre-mRNA splicing defects in a large number of genes, many of which regulate light signaling and the circadian clock. Thus, light might control pre-mRNA splicing in addition to transcription of many genes through SFPS to promote photomorphogenesis.
Keywords: pre-mRNA splicing, photomorphogenesis, phytochrome signaling, splicing factor, Arabidopsis
Abstract
Light signals regulate plant growth and development by controlling a plethora of gene expression changes. Posttranscriptional regulation, especially pre-mRNA processing, is a key modulator of gene expression; however, the molecular mechanisms linking pre-mRNA processing and light signaling are not well understood. Here we report a protein related to the human splicing factor 45 (SPF45) named splicing factor for phytochrome signaling (SFPS), which directly interacts with the photoreceptor phytochrome B (phyB). In response to light, SFPS-RFP (red fluorescent protein) colocalizes with phyB-GFP in photobodies. sfps loss-of-function plants are hyposensitive to red, far-red, and blue light, and flower precociously. SFPS colocalizes with U2 small nuclear ribonucleoprotein-associated factors including U2AF65B, U2A′, and U2AF35A in nuclear speckles, suggesting SFPS might be involved in the 3′ splice site determination. SFPS regulates pre-mRNA splicing of a large number of genes, of which many are involved in regulating light signaling, photosynthesis, and the circadian clock under both dark and light conditions. In vivo RNA immunoprecipitation (RIP) assays revealed that SFPS associates with EARLY FLOWERING 3 (ELF3) mRNA, a critical link between light signaling and the circadian clock. Moreover, PHYTOCHROME INTERACTING FACTORS (PIFs) transcription factor genes act downstream of SFPS, as the quadruple pif mutant pifq suppresses defects of sfps mutants. Taken together, these data strongly suggest SFPS modulates light-regulated developmental processes by controlling pre-mRNA splicing of light signaling and circadian clock genes.
All organisms perceive and respond to environmental light signals, and plants are no exception. Plants can perceive minute changes in light quantity, quality, and direction, and integrate this information to modulate growth and development. At the young seedling stage, the perception of light enables plants to switch from skotomorphogenesis (a dark-adapted developmental program characterized by long hypocotyl, short root, and small and unopened cotyledons) to photomorphogenesis (a light-adapted developmental program characterized by short hypocotyl, open expanded and green cotyledons, and elongated root suitable for photosynthetic growth). At later stages of growth, light plays a crucial role in regulating shade avoidance, flowering time, and eventually senescence. Plants have several classes of photoreceptors sensing and responding to major bandwidths of the visible light spectrum (1, 2). Among these, phytochromes (phys) perceive and respond to the red/far-red region of the spectrum. Phytochromes consist of a small multigene family (designated PHYA to PHYE in Arabidopsis thaliana) encoding ∼125-kDa soluble proteins that can form selective homo- and heterodimers between family members (3–5). They exist as two spectrally distinct light-switchable forms: a red light-absorbing Pr (inactive) form, which can be converted to the active Pfr form by exposure to red light; the far-red light–absorbing Pfr form can in turn be converted back to the Pr form by exposure to far-red light. Subcellular localization studies show that the Pr forms of plant phytochromes are mainly present in the cytosol. However, the Pfr forms of all family members are translocated into the nucleus on light activation with differential kinetics (6, 7). Within the nucleus, phytochromes form nuclear photobodies in response to light; the size and number of photobodies for phyB correlates with phyB activity (7, 8).
As a pivotal light sensor, phytochromes regulate plant growth and development by controlling protein translocation, gene expression, translation, and proteolysis. Phytochromes also fine-tune downstream light responses through posttranslational modifications, chromatin remodeling, and posttranscriptional regulation of gene expression (9). Extensive work describes the role of phytochromes in global regulation of gene expression in response to light, especially through modulation of the stability of the transcription factors LONG HYPOCOTYL 5 (HY5, an activator) and PHYTOCHROME-INTERACTING FACTORS (PIFs, repressors) (10). However, much less is known about the roles of phytochromes in controlling pre-mRNA splicing in response to light.
Pre-mRNA splicing involves removal of introns from pre-mRNAs transcribed from eukaryotic genes by large ribonucleoprotein complexes called spliceosomes (11, 12). Five small nuclear ribonucleoprotein complexes (U1, U2, U4, U5, and U6) recognize and assemble sequentially on each intron and catalyze the removal of introns (12). Introns can be either constitutively or alternatively spliced. Alternative splicing (AS) can lead to intron removal or retention, or the use of alternative 5′ and 3′ splice sites (SS) of an exon. AS is a key posttranscriptional regulatory process to adjust transcript abundance and enhance the genome’s diversity without increasing the number of genes (11–13). Introns have at least four loosely conserved sequence determinants for recognition and removal by spliceosome. These include the 5′ SS with a conserved GU nucleotide residue, 3′ SS with a conserved AG nucleotide residue, a branched point adenine (A) 18–40 nucleotides upstream of the 3′ SS, and a poly-pyrimidine tract following the branched point. In addition to the above sequence determinants in all introns, there are splicing regulatory elements that confer gene-specific regulation of splicing. These include exonic or intronic splicing enhancers and exonic or intronic splicing silencers. These cis-acting sequences act as binding sites for transacting splicing factors such as serine/arginine-rich (SR) proteins to regulate either AS or splicing efficiency (12, 14).
Recent reports showed that phytochromes as well as other photoreceptors are involved in regulating pre-mRNA splicing of light response genes functioning in photomorphogenesis and the circadian clock in Arabidopsis (15). For example, RNA-sequencing data showed that 2,230 genes undergo AS within 1–3 h of red light exposure in a phytochrome-dependent manner (16). The Arabidopsis protein reduced red light responses in cry1cry2 background 1 (RRC1), an orthologous protein of the potential human splicing factor SR140, are involved in phyB-mediated AS of light response genes. The rrc1 seedlings show hyposensitive phenotype under red light conditions (17). An acute light pulse in the middle of night, as well as a retrograde signal emanating from the chloroplast regulate splicing of light- and circadian clock-regulated genes (18, 19). In addition, five mutants (skip, prmt5, stipl1, gemin2, and sickle) with global defects in splicing have been characterized with altered circadian clock function (20–25). Among these, SKIP, STIPL1, and GEMIN2 encode putative splicing factors, and skip and prmt5 displayed defects in photomorphogenesis (20, 26). Pre-mRNA splicing is therefore an important regulatory mechanism in light signal transduction. Here we describe the isolation and cloning of a mutant encoding a splicing factor called splicing factor for phytochrome signaling (SFPS). SFPS colocalizes and physically interacts with phyB in vivo, suggesting SFPS might function in the phyB signaling pathway to promote photomorphogenesis by regulating pre-mRNA splicing of many light signaling and circadian clock genes.
Results
SFPS Required for red Light Responses.
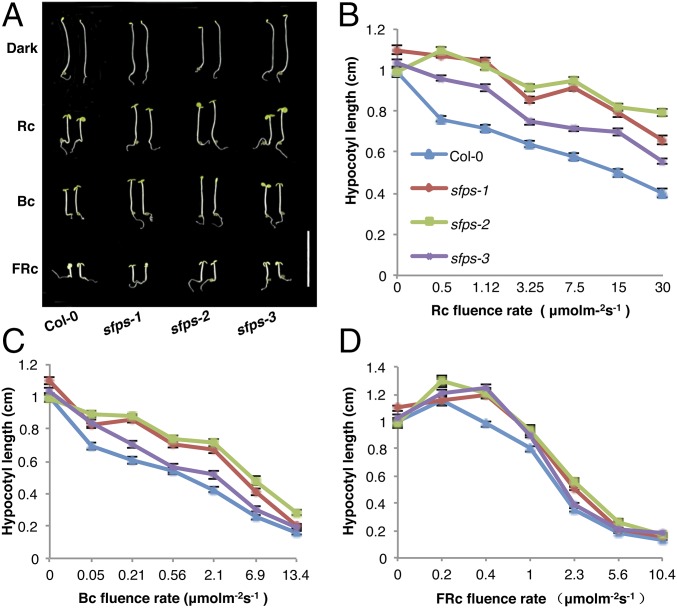
Hypocotyl growth provides a sensitive assay to measure responses to light signals and has been widely used to isolate mutants in this pathway (27). In an ethyl methanesulfonate mutagenesis screen, we identified an sfps mutant with a long hypocotyl under red light, indicative of reduced red light sensitivity. We mapped the causal locus to the upper arm of chromosome 1 by whole-genome sequencing (28). Of the five mutations relative to the published reference genome sequence within the mapping interval, we focused on one in At1g30480, which contained an ethyl methanesulfonate-diagnostic G-to-A substitution mutation only in the mutant background, affecting the start codon (M1I) (SI Appendix, Fig. S1A). We isolated two independent T-DNA insertion lines for At1g30480. Our initial ethyl methanesulfonate allele will be referred to as sfps-1, whereas the T-DNA insertion lines are sfps-2 (SALK_001489) and sfps-3 (SALK_066706; SI Appendix, Fig. S1A and Fig. 1). Examination of the mRNA levels by qPCR showed that the sfps-2 allele had the lowest mRNA level among all three alleles (SI Appendix, Fig. S1B). Both T-DNA insertion lines, as well as the original mutant, displayed a long hypocotyl specifically under continuous red, far-red, and blue light conditions, but not in constant darkness (Fig. 1 A–D and SI Appendix, Fig. S2F). Other phenotypes recapitulated by the T-DNA insertion lines included much reduced red, far-red, and blue light-induced cotyledon opening (SI Appendix, Fig. S2 A–C), and lower chlorophyll and anthocyanin levels in white light and far-red light, respectively (SI Appendix, Fig. S2 D and E). These results demonstrate that the causal mutation responsible for hyposensitivity to red light in our mutant lies in At1g30480.
