Significance
Repair of the majority of UV-induced DNA damage in mammalian cells by the nucleotide excision repair pathway starts with rapid recruitment of Xeroderma pigmentosum C (XPC) protein to the lesion. However, rapidity of XPC recruitment to the lesion site in a genomic context cannot be fully explained by the known properties of XPC or its partner protein DDB2. Here, we show that the DNA damage-detecting nuclear protein PARP1 forms a stable complex with XPC before DNA damage and transfers it very rapidly to the DNA lesion site if other repair conditions are present. Since PARP1 is known to interact with many proteins under steady-state conditions, our results reveal a paradigm that an association with PARP1 could confer a functional advantage to these proteins.
Keywords: PARP1, XPC, NER, DDB2, UV
Abstract
Xeroderma pigmentosum C (XPC) protein initiates the global genomic subpathway of nucleotide excision repair (GG-NER) for removal of UV-induced direct photolesions from genomic DNA. The XPC has an inherent capacity to identify and stabilize at the DNA lesion sites, and this function is facilitated in the genomic context by UV-damaged DNA-binding protein 2 (DDB2), which is part of a multiprotein UV–DDB ubiquitin ligase complex. The nuclear enzyme poly(ADP-ribose) polymerase 1 (PARP1) has been shown to facilitate the lesion recognition step of GG-NER via its interaction with DDB2 at the lesion site. Here, we show that PARP1 plays an additional DDB2-independent direct role in recruitment and stabilization of XPC at the UV-induced DNA lesions to promote GG-NER. It forms a stable complex with XPC in the nucleoplasm under steady-state conditions before irradiation and rapidly escorts it to the damaged DNA after UV irradiation in a DDB2-independent manner. The catalytic activity of PARP1 is not required for the initial complex formation with XPC in the nucleoplasm but it enhances the recruitment of XPC to the DNA lesion site after irradiation. Using purified proteins, we also show that the PARP1–XPC complex facilitates the handover of XPC to the UV-lesion site in the presence of the UV–DDB ligase complex. Thus, the lesion search function of XPC in the genomic context is controlled by XPC itself, DDB2, and PARP1. Our results reveal a paradigm that the known interaction of many proteins with PARP1 under steady-state conditions could have functional significance for these proteins.
Nucleotide excision repair (NER) is a versatile pathway that removes a wide variety of DNA lesions including UV radiation (UV)-induced cyclobutane pyrimidine dimers (CPD) and 6–4 pyrimidine-pyrimidone photoproducts (6-4PP). There are two subpathways of NER: the global genomic NER (GG-NER) that removes the majority of lesions from the entire genome and the transcription-coupled NER that repairs the minority of total lesions that occur on the transcribed strand (1). The GG-NER process is dependent on the Xeroderma pigmentosum C (XPC) protein, the arrival and stabilization of which at the lesion site, followed by its timely departure, are crucial for permitting the downstream NER events (2). XPC accomplishes some of these tasks on its own or with the help of processes initiated by two proteins that independently reach the lesion site very rapidly, namely UV-damaged DNA-binding protein 2 (DDB2) and poly(ADP-ribose) polymerase 1 (PARP1). It is known that once XPC reaches the vicinity of the DNA lesion site, the UV–DDB ubiquitin ligase complex containing DDB1, DDB2, Cul4A, and Rbx1 regulates its specific binding and stabilization at the site (2). However, we have the least understanding of the ability of XPC to rapidly search for and arrive at the very few lesions surrounded by a vast majority of undamaged bases in the chromatin structure (3).
There are three proposed mechanisms by which XPC could be rapidly recruited to the lesion site in the genomic context. The first mechanism is based on XPC’s inherent capacity to interrogate the DNA for verifying the presence or absence of a lesion. It has been suggested that XPC searches for the lesion-induced helical distortion in the genome (1) by association and quick dissociation until it encounters the lesion site where it stabilizes due to a stronger association (4). Since the yeast ortholog of XPC forms identical crystal structures with normal and UV-damaged DNA (5), it is proposed that the discrimination between normal and damaged DNA depends on the difference in the kinetics of twisting and opening of the helix by XPC at these two sites (5, 6). Although this physical verification mechanism could work rapidly in vitro with small and naked DNA, it would be too slow to explain the rapid recruitment of XPC that is known to occur within minutes at UV-lesion sites in a complex eukaryotic chromatin. Therefore, the second proposed mechanism is that DDB2 helps XPC in finding the UV lesions in the genomic context. This is based on the observation that the Xeroderma pigmentosum group E (XP-E) cells, which are deficient in DDB2 but proficient in XPC, can slowly repair 6–4PP lesions but fail to remove CPD lesions, which constitute the majority of the lesions formed by UV radiation (7, 8). Based on the phenotype of XP-E cells and the biochemical studies of XPC with these two types of lesions (2), it is proposed that, although XPC can directly recognize the greater degree of helical distortion in DNA induced by 6–4PP lesions, it cannot recognize the smaller degree of distortion caused by CPD lesions until DDB2 binds to these lesions and increases the degree of helical distortion at the site (3). Finally, the third proposed mechanism is that XPC recognizes the remodeled chromatin at the lesion site, which is the result of events initiated by ubiquitination of histones by the UV–DDB ligase complex (9). This is supported by the observations that decreased histone ubiquitination impairs the eviction of the UV–DDB ligase complex (10) and recruitment of XPC to the lesions site (11). Thus, apart from the direct recognition of the DNA damage by XPC, all other mechanisms to explain XPC’s ability to rapidly reach most of the UV lesions in the genomic context depend on DDB2. However, XPC must have some DDB2-independent mechanism of recruitment to lesions other than 6–4PP. For example, CPD are recognized and repaired even after DDB2 is degraded (12) or when the number of lesion sites exceeds the number of DDB2 molecules in the cell (13). In addition, XPC recognizes and starts NER at other types of DNA damages, such as bulky adducts and cross-links that are not likely to be recognized by DDB2, because these lesions cannot be accommodated in DDB2′s recognition pocket (14–16).
Earlier, we showed that PARP1, an abundant nuclear enzyme in higher eukaryotes, is recruited very early to the UV-lesion site and catalytically activated to form polymers of ADP-ribose (PAR) (17, 18). It has been shown by others and our team that PARP1 and DDB2 reach the lesion site in the same time frame and cooperate with each other to increase the efficiency of GG-NER. More specifically, DDB2 has been shown to stimulate catalytic activity of PARP1, which in turn PARylates DDB2 and DDB1 (18, 19). The inhibition of PARylation has been shown to block the turnover of DDB2 at UV-damaged chromatin (18) and to decrease total cellular levels of DDB2 (19). Additionally, the activated PARP1 promotes chromatin decondensation by DDB2 (20) via recruitment of the remodeler protein ALC1 at the UV-lesion site (19). Together, these studies suggest that the interplay of PARP1 and DDB2 at UV-damaged DNA could be a mechanism for recruitment and stabilization of XPC at UV-damaged chromatin (18, 20). Recent studies have shown PARylation of XPC in vitro (21) and in the cells responding to oxidative DNA damage (22). However, the significance of the PARylation of XPC in NER of UV-induced DNA damage is not clear since the higher affinity of XPC for larger PAR chains shown in vitro (21) would repel XPC from DNA, and in vivo PARylation of XPC was not observed in UV-irradiated cells (22). Thus, studies to date have not shown an unequivocal direct DDB2-independent role for PARP1 and PARylation in recruitment of XPC in GG-NER.
Here, we show that PARP1 stably interacts with XPC in the nucleoplasm under unchallenged conditions, i.e., in the absence of any type of exogenously induced DNA damage. The functionally important DNA-binding region of XPC is involved in its interaction with PARP1. Using various cellular models and in vitro assays with purified proteins, we show that, after UV irradiation, PARP1 rapidly escorts XPC to the UV-lesion site and facilitates its handover to the damaged DNA in the presence of the UV–DDB ligase complex. We also show that, although the PARP1 catalytic function does not influence the initial interaction between these two proteins in the nucleoplasm, it is required for efficient recruitment of their complex to the lesion site. Our results reveal that the interaction of XPC with PARP1 in nucleoplasm under steady-state conditions facilitates the search function of XPC for DNA damage in the genomic context.
Results
DDB2-Independent Nucleoplasmic Interaction of PARP1 with XPC Before and After DNA Damage.
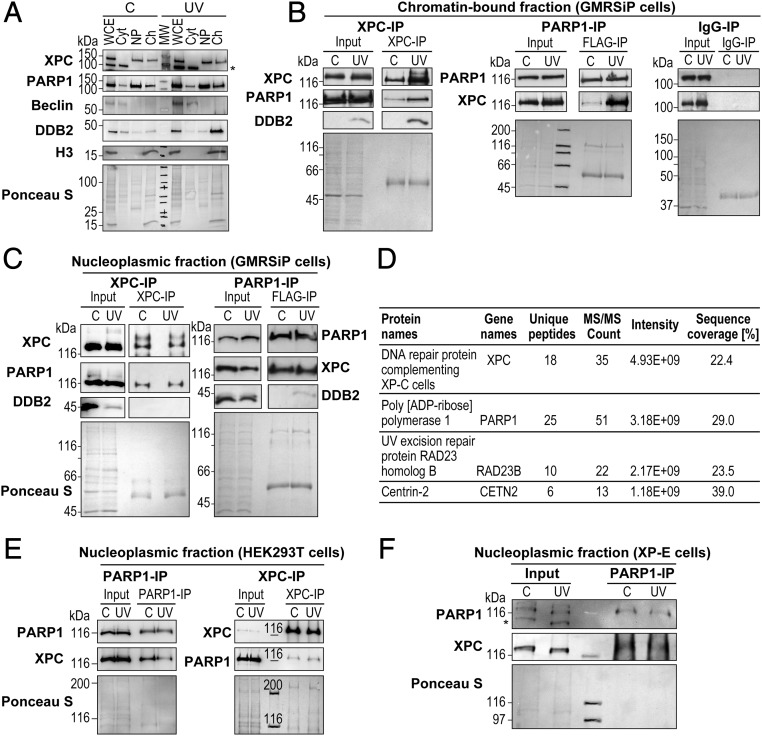
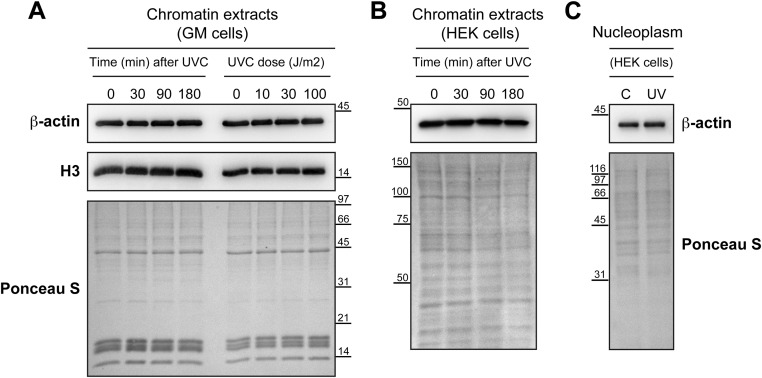
Within minutes after UV irradiation, XPC, DDB2, and PARP1 are present at the UV-induced DNA lesions in cells. Although independent interactions of DDB2 with XPC (18, 23) or PARP1 (18, 19) at the lesion site are known, here we examined whether XPC interacts directly with PARP1 on the UV-damaged chromatin. The FLAG-PARP1–expressing human skin fibroblasts (GMRSiP) were fractionated before and after UV irradiation to prepare cytoplasm, nucleoplasm, and chromatin-bound protein fractions (Fig. 1A), as described earlier (18). In this protocol, the chromatin proteins such as histone H3 do not leak in the nucleoplasmic fraction but are extracted in the chromatin-bound protein fraction. Moreover, we further validated this fractionation protocol by confirming our earlier observation (18) that DDB2 accumulates in the chromatin fraction after UV irradiation (Fig. 1A). The XPC-immunoprecipitation (IP) of chromatin extracts with equal protein content from control and UV-irradiated cells revealed a significant increase in UV-induced association of PARP1 with XPC on the chromatin (Fig. 1B, Left). We also observed a UV-induced increase in the interaction of DDB2 with XPC at the chromatin, which is in agreement with our previous observation of this interaction identified by DDB2-IP (18). The reciprocal PARP1-IP using FLAG antibody revealed a strong UV-induced association of XPC with PARP1 in the chromatin-bound fraction (Fig. 1B, Middle). Since PARP1 and XPC are present in the nucleoplasm before and after irradiation (Fig. 1A), we examined whether they also interact with each other in this subnuclear fraction. The reciprocal IP for XPC and PARP1 in the nucleoplasmic fraction revealed a strong association of these two proteins not just after UV irradiation but also under control conditions before irradiation (Fig. 1C). Both the IPs failed to pull down DDB2 from control nucleoplasm, indicating a DDB2-independent nature of PARP1–XPC nucleoplasmic interaction before irradiation. Even after UV irradiation, XPC-IP confirmed its lack of interaction with DDB2 in nucleoplasm, whereas PARP1-IP revealed a feeble interaction of PARP1 with DDB2 (Fig. 1C), reflecting some turnover of the PARP1–DDB2 complex from postirradiation chromatin to nucleoplasm.
Fig. 1.
PARP1 interacts with XPC in the nucleoplasmic and chromatin fractions. (A) The GMRSiP cells expressing FLAG-tagged PARP1 were irradiated with 30 J/m2 UVC (or control), and whole-cell extracts (WCE) were fractionated to obtain cytoplasm (Cyt), nucleoplasm (Np), and chromatin-bound (Ch) fractions. The proteins from each fraction were immunoblotted for XPC, PARP1, and DDB2. Beclin and histone H3 were used as cytoplasmic and chromatin markers, respectively. The asterisk indicates a nonspecific band. The Ponceau S staining reflects the protein content at the end of each fractionation step. (B and C) The chromatin (B) and nucleoplasm fractions (C) of GMRSiP cells prepared as described above were subjected to IP for XPC, FLAG (PARP1), and mouse IgG (negative control), followed by the detection of PARP1, XPC, and DDB2. (D) Table showing the XPC-interacting proteins identified after XPC-IP of HEK293T cells followed by mass spectrometry analysis. (E) PARP1-IP and XPC-IP were performed in the Np fractions of control and UVC-treated HEK293T cells as shown in C. The Input and IP eluates were probed for XPC and PARP1. (F) PARP1-IP was performed in the Np fractions of control, and UVC-treated XP-E cells as described above. The Input and IP eluates were probed for XPC and PARP1. For B, C, E, and F the Ponceau S staining was used as loading control, and results shown here are representative of results from two to four experiments.
The interaction between PARP1 and XPC in UV-irradiated chromatin was expected, as both are known to bind to UV-damaged DNA. However, their interaction in nucleoplasm under steady-state conditions before UV challenge was unexpected; therefore, we examined this interaction in further detail using multiple cellular and in vitro models. To exclude the possibility that this could be an artifact of the expression of exogenous FLAG-tagged PARP1 in the above model, we examined this interaction in HEK293T cells that express endogenous XPC, PARP1, and DDB2. The mass spectrometry of the proteins that coimmunoprecipitate (co-IP) with XPC from the lysates of unirradiated HEK293T cells confirmed the presence of PARP1 as well as two known partners of XPC, namely HR23B and Centrin2 (Fig. 1D). The reciprocal IP for PARP1 and XPC validated the interaction of even the endogenous PARP1 with XPC in this fraction (Fig. 1E). Interestingly, the mass spectrometry of XPC eluate did not reveal the presence of DDB2, confirming its lack of interaction with XPC under control conditions. The DDB2-independent interaction between endogenous PARP1 and endogenous XPC was further confirmed in the nucleoplasm of DDB2-deficient XP-E cells (Fig. 1F). Thus, using different cellular models expressing endogenous or exogenous PARP1, and the variable status of DDB2, we consistently observed a DDB2-independent interaction between XPC and PARP1 in the nucleoplasm before and after irradiation and an increased interaction at the chromatin after irradiation.
Direct Interaction of XPC and PARP1.
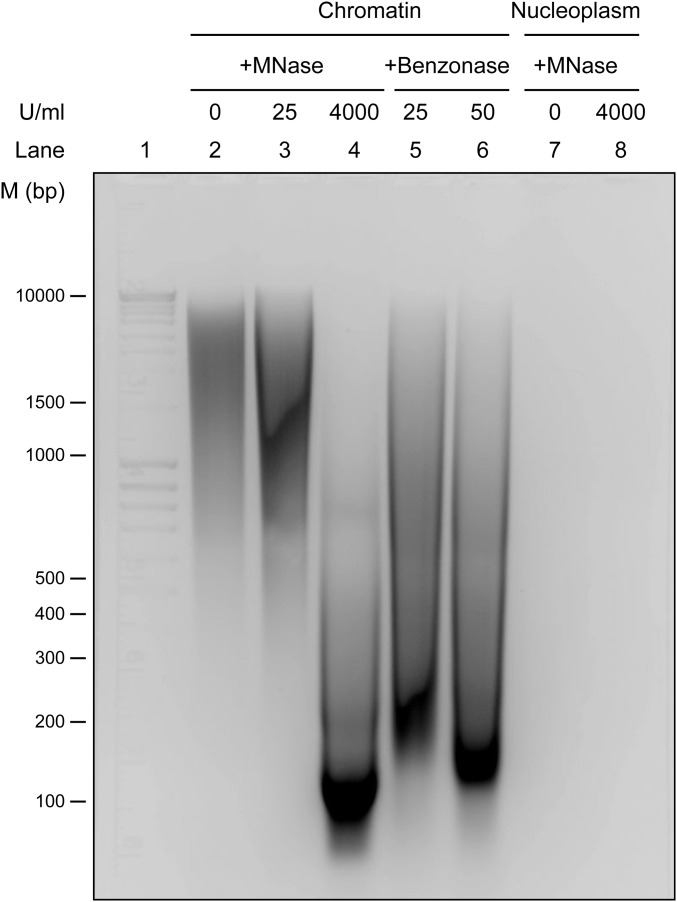
PARP1 and XPC are DNA-binding proteins; therefore, we examined the possibility that DNA could be mediating their interaction in the nucleoplasm. Unlike chromatin fraction, the nucleoplasmic fraction of the unirradiated HEK293T cells contained undetectable levels of DNA (Fig. S1). Very high doses of micrococcal nuclease (MNase) or benzonase are often used for digesting DNA from chromatin preparations to demonstrate direct interactions of proteins. In our IP studies, we prepared a chromatin-bound protein fraction after treatment of chromatin pellets with very low-dose MNase (25 U/mL). Since treatment of HeLa cell extracts with high-dose MNase was shown to digest the DNA to mononucleosomes (23), we treated the chromatin fraction of HEK293T cells with different doses of MNase up to 4,000 U/mL or benzonase up to 50 U/mL to digest it down to mononucleosomal DNA at 147 bp (Fig. S1). Although there was no detectable DNA in the nucleoplasmic fraction, we still treated this fraction from control or UV-treated HEK293T cells with 4,000 U/mL MNase before subjecting it to XPC-IP and observed that MNase did not break the association of XPC with PARP1 (Fig. 2A), confirming DNA-independent interaction of these two proteins in the nucleoplasm.
Fig. S1.
DNA profile after nuclease activity. The chromatin and nucleoplasmic fraction of HEK293T cells were treated with an increasing concentration of MNase or Benzonase, as indicated. The DNA was purified with phenol–chloroform and resolved on 2% agarose gel.
Fig. 2.
Identification of the domains implicated in the interaction between PARP1 and XPC. (A, Left) IP for XPC and rabbit IgG were performed in the nucleoplasm of HEK293T cells prepared as described in Fig. 1E. (A, Right) Nucleoplasmic extracts were also treated with 4,000 U/mL MNase and subjected to XPC-IP. The Input and eluates were probed for XPC and PARP1. Ponceau S was used as loading control. (B, Top) Pictogram of GFP-tagged full-length XPC and its five fragments used in the study. The domains marked as TGD and BHD1-3 represent the transglutaminase homology domain and β hairpin domains 1–3. (B, Lower) The HEK293T cells were transiently transfected with GFP-tagged full-length XPC or its fragments for 48 h, and cell extracts were subjected to PARP1-IP. The Input and the IP eluates were analyzed for GFP (XPC) and PARP1. The relative intensity of the IP band was measured as a fraction of the total input protein. The strength of the interaction between XPC fragments and PARP1 was expressed as relative to the interaction of full-length XPC with PARP1. (C, Top) The pictogram of the full-length XPC and its fragments used in this study. (C, Lower) The XPC fragments were reacted with pure PARP1 for 30 min at 25 °C, followed by PARP1-IP on magnetic beads. The bead elutes were probed for PARP1, GST (XPC-N), and histidine (XPC-C1 and XPC-C2). (D, Top) Pictogram showing the domains of full-length PARP1 and its N- or C-terminal fragments used in this study. The DBD, AMD, and CAT are the DNA-binding, automodification, and catalytic domains, respectively. (D, Lower) The PARP1-depleted GMSiPs were transiently transfected with the full-length FLAG-PARP1 or its N-terminal fragment (GFP-DBD) and the PARP1−/− A1 cells with the C-terminal fragment for 48 h. The cell extracts were subjected to XPC-IP, and the bead eluates were analyzed for XPC and PARP1 or its fragments (F1-23 and C2-10). The data represent similar results observed in two experiments.
To identify the domains of XPC involved in interaction with PARP1, we expressed GFP-tagged XPC and its five different partially overlapping fragments in HEK293T cells (Fig. 2B, Top). The extracts from these cells under control conditions were subjected to PARP1-IP followed by immunoblotting for GFP. Since the expression levels of full-length XPC and its fragments varied greatly after transfection (Fig. 2B, Input lanes), the strength of interaction of each XPC fragment with PARP1 was measured as a fraction of total input protein that coimmunoprecipitated with PARP1 and expressed relative to the interaction of full-length XPC with PARP1 (Fig. 2B, Lower). While control GFP protein did not interact with PARP1, four of the XPC fragments that span from 427 to 940 aa showed an interaction with PARP1 similar to that seen with full-length XPC, whereas a comparatively weaker interaction was observed with an N-terminal fragment (1–495 aa).
The interaction observed above between XPC or its fragments and PARP1 after PARP1-IP could not exclude the possibility that PARP1-interacting proteins may be indirectly mediating their interaction. Therefore, we examined the interaction of purified PARP1 in vitro with equimolar amounts of three purified fragments of XPC, namely GST-tagged 141–250 aa (XPC-N), His-tagged 496–734 aa (XPC-C1), and His-tagged 734–933 aa (XPC-C2) (Fig. 2C, Top). The PARP1-IP of above reaction mixtures revealed a strong interaction of PARP1 with XPC-C1 fragment, a weak interaction with XPC-C2 fragment and no interaction with XPC-N fragment, indicating that 496–734 aa is key region of XPC that interacts with PARP1 (Fig. 2C, Lower). These results are in agreement with the above in vivo data (Fig. 2B) showing a comparatively weaker interaction with PARP1 for the N-terminal XPC fragment (1–495 aa) compared with a strong interaction seen with full-length XPC or four of its fragments that contain the central region of XPC. The results with purified proteins and in vivo data with cells expressing XPC fragments strongly indicate an interaction of the central portion of the XPC protein spanning 496 to 734 aa with PARP1 without intervention of other proteins or DNA.
To identify the domains of PARP1 implicated in this interaction, its N-terminal (1–232 aa) fragment was expressed in GMSiP cells in which endogenous PARP1 was depleted by shRNA, whereas the C-terminal (232–1014 aa) fragment of PARP1 was expressed in PARP1−/− A1 cells. For control, we used GMRSiP cells that express FLAG-tagged full-length PARP1 in the GMSiP cells (Fig. 2D, Top). The XPC-IP of the extracts of these cells revealed that the C-terminal fragment of PARP1 is implicated in its interaction with XPC (Fig. 2D, Lower). Collectively, these results demonstrate that the region of XPC that is involved in its interaction with DNA, DDB2, and HR23B (2) is also involved in its interaction with PARP1.
Roles of the PARP1–XPC Complex and Catalytic Activity of PARP1 in Recruitment of XPC to UV-Damaged DNA.
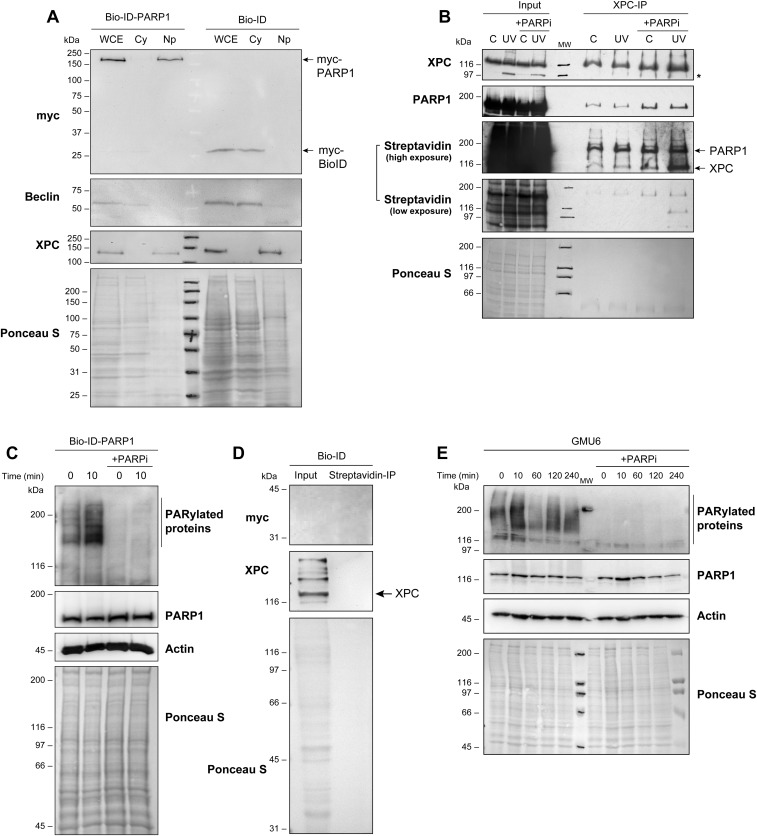
PARP1 is known to rapidly reach damaged DNA in the chromatin context; hence, the PARP1–XPC complex formed in the nucleoplasm before irradiation could have a role in rapidly escorting XPC to the DNA lesion site after UV irradiation. To trace the intranuclear movement of PARP1-bound XPC, we used the proximity-dependent biotin identification (Bio-ID) technique (24). The FLAG-PARP1 was cloned in the myc-tagged biotin ligase BirA vector to create a new Bio-ID-PARP1 vector. The expression of Bio-ID-PARP1 in GMSiP cells depleted of endogenous PARP1 in the nuclear fraction (Fig. S2A) ensured that any nuclear protein that stably associates with or stays within 10 nm of the cloned PARP1 would be stably biotinylated. The absence of endogenous PARP1 in these cells eliminated the possibility that any PARP1-associated protein would not be biotinylated. For optimal biotinylation, we incubated these cells for 16–18 h with biotin before irradiation (24) and confirmed biotinylation of XPC and auto-biotinylation of PARP1 in XPC-IP eluates of the nucleoplasm of control and UV-irradiated cells (Fig. S2B). To examine the role of the catalytic activity of PARP1 in this interaction, we also treated the cells with PARPi PJ-34 under conditions that abolished the signal for PAR-modified proteins in control or UV-irradiated cells (Fig. S2C). Interestingly, the presence of PARPi did not abolish the interaction between XPC and PARP1, indicating that catalytic function of PARP1 is not required for this interaction in the nucleoplasm (Fig. S2B). Using this model, we tracked UV-induced movement of biotinylated XPC and PARP1 from the nucleoplasmic to chromatin fractions of control or UV-treated cells (Fig. 3A). In the streptavidin-IP of nucleoplasm, both PARP1 and XPC were detectable before and after irradiation, confirming biotinylation of XPC and auto-biotinylation of Bio-ID-PARP1 (Fig. 3A, Left, and Fig. S2D). The decrease in signal for biotinylated XPC after UV irradiation suggested a movement of XPC away from this fraction. Interestingly, PARPi blocked the reduction in the signal for XPC after UV irradiation, indicating a potential role of catalytic activity of PARP1 in the movement of XPC from the nucleoplasmic fraction to damaged DNA. The absence of biotinylated DDB2 in streptavidin-IP (Fig. 3A, Left), once again confirmed that DDB2 did not interact with Bio-ID-PARP1 or its complex with XPC in the nucleoplasm.
Fig. S2.
Characterization of the Bio-ID and Bio-ID-PARP1 cells. (A) The control Bio-ID and Bio-ID-PARP1 whole-cell extracts (WCE) were fractionated to obtain cytoplasm (Cy) and nucleoplasm (Np) fractions. The proteins in each fraction were resolved on SDS/PAGE and immunoblotted for myc. The cytoplasm and nuclear fraction were verified by probing for Beclin and XPC, respectively. (B) Bio-ID-PARP1 cells were treated with 50 µM biotin 16 h before irradiation with 30 J/m2 UVC (or control). The PARPi PJ-34 was used at 10 µM to inhibit PARP1 catalytic activity. Cells were fractionated 10 min after irradiation to obtain nucleoplasm that was subjected to XPC-IP. The eluates were analyzed for PARP1, XPC, and biotin using streptavidin. The data represent identical results observed in two experiments. (C) The Bio-ID-PARP1 cells treated with 10 µM PJ-34 (or control) were irradiated with 30 J/m2 UVC. The total cell extracts prepared 10 min after UVC irradiation were resolved on SDS/PAGE and immunoblotted for PARylated proteins (LP-96-10), PARP1, and β-actin. (D) The pcDNA3.1 mycBioID plasmid-expressing cells were fractionated and streptavidin-IP was performed in the nucleoplasmic extract. Input and eluates were probed for myc and XPC. (E) The GMU6 cells were pretreated with 10 µM PJ-34 (or control) and irradiated 30 min after with 30 J/m2 UVC. Total cell extract collected at different times after UVC irradiation as indicated were resolved on SDS/PAGE and probed for the PARylated proteins (LP-96-10) PARP1 and actin. For A–E, Ponceau S staining was used as loading control.
Fig. 3.
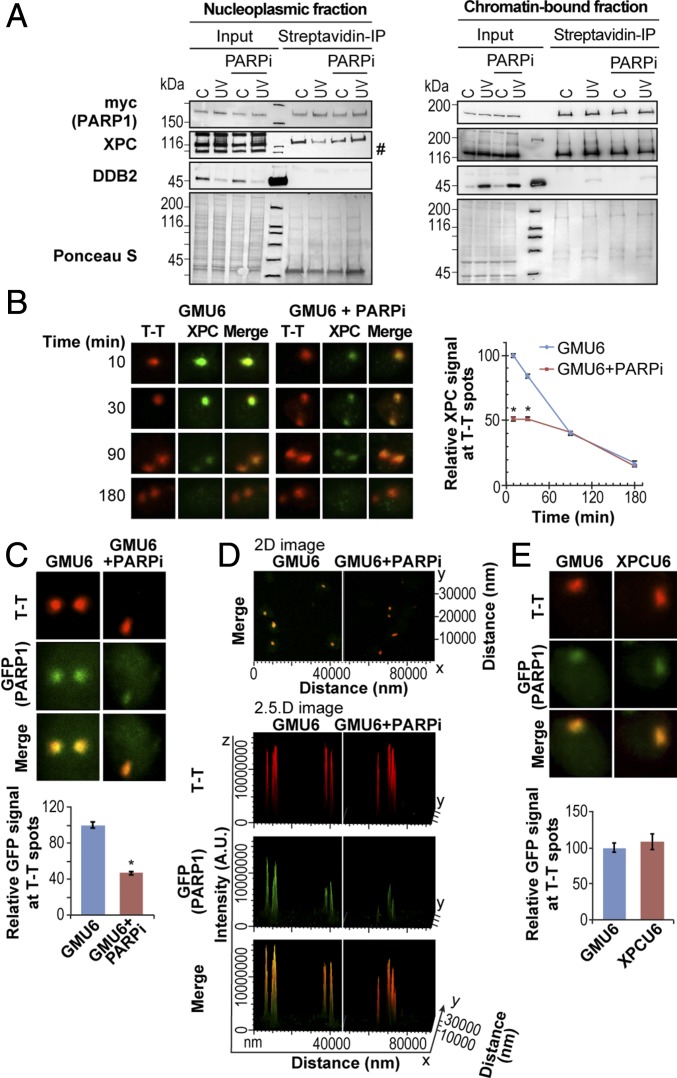
Efficient recruitment of PARP1 and XPC to the UV lesion requires PARP1 catalytic activity. (A) Bio-ID-PARP1 cells expressing myc-Bio-ID-FLAG-PARP1 were irradiated (30 J/m2 UVC or control) with or without PARPi (PJ-34). The nucleoplasm (Left) and chromatin-bound (Right) fractions were subjected to streptavidin-IP. The eluates were analyzed for myc (PARP1), XPC, and DDB2. The data represent similar results observed in three experiments, and Ponceau staining provides loading control. The “#” refers to a nonspecific band. (B) The XPC kinetics at the UV lesions were monitored in GMU6 cells up to 180 min after local UVC irradiation with 100 J/m2 UVC through 5-µm pores of a polycarbonate filter with or without PARPi PJ-34. The background-corrected signal for XPC (green) at T-T spots (red) relative to the 10-min signal is represented as mean ± SEM (200–500 spots from three experiments). Note: In all panels of this and subsequent figures, an asterisk (*) denotes a statistically significant difference with P value < 0.05 with unpaired two-tailed t test. (C and D) The GMU6 cells were transiently transfected with GFP-PARP1 for 24 h and locally irradiated with 100 J/m2 UVC in the absence or presence of PARPi PJ-34. (C) The GFP (PARP1) signals at local T-T spots (red) after background correction were pooled from 200 to 300 spots derived from three experiments and expressed relative to the signal observed in cells not treated with PARPi. (D) A representative 2D-merged image for GFP (PARP1) and T-T (red) colocalization and the orthogonal view (2.5D image) for the same field is shown to visualize signal intensity of T-T and GFP. The x and y axis represent distance in nanometers, and the z axis represents fluorescence intensity in arbitrary units. (E) The accumulation of the GFP (PARP1) at T-T lesions was monitored in GMU6 and XPCU6 cells 10 min after irradiation with UVC at 100 J/m2. The background-corrected GFP signal at lesion sites relative to the signal observed in GMU6 cells is expressed as mean ± SEM derived from ≥150 spots from two experiments.
To examine whether the biotinylated nucleoplasmic XPC reaches the UV-damaged DNA, we performed the streptavidin-IP of corresponding chromatin fractions from these cells (Fig. 3A, Right). There was some signal for biotinylated XPC under steady-state conditions, which represents a basal level of interrogation of DNA at all times by the multifunctional XPC protein and an increase in this signal in UV-treated cells, which was suppressed by PARPi (Fig. 3A, Right). Our results are in agreement with a previous report that XPC constantly associates–dissociates with chromatin under steady-state conditions; an additional association of XPC is seen with chromatin after UV-induced DNA damage (4). The UV-mediated increase in biotinylated XPC at chromatin with a corresponding decrease in nucleoplasm suggests that either PARP1-bound XPC moved from nucleoplasm to UV-damaged chromatin or a completely independent XPC arrived at the lesion site and was biotinylated by PARP1 within 30 min after irradiation when the samples were harvested. However, within the same time period of 30 min postirradiation, DDB2, which is known to closely interact with PARP1 at the chromatin immediately after UV irradiation (18, 19), was not strongly biotinylated by Bio-ID-PARP1 (Fig. 3A, Right). Thus, the biotinylated XPC that is deposited on the UV-damaged chromatin must have originated from nucleoplasmic PARP1–XPC complexes formed before UV damage, and PARPi suppressed this movement.
To validate the inhibitory effect of PARPi on the movement of XPC from nucleoplasm to chromatin, we used immunocytological methods to visualize XPC at the site of local ultraviolet C (UVC) irradiation up to 3 h in the NER-proficient GMU6 human skin fibroblasts with or without treatment with PARPi (Fig. 3B). The GMU6 fibroblasts were locally irradiated with UVC through 5-µm pores in a polycarbonate filter, which produces distinct subnuclear areas of irradiation that are surrounded by unirradiated zones in the nucleus (25). Unlike Western blot data that reveal total XPC molecules that are bound to both UV-damaged and undamaged portions of chromatin, the immunocytological data with local irradiation reflect the status of XPC only in the irradiated subnuclear zones, thus excluding the “noise” of XPC signals from the unirradiated portions of the nucleus. In the subnuclear irradiated zones identified by staining with thymine dimer (T-T) CPD-specific antibody, the endogenous XPC followed a normal kinetics, i.e., initial strong accumulation at 10 min followed by a steady decline to 40% of initial levels in 90 min. The treatment of cells with PARPi, which could abolish the signal for PAR-modified proteins in control or UV-irradiated cells (Fig. S2E), also suppressed the initial recruitment of XPC at 10 min by 50%. Moreover, PARPi slowed down the departure of XPC from the lesion site in the first 90 min, compared with the rapid turnover of XPC in cells not treated with PARPi (Fig. 3B, Right). Thus, the major impact of PARP inhibition was in partially suppressing the initial recruitment of XPC to UV-lesion sites.
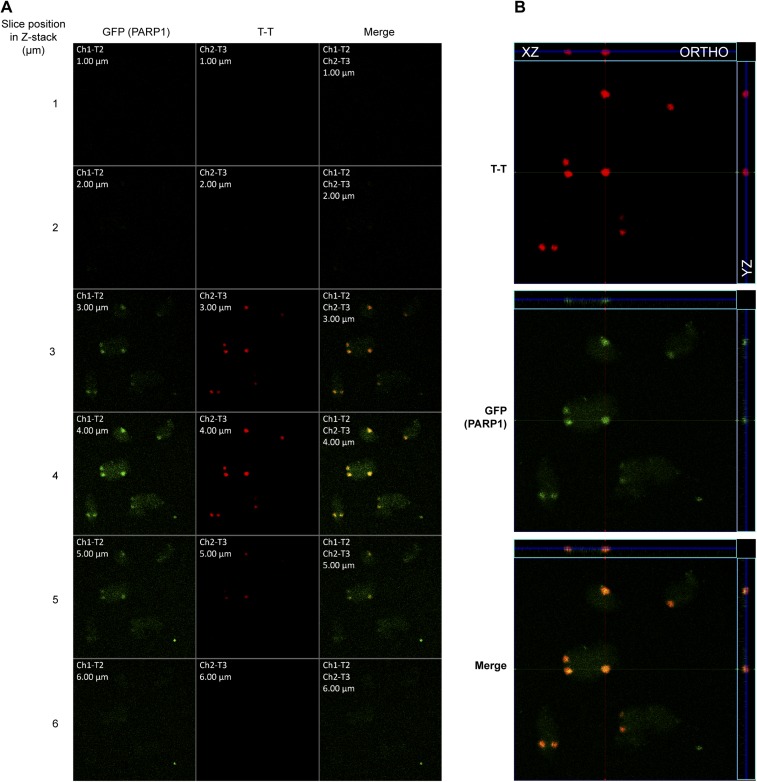
Since XPC is in a complex with PARP1 in the nucleoplasm, we examined the effect of PARPi on the recruitment of PARP1 itself to UV-damaged chromatin. Using cells expressing GFP-PARP1 and a recently developed in situ extraction technique that can selectively identify DNA-bound PARP1 (26), we observed that PARPi PJ-34 suppressed by 50% the initial recruitment of GFP-PARP1 to local UV-irradiated subnuclear zones, which were identified by immunostaining for T-T (Fig. 3C, Lower). The Z-stack images (Fig. S3A) and their orthogonal view (Fig. S3B) of the locally irradiated GMU6 cells confirmed the spatial colocalization of the GFP-PARP1 with T-T (Fig. 3C). The immunofluorescence image with multiple locally irradiated cells presented in 2D and 2.5D format revealed that PARPi treatment significantly reduced the intensity of colocalized signal for GFP-PARP1 at the site of DNA damage (Fig. 3D). Interestingly, the recruitment of PARP1 itself to a UV-lesion site is not dependent on XPC because it occurs to an identical extent in both XPC-proficient (GMU6) and -deficient (XP-C) cells (Fig. 3E). Collectively, our results indicate that the initial phase of the basal level of recruitment of PARP1 and XPC to UV-damaged DNA does not require catalytic activity of PARP1, whereas the second phase occurs in response to PARP1 activation.
Fig. S3.
PARP1 colocalizes with UV-damaged DNA in vivo. (A) The GMU6 cells were transiently transfected with GFP-PARP1, and 24 h later were locally irradiated with 100 J/m2 UVC. The cells were subjected to the in situ extraction protocol with detergent and salt as described in SI Materials and Methods. Representative Z-stack images collected at 1-µm sections show the colocalization of the GFP (PARP1) with the T-T lesions (slices 3–5). Each boxed image collected at 63× magnification represents the field size of 100 μm × 100 μm. (B) The Z-stack presented in A is shown in the orthogonal view to observe the GFP colocalization with T-T. The green and red lines indicate the orthogonal planes of the X-Z and Y-Z projection, respectively, and the blue lines, the Z-depth of the 4-µm optical slice.
PARP1-Mediated Recruitment of XPC to UV-Lesion Sites Is Independent of DDB2.
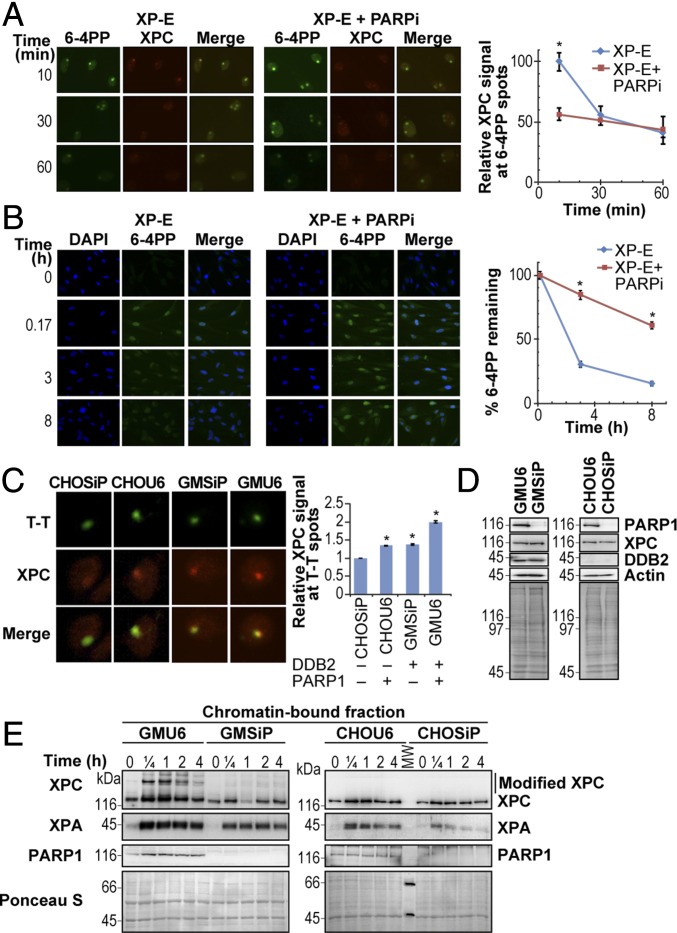
DDB2 is known to recruit XPC to the UV-lesion site, and since DDB2 and PARP1 interact with each other to facilitate NER (27), our results of suppressed XPC recruitment to the UV-lesion site could also be an indirect effect of PARPi on the role of DDB2 in recruiting XPC. To exclude this possibility, we used DDB2-deficient XP-E cells to examine the effect of PARPi on colocalization of XPC with 6–4PP lesions after local UV irradiation. At the lesion sites, the signal for XPC declined rapidly by 50–60% from 10 to 60 min (Fig. 4A). In contrast, PARPi treatment not only reduced the initial recruitment of XPC at lesion site by 50%, but also slowed down XPC turnover up to 60 min, a trend that was also observed in DDB2-proficient GM cells (Fig. 3B). An identical profile of suppression of XPC recruitment and turnover in DDB2-proficient and -deficient cells indicates that the suppression of XPC recruitment by PARPi is not mediated via DDB2. The biological end-point of XPC recruitment to the lesion site is the repair of UV-damaged DNA. Therefore, we measured the kinetics of removal of 6–4PP lesions up to 8 h following global UV irradiation in XP-E cells. As expected, almost all 6–4PP lesions were removed in XP-E cells by 8 h, but the treatment with PARPi significantly slowed down this repair process (Fig. 4B). Since the depletion of PARP1 completely suppressed any PAR formation in response to UV irradiation without affecting PARP2 expression (18, 28, 29), these results indicate that XPC recruitment is partially controlled by the catalytic activity of PARP1 in a DDB2-independent manner. Collectively, our findings demonstrate that the inhibition of PARP1 activity reduces recruitment of XPC to UV damage with a direct negative consequence on the repair of UV-induced lesions.
Fig. 4.
XPC recruitment to UV lesions by PARP1 is DDB2-independent. (A) XP-E cells with or without PARPi (ABT-888) were locally irradiated with 100 J/m2 UVC and probed for XPC (red) and 6–4PP sites (green) at different times. The background-corrected signal for XPC at 6–4PP spots is represented as mean ± SEM (300 spots derived from three experiments). (B) The DDB2-deficient XP-E cells, with or without PARPi ABT-888, were globally irradiated with 10 J/m2 UVC and immunostained for 6–4PP lesions to determine its repair kinetics up to 8 h. The data are presented as signal intensity relative to the maximum signal at 10 min (mean ± SEM, n ≥ 300 nuclei from three experiments). (C) The four cell lines with differing status of PARP1 and DDB2, as shown at the Right, were locally irradiated with 100 J/m2 and probed for T-T and XPC at 10 min after irradiation. Background-corrected signal for XPC at the T-T under identical exposure conditions was calculated as mean ± SEM derived from 300 to 700 spots from three experiments. The accumulation of the XPC at the T-T lesion in CHOU6, GMSiP, and GMU6 is expressed as fold increase over that observed in CHOSiP. (D) The total cell extracts of the four indicated cell lines were separated on the SDS/PAGE and probed for PARP1, XPC, and DDB2. Actin and Ponceau S were used as loading controls. (E) The four cell lines from above were globally irradiated with 30 J/m2 UVC and fractionated after the time indicated. The chromatin extracts with equal protein content were separated on SDS/PAGE and probed for XPC, XPA, and PARP1. Ponceau S staining was used as loading control.
To determine the extent of the contribution of DDB2 and PARP1 in the recruitment of XPC to DNA lesions, we used four different cell lines described earlier (29) (Fig. 4C): CHOSiP cells (deficient in both PARP1 and DDB2); CHOU6 cells (deficient only in DDB2); GMSiP cells (deficient only in PARP1); and GMU6 (proficient in both DDB2 and PARP1). In each cell type, we examined the early accumulation of XPC at the local UV-lesion site (Fig. 4C, Right). The CHOSiP cells displayed a basal level of accumulation of XPC at the UV-lesion site, which represents the inherent capacity of XPC to reach the lesions without the help of DDB2 or PARP1. Relative to this basal level, the presence of only PARP1 (CHOU6) or only DDB2 (GMSiP) increased XPC recruitment by about 40%. However, the presence of both DDB2 and PARP1 in GMU6 cells nearly doubled the XPC accumulation compared with the CHOSiP model, strongly supporting independent roles of DDB2 and PARP1 in this process.
The total cellular levels of XPC in PARP1-depleted GMSiP and CHOSiP cell lines were similar to PARP1-proficient GMU6 and CHOU6 cells (Fig. 4D); therefore, the suppression of the recruitment of XPC to the damaged site in PARP1-depleted cells was not an artifact of reduced XPC expression in these cells. Since the recruitment of the downstream NER proteins depends on XPC loading and stabilization at the damage site (1), we measured the accumulation of Xeroderma pigmentosum A (XPA) protein on the chromatin-bound fraction of these cells up to 4 h after damage (Fig. 4E). In each of these cellular models, the kinetics of XPA recruitment reflected the status of XPC at the UV-damaged chromatin. The XPA accumulation was robust in GMU6 cells and reduced in the absence of PARP1 (GMSiP cells) or DDB2 (CHO cells), whereas the weakest accumulation and a rapid turnover of XPA were seen in the CHOSiP cells devoid of both DDB2 and PARP1. Collectively, our results show that there are three components of recruitment of XPC to a DNA lesion site: the first is a basal level of recruitment controlled by XPC itself; the second is dependent on PARP1 or DDB2 proteins; and the last component depends on the catalytic activation of PARP1.
Characterization of the Handover of XPC from Its Complex with PARP1 to UV-Damaged DNA.
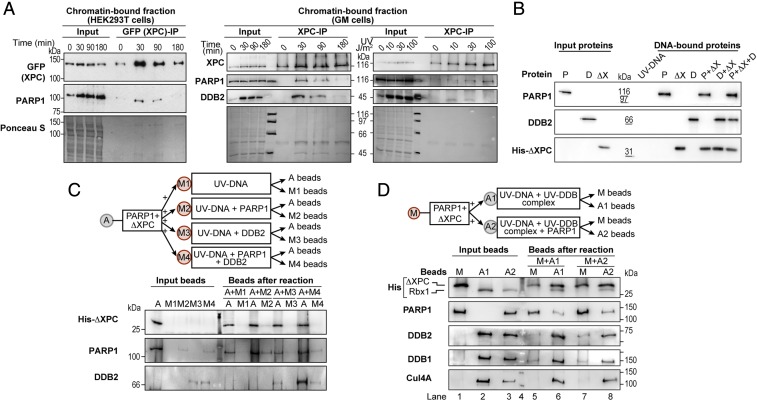
Our results show that PARP1 and XPC form a complex in the nucleoplasm, and the biotin tag on XPC at the UV-lesion site indicates a physical handover of XPC from its complex with PARP1 to UV-damaged DNA. To explore the mechanistic aspect of this transfer in vivo, we carried out GFP (XPC)-IP of chromatin fractions from HEK293T cells expressing GFP-tagged XPC up to 3 h after irradiation and examined the state of association of PARP1 with XPC (Fig. 5A, Left). The IP revealed a normal kinetics of recruitment and departure of XPC at the UV-damaged chromatin with a strong accumulation at 30 min and a significant reduction by 3 h. In contrast, the amount of PARP1 that is associated with GFP-XPC decreased rapidly from 30 to 90 min, and no signal was detected at 180 min. It is noteworthy that, although a significant amount of PARP1 was still present in the chromatin fraction from 30 to 180 min (as seen in the Input samples in Fig. 5A, Left), it was not associated with XPC after the peak period of recruitment of XPC to the lesion. A similar kinetics of association and dissociation of XPC and PARP1 was observed in chromatin over 3-h period after exposure to 30 J/m2 UVC in GM cells that express endogenous XPC and PARP1 (Fig. 5A, Middle), demonstrating the general nature of this observation. Moreover, XPC-IP of the chromatin-bound fraction of the same GM cells 10 min after exposure to various UVC doses up to 100 J/m2 revealed a dose-dependent increase in the interaction between XPC and PARP1 at the chromatin (Fig. 5A, Right). On the other hand, despite an abundance of DDB2 in the input chromatin fraction at all doses, there was a dose-dependent decrease in association of DDB2 with XPC (Fig. 5A, Right). The dose-dependent increase in association of XPC and PARP1 at an early time point and their dissociation at a later time support a model that early recruitment of XPC occurs as a complex with PARP1 but, having reached the lesion site, XPC gradually dissociates from PARP1 to continue with its functions in NER.
Fig. 5.
Role of PARP1 and the UV–DDB ligase complex in the handover of XPC to the lesion site. (A, Left) HEK293T cells transfected with GFP-XPC plasmid were irradiated 48 h later with 30 J/m2 UVC (or control) and fractionated to isolate chromatin-bound fraction, which was subjected to IP with GFP-trap beads. The eluates were probed with PARP1 and GFP antibody. (A, Middle and Right) GMU6 cells were irradiated with either 30 J/m2 UVC (Middle) or different UVC doses as indicated (Right) and fractionated after the time indicated for the Middle panel or at 10 min for the Right panel. The chromatin-bound protein fraction was used for immunoprecipitation of XPC. The IP eluates were resolved on the SDS/PAGE and probed for XPC, PARP1, and DDB2. The Ponceau S staining was used as loading control, and each panel is representative of results from two experiments. (B) UVC-DNA was bound to magnetic beads via T-T antibody and reacted with the pure proteins PARP1 (P), XPC fragment (ΔX), DDB2 (D), and different protein combinations for 15 min at 25 °C. The beads were washed, and the bound proteins were eluted, separated on SDS/PAGE, and probed with their respective antibodies. (C and D, Top) The Top panels represent schematic of the different conditions used in the in vitro assays for examining the separation of XPC from the PARP1–XPC complex and its handover to UV-DNA. The gray and red circles represent agarose and magnetic beads, respectively. The bead-bound PARP1–ΔXPC complex was prepared either on magnetic or agarose beads as described in Fig. 2C. The representative results for each model from two to three experiments are shown. (C, Lower) The PARP1–ΔXPC complex was prepared on agarose beads (A), and magnetic bead-bound UV-DNA (M1) was reacted with PARP1 (M2), DDB2 (M3), or both (M4), as described for B. The beads were mixed as indicated in the Top panel, reacted for 15 min at 25 °C in buffer, separated, washed, and eluted, and the elution was separated on SDS/PAGE and probed with specified antibodies. (D, Lower) The PARP1–ΔXPC complex was prepared on magnetic beads (M), and the agarose bead-bound UV–DDB ligase complex without (A1) or with (A2) second PARP1 was prepared by prereacting the agarose bead-bound Cul4A–Rbx1 complex with free UV-DNA, DDB1, and DDB2 for 15 min at 25 °C. The beads were mixed, reacted for 15 min at 25 °C in buffer, separated, washed, and eluted, and the elution was separated on SDS/PAGE and probed with specified antibodies.
To explore the conditions required for XPC to dissociate from its complex with PARP1 and bind to UV-damaged DNA, we designed in vitro assays using the factors prevalent at the lesion site in vivo, namely UV-damaged DNA, DDB2, PARP1, and XPC represented by its key fragment 496–734 aa (ΔXPC) that interacts with PARP1 (Figs. 5 B–D). To examine the endogenous properties of these three proteins to bind to UV-damaged DNA, we reacted them singly or in combinations with UVC-irradiated plasmid DNA, which was immobilized on the magnetic beads via T-T antibody (Fig. S4). All of the proteins could bind to UV-damaged DNA on their own and even in combination with other proteins (Fig. 5B). Although this assay confirms the inherent capacity of XPC, DDB2, and PARP1 to bind to UV-damaged DNA, it does not reveal how DDB2 or PARP1, which are recruited before XPC, could participate in the loading of XPC to the lesion site.
Fig. S4.
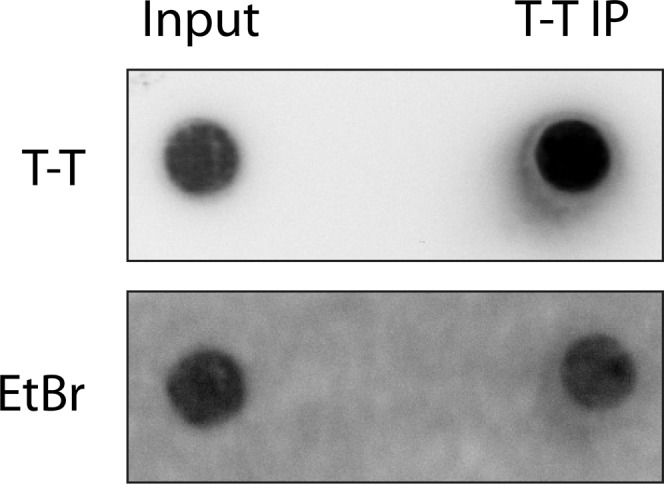
Magnetic bead-bound UVC-DNA used in the in vitro assays. The pPBS-U6 plasmid was irradiated with 5,000 J/m2 UVC and bound to magnetic beads via T-T antibody. The IP was validated by dot-blot assay using specific antibody. Ethidium bromide staining was used as loading control.
Since XPC exists as a complex with PARP1 in the nucleoplasm before reaching UV-damaged chromatin, we examined whether this PARP1–XPC complex would simulate conditions for in vivo loading of XPC to UV-DNA. We immobilized the PARP1–ΔXPC complex on the agarose beads with PARP1 antibody (A beads) and reacted it with the above-described UV-damaged DNA bound to magnetic beads (M1) in the presence of either PARP1 (M2 beads) or DDB2 (M3 beads) or both PARP1 and DDB2 (M4) (Fig. 5C, Top). After each reaction, the agarose (A beads) and magnetic beads (M1–M4) were separated and washed, and the proteins present in each of these beads were examined by immunoblotting of bead eluates. The immunoprobing for ΔXPC in the agarose and magnetic beads after the reaction revealed that none of the above conditions could dissociate ΔXPC from its complex with PARP1 (Fig. 5C).
At the UV-lesion site on DNA, DDB2 is not recruited alone but as a UV–DDB ligase complex containing DDB2, DDB1, Cul4A, and Rbx1. In addition, DDB2 interacts with PARP1 at the lesion site. Therefore, we examined whether the entire UV–DDB complex as well as PARP1 are required for loading of XPC to UV-DNA (Fig. 5D, Top). We recreated this complex by loading purified Cul4A–Rbx1 on agarose beads and reacted it with purified DDB1, DDB2, and UV-DNA in the absence (A1) or presence of PARP1 (A2). On the other hand, the PARP1–ΔXPC complex was immobilized via PARP1 antibody on the magnetic beads (M). The immunoblotting confirmed the presence or absence of each of the six designated proteins in the input beads M, A1, and A2 (Fig. 5D, Input lanes). The PARP1–ΔXPC (M) beads were reacted with A1 or A2 agarose beads, and the beads were separated and washed followed by immunoblotting of each of the beads for detection of the six proteins. The immunoprobing for His-ΔXPC in the magnetic and agarose beads revealed that the UV–DDB ligase complex with UV-DNA provided favorable conditions for promoting the dissociation of ΔXPC from its complex with PARP1 on magnetic beads (Fig. 5D, compare lane 1 with lane 5) as well as loading of ΔXPC onto the UV-DNA on A1 agarose beads (Fig. 5D, compare lane 2 with lane 6). Addition of PARP1 to the UV–DDB complex on A2 agarose beads in the above reaction did not confer any additional movement of ΔXPC to UV-DNA on agarose beads (Fig. 5D, compare lane 6 with lane 8). Collectively, these in vitro assays with purified proteins reveal that, although free XPC has an inherent capacity to efficiently bind to UV-DNA, its presence as a complex with PARP1 before irradiation ensures that XPC is preferably deposited at the UV-damaged sites that contain the UV–DDB ligase complex.
Discussion
For the last 15 years, focused efforts have been made to understand how XPC, with or without the help of other proteins, rapidly searches for its target lesions scattered across the entire genome in higher-order chromatin structure. Many studies indicated a role for DDB2 in the proper functioning of XPC with an indirect role for PARP1 via its ability to participate in chromatin remodeling (20, 27). The present study reveals a paradigm for the functional role of physical interaction of PARP1 with XPC before DNA damage in the initial recruitment and handover of XPC at UV-induced DNA lesions. Using various cell lines with exogenous or endogenous PARP1 and XPC, we show that PARP1 and XPC interact in the nucleoplasmic fraction of the cells even in the absence of DNA damage and that this interaction is independent of DDB2 and catalytic activation of PARP1. By using the PARP1 proximity-mediated biotinylation model in vivo, we also show that XPC from the nucleoplasmic PARP1–XPC complex is deposited at the DNA lesion site after UV irradiation. Using PJ-34 as PARPi, we observed that PARP inhibition partially suppresses the initial recruitment of XPC and PARP1 to the UV-lesion site, which is in agreement with earlier reports showing decreased recruitment of PARP1 to the site of microirradiation-induced DNA damage in the presence of PARPi such as PJ-34 (30) and NU-1025 (31). Another study reported an increased signal for PARP1 at damaged DNA after treatment with the PARPi 4-amino-1,8-naphthalimide (32). The difference in the end results among these studies could be attributed to the time of harvesting of the samples after treatment and the capacity of different PARPi to immobilize PARP1 on the DNA lesion sites (30, 33). Since PARP1 depletion reduces XPC recruitment to the lesion site, and PARPi reduces the rapid colocalization of PARP1 and XPC to the lesion site in vivo, our results indicate that both PARP1 and its catalytic function determine the movement of the PARP1–XPC complex from nucleoplasm to chromatin after irradiation.
In XP-E cells deficient in DDB2 function, the repair of 6–4PP is attributed to the inherent property of XPC to recognize 6–4PP lesions. Nonetheless, some studies demonstrated a reduced level of recruitment of XPC to UV damage in these cells compared with DDB2-proficient cells (7, 8). Additionally, we show that PARPi not only causes further reduction in initial recruitment of XPC to local spots of UV-induced DNA lesions but also significantly hampers the repair of 6–4PP lesions in these cells. Thus, in the XP-E model, our results clearly reveal a DDB2-independent role of PARP1 in facilitating XPC recruitment to the UV lesions and repair of 6–4PP by NER. It has been shown that XP-E cells have very low levels of PAR and that both ubiquitination and ALC1-mediated chromatin remodeling are absent in these cells (19). Hence, the decrease in recruitment of XPC by PARPi could not be related to ubiquitination or chromatin remodeling at the damaged site but instead due mainly to the suppression of movement of the PARP1–XPC complex from nucleoplasm to the lesion site on chromatin in XP-E cells. Using cells that are DDB2-deficient, PARP1-depleted, or treated with PARPi, we identified that the recruitment of XPC to the UV-lesion site in the genomic context is the sum of efforts by multiple factors including XPC itself, DDB2, PARP1, and the catalytic activity of PARP1. In a cell line that is devoid of functional DDB2 and PARP1 (CHO-SiP), there is a basal level of recruitment of XPC to the lesion site, indicating that XPC has some inherent capacity to reach the DNA lesion site that is not dependent on DDB2 or PARP1 or its activation. The reduced level of XPC translates to an impaired accumulation of XPA at the lesion site. Nonetheless, adding PARP1 alone in this DDB2-deficient background (CHOU6 cells) or DDB2 alone in a PARP1-deficient background (GMSiP) improves the recruitment of XPC above the basal level, indicating that each of these two proteins independently participates in XPC recruitment and stabilization. Finally, in the cells with PARP1 and DDB2, it is the DDB2-stimulated catalytic activation of PARP1 (18) that provides the last boost for recruitment of XPC to the lesion.
PARP1 has many characteristics that would facilitate the search function of XPC in NER: (i) PARP1 is an abundant protein in the mammalian nucleus that is rapidly recruited to all types of DNA damages (34) including UV-induced DNA lesions (26). Hence, an association with PARP1 will allow XPC to be quickly recruited to different types of DNA lesions anywhere in the genome. (ii) PARP1 can detect lesions and become activated to form PAR and create a protein-recruiting PAR platform (35), which in turn can bring in more PARP1 with XPC and other PAR-seeking proteins to the site. (iii) Like XPC, the binding of PARP1 to damaged DNA is independent of the sequence or the chemical nature of DNA damage (1, 36). Moreover, both XPC and PARP1 have affinity for unusual DNA structures with nonhydrogen-bonded bases, such as hairpins, stem loops, bubbles, and overhangs (1, 37). Thus, PARP1 could rapidly recruit XPC to all types of damages that are repaired by NER irrespective of their recognition by DDB2 (38). (iv) PARP1 is a part of the chromatin structure with preference for binding to the internucleosomal linker region (39). The chromatin-bound PARP1 can bind rapidly to lesions in this region and help recruit the nucleoplasmic PARP1–XPC complex. Additionally, the presence of the UV–DDB ligase complex in the linker (23) will allow handover and stabilization of XPC at this site. (v) Finally, the role of PARP1 activation in chromatin remodeling at the lesion site via recruitment of ALC1 (19) and PARylation of histones (34) would subsequently permit XPC to repair less accessible intranucleosomal lesions.
It has been shown that the handover of the UV lesion from the UV–DDB complex to XPC requires a transient physical interaction between DDB2 and the central region of XPC (496–679 aa) containing the domains required for its interaction with DNA (23). Interestingly, our in vitro studies have identified that the same central domain of XPC (496–734 aa) mediates its interaction with PARP1. This indicates a dynamic and cooperative process in which XPC is released from its complex with PARP1 and transferred to the lesion site containing the UV–DDB complex. Thus, our in vitro model faithfully replicates the sequence of events surrounding efficient stabilization of XPC to the lesion site, which starts with its dissociation from the complex formed with PARP1, followed by the formation of a new complex with UV–DDB. The requirement of the UV–DDB ligase complex at the lesion site ensures that PARP1-escorted XPC is preferably released at a site that is primed for GG-NER due to ubiquitination and chromatin-remodeling events initiated by the UV–DDB ligase complex. Our data do not exclude the role of the PARylation of proteins as well as changes in the structure of DNA at the lesion site toward dissociation of the PARP1–XPC complex and stabilization of XPC. These factors could play a key role in delivering XPC from its complex with PARP1 to the damage site in the absence of DDB2. Such events have been shown to play a role in dissociation of PARP1 from XRCC1 or APE1 during base excision repair (40, 41).
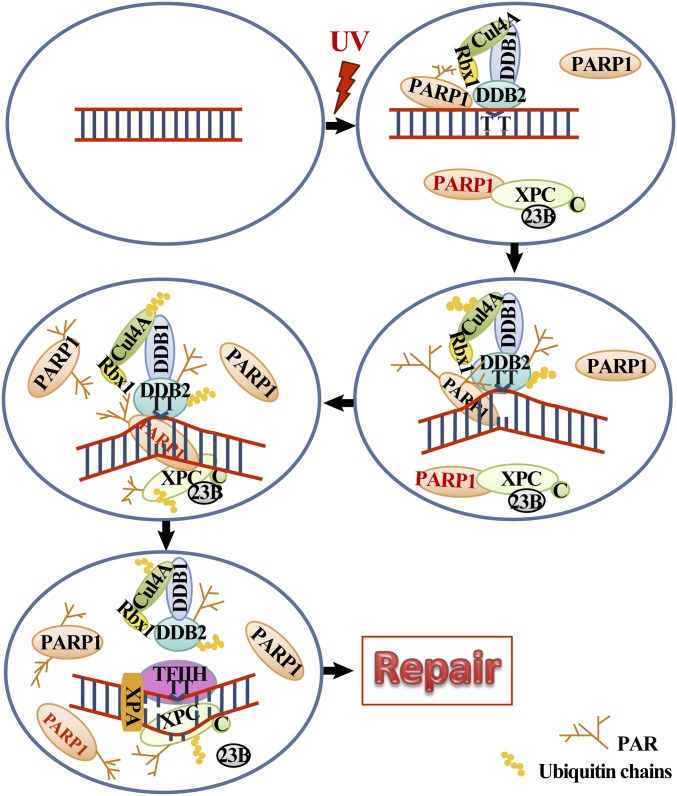
We propose a model for the roles of PARP1 in the lesion recognition function of XPC (Fig. 6). Before UV irradiation, PARP1 and XPC coexist as a complex in the nucleoplasm, and DDB2 is not part of this complex. Since PARP1 is a more abundant protein compared with XPC (9, 42), there will still be sufficient free PARP1 molecules to separately interact with DDB2 at the lesion site. The free PARP1 molecules as well as the PARP1–XPC complex will scan the intact DNA due to the affinity of PARP1 and XPC for DNA, which explains the presence of basal levels of biotinylated XPC and PARP1 in chromatin-bound protein fractions from control cells. However, the transient binding of the complex to control DNA will not result in separation of XPC from PARP1 because the UV–DDB ligase complex is not recruited to chromatin until DNA damage occurs. Upon UV irradiation, free PARP1 as well as XPC-bound PARP1 molecules will reach the lesion site and may deposit XPC from the complex to the lesion site with the help of other factors. However, the optimum deposition of XPC from its complex with PARP1 to the UV-lesion site would occur when the UV–DDB ligase complex is present at the lesion site, a condition that would be observed in normal DDB2-competent cells. The formation of PAR by DDB2-stimulated PARP1 (18) provides a platform for additional PAR-seeking molecules to accumulate at the damaged site, including more of the nucleoplasmic PARP1–XPC complex. The presence of the UV–DDB complex at the lesion site in the linker region (23) or in the core region (43) would facilitate prioritization of these sites for initial recruitment of XPC and the repair. Additionally, the PARylation, DDB2, and UV–DDB ubiquitin ligase complex-mediated chromatin-remodeling events opens the nucleosomal structure to allow the arrival of downstream proteins to complete the process of NER at all of the remaining lesions in the genome.
Fig. 6.
Model for the role of PARP1 in recruitment and stabilization of XPC in NER. See details in Discussion.
Much effort has gone into understanding the interaction of PAR and PARP1 with different proteins in the cells after DNA damage. However, not much is known about the importance of the interaction of PARP1 with multiple cellular proteins in steady-state conditions before DNA damage, which has been reported in the proteomics study (44). Here, we clearly show that the interaction of PARP1 with XPC before DNA damage is not a random phenomenon, but serves a definitive purpose of delivering XPC to the site of DNA damage within minutes after irradiation for efficient NER-mediated repair by XPC. We suggest that similar functional roles are possible for the steady-state interaction of other proteins with PARP1. Our study also highlights the fact that proteins move from one subnuclear compartment to another and thus that they may carry old partners into a new compartment or make new partners in the new location. Hence, the proteomic studies of PARP1 interactors would be much more informative if these analyses were performed before and after DNA damage and in different subnuclear compartments.
Materials and Methods
Full details are provided in SI Materials and Methods.
Cell Lines, Clones, Plasmids, and Cloning.
The SV-40 immortalized GM637 and primary XP-E (GM01389) human skin fibroblasts were obtained from Coriell. HEK293T and CHO cells were from ATCC. The clones PARP1-replete (GMU6, CHOU6), PARP1-depleted (GMSiP, CHOSiP), and FLAG-tagged PARP1-expressing (GMRSiP) were described earlier (18, 29). The Bio-ID-PARP1–expressing plasmid was generated in pcDNA3.1 mycBioID backbone from Addgene (24) and used for creating Bio-ID-PARP1 cell lines in the PARP1-depleted GMSiP cells. The creation of the pGFP-DBD vector expressing the N-terminal fragment of PARP1 (1–232 aa) was described earlier (26). The vector expressing the C-terminal fragment of PARP1 (232–1,014 aa) was cloned from PARP31 vector.
UV Irradiation and Immunofluorescence Microscopy Studies.
The local UVC irradiation using a 5-µm polycarbonate filter (Millipore), global UVC irradiation, the repair kinetics assays for 6–4PP, recruitment of NER proteins and GFP-PARP1 to local UVC-induced DNA damage, the image acquisition and analyses and software used for analyses of images, and full details of the statistical analyses of images are described in SI Materials and Methods.
Cell Fractionation and co-IP of Proteins in the Cell Fractions.
Cell fractionation to obtain nucleoplasmic and chromatin-bound protein fractions and the IP protocols followed by immunoprobing for proteins in these fractions were described earlier (18) and are explained in SI Materials and Methods. For streptavidin-IP, cells were incubated with 50 µM biotin in the medium for 16–18 h before UV treatment.
Identification of XPC-Interacting Proteins by Mass Spectrometry.
The preparation of the cell extracts from HEK293T cells, the XPC-IP, the identification XPC-interacting proteins using LC-MS/MS, along with their quantification using the appropriate software and the threshold limits, were described earlier (45) and also in SI Materials and Methods.
In Vitro Studies to Examine the Handover of XPC from PARP1 to UV-Damaged DNA.
Use of purified bovine PARP1 (Apartosis), XPC fragments (Antibodies online), GST-DDB1 and GST-DDB2 (Abnova), purification of full-length human Cul4A, binding of UVC-DNA and proteins to magnetic or agarose beads through their respective antibodies, and the handover assays are described in SI Materials and Methods.
SI Materials and Methods
Cell Lines, Clones, and Transient Transfection.
The SV-40–immortalized GM637 and primary XP-E (GM01389) human skin fibroblasts were obtained from Coriell and HEK293T and CHO cells from ATCC. The different clones PARP1-replete GMU6, PARP1-depleted GMSiP, PARP1-replete CHOU6, PARP1-depleted CHOSiP, and the FLAG-tagged PARP1-expressing GMRSiP have been described (18, 29). For creating Bio-ID clones, the GMSiP cells were transfected with Bio-ID-PARP1 or mycBioID plasmids (see below), and clones were selected with 800 µg/mL G418.
Plasmids and Cloning.
The pcDNA3.1 mycBioID plasmid expressing the promiscuous Escherichia coli biotin protein ligase BirA with N-terminal myc tag was obtained from Addgene (24). To create the Bio-ID-PARP1 plasmid, the EcoICRI/SmaI fragment of RNAi-resistant FLAG-PARP1 from the p3X FLAG-hPARP1-RSiP-CMV 7.1 plasmid (18) was ligated to the EcoRV-linearized pcDNA3.1 mycBioID plasmid. The pGFP-DBD vector expressing an N-terminal fragment of PARP1 (1–232 aa) was described earlier (26). The vector expressing the C-terminal fragment of PARP1 (232–1,014 aa) was created by subcloning the HindIII-XbaI fragment of the PARP1 expression construct PARP31 into the pcDNA-neo vector. The GFP-PARP1 expression vector was a gift from Valerie Schreiber, National Institute for Scientific Research, Strasbourg, France. Plasmid transfection was performed using TurboFect reagent according to the manufacturer’s protocol (Thermo Fisher Scientific).
UV Irradiation and Immunofluorescence Microscopy Studies.
Global and local UVC (lamp with 254-nm peak emission) irradiation of cells through a 5-µm polycarbonate filter (Millipore) was carried out in PBS or dye-free medium. For the immunofluorescence studies, cells were grown on circular coverslips in 35-mm dishes to 80% confluence. The repair kinetics assays for 6–4PP and recruitment of NER proteins to local UVC-induced DNA damage were performed as described (18). For GFP-PARP1 kinetics, cells were transfected using TurboFect reagent 24 h before irradiation. The in vivo recruitment of GFP-PARP1 to local the UV-lesion site in the nuclei was performed by selective in situ extraction protocol as described (26). Microscopic images were taken with Axiovert 200 at 40× or 63× using the AxioCam MRm. The Z-stack images were taken using a 63× oil objective of the LSM 700 confocal laser-scanning microscope (Zeiss), equipped with ZEN 2009 software for image acquisition and for preparing 2.5D images.
Cell Fractionation and co-IP of Proteins in the Cell Fractions.
Cell fractionation to obtain nucleoplasm and chromatin extract and the co-IP studies were carried out as described (18). The protein content in each fraction was estimated by Bradford assay, and samples with an equal amount of protein were subjected to co-IP, as confirmed by Ponceau S staining for all of the blots. The Ponceau S staining as a measure of equal loading was validated by the immunoprobing for housekeeping gene β-actin or for histone H3 in some of these fractions on representative blots of GM or HEK293T cells (Fig. S5). For streptavidin-IP, cells were incubated with 50 µM biotin in the medium for 16–18 h before UV treatment. The IP with M280 streptavidin beads was carried out for 30 min at ambient temperature, and washing of beads was carried out as suggested by the supplier (Life Technologies).
Fig. S5.
Loading controls for the nucleoplasmic and chromatin extracts of GMU6 and HEK293T cells. The chromatin extract of GMU6 cells (A) or HEK293T cells (B), and nucleoplasmic extract of HEK293T (C) prepared for IP from these cells were separated on SDS/PAGE and probed for β-actin and H3 to validate the equality of the proteins as estimated by Bradford. Ponceau S staining is shown as an additional loading control.
Identification of XPC-Interacting Proteins by Mass Spectrometry.
The 50–60 × 106 HEK293T cells were suspended in 2 mL of 50 mM Tris⋅HCl buffer, pH 7.5, containing 5 mM EDTA, 0.2% IGEPAL, 300 mM NaCl, 0.5 mM PMSF, and protease inhibitors. After incubation at 4 °C for 20 min, the cell lysate was sonicated for 20 s at lowest amplitude (11%, Fisher Ultrasonic Dismembrator, model 500). The extract was diluted with an equal volume of the same buffer without NaCl to dilute the NaCl to 150 mM and centrifuged at 13,000 × g for 10 min. The supernatant was subjected to XPC-IP at 4 °C for 16 h using XPC polyclonal antibody (1:500) followed by recovery of protein-antibody complex with magnetic Protein G beads (Life Technologies). The IP’s beads were extensively washed and suspended in 2× volume of TBS containing 30% glycerol and protease inhibitors and stored at −30 °C. The XPC-interacting proteins were identified using LC-MS/MS and quantified on a Dionex Ultimate 3000 nano HPLC system as previously described (45). Briefly, acetonitrile/formic acid extract of tryptic digest of the IP beads was separated on a PepMap100 C18 column and loaded on a PepMap C18 nano column. The peptides were then electro-sprayed on a OrbiTrap QExactive mass spectrometer to obtain a survey spectra at resolution of 70,000, using 106 ions. The 10 highest intensity peptides were selected for collision-induced dissociation at 35% energy and resolution of 17,500 for 50,000 ions. Dynamic exclusion was set to retain 500 entries with a maximum retention time of 40 s using a 10-ppm mass window and an enabled lock mass option. Data acquisition was done using the Xcalibur version 2.2 SP1.48. Protein identification and quantification was performed using the MaxQuant software package version 1.5.2.8 with the protein database from Uniprot (Homo sapiens, 16/07/2013, 88,354 entries). Maximum mass tolerance was set at 7 ppm and 20 ppm for precursor and fragment ions, respectively. The threshold for the false discovery rate was 5%, and protein quantification was set at a minimum of two peptides identified for each protein.
In Vitro Studies to Examine the Handover of XPC from PARP1 to UV-Damaged DNA.
The purified proteins used in the assay were bovine PARP1 (Apartosis), XPC fragments (Antibodies online), and GST-DDB1 and GST-DDB2 (Abnova). Full-length human Cul4A was coexpressed as a GST-fusion protein with His6-tagged Rbx1 in E. coli, affinity-purified by glutathione affinity chromatography, and subsequently subjected to anion exchange and gel filtration chromatography. The purified GST–Cul4A/His–Rbx1 complex was concentrated by ultrafiltration and suspended in 20 mM Tris⋅HCl, pH 8.0, 200 mM NaCl, and 5 mM DTT. Antibodies were bound to magnetic beads at ambient temperature (45 min) and to agarose beads at 4 °C (1 h) in 400–500 µL of TBS-T (50 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 0.05% Tween-20). The beads were washed three times with TBS-T and suspended in Tris buffer containing 100 mM Tris⋅HCl, pH 8.0, 10 mM MgCl2, 10% glycerol, 1.5 mM DTT, 150 mM NaCl. To prepare beads bound with UVC-DNA, a 3.2-kb PBS-U6 plasmid DNA was irradiated with 5,000 J/m2 UVC and subjected to IP with bead-bound T-T antibody at 4 °C for 2 h. To determine binding affinity of proteins to UV-DNA, free purified proteins were reacted with bead-bound UVC-DNA in the above Tris buffer for 15 min at 25 °C and washed to remove the unbound proteins before elution and detection of UV-DNA–bound proteins. PARP1–XPC fragment complexes were prepared by reacting PARP1 and the XPC fragments at equimolar ratio in the above-mentioned Tris buffer at 25 °C for 30 min, followed by PARP1-IP at 4 °C for 1.5 h using the bead-bound antibody. The bead-bound UV–DDB ligase complex was prepared by incubating agarose bead-bound Cul4A/Rbx1 for 15 min at 25 °C with free UVC-DNA, DDB1, and DDB2 in the reaction buffer with or without PARP1. The unreacted proteins were removed by washing. In the experiments that involved mixing of magnetic and agarose beads loaded with proteins or DNA as specified, these two beads were incubated together, washed, and separated using the magnetic stand. The bead-bound proteins were eluted, separated on SDS/PAGE, and probed with their respective antibodies.
Statistics.
For the immunofluorescence studies, three independent experiments were performed, and the intensity of fluorescence from at least 200–400 nuclei was analyzed by Axiovert 200 (Zeiss) using the program AutoMeasure Plus of the software Axiovision version 4.7 and expressed as pixel per area. To calculate the amount of accumulation of proteins at the UV lesion, the fluorescence intensity at the damage site was measured as described above followed by the subtraction of the background intensity detected outside of the spots from an identical area in the nucleus. Results are expressed as mean ± SEM. The statistical significance of difference was determined by unpaired two-tailed t tests, and an asterisk refers to significant P values < 0.05.
Acknowledgments
We thank V. Schreiber for GFP-PARP1 and M. Miwa for permission to receive 10H hybridoma and F. M. Boisvert and D. Levesque for interpretation of the proteomics data generated at Université de Sherbrooke. This work was supported by Discovery Grant RGPIN-2016-05868 and Discovery Accelerator Grant RGPAS-492875-2016 (to G.M.S.) from the Natural Sciences and Engineering Research Council of Canada. N.K.P. received a foreign student fee-waiver scholarship from the Québec Government and Shastri Indo-Canadian Institute. M.R. and N.K.P. were recipients of graduate scholarships from the Fonds de Recherche du Québec-Santé and Neuroscience Axis of Centre Hospitalier Universitaire de Québec Research Center-Université Laval, respectively.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706981114/-/DCSupplemental.
References
- 1.Schärer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5:a012609. doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puumalainen MR, Rüthemann P, Min JH, Naegeli H. Xeroderma pigmentosum group C sensor: Unprecedented recognition strategy and tight spatiotemporal regulation. Cell Mol Life Sci. 2016;73:547–566. doi: 10.1007/s00018-015-2075-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sugasawa K. Molecular mechanisms of DNA damage recognition for mammalian nucleotide excision repair. DNA Repair (Amst) 2016;44:110–117. doi: 10.1016/j.dnarep.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 4.Hoogstraten D, et al. Versatile DNA damage detection by the global genome nucleotide excision repair protein XPC. J Cell Sci. 2008;121:2850–2859. doi: 10.1242/jcs.031708. [DOI] [PubMed] [Google Scholar]
- 5.Chen X, et al. Kinetic gating mechanism of DNA damage recognition by Rad4/XPC. Nat Commun. 2015;6:5849. doi: 10.1038/ncomms6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Velmurugu Y, Chen X, Slogoff Sevilla P, Min JH, Ansari A. Twist-open mechanism of DNA damage recognition by the Rad4/XPC nucleotide excision repair complex. Proc Natl Acad Sci USA. 2016;113:E2296–E2305. doi: 10.1073/pnas.1514666113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moser J, et al. The UV-damaged DNA binding protein mediates efficient targeting of the nucleotide excision repair complex to UV-induced photo lesions. DNA Repair (Amst) 2005;4:571–582. doi: 10.1016/j.dnarep.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Oh KS, et al. Nucleotide excision repair proteins rapidly accumulate but fail to persist in human XP-E (DDB2 mutant) cells. Photochem Photobiol. 2011;87:729–733. doi: 10.1111/j.1751-1097.2011.00909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luijsterburg MS, et al. Dynamic in vivo interaction of DDB2 E3 ubiquitin ligase with UV-damaged DNA is independent of damage-recognition protein XPC. J Cell Sci. 2007;120:2706–2716. doi: 10.1242/jcs.008367. [DOI] [PubMed] [Google Scholar]
- 10.Lan L, et al. Monoubiquitinated histone H2A destabilizes photolesion-containing nucleosomes with concomitant release of UV-damaged DNA-binding protein E3 ligase. J Biol Chem. 2012;287:12036–12049. doi: 10.1074/jbc.M111.307058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, et al. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell. 2006;22:383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 12.El-Mahdy MA, et al. Cullin 4A-mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC. J Biol Chem. 2006;281:13404–13411. doi: 10.1074/jbc.M511834200. [DOI] [PubMed] [Google Scholar]
- 13.Nishi R, et al. UV-DDB-dependent regulation of nucleotide excision repair kinetics in living cells. DNA Repair (Amst) 2009;8:767–776. doi: 10.1016/j.dnarep.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Shell SM, et al. Xeroderma pigmentosum complementation group C protein (XPC) serves as a general sensor of damaged DNA. DNA Repair (Amst) 2013;12:947–953. doi: 10.1016/j.dnarep.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Payne A, Chu G. Xeroderma pigmentosum group E binding factor recognizes a broad spectrum of DNA damage. Mutat Res. 1994;310:89–102. doi: 10.1016/0027-5107(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 16.Scrima A, et al. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell. 2008;135:1213–1223. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vodenicharov MD, Ghodgaonkar MM, Halappanavar SS, Shah RG, Shah GM. Mechanism of early biphasic activation of poly(ADP-ribose) polymerase-1 in response to ultraviolet B radiation. J Cell Sci. 2005;118:589–599. doi: 10.1242/jcs.01636. [DOI] [PubMed] [Google Scholar]
- 18.Robu M, et al. Role of poly(ADP-ribose) polymerase-1 in the removal of UV-induced DNA lesions by nucleotide excision repair. Proc Natl Acad Sci USA. 2013;110:1658–1663. doi: 10.1073/pnas.1209507110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pines A, et al. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J Cell Biol. 2012;199:235–249. doi: 10.1083/jcb.201112132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luijsterburg MS, et al. DDB2 promotes chromatin decondensation at UV-induced DNA damage. J Cell Biol. 2012;197:267–281. doi: 10.1083/jcb.201106074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maltseva EA, Rechkunova NI, Sukhanova MV, Lavrik OI. Poly(ADP-ribose)polymerase 1 modulates interaction of the nucleotide excision repair factor XPC-RAD23B with DNA via poly(ADP-ribosyl)ation. J Biol Chem. 2015;290:21811–21820. doi: 10.1074/jbc.M115.646638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jungmichel S, et al. Proteome-wide identification of poly(ADP-ribosyl)ation targets in different genotoxic stress responses. Mol Cell. 2013;52:272–285. doi: 10.1016/j.molcel.2013.08.026. [DOI] [PubMed] [Google Scholar]
- 23.Fei J, et al. Regulation of nucleotide excision repair by UV-DDB: Prioritization of damage recognition to internucleosomal DNA. PLoS Biol. 2011;9:e1001183. doi: 10.1371/journal.pbio.1001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moné MJ, et al. Local UV-induced DNA damage in cell nuclei results in local transcription inhibition. EMBO Rep. 2001;2:1013–1017. doi: 10.1093/embo-reports/kve224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purohit NK, Robu M, Shah RG, Geacintov NE, Shah GM. Characterization of the interactions of PARP-1 with UV-damaged DNA in vivo and in vitro. Sci Rep. 2016;6:19020. doi: 10.1038/srep19020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pines A, Mullenders LH, van Attikum H, Luijsterburg MS. Touching base with PARPs: Moonlighting in the repair of UV lesions and double-strand breaks. Trends Biochem Sci. 2013;38:321–330. doi: 10.1016/j.tibs.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Ghodgaonkar MM, Zacal N, Kassam S, Rainbow AJ, Shah GM. Depletion of poly(ADP-ribose) polymerase-1 reduces host cell reactivation of a UV-damaged adenovirus-encoded reporter gene in human dermal fibroblasts. DNA Repair (Amst) 2008;7:617–632. doi: 10.1016/j.dnarep.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Shah RG, Ghodgaonkar MM, Affar B, Shah GM. DNA vector-based RNAi approach for stable depletion of poly(ADP-ribose) polymerase-1. Biochem Biophys Res Commun. 2005;331:167–174. doi: 10.1016/j.bbrc.2005.03.135. [DOI] [PubMed] [Google Scholar]
- 30.Hanssen-Bauer A, et al. XRCC1 coordinates disparate responses and multiprotein repair complexes depending on the nature and context of the DNA damage. Environ Mol Mutagen. 2011;52:623–635. doi: 10.1002/em.20663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortusewicz O, Amé JC, Schreiber V, Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Godon C, et al. PARP inhibition versus PARP-1 silencing: Different outcomes in terms of single-strand break repair and radiation susceptibility. Nucleic Acids Res. 2008;36:4454–4464. doi: 10.1093/nar/gkn403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gassman NR, Wilson SH. Micro-irradiation tools to visualize base excision repair and single-strand break repair. DNA Repair (Amst) 2015;31:52–63. doi: 10.1016/j.dnarep.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo X, Kraus WL. On PAR with PARP: Cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26:417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pascal JM, Ellenberger T. The rise and fall of poly(ADP-ribose): An enzymatic perspective. DNA Repair (Amst) 2015;32:10–16. doi: 10.1016/j.dnarep.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langelier MF, Pascal JM. PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis. Curr Opin Struct Biol. 2013;23:134–143. doi: 10.1016/j.sbi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lonskaya I, et al. Regulation of poly(ADP-ribose) polymerase-1 by DNA structure-specific binding. J Biol Chem. 2005;280:17076–17083. doi: 10.1074/jbc.M413483200. [DOI] [PubMed] [Google Scholar]
- 38.Sugasawa K, et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 39.Clark NJ, Kramer M, Muthurajan UM, Luger K. Alternative modes of binding of poly(ADP-ribose) polymerase 1 to free DNA and nucleosomes. J Biol Chem. 2012;287:32430–32439. doi: 10.1074/jbc.M112.397067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moor NA, Vasil’eva IA, Anarbaev RO, Antson AA, Lavrik OI. Quantitative characterization of protein-protein complexes involved in base excision DNA repair. Nucleic Acids Res. 2015;43:6009–6022. doi: 10.1093/nar/gkv569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei L, et al. Damage response of XRCC1 at sites of DNA single strand breaks is regulated by phosphorylation and ubiquitylation after degradation of poly(ADP-ribose) J Cell Sci. 2013;126:4414–4423. doi: 10.1242/jcs.128272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kraus WL, Lis JT. PARP goes transcription. Cell. 2003;113:677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- 43.Osakabe A, et al. Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome. Sci Rep. 2015;5:16330. doi: 10.1038/srep16330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Isabelle M, et al. Investigation of PARP-1, PARP-2, and PARG interactomes by affinity-purification mass spectrometry. Proteome Sci. 2010;8:22. doi: 10.1186/1477-5956-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dubois ML, Bastin C, Lévesque D, Boisvert FM. Comprehensive characterization of minichromosome maintenance complex (MCM) protein interactions using affinity and proximity purifications coupled to mass spectrometry. J Proteome Res. 2016;15:2924–2934. doi: 10.1021/acs.jproteome.5b01081. [DOI] [PubMed] [Google Scholar]