Significance
Cdc48/TERA/p97/VCP is an enzyme that utilizes energy stored in ATP to coordinate protein degradation and recycling in eukaryotic cells. Point mutations in p97 cause a degenerative disease in humans that affects the central nervous system, bone, and muscle. These mutations deregulate the structure and dynamics of p97 with implications for binding of a cofactor that recruits p97 to the lysosomal degradation pathway. Here we probe the plasticity of p97, searching for hotspots in this enzyme to revert the effect of disease mutations. We demonstrate that mutations at secondary sites can shift the dynamics and structure toward wild type, highlighting the potential of exploiting the plasticity of p97 in the rational design of compounds that restore function in disease mutants.
Keywords: p97/VCP, conformational plasticity, domain equilibrium, methyl-TROSY NMR, IBMPFD disease mutants
Abstract
p97/VCP, a member of the AAA+ (ATPases associated with diverse cellular activities) family of proteins, is implicated in the etiology of a group of degenerative diseases affecting bone and muscle tissue as well as the central nervous system. Methyl-TROSY–based NMR studies have previously revealed how disease-causing mutations deregulate a subtle dynamic conformational equilibrium involving the N-terminal domain (NTD) with implications for the binding of certain adaptors, providing insight into how disease mutations lead to abnormal function. Herein the conformational plasticity of the p97 system is explored in an attempt to identify hotspots that can serve as targets for restoring function in disease mutants by shifting the position of the NTD back to its wild-type location. Although p97 is overall robust with respect to extensive mutagenesis throughout the protein involving conservative substitutions of hydrophobic residues, key positions have been identified that alter the NTD equilibrium; these lie in specific regions that localize to the interface between the NTD and the D1 nucleotide-binding domain of the complex. Notably, for a severe disease mutant involving an R155C substitution the NTD equilibrium can be shifted back to its wild-type position by mutation at a secondary site with restoration of wild-type two-pronged binding of the UBXD1 adaptor protein that is impaired in disease; this underlies the potential for recovering function by targeting p97 disease mutants with drug molecules.
The highly conserved valosin containing protein (VCP), also known as p97 in mammals or Cdc48 in yeast, plays a critical role in a wide range of cellular functions (1). These include segregation of proteins from complexes or membranes (2), membrane fusion (3), progression of the cell cycle (4), and protein degradation via either the proteasomal (5) or lysosomal pathways (6). p97 is directed to its various functions through binding to one of over 40 adaptors that have been discovered in mammalian cells and the fidelity of binding is critical for proper function (7). Detailed structural studies that include both X-ray (8, 9) and cryo-EM–based (10) approaches have established that p97 has a homohexameric double-ring structure with each 89 kDa protomer consisting of an N-terminal domain (NTD) followed by two nucleotide-binding domains (D1 and D2). These studies clearly show that the NTD of p97 can assume one of two positions: in the plane formed by the D1 ring (referred to as the down position) or above the plane of the ring (up) depending on whether the nucleotide in D1 is ADP or ATP, respectively (Fig. 1A).
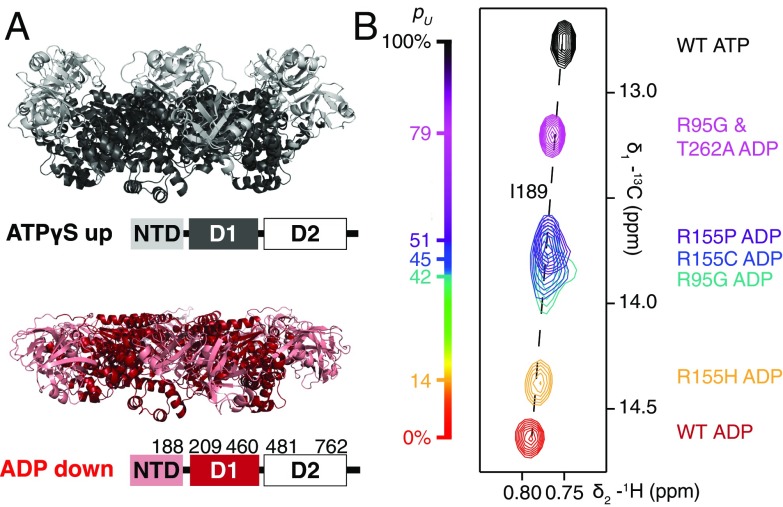
Fig. 1.
(A) Ribbon diagram representations of X-ray structures of ND1Lp97 (residues 1–480) with NTD in the up (ATPγS, black) and down (ADP, red) states [PDB ID code 4KO8 (29) for R115H p97-ATPγS; 1E32 (33) for p97-ADP]. The domain organization of full-length p97 is shown; note that D2 has been removed in this study. NTDs of disease mutants of ND1Lp97-ADP interconvert between these two conformations with a rate that is fast on the NMR chemical shift timescale, >15,000 s−1. The chemical shifts of key residues, such as I189 located in the NTD–D1 linker, thus encode the fractional populations of up (pU) and down states. (B) Superpositions of small regions of 13C-1H HMQC spectra focusing on the I189 peaks from a series of WT and disease mutants of Iδ1-13CH3, proR L,V-13CH3, Mε1-13CH3–labeled p97 recorded at 18.8 T, 50 °C.
In humans, p97 is implicated in a dominantly inherited and ultimately lethal degenerative disorder affecting muscle, bone, and the central nervous system that is referred to as inclusion body myopathy associated with Paget disease of the bone and frontotemporal dementia (IBMPFD) (11) or as multisystem proteinopathy type 1 (MSP1) (12). Mutations identified in patients have been reported at various locations clustered at the interface between the N-terminal and D1 domains or in the linker region between these domains (12). Using solution-state NMR spectroscopy and focusing on methyl group probes of structure and dynamics, we have previously shown that in the p97-ADP state the NTD is in equilibrium between up/down conformations and that as a function of disease mutant severity the equilibrium becomes progressively shifted from the down [wild type (WT)] to the up conformation, the latter characterizing the WT ATP state (13). This picture is distinct from that obtained via X-ray and EM studies of both WT and disease mutants of p97 where the NTD conformation, either up or down, appears to be dictated exclusively by the nature of the bound nucleotide (10, 14). However, the dynamic model that emerges from NMR studies provides an important link between mutation and p97 function. Notably, as the equilibrium is shifted, binding of the adaptor UBXD1, which recruits p97 to the lysosomal degradation pathway (6, 15), becomes increasingly impaired. Because a complex of UBXD1-p97 is required for recruitment of ubiquitylated caveolin-1 molecules to the lysosome for degradation this leads to the accumulation of caveolin-1–positive endolysosomes in IBMPFD patients (6). In contrast to the negative effects on the lysosomal pathway caused by impaired UBXD1 binding, interactions with other adaptors, such as p47 that targets p97 for membrane remodeling, are little affected by the IBMPFD mutations (16–18).
The underlying energetics of the disease mutations that lead to the shift of the up/down equilibrium are small. For example, in the case of weak (R155H) and strong (R95G) disease mutants, the difference in free energy between the up and down conformations for p97-ADP is estimated to be on the order of ∼1200 cal/mol and 200 cal/mol, respectively, based on the relative populations of the up and down states (13). The fact that subtle differences in up/down conformational energetics are related to large differences in structure and, importantly, are correlated strongly with function suggested to us that it may be possible to manipulate the up/down equilibrium through the introduction of mutations, so that WT activity could be restored, at least in vitro, in severe disease mutants. Here we have explored the p97 energy landscape by carrying out a series of NMR spin relaxation experiments on disease mutants that clearly establish that mutant p97 molecules can be dynamic over a range of timescales. In an effort to exploit this inherent plasticity, a series of mutations were designed to either destabilize the up or stabilize the down states as a first step toward manipulation of function by controlling the NTD equilibrium. Although the equilibrium was resistant to conservative substitutions involving hydrophobic residues, key sites at the interface of the NTD and D1 domains were identified as hotspots, where mutations lead to changes in relative populations of the up/down conformers. Notably, the perturbed up/down equilibrium in a severe R155C disease mutant could be shifted back to the WT state by a single compensating mutation and WT binding of the UBXD1 adaptor protein was restored; this highlights the potential of exploiting the plasticity of p97 in the design of rational therapeutics for regenerating WT function in IBMPFD disease mutants.
Results
Disease Mutations Induce Dynamics on Multiple Timescales.
