Significance
Elevated leaf vein density is a key step in the evolution from C3 to C4 plants. We hypothesized that high vein density in C4 leaves is due to elevated auxin biosynthesis and transport in developing leaves. We found higher expression levels of genes promoting auxin biosynthesis and higher auxin content in developing C4 leaves than in developing C3 leaves. We also found higher auxin content and vein density in loss-of-function mutants of MYC2, an auxin biosynthesis suppressor. Moreover, treatment with an auxin biosynthesis or transport inhibitor reduced vein density in new leaves. Finally, mutations that reduce auxin efflux or influx reduce vein density. These observations support our hypothesis and provide a molecular basis for high vein density in C4 leaves.
Keywords: vein density, auxin biosynthesis, auxin transport, C4 plants
Abstract
High vein density, a distinctive trait of C4 leaves, is central to both C3-to-C4 evolution and conversion of C3 to C4-like crops. We tested the hypothesis that high vein density in C4 leaves is due to elevated auxin biosynthesis and transport in developing leaves. Up-regulation of genes in auxin biosynthesis pathways and higher auxin content were found in developing C4 leaves compared with developing C3 leaves. The same observation held for maize foliar (C4) and husk (C3) leaf primordia. Moreover, auxin content and vein density were increased in loss-of-function mutants of Arabidopsis MYC2, a suppressor of auxin biosynthesis. Treatment with an auxin biosynthesis inhibitor or an auxin transport inhibitor led to much fewer veins in new leaves. Finally, both Arabidopsis thaliana auxin efflux transporter pin1 and influx transporter lax2 mutants showed reduced vein numbers. Thus, development of high leaf vein density requires elevated auxin biosynthesis and transport.
Photosynthesis efficiency is higher in C4 than in C3 leaves mainly due to the presence of Kranz anatomy and high vein density, which are central to both C3–C4 evolution (1–5) and conversion of C3 to C4-like crops (6). High vein density may confer an adaptive advantage in arid or/and high light environments (5, 7, 8). The importance of auxin in leaf vein patterning is well documented (9–14). The auxin canalization model postulates that, in developing leaves, auxin is transported to precursor cells to initiate development of veins (9, 15). This model was supported by the observations that PIN-FORMED1 (PIN1) directs auxin to form local auxin maxima in procambial cells to initiate vein development (10, 13). Moreover, exogenous application of auxin led to dramatic expansion of PIN1 expression in the ground meristem, leading to formation of additional veins (9, 15). However, the molecular basis for high vein density in C4 leaves is not well understood.
We hypothesized that high vein density in C4 leaves is due to elevated auxin biosynthesis and transport. To test this hypothesis, we conducted the following studies. First, we compared transcriptomes of developing leaves of C3 Tarenaya hassleriana and C4 Gynandropsis gynandra in Cleomaceae to see whether genes promoting auxin biosynthesis tend to be up-regulated in G. gynandra, while the basic helix–loop–helix transcription factor MYC2, which is a suppressor of auxin biosynthesis, is down-regulated. For the same purpose, we compared the transcriptomes of maize foliar (Kranz, high vein density) and husk (non-Kranz, low vein density) leaf primordia. Second, we examined whether the auxin content is higher in G. gynandra developing leaves and in maize foliar leaf primordia. Third, we examined the vein density in two myc2 mutant lines of Arabidopsis thaliana to see whether they showed increased vein density, because, as mentioned above, MYC2 is a suppressor of auxin biosynthesis. Fourth, we treated G. gynandra developing leaves with an auxin biosynthesis inhibitor to see whether it reduces vein density. Fifth, we treated the developing leaves of both species with an auxin transport inhibitor to see its effect on vein density. Sixth, we studied the A. thaliana auxin efflux transporter pin1 mutant and auxin influx transporter lax2 mutant to see whether they both showed reduced vein numbers. The results of these studies all supported our hypothesis and have implications for the C4 rice project, which is to transform C3 rice to a C4 crop (16, 17).
Results
Up-Regulation of Genes in Auxin Biosynthesis Pathways in Developing C4 Leaves.