Fig. 1.
SFPS acts positively in phytochrome signaling. (A) Photographs of wild-type Col-0 and different sfps mutant alleles grown for 4 d under continuous red (1.2 μmol⋅m−2⋅s−1), far-red (0.56 μmol⋅m−2⋅s−1), and blue light (0.73 μmo⋅m−2⋅s−1) conditions. White bar, 1 cm. Fluence rate response curves for hypocotyl elongation of wild-type Col-0 and different alleles of sfps under constant red (B), blue (C), and far-red (D) light conditions. Seedlings were grown under different fluences of red, far-red, and blue light conditions for 4 d. Error bars indicate SEM (n > 30). Bc, continuous blue light; D, dark; FRc, continuous far-red; Rc, continuous red.
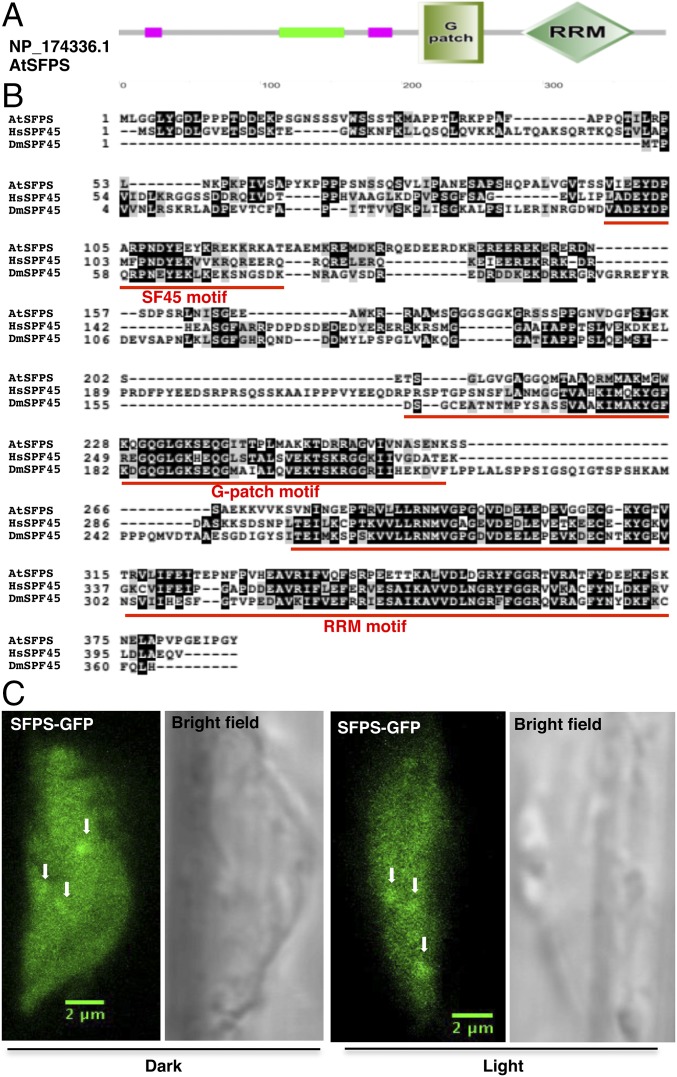
The predicted SFPS protein contains a conserved SF45 motif, a glycine-rich (G-patch) motif, and an RNA recognition motif at the C terminus, as in human and Drosophila SPF45 (Fig. 2A). SFPS is a potential ortholog to the human and Drosophila splicing factor SPF45, as it displays a high degree of sequence similarity (Fig. 2B) (29, 30). SFPS from A. thaliana was first described in 1993 as DNA damage response tolerance 111 because it partially restored, by an unknown mechanism, DNA damage resistance to Escherichia coli mutants compromised in recombination (31). We have renamed this locus SFPS (SPLICING FACTOR FOR PHYTOCHROME SIGNALING), based on its role in pre-mRNA splicing. We expressed a fusion protein between SFPS and the GFP, using the SFPS promoter in the sfps-1 background to examine its subcellular localization. The construct rescued the long hypocotyl phenotype of the sfps-1 mutant to wild type, confirming the identity of the causal locus (SI Appendix, Fig. S1 C and D). SFPS-GFP localized to the nucleoplasm as both diffuse signal and nuclear speckles under both dark- and light-exposed seedlings, characteristic of splicing factors in many eukaryotes (Fig. 2C) (32). Thus, SFPS might function as a splicing factor in light signaling pathways.
Fig. 2.
SFPS is a nuclear-localized splicing factor in Arabidopsis. (A) Domain structure of SFPS protein. Protein sequences are drawn to scale. SF45, G-patch, and RNA recognition motifs are shown boxed. (B) Alignment of SPF45 family protein sequences. SF45, G-patch, and RNA recognition motifs are underlined by the red line. NP_174336.1 (SFPS in Arabidopsis); NP_116294.1 (splicing factor 45 in Homo sapiens); NP_001036424.1 (Sfp45, isoform C in Drosophila melanogaster). (C) Confocal microscopy of pSFPS:SFPS-GFP/sfps-1 transgenic seedlings shows that SFPS protein is both diffuse in the nucleoplasm and forms speckles (white arrowheads) in the nucleus under both dark and light conditions. (Scale bar, 2 μm.) RRM, RNA recognition motif.
sfps Mutants Flower Early Under Short and Long Days.
To examine whether SFPS also plays a role in transition from vegetative to reproductive growth, we grew plants under both short days (SD) and long days (LD) and measured flowering time. The two strong alleles sfps-1 and sfps-2 flowered earlier than wild type, as observed by number of rosette leaves and days to flower under both SD and LD (SI Appendix, Fig. S3 A–D). Expression of CONSTANS (CO) was marginally changed in SD (SI Appendix, Fig. S4A), whereas FLOWERING LOCUS C (FLC) was down-regulated and FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CO (SOC1) were up-regulated in the same conditions (SI Appendix, Fig. S4 B–D), consistent with early flowering. In LD, CO and FT expression was unaffected, whereas FLC expression was strongly down-regulated, and SOC1 expression is up-regulated in the sfps-2 mutant (SI Appendix, Fig. S4 E–H). The flowering time phenotypes might be a result of a defect in circadian clock function, as sfps-2 had slightly shorter period length compared with wild type (SI Appendix, Fig. S5 A–F). Under continuous white light conditions at 22 °C, sfps alleles did not display any other obvious morphological defects during either juvenile or adult stages of growth (SI Appendix, Fig. S6 A and B). Overall, these data suggest SFPS promotes flowering under both SD and LD.
SFPS Colocalizes with U2-Associated Splicing Factors to Nuclear Speckles.
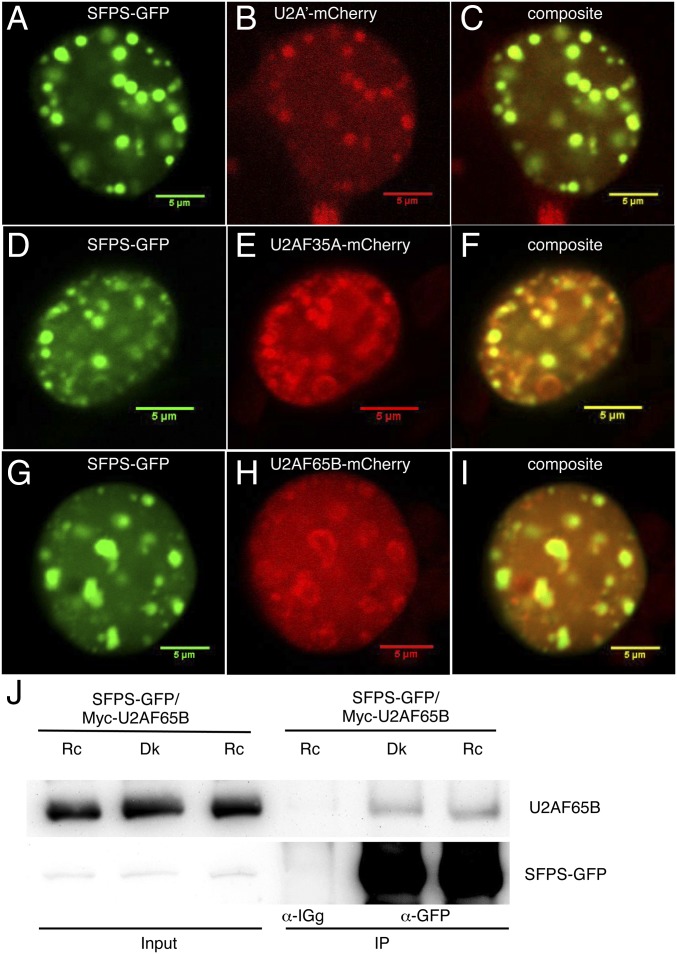
The human and Drosophila homologs of SFPS associate with U2 small nuclear ribonucleoprotein-associated complexes and function in 3′ SS determination. We tested whether SFPS colocalizes with U2-associated factors in Arabidopsis by coexpressing SFPS-GFP along with U2A′-mCherry, U2AF35A-mCherry, or U2AF65B-mCherry by transient expression in tobacco epidermal cells. SFPS-GFP and all three U2-associated factors formed nuclear speckles (Fig. 3 A–H). Importantly, SFPS colocalized with all three U2-associated factors within the nucleus (Fig. 3 C, F, and I).
Fig. 3.
SFPS colocalizes with U2 small nuclear ribonucleoprotein (snRNP) components in discrete nuclear foci. (A, D, and G) Subcellular localization of SFPS-GFP in tobacco cells. Subcellular localization of U2A′-mCherry (B), U2AF35A-mCherry (E), and U2AF65B-mCherry (H) in tobacco cells. (C, F, and I) Overlay panels from GFP and mCherry channels. (Scale bar, 5 μm.) (J) SFPS-GFP and MYC-U2AF65B proteins coimmunoprecipitate in Arabidopsis. Total proteins are extracted from double-transgenic Arabidopsis seedlings coexpressing SFPS-GFP and MYC-U2AF65B proteins. IP samples were pulled down with anti-GFP antibody and probed with anti-MYC and anti-GFP antibodies, respectively. Anti-IgG was used as a negative control. Dk, dark; Rc, continuous red.
As an independent confirmation, we tested for coimmunoprecipitation between U2AF65B and SFPS-GFP in transient assays in tobacco and stable Arabidopsis transgenic plants. We found that SFPS immunoprecipitated U2AF65B in both assays (Fig. 3J and SI Appendix, Fig. S7), indicating SFPS associates with U2 components and may be involved in 3′ SS determination.
SFPS Interacts with the Photoreceptor phyB.
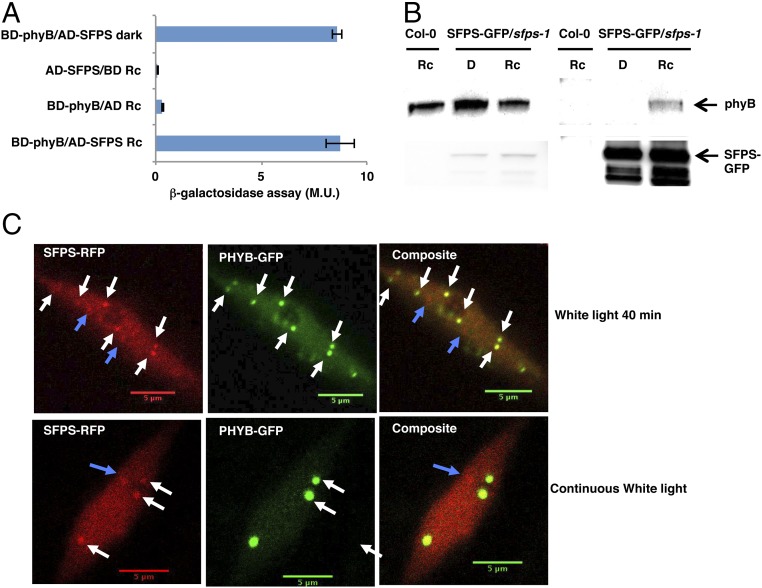
Because sfps mutant alleles showed defects under red light, we examined whether SFPS directly interacted with phytochromes. We first performed yeast-two-hybrid assays using full-length phyB as a bait and SFPS as a prey. SFPS interacted with full-length phyB both in the dark and in red light (Fig. 4A). We then sought to confirm this interaction in vivo by immunoprecipitating SFPS-GFP, using the anti-GFP antibody and probing for native phyB. Fig. 4B shows that SFPS interacts with phyB in a red light-dependent manner in vivo. The difference between the yeast two-hybrid and in vivo coimmunoprecipitation (Co-IP) assays might be a result of the light-induced translocation of phyB into the nucleus in plants, whereas SFPS is located in the nucleus constitutively in both yeast and plant.
Fig. 4.
phyB interacts with SFPS in a yeast two-hybrid assay and in vivo. (A) phyB interacts with SFPS in the yeast-two hybrid assay in a light-independent manner. Full-length PHYB protein was fused to the GAL4 DNA binding domain; full-length SFPS was fused to the GAL4 activation domain. β-galactosidase activity was measured to quantify the interaction between phyB and SFPS. Error bar, SEM (n > 3). (B) phyB and SFPS coimmunoprecipitate in a light-dependent manner in vivo. Four-day-old dark-grown seedlings were kept in the darkness or transferred to red light conditions (7 μmol m−2 ⋅ s−1) for 6 h. IP samples were pulled down with anti-GFP antibody and probed with anti-phyB, and anti-GFP antibodies, respectively. (C) Colocalization of phyB-GFP and SFPS-RFP in Arabidopsis seedlings. 35S:PHYB-GFP/35S:SFPS-RFP in Col-0 seedlings were grown under continuous white light for 4 d and imaged (Bottom). A subset of these seedlings were exposed to saturated far-red light and kept in darkness for 24 h followed by exposure to additional 40 min white light before the images were taken (Upper). (Scale bar, 5 mm.) Broken arrowheads indicate SFPS-specific speckles; white arrowheads indicate colocalized photobodies. AD, activation domain; BD, binding domain; D, dark; M.U., miller unit; Rc, continuous red.
To map the interaction domains between phyB and SFPS, we tested several fragments of SFPS. Quantitative β-galactosidase assays showed that the G-patch domain of SFPS is necessary for interaction with full-length phyB (SI Appendix, Fig. S8A). In addition, the C-terminal domain of phyB interacts with full-length SFPS (SI Appendix, Fig. S8B). Overall, these data suggest SFPS and phyB physically interact through the G-patch domain of SFPS and the C-terminal domain of phyB. phyB may regulate SFPS function in vivo via direct interaction.
Previously, phyB was shown to translocate to the nucleus in response to light, where it then forms photobodies (7, 8). To examine whether SFPS colocalizes with phyB photobodies, we generated SFPS-RFP and phyB-GFP double transgenic lines, using the 35S promoter. Examination of fluorescence using confocal microscopy showed that a large proportion of SFPS nuclear speckles colocalized with phyB photobodies (Fig. 4C and SI Appendix, Fig. S8C). However, SFPS also formed nuclear speckles that did not colocalize with phyB photobodies. Thus, the overlap in localization of these proteins in photobodies suggests phytochrome might regulate the splicing machinery through the recruitment of splicing factors to photobodies.
SFPS Transcription and Protein Stability Is Not Regulated by Light.
To examine whether SFPS activity might be light-dependent, we first asked whether SFPS RNA expression is regulated by light, using RT-qPCR with RNA from wild-type seedlings grown in darkness for 4 d or dark-grown seedlings exposed to red light for 30 min or 3 h. SFPS was expressed equally in dark- and light-exposed samples (SI Appendix, Fig. S9A). We also examined SFPS protein accumulation in 4-d-old seedlings grown under continuous red, far-red, and blue light conditions, or maintained in darkness, using the SFPSp:SFPS-GFP/sfps-1 transgenic plants expressing SFPS from its native promoter. SFPS-GFP protein levels were comparable in all four samples (SI Appendix, Fig. S9B), suggesting light signals do not regulate SFPS protein stability under these conditions. Thus, light may regulate the activity of SFPS either by direct physical interaction with the photoreceptor phyB and/or by posttranslational modifications.
sfps Regulates Gene Expression in Arabidopsis.
To further characterize the range of phenotypes of the sfps mutants, we performed RNA deep sequencing to identify genome-wide defects in gene expression and pre-mRNA splicing in 4-d-old dark-grown wild-type and sfps-2 mutant seedlings with or without exposure to a 3-h red light pulse. With a false-discovery rate (FDR) of 0.05, we identified 1,494 genes as differentially expressed in the light and 1,361 in the dark between Col-0 and sfps-2 mutant (|log2 fold-change| >0.58; FDR <0.05) (SI Appendix, Fig. S10; Dataset S1, III and IV). These data highlight the significant effect of SFPS on gene expression. GO-term analyses revealed that 246 GO terms are enriched in the dark and 195 in the light, pointing to the involvement of various plant growth and developmental processes (SI Appendix, Fig. S11; Dataset S1, V and VI). The GO terms “photosynthesis,” “response to light-stimulus,” and “response to red and far-red light” were significantly enriched in the differentially expressed gene list (SI Appendix, Fig. S11; Dataset S1, V and VI).
SFPS Controls pre-mRNA Splicing in Arabidopsis Transcriptome.
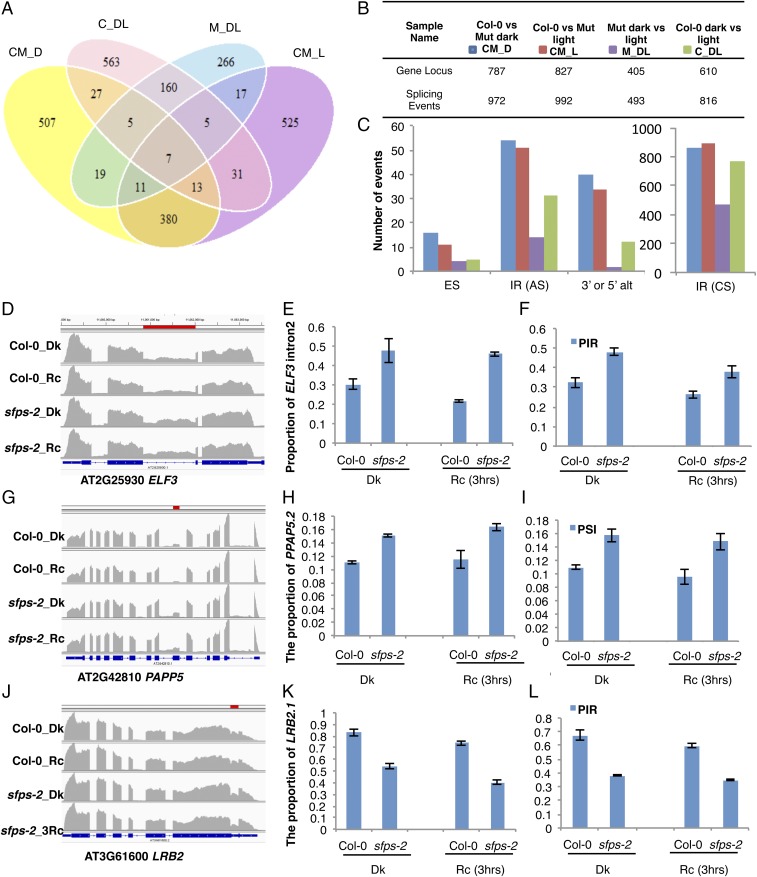
Given that SFPS has sequence homology to splicing factor SPF45 and colocalizes with U2-associated factors, we investigated whether SFPS regulates pre-mRNA splicing. To this end, we looked for changes in pre-mRNA splicing pattern between wild type and sfps-2 by mapping reads to exonic and intronic as well as the junction regions of all genes of the Arabidopsis genome. We then evaluated splicing defects resulting from sfps mutation by comparing the percentage of inclusion of exons (percentage spliced-in, PSI) and introns (percentage intron retention, PIR) in four different comparisons: Col-0 vs. Mut dark (CM_D), Col-0 vs. Mut 3 h Rc (CM_L), Col-0 dark vs. 3 h Rc (C_DL), and Mut dark vs. 3 h Rc (M_DL), respectively, as previously described (18). We identified 972 (FDR, <0.05 and Delta PSI or Delta PIR, >3%) pre-mRNA splicing defects between wild type and sfps-2 in the dark covering 787 genes (Fig. 5 A and B; Dataset S2, III). We also identified 992 (FDR, <0.05 and Delta PSI or PIR, >3%) splicing defects under light condition, which mapped to 827 genes in the genome (Fig. 5 A and B; Dataset S2, IV). Among those, 411 splicing changes overlapped between light and dark conditions (dark, ∼52%; light, ∼41%), which indicates SFPS regulates pre-mRNA splicing for a different subset of genes in the dark vs. light conditions. Several of the splicing defects detected in the mutant corresponded to changes in AS, which include intron retention (IR), exon skipping, and AS donor or acceptor sites (3′ or 5′ alt; Fig. 5C; Dataset S2, VI). Although all four categories were affected in the mutant, there was a significant enrichment in defects in IR events (Dataset S2, VI). Indeed, most of the splicing events identified as affected in the mutant actually correspond to IR events not previously annotated as AS events (Dataset S2, III, IV, and VI). Although some of these may represent not previously annotated AS events, many of them may simply be constitutively spliced introns in wild-type plants. Taken together, these data indicate a crucial regulatory role of SFPS during pre-mRNA splicing in Arabidopsis.
Fig. 5.
SFPS controls pre-mRNA splicing of the Arabidopsis transcriptome. (A) Venn diagram of the number of genes that displayed altered AS pattern in Col-0 vs. Mut dark (CM_D), Col-0 vs. Mut 3 h Rc (CM_L), Col-0 dark vs. 3 h Rc (C_DL), and Mut dark vs. 3 h Rc (M_DL) respectively. P < 0.05, FDR <0.05 and PSI or PIR change is larger than 3%. (B) Summary table giving number of genes with altered AS patterns. (C) Bar graph shows the number of different types of AS (alternative splicing) events in four groups of comparisons. (D–L) Confirmation of RNA-seq data by RT-qPCR for selected light signaling genes. (D, G, and J) Integrative Genomics Viewer shows alternative splicing defects detected by RNA-seq between Col-0 and sfps-2. Red bar indicates the position of AS events in the genes. (E, H, and K) RT-qPCR validation of RNA-seq data. Total RNA was extracted from Col-0 and sfps-2 seedlings grown in the dark or under Rc (3 h, 7 μmol⋅m−2⋅s−1) light, respectively. AS patterns of the light signaling genes, ELF3, PAPP5, and LRB2 are expressed as the proportion of specific isoforms in the total transcripts of the same gene. PP2A was used as an internal control. Each bar is the mean ± SEM (n = 3 independent biological repeats, and each biological repeat include three technical repeats). (F, I, and L) AS patterns indicated by RNA-seq. PSI and PIR are calculated according to formulas in Materials and Methods (n = 3 independent biological repeats). CS, constitutive splicing; Dk, dark; ES, exon skipping; IR, intron retention; Rc, continuous red.
We selected 10 genes with AS patterns for independent verification (Fig. 5 D, G, and J, Left; SI Appendix, Fig. S15 A, D, G, J, and M). We isolated RNA from three biological replicates and performed RT-qPCR, using primers from the junction regions of these genes. RNAseq data are shown on the right in Fig. 5 F, I, and K (SI Appendix, Fig. S15 C, F, I, L, and O), and qPCR data are shown in the middle column of Fig. 5 E, H, and K (SI Appendix, Fig. S15 B, E, H, K, and N). These results show that the RNAseq data can be reproduced by an independent method. In addition, semiquantitative RT-PCR analyses show that pre-mRNA splicing of the splicing regulator, SR genes is altered in the mutant compared with wild type (SI Appendix, Fig. S16), suggesting at least some of the AS events we detected in the sfps mutant might be indirect.
SFPS Is Involved in Light-Regulated Pre-mRNA Splicing.
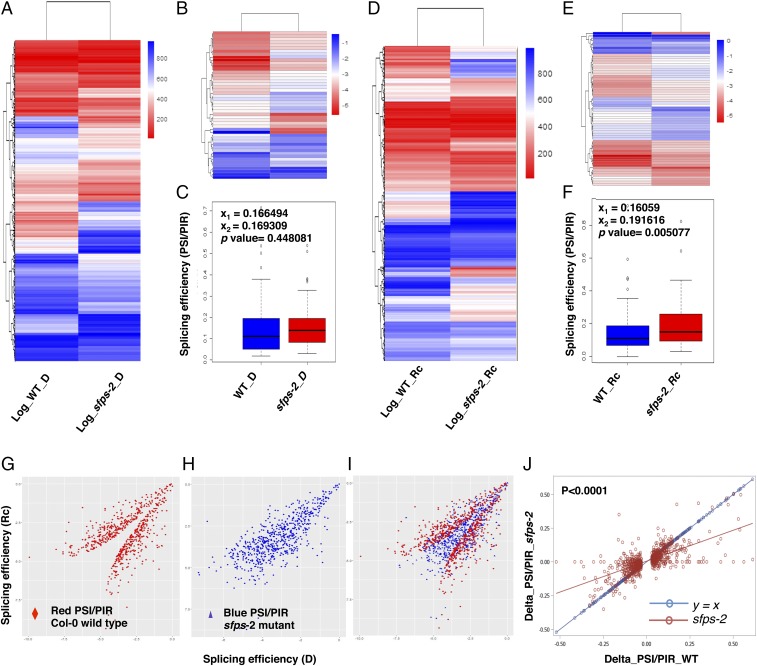
To further quantify differential splicing efficiency in the wild type and sfps-2 mutant, we calculated log2PSI,PIR, which normalizes the variance of the data. Heat maps show that splicing patterns are significantly altered in the sfps-2 mutant compared with in wild type (Fig. 6 A and D). We conducted GO analysis for the genes that displayed splicing defects between the wild type and sfps-2 to identify pathways regulated by SFPS. In the dark and light conditions, respectively, 213 and 239 GO categories (FDR <0.05) were overrepresented (SI Appendix, Fig. S12; Dataset S2, VII and VIII). Several GO terms related to “light stimulus,” “photosynthesis,” and “circadian clock” were significantly enriched (P < 0.05) in the dark and light conditions, respectively (SI Appendix, Fig. S12). The Kyoto Encyclopedia of Genes and Genomes pathway analysis also indicated that the “circadian rhythm” pathway is significantly enriched in the sfps mutant (SI Appendix, Fig. S13; Dataset S2, IX).
Fig. 6.
SFPS mediates light regulation of pre-mRNA splicing. Heat maps of genome-wide pre-mRNA splicing profiling, based on log2 PSI, PIR of Col-0 and sfps-2 dark (A) and light samples (D), respectively. Hierarchical clustering was performed on genes with altered pre-mRNA splicing pattern between Col-0 and sfps-2. Blue color, higher log2 PSI, PIR; red color, lower log2 PSI, PIR. red light intensity used was 7 μmol⋅m−2⋅s−1. (B and E) Heat maps of quantified pre-mRNA splicing profile of the genes related to the light-induced phenotype of sfps mutant. Gene lists are included in Dataset S2, I. (C and F) Box plots of PSI and PIR for the genes related to the phenotype in dark and Rc conditions. (G and H) Scatter plots of splicing efficiency for light-regulated splicing events in wild type (G) and sfps-2 (H). X-axis is the splicing efficiency (log2 PSI, PIR) in the dark. y axis is the splicing efficiency (log2 PSI, PIR) after light treatment. (I) Overlay of G and H. The scatter plot was generated on the basis of the 816 differentially spliced events detected in wild type. (J) Linear regression analyses of the splicing efficiency changes (Delta_PSI, PIR = PSI, PIR_Dark – PSI, PIR_Light) between the wild type and sfps-2 mutant in response to light. The differentially spliced events that were used to generate this analysis are described in G–I. The linear regression model for sfps-2 P < 0.0001, slope= 0.46, adjusted R2 = 0.45. Red line indicates linear regression for sfps-2; blue line indicates the control y = x. The splicing efficiency changes in sfps-2 are significantly different from y = x (P < 0.0001). D, dark; Rc, continuous red.
Based on the GO term annotations, we zeroed in on genes related to the light-regulated phenotype of sfps mutant (Dataset S2, X). GO analysis for molecular function indicates the “transcription factor activity” term was enriched (SI Appendix, Fig. S14), suggesting SFPS might regulate the activity of transcription factors, which in turn regulate transcription of many downstream target genes. Among these genes, we observed key regulatory genes (e.g., ELF3, LRB2, PIF3, FLC, PAPP5 and others) in light signaling pathways, consistent with the photomorphogenesis and flowering time phenotypes of the sfps mutant. Heat maps for splicing these genes show that pre-mRNA splicing efficiency was altered in the sfps-2 mutant compared with wild type (Fig. 6 B and E). We also generated box plots for these genes to display splicing efficiency (PSI, PIR) (Figs. 6 C and F). Interestingly, the average distribution of splicing efficiency was significantly different between the wild type and sfps-2 mutant only under Rc conditions, consistent with the light-dependent phenotype of sfps mutant.
To determine whether SFPS is involved in light-regulated pre-mRNA splicing, we examined pre-mRNA splicing efficiency after a 3-h pulse of red light. We found that 816 splicing events (corresponding to 610 gene locus) underwent changes in pre-mRNA splicing after 3-h red light pulse in wild type (Dataset S2, I); however, this number drops to 493 splicing events (corresponding to 405 gene locus) in sfps-2 (Fig. 5B; Dataset S2, II). To examine the role of light in regulating pre-mRNA splicing, we calculated log2PSI,PIR for these 816 light-regulated splicing events in wild type and for the same set of events in sfps mutant (Dataset S2, I). Scatter plots show that the splicing efficiency of those light-regulated events are either up-regulated or down-regulated in wild type on light exposure (Fig. 6 G and I); however, light-induced splicing efficiency changes of these events become much less dramatic in sfps-2 mutant (Fig. 6 H and I). Moreover, a simple linear regression analysis shows that splicing efficiency changes in response to light irradiation are significantly reduced in the sfps-2 mutant compared with wild type (Fig. 6J). Overall, these data support our hypothesis that SFPS might function in light-regulated pre-mRNA splicing and fine-tune the abundance and diversity of light response genes in addition to the transcriptional regulation in Arabidopsis.
SFPS Associates with ELF3 Transcript in Vivo.
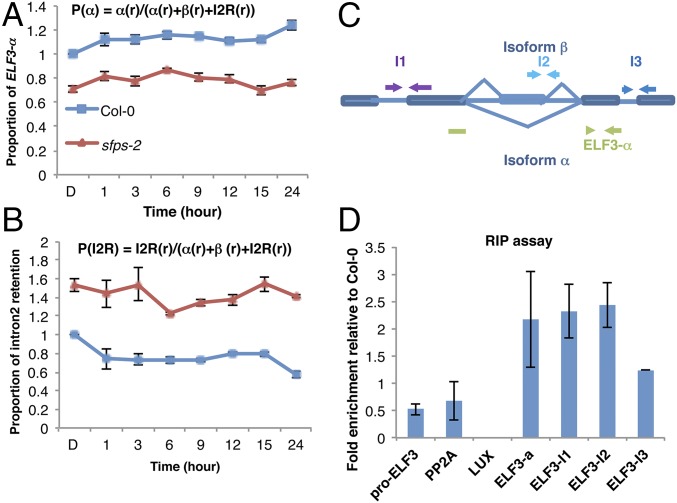
An excellent candidate for understanding the role of SFPS in light-dependent splicing is ELF3, which interacts with phyB to regulate photomorphogenesis and flowering time. ELF3 is also a key component in the evening complex, which is necessary for circadian clock regulation (33, 34). Our phenotypic analyses show that sfps and elf3 display similar defects, including long hypocotyl and early flowering, although elf3 phenotypes are much stronger that those of sfps. ELF3 transcript itself is a known target of alternative splicing, making it a prime candidate for control of photomorphogenesis by SFPS (35). Thus, we performed a time course analysis for spliced and prespliced forms of the ELF3 transcript in the sfps-2 mutant compared with wild type (Fig. 7 A and B; SI Appendix, Fig. S17). We also calculated total transcript levels, including light-induced transcriptional changes in ELF3, and determined the proportion of splice variants over time. sfps mutants produced much reduced level of the fully spliced and functional form of the ELF3 transcript (Fig. 7A; SI Appendix, Fig. S17 A, C, and E). In contrast, sfps mutants had much higher levels of the prespliced form of ELF3 transcript, which leads to a nonfunctional protein because of intron retention (Fig. 7B; SI Appendix, Fig. S17 B, D, and F). These data show that SFPS controls pre-mRNA splicing of ELF3 under both dark and light conditions to regulate photomorphogenesis and flowering time.
Fig. 7.
SFPS regulates ELF3 pre-mRNA splicing through the direct association of its transcripts in vivo. RT-qPCR to detect the proportion of ELF3 functional isoform α (A) and intron 2 retention (I2R) (B) in vivo over the course of 24 h red light irradiation (7 μmol⋅m−2⋅s−1). Total RNA was extracted from the Col-0 and sfps-2 mutants grown in the dark or transferred to red light (6, 9, 12, 15, and 24 h). PP2A was used as an internal control. Each bar is the mean ± SEM (n = 3 independent biological repeats, and each biological repeat includes three technical repeats). (C) Schematic diagram indicates the positions of primers used for the RNA immunoprecipitation assay. (D) RIP assay shows association of SFPS protein to ELF3 transcripts in vivo. The RNA–protein complexes were extracted from SFPS-GFP/sfps-1 seedling with Col-0 as control and immunoprecipitated by anti-GFP antibody. RNA was purified from IP products and then reverse-transcribed into cDNA. The abundance of each gene was quantified by RT-qPCR. The results were normalized to input of each sample, and then normalized to Col-0 wild type to calculate fold-enrichment. Each bar is the mean ± SEM (n = 3 independent biological repeats).
Our hypothesis posits that SFPS associates with the ELF3 transcript in vivo to regulate its pre-mRNA splicing. We therefore performed a RIP assay using SFPS-GFP as a bait. We grew SFPSp:SFPS-GFP/sfps-1 and wild type as a control under dark/light cycles (12 h dark:12 h red light) and isolated nuclear RNA species associated with SFPS-GFP. RT-qPCR was used to quantify various regions of the ELF3 transcript as well as two control genes [LUX ARRHYTHMO (LUX) and PROTEIN PHOSPHATASE 2A (PP2A)] and the promoter region of ELF3 as controls. SFPS-GFP efficiently coimmunoprecipitated the ELF3 transcript, but not LUX or PP2A, as expected (Fig. 7 C and D). These data suggest SFPS associates with ELF3 pre-mRNA in vivo to promote proper splicing of ELF3 transcripts.
Because ELF3 pre-mRNA splicing is altered in sfps and SFPS associates with ELF3 transcript in vivo, we examined whether ectopic expression of the functional, mature form of ELF3 can rescue the sfps phenotype. We placed the ELF3 ORF (fused to myc tag) under the control of constitutively active Cauliflower Mosaic Virus 35S promoter and transformed sfps-2. We selected homozygous transgenic lines expressing different amounts of ELF3 transcript (SI Appendix, Fig. S18A) and then tested their hypocotyl length phenotype when grown in darkness and continuous red light conditions. We found that fivefold higher expression of Myc-ELF3 compared with wild type was insufficient to rescue the sfps phenotype under red light. However, a 30-fold higher expression of Myc-ELF3 compared with wild type rescued the sfps phenotype under red light, probably because of very high overexpression of ELF3. These data suggest ELF3 is not the only downstream target responsible for the sfps phenotypes. Consistent with these data, elf3 spfs double mutant also displayed additive phenotypes under red and blue light conditions (SI Appendix, Fig. S18B).
SFPS Acts Upstream of Phytochrome Interacting Factors.
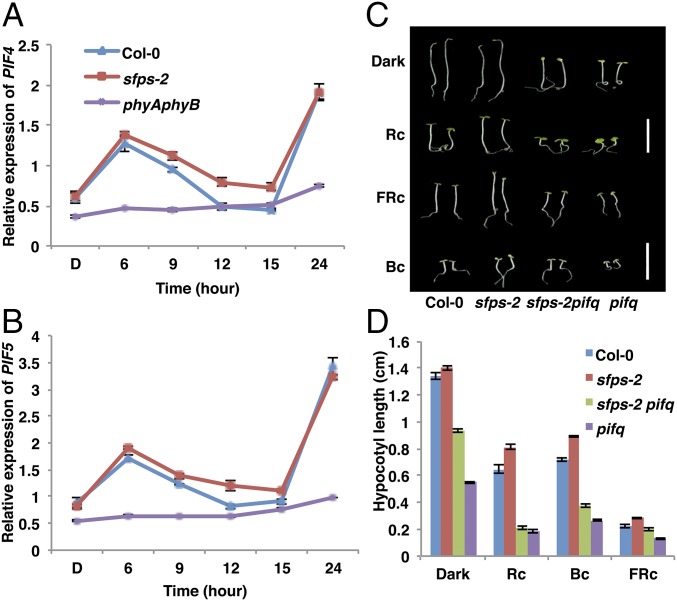
PIFs act as negative regulators downstream of phytochromes to promote hypocotyl elongation under constant dark and diurnal conditions (34, 36, 37). The expression of PIF4 and PIF5 is repressed by the evening complex in the evening, but increased later at night when PIF4/PIF5 promote hypocotyl elongation (34, 37). To examine whether the expression of PIF4/PIF5 is altered in the sfps mutant, we performed RT-qPCR assays to measure the abundance of PIF4 and PIF5. We noticed an increased transcript abundance of PIF4 and PIF5 during the evening, when the evening complex is most functional (Fig. 8 A and B), consistent with the reduced expression of ELF3 in the sfps mutant. To examine the genetic relationship between SFPS and PIFs, we crossed sfps-2 to the quadruple pif1, pif3, pif4, pif5 mutant (pifq) to generate a pifq sfps-2 quintuple mutant. Hypocotyl lengths of pifq sfps-2 seedlings were similar to pifq seedlings under red light (Fig. 8 C and D), indicating pifq is epistatic to sfps-2. SFPS therefore acts upstream of the PIF family of transcription factors to regulate photomorphogenesis.
Fig. 8.
PIFs act downstream of the genome-wide splicing defects in sfps regulating photomorphogenesis. Relative expression of PIF4 (A) and PIF5 (B) is up-regulated in the sfps-2 mutant compared with Col-0 wild type over the course of 24 h red light irradiation (7 μmol⋅m−2⋅s−1). Total RNA was extracted from the Col-0, sfps-2, and phyAphyB mutants grown in the dark or transferred to red light irradiation for 6, 9, 12, 15, and 24 h. PP2A was used as an internal control. Each bar is the mean ± SEM (n = 3 independent biological repeats, and each biological repeat include three technique repeats). (C) Representative photographs of wild-type Col-0, sfps-2, pifq, and sfps-2 pifq seedlings grown for 4 d under continuous red (1.2 μmol⋅m−2⋅s−1), far-red (0.56 μmol⋅m−2⋅s−1), and blue light (0.73 μmol⋅m−2⋅s−1) conditions. White bar, 1 cm. (D) Quantification of the hypocotyl length of different seedlings grown under light conditions indicated above. Error bar, SEM (n > 40). Bc, continuous blue light; FRc, continuous far-red; Rc, continuous red.
Discussion
This study provides strong genetic, molecular, and photobiological evidence in support of a role for SFPS as a positive regulator of phytochrome signaling pathways. To date, of the many genes with ascribed roles in phytochrome signaling, only RRC1 encodes a splicing factor with an SR domain (17, 27). rrc1 and sfps display similar phenotypes, including long hypocotyl under red light and early flowering, although sfps phenotypes appear stronger than rrc1 (17). In addition, sfps also displays long hypocotyl under blue light conditions, suggesting that, unlike RRC1, SFPS might play a broad role in photomorphogenesis. Moreover, two of the five mutants impaired in circadian clock and splicing (e.g., skip and prmt5) display photomorphogenic defects (20–22, 24–26). sfps displays a marginally short period phenotype based on luciferase reporter assays under free-running conditions, indicating SFPS plays a much stronger role in regulating photomorphogenesis than the circadian clock. Thus, it is possible that SFPS and RRC1 splicing factors function in the same pathway, or in parallel to regulate photomorphogenesis and transition from vegetative to reproductive growth. Future analyses of single and double mutants will help understand whether SFPS and RRC1 function in the same pathway.
SFPS is very similar in sequence to human and Drosophila splicing factors SPF45 (29, 30). SPF45-like proteins are encoded by all animal and plant genomes analyzed so far, suggesting a conserved function in regulating splicing. SFPS-GFP shows a characteristic localization within nuclear speckles, where it is found together with nuclear splicing factors including U2AF65B, U2A′, and U2AF35A. These factors form a complex with U2 small nuclear ribonucleoprotein and function in 3′ SS determination in animals. SFPS-GFP associates with ELF3 pre-mRNA in a RIP assay, demonstrating that SFPS can bind specific mRNAs in vivo. Overall, these data strongly suggest SFPS functions as a splicing factor, likely in 3′ SS determination in Arabidopsis.
We surprisingly found that SFPS regulates pre-mRNA splicing of as many genes in the dark as it does in the light (787 genes in dark vs. 827 genes under light), which is at first glance inconsistent with the light-dependent phenotypes of sfps. However, GO analyses revealed that both dark- and light-regulated AS pattern genes are enriched in genes regulating responses to light stimuli, photosynthesis, and the circadian clock. Thus, SFPS does not control the pre-mRNA splicing only under light conditions, but also under darkness to regulate photomorphogenesis. SFPS modulates the transcriptome both in the dark and light in a way that properly responds to the ambient light signal.
Splicing of the early flowering 3 (ELF3) pre-mRNA is under the control of SFPS and likely one of its main targets. elf3 and sfps share qualitatively similar phenotypes including long hypocotyl under light and early flowering. ELF3 functions as part of the evening complex that regulates the expression of PIF4 and PIF5 under diurnal conditions. This regulation contributes to rhythmic diurnal hypocotyl elongation, with maximal growth coinciding with PIF4 and PIF5 peak expression at the end of the night. However, despite strong defects in pre-mRNA splicing of ELF3 transcript in the sfps mutant and direct binding of SFPS to ELF3 pre-mRNA in vivo, transgenic expression of ELF3 in the sfps mutant closer to a wild-type level failed to rescue the sfps phenotypes in the light. In contrast, a simple epistasis analysis between pifq and sfps clearly places PIFs downstream of SFPS in the control of photomorphogenesis. These data suggest the regulation of pre-mRNA splicing of ELF3 alone may not be sufficient to account for the phenotypes of sfps; rather, the defects in pre-mRNA splicing of multiple genes in the light signaling and circadian clock pathways contribute to the phenotypes of sfps.
Although SFPS affects pre-mRNA splicing of many light-signaling genes to promote photomorphogenesis, the biochemical mechanisms by which SFPS regulates this process remain unknown. Phytochromes physically interact with diverse groups of factors, including PIFs, and regulate their abundance and/or activity (27). In fact, phytochromes regulate PIF function in two distinct ways (10). First, direct physical interactions between phytochromes and PIFs result in rapid light-induced phosphorylation, poly-ubiquitination, and subsequent degradation of PIFs in part through CUL4COP1-SPA and CUL3LRB E3 ubiquitin ligases (38, 39). Alternatively, phyB directly interacts with PIFs and sequesters PIFs from their function (40). Although we observed direct physical interaction between SFPS with phyB in vivo, and SFPS and phyB colocalizes to photobodies, immunoblots and qPCR assays showed that SFPS protein abundance and transcription are not regulated by light. Thus, phytochromes might regulate SFPS activity by sequestration and/or by posttranslational modification. The animal ortholog to SFPS is phosphorylated (41, 42). In addition, phytochromes possess kinase activity (43, 44). Therefore, phytochromes might regulate the activity of SFPS by phosphorylation in response to light. Further experiments are necessary to examine this hypothesis.
Various models have been proposed to explain the function of photobody formation by phytochromes in response to light (7, 8). These include storage of active phytochromes, site of degradation of phytochrome signaling components (e.g., PIFs), and site of transcription complex for rapid gene expression in response to light signals. Our data showing the colocalization of a splicing factor with phyB in nuclear photobodies suggest the photobodies might also be a site for storage and/or modification of splicing factors to regulate pre-mRNA splicing in response to light. Thus, phytochromes might form different types of photobodies specialized for each function. The size, timing, and overall dynamics of photobody formation in response to light support this hypothesis.
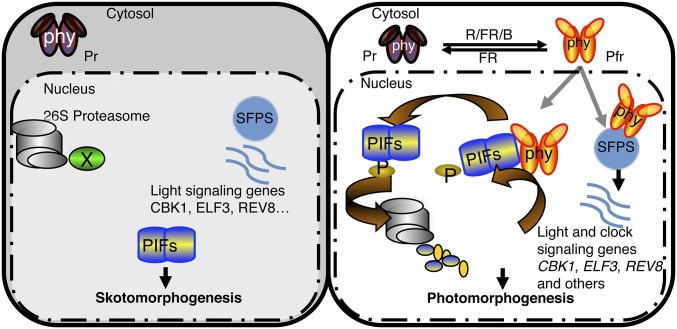
Phytochromes control the transcription of a large number of genes by regulating the activity and/or abundance of a diverse group of transcription factors including PIFs and HY5 (Fig. 9). In addition, we have shown here that phytochromes also regulate splicing of many light-signaling genes by directly affecting the activity of splicing factors such as SFPS. The coordinated control of transcription as well as splicing of light-regulated genes promotes photomorphogenesis.
Fig. 9.
Model of SFPS function in regulating photomorphogenesis. (Left) PIFs are constitutively nuclear localized, whereas phytochromes are in the cytosol in the dark. PIFs regulate photomorphogenesis in the dark and light. (Right) In response to light, phytochromes migrate into the nucleus, interact with PIFs, and induce rapid degradation of PIFs to promote photomorphogenesis. In addition, SFPS regulates pre-mRNA splicing of light signaling genes (e.g., CKB1, ELF3, REV8, and others) to fine-tune the light-regulated developmental processes in Arabidopsis. Phytochromes also interact with SFPS in vivo and regulate AS in response to light to promote photomorphogenesis. B, blue; FR, far-red; phy, phytochrome; R, red.
Materials and Methods
Detailed information is provided in SI Materials and Methods.
Plant Materials, Growth Conditions, and Measurements.
Wild-type Col-0 and various mutant seeds in the Col-0 background were used. We grew plants in Metro-Mix 200 soil (Sun Gro Horticulture) under 24-h light at 22 ± 0.5 °C for seed propagation. Wild-type and mutant seeds were surface-sterilized and plated on Murashige and Skoog growth medium containing 0.9% agar without sucrose, as described (45). After 4 d of chilling at 4 °C in the dark for stratification, seeds were exposed to 3 h of white light at room temperature to induce germination before placing them in the dark.
To measure hypocotyl lengths, cotyledon areas, and cotyledon angles, we took digital photographs of 4-d-old dark- and light-grown seedlings; at least 30 seedlings were processed in ImageJ (https://imagej.nih.gov/ij/). For flowering time measurements, plants were grown under SD (16 h dark/8 h light) or LD (8 h dark/16 h light) conditions. When the flower buds were ∼1 cm long after emergence, we counted the rosette leaf numbers as well as days to flower. These experiments were repeated at least three times.
RNA Extraction and RNAseq Analyses.
For RNA deep sequencing, we grew wild-type and sfps-2 seedlings for 4 d in the dark, and then one batch of seedlings was exposed to continuous red light for 3 h. Total RNA was isolated from dark-grown and light-exposed samples in triplicate biological repeats for each sample. Five micrograms total RNA was used to generate the RNA library for sequencing. The RNA library generation process followed the manufacturer’s protocol from the Illumina kit TruSeq Stranded Total RNA with Ribo-Zero Plant. The average RNA fragment is about 350 bp, followed by a 12-cycle PCR amplification with the PCR primer mixture provided in the kit. Library concentration was estimated by fluorescence measurement by Qubit fluorometer (Life Technologies), and quality was assessed on a Bioanalyzer DNA chip (Agilent Technologies). Sequencing was performed on an Illumina Hiseq4000 instrument with 2 × 150-bp paired-ends reads. The sequencing yield is about 120 million reads per sample. For each library, more than 90% of the reads were mapped to the unique loci of Arabidopsis TAIR10 genome with the Tophat2 pipeline (46).
Differential Gene Expression and Differential Splicing Analysis.
Differential gene expression was analyzed through Cufflink and cuffdiff pipelines (47). Genes with FDR values lower than 0.05 and absolute log twofold change greater than 0.58 (1.5-fold) were considered as differentially expressed. AS is analyzed through ASpli_1.9 (18), which makes use of junction reads information and quantifies the pre-mRNA splicing events through calculating PSI and PIR matrix {formulas: PSI(Altss) = #Jinclusion/(#Jinclusion + #Jexclusion); PSI(exon skipping) = (#Jstart + #Jend)/(#Jstart + #Jend + 2#Jexclusion); PIR(IR) = (#E1l + #IE2)/(#E1l + #IE2 + 2#jE1E2)}. The AS events with an absolute FDR <5% and Delta PSI_PIR >3% were deemed differentially spliced. GO term enrichments are analyzed by the agriGO (bioinfo.cau.edu.cn/agriGO/), using TAIR9 as the reference. For differentially expressed genes or differentially spliced genes, we compared Col-0 vs. sfps-2 mutant dark and Col-0 vs. sfps-2 mutant light samples. GO categories belonging to biological processes, molecular functions, and cellular components were analyzed. GO terms with P value < 0.05 and FDR <0.05 were considered as significantly enriched. Raw sequences (fastq files) and count tables at gene, exon, intron, AS bin and junction levels used in this paper have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE85883).
Quantitative RT-PCR Analyses.
Two micrograms total RNA were reverse transcribed using M-MLV (Invitrogen), as per manufacturer’s protocol. For RT-PCR, gene-specific primers listed in Dataset S3 were used to detect mRNA levels. The housekeeping gene PP2A (At1g13320) was used as an internal control for all RT-PCR assays. The RT-qPCR assays used the Power SYBR Green RT-PCR Reagents Kit (Applied Biosystems Inc.). Primer sequences used for RT-qPCR and RT-PCR assays are listed in Dataset S3. The cycle threshold (Ct) values were used for calculation of the levels of expression of different genes relative to PP2A as follows: 2∆CT, where ∆CT = CT (PP2A) − CT (specific gene).
RNA Immunoprecipitation Assays.
The RNA-IP assay was conducted as described previously (48), with slight modifications. Ten-d-old pSFPS:SFPS-GFP/sfps-1 seedlings grown under 12 h light/12 h dark conditions were harvested at Zeitgeber time 12 to maximize the association of SFPS with ELF3 pre-mRNA. Cross-linking is accomplished by vacuum infiltration in 0.5% formaldehyde for 10 min, and later quenched by vacuum infiltration with 0.125 M glycine for 5 min. Samples were washed with large amounts of de-ionized water, dried on filter papers, and ground into powder in liquid nitrogen. Three volumes of nuclei isolation buffer (0.25 M sucrose, 15 mM pipes at pH 6.8, 5 mM MgCl2, 60 mM KCl, 15 mM NaCl, 0.9% Triton X-100, 1 mM PMSF, and 1× Protease 591 inhibitor mixture (Sigma–Aldrich; catalog No. P9599), 1× SUPERase·In (Thermo Fisher Scientific Inc., AM269) was added to the powder and incubated on ice for 15 min. Samples were filtered with 2 layers of miracloth and then centrifuged at 16,000 × g for 10 min at 4 °C. The pellets were resuspended with 600 μL lysis buffer (50 mM Hepes at pH 7.5, 150 mM 594 NaCl, 10 mM EDTA, 1% Triton X-100, 0.1% Na Deoxycholate, 0.1% SDS, 1 mM PMSF, and 595 1× protease inhibitor mixture, SUPERase·In) before sonication. Sonicated samples were clarified by centrifugation at 16,000 × g at 4 °C for 5 min. Forty microliters Dynabead protein A and 5 microliters Anti-GFP antibody (Abcam; catalog no. ab290) was used for immunoprecipitation at 4 °C for 3 h. Immunoprecipitated samples were sequentially washed by low-salt wash buffer (150 mM NaCl, 600 0.2% SDS, 0.5% Triton X-100, 2 mM EDTA, and 20 mM Tris·Cl at pH 8.0, SUPERase·In), high-salt wash buffer (500 mM NaCl, 0.2% SDS, 0.5% Triton X-100, 2 mM EDTA, and 20 mM Tris·Cl at pH 8.0, SUPERase·In), LiCl wash buffer (0.25 M LiCl, 0.5% Nonidet P-40, 0.5% deoxycholate sodium salt, 1 mM EDTA, and 10,603 mM Tris·Cl at pH 8.0, SUPERase·In) and TE buffer (10 mM Tris·Cl at pH 8.0 and 1 mM EDTA, SUPERase·In). One milliliter buffer per sample was used for all washes; each wash requires 5 min rotation at 4 °C. Immune complexes were eluted in 125 µL elution buffer (1% SDS and 0.1 M NaHCO3, SUPERase·In) twice. The total 250-µL eluted sample was incubated with 10 µL 5 M NaCl at 65 °C for 1 h for de-cross-linking. Then add 5 µL 0.5M EDTA, 10 µL Tris⋅HCl at pH 6.5, and 1 µL 2 μg/μL proteinase 45 degree incubate for 1 h. Then add equal volume phenol:chloroform to remove protein, the RNA was precipitated in 2.5× volume 100% ethanol at −20 °C overnight. The associated RNAs were quantified by RT-qPCR with primer pairs crossing the intron–exon junctions of ELF3 pre-mRNAs.
Supplementary Material
Acknowledgments
We thank the members of the E.H. laboratory for critical reading of the manuscript. This work was supported by National Institutes of Health Grant 1R01 GM-114297 and National Science Foundation Grant MCB-1543813 (to E.H.), and a grant from the Max Planck Society (to D.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE85883).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706379114/-/DCSupplemental.
References
- 1.Bae G, Choi G. Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol. 2008;59:281–311. doi: 10.1146/annurev.arplant.59.032607.092859. [DOI] [PubMed] [Google Scholar]
- 2.Rizzini L, et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science. 2011;332:103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- 3.Mathews S, Sharrock RA. Phytochrome gene diversity. Plant Cell Environ. 1997;20:666–671. [Google Scholar]
- 4.Clack T, et al. 2009. Obligate heterodimerization of Arabidopsis phytochromes C and E and interaction with the PIF3 bHLH protein. Plant Cell 2009;21:786–799.
- 5.Sharrock RA, Clack T. Heterodimerization of type II phytochromes in Arabidopsis. Proc Natl Acad Sci USA. 2004;101:11500–11505. doi: 10.1073/pnas.0404286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fankhauser C, Chen M. Transposing phytochrome into the nucleus. Trends Plant Sci. 2008;13:596–601. doi: 10.1016/j.tplants.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Klose C, Viczián A, Kircher S, Schäfer E, Nagy F. Molecular mechanisms for mediating light-dependent nucleo/cytoplasmic partitioning of phytochrome photoreceptors. New Phytol. 2015;206:965–971. doi: 10.1111/nph.13207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Buskirk EK, Decker PV, Chen M. Photobodies in light signaling. Plant Physiol. 2012;158:52–60. doi: 10.1104/pp.111.186411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu S-H. Gene expression regulation in photomorphogenesis from the perspective of the central dogma. Annu Rev Plant Biol. 2014;65:311–333. doi: 10.1146/annurev-arplant-050213-040337. [DOI] [PubMed] [Google Scholar]
- 10.Xu X, Paik I, Zhu L, Huq E. Illuminating progress in phytochrome-mediated light signaling pathways. Trends Plant Sci. 2015;20:641–650. doi: 10.1016/j.tplants.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 11.Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014;15:108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee Y, Rio DC. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reddy ASN, Marquez Y, Kalyna M, Barta A. Complexity of the alternative splicing landscape in plants. Plant Cell. 2013;25:3657–3683. doi: 10.1105/tpc.113.117523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu X-D, Ares M., Jr Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Lin C, Gu L. Light regulation of alternative pre-mRNA splicing in plants. Photochem Photobiol. 2017;93:159–165. doi: 10.1111/php.12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shikata H, et al. Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proc Natl Acad Sci USA. 2014;111:18781–18786. doi: 10.1073/pnas.1407147112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shikata H, et al. The RS domain of Arabidopsis splicing factor RRC1 is required for phytochrome B signal transduction. Plant J. 2012;70:727–738. doi: 10.1111/j.1365-313X.2012.04937.x. [DOI] [PubMed] [Google Scholar]
- 18.Mancini E, et al. Acute effects of light on alternative splicing in light-grown plants. Photochem Photobiol. 2016;92:126–133. doi: 10.1111/php.12550. [DOI] [PubMed] [Google Scholar]
- 19.Petrillo E, et al. A chloroplast retrograde signal regulates nuclear alternative splicing. Science. 2014;344:427–430. doi: 10.1126/science.1250322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, et al. SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell. 2012;24:3278–3295. doi: 10.1105/tpc.112.100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlaen RG, et al. The spliceosome assembly factor GEMIN2 attenuates the effects of temperature on alternative splicing and circadian rhythms. Proc Natl Acad Sci USA. 2015;112:9382–9387. doi: 10.1073/pnas.1504541112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones MA, et al. Mutation of Arabidopsis spliceosomal timekeeper locus1 causes circadian clock defects. Plant Cell. 2012;24:4066–4082. doi: 10.1105/tpc.112.104828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marshall CM, Tartaglio V, Duarte M, Harmon FG. 2016. The Arabidopsis sickle mutant exhibits altered circadian clock responses to cool temperatures and temperature-dependent alternative splicing. Plant Cell 28:2560–2575 10.1105/tpc.16.00223. [DOI] [PMC free article] [PubMed]
- 24.Hong S, et al. Type II protein arginine methyltransferase 5 (PRMT5) is required for circadian period determination in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107:21211–21216. doi: 10.1073/pnas.1011987107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez SE, et al. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature. 2010;468:112–116. doi: 10.1038/nature09470. [DOI] [PubMed] [Google Scholar]
- 26.Hernando CE, Sanchez SE, Mancini E, Yanovsky MJ. Genome wide comparative analysis of the effects of PRMT5 and PRMT4/CARM1 arginine methyltransferases on the Arabidopsis thaliana transcriptome. BMC Genomics. 2015;16:192. doi: 10.1186/s12864-015-1399-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huq E, Quail PH. Phytochrome signaling. In: Briggs WR, Spudich JL, editors. Handbook of Photosensory Receptors. Wiley-VCH; Weinheim, Germany: 2005. pp. 151–170. [Google Scholar]
- 28.Schneeberger K, et al. SHOREmap: Simultaneous mapping and mutation identification by deep sequencing. Nat Methods. 2009;6:550–551. doi: 10.1038/nmeth0809-550. [DOI] [PubMed] [Google Scholar]
- 29.Lallena MJ, Chalmers KJ, Llamazares S, Lamond AI, Valcárcel J. Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell. 2002;109:285–296. doi: 10.1016/s0092-8674(02)00730-4. [DOI] [PubMed] [Google Scholar]
- 30.Neubauer G, et al. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat Genet. 1998;20:46–50. doi: 10.1038/1700. [DOI] [PubMed] [Google Scholar]
- 31.Pang Q, Hays JB, Rajagopal I. Two cDNAs from the plant Arabidopsis thaliana that partially restore recombination proficiency and DNA-damage resistance to E. coli mutants lacking recombination-intermediate-resolution activities. Nucleic Acids Res. 1993;21:1647–1653. doi: 10.1093/nar/21.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddy AS, Day IS, Göhring J, Barta A. Localization and dynamics of nuclear speckles in plants. Plant Physiol. 2012;158:67–77. doi: 10.1104/pp.111.186700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR. ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell. 2001;13:1293–1304. doi: 10.1105/tpc.13.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nusinow DA, et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwon Y-J, Park M-J, Kim S-G, Baldwin IT, Park C-M. Alternative splicing and nonsense-mediated decay of circadian clock genes under environmental stress conditions in Arabidopsis. BMC Plant Biol. 2014;14:136. doi: 10.1186/1471-2229-14-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leivar P, Monte E. PIFs: Systems integrators in plant development. Plant Cell. 2014;26:56–78. doi: 10.1105/tpc.113.120857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nozue K, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 38.Ni W, et al. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science. 2014;344:1160–1164. doi: 10.1126/science.1250778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu L, et al. CUL4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nat Commun. 2015;6:7245. doi: 10.1038/ncomms8245. [DOI] [PubMed] [Google Scholar]
- 40.Park E, et al. Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 2012;72:537–546. doi: 10.1111/j.1365-313X.2012.05114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Al-Ayoubi AM, Zheng H, Liu Y, Bai T, Eblen ST. Mitogen-activated protein kinase phosphorylation of splicing factor 45 (SPF45) regulates SPF45 alternative splicing site utilization, proliferation, and cell adhesion. Mol Cell Biol. 2012;32:2880–2893. doi: 10.1128/MCB.06327-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, et al. Phosphorylation of the alternative mRNA splicing factor 45 (SPF45) by Clk1 regulates its splice site utilization, cell migration and invasion. Nucleic Acids Res. 2013;41:4949–4962. doi: 10.1093/nar/gkt170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin A-Y, et al. Evidence that phytochrome functions as a protein kinase in plant light signalling. Nat Commun. 2016;7:11545. doi: 10.1038/ncomms11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeh KC, Lagarias JC. Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen H, Moon J, Huq E. PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 2005;44:1023–1035. doi: 10.1111/j.1365-313X.2005.02606.x. [DOI] [PubMed] [Google Scholar]
- 46.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.