Hexameric p97 is a 540-kDa complex whose large size challenges traditional solution-state NMR approaches for studies of protein structure and dynamics. Herein, as in our previous study of p97 (13), we have exploited stereospecific methyl labeling of highly deuterated protein complexes (19) in concert with a methyl-TROSY (20)–based NMR approach to obtain well-resolved, high-quality spectra of this large system. Further, to reduce the aggregate size of p97, we focused on a 320-kDa construct of p97 containing the NTD, D1, and the linker between D1 and D2 (ND1Lp97, 6 × 53 kDa, residues 1–480) (9) that was prepared as a highly deuterated, selectively Iδ1-13CH3, proR L,V-13CH3, Mε1-13CH3–labeled protein (19, 21, 22). ND1Lp97 has previously been shown to be a good model system that faithfully reports on the NTD equilibrium in the same way as the full-length protein (13). Fig. 1B shows a superposition of small regions of 13C-1H HMQC spectra recorded of a number of p97 variants, including WT ND1Lp97-ADP, several disease mutants of ND1Lp97-ADP, and WT ND1Lp97-ATP, focusing on the I189 correlation. I189 lies at the N-terminal side of the linker connecting the NTD to the D1 domain and thus serves as a probe of the linker helix to coil transition that accompanies the hydrolysis of ATP (9). As a result, the position of the I189δ1 cross-peak in spectra serves as a proxy for the up/down NTD equilibrium. Notably, as observed with many other methyl probes (13), the peak position of I189 in p97-ADP variants titrates as a function of mutation in a near linear manner; this is most simply explained as a shift in an equilibrium that involves rapid interconversion between down (ADP-bound like) and up (ATP-bound like) conformers (Fig. 1A) from an all down state for the WT protein (pU = 0, where pU is the fraction of up NTD population) to increasing amounts of up conformer as a function of disease mutant severity (13). As described previously, pU for a given disease mutant can then be simply calculated from the relative position of the cross-peak in relation to the limiting corresponding chemical shifts in spectra of the WT p97-ADP and WT p97-ATP states. NTDs in the weak disease mutant R155H are mostly down (pU ∼0.15), whereas the up/down equilibrium in the case of the strong disease mutants R155C, R155P, and R95G is shifted significantly more to the up state, pU ∼0.45. Notably, when the two disease-causing mutations, R95G and T262A, were combined, pU increases further to 0.79, suggesting that the energy landscape is plastic and that this plasticity can be exploited to manipulate the up/down equilibrium (see below).
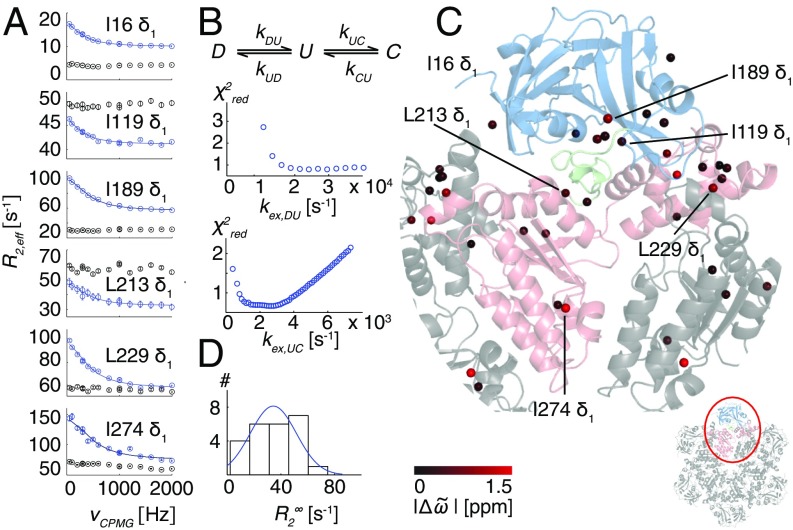
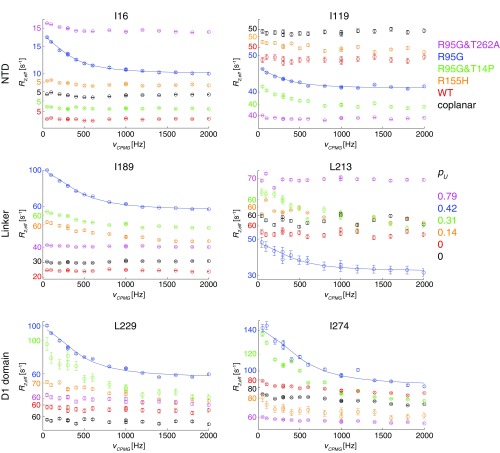
A qualitative estimate of the lower bound for the up/down exchange rate, ∼2,000 s−1, is obtained from the 2 ppm I189 13Cδ1 chemical shift difference between up and down states (Fig. 1), assuming that their interconversion is fast on the NMR chemical shift timescale (see below). Because it is known that molecular motions of the N-terminal domain are of central importance for the ATPase activity of p97, with cross-linking of NTD to D1 suppressing activity in both nucleotide binding domains (23), we were interested in obtaining further insight into the dynamics of p97 beyond the qualitative measures that are obtained from changes in chemical shifts with mutation. We therefore characterized microsecond-to-millisecond timescale dynamics in the ADP state of p97 by performing methyl-TROSY–based CPMG relaxation dispersion experiments (24) that are sensitive to stochastic fluctuations of chemical shifts resulting from exchange between different protein conformational states (25). The experiment measures the effective decay of 1H-13C multiple quantum coherences during a fixed interval where the effective chemical shift difference between exchanging states is modulated by the application of 13C chemical shift refocusing pulses (24). As the number of pulses increases (increasing νCPMG), the effective shift differences decrease, leading to a decrease in the attenuation of signal. Hence the effective relaxation rate, R2,eff, becomes smaller. Dispersion profiles, R2,eff vs. νCPMG, where R2,eff decreases with νCPMG, are the hallmark of chemical exchange and when fit to an appropriate model of exchange estimates of populations of interconverting states and rates of interconversion are obtained. In contrast, flat dispersion profiles are consistent with an absence of millisecond timescale dynamics or with processes for which there are no chemical shift changes between states. A challenge in the analysis of relaxation dispersion data in general often relates to the choice of a proper model, because a priori little insight into the underlying structural rearrangements is obtained from the raw dispersion profiles. Fig. 2A illustrates a number of the larger dispersion profiles obtained for R95G ND1Lp97-ADP (blue circles) along with the corresponding curves generated from measurements on a coplanar variant of the R95G mutant in which the NTD is covalently tethered to D1 via a disulfide bond connecting residues 155 and 387 that have been converted to cysteines (black circles). Dispersions are found for methyl groups that are distributed throughout the structure, as illustrated by the examples in Fig. 2 that were selected from the NTD, the D1 domain, and the linker region connecting these two domains. Notably, flat dispersion profiles are measured upon cross-linking indicating that the exchange process becomes quenched. In addition to recording dispersion experiments on the R95G mutant (pU = 0.42) and the coplanar R95G variant we obtained datasets for WT ND1Lp97-ADP and additional mutants with pU = 0.14 (R155H ND1Lp97-ADP), 0.31 (R95G&T14P ND1Lp97-ADP), and 0.79 (R95G&T262A ND1Lp97-ADP; Fig. S1). We find that relaxation dispersion profiles are maximal when pU is ∼0.5, as for the R95G mutant (Fig. S1), and that motional freedom of the NTD is necessary for conformation exchange, because the exchange process is eliminated upon cross-linking (see above; Fig. 2A).
Fig. 2.
R95G p97-ADP disease mutant is plastic. (A) 13C-1H multiple quantum relaxation dispersion profiles (50 °C, 18.8T) for selected residues in the NTD (I16, I119), NTD–D1 linker (I189, L213), and the D1 domain (L229, I274) for R95G ND1Lp97-ADP (blue) and the corresponding molecule with the NTD domain covalently tethered to D1 (black). A global fit is included for the R95G profiles based on an analysis of 22 residues (Combined Fits of Chemical Shift and CPMG Data). (B) Three-state exchange model that takes into account both the 13C chemical shift data (Fig. 1B) and CPMG dispersion data that consists of the NTD up (U)/down (D) equilibrium and an additional process involving a third state, C. Reduced χ2 surfaces for rates for the first and second processes for the R95G mutant. (C) Positions of methyl groups from which CPMG dispersion profiles were analyzed via a global fit of the data using the model in B. Methyls are shown as spheres on the X-ray structure of ND1Lp97-ADP color-coded by the size of differences between 13C chemical shifts measured in spectra of R95G ND1Lp97-ADP and for the corresponding probes in state C (Combined Fits of Chemical Shift and CPMG Data), |Δϖ|. (D) Histogram of fitted rates for R95G ND1Lp97-ADP.
Fig. S1.
13C-1H multiple-quantum relaxation dispersion profiles from experiments recorded on WT ND1Lp97-ADP (red) and the following mutants: R95G&T262A (magenta), R95G (blue), R95G&T14P (green), R155H (orange), and R95G-coplanar (black). All p97 constructs were in the ADP loaded state. Selected residues are located in the NTD (I16, I119), in the NTD–D1 linker (I189, L213), and in the D1 domain (L229, I274). Notably, dispersion profiles are largest for R95G ND1Lp97-ADP and decrease as pU becomes either larger or smaller than ∼0.5. Nonflat dispersion profiles were obtained for a small number of residues in R155H ND1Lp97-ADP (6) and for 15 residues in T14P,R95G ND1Lp97-ADP where the interface between the N terminus of the NTD and the D1 domain of an adjacent protomer has been modified by removal of the N-terminal NTD helix (T14P; see text). For all other variants considered, profiles were essentially flat. All residue-specific dispersion curves from different mutants have been offset slightly for ease of visualization; y axis values are color-coded differently for each mutant, as indicated. Note the different plateau values for dispersion profiles (taking into account the different offsets), which likely indicates a diversity of fast timescale dynamics (faster than νCPMG) both as a function of position within a given p97 variant and between variants. A case in point is provided by I16, located in a flexible loop region in the down state that undergoes a secondary structure change to a helix that folds back onto the D1 hexamer, in the up state. Plateau values of ≈5 s−1 are obtained for profiles from this residue when the NTDs are in the down (WT, disulfide linked) or predominantly down (R155H) conformations, or when the helix cannot form due to mutation (R95G&T14P), yet increase to 10–15 s−1 when the NTDs are partially up (R95G, R95G&T262A). Trends for other residues can sometimes be more difficult to rationalize, but it is worth noting that, as discussed in the text, the position of the plateau is influenced not only by dynamics but also by the magnitude (and sign) of 1H Δϖ in multiple quantum-based CPMG experiments (24).