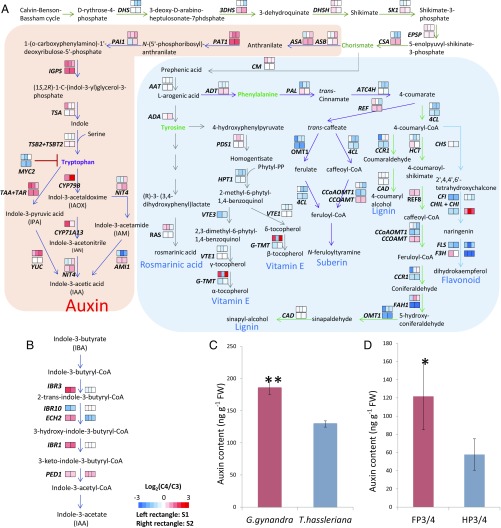
We obtained leaf transcriptomes of C3 Tarenaya hassleriana and C4 Gynandropsis gynandra, both in Cleomaceae (Materials and Methods). Because veins develop early during leaf development (18), we chose two early developmental stages, stage 1 (S1) (0.5–0.8 mm) and stage 2 (S2) (∼2 mm), of young fifth leaves that may better pinpoint key transcriptional events in vein density control than an earlier study using mature leaves (19). At both S1 and S2 stages, genes in the auxin biosynthesis pathways tend to have a higher transcript level in C4 G. gynandra than in C3 T. hassleriana (Fig. 1). In the tryptophan (Trp)-dependent auxin biosynthesis pathway, no significant expression differences between the two species was found for the six genes in the pathway before the biosynthesis of chorismate, a key precursor of auxin and several other essential metabolites (Fig. 1A). In contrast, more than one-half of the genes in the subpathway from chorismate to auxin showed higher expression levels in G. gynandra. Similarly, four out of the five genes in the Trp-independent auxin biosynthesis pathway showed a higher expression level in G. gynandra (Fig. 1B). Note that none of the genes involved in the biosynthesis of nonauxin metabolites from chorismate showed a significantly higher expression level in G. gynandra (Fig. 1A), indicating specific up-regulation of auxin biosynthesis genes in C4 G. gynandra. This finding is further supported by the 1.3-fold higher auxin content in young fifth leaves (∼3 mm) of G. gynandra compared with T. hassleriana (P < 0.001; Fig. 1C).
Fig. 1.
Expression ratios of genes involved in auxin biosynthesis and differences in auxin content between C4 G. gynandra and C3 T. hassleriana developing leaves or between maize foliar and husk leaf primordia. (A) The Trp-dependent auxin (shaded in pink) and other secondary metabolite (shaded in light blue) biosynthesis pathways, including key intermediates and enzymes. (B) The Trp-independent auxin biosynthesis pathway. (A and B) The Trp-dependent and -independent auxin biosynthesis pathways were based on the pathways in A. thaliana (33–36). We could not find maize genes that are clearly orthologous to CYP79B and CYP71A13 in A. thaliana. In A and B, each enzymatic reaction is represented by an arrow. In each step, the two colored rectangles indicate the fold differences (log2 ratios) of gene expression between G. gynandra and T. hassleriana at the S1 (left rectangle) and S2 (right rectangle) developmental stages, respectively. The three colored rectangles indicate the fold differences of gene expression between maize foliar (FP) and husk leaf (HP) primordia during the midvein initiated stage (Left), midvein lateral initiated stage (Middle), and the midvein lateral and intermediate initiated stage (Right), respectively. (For definitions of developmental stages of maize foliar and husk leaf primordia, see ref. 20.) Colors represent log2 ratios of gene expression levels according to the color bar at the Bottom of the figure. The G. gynandra and T. hassleriana gene RPKM values are from Dataset S3, and the maize data are from Wang et al. (20). (C) The auxin contents in developing leaves of G. gynandra and T. hassleriana. The fifth young developing leaves (∼3 mm long) were selected to analyze auxin content (Dataset S4). (D) The auxin contents in maize foliar and husk leaf primordia. The maize foliar (FP3/4) and husk (HP3/4) leaf primordia (∼2 mm long) were selected to analyze auxin contents. (C and D) Error bar indicates the SD of the mean auxin contents (nanograms per gram fresh weight) in three biological replicates (Dataset S4). FW, fresh weight. *P < 0.05 and **P < 0.001 (Student’s one-tailed t test).
