Significance
Despite decades of studies, the development of competitive glutamate-site antagonists that can distinguish between NMDA receptor subtypes based on GluN2 subunits has been unsuccessful. The resulting lack of subunit-selective NMDA receptor ligands has led to the widespread use of competitive antagonists with only modest subunit preference in neurophysiological and behavioral studies. This study describes competitive glutamate-site antagonists with a binding mode in the GluN2A agonist binding domain that enables indirect engagement between ligands and nonconserved residues to achieve preferential binding to GluN1/2A over GluN1/2B. These findings are required for rational drug design and suggest that glutamate-site competitive antagonists with considerable subunit selectivity can be developed, despite the highly conserved nature of the glutamate binding site.
Keywords: synaptic transmission, Schild analysis, kinetic modeling, X-ray crystallography, PEAQX
Abstract
NMDA-type glutamate receptors are ligand-gated ion channels that contribute to excitatory neurotransmission in the central nervous system (CNS). Most NMDA receptors comprise two glycine-binding GluN1 and two glutamate-binding GluN2 subunits (GluN2A–D). We describe highly potent (S)-5-[(R)-2-amino-2-carboxyethyl]-4,5-dihydro-1H-pyrazole-3-carboxylic acid (ACEPC) competitive GluN2 antagonists, of which ST3 has a binding affinity of 52 nM at GluN1/2A and 782 nM at GluN1/2B receptors. This 15-fold preference of ST3 for GluN1/2A over GluN1/2B is improved compared with NVP-AAM077, a widely used GluN2A-selective antagonist, which we show has 11-fold preference for GluN1/2A over GluN1/2B. Crystal structures of the GluN1/2A agonist binding domain (ABD) heterodimer with bound ACEPC antagonists reveal a binding mode in which the ligands occupy a cavity that extends toward the subunit interface between GluN1 and GluN2A ABDs. Mutational analyses show that the GluN2A preference of ST3 is primarily mediated by four nonconserved residues that are not directly contacting the ligand, but positioned within 12 Å of the glutamate binding site. Two of these residues influence the cavity occupied by ST3 in a manner that results in favorable binding to GluN2A, but occludes binding to GluN2B. Thus, we reveal opportunities for the design of subunit-selective competitive NMDA receptor antagonists by identifying a cavity for ligand binding in which variations exist between GluN2A and GluN2B subunits. This structural insight suggests that subunit selectivity of glutamate-site antagonists can be mediated by mechanisms in addition to direct contributions of contact residues to binding affinity.
Glutamate mediates fast excitatory neurotransmission in the mammalian CNS by binding to AMPA, kainate, and NMDA receptors, which are ligand-gated ion channels involved in critical processes ranging from neuronal development to learning and memory (1, 2). In particular, dysfunction or dysregulation of NMDA receptors has been implicated in numerous neurological and psychiatric disorders (1, 2). NMDA receptors are heterotetrameric subunit assemblies containing two GluN1 subunits that bind glycine or d-serine and two GluN2 subunits that bind glutamate (3, 4). Four different GluN2 subunits (GluN2A–D) exist that have distinct regional and developmental expression patterns and endow NMDA receptor subtypes with strikingly different biophysical and pharmacological properties (5, 6). The GluN2 subunits therefore determine the physiological roles of NMDA receptor subtypes, and, for this reason, they have received considerable interest as potential therapeutic targets. To this end, ligands that distinguish NMDA receptor subtypes based on GluN2 subunits are desirable due to their obvious utility as pharmacological tools and as potential therapeutic agents for the treatment of CNS disorders (7, 8).
Considerable progress has been made in the development of subunit-selective allosteric modulators (7–13), but the development of subtype-selective competitive NMDA receptor antagonists has been less successful. The competitive glutamate-site antagonist NVP-AAM077 (hereafter NVP) was originally reported to have 100-fold preference for GluN1/2A over GluN1/2B (14). As such, this compound has been extensively used to investigate the role of GluN2A-containing receptors in different brain regions and cellular processes. Subsequent studies using Schild analysis suggested that NVP only has a 5.4-fold preference for GluN1/2A over GluN1/2B (15). The modest 5.4-fold GluN2A preference of NVP has not discouraged its use in numerous published studies as a pharmacological tool compound to dissect the relative contributions of GluN2A- and GluN2B-containing NMDA receptors to synaptic responses. The widespread use of competitive antagonists with only modest subunit preference as tool compounds highlights a broad interest in GluN2A-selective antagonists and suggests a lack of structural and pharmacological understanding of competitive antagonism at the glutamate site in NMDA receptors. Until this study, crystal structures of NMDA receptor agonist binding domains (ABDs) in complex with competitive antagonists have been limited to the glycine site ligands—DCKA, cycloleucine, and TK40 (16-20) —and the glutamate site ligands—2-amino-5-phosphonopentanoic acid (d-AP5), (−)-PPDA, and NVP (18, 21).
In this study, we explore the structural and pharmacological properties of a series of ligands based on (S)-5-[(R)-2-amino-2-carboxyethyl]-4,5-dihydro-1H-pyrazole-3-carboxylic acid (ACEPC) (22). Competitive NMDA receptor antagonists in this series have been evaluated as potential neuroprotective and radioligand imaging agents (22, 23). Furthermore, preliminary functional results suggested that addition of halogen substituents to one of these ligands, FRA-19 {(S)-5-[(R)-2-amino-2-carboxyethyl]-1-phenyl-4,5-dihydro-1H-pyrazole-3-carboxylic acid}, resulted in modest preference for GluN1/2A over GluN1/2B receptors, as seen for compounds ST1 {(S)-5-[(R)-2-amino-2-carboxyethyl]-1-(4-fluorophenyl)-4,5-dihydro-1H-pyrazole-3-carboxylic acid} and ST6 {(S)-5-[(R)-2-amino-2-carboxyethyl)-1-(4-bromophenyl)-4,5-dihydro-1H-pyrazole-3-carboxylic acid} (23). The GluN2A preference prompted the synthesis of additional analogs, and we show here that a compound in the ACEPC series of competitive antagonists, ST3 {(S)-5-[(R)-2-amino-2-carboxyethyl]-1-[4-(3-fluoropropyl)phenyl]-4,5-dihydro-1H-pyrazole-3-carboxylic acid}, has a 15-fold preference for GluN1/2A over GluN1/2B. To facilitate the design of novel competitive antagonists, we use a combination of pharmacological, crystallographic, and mutational experiments to describe the structural determinants of binding and subunit selectivity for the ACEPC ligands.
Results
Pharmacology of ACEPC Compounds at NMDA Receptors.
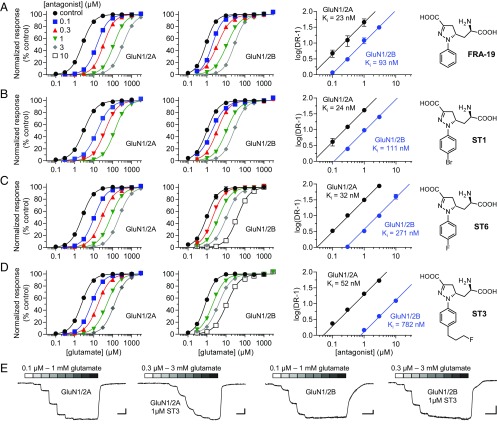
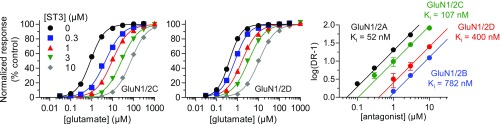
We performed Schild analyses to determine binding affinities for the ACEPC ligands at GluN1/2A and GluN1/2B receptors (Materials and Methods). Schild analyses of glutamate concentration–response relationships in the absence and presence of FRA-19, ST1, ST6, or ST3 revealed variation in selectivity between GluN1/2A and GluN1/2B depending on the substituents on the ligand phenyl ring. FRA-19 (phenyl-ACEPC) displayed 4.0-fold preference for GluN1/2A over GluN1/2B with binding affinities of 23 and 93 nM, respectively (Fig. 1A and Table 1). ST1 (4-bromophenyl-ACEPC) and ST6 (4-fluorophenyl-ACEPC) displayed 4.6-fold (24 vs. 111 nM) and 8.5-fold (32 vs. 271 nM) preference for GluN1/2A over GluN1/2B receptors, respectively (Fig. 1 B and C and Table 1). Most notably, ST3 [4-(3-fluoropropyl)phenyl-ACEPC] was 15-fold selective for GluN1/2A over GluN1/2B receptors (52 vs. 782 nM) (Fig. 1 D and E and Table 1). To further evaluate the subunit selectivity of ST3, we also determined binding affinities at GluN1/2C and GluN1/2D using Schild analyses. The binding affinity of ST3 was 107 and 400 nM at GluN1/2C and GluN1/2D, respectively (Fig. S1). ST3 therefore displays a marked preference for the glutamate binding sites in GluN2A/C over GluN2B/D subunits. The Ki values determined using Schild analyses are generally consistent with corresponding Ki values estimated using the Cheng–Prusoff relationship (Fig. S2 and Table S1). However, some differences are expected due to limitations of the Cheng–Prusoff relationship (24), highlighting the requirement of Schild analysis for more robust determination of binding affinities (25). Thus, we demonstrate that ST3 is a competitive antagonist with 15-fold preference for the glutamate binding site in GluN2A over that in GluN2B.
Fig. 1.
Schild analysis of NMDA receptor inhibition by competitive antagonists. (A–D) Glutamate concentration–response data in the absence and presence of FRA-19 (A), ST1 (B), ST6 (C), or ST3 (D) and corresponding Schild plots for GluN1/2A and GluN1/2B determined using two-electrode voltage-clamp recordings. Glutamate concentration–response data in the absence and presence of antagonist are analyzed using global nonlinear regression to determine inhibition binding affinity (Ki). The dose ratio (DR) is the ratio of glutamate EC50 values in the presence and absence of antagonist. See SI Materials and Methods for details and Table 1 for values. (E) Representative two-electrode voltage-clamp recordings of responses from GluN1/2A and GluN1/2B in the absence and presence of ST3. [Scale bars: 200 nA (vertical) and 1 min (horizontal).]
Table 1.
Schild analysis of NMDA receptor inhibition by competitive antagonists
| Antagonist | GluN1/2A | GluN1/2B | ||
| Ki, nM (95% CI) | n | Ki, nM (95% CI) | n | |
| FRA-19 | 22.6 (21.2–24.0) | 39 | 93.3 (87.2–100) | 38 |
| ST1 | 24.3 (22.6–26.1) | 24 | 111 (104–119) | 33 |
| ST6 | 32.2 (29.9–34.6) | 35 | 271 (245–299) | 34 |
| ST3 | 51.8 (49.0–54.7) | 30 | 782 (730–837) | 24 |
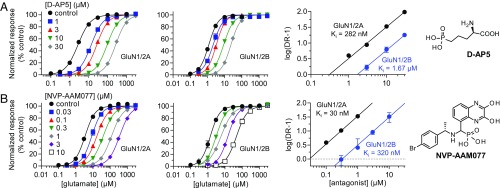
| d-AP5 | 282 (257–309) | 25 | 1,670 (1,520–1,830) | 26 |
| NVP (rat) | 29.7 (27.4–32.2) | 47 | 320 (282–363) | 34 |
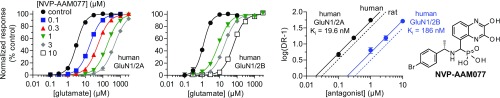
| NVP (human) | 19.6 (17.9–21.3) | 32 | 186 (168–207) | 28 |
Glutamate concentration–response data in the absence or presence of antagonist were analyzed by simultaneously fitting all data to both the Schild and Hill equations using global nonlinear regression (SI Materials and Methods). This analysis yields values for glutamate EC50 (in the absence of antagonist) and antagonist binding affinity (Ki) that best describe all of the experimental data. Ki values are listed with their 95% CI from the global nonlinear least-squares fits. n is the total number of oocytes used to generate all glutamate concentration–response data in the absence or presence of antagonist.
Fig. S1.
Schild analysis of NMDA receptor inhibition by ST3. Glutamate concentration–response data for rat GluN1/2C and rat GluN1/2D in the absence and presence of ST3 and corresponding Schild plots determined using two-electrode voltage-clamp recordings are shown. Schild plots for rat GluN1/2A and rat GluN1/2B are included for comparison, but are also shown in Fig. 1. The binding affinity (Ki) for ST3 is 107 nM at GluN1/2C (95% CI, 99–115 nM; determined using 30 oocytes) and 400 nM at GluN1/2D (95% CI, 372–431 nM; determined using 32 oocytes). Data are presented as mean ± SEM.
Fig. S2.
Inhibition of NMDA receptor subtypes by competitive antagonists. Concentration–inhibition data for FRA-19 (A), ST1 (B), ST6 (C), and ST3 (D) at recombinant GluN1/2A-D receptors expressed in Xenopus oocytes and measured using two-electrode voltage-clamp electrophysiology. Responses were activated by 100 µM glycine plus 10 µM (GluN1/2A), 3 µM (GluN1/2B and GluN1/2C), or 1 µM (GluN1/2D) glutamate. Data are normalized to the response in the absence of antagonist and are presented as mean ± SEM from five to eight oocytes. See Table S1 for values.
Table S1.
Estimated binding affinities for competitive antagonists at NMDA receptors
| Receptor | Antagonist | Glutamate EC50, µM | Hill slope | n | [Glutamate], µM | Antagonist IC50, nM | Hill slope | Estimated Ki, nM | n |
| GluN1/2A | FRA-19 | 3.00 ± 0.10 | 1.4 | 36 | 10 | 317 ± 14* | 1.1 | 73 ± 3 | 6 |
| ST1 | 10 | 91 ± 4* | 1.3 | 21 ± 1 | 6 | ||||
| ST3 | 1 | 49 ± 1 | 1.1 | 35 ± 1 | 42 | ||||
| ST3 | 10 | 282 ± 3* | 1.4 | 65 ± 1 | 7 | ||||
| ST6 | 10 | 159 ± 7* | 1.3 | 37 ± 2 | 8 | ||||
| d-AP5 | 1 | 214 ± 9 | 1.2 | 160 ± 6 | 5 | ||||
| NVP | 1 | 25 ± 2 | 1.3 | 19 ± 1 | 5 | ||||
| GluN1/2B | FRA-19 | 1.06 ± 0.03 | 1.5 | 37 | 3 | 209 ± 38* | 1.3 | 54 ± 10 | 5 |
| ST1 | 3 | 247 ± 17* | 1.2 | 65 ± 4 | 6 | ||||
| ST3 | 1 | 1,390 ± 43 | 1.2 | 647 ± 20 | 39 | ||||
| ST3 | 3 | 1,980 ± 80* | 1.2 | 516 ± 21 | 6 | ||||
| ST6 | 3 | 856 ± 108* | 1.2 | 224 ± 28 | 6 | ||||
| d-AP5 | 1 | 1,420 ± 130 | 1.1 | 722 ± 57 | 5 | ||||
| NVP | 1 | 269 ± 11 | 1.2 | 136 ± 5 | 6 | ||||
| GluN1/2C | FRA-19 | 0.91 ± 0.08 | 1.3 | 6 | 3 | 196 ± 8* | 1.3 | 46 ± 2 | 6 |
| ST1 | 3 | 189 ± 12* | 1.3 | 44 ± 3 | 6 | ||||
| ST3 | 3 | 503 ± 9* | 1.3 | 117 ± 2 | 5 | ||||
| ST6 | 3 | 918 ± 20* | 1.2 | 214 ± 5 | 8 | ||||
| GluN1/2D | FRA-19 | 0.41 ± 0.02 | 1.5 | 6 | 1 | 390 ± 82* | 1.1 | 113 ± 24 | 6 |
| ST1 | 1 | 627 ± 37* | 1.2 | 182 ± 11 | 6 | ||||
| ST3 | 1 | 1,190 ± 60* | 1.1 | 345 ± 17 | 6 | ||||
| ST6 | 1 | 918 ± 20* | 1.2 | 267 ± 6 | 8 | ||||
| GluN1/2A E714D | ST3 | 3.14 ± 0.11 | 1.3 | 6 | 1 | 48 ± 1 | 1.2 | 36 ± 1 | 6 |
| GluN1/2A V529I | ST3 | 4.17 ± 0.08 | 1.3 | 4 | 1 | 44 ± 2 | 1.2 | 36 ± 1 | 5 |
| GluN1/2A K738M | ST3 | 2.55 ± 0.10 | 1.2 | 12 | 1 | 80 ± 2 | 1.2 | 57 ± 1 | 13 |
| d-AP5 | 1 | 341 ± 24 | 1.1 | 244 ± 15 | 5 | ||||
| NVP | 1 | 45 ± 4 | 1.2 | 33 ± 2 | 5 | ||||
| GluN1/2A Y754K | ST3 | 1.33 ± 0.10 | 1.3 | 7 | 1 | 151 ± 9 | 1.3 | 86 ± 5 | 4 |
| d-AP5 | 1 | 482 ± 5 | 1.1 | 272 ± 14 | 5 | ||||
| NVP | 1 | 62 ± 4 | 1.3 | 35 ± 2 | 6 | ||||
| GluN1/2A I755V | ST3 | 3.55 ± 0.17 | 1.3 | 14 | 1 | 54 ± 1 | 1.2 | 42 ± 1 | 8 |
| d-AP5 | 1 | 225 ± 19 | 1.2 | 174 ± 13 | 4 | ||||
| NVP | 1 | 24 ± 1 | 1.1 | 19 ± 1 | 8 | ||||
| GluN1/2A T758S | ST3 | 2.46 ± 0.08 | 1.4 | 6 | 1 | 105 ± 2 | 1.2 | 74 ± 1 | 6 |
| d-AP5 | 1 | 239 ± 18 | 1.2 | 170 ± 11 | 6 | ||||
| NVP | 1 | 35 ± 1 | 1.2 | 25 ± 1 | 5 | ||||
| GluN1/2A-(2B ATD) | ST3 | 2.19 ± 0.28 | 1.5 | 5 | 1 | 88 ± 4 | 1.1 | 60 ± 2 | 6 |
| GluN1/2A-(2B S1) | ST3 | 2.75 ± 0.09 | 1.2 | 5 | 1 | 48 ± 4 | 1.2 | 35 ± 3 | 8 |
| GluN1/2A-(2B S2) | ST3 | 2.18 ± 0.14 | 1.2 | 6 | 1 | 606 ± 42 | 1.2 | 413 ± 27 | 8 |
| GluN1/2A-(2B S1+S2) | ST3 | 1.55 ± 0.09 | 1.1 | 5 | 1 | 782 ± 58 | 1.2 | 502 ± 34 | 6 |
| GluN1/2A K738M+Y754K | ST3 | 0.94 ± 0.08 | 1.2 | 8 | 1 | 253 ± 12 | 1.2 | 121 ± 5 | 7 |
| GluN1/2A Y754K+T758S | ST3 | 1.26 ± 0.13 | 1.1 | 7 | 1 | 231 ± 19 | 1.1 | 127 ± 9 | 7 |
| GluN1/2A K738M+T758S | ST3 | 1.92 ± 0.06 | 1.3 | 6 | 1 | 186 ± 28 | 1.1 | 122 ± 17 | 7 |
| GluN1/2A K/M+Y/K+T/S | ST3 | 1.05 ± 0.05 | 1.0 | 6 | 1 | 522 ± 58 | 1.2 | 267 ± 27 | 6 |
| GluN1/2A K/M+Y/K+I/V+T/S | ST3 | 1.70 ± 0.15 | 1.2 | 6 | 1 | 501 ± 31 | 1.2 | 312 ± 18 | 8 |
Glutamate concentration–response and –inhibition data for competitive antagonists measured using two-electrode voltage-clamp recordings at NMDA receptors expressed in Xenopus oocytes. Inhibition IC50 and Hill slope values were determined for responses activated by the indicated concentration of glutamate plus 100 µM glycine, and the estimated binding affinity (Ki) was calculated using the Cheng–Prusoff relationship (24). Ki values are mean ± SEM of the estimated Ki from individual oocytes calculated using the Cheng–Prusoff relationship and average glutamate EC50 for each receptor. The 2A-(2B ATD), 2A-(2B S1), 2A-(2B S2), and 2A-(2B S1+S2) are chimeric subunits with regions in GluN2A exchanged with the corresponding region in GluN2B. K/M+Y/K+T/S and K/M+Y/K+I/V+T/S indicate triple and quadruple GluN2A mutants, respectively. Data are presented as mean ± SEM of values from individual oocytes, and n is the number of oocytes used to generate the data.
Concentration–inhibition data from ref. 23. The estimated Ki values were recalculated using the indicated EC50 values for each receptor.
Binding Affinities of d-AP5 and NVP at GluN1/2A and GluN1/2B.
To compare binding affinities of ACEPC ligands to well-known competitive NMDA receptor antagonists, we evaluated the pharmacology of d-AP5 and NVP (Fig. 2). d-AP5 is commonly applied for inhibition of neuronal NMDA receptors without consideration of selectivity among receptor subtypes (i.e., d-AP5 is considered a nonselective NMDA receptor antagonist) (26). Using Schild analysis, we found that d-AP5 has a relatively low binding affinity compared with the ACEPC ligands and displayed 5.9-fold preference for GluN1/2A over GluN1/2B (Fig. 2A and Table 1). NVP has been used in numerous studies as a GluN2A-selective antagonist to differentiate the roles of GluN2A- and GluN2B-containing NMDA receptors in synaptic transmission. Using Schild analysis, we determined Ki values for NVP of 30 nM at GluN1/2A and 320 nM at GluN1/2B, corresponding to 11-fold preference for GluN1/2A over GluN1/2B (Fig. 2 B and C and Table 1). The original report evaluated NVP at human GluN1/2A and human GluN1/2B (14), and discrepancies among studies regarding the subunit selectivity of NVP could be attributed to differences between human and rat NMDA receptors. Here, we determined 9.5-fold preference for human GluN1/2A over human GluN1/2B, with Ki values of 20 and 186 nM, respectively (Fig. S3 and Table 1). Thus, NVP binding is not noticeably different between rat and human NMDA receptors. The ACEPC ligands bind GluN1/2A with higher affinity (23–52 nM) compared with the nonselective antagonist d-AP5 (282 nM at GluN1/2A), and the 15-fold preference of ST3 for GluN1/2A over GluN1/2B is modestly improved compared with NVP.
Fig. 2.
Schild analysis of NMDA receptor inhibition by d-AP5 and NVP. Glutamate concentration–response data in the absence and presence of d-AP5 (A) and NVP (B) and corresponding Schild plots for GluN1/2A and GluN1/2B determined using two-electrode voltage-clamp recordings. See Table 1 for values.
Fig. S3.
Inhibition of human NMDA receptors by NVP. Glutamate concentration–response data in the absence and presence of NVP and corresponding Schild plots for human GluN1/2A and human GluN1/2B determined using two-electrode voltage-clamp recordings. Schild plots for rat GluN1/2A and rat GluN1/2B are shown as dashed lines for comparison. See Table 1 for values.
NMDA Receptor Inhibition Under Nonequilibrium Conditions.
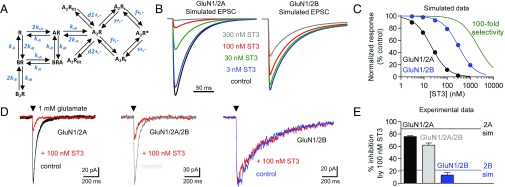
During excitatory synaptic transmission, NMDA receptors are activated by exposure to a high peak concentration of glutamate (∼1 mM) for a few milliseconds (27). The presence of competitive antagonist will reduce the fraction of NMDA receptors that activate during synaptic release of glutamate. That is, the antagonist will bind a subset of NMDA receptors, depending on antagonist concentration and binding affinity, and prevent agonist binding and subsequent gating of these receptors after synaptic release of glutamate; the mean lifetimes of antagonist–receptor complexes are typically much longer than the few milliseconds that glutamate is available for binding (28, 29). Frizelle et al. (15) used kinetic modeling to predict that, under nonequilibrium conditions relevant to those encountered during rapid release and removal of synaptic glutamate, NVP is incapable of fully inhibiting GluN1/2A without markedly reducing GluN1/2B responses. We modeled inhibition of GluN1/2A and GluN1/2B by ST3 under the same nonequilibrium conditions using a published kinetic scheme (30). In this kinetic scheme (Fig. 3A; see Table S2 for rate constants), the receptor contains two equivalent glutamate binding sites (i.e., GluN2 subunits) and requires agonist occupancy at both of these sites for activation. We assume that the association rate (k+B) of the antagonist ST3 is 10 µM−1⋅s−1, consistent with values for other competitive NMDA receptor antagonists (28, 29). The antagonist dissociation rates (k-B) for ST3 are therefore 0.52 s−1 at GluN1/2A and 7.8 s−1 at GluN1/2B, according to the experimentally determined Ki values (Ki = k-B/k+B). Using these parameters, we modeled inhibition of GluN1/2A and GluN1/2B by ST3 for responses activated by exposure to 1 mM glutamate for a duration of 1 ms (Fig. 3B). This modeling suggests that 300 nM ST3 fully blocks peak responses from GluN1/2A, but also inhibits peak responses from GluN1/2B by 48%. Simulated concentration–response data under these nonequilibrium conditions predict that, despite the 15-fold preference of ST3 for GluN1/2A over GluN1/2B, it is not possible to achieve full block of GluN1/2A without accompanying inhibition of GluN1/2B responses (Fig. 3C).
Fig. 3.
Competitive antagonism under nonequilibrium conditions. (A) Kinetic scheme used to simulate NMDA receptor-mediated responses. The kinetic scheme and rate constants are published (30). See Table S2 for rate constants. (B) Simulated responses mediated by GluN1/2A and GluN1/2B receptors to a 1-ms exposure to 1 mM glutamate in the absence (control) or continuous presence of increasing concentrations of ST3. EPSC, excitatory postsynaptic current. (C) Simulated concentration–response data for inhibition of peak responses by ST3. The theoretical concentration–response data for a compound that inhibits with 100-fold selectivity is shown for comparison. (D) Representative whole-cell voltage-clamp recordings of responses from recombinant GluN1/2A, triheteromeric GluN1/2A/2B, and GluN1/2B receptors expressed in HEK-293 cells. Responses were activated by fast-application (1–3 ms) of 1 mM glutamate in the absence (control) and presence (red) of 100 nM ST3 in the continuous presence of 100 µM glycine. (E) Bar graph of the average percent inhibition by 100 nM ST3 at GluN1/2A, triheteromeric GluN1/2A/2B, and GluN1/2B receptors. Data are from five or six cells per NMDA receptor subtype. Simulated values for inhibition of GluN1/2A (2A sim) and GluN1/2B (2B sim) are indicated by lines.
Table S2.
Rate constants for glutamate activation and ST3 inhibition
| Rate constant | GluN1/2A | GluN1/2B |
| k+A, µM−1⋅s−1 | 31.6 | 2.83 |
| k-A, s−1 | 1,010 | 38.1 |
| kd1+, s−1 | 85.1 | 550 |
| kd1-, s−1 | 29.7 | 81.4 |
| kd2+, s−1 | 230 | 112 |
| kd2-, s−1 | 1.01 | 0.91 |
| ks+, s−1 | 230 | 48 |
| ks-, s−1 | 178 | 230 |
| kf+, s−1 | 3,140 | 2,836 |
| kf-, s−1 | 174 | 175 |
| k+B, µM−1⋅s−1 | 10 | 10 |
| k-B, s−1 | 0.52 | 7.8 |
All rate constants are from ref. 30, except k+B and k-B for binding and unbinding of ST3, respectively. We assume the association rate (k+B) of ST3 is 10 µM−1⋅s−1, consistent with values for other competitive antagonists at NMDA receptors (28, 29). The calculated antagonist dissociation rates (k-B) for ST3 are 0.52 s−1 at GluN1/2A and 7.8 s−1 at GluN1/2B according to the experimentally determined Ki values of 52 and 780 nM at GluN1/2A and GluN1/2B, respectively (Ki = k-B/k+B). See Fig. 3A for kinetic scheme.
To provide experimental data to support the simulations, we used fast-application whole-cell patch-clamp recordings of responses from recombinant GluN1/2A, GluN1/2B, and triheteromeric GluN1/2A/2B receptors expressed in HEK-293 cells. We measured responses to a brief 1- to 3-ms exposure to 1 mM glutamate in the continuous presence of 100 µM glycine and in the absence or presence of 100 nM ST3 (Fig. 3D). Under these conditions, the presence of ST3 inhibited peak responses from GluN1/2A and triheteromeric GluN1/2A/2B by 76 ± 1% (n = 5) and 62 ± 4% (n = 5), respectively, whereas peak responses from GluN1/2B were reduced by 13 ± 4% (n = 6) (Fig. 3E). These results are consistent with the simulated data and demonstrate that 100 nM ST3 blocks a significant portion of responses from GluN2A-containing NMDA receptors, but also produces some inhibition of GluN1/2B responses. In theory, competitive antagonists with >100-fold selectivity are required to produce full inhibition of GluN1/2A without affecting GluN1/2B receptors under the conditions simulated here (Fig. 3C). However, a competitive antagonist with 100-fold selectivity for GluN1/2A over GluN1/2B is still expected to produce robust inhibition of triheteromeric GluN1/2A/2B receptors, which are the primary GluN2B-containing NMDA receptors in the adult hippocampus and cortex (31–33).
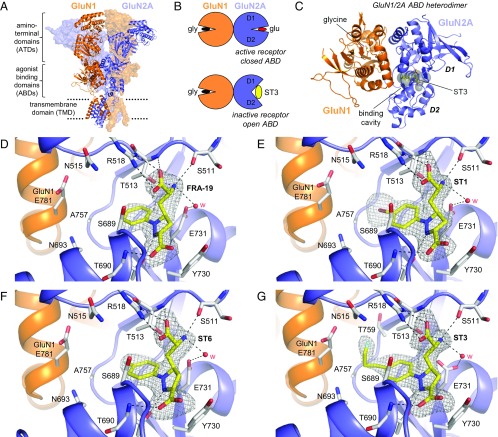
Crystal Structures of the GluN1/2A ABD Heterodimer in Complex with ACEPC Ligands.
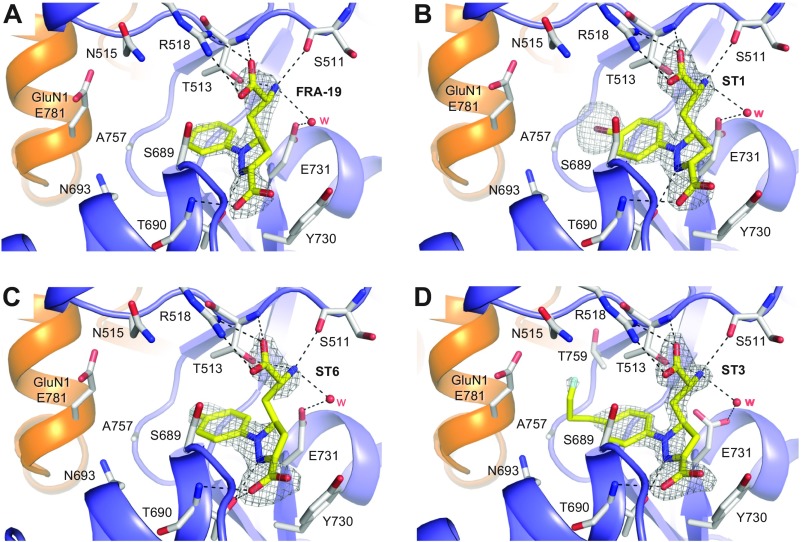
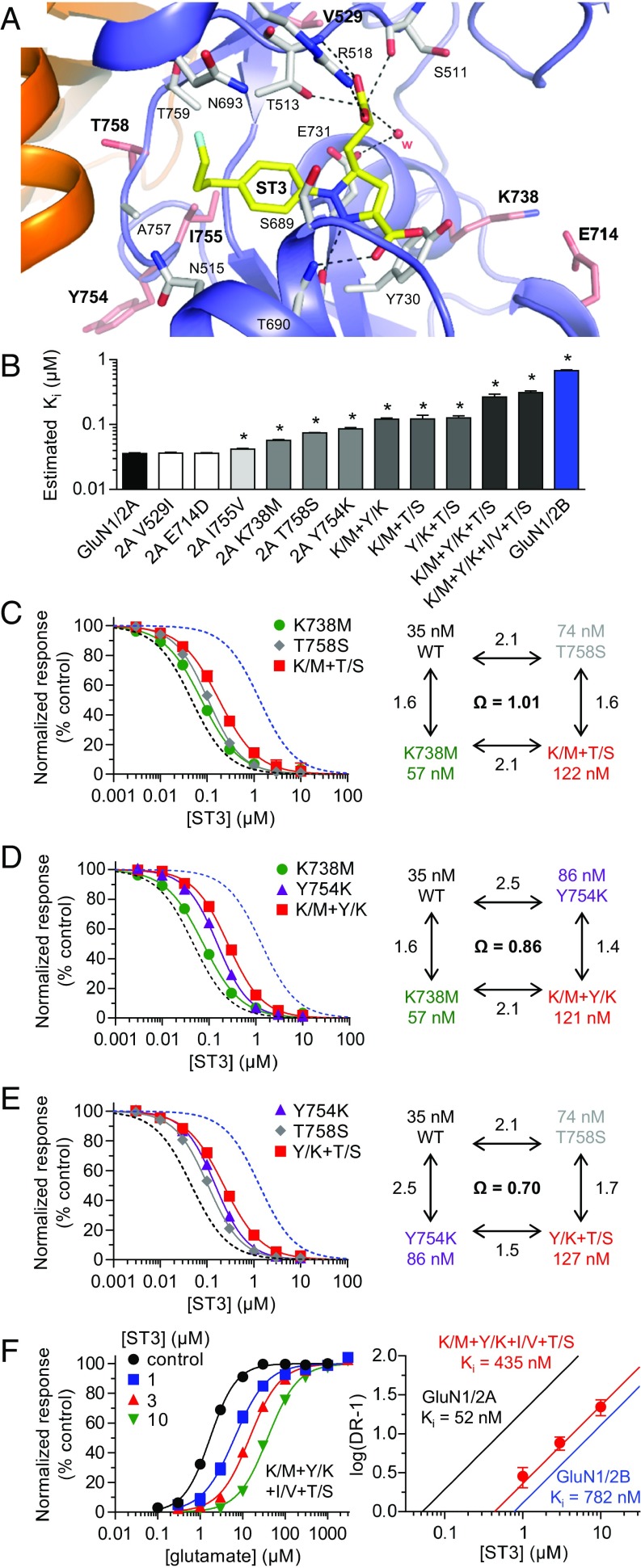
To define the structural basis for selectivity of the ACEPC compounds, we determined crystal structures of GluN1/2A ABD heterodimers in complex with glycine and FRA-19 (2.4 Å), ST1 (2.11 Å), ST6 (2.3 Å), and ST3 (1.95 Å) (Fig. 4 and Table S3). In these structures, the ACEPC ligands occupy the glutamate binding site and stabilize a more open conformation of the GluN2A ABD compared with agonists, consistent with their mechanism of action as competitive antagonists (Fig. 4B) (18). The degree of GluN2A ABD opening by the ACEPC ligands is similar to published GluN1/2A ABD heterodimer structures in complex with the antagonists d-AP5, (−)-PPDA, and NVP (18, 21), albeit (−)-PPDA stabilizes the GluN2A ABD in a slightly more open conformation compared with the other ligands. The degree of GluN2A ABD opening compared with the glutamate-bound structure is ∼16° for d-AP5, ∼20° for (−)-PPDA, ∼16° for NVP, ∼14° for ST1, ∼13° for ST3, ∼13° for ST6, and ∼14° for FRA-19 (Materials and Methods). Polar interactions between the ligand amino acid moiety and residues in the upper lobe of the GluN2A ABD (D1) are conserved among ACEPC compounds and d-AP5, but polar interactions between ligands and residues in the lower lobe of the GluN2A ABD (D2) are different for ACEPC ligands and d-AP5 (Fig. 4) (18). Polar interactions are formed between the pyrazoline 2-nitrogen atoms of the ACEPC ligands and the side-chain hydroxyl of GluN2A Thr-690, whereas the distal carboxyl groups of the ACEPC ligands interact with the backbone NH of GluN2A Thr-690.
Fig. 4.
Structures of antagonist-bound GluN1/2A ABD heterodimers. (A) Structure of the NMDA receptor composed of two GluN1 and two GluN2 subunits (PDB ID code 4PE5) (3). Soluble NMDA receptor ABDs are expressed by deleting the ATD and replacing the transmembrane domain (TMD) with a dipeptide linker. (B) Cartoon illustrating the GluN1/2A ABD heterodimer with glycine bound in GluN1 and glutamate or ST3 bound in GluN2A. Binding of competitive antagonists stabilizes the cleft formed between the upper lobe (D1) and the lower lobe (D2) of the ABD in a more open conformation compared with binding of agonists. (C) Crystal structure of the GluN1/2A ABD heterodimer with bound glycine and ST3. The cavity (shown in gray) can be occupied by ACEPC ligands in the open ABD conformation and extends from the glutamate binding site toward the ABD dimer interface. The cavity was detected using CAVER (SI Materials and Methods). (D–G) Views of glutamate binding sites in GluN1/2A ABD heterodimer structures with bound FRA-19 (2.4 Å; D), ST1 (2.11 Å; E), ST6 (2.3 Å; F), and ST3 (1.95 Å; G). Select residues (gray carbon) within 4 Å of the ST3 ligand and potential polar interactions (dashed lines) are shown. Residue GluN2A H485 and backbone atoms for some residues are omitted for clarity. The antagonists could be unambiguously fitted to their electron densities shown as gray mesh (mFo − DFc omit map contoured at 3.0 σ; see Fig. S5 for contour level at 5.0 σ). See Table S3 for data collection and refinement statistics.
Table S3.
Crystallography data collection and refinement statistics
| Data collection | ST1 | ST3 | ST6 | FRA-19 |
| Beamline | APS 19ID | APS 19ID | APS 19ID | APS 19ID |
| Wavelength, Å | 0.98 | 0.98 | 0.98 | 0.98 |
| PDB ID code | 5VIJ | 5VII | 5VIH | 5DEX |
| Ligand | ST1 | ST3 | ST6 | FRA-19 |
| Resolution range, Å* | 33.00–2.11 (2.18–2.11) | 33.01–1.95 (2.02–1.95) | 33.31–2.30 (2.39–2.30) | 34.13–2.40 (2.49–2.40) |
| Space group | P 21 21 21 | P 21 21 21 | P 21 21 21 | P 21 21 21 |
| Unit cell dimensions | ||||
| a, b, c, Å | 54.9, 87.2, 122.3 | 54.6, 87.3, 122.6 | 54.7, 87.2, 122.3 | 54.6, 87.3, 122.6 |
| α, β, γ, ◦ | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Unique reflections* | 34,022 (2,941) | 42,271 (3,721) | 25,693 (2,187) | 25,970 (2,394) |
| Redundancy* | 3.0 (1.9) | 4.6 (2.3) | 4.6 (2.3) | 4.4 (3.0) |
| Completeness,* % | 98.1 (86.4) | 97.4 (87.0) | 96.6 (83.0) | 97.7 (90.6) |
| Mean I/σ,* I | 8.9 (1.9) | 7.6 (1.9) | 7.0 (2.0) | 6.7 (1.6) |
| Wilson B-factor | 32.64 | 25.41 | 26.59 | 29.79 |
| Rsym*,† | 0.12 (0.65) | 0.11 (0.48) | 0.14 (0.69) | 0.15 (0.73) |
| Refinement | ||||
| Resolution range, * Å | 20.00–2.11 (2.18–2.11) | 20.00–1.95 (2.02–1.95) | 20.00–2.40 (2.46–2.40) | 20.00–2.40 (2.49–2.40) |
| Rwork*,‡ | 0.192 (0.291) | 0.200 (0.347) | 0.200 (0.283) | 0.224 (0.406) |
| Rfree*,‡ | 0.246 (0.333) | 0.259 (0.376) | 0.271 (0.330) | 0.278 (0.365) |
| No. of total atoms | 4,678 | 4,813 | 4,610 | 4,464 |
| Protein | 4,387 | 4,352 | 4,361 | 4,375 |
| ACEPC ligand | 21 | 24 | 28 | 20 |
| Water | 270 | 422 | 221 | 169 |
| Total protein residues | 551 | 547 | 548 | 550 |
| rms deviations | ||||
| Bond lengths, Å | 0.009 | 0.008 | 0.003 | 0.008 |
| Bond angles, ◦ | 1.23 | 0.87 | 0.63 | 1.23 |
| Ramachandran favored, § % | 96 | 95 | 95 | 94 |
| Ramachandran outlier, § % | 0.4 | 0.2 | 0.6 | 0.0 |
| Average B-factor | 37.70 | 32.43 | 32.58 | 28.00 |
| Macromolecules | 37.50 | 32.18 | 32.67 | 27.90 |
| ACEPC ligand | 28.00 | 23.07 | 23.30 | 19.50 |
| Water | 41.20 | 35.83 | 31.87 | 31.90 |
Data for highest resolution shell are given in brackets.
Rsym = ∑hkl∑i|Ii(hkl) −〈I(hkl)〉|/∑hkl∑iIi(hkl), where Ii(hkl) is the ith observation of the intensity of the reflection hkl.
Rwork = ∑hkl ||Fobs| − |Fcalc||/∑hkl |Fobs|, where Fobs and Fcalc are the observed and calculated structure-factor amplitudes for each reflection hkl. Rfree was calculated with 5% of the diffraction data that were selected randomly and excluded from refinement.
Calculated using MolProbity (55).
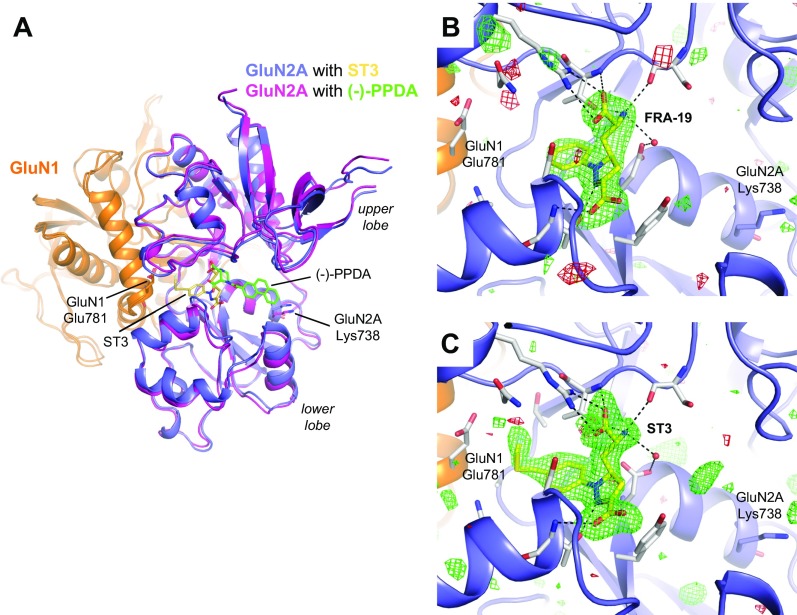
The published structure of the GluN1/2A ABD heterodimer in complex with glycine and (‒)-PPDA revealed a binding mode in which the phenanthrene group of (‒)-PPDA is oriented along the cleft between the upper and lower lobes of the GluN2A ABD and forms a hydrophobic contact with the nonconserved residue GluN2A Lys-738 (Fig. S4) (18). Interestingly, the ACEPC ligands adopt a strikingly different binding mode compared with (‒)-PPDA, with the phenyl group and its substituents directed into a cavity of the glutamate binding site toward the subunit interface between GluN1 and GluN2A ABDs (Fig. 4; see also Fig. S4 for additional justification of the binding pose). Recently, the 4-bromophenyl substituent of NVP was shown to occupy the same cavity in the GluN2A ABD (21), albeit the lack of an amino acid moiety in NVP (Fig. 2B) results in a distinct binding mode for this ligand compared with d-AP5, (‒)-PPDA, and ACEPC ligands.
Fig. S4.
Binding mode of ACEPC ligands in the GluN2A ABD. (A) Structural alignment of GluN1/2A ABD heterodimer structures in complex with glycine/ST3 (PDB ID code 5VII) and glycine/(−)-PPDA [PDB ID code 4NF6 (18)]. ACEPC ligands (e.g., ST3) and (−)-PPDA adopt strikingly different binding modes. The phenanthrene rings of (−)-PPDA are oriented along the cleft formed by the upper and lower lobes of the GluN2A ABD and form a hydrophobic contact with the nonconserved residue GluN2A Lys-738 (18). For ACEPC ligands, the phenyl groups with substituents are directed into a cavity of the GluN2A agonist binding site toward the subunit interface between GluN1 and GluN2A ABDs, and, in the case of ST3, come in close vicinity to GluN1 Glu-781. (B and C) Views of glutamate binding sites in GluN1/2A ABD heterodimer structures with bound FRA-19 (2.4 Å; B) and ST3 (1.95 Å; C). Select residues (gray carbon) within 4 Å of the ST3 ligand and potential polar interactions (dashed lines) are shown. Residue GluN2A H485 and backbone atoms for some residues are omitted for clarity. The antagonists could be unambiguously fitted to their electron densities with no residual positive (green mesh) or negative (red mesh) density in the region occupied by the (−)-PPDA ligand (mFo − DFc omit map contoured at ±3.0 σ).
The cavity occupied by the substituted phenyl groups of the ACEPC ligands extends from the central glutamate binding site in GluN2A through an opening between the backbone and side chain of Ser-689, the side chains of Thr-513 and Ile-533, and Gly-532. The cavity is then lined by the side chains of Asn-693 and Ile-533 at the bottom and by the side chains of Thr-513, Asn-515, and Thr-759 at the top. The back wall of the cavity is lined by Ala-757 from GluN2A and the side chain of GluN1 Glu-781. The cavity is solvent-exposed through an opening between side chains of GluN1 Glu-781 and GluN2A Asn-515, -693, and Ser-689. The ACEPC ligands could be unambiguously fitted to their electron densities and are not forming obvious polar contacts in this cavity (Fig. 4; see also Fig. S5). The ACEPC ligands were refined with an occupancy of 1.0, which yielded B factors for ligand atoms that are similar surrounding protein atoms (Table S3 for average ligand B factors). The fluorine atom of ST3 is located 3.8 Å from the side chain carboxylate of GluN1 Glu-781 and 3.5 Å from the backbone carbonyl of GluN2A Ile-514. Thus, the fluorine atom of ST3 could potentially form weak interactions with either of these residues (34). This finding is similar to the bromine atom of NVP in a recently described structure, in which the bromine atom is located 3.5 Å from the side chain carboxylate of GluN1 Glu-781 and could form a weak bromine–oxygen interaction (21). In summary, all residues that line the binding pocket for the ACEPC ligands, including the cavity occupied by the substituted phenyl groups, are fully conserved among GluN2 subunits (Fig. S6), suggesting that the 15-fold GluN2A binding preference of ST3 is not mediated by nonconserved residues in direct contact with the ligand.
Fig. S5.
Ligand densities for ACEPC ligands in GluN1/2A ABD heterodimer structures. Views of glutamate binding sites in GluN1/2A ABD heterodimer structures with bound FRA-19 (2.4 Å; A), ST1 (2.11 Å; B), ST6 (2.3 Å; C), and ST3 (1.95 Å; D). Select residues (gray carbon) within 4 Å of the ST3 ligand and potential polar interactions (dashed lines) are shown. Residue GluN2A H485 and backbone atoms for some residues are omitted for clarity. The antagonists could be unambiguously fitted to their electron densities shown as gray mesh (mFo − DFc omit map contoured at 5.0 σ). See Table S3 for data collection and refinement statistics.
Fig. S6.
Amino acid sequence alignment of GluN2 ABDs. The S1 and S2 segments in glutamate-binding GluN2 subunits form the ABD and correspond to residues included in the GluN2A-S1S2 protein used for X-ray crystallography (SI Materials and Methods). The α-helices and β-sheets in the GluN2A ABD structure are shown above the alignment in red and blue, respectively. The α-helix following β-sheet 13 in the S2 segment is named αX to keep helix assignments consistent with previous studies (18). Nonconserved residues are highlighted in yellow, and the subset of these residues that are conserved between GluN2A and GluN2B are highlighted in green. Amino acid numbering is according to full-length GluN2 subunits. # indicates nonconserved residues within 12 Å of ST3 in the GluN1/2A ABD heterodimer structure that mediate the GluN2A preference of ST3. − indicates nonconserved residues within 12 Å of ST3 in the GluN1/2A ABD heterodimer structure that do not affect ST3 binding affinity. * indicates residues that line the cavity that is occupied by the phenyl groups of the ACEPC ligands and extends from the glutamate binding site toward the interface between GluN1 and GluN2A subunits.
Identification of NMDA Receptor Regions That Mediate Subunit Selectivity.
The crystal structures provide insight to binding of ACEPC ligands, but do not explicitly reveal the structural determinants for the GluN2A preference of ST3. To identify regions of GluN2A that mediate subunit selectivity of the ACEPC compounds, we created chimeric GluN2 subunits by replacing regions in GluN2A with corresponding regions from GluN2B (Fig. 5A). First, the effects of swapping individual regions from GluN2B into GluN2A on glutamate EC50 and ST3 IC50 were evaluated using two-electrode voltage-clamp recordings (Fig. 5B and Table S1). These values were then used to estimate the ST3 binding affinity (Ki) using the Cheng–Prusoff relationship. The amino-terminal domain (ATD) has a small, but significant, influence on the GluN2A preference of ST3, because replacing the GluN2A ATD with the GluN2B ATD reduced the estimated ST3 binding affinity from 35 ± 1 nM (n = 42) at GluN1/2A to 60 ± 2 nM (n = 6) at GluN1/2A-(2B ATD) (Fig. 5 B–D and Table S1). Swapping the GluN2B S1 segment, which forms approximately half of the ABD, into GluN2A did not change the estimated binding affinity of ST3. By contrast, a strong effect was observed with the introduction of the GluN2B S2 segment, which forms roughly the other half of the ABD, into GluN2A. Schild analysis determined Ki values of ST3 to be 687 nM [95% confidence interval (CI) 612–772 nM, n = 23] and 588 nM (95% CI 504–686 nM, n = 20) at GluN1/2A-(2B S2) and GluN1/2A-(2B S1+S2), respectively, which are markedly reduced compared with GluN1/2A (52 nM) and more similar to the binding affinity at GluN1/2B (782 nM) (Fig. 5E). These results demonstrate that the S2 segment of the GluN2 ABD is the primary region in the NMDA receptor that mediates the 15-fold preference of ST3 for GluN1/2A over GluN1/2B.
Fig. 5.
NMDA receptor regions that mediate ST3 subunit selectivity. (A) Linear representations of GluN2 polypeptide chains showing the ATD, S1, and S2 segments that together form the ABD, transmembrane helices (M1, M3, and M4), and the reentrant pore loop (M2) for GluN2A (black), GluN2B (blue), and chimeric GluN2A–GluN2B subunits. (B) ST3 concentration–inhibition data for wild-type and chimeric NMDA receptors. Responses were activated by 1 µM glutamate plus 100 µM glycine and measured using two-electrode voltage-clamp electrophysiology. Data are mean ± SEM from 4 to 38 oocytes. (C) Bar graph of ST3 binding affinities (Ki) estimated using the Cheng–Prusoff relationship (Table S1). *P < 0.05 (significantly different from GluN1/2A; one-way ANOVA with Tukey–Kramer posttest). (D) Representative two-electrode voltage-clamp recordings of ST3 inhibition in the continuous presence of 1 µM glutamate plus 100 µM glycine. (E) Glutamate concentration–response data in the absence and presence of ST3 and corresponding Schild plots. Data for wild-type GluN1/2A (black) and GluN1/2B (blue) are shown for comparison. See also Table 1.
Mutational Analysis of the GluN2A Preference for Competitive Antagonists.
We used site-directed mutagenesis to identify individual residues that may contribute to the preference of ST3 for GluN1/2A over GluN1/2B. We selected nonconserved residues in GluN2A within 12 Å of ST3 in the crystal structure and substituted these residues to the corresponding residues in GluN2B (Fig. 6A and Fig. S6). The effects of substituting each of these residues (GluN2A V529I, E714D, K738M, Y754K, I755V, and T758S) on glutamate EC50 and ST3 IC50 were evaluated using two-electrode voltage-clamp recordings (Fig. 6B and Table S1). The V529I and Y754K substitutions had the greatest effect on glutamate EC50: 4.2 and 1.3 µM, respectively, compared with 3.0 µM at wild-type GluN1/2A and 1.1 µM at GluN1/2B. We then measured the IC50 of ST3 and used the Cheng–Prusoff relationship to estimate Ki at each mutant receptor (Fig. 6B and Table S1). Four of the individual mutations significantly increased the estimated Ki of ST3 compared with GluN1/2A (35 ± 1 nM, n = 42) —namely, K738M (57 ± 1 nM, n = 13), Y754K (86 ± 5 nM, n = 4), I755V (42 ± 1 nM, n = 8), and T758S (74 ± 1 nM, n = 6). To explore potential synergistic or additive effects of these residues in mediating the GluN2A preference of ST3, we performed mutant cycle analysis (35–37) by calculating coupling coefficients for GluN2A K738M + Y754K (denoted K/M+Y/K), K738M + T758S (K/M+T/S), and Y754K + T758S (Y/K+T/S) double substitutions (Fig. 6 C–E and Table S1). This mutant cycle analysis compares the effects of mutation 1 when introduced into receptors with mutation 2 or into wild-type receptors. A coupling coefficient Ω of 1.0 indicates that mutation 1 has the same effect in wild-type receptors as in the presence of mutation 2 and that the two mutations are independent and additive (i.e., energetically uncoupled; the coupling energy −RTlnΩ is zero). Coupling coefficients Ω different from 1.0 indicate that the effect of mutation 1 is modified by mutation 2 and that the two mutations are nonadditive (i.e., energetically coupled). The coupling coefficient of Ω = 1.01 for K/M+T/S suggests that the effects of K738M and T758S mutations are additive and independent. Coupling coefficients of Ω = 0.86 and 0.70 for K/M+Y/K and Y/K+T/S, respectively, suggest that the influence of K738M and T758S substitutions on ST3 binding affinity is slightly modified by the Y754K substitution. More specifically, the Y754K substitution reduces the effects of the K738M and T758S and vice versa. The Y754K substitution also increases glutamate potency (Table S1), suggesting that this mutation has a global effect on the GluN2A ABD, perhaps by altering the dynamic behavior of the glutamate binding site. We also evaluated the triple substitution GluN2A K738M + Y754K + T758S (K/M+Y/K+T/S) and the quadruple substitution K/M+Y/K+I/V+T/S, which combines all four mutations with significant effects on the estimated affinity of ST3. Estimation of ST3 binding affinities for the triple and quadruple substitutions suggested that the four residues mediate a majority, but not all, of the GluN2A preference of ST3 (Fig. 6B and Table S1). We supported this result using Schild analysis to determine the binding affinity of ST3 at K/M+Y/K+I/V+T/S (435 nM, 95% CI 307–505 nM, n = 29), which was markedly reduced compared with wild-type GluN1/2A (52 nM) and closer to GluN1/2B (782 nM) (Fig. 6F and Table 1).
Fig. 6.
Identification of nonconserved residues that mediate ST3 subunit selectivity. (A) Structure of the GluN1/2A ABD heterodimer with bound ST3. Residues that are nonconserved between GluN2A and GluN2B and within 12 Å of ST3 are shown as salmon carbon. (B) Bar graph of ST3 binding affinities (Ki) for wild-type and mutant NMDA receptors estimated using the Cheng–Prusoff relationship. Mutations are made in GluN2A to the corresponding residues in GluN2B (Fig. S6). Double (e.g., K/M+Y/K), triple (K/M+Y/K+T/S), and quadruple (K/M+Y/K+I/V+T/S) substitutions are abbreviated. *P < 0.05 (significantly different from GluN1/2A; one-way ANOVA with Tukey–Kramer posttest). See Table S1 for values. (C–E) ST3 concentration–inhibition data and mutant cycle analysis for wild-type, single-, and double-mutant NMDA receptors. Coupling coefficients (Ω = Ki,WT × Ki,M1+M2/Ki,M1 × Ki,M2) were calculated using estimated binding affinities from Table S1. (F) Glutamate concentration–response data in the absence and presence of ST3 and corresponding Schild plot for GluN1/2A K/M+Y/K+I/V+T/S. Data for wild-type GluN1/2A (black) and GluN1/2B (blue) are shown for comparison. See also Table S1.
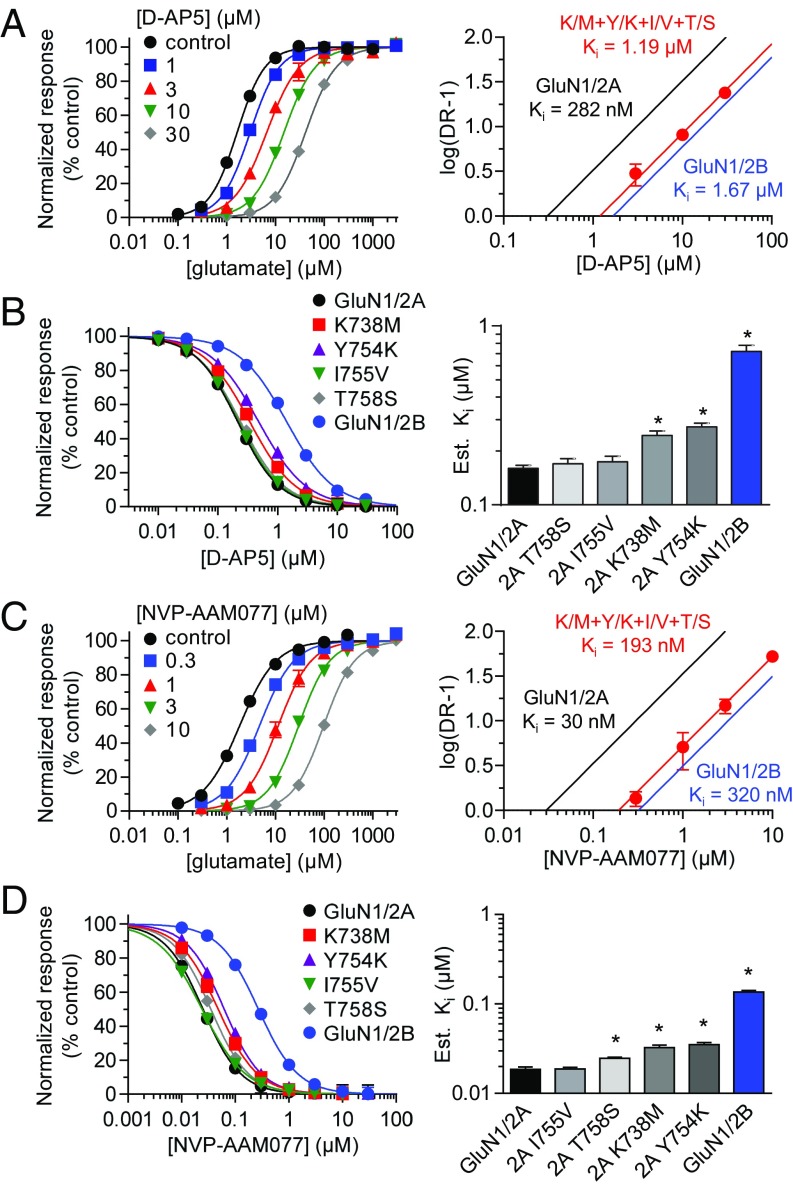
To address whether the effects of the identified residues are specific to ST3, we used Schild analysis to determine the binding affinity of d-AP5 at wild-type GluN1/2A and GluN1/2B, as well as K/M+Y/K+I/V+T/S. We found that the binding affinity of d-AP5 was significantly reduced to 1.19 µM (95% CI 1.08–1.32 µM, n = 31) by the quadruple substitution compared with 282 nM at GluN1/2A and 1.67 µM at GluN1/2B (Fig. 7A and Table 1). To identify the individual residues that affect d-AP5 binding, we estimated the affinity of d-AP5 at each of the four single mutants using the Cheng–Prusoff relationship (Fig. 7B and Table S1). Two of these substitutions, K738M and Y754K, significantly reduced the estimated binding affinity of d-AP5 compared with wild-type GluN1/2A, whereas the other two substitutions, T758S and I755V, had no effect on d-AP5 binding. To determine whether the identified residues similarly affect NVP and ST3, we used Schild analysis to determine the binding affinity of NVP at wild-type GluN1/2A and GluN1/2B, as well as K/M+Y/K+I/V+T/S. We found that the binding affinity of NVP was significantly reduced to 193 µM (95% CI 174–215 nM, n = 31) by the quadruple substitution compared with 30 nM at GluN1/2A and 320 µM at GluN1/2B (Fig. 7C and Table 1). Estimation of NVP affinity using the Cheng–Prusoff relationship showed that three of the substitutions, K738M, Y754K, and T758S, significantly reduced the estimated binding affinity of NVP compared with wild-type GluN1/2A, whereas I755V had no effect on NVP binding (Fig. 7D and Table S1).
Fig. 7.
Mutational analysis of inhibition by d-AP5. (A) Glutamate concentration–response data in the absence and presence of d-AP5 and corresponding Schild plot for GluN1/2A K/M+Y/K+I/V+T/S. Data for wild-type GluN1/2A (black) and GluN1/2B (blue) are shown for comparison. (B) d-AP5 concentration–inhibition data and bar graph of d-AP5 binding affinities estimated using the Cheng–Prusoff relationship. (C) Glutamate concentration–response data in the absence and presence of NVP and corresponding Schild plot for GluN1/2A K/M+Y/K+I/V+T/S. (D) NVP concentration–inhibition data and bar graph of NVP binding affinities estimated using the Cheng–Prusoff relationship. Responses for antagonist concentration–inhibition data were activated by 1 µM glutamate plus 100 µM glycine. *P < 0.05 (significantly different from GluN1/2A; one-way ANOVA with Tukey–Kramer posttest). See also Table 1 and Table S1.
In summary, GluN2A Lys-738 and Tyr-754 residues appear to have nonspecific effects on binding of competitive antagonists at the glutamate binding site, whereas ST3 binding is also influenced by GluN2A Ile-755 and Thr-758 residues. NVP binding is influenced by GluN2A Thr-758, but not Ile-755, consistent with the 4-bromophenyl substituent of NVP occupying the same cavity as the substituted phenyl group of ST3. These results also suggest that the 11-fold preference of NVP for GluN2A over GluN2B is mediated, in part, by indirect engagement of the 4-bromophenyl substituent with the nonconserved GluN2A Thr-758 residue, whereas the 15-fold GluN2A preference of ST3 is achieved by engaging both GluN2A Ile-755 and Thr-758.
Discussion
In this study, we have provided structural and functional insight into binding of ACEPC competitive antagonists at the NMDA receptor glutamate binding site. The ACEPC ligands have a binding mode in which they occupy an extension of the glutamate binding site near the subunit interface between the GluN1 and GluN2A ABDs. One of the compounds, ST3, which extends into this cavity, has 15-fold preference for GluN1/2A over GluN1/2B. We found that the GluN2A preference of ST3 was mediated by the S2 region of the GluN2A ABD with four nonconserved residues (GluN2A Lys-738, Tyr-754, Ile-755, and Thr-758) being responsible for the majority of the difference in antagonist binding between GluN2A and GluN2B subunits (Figs. 5 and 6). Two of these residues, GluN2A Lys-738 and Tyr-754, did not selectively influence ST3 binding, because they also shaped the modest 5.9- and 11-fold preference of d-AP5 and NVP, respectively, for GluN1/2A over GluN1/2B (Fig. 7). GluN2A Lys-738 and Tyr-754 were previously found to play critical roles in the binding preference of conantokin-G, a GluN1/2B-selective peptide toxin (38). In addition, GluN2A Lys-738 was shown to mediate the preferential binding of (‒)-PPDA to GluN1/2B-D receptors, consistent with the distinct binding mode adopted by this ligand and its direct contact with this residue (Fig. S4) (18). GluN2A Lys-738 and Tyr-754 have also been shown to influence glycine potency, albeit the mechanism by which these residues affect agonist activity at the GluN1 subunit are unclear (39). Finally, GluN2A Lys-738 influences inhibition by the GluN2D-selective negative allosteric modulator, QNZ-46, which requires glutamate binding for its activity (40). Thus, GluN2A Lys-738 and Tyr-754 appear to have global effects on ligand binding to the GluN2 ABD. We suggest that these residues may indirectly affect binding of competitive antagonists by changing the relative stabilities of open and closed conformations (i.e., the dynamic behavior of the ABD) and/or by influencing the orientation of the upper and lower lobes of the ABD. Given the location of GluN2A Tyr-754 (Fig. 6A), it is possible that the residue at this position is implicated in interactions between GluN1 and GluN2A ABDs that change during transitions between open and closed ABD conformations (i.e., agonist- and antagonist-bound conformations). GluN2A Lys-738 is positioned to influence the hydrogen-bonding network surrounding the glutamate binding site and could also affect cross-cleft interactions between the upper and lower lobes of the GluN2A ABD (Fig. 6A). In this way, GluN2A Lys-738 and Tyr-754 may enhance antagonist binding in GluN2A compared with in GluN2B-D, where these residues are methionine and lysine, respectively. Nonetheless, the mechanisms by which GluN2A Lys-738 and Tyr-754 influence ligand binding to the GluN2A ABD remain elusive.
Conversely, GluN2A Ile-755 and Thr-758 specifically influence ST3 binding and are located closer to the 4-(3-fluoropropyl)phenyl group of ST3 compared with GluN2A Lys-738 and Tyr-754 (Fig. 6A). The side chains of GluN2A Ile-755 and Thr-758 are in close proximity to each other as part of a larger hydrophobic cluster of residues at the interface between GluN1 and GluN2A ABDs. Small perturbations to the side chains of GluN2A Ile-755 and Thr-758, as with I755V and T758S mutations, could alter the positions of neighboring residues, three of which (GluN2A Gly-532, Ala-757, and Thr-759) line the cavity occupied by the 4-(3-fluoropropyl)phenyl group of ST3. We therefore suggest that GluN2A Ile-755 and Thr-758 influence the shape and volume of the cavity favorably for ST3 binding, whereas valine and serine residues at these positions in GluN2B result in a less optimal cavity for ST3 binding. This idea is supported by a recent structure of the GluN1/2A ABD dimer in complex with NVP, which revealed that the 4-bromophenyl group of NVP extends into the same cavity as the substituted phenyl groups of the ACEPC ligands (21). We show that binding affinities of NVP and ST1, which also has a 4-bromophenyl group, are very similar for the GluN2A ABD (30 and 24 nM, respectively), but NVP has a lower affinity for the GluN2B ABD compared with ST1 (320 and 111 nM, respectively) (Table 1). The 4-bromophenyl group of NVP is located deeper in the cavity compared with the 4-bromophenyl group of ST1 (i.e., the distance between the bromine and the carboxylate of GluN1 Glu-781 is 3.5 Å for NVP and 6.7 Å for ST1). This finding demonstrates that the same substituent (4-bromophenyl) positioned differently in the cavity does not change affinity at GluN2A, but, rather, reduces binding to GluN2B. Thus, the GluN2A preference for the ACEPC ligands, and likely NVP, is not mediated by increased binding to GluN2A, but, rather, occlusion of binding to GluN2B. Because residues lining the cavity are conserved between GluN2A and GluN2B, the occlusion from GluN2B binding is presumably mediated by steric differences related to the shape and/or volume of the cavity. In this regard, the relatively high binding affinity of ST3 at GluN1/2C receptors (107 nM) is surprising (Fig. S1), because sequence analysis suggests that ST3 binding to GluN2C should be similar to GluN2B and GluN2D (Fig. S6). However, GluN1/2C receptors may have additional, unique structural determinants that facilitate binding of ST3 and other ligands at the glutamate site.
The 15-fold preference of ST3 for inhibition of GluN1/2A over GluN1/2B receptors is improved compared with described GluN2A-preferring competitive antagonists (15, 41–43). However, our kinetic modeling and fast-application patch-clamp recordings suggest that 15-fold subunit preference is not sufficient to fully distinguish between GluN1/2A and GluN1/2B receptors under conditions relevant to synaptic transmission (Fig. 3). This issue is further complicated when triheteromeric GluN1/2A/2B receptors are considered, because these receptors are inhibited by ST3 to a similar level as GluN1/2A receptors (Fig. 3 D and E). NMDA receptors require simultaneous occupancy at both agonist binding sites in the two GluN2 subunits for channel gating, and binding of one competitive antagonist will therefore prevent activation (44). Thus, competitive antagonists with high GluN2A selectivity would result in full inhibition of GluN1/2A and GluN1/2A/2B receptors, thereby enabling pharmacological isolation of GluN1/2B receptors. By contrast, some highly subunit-selective negative allosteric modulators are less effective inhibitors of triheteromeric GluN1/2A/2B receptors and therefore primarily inhibit either GluN1/2A or GluN1/2B receptors [e.g., TCN-201 or ifenprodil, respectively (20, 45)]. Both subunit selectivity and mechanism of action are therefore important considerations for the application of a pharmacological tool compound in neurophysiological studies, because many central synapses in the adult hippocampus and cortex contain a mixed population of GluN1/2A, GluN1/2B, and triheteromeric GluN1/2A/2B receptors (31).
The development of highly subunit-selective competitive glutamate-site antagonists that can distinguish between NMDA receptor subtypes based on GluN2 subunits has so far been unsuccessful. The insights into binding of ACEPC competitive antagonists revealed in this study provide a starting point for manipulations of existing ligands and for additional medicinal chemistry. The structural insight provided here is a prerequisite for rational drug design and suggests that glutamate-site competitive antagonists with considerable subunit selectivity can be developed, despite the highly conserved nature of the glutamate binding site. This work, therefore, facilitates the development of highly subunit-selective competitive NMDA receptor antagonists, which are critical to advancing our understanding of NMDA receptor subtypes in the CNS, as well as for the development of new therapeutic agents.
Materials and Methods
DNA Constructs and Ligands.
Rat cDNAs for GluN1-1a (GenBank accession no. U08261; hereafter GluN1), GluN2A (D13211), GluN2B (U11419), GluN2C (M91563), and GluN2D (L31611) were provided by S. Heinemann, Salk Institute, La Jolla, CA; S. Nakanishi, Osaka Bioscience Institute, Osaka; and P. Seeburg, University of Heidelberg, Heidelberg. DNA constructs for human GluN1-1a, GluN2A, and GluN2B subunits have been described (46). DNA constructs for expression of rat triheteromeric GluN1/2A/2B receptors have also been described (45). Additional details on the DNA constructs and the chimeric GluN2A-GluN2B subunits are described in SI Materials and Methods. The amino acids are numbered according to the full-length protein, including the signal peptide. See also SI Materials and Methods and Fig. S7 for details on ligands and synthesis of ST3.
Fig. S7.
Synthesis of ST3. a: NaHCO3, EtOAc, Δ; b: XtalFluor-E, TEA*3HF, CH2Cl2, –78 °C; c: 1 M NaOH, MeOH (i) and 30% TFA, CH2Cl2 (ii).
Two-Electrode Voltage-Clamp Recordings.
See SI Materials and Methods for details on NMDA receptor expression in Xenopus oocytes. Electrophysiological recordings were performed at room temperature (23 °C) as described (47). The oocytes were voltage-clamped at −40 mV during recordings. The extracellular recording solution contained (in mM) 90 NaCl, 1 KCl, 10 Hepes, 0.5 BaCl2, and 0.01 EDTA (pH 7.4 with NaOH), and recording electrodes were filled with 3 M KCl. Approximately 10–30 min before recordings, oocytes expressing GluN1/2A and GluN1/2B receptors were injected with 20–50 nL of 50 mM BAPTA to prevent activity-dependent increases in response amplitude (48).
Whole-Cell Patch-Clamp Recordings.
Cell culturing and expression of NMDA receptors in HEK-293 cells are described in SI Materials and Methods. Recordings were performed using an Axopatch 200B amplifier (Molecular Devices) at room temperature (23 °C) with the holding potential at −60 mV. Electrodes were filled with internal solution containing (in mM) 110 d-gluconate, 110 CsOH, 30 CsCl, 5 Hepes, 4 NaCl, 0.5 CaCl2, 2 MgCl2, 5 BAPTA, 2 NaATP, and 0.3 NaGTP (pH 7.35 with CsOH), and the extracellular recording solution was composed of (in mM) 150 NaCl, 10 Hepes, 3 KCl, 0.5 CaCl2, 0.01 EDTA, and 20 mM d-mannitol (pH 7.4 with NaOH). The speed of solution exchange was measured at 0.4–0.8 ms (10–90% rise times) from the open tip junctional potential and was achieved using a two-barrel theta-glass pipette controlled by a piezoelectric translator.
Kinetic Modeling of NMDA Receptor Current Responses.
Simulations of GluN1/2A and GluN1/2B synaptic currents were performed using Kinetic Model Builder (Version 2.0) (49). Published kinetic schemes and rate constants for activation of GluN1/2A and GluN1/2B (30) can be found in Table S2, along with ST3 binding and unbinding rates. Synaptic-like NMDA receptor responses were simulated by an instantaneous step change from 0 to 1 mM glutamate for a duration of 1 ms.
Data Analysis.
See SI Materials and Methods for details on estimation of Ki values using the Cheng–Prussoff relationship, Schild analysis, and mutant cycle analysis.
Crystallography of GluN1 and GluN2A ABDs.
Details on expression, purification, and crystallization of GluN1 and GluN2A ABDs are described in SI Materials and Methods. Diffraction data were collected at the Advanced Photon Source SBC-CAT 19-ID beamline. Images were processed using HKL2000 (50). The initial phasing maps were determined by molecular replacement in PHASER (51) using the published glycine/glutamate-bound GluN1/2A ABD structure [PDB ID code 4NF8 (18)] as search model. The initial models of antagonist-bound structures were built into a 2mFo − DFc map with COOT (52) and subjected to one cycle of rigid-body refinement using PHENIX (53). ACEPC ligands were then located and fitted in a mFo − DFc omit map for each structure (54). The models were then further refined by iterative model rebuilding into a sigma-weighted 2|Fo| − |Fc| map in COOT and refinement cycles in PHENIX. Data collection and refinement statistics are shown in Table S3. The coordinates and structure factors have been deposited to the PDB with ID codes 5VIJ (ST1), 5VII (ST3), 5VIH (ST6), and 5DEX (FRA-19). See SI Materials and Methods for details on calculation of the degree of opening of the GluN2A ABD by the antagonists compared with the glutamate-bound GluN1/2A ABD dimer structure and for details on detection of the cavity extending from the glutamate binding site in GluN2A toward the interface between GluN1 and GluN2A ABDs.
SI Materials and Methods
DNA Constructs and Ligands.
Rat cDNAs for GluN1-1a (GenBank accession no. U08261; hereafter GluN1), GluN2A (D13211), GluN2B (U11419), GluN2C (M91563), and GluN2D (L31611) were provided by S. Heinemann, S. Nakanishi, and P. Seeburg. The cDNA encoding rat GluN2B was modified without changing the amino acid sequence to remove a T7 RNA polymerase termination site located in the C-terminal domain (45). DNA constructs for human GluN1-1a, GluN2A, and GluN2B subunits have been described (46). DNA constructs for expression of rat triheteromeric GluN1/2A/2B receptors have also been described (45). Briefly, peptide tags C1 and C2 that prevent surface expression of diheteromeric GluN1/2A and GluN1/2B receptors were inserted in place of the stop codon in the ORF of GluN2A to generate 2AC1 and 2AC2. The C-terminal domain of GluN2B was then replaced by the C-terminal domain of 2AC2, resulting in the chimeric subunit denoted 2BAC2. Chimeric GluN2 subunits were generated using the Gibson assembly method (New England Biolabs), and site-directed mutagenesis was performed using standard QuikChange protocols (Agilent Technologies). The composition of the chimeric subunits was as follows (numbers refer to residues): 2A-(2B S1), 1–401 + 540–1464 from GluN2A, 402–540 from GluN2B; 2A-(2B S2), 1–655 + 803–1464 from GluN2A, 657–803 from GluN2B; 2A-(2B S1+S2), 1–401 + 540–655 + 803–1464 from GluN2A, 402–540 + 657–803 from GluN2B. The 2A-(2B ATD) contained residues 402–1464 from GluN2A and 1–402 from GluN2B and has been published (56). The amino acids are numbered according to the full-length protein, including the signal peptide.
Glutamate (l-glutamic acid) and glycine were purchased from Sigma-Aldrich. d-AP5 and NVP (PEAQX) were from Hello Bio. FRA-19, ST1, and ST6 were provided by Dr. Andrea Pinto (University of Milan) and synthesized as described (22, 23). ST3 was synthesized according to the procedure reported below. Stock solutions of all ligands were made by dissolving the compound in extracellular recording solution.
Two-Electrode Voltage-Clamp Recordings.
Xenopus oocytes were purchased from Rob Weymouth (Xenopus 1) and prepared essentially as described (47). For NMDA receptor expression in Xenopus oocytes, DNA constructs were linearized by restriction enzymes to produce cRNAs using the mMessage mMachine kit (Thermo Fisher Scientific). The cRNAs were combined at a ratio of 1:2 for GluN1 and GluN2 and diluted with RNase-free water before injection to achieve desired expression levels for subsequent recordings. Injected oocytes were incubated at 17 °C in modified Barth’s solution containing (in mM) 88 NaCl, 2.4 NaHCO3, 1 KCl, 0.33 Ca(NO3)2, 0.41 CaCl2, 0.82 MgSO4, and 5 Tris⋅HCl (pH 7.5) with NaOH supplemented with 100 U/mL penicillin and 100 µg/mL streptomycin (Invitrogen) as well as 50 mg/mL gentamicin (Fisher Scientific). Electrophysiological recordings were performed 2–4 d after cRNA injection at room temperature (23 °C) as described (47).
Whole-Cell Patch-Clamp Recordings.
HEK-293 cells were cultured in Dulbecco’s Modified Eagle's Medium with GlutaMax-I (Invitrogen) supplemented with 10% dialyzed FBS, 1 mM sodium pyruvate, 10 U/mL penicillin, and 10 µg/mL streptomycin and plated in 24-well plates on glass coverslips coated with 0.1 mg/mL poly-d-lysine ∼48 h before the experiments. For expression of GluN1/2A and GluN1/2B, cells were transfected with plasmid cDNAs encoding GluN1 and GluN2 subunits at a ratio of 1:2 using the calcium phosphate precipitation method as described (47). Triheteromeric GluN1/2A/2B receptors were expressed in HEK-293 cells as described (45). To minimize cytotoxicity, the culture medium was replaced 4–5 h after transfection with medium containing NMDA receptor antagonists d,l-AP5 and 7-CKA (200 µM each). Experiments were performed ∼24 h after transfection.
Data Analysis.
Concentration–response data were analyzed using GraphPad Prism (GraphPad Software). Data from individual oocytes were fitted to the Hill equation using variable slope and fitted EC50 or IC50 values, and Hill slopes from individual oocytes were used to calculate the mean and SEM. For graphing, data points were normalized to the fitted maximum response from the same recording, and the averaged data were then fitted to the Hill equation and plotted together. Antagonist binding affinities were estimated using average EC50 values, IC50 values for individual oocytes, the agonist concentration [A], and the Cheng–Prusoff relationship (24): Estimated Ki = IC50/(1 + [A]/EC50). For compounds evaluated using Schild analysis (57), the glutamate EC50 was measured in the absence of antagonist and EC50′ measured in the presence of increasing concentrations of antagonist. These values were used to calculate a dose ratio (DR = EC50′/EC50) for each antagonist concentration and log(DR − 1) was plotted vs. log(antagonist concentration). The slope of the Schild plot is 1 for a competitive antagonist according to the Schild equation: log(DR − 1) = pKi + log[B], where [B] is the antagonist concentration and pKi is the negative logarithm of the antagonist concentration that produces a twofold shift of the agonist EC50 (i.e., Ki is the antagonist binding affinity). Importantly, Ki values were not determined by fitting a straight line to the DR values, but, rather, determined using global nonlinear least-squares fitting. That is, all of the agonist concentration–response data obtained at different antagonist concentrations were simultaneously fit to the following equations (58):
where [A] is the agonist concentration, [B] is the antagonist concentration, EC50 is for the agonist in the absence antagonist, EC50′ is for agonist in the presence of different concentrations of antagonist, nH is the Hill slope, and Ki is the antagonist binding affinity. This method will fit the data using EC50 in the absence of antagonist and Ki as global parameters (i.e., shared among all concentration–response curves), whereas nH and EC50′ values are different for each concentration–response curve. Unlike conventional Schild analysis that fits a straight line to the DR values, the global nonlinear regression approach does not overemphasize the EC50 value in the absence of antagonist, thereby providing a more robust determination of Ki with stronger theoretical basis (58). Mutant cycle analysis (35–37) was performed using estimated binding affinities for wild-type GluN1/2A (Ki,WT), mutant 1 (Ki,M1), mutant 2 (Ki,M2), and the double mutant combining mutant 1 and 2 (Ki,M1+M2) to calculate the coupling coefficient (Ω = Ki,WT × Ki,M1+M2/Ki,M1 × Ki,M2). All data are presented as mean ± SEM. One-way ANOVA with Tukey–Kramer posttest were used for statistical comparisons as indicated, and P < 0.05 was considered significant.
Crystallography of GluN1 and GluN2A ABDs.
The GluN1 ABD contains residues 394–544 and 663–800 from full-length GluN1 joined by a Gly–Thr dipeptide linker and the GluN2A ABD contains residues 402–539 and 661–802 from full-length GluN2A joined by a Gly–Thr dipeptide linker as described (16, 18). Constructs for the GluN1 ABD and the fusion protein of the small ubiquitin-like modifier (SUMO) and the GluN2A ABD (SUMO-GluN2A ABD) were provided by Hiro Furukawa, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, and expressed in Escherichia coli OrigamiB (DE3) cells essentially as described (16, 18). These constructs, which contain an amino-terminal 6×His-tag, were captured from cell lysates and purified as described (20).
The isolated GluN1 and GluN2A ABD proteins were mixed at a 1:1 molar ratio and dialyzed against a buffer of (in mM) 100 NaCl, 10 Hepes (pH 7.0), 0.01 glycine, and 1 l-glutamate for 14–18 h and concentrated to 4–7 mg/mL for crystallization. Crystallization of GluN1/2A ABD heterodimers in complex with glycine and glutamate was done by mixing protein samples with equal volumes of reservoir solution [0.2 M ammonium acetate and 16–22% PEG 4000 (final pH 6.2)] in VDX 24-well hanging drop plates. Antagonist-bound crystals were obtained by soaking glycine/glutamate-bound crystals against the reservoir solution plus 10 µM glycine and 100 µM different ACEPC ligands for 48 h, and then crystals were transferred to same soaking solution with 1 mM respective ACEPC ligands for additional 48 h. Crystals were cryoprotected in reservoir solution plus 20% (vol/vol) glycerol and flash-frozen in a 100 K nitrogen gas stream.
The degree of opening of the GluN2A ABD compared with the glutamate-bound GluN1/2A ABD dimer structure (PDB ID code 5I57) (20) was calculated for the different competitive antagonists using RotationAxis (draw_rotation_axis.py; python script by Pablo Guardado Calvo and available from PyMOLWiki) in the PyMOL Molecular Graphics System (Version 1.8; Schrödinger, LLC). The cavity extending from the glutamate binding site in GluN2A toward the interface between GluN1 and GluN2A ABDs was detected using the CAVER (Version 3.0.1) PyMOL plugin (59). Briefly, ST3 bound in the GluN1/2A ABD dimer crystal structure (PDB ID code 5VII) was selected as starting point, and the cavity was detected using default parameters. Preparation of figures depicting crystal structures were performed using the PyMOL Molecular Graphics System (Version 1.8).
Synthesis of ST3.
Solvents and chemicals purchased from commercial sources were of analytical grade or better and used without further purification. The 1H NMR spectra were recorded on a Varian Mercury 300 (300 MHz) spectrometer. Chemical shifts (δ) are expressed in ppm, and coupling constants (J) are expressed in Hz. Rotary power determinations were carried out using a Jasco P-1010 spectropolarimeter, coupled with a Haake N3-B thermostat. TLC analyses were performed on commercial silica gel 60 F254 aluminum sheets; spots were further evidenced by spraying with a dilute alkaline potassium permanganate solution. Melting points were determined on a model B 540 Büchi apparatus and are uncorrected. MS analyses were performed on a Varian 320-MS triple quadrupole mass spectrometer with ESI source. Compounds 1 (60) and 4 (61) shown in Fig. S7 were synthesized as reported in the literature.
Synthesis of Methyl 2-chloro-2-(2-(4-(3-hydroxypropyl)phenyl)hydrazono)acetate (3).
Chloro-hydrazone 3 was synthesized in a 78% yield starting from compound 1 following a literature procedure (62). 1H NMR (CDCl3, 300 MHz): 1.50 (1H, bs), 1.80–1.92 (2H, m); 2.67 (2H, 7, J = 6.5); 3.64 (2H, t, J = 6.5); 3.95 (3H, s); 7.10–7.20 (4H, m); 8.25 (1H, bs).
Synthesis of (S)-Methyl 5-((R)-2-(tert-butoxycarbonyl)-3-methoxy-3-oxopropyl)-1-(4-(3-hydroxypropyl)phenyl)-4,5-dihydro-1H-pyrazole-3-carboxylate (5a).
To a solution of (R)-methyl 2-(tert-butoxycarbonylamino)pent-4-enoate 4 (1.50 g, 6.54 mmol, 1.0 eq) in EtOAc (25 mL), chloro-hydrazone 3 (9.81 mmol, 1.5 eq) and solid NaHCO3 (2.75 g, 32.7 mmol, 5 eq) were added. The mixture was vigorously stirred for 24 h at 80 °C, and the reaction progress was followed by TLC (cyclohexane/EtOAc 1:1). Water (5 mL) was added, and the organic layer was separated and dried over anhydrous Na2SO4. The crude material obtained after evaporation of the solvent was purified by column chromatography on silica gel (cyclohexane/EtOAc 8:2) to isolate the single diastereoisomers 5a and 5b (overall yield: 69%).
5a: Rf: 0.52 (cyclohexane/EtOAc 1:1); [α]D20 = + 222.2 (c = 0.25; CHCl3); [M+H]+ = 464.4; 1H-NMR (CDCl3, 300 MHz): 1.47 (9H, s); 1.50 (1H, bs); 1.80–1.92 (3H, m); 1.98–2.10 (1H, m); 2.65 (2H, t, J = 7.3); 3.08 (1H, dd, J = 4.7, 17.9); 3.38 (1H, dd, J = 12.0, 17.9); 3.67 (2H, t, J = 6.5); 3.73 (3H, s); 3.87 (3H, s); 4.30–4.44 (1H, m); 4.58–4.72 (1H, m); 5.18–5.26 (1H, m); 7.05 (2H, d, J = 8.5); 7.15 (2H, d, J = 8.5).
Synthesis of (S)-Methyl 5-((R)-2-(tert-butoxycarbonyl)-3-methoxy-3-oxopropyl)-1-(4-(3-fluoropropyl)phenyl)-4,5-dihydro-1H-pyrazole-3-carboxylate (6a).
Under a N2 atmosphere, a solution of TEA*3HF (4.32 mmol, 5.0 eq) and XtalFluor-E (3.24 mmol, 3.8 eq) was prepared in anhydrous CH2Cl2 (6 mL). A solution of compound 5a (0.86 mmol, 1 eq) in anhydrous CH2Cl2 (6.5 mL) was added dropwise to the mixture cooled at –78 °C. The reaction was stirred 30 min at –78 °C, followed by 1 h at room temperature. The reaction was monitored by TLC (cyclohexane/EtOAc 1:1). After completion, a 5% aqueous solution of NaHCO3 (5 mL) was added, and the aqueous phase was extracted with CH2Cl2 (3 × 5 mL). The organic layer was dried over anhydrous Na2SO4, and the solvent was evaporated under reduced pressure. The crude material obtained was purified by column chromatography on silica gel (cyclohexane/EtOAc 8:2) to yield compound 6a as a yellow oil (yield: 63%).
6a: Rf:0.53 (cyclohexane/EtOAc 6:4); [α]D20 = + 244.6 (c = 0.5; CHCl3); [M+H]+ = 466.4; 1H-NMR (CDCl3, 300 MHz): 1.47 (9H, s); 1.84–2.10 (4H, m); 2.69 (2H, t, J = 7.3); 3.08 (1H, dd, J = 4.9, 17.9); 3.38 (1H, dd, J = 12.3, 17.9); 3.72 (3H, s); 3.85 (3H, s); 4.45 (2H, dt, J = 5.9, 47.2); 4.59–4.70 (1H, m); 5.20 (1H, d, J = 7.3); 7.06 (2H, d, J = 8.5); 7.15 (2H, d, J = 8.5).
Synthesis of (S)-5-((R)-2-Amino-2-carboxyethyl)-1-(4-(3-fluoropropyl)phenyl)-4,5-dihydro-1H-pyrazole-3-carboxylic acid (ST3).
(a) Derivative 6a (0.6 mmol, 1.0 eq) was dissolved in MeOH (1.8 mL) and treated with 1 M aqueous NaOH (1.8 mL, 3.0 eq). The disappearance of the starting material was monitored by TLC (cyclohexane/EtOAc 8:2). After evaporation of MeOH, the aqueous layer was washed with Et2O (1 × 3 mL), made acidic with 2 M aqueous HCl, and extracted with EtOAc (3 × 5 mL). The organic layer was dried over anhydrous Na2SO4, and, after evaporation of the solvent, the diacid derivative was obtained as a yellow solid. (b) The diacid derivative (0.56 mmol, 1.0 eq) was treated with a 30% CH2Cl2 solution of trifluoroacetic acid (5.6 mmol, 10 eq) at 0 °C. The solution was stirred at room temperature for 3 h, and the reaction was followed by TLC (CH2Cl2/MeOH 9:1 + 1% AcOH). The volatiles were removed under reduced pressure, and the residue was crystallized from EtOH to yield the final amino acid ST3 as a pale green solid (overall yield: 68%).
ST3: Rf: 0.44 (n-butanolo/H2O/AcOH 4:2:1); [α]D20 = + 250.2 (c = 0.25; DMSO); [M+H]+ = 338.1; M.p. = 175.5–176.6 °C; 1H-NMR (DMSO-d6, 300MHz): 1.69–2.08 (4H, m); 2.59 (2H, t, J = 6.9); 2.98 (1H, dd, J = 3.9, 18.4); 3.13 (1H, dd, J = 11.3, 18.4); 3.53 (1H, dd, J = 3.3, 9.9); 4.42 (2H, dt, J = 5.8, 47.3); 4.58–4.72 (1H, m); 7.08–7.18 (4H, m).
Acknowledgments
We thank the staff at the Advanced Photon Source, Argonne National Laboratory for excellent beamline support. Argonne is operated by UChicago Argonne, LLC, for the US Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357. We thank Dr. Hiro Furukawa (Cold Spring Harbor Laboratory) for providing DNA constructs for expression of GluN1 and GluN2A ABDs; Gina C. Bullard, Marlene E. Woldstad, and Cindee K. Yates-Hansen for excellent technical assistance; and Dr. Stephen R. Sprang (University of Montana) for helpful discussions and comments on the manuscript. A.P. was supported by University of Milan Grant Piano di Sostegno alla Ricerca 2015/2017–Linea 2A. This work was supported by National Institutes of Health Grants P20 GM103546 and R01 NS097536.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 5VIJ (ST1), 5VII (ST3), 5VIH (ST6), and 5DEX (FRA-19)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707752114/-/DCSupplemental.
References
- 1.Traynelis SF, et al. Glutamate receptor ion channels: Structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: Impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci. 2013;14:383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- 3.Karakas E, Furukawa H. Crystal structure of a heterotetrameric NMDA receptor ion channel. Science. 2014;344:992–997. doi: 10.1126/science.1251915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee CH, et al. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature. 2014;511:191–197. doi: 10.1038/nature13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 6.Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF. Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J Neurosci. 2009;29:12045–12058. doi: 10.1523/JNEUROSCI.1365-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strong KL, Jing Y, Prosser AR, Traynelis SF, Liotta DC. NMDA receptor modulators: An updated patent review (2013-2014) Expert Opin Ther Pat. 2014;24:1349–1366. doi: 10.1517/13543776.2014.972938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogden KK, Traynelis SF. New advances in NMDA receptor pharmacology. Trends Pharmacol Sci. 2011;32:726–733. doi: 10.1016/j.tips.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khatri A, et al. Structural determinants and mechanism of action of a GluN2C-selective NMDA receptor positive allosteric modulator. Mol Pharmacol. 2014;86:548–560. doi: 10.1124/mol.114.094516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Volkmann RA, et al. MPX-004 and MPX-007: New pharmacological tools to study the physiology of NMDA receptors containing the GluN2A subunit. PLoS One. 2016;11:e0148129. doi: 10.1371/journal.pone.0148129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hackos DH, et al. Positive allosteric modulators of GluN2A-containing NMDARs with distinct modes of action and impacts on circuit function. Neuron. 2016;89:983–999. doi: 10.1016/j.neuron.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 12.Costa BM, et al. A novel family of negative and positive allosteric modulators of NMDA receptors. J Pharmacol Exp Ther. 2010;335:614–621. doi: 10.1124/jpet.110.174144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monaghan DT, Irvine MW, Costa BM, Fang G, Jane DE. Pharmacological modulation of NMDA receptor activity and the advent of negative and positive allosteric modulators. Neurochem Int. 2012;61:581–592. doi: 10.1016/j.neuint.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auberson YP, et al. 5-Phosphonomethylquinoxalinediones as competitive NMDA receptor antagonists with a preference for the human 1A/2A, rather than 1A/2B receptor composition. Bioorg Med Chem Lett. 2002;12:1099–1102. doi: 10.1016/s0960-894x(02)00074-4. [DOI] [PubMed] [Google Scholar]
- 15.Frizelle PA, Chen PE, Wyllie DJ. Equilibrium constants for (R)-[(S)-1-(4-bromo-phenyl)-ethylamino]-(2,3-dioxo-1,2,3,4-tetrahydroquinoxalin-5-yl)-methyl]-phosphonic acid (NVP-AAM077) acting at recombinant NR1/NR2A and NR1/NR2B N-methyl-D-aspartate receptors: Implications for studies of synaptic transmission. Mol Pharmacol. 2006;70:1022–1032. doi: 10.1124/mol.106.024042. [DOI] [PubMed] [Google Scholar]
- 16.Furukawa H, Gouaux E. Mechanisms of activation, inhibition and specificity: Crystal structures of the NMDA receptor NR1 ligand-binding core. EMBO J. 2003;22:2873–2885. doi: 10.1093/emboj/cdg303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inanobe A, Furukawa H, Gouaux E. Mechanism of partial agonist action at the NR1 subunit of NMDA receptors. Neuron. 2005;47:71–84. doi: 10.1016/j.neuron.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 18.Jespersen A, Tajima N, Fernandez-Cuervo G, Garnier-Amblard EC, Furukawa H. Structural insights into competitive antagonism in NMDA receptors. Neuron. 2014;81:366–378. doi: 10.1016/j.neuron.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kvist T, et al. Crystal structure and pharmacological characterization of a novel N-methyl-D-aspartate (NMDA) receptor antagonist at the GluN1 glycine binding site. J Biol Chem. 2013;288:33124–33135. doi: 10.1074/jbc.M113.480210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yi F, et al. Structural basis for negative allosteric modulation of GluN2A-containing NMDA receptors. Neuron. 2016;91:1316–1329. doi: 10.1016/j.neuron.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romero-Hernandez A, Furukawa H. Novel mode of antagonist binding in NMDA receptors revealed by the crystal structure of the GluN1-GluN2A ligand-binding domain complexed to NVP-AAM077. Mol Pharmacol. 2017;92:22–29. doi: 10.1124/mol.116.107912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conti P, et al. Novel 3-carboxy- and 3-phosphonopyrazoline amino acids as potent and selective NMDA receptor antagonists: Design, synthesis, and pharmacological characterization. ChemMedChem. 2010;5:1465–1475. doi: 10.1002/cmdc.201000184. [DOI] [PubMed] [Google Scholar]
- 23.Tamborini L, et al. Development of radiolabeled ligands targeting the glutamate binding site of the N-methyl-d-aspartate receptor as potential imaging agents for brain. J Med Chem. 2016;59:11110–11119. doi: 10.1021/acs.jmedchem.6b01344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 25.Wyllie DJ, Chen PE. Taking the time to study competitive antagonism. Br J Pharmacol. 2007;150:541–551. doi: 10.1038/sj.bjp.0706997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans RH, Francis AA, Jones AW, Smith DA, Watkins JC. The effects of a series of omega-phosphonic alpha-carboxylic amino acids on electrically evoked and excitant amino acid-induced responses in isolated spinal cord preparations. Br J Pharmacol. 1982;75:65–75. doi: 10.1111/j.1476-5381.1982.tb08758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clements JD, Lester RA, Tong G, Jahr CE, Westbrook GL. The time course of glutamate in the synaptic cleft. Science. 1992;258:1498–1501. doi: 10.1126/science.1359647. [DOI] [PubMed] [Google Scholar]
- 28.Benveniste M, Mayer ML. Structure-activity analysis of binding kinetics for NMDA receptor competitive antagonists: The influence of conformational restriction. Br J Pharmacol. 1991;104:207–221. doi: 10.1111/j.1476-5381.1991.tb12409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benveniste M, Mienville JM, Sernagor E, Mayer ML. Concentration-jump experiments with NMDA antagonists in mouse cultured hippocampal neurons. J Neurophysiol. 1990;63:1373–1384. doi: 10.1152/jn.1990.63.6.1373. [DOI] [PubMed] [Google Scholar]
- 30.Erreger K, Dravid SM, Banke TG, Wyllie DJ, Traynelis SF. Subunit-specific gating controls rat NR1/NR2A and NR1/NR2B NMDA channel kinetics and synaptic signalling profiles. J Physiol. 2005;563:345–358. doi: 10.1113/jphysiol.2004.080028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rauner C, Köhr G. Triheteromeric NR1/NR2A/NR2B receptors constitute the major N-methyl-D-aspartate receptor population in adult hippocampal synapses. J Biol Chem. 2011;286:7558–7566. doi: 10.1074/jbc.M110.182600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo J, Wang Y, Yasuda RP, Dunah AW, Wolfe BB. The majority of N-methyl-D-aspartate receptor complexes in adult rat cerebral cortex contain at least three different subunits (NR1/NR2A/NR2B) Mol Pharmacol. 1997;51:79–86. doi: 10.1124/mol.51.1.79. [DOI] [PubMed] [Google Scholar]
- 33.Sheng M, Cummings J, Roldan LA, Jan YN, Jan LY. Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature. 1994;368:144–147. doi: 10.1038/368144a0. [DOI] [PubMed] [Google Scholar]
- 34.Bissantz C, Kuhn B, Stahl M. A medicinal chemist’s guide to molecular interactions. J Med Chem. 2010;53:5061–5084. doi: 10.1021/jm100112j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ranganathan R, Lewis JH, MacKinnon R. Spatial localization of the K+ channel selectivity filter by mutant cycle-based structure analysis. Neuron. 1996;16:131–139. doi: 10.1016/s0896-6273(00)80030-6. [DOI] [PubMed] [Google Scholar]
- 36.Horovitz A, Fersht AR. Strategy for analysing the co-operativity of intramolecular interactions in peptides and proteins. J Mol Biol. 1990;214:613–617. doi: 10.1016/0022-2836(90)90275-Q. [DOI] [PubMed] [Google Scholar]
- 37.Carter PJ, Winter G, Wilkinson AJ, Fersht AR. The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus) Cell. 1984;38:835–840. doi: 10.1016/0092-8674(84)90278-2. [DOI] [PubMed] [Google Scholar]
- 38.Sheng Z, Liang Z, Geiger JH, Prorok M, Castellino FJ. The selectivity of conantokin-G for ion channel inhibition of NR2B subunit-containing NMDA receptors is regulated by amino acid residues in the S2 region of NR2B. Neuropharmacology. 2009;57:127–136. doi: 10.1016/j.neuropharm.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen PE, et al. Modulation of glycine potency in rat recombinant NMDA receptors containing chimeric NR2A/2D subunits expressed in Xenopus laevis oocytes. J Physiol. 2008;586:227–245. doi: 10.1113/jphysiol.2007.143172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansen KB, Traynelis SF. Structural and mechanistic determinants of a novel site for noncompetitive inhibition of GluN2D-containing NMDA receptors. J Neurosci. 2011;31:3650–3661. doi: 10.1523/JNEUROSCI.5565-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costa BM, et al. N-methyl-D-aspartate (NMDA) receptor NR2 subunit selectivity of a series of novel piperazine-2,3-dicarboxylate derivatives: Preferential blockade of extrasynaptic NMDA receptors in the rat hippocampal CA3-CA1 synapse. J Pharmacol Exp Ther. 2009;331:618–626. doi: 10.1124/jpet.109.156752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kinarsky L, et al. Identification of subunit- and antagonist-specific amino acid residues in the N-methyl-D-aspartate receptor glutamate-binding pocket. J Pharmacol Exp Ther. 2005;313:1066–1074. doi: 10.1124/jpet.104.082990. [DOI] [PubMed] [Google Scholar]
- 43.Feng B, Morley RM, Jane DE, Monaghan DT. The effect of competitive antagonist chain length on NMDA receptor subunit selectivity. Neuropharmacology. 2005;48:354–359. doi: 10.1016/j.neuropharm.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 44.Benveniste M, Mayer ML. Kinetic analysis of antagonist action at N-methyl-D-aspartic acid receptors. Two binding sites each for glutamate and glycine. Biophys J. 1991;59:560–573. doi: 10.1016/S0006-3495(91)82272-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hansen KB, Ogden KK, Yuan H, Traynelis SF. Distinct functional and pharmacological properties of Triheteromeric GluN1/GluN2A/GluN2B NMDA receptors. Neuron. 2014;81:1084–1096. doi: 10.1016/j.neuron.2014.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hedegaard M, Hansen KB, Andersen KT, Bräuner-Osborne H, Traynelis SF. Molecular pharmacology of human NMDA receptors. Neurochem Int. 2012;61:601–609. doi: 10.1016/j.neuint.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hansen KB, et al. Structural determinants of agonist efficacy at the glutamate binding site of N-methyl-D-aspartate receptors. Mol Pharmacol. 2013;84:114–127. doi: 10.1124/mol.113.085803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams K. Ifenprodil discriminates subtypes of the N-methyl-D-aspartate receptor: Selectivity and mechanisms at recombinant heteromeric receptors. Mol Pharmacol. 1993;44:851–859. [PubMed] [Google Scholar]
- 49.Goldschen-Ohm MP, Haroldson A, Jones MV, Pearce RA. A nonequilibrium binary elements-based kinetic model for benzodiazepine regulation of GABAA receptors. J Gen Physiol. 2014;144:27–39. doi: 10.1085/jgp.201411183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otwinowski Z, Minor W. [20] Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 51.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Read RJ. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr A. 1986;42:140–149. [Google Scholar]
- 55.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun W, Hansen KB, Jahr CE. Allosteric interactions between NMDA receptor subunits shape the developmental shift in channel properties. Neuron. 2017;94:58–64 e53. doi: 10.1016/j.neuron.2017.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arunlakshana O, Schild HO. Some quantitative uses of drug antagonists. Br Pharmacol Chemother. 1959;14:48–58. doi: 10.1111/j.1476-5381.1959.tb00928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lew MJ, Angus JA. Analysis of competitive agonist-antagonist interactions by nonlinear regression. Trends Pharmacol Sci. 1995;16:328–337. doi: 10.1016/s0165-6147(00)89066-5. [DOI] [PubMed] [Google Scholar]
- 59.Chovancova E, et al. CAVER 3.0: A tool for the analysis of transport pathways in dynamic protein structures. PLOS Comput Biol. 2012;8:e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moreau E, et al. Optimized N-phenyl-N'-(2-chloroethyl)ureas as potential antineoplastic agents: Synthesis and growth inhibition activity. Bioorg Med Chem. 2005;13:6703–6712. doi: 10.1016/j.bmc.2005.07.048. [DOI] [PubMed] [Google Scholar]
- 61.Busscher GF, Rutjes FPJT, van Delft FL. Synthesis of a protected enantiomerically pure 2-deoxystreptamine derivative from d-allylglycine. Tetrahedron Lett. 2004;45:3629–3632. [Google Scholar]
- 62.Pfefferkorn JA, et al. Substituted pyrazoles as hepatoselective HMG-CoA reductase inhibitors: Discovery of (3R,5R)-7-[2-(4-fluoro-phenyl)-4-isopropyl-5-(4-methyl-benzylcarbamoyl)-2H-pyrazol-3-yl]-3,5-dihydroxyheptanoic acid (PF-3052334) as a candidate for the treatment of hypercholesterolemia. J Med Chem. 2008;51:31–45. doi: 10.1021/jm070849r. [DOI] [PubMed] [Google Scholar]