Because dispersion profiles were largest for the R95G ND1Lp97-ADP variant, we analyzed these profiles in detail. Dispersion curves (24 profiles) could be well fit to a simple two-site exchange model (χ2red < 1), , where G and E are the populated, ground and sparse, invisible states, respectively, with pE = 10% and kex = kGE + kEG = 2,700 s−1, 50 °C. However, the resulting chemical shift differences between states, ΔϖGE = ϖE – ϖG, did not match those measured between NTD up and down states from spectra of WT ND1Lp97-ADP and WT ND1Lp97-ATP, although results from cross-linking (see above) clearly argue that the up/down exchange is integral to the observation of nonflat dispersion profiles. Moreover, if the ΔϖGE values were enforced to be those that correspond to the up/down shifts, the dispersions could not be well fit, with reduced χ2 values increasing from 1 to over 6. Thus, although the dispersion data in isolation could be fit to a two-site model of chemical exchange, the use of a more complex model that could explicitly account for both the chemical shift titration (vs. mutation) and relaxation dispersion data is justified, as described in general terms recently by Chao and Byrd (26).
Fig. 2B shows the three-state model that we have used to explain the chemical shift and relaxation data. Here we explicitly include the up (U)/down (D) NTD equilibrium, which is strongly suggested by the chemical shift data (Fig. 1B), with U and D corresponding to the pure up and down states, respectively, as well as an additional exchange process to a state designated as C. In the fits, ΔϖDU values are fixed to shift differences obtained from spectra of WT p97-ADP and WT p97-ATP states for methyl groups from five residues (I189 Cδ1, L213Cδ1, L229Cδ1, M388Cε1, and M442Cε1) that have large shift differences and that are far removed from bound nucleotide and from the position of mutation (residue 95). In addition, the 13C chemical shifts of these five residues that inform on the up/down equilibrium are also included in the fits. The populations of states U and C are fitting parameters, as are kex,DU = kDU + kUD and kex,UC, along with ΔϖDU values for residues with dispersions whose shifts are not fixed and ΔϖCD values for all residues for which dispersion curves are analyzed. Fits are shown by solid curves for the residues highlighted in Fig. 2A, along with the resulting χ2red surfaces for fits obtained as a function of a series of kex,DU and kex,UC values. Notably, the up/down equilibrium is fast (kex,DU), greater than ∼15,000 s−1, whereas kex,UC is between ∼1,500 and 4,000 s−1 (Fig. 2B), pU = 0.41 (as expected from the chemical shift data), and pC = 0.09. Fig. 2C plots the differences between the 13C chemical shifts of methyl probes as measured in spectra of R95G ND1Lp97-ADP and the chemical shifts of the corresponding probes in state C (Combined Fits of Chemical Shift and CPMG Data) on the structure of WT ND1Lp97-ADP, color-coded according to the size of the shift differences. Large shift differences are localized to interfaces between domains, including between adjacent protomers (e.g., L229; pink and gray protomers in Fig. 2C); this suggests that the three-state nature of the exchange process may arise from relative dynamics between adjacent protomers that become prominent in the R95G mutant. It is worth noting that in the scheme of Fig. 2B the positioning of state C in relation to U and D is arbitrary given that the exchange between D and U is fast; fits that have been performed using a model in which C is placed before D (Fig. 2B) produce the same kDU, kUD, and ΔϖCD values, with kUC and kCU rates replaced by kDC and kCD, respectively.
Additional, albeit qualitative, information is available from the intrinsic relaxation rates, , that are extracted from analyses of dispersion profiles, such as those in Fig. 2A. In this regard, it is worth noting that for the 13C-1H multiple quantum-based CPMG experiments considered here, plateau values of R2,eff cannot be used as a proxy for rates without caution because the former are influenced by changes in 1H chemical shifts of spins between ground and excited states that are not pulsed out using the 13C chemical shift refocusing scheme (24). In the case here, however, it seems likely that the differences in R2,eff plateau values for some of the black and blue curves (Fig. 2) reflect differences in fast time-scale dynamics that result from the disulfide (Fig. S1). A large distribution of rates is noted for R95G ND1Lp97-ADP, varying from ∼10 s−1 (I16δ1) to 80 s−1 (I274 δ1) (Fig. 2D). The large values are consistent with additional motional processes that occur with rates faster than νCPMG,max that can therefore not be modulated through the application of pulses. Taken together, our data provides strong evidence for a range of dynamics over a wide spectrum of timescales in the severe R95G p97-ADP disease mutant.
Key Hotspots Regulate p97 Dynamics.
Having shown that the p97-ADP up/down equilibrium is significantly affected by disease mutations that localize to the interface between domains (13) (Fig. S2A), we were interested in establishing whether perturbation of other sites in the protein might also trigger changes. We therefore carried out an extensive mutagenesis of ND1Lp97-ADP that involved over 50 different sites in the molecule, shown in Fig. S2B and listed in Table S1. The mutations were distributed all over ND1Lp97, and in general were highly conservative where one methyl-bearing amino acid side chain was converted into another. None of these mutants produced changes to the up/down equilibrium in the ADP bound state, as judged by the positions of cross-peaks in NMR spectra. Thus, it appears that there are a few key hotspots that control the NTD dynamics (Fig. 1B and Fig. S2A), while the majority of the protein is robust with respect to localized structural disturbances. We decided, therefore, to further explore the plasticity of p97 by making mutations that targeted specific structural features that are unique to either the up or down states, localized to the interface regions that appear critical in regulating the position of the NTD, in the hope that these manipulations might restore the WT equilibrium in the disease mutants.
Fig. S2.
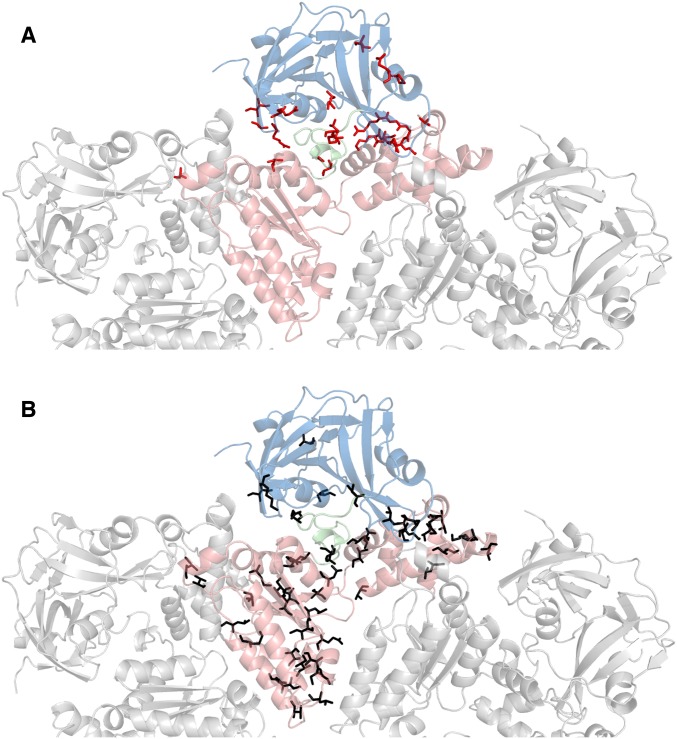
Location of disease-associated mutations (A) (12) and predominantly hydrophobic substitutions that have been tested (B) in ND1Lp97. Top view of a half-hexamer of ND1Lp97-ADP in cartoon representation with the central protomer color-coded by domain (NTD-linker-D1 in blue, green, red). (A) The 22 residues in ND1Lp97 that are mutated in disease patients (12) are shown in red (sticks) on the structure of the ADP state [PDB ID code 1E32 (33)]. (B) Black sticks denote the positions of 56 point mutations that do not shift the NTD up/down equilibrium in p97-ADP.
Table S1.
Point mutations that do not perturb the up/down NTD equilibrium in ND1Lp97-ADP
| Mutation at | Mutated to | CSP* | RD† | Other observations | Mutation at | Mutation to | CSP* | RD† |
| L17 | I | Located in N-terminal helix that is formed in the ATP state only | V320 | I | ||||
| I27 | V | x | Associated with disease (38) but also found in healthy control patients (12) | I324 | V | |||
| I32 | V | x | V325 | I | ||||
| K63 | A | Affects salt bridge formation in ATP state | M332 | L | ||||
| E | ||||||||
| V71 | I | L335 | M | |||||
| V123 | I | x | V341 | I | ||||
| I126 | L | M344 | I | x | ||||
| M158 | L | x | Location of disease mutation M158V | I353 | V | x | ||
| I189 | V | x | x | Located at the beginning of linker helix | V367 | I | ||
| E194 | P | Located in the middle of linker helix; affects helix formation in ATP state | I369 | V | ||||
| V201 | I | x | Located at the end of linker helix | L378 | M | |||
| I206 | V | Location of disease mutation I206F | I380 | V | ||||
| L213 | M | x | x | I383 | V | |||
| M219 | L | x | M388 | L | x | x | ||
| I233 | L | V407 | T | |||||
| V235 | I | L420 | M | |||||
| L253 | I | I423 | V | |||||
| V258 | I | M427 | L | x | ||||
| L268 | M | L429 | M | |||||
| I269 | V | L432 | I | |||||
| I274 | V | x | x | M442 | L | x | ||
| M275 | L | L445 | M | x | ||||
| L278 | M | V447 | I | |||||
| I300 | V | L464 | M | |||||
| E305 | Q | Walker B position | V468 | I | ||||
| I309 | V | V471 | L |
For each residue, it is noted whether the δ1 (Ile), γ1 (Val), or δ1 (Leu) methyl peak position is sensitive to the NTD up/down equilibrium [x, combined 13C and 1H chemical shift differences between ND1Lp97-ADP and R95G mutant exceeds 0.25 ppm, > 0.25 ppm, where Δϖ is in ppm].
The x indicates that ΔR2,eff > 1.5 s−1, where ΔR2,eff = R2,eff (νCPMG = 50 Hz) – R2,eff (νCPMG = 2,000 Hz) in relaxation dispersion profiles recorded on R95G ND1Lp97-ADP.
Destabilizing the Up State of ND1Lp97.
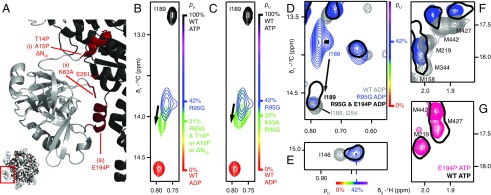
The X-ray structure of ND1Lp97-ATPγS (ATPγS is a slowly hydrolyzing analog of ATP) provides a number of clues as to how one might destabilize the up state that becomes overpopulated in p97-ADP disease mutants. Fig. 3A focuses on the three regions of the structure labeled i–iii that were explored. In i, the N-terminal NTD helix (in red) makes contacts with the D1 domain of an adjacent protomer. Notably, this helix is not formed in p97-ADP, where the N terminus of the NTD is disordered. We reasoned, therefore, that elimination of the helix might shift the equilibrium in disease mutants such that the NTD becomes positioned in the down conformation. Insertion of a proline residue in the middle of an α-helix is known to destabilize it by over 3 kcal/mol (27), effectively disrupting the helix. Therefore, we considered a number of mutants, including T14P and A15P, where residues 14 and 15 are located in the center of the helix, as well as a deletion that eliminates the first 20 amino-terminal residues that includes the helix, ΔN20. These mutants were inserted into the R95G disease mutant background and the effect of the substitutions evaluated by monitoring the position of the I189 cross-peak that reports on the NTD up/down equilibrium. Fig. 3B (Fig. S3A) shows that pU decreases from 0.42 for R95G ND1Lp97-ADP to 0.31 when any of the above-mentioned mutations is inserted. As a second manipulation (ii) we considered the elimination of favorable electrostatic interactions that are formed in the up state (ATP-p97) but not in the WT down conformation involving the side chains of K63 and E200/E261. There is a decrease in pU (to 0.33) for K63A,R95G ND1Lp97-ADP (Fig. 3C).
Fig. 3.
Destabilization of the NTD up state. (A) Three sites were targeted at the interface between the NTD and D1, as highlighted in red on the X-ray structure of WT ND1Lp97-ATPγS (29) and denoted by i–iii. (B) The N-terminal helix (site i) is destabilized through the substitution of a proline residue at positions 14 or 15 or removed by truncation, ΔN20. Superposition of small regions of 13C-1H HMQC spectra (18.8T, 50 °C) focusing on I189, with the I189δ1 cross-peak for ΔN20 R95G ND1Lp97-ADP in green (note that I189δ1 chemical shifts are identical for each of T14P, A15P, or ΔN20 substitutions), blue for R95G ND1Lp97-ADP, red for WT ND1Lp97-ADP, and black for WT ND1Lp97-ATP. Relevant mutations are placed in the R95G background because it is straightforward to evaluate changes to the up/down equilibrium in a system where pU ∼0.5. (C) As in B but for K63A,R95G ND1Lp97-ADP where an electrostatic interaction that stabilizes the up state has been removed (site ii). (D–F) Effect of removing the helix in the linker region connecting the NTD and D1 domains via the E194P mutation (site iii). The I189δ1 correlation in R95G,E194P ND1Lp97-ADP shifts (arrow) close to its position in WT ND1Lp97-ADP, indicative of an unfolding of the helix (D). Movement of the NTD into the down state does not occur as established by the positions of methyl correlations for I146 (E) and a number of methionines (F) that report on the up/down equilibrium that do not change with the E194P mutation (compare black single contour and blue contours in E and F). (G) Spectra of methionine methyls from WT ND1Lp97-ATPγS and E194P ND1Lp97-ATPγS superimpose, confirming that the helix in the NTD–D1 linker is not required for the NTD up state. pU values are highlighted on the sides of spectra in B–E, using the same color code as in Fig. 1.
Fig. S3.
Effect of mutations on the NTD equilibrium in R95G ND1Lp97-ADP. Superposition of the isoleucine region from 13C-1H HMQC spectra (18.8T, 50 °C) focusing on I146δ1 and I189δ1 cross-peaks that report on the NTD up/down equilibrium. (A) The ΔN20 deletion in the R95G context (black contours) partially restores the equilibrium to the down position (blue to black). (B) Position 95 is distal from the N387C mutation, and thus, unlike the case for R155C ND1Lp97-ADP (see text), the N-to-C substitution at position 387 has no effect on the NTD equilibrium in R95G ND1Lp97-ADP (as evidenced by the superposition of blue and black peaks).
Next we examined whether the helix that is formed in the NTD–D1 linker in the p97-ATP state (site iii) is essential for supporting the up conformation, and thus whether this region might serve as a further point for controlling the up/down equilibrium. Notably, this helix is thought to play a role in “pushing” the NTD to the up state in the p97-ATP form and becomes unstructured in p97-ADP (9). A proline was introduced at position 194, with a predicted destabilization of 2.8 kcal/mol based on ROSETTA calculations (28), which further establish that the proline substitution at 194 is the most destabilizing of all 20 amino acid replacements based on the structure of the up state of ND1Lp97 (PDB ID code 4K08) (29). Indeed, the E194P mutation fully abolished the helix, as evidenced by the 13Cδ1 chemical shift of residue I189, which is located at the beginning of the helix. The Iδ1 cross-peak for this residue shifts almost fully to its position in WT ND1Lp97-ADP (Fig. 3D), where the linker is in a loop conformation. However, the up/down equilibrium is not perturbed, as evidenced by the fact that cross-peaks for I146 (Fig. 3E) and for many of the methionines (Fig. 3F), which are all sensitive to the up/down states, do not revert back to their positions in WT p97-ADP (down) but rather remain superimposable in spectra of R95G p97-ADP and E194P,R95G p97-ADP (compare black single contour with blue contours). Thus, in the context of the E194P mutation, or for that matter any mutation that disrupts formation of the linker helix, the position of the δ1 methyl of I189 is not a good proxy of the up/down equilibrium because, as noted, it senses precisely the formation of the linker helix which, in general, accompanies the up transition. All other mutations that we have studied do not affect the linker helix, and the pU values obtained from the resonance position of I189 are in good agreement with those from other probes of the equilibrium (i.e., I189 is an excellent probe).
We also tested whether the linker helix is required for stabilization of the pure up state in the ATPγS-loaded form of the protein. In E194P ND1Lp97-ATPγS, the NTDs are in the up conformation even in the absence of the helix, as evidenced by the superposition of methionine peaks in 13C-1H HMQC spectra of E194 p97-ATPγS and WT p97-ATPγS (Fig. 3G, black and red peaks), confirming that this helix is not required for stabilizing the up state. In summary, our mutagenesis study identified two sites (i and ii; Fig. 3A) as potential hotspots for manipulation of the aberrant NTD position in a severe disease mutant and established that the coil-to-helix transition that accompanies ATP binding in the WT protein is not essential for populating the up conformation.
Stabilizing the Down State of ND1Lp97.
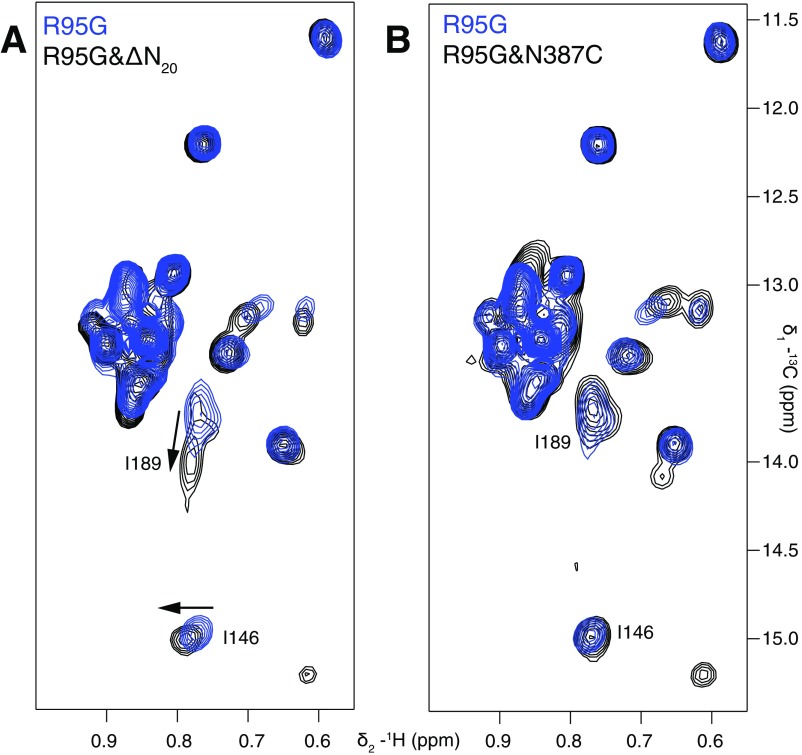
The above experiments provide a clear demonstration that it is possible to manipulate the NTD equilibrium in a rational way by modifying structural features that appear to stabilize one state over the other. With this in mind, we focused on stabilizing the down state of ND1Lp97-ADP by targeting a region of the NTD–D1 interface formed by residues 155–159 and 386–387 in the NTD and in D1, respectively (Fig. 4A). Notably, 16 of the 41 reported disease mutations are localized to this small section of the complex with residue 387 the site of a number of disease mutations formed by the substitution of His, Thr, or Ser for the WT Asn (12). Because residues 155 and 387 are within 3 Å in p97-ADP and R155 is mutated to either cysteine or proline in strong disease mutants (Fig. 4A), we considered amino acid substitutions for N387 that might lead to energetically favorable interactions with C155 or P155 and hence stabilization of the down conformation in these disease mutants (pU = 0.45 and 0.51 for R155C ND1Lp97-ADP and R155P ND1Lp97-ADP, respectively). Fig. 4 B and C shows that the N387C mutation is one possibility; the position of the NTDs are shifted toward the down state in both of the double mutants R155C,N387C ND1Lp97-ADP and R155P,N387C ND1Lp97-ADP, to different degrees, with the N387C substitution restoring the down state of the R155C mutant to nearly WT levels (pU = 0.06). It is worth noting that the NMR samples were maintained under reducing conditions using the reducing agent Tris(2-carboxyethyl)phosphine (TCEP), and that the reduced state of the cysteines at positions 155 and 387 in R155C,N387C ND1Lp97-ADP could be confirmed by NMR spectra (see below; Fig. S4). The fact that the R155C and R155P mutants are restored to different degrees argues that the effect, at least in part, is local, involving the interplay between sidechains of residues 155 and 387. In contrast, if the change in equilibrium from the N387C substitution was principally due to destabilization of the up state (where positions 155 and 387 are >20 Å from each other), it might be expected that the effects on the R155C and R155P mutants would be more uniform. We also tested R95G,N387C ND1Lp97-ADP to establish whether the N387C mutation might revert the aberrant NTD equilibrium in the R95G case as well; Fig. S3B shows that this is not the case, with no change in the up/down equilibrium. Because positions 95 and 387 are distal in the p97-ADP conformation, this result argues, as above, that a local effect underlies the stabilization of the down state in the R155C/P mutants.
Fig. 4.
Stabilizing the NTD down state. (A) Positions 155 and 387 (orange sticks) are proximal in the NTD down state, as highlighted by the X-ray structure of WT ND1Lp97-ADP (33). (B and C) The N387C mutation shifts the up/down equilibrium in R155C ND1Lp97-ADP (B) and R155P ND1Lp97-ADP (C) toward the down state, with the NTDs almost in the WT p97-ADP position in the case of R155C,N387C ND1Lp97-ADP. Position 95 is distal from 155 and the N387C mutation has no effect on the location of the NTDs in R95G ND1Lp97-ADP (Fig. S3B). Note that for both of the R155C and R155P mutants considered in B and C, position 95 was WT.
Fig. S4.
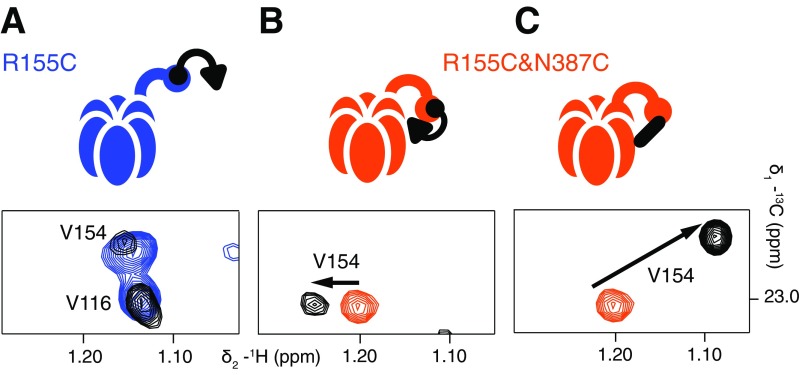
V154 is a proxy for disulfide bond formation between proximal C155 and C387 residues in R155C,N387C ND1Lp97-ADP. Selected regions of 13C-1H HMQC spectra are shown highlighting the resonance position of the δ1 methyl group of V154. (A) R155C ND1Lp97-ADP (blue, no UBXD1-N; black, threefold excess UBXD1-N over p97 protomers). As indicated schematically above the spectrum, binding of UBXD1-N to the NTD occurs via the VIM domain (black circle) but the H1/H2 motif (black triangle) is not able to interact, and there is no change in the peak position of V154. (B) In the R155C,N387C ND1Lp97-ADP variant, the up/down equilibrium is restored to the WT situation and WT two-pronged binding of UBXD1-N is observed (see main text and schematic). The position of V154 shifts slightly to reflect this, as indicated by the arrow. Sample is prepared with 5 mM TCEP. (C) A large chemical shift change is observed for V154 δ1 that reflects formation of the 155–387 disulfide in R155C,N387C ND1Lp97-ADP under oxidizing conditions (orange, 5 mM TCEP; black, oxidizing conditions). UBXD1-N is not added in this case.
UBXD1 Binding Is Restored to R155C,N387C ND1Lp97-ADP.
The UBXD1 adaptor–p97 complex plays an important role in regulating the transport of ubiquitylated cargo to the lysosome for degradation (30), with interactions between UBXD1 and p97 specifically disrupted in the IBMPFD disease mutants considered in this study (6). We have previously shown (13) that an N-terminal fragment of UBXD1 (UBXD1-N, reported in ref. 15), consisting of the first 133 residues of the adaptor and including the H1/H2 and VCP-interacting motif (VIM) regions, binds to p97-ADP via a two-pronged mechanism with the VIM and H1/H2 motifs contacting the canonical VIM binding region of the NTD and the NTD–D1 interface, respectively. This model is supported by additional NMR studies on UBXD1-N and the isolated NTD domain (15). The effective binding of UBXD1-N to p97-ADP decreases with increasing disease severity because the H1/H2 interactions are weakened both by the disruption of the NTD–D1 interface and by additional structural changes that occur in this region upon mutation (13).
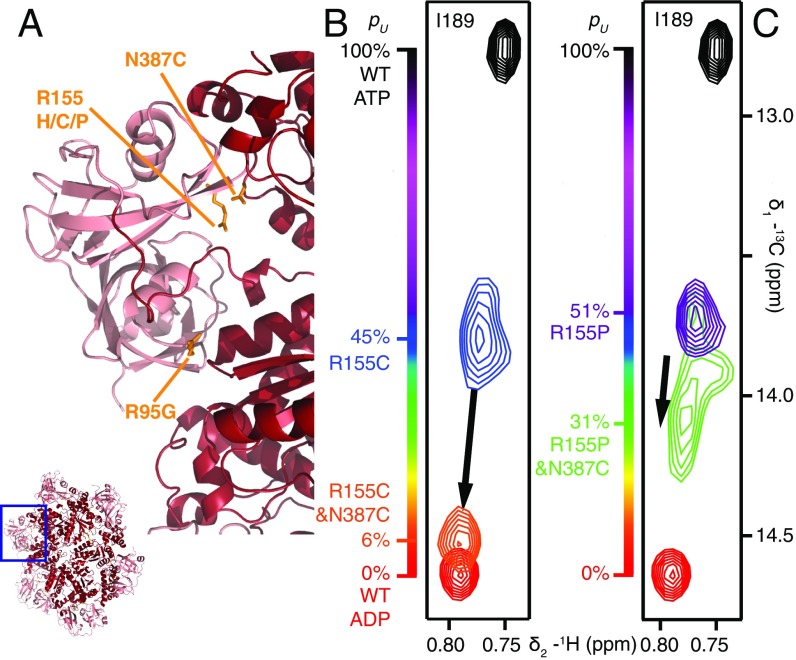
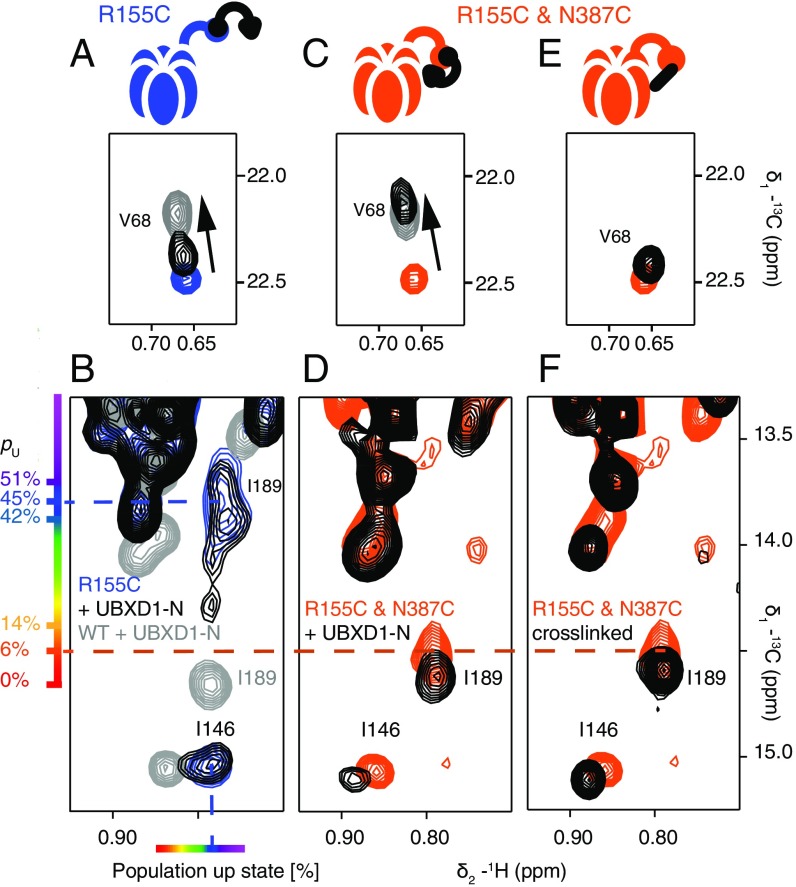
Fig. 5 A and B shows selected regions of 13C-1H HMQC spectra highlighting the binding of threefold excess UBXD1-N to WT ND1Lp97-ADP (gray bound) and to R155C ND1Lp97-ADP (blue free; black bound). Here we focus on a small number of residues that serve as a readout of binding, with V68 of the NTD reporting on the VIM–NTD interaction (31, 32) and residues I146 (NTD) and I189 (NTD–D1 linker) probing binding of the H1/H2 motif to the p97 NTD–D1 interface (13). Fig. 5A shows that for the same excess of UBXD1-N, the position of the V68 methyl resonance for R155C ND1Lp97-ADP shifts only partially to the fully bound WT location (compare black vs. gray peaks) that implies a weaker UBXD1-N binding affinity for the R155C mutant relative to the WT protein [Kd,macro = 20 μM (13)]. An estimated Kd,macro = 200 μM is calculated (based on the relative shift; see ref. 13 for details) that is the same as obtained for the R95G disease mutant. Notably, the I146 and I189 cross-peaks do not shift upon addition of excess UBXD1-N (compare black vs. blue peaks in Fig. 5B), as reported previously for the R95G and R155P strong disease mutants (13); this clearly indicates that the second binding interaction involving the H1/H2 domain of the adaptor is eliminated, accounting for the increased Kd,macro value. A schematic of the complex is indicated in Fig. 5A, illustrating that only one of the two binding prongs is engaged. In contrast, the R155C,N387C ND1Lp97-ADP double mutant displays WT-like binding behavior (schematic in Fig. 5C), with V68 shifting to its location in the WT complex (orange free; black bound; gray WT complex). Cross-peaks for I146 and I189 in the unbound/free form of R155C,N387C ND1Lp97-ADP are much closer to the WT positions than for the R155C single mutant because, as discussed in the previous section, the double mutant is essentially in the all-down position. Binding of the adaptor does, however, cause small shifts that correspond to locking of the structure in the down conformation (compare orange and black in Fig. 5D). That a down, locked state has been achieved can be verified by recording spectra of unbound/free R155C,N387C ND1Lp97-ADP (Fig. 5E) under oxidizing conditions where the cysteines at positions 155 and 387 become cross-linked (schematic in Fig. 5E). Formation of the disulfide produces a down, locked conformation that is essentially identical to that for the adaptor bound state under reducing conditions (Fig. 5F, black peaks). We have established that the R155C,N387C ND1Lp97-ADP sample (containing the TCEP reducing agent) used to recorded the spectra in Fig. 5 C and D is not oxidized by monitoring the position of V154 that is sensitive to formation of the 155–387 disulfide. Formation of the disulfide bond leads to a significant chemical shift perturbation for the γ1-methyl of V154 that is much larger and very different from the small shift change observed for V154 upon binding of UBXD1-N (i.e., the γ1-methyl group of V154 resonates at a very different position in adaptor bound reduced and in unbound oxidized p97 states; Fig. S4).
Fig. 5.
The N387C mutation restores WT UBXD1-N binding to R155C ND1Lp97-ADP. (A–D) Selected regions of 13C-1H HMQC spectra of mutant ND1Lp97-ADP in the free form (color) and upon addition of three equivalents UBXD1-N (15) per protomer (black). The corresponding cross-peaks from spectra of WT ND1Lp97-ADP are indicated as a reference for fully bound and down, locked p97 in gray. Binding of UBXD1-N to R155C ND1Lp97-ADP is weakened (A, compare black and gray contours for V68), with no contacts of the H1/H2 motif of UBXD1-N to p97 as established by the identical shifts for I146 and I189 with and without addition of adaptor (B). R155C,N387C ND1Lp97-ADP binds UBXD1-N with WT affinity (C) and with NTDs in the down, locked conformation under reducing conditions (D). The positions of cross-peaks for I146 and I189 are as in spectra obtained of a covalent cross-linked version of R155C,N387C ND1Lp97-ADP (F) in the absence of adaptor (E). Under reducing conditions the disulfide between C155 and C387 is not formed (Fig. S4) so that the down, locked conformation that is reported in D is the result of UBXD1-N binding and not oxidation. Schematics of the structures that are formed via UBXD1-N binding or disulfide oxidation are shown above each of the spectra in A, C, and E.
Combined Fits of Chemical Shift and CPMG Data
As described in the main text, we have analyzed both chemical shift and CPMG data simultaneously, focusing on R95G ND1Lp97-ADP that has significantly larger dispersion profiles than the other disease mutants. Our goal is to use a simple model that explicitly takes into account the up/down equilibrium (see below) in a description of the NTD dynamics. One possible scheme is
| [S1] |
where D and U denote the NTD down (WT p97-ADP) and up (WT p97-ATP) states (Fig. 1A), respectively, and C is an additional state. States D and U are in fast exchange, as evidenced by the linear titration of chemical shifts as a function of disease mutant severity (Fig. 1B) (13) and a lower limit of the D–U exchange rate (2,000 s−1) could be established from the chemical shifts of pure up and pure down states that were measured in spectra of WT p97 (see main text). The combined analysis of dispersion and chemical shift data allows us to refine this estimate. Consistent with the rapid exchange between D and U (see below), we could fit our data equally well to a model where the positions of D and U are reversed in Eq. S1 above, and, of course, to a more complex triangular model.
To link states D and U to the pure down and pure up NTD conformations, respectively, ΔϖDU = ϖU – ϖD values (ppm) were equated to chemical shift differences observed experimentally in spectra of p97-ATP (U) and p97-ADP (D) based on five methyl groups (Ile189 δ1, Leu213 δ1, Leu229 δ1, Met388 ε1, and Met442 ε1), and the chemical shift values of carbons of these methyls were included in the fits (see text and below). These five methyl groups were chosen based on the following criteria: (i) they have the largest 13C chemical shift differences between the p97-ATP and p97-ADP states; (ii) they are not close in space to position 95; and (iii) they are not proximal to the terminal phosphate of ATP, so that although they are sensitive reporters of the NTD up/down equilibrium, they are not influenced directly by bound nucleotide.
Twenty-two dispersion profiles were identified as nonflat [ΔR2,eff > 1.5 s−1, where ΔR2,eff = R2,eff (νCPMG = 50 Hz) – R2,eff (νCPMG = 2,000 Hz)] and used in the three-state analysis. Methyl-TROSY–based profiles were fit as described previously (24) and exchange parameters extracted by minimization of a χ2 function (Eq. S2) that also included a term to enforce the 13C chemical shift values of the five residues indicated above,
| [S2] |
In Eq. S2, the first and second terms are fitting functions for dispersion profiles and chemical shifts, respectively; j and k denote the 22 (j) dispersion profiles and the five (k) residues that are included in the fit; Ij(νCPMG) is the intensity of a peak in the dispersion data set recorded with a CPMG frequency of νCPMG; ωk is the 13C shift (rad/s) of residue k measured in 13C-1H HMQC spectra of R95G ND1Lp97-ADP; and σCPMG, σω are errors in , as evaluated from repeat measurements, and in ωk (6 Hz = 2π*6 rad/s), respectively, based on measured positions of cross-peaks in separate samples of R95G ND1Lp97-ADP.
Values of were obtained by evaluating the time evolution of magnetization using the following set of equations:
| [S3] |
where Aj denotes the j (x, y, z) component of spin A (D,U,C) and ωA is the 13C resonance frequency of spin A with starting magnetization values of [pD, 0, pU, 0, pC, 0], where pA is the fractional population of state A. Values of each of the six elements (Dx…Cy) were evaluated over an interval of 70 ms, starting from 0, to produce numerical values for the total magnetization, M+(t) = Mx(t) + iMy(t), (i = √ − 1). M+(t) was apodized and then Fourier transformed; the position of the peak in the simulated spectrum was fitted to estimate . The parameters that were fixed/fit in our analysis (using the program ChemEx and a new fitting module that is available upon request) are summarized below:
| Parameter name | Description | Floating/fixed |
| Chemical shifts of down state | Fixed for five selected residues (13C only) and used in Eq. S3 | |
| Difference in chemical shifts between up and down states | Fixed for five selected residues (13C and 1H) and floating for others | |
| Difference in chemical shifts between states C and D | Floating | |
| Multiple quantum (MQ) relaxation rates for states D, U, C | Floating, but assumed the same for a given methyl | |
| Exchange rates and populations | Floating global parameters |
It is worth noting that 13C-1H HMQC dispersion profiles are sensitive to differences in both methyl 1H and 13C chemical shifts (24) and both of these differences are either fit or fixed as described above. Fig. 2B presents 1D surfaces for kex,DU and kex,UC from which it is clear that kex,DU > 15,000 s−1 and 1,500 s−1 ≤ kex,UC < 4,000 s−1. Values of pU = 0.41 and pC = 0.09 are obtained from fits.
As described in the main text, dispersion profiles could be well fit to a two-state model, ; however, the extracted chemical shift differences, ΔϖGE, did not correspond to ΔϖDU. Because the interconversion between states D and U is fast (>15,000 s−1; see above), it is possible to derive a simple relation between ΔϖGE and the Δϖ values that are extracted from the three-state fit by noting that the up/down exchange averaged positions of resonances in spectra are given by
| [S4] |
and thus the fitted shift difference assuming a two-state model, ΔϖGE, can be expressed as
| [S5] |
We have verified that Eq. S5 holds. An advantage of the three-state fits is that it becomes possible to quantify the lower bound for the D–U exchange rate.
Discussion
p97 plays an important role in cellular proteostasis by interacting with a variety of different adaptor molecules that target it for specific biological functions (7). Impairment of adaptor binding leads to disease, as is the case for the IBMPFD disease mutants that affect p97-UBXD1 interactions (6) and, hence, recruitment of p97 to the lysosomal degradation pathway (30). Although high-resolution X-ray images of p97 are consistent with the enzyme adopting either NTD up or down conformations, that, in turn, are a function of either bound ATP or ADP in D1 (9, 33), recent NMR studies show that the NTDs are dynamic and that disease mutations at the interface between the NTD and D1 domains lead to subtle changes in the energy landscape and significant perturbations to the up/down equilibrium (13). In the case of the IBMPFD mutants considered here, the down NTD conformation in WT p97-ADP is increasingly skewed upward as a function of disease severity and proper binding of UBXD1 becomes progressively compromised in the case of severe disease mutants (13).
The energetic differences between moderate and severe disease mutants are subtle, on the order of 1 kcal/mol, whereas the Kd for binding of the VIM domain of UBXD1-N to the NTD of p97 has been measured to be 200 μM (50 °C), corresponding to a binding free energy of 5.5 kcal/mol. Why is it then that binding of VIM cannot overcome the impaired up/down equilibrium? The answer lies in the two-pronged nature of the interaction with the macroscopic binding affinity to p97 a function of both VIM and H1/H2 contacts. Chemical shift changes that report on the binding of UBXD1-N show that the impairment in binding with disease severity can only be explained by a significant lowering of the affinity of the H1/H2 domain of UBXD1-N for the interface between the NTD/D1 domains of p97 (Fig. 5B), whereas the microscopic binding of the VIM domain to the NTD of p97 is much less affected. Thus, binding of VIM still occurs, but the second prong involving the H1/H2 domain is effectively no longer able to interact and hence stabilize the down state. Shifting of the up/down NTD equilibrium upon the addition of UBXD1-N thus serves as a proxy for two-pronged adaptor binding. A detailed description of our binding model has been published (13).
The results of the present study make it clear that p97 is highly dynamic with motion on a range of timescales that extend from the millisecond processes characterized here using relaxation dispersion NMR spectroscopy (Fig. 2) to the faster up/down equilibrium for which a lower bound of 15,000 s−1 is obtained. Additional processes that are beyond the CPMG window are also detected via elevated relaxation rates. Notably, the dynamics localize to regions at interfaces between domains, including predominantly between the NTD and D1 domains. The plasticity of the p97 molecule, coupled with the subtle changes in energies between weak and strong disease mutants, suggested to us that it might be possible to manipulate the up/down equilibrium by small changes to structure at key sites; if possible, this would constitute a first demonstration of the feasibility of shifting the aberrant NTD equilibrium in disease mutants to that of the WT state.
A pair of sites was identified by inspection of the X-ray structure of p97-ATPγS where stabilizing features of the up state could be removed via mutagenesis that would be predicted to shift the aberrant R95G ND1Lp97-ADP equilibrium to the down conformation. Shifts from pU = 0.42 (R95G ND1Lp97-ADP) to ∼0.3 [R95G and an additional mutation (Fig. 3A), i or ii] were obtained that involved destabilization of either the amino terminal helix of the NTD or an electrostatic interaction, both of which were present in the p97-ATP structure but not in p97-ADP. Having established that the p97 energy landscape is malleable, we next tried to stabilize the down state by mutating position 387 that is proximal to the site of the strong disease mutations R155P and R155C in the p97-ADP structure. Notably, the NTDs of R155C ND1Lp97-ADP are restored effectively to their WT positions (pU = 0.06) via the N387C substitution. Importantly, the functional properties of R155C,N387C ND1Lp97-ADP are also restored, at least as measured by UBXD1 binding.
The present study provides a proof of principle that severe p97 disease mutants can be moderated, or in the case of R155C ND1Lp97-ADP rescued, by exploiting the inherent plasticity of this protein. Although a number of hotspots have been identified here (summarized in Table S2) that may be of interest for further studies, it seems likely that additional sites for manipulation will be discovered by focusing on allosteric pathways that have been identified in a previous NMR study (13). Notably, it may be possible to work with multiple sites simultaneously to better control changes to the free-energy landscape. In this regard, as discussed above, the combination of the two disease mutants, T262A and R95G, that both individually increase pU in the p97-ADP state, collectively lead to an even larger pU for the T262A&R95G double mutant (pU = 0.79), whereas a similar situation is observed for the combination of two weak disease mutants (R155H and N387H) that together give pU = 0.45. Manipulating pairs of sites, where each change decreases pU, may be a useful strategy for achieving a more complete down conformation for disease mutants of p97-ADP. Our present work has focused on mutational studies. As a next step, the identification of drug molecules that shift the NTD equilibrium in desired ways would constitute a potential route to restoring proper function to IBMPFD disease mutants of p97.
Table S2.
Point mutations that perturb the up/down equilibrium in ND1Lp97-ADP
| Mutation* | Mutation to | Class† | Background** | pU (from background)‡ | Observations |
| ΔN20 | Deletion of residues 1–20 | A | R95G ND1Lp97-ADP | 0.31 (0.42) | |
| T14 | P | A | R95G ND1Lp97-ADP | 0.31 (0.42) | I16 uncoupled from CPMG dynamics |
| A15 | P | A | R95G ND1Lp97-ADP | 0.31 (0.42) | I16 uncoupled from CPMG dynamics |
| K63 | A,E | A | WT ND1Lp97-ADP | 0 (0) | |
| A | A | R95G ND1Lp97-ADP | 0.33 (0.42) | ||
| R95 | G | D | WT ND1Lp97-ADP | 0.42 (0) | |
| R155 | H | D | WT ND1Lp97-ADP | 0.14 (0) | |
| C | D | WT ND1Lp97-ADP | 0.45 (0) | ||
| P | D | WT ND1Lp97-ADP | 0.51 (0) | ||
| R191 | Q | D | WT ND1Lp97-ADP | 0.39 (0) | |
| E194 | P | A | WT ND1Lp97-ADP | 0 (0) | NTD/D1 linker helix broken but NTD equilibrium not shifted as evident from other residues (e.g., I146, M388, M442) |
| P | A | R95G ND1Lp97-ADP | 0.42 (0.42) | ||
| P | A | WT ND1Lp97-ATPgS | 1.0 (1.0) | ||
| L198 | W | D | WT ND1Lp97-ADP | 0.27 (0) | |
| A232 | E | D | WT ND1Lp97-ADP | 0.18 (0) | |
| N387 | C | A | WT ND1Lp97-ADP | 0 (0) | |
| C | A | R95G ND1Lp97-ADP | 0.42 (0.42) | ||
| C | A | R155C ND1Lp97-ADP | 0.06 (0.45) | Under reducing conditions | |
| C | A | R155C ND1Lp97-ADP | 0.00 (0.45) | Under oxidizing condition, cross-linked | |
| C | A | R155P ND1Lp97-ADP | 0.31 (0.51) | ||
| H | D | WT ND1Lp97-ADP | 0.14 |
Data from our previous study (13) and the present work were compiled.
pU values for the construct listed under Background are given in parentheses.
Mutations are classified as artificial (A) or naturally occurring, disease causing (D).
The population of the up state is given, pU, as determined from the position of the I189 correlation (except for E194P mutants; see text).
Materials and Methods
Cloning, Protein Expression, Purification, and NMR Sample Preparation.
p97 protein from Mus musculus (identical to Homo sapiens) with an N-terminal 6xHis tag and TEV cleavage site was expressed in Escherichia coli BL21(DE3) cells in minimal M9 D2O media supplemented with 15NH4Cl and [2H,12C]-glucose as the only nitrogen and carbon sources (Cambridge Isotope Laboratories) and purified as described before (13). Point mutations and the N-terminal deletion, ΔN20, were introduced using QuikChange site-directed mutagenesis (Agilent). Disulfide cross-linking, connecting positions 155 and 387 that were first mutated to cysteine, was accomplished as detailed previously (13). Selective methyl labeling with Iδ1-[13CH3],V/L-γ1/δ1(proR)-[13CH3,12CD3] and M-ε1[13CH3] was achieved by the addition of appropriate precursors (Cambridge Isotope Laboratories), as detailed previously (19, 22, 34). The UBXD1-N fragment (15) from Mus musculus with an N-terminal 6xHis tag and tobacco etch virus protease (TEV) cleavage site was expressed and purified as described (13). NMR samples were buffer-exchanged into 25 mM Hepes, pH 7.0, 50 mM NaCl, 10 mM ADP, 5 mM TCEP in 100% D2O at protein concentrations between 50 μM (protomer concentration; for HMQC spectra), and 600 μM (relaxation dispersion experiments). Monomer concentrations in UBXD1-N binding experiments were perdeuterated UBXD1-N 150 μM, wt/R155C/R155C,N387C ND1Lp97-ADP 50 μM.
Data Acquisition and Analysis.
NMR experiments were performed using either Varian or Bruker 18.8 T spectrometers, 50 °C, the latter equipped with a cryogenically cooled probe. Methyl-TROSY–based CPMG data sets (24) were acquired as pseudo 3D experiments with a constant-time CPMG evolution period (35) of 20 ms, 14 νCPMG values [νCPMG = 1/(2δ), where δ is the separation between successive 180° refocusing pulses] extending from 50 to 2,000 Hz, with two duplicates for error analysis and a recycle delay of 2.5 s. The total acquisition time was 68 h for each experiment. A reference spectrum was recorded with the CPMG evolution period removed (24). Peak fitting was performed using FuDA (www.biochem.ucl.ac.uk/hansen/fuda/) and the resulting dispersion profiles analyzed using ChemEx software. CPMG datasets were collected on samples of WT ND1Lp97-ADP, coplanar R95G ND1Lp97-ADP, and mutants of ND1Lp97-ADP that included the following substitutions: R155H, R95G, R95G&T14P, and R95G&T262A. Details of the fitting procedure are given in Combined Fits of Chemical Shift and CPMG Data. All spectra were processed in NMRPipe (36) and analyzed with the CcpNmr (37) program with additional data analysis (i.e., excluding CPMG dispersions) achieved using home-written MATLAB scripts.
Acknowledgments
Funding for this work was provided by Swiss National Science Foundation Grant P2EZP2_148754 (to A.K.S.); European Molecular Biology Organization Grant ALTF 100-2013; Leukemia and Lymphoma Society Grant 3367-17; and the Canadian Institutes of Health Research. L.E.K. holds a Canada Research Chair in Biochemistry.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707974114/-/DCSupplemental.
References
- 1.Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol. 2012;14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- 2.Rabinovich E, Kerem A, Fröhlich KU, Diamant N, Bar-Nun S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol. 2002;22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rabouille C, Levine TP, Peters JM, Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell. 1995;82:905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- 4.Cao K, Nakajima R, Meyer HH, Zheng Y. The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell. 2003;115:355–367. doi: 10.1016/s0092-8674(03)00815-8. [DOI] [PubMed] [Google Scholar]
- 5.Richly H, et al. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 6.Ritz D, et al. Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nat Cell Biol. 2011;13:1116–1123. doi: 10.1038/ncb2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchberger A, Schindelin H, Hänzelmann P. Control of p97 function by cofactor binding. FEBS Lett. 2015;589:2578–2589. doi: 10.1016/j.febslet.2015.08.028. [DOI] [PubMed] [Google Scholar]
- 8.DeLaBarre B, Brunger AT. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat Struct Biol. 2003;10:856–863. doi: 10.1038/nsb972. [DOI] [PubMed] [Google Scholar]
- 9.Tang WK, et al. A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants. EMBO J. 2010;29:2217–2229. doi: 10.1038/emboj.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banerjee S, et al. 2.3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science. 2016;351:871–875. doi: 10.1126/science.aad7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watts GD, et al. Inclusion body myopathy–Paget bone disease–frontotemporal dementia syndrome caused by mutated valosin containing protein. Nat Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 12. Evangelista T, Weihl CC, Kimonis V, Lochmüller H, VCP Related Diseases Consortium (2016) 215th ENMC International Workshop VCP-related multi-system proteinopathy (IBMPFD) 13–15 November 2015, Heemskerk, The Netherlands. Neuromuscul Disord 26(8):535–547. [DOI] [PMC free article] [PubMed]
- 13.Schuetz AK, Kay LE. A dynamic molecular basis for malfunction in disease mutants of p97/VCP. eLife. 2016;5:e20143. doi: 10.7554/eLife.20143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang WK, Xia D. Role of the D1-D2 linker of human VCP/p97 in the asymmetry and ATPase activity of the D1-domain. Sci Rep. 2016;6:20037. doi: 10.1038/srep20037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trusch F, et al. The N-terminal region of the ubiquitin regulatory X (UBX) domain-containing protein 1 (UBXD1) modulates interdomain communication within the valosin-containing protein p97. J Biol Chem. 2015;290:29414–29427. doi: 10.1074/jbc.M115.680686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tresse E, et al. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy. 2010;6:217–227. doi: 10.4161/auto.6.2.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer H, Weihl CC. The VCP/p97 system at a glance: Connecting cellular function to disease pathogenesis. J Cell Sci. 2014;127:3877–3883. doi: 10.1242/jcs.093831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson AE, Shu H, Hauswirth AG, Tong A, Davis GW. VCP-dependent muscle degeneration is linked to defects in a dynamic tubular lysosomal network in vivo. eLife. 2015;4:e07366. doi: 10.7554/eLife.07366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gans P, et al. Stereospecific isotopic labeling of methyl groups for NMR spectroscopic studies of high-molecular-weight proteins. Angew Chem Int Ed Engl. 2010;49:1958–1962. doi: 10.1002/anie.200905660. [DOI] [PubMed] [Google Scholar]
- 20.Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE. Cross-correlated relaxation enhanced 1H[bond]13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J Am Chem Soc. 2003;125:10420–10428. doi: 10.1021/ja030153x. [DOI] [PubMed] [Google Scholar]
- 21.Tugarinov V, Kay LE. An isotope labeling strategy for methyl TROSY spectroscopy. J Biomol NMR. 2004;28:165–172. doi: 10.1023/B:JNMR.0000013824.93994.1f. [DOI] [PubMed] [Google Scholar]
- 22.Gelis I, et al. Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell. 2007;131:756–769. doi: 10.1016/j.cell.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niwa H, et al. The role of the N-domain in the ATPase activity of the mammalian AAA ATPase p97/VCP. J Biol Chem. 2012;287:8561–8570. doi: 10.1074/jbc.M111.302778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korzhnev DM, Kloiber K, Kanelis V, Tugarinov V, Kay LE. Probing slow dynamics in high molecular weight proteins by methyl-TROSY NMR spectroscopy: Application to a 723-residue enzyme. J Am Chem Soc. 2004;126:3964–3973. doi: 10.1021/ja039587i. [DOI] [PubMed] [Google Scholar]
- 25.Palmer AG, 3rd, Kroenke CD, Loria JP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 26.Chao F-A, Byrd RA. Application of geometric approximation to the CPMG experiment: Two- and three-site exchange. J Magn Reson. 2017;277:8–14. doi: 10.1016/j.jmr.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fersht AR. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. Freeman; New York: 1998. [Google Scholar]
- 28.Kellogg EH, Leaver-Fay A, Baker D. Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins. 2011;79:830–838. doi: 10.1002/prot.22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang WK, Xia D. Altered intersubunit communication is the molecular basis for functional defects of pathogenic p97 mutants. J Biol Chem. 2013;288:36624–36635. doi: 10.1074/jbc.M113.488924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bug M, Meyer H. Expanding into new markets–VCP/p97 in endocytosis and autophagy. J Struct Biol. 2012;179:78–82. doi: 10.1016/j.jsb.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Hänzelmann P, Schindelin H. The structural and functional basis of the p97/valosin-containing protein (VCP)-interacting motif (VIM): Mutually exclusive binding of cofactors to the N-terminal domain of p97. J Biol Chem. 2011;286:38679–38690. doi: 10.1074/jbc.M111.274506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stapf C, Cartwright E, Bycroft M, Hofmann K, Buchberger A. The general definition of the p97/valosin-containing protein (VCP)-interacting motif (VIM) delineates a new family of p97 cofactors. J Biol Chem. 2011;286:38670–38678. doi: 10.1074/jbc.M111.274472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, et al. Structure of the AAA ATPase p97. Mol Cell. 2000;6:1473–1484. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 34.Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat Protoc. 2006;1:749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- 35.Mulder FA, Skrynnikov NR, Hon B, Dahlquist FW, Kay LE. Measurement of slow (micros-ms) time scale dynamics in protein side chains by (15)N relaxation dispersion NMR spectroscopy: Application to Asn and Gln residues in a cavity mutant of T4 lysozyme. J Am Chem Soc. 2001;123:967–975. doi: 10.1021/ja003447g. [DOI] [PubMed] [Google Scholar]
- 36.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 37.Vranken WF, et al. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 38.Rohrer JD, et al. A novel exon 2 I27V VCP variant is associated with dissimilar clinical syndromes. J Neurol. 2011;258:1494–1496. doi: 10.1007/s00415-011-5966-4. [DOI] [PMC free article] [PubMed] [Google Scholar]