To assess whether this specific up-regulation of Trp-dependent auxin biosynthesis also occurs in other independently evolved C4 species, we examined transcriptome data of maize foliar (Kranz, high vein density) and husk (non-Kranz, low vein density) leaf primordia (20). Similar to our comparison between G. gynandra and T. hassleriana, more than one-half of the genes in the subpathway from chorismate to auxin biosynthesis were up-regulated in foliar leaf primordia (Fig. 1A). Moreover, we found higher auxin content in developing foliar leaf primordia than in developing husk leaf primordia (P < 0.02; Fig. 1D). Thus, the up-regulation of the Trp-dependent auxin biosynthesis pathway is likely a common feature among C4 plants.
Elevated Auxin Content and Vein Density in T-DNA Insertion myc2 Mutants.
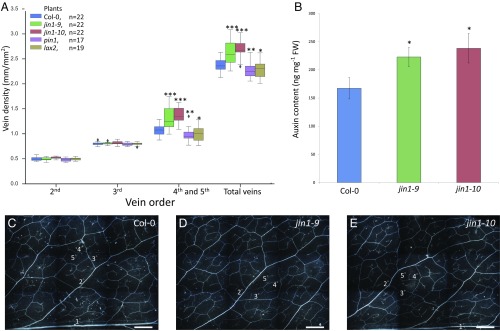
MYC2, a basic helix–loop–helix family transcription factor, was significantly down-regulated in G. gynandra developing leaves (Fig. 1A). Importantly, MYC2 negatively regulates the biosynthesis of Trp, a key substrate for auxin biosynthesis (21). To assess MYC2’s role in auxin biosynthesis and vein density control, we compared the auxin content and vein density in two A. thaliana T-DNA insertion myc2 mutants, jin1-9 (SALK_017005) and jin1-10 (SALK_083483) (22), to those of the wild-type plants. The vein densities in the two myc2 mutants were comparable, but both were significantly higher than that in the wild-type plants (P < 0.0005), mainly due to the increases in the 4° (fourth order) and 5° veins (P < 0.0005) (Fig. 2A). In addition, the auxin contents in the developing leaves of jin1-10 and jin1-9 were 40% and 30% higher than that in the wild-type control, respectively (Fig. 2B), similar to the leaf auxin content ratio between G. gynandra and T. hassleriana (Fig. 1C). Thus, a modest increase in auxin content in developing leaves may significantly increase vein density.
Fig. 2.
Leaf vein densities in A. thaliana wild type (Col-0), jin1-9 and jin1-10 homozygotes, pin1 heterozygotes, and lax2 homozygotes, and auxin contents in developing leaves of the wild-type and jin1-9 and jin1-10 homozygotes. (A) The box plots show the vein densities (in millimeters per square millimeter) in the seventh mature leaves (∼2 cm long) (n = the number of biological replicates; Dataset S5). The statistical significance of the difference between the wild type and a mutant was determined by Student's one-tailed t test (*P < 0.05, **P < 0.01, and ***P < 0.0005). (B) Auxin contents in the seventh developing leaves (∼3 mm long). Error bar: SD of the auxin contents (nanograms per gram fresh weight) in three biological replicates (Dataset S4). (C–E) Leaf vein patterns and orders of Col-0, jin1-9, and jin1-10. (Scale bar: 100 mm.) Wild type: Col-0; jin1-9: SALK_017705; jin1-10: SALK_083483; pin1: SALK_047613; and lax2: SAIL_178_C02.
Reduced Leaf Vein Density in Yucasin-Treated Plants.
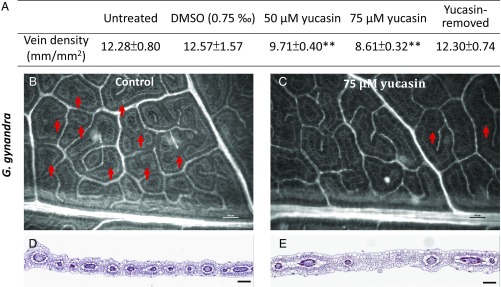
To investigate whether the leaf vein density in a C4 plant could be reduced by a reduction in auxin content, G. gynandra plants were treated with yucasin [5-(4-chlorophenyl)-4H-1,2,4-triazole-3-thiol]. Yucasin is an inhibitor of the YUCCA (YUC) enzyme that catalyzes the final step of the Trp-dependent auxin biosynthesis pathway (Fig. 1A), and its application reduces the complexity of A. thaliana vascular systems (23). We found a yucasin concentration-dependent reduction in the vein density in newly developed leaves of yucasin-treated G. gynandra (Fig. 3A), mainly due to reduced numbers of higher-order veins (Fig. 3 B–E). Note that, after removal of the yucasin treatment, the newly developed leaves exhibited a vein density comparable to that of the control (Fig. 3A). In conclusion, despite the existence of two other subpathways from Trp to auxin (Fig. 1A), blocking the YUC subpathway alone is sufficient to reduce vein density.
Fig. 3.
Effects of yucasin, an auxin biosynthesis inhibitor, on leaf vein development in G. gynandra. Plants were grown in half-strength Kimura B nutrient solution with or without yucasin. (A) Vein densities in untreated, DMSO-treated, and yucasin-treated leaves. DMSO was used to dissolve yucasin. Each vein density is given in mean value ± SD (four biological replicates; Dataset S6). **P < 0.001, Student’s t test. (B) Vein pattern of the fifth newly developed leaf (∼2.5 cm long) from a plant grown in a control (untreated) solution. (C) Vein pattern of the fifth newly developed leaf (∼2.5 cm long) from a plant grown in a solution containing 75 μM yucasin. (B and C) Red arrows indicate the highest-order veins. (D and E) Cross-sections of the fifth leaf (∼2.5 cm long) grown in a control solution (D) or in a solution containing 75 μM yucasin (E) were stained by toluidine blue stain. Yucasin reduced the number of higher-order veins. (Scale bar: 100 μm.)
Reduced Leaf Vein Density in 1-N-Naphthylphthalamic Acid-Treated Plants.
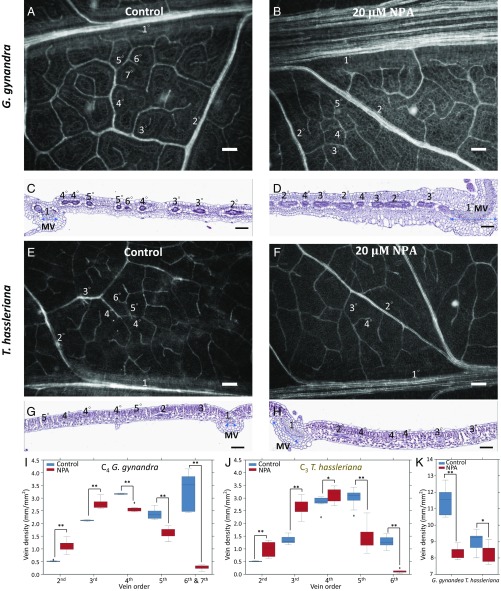
To see whether decreased auxin transport reduces vein density, we applied 1-N-naphthylphthalamic acid (NPA), an inhibitor of auxin efflux (24), to G. gynandra and T. hassleriana plants. Previous studies showed that A. thaliana treated with NPA during early leaf development displayed larger mid (1°) veins and an increased number of 2° veins (11, 12). This is because NPA reduces auxin transport, so that auxin accumulates in and near auxin sources, increasing both the size of mid veins and the formation of 2° veins (12, 15). Consistent with these studies in A. thaliana, NPA-treated G. gynandra and T. hassleriana showed larger 1° veins and more 2° veins than their controls (Fig. 4 A vs. B, E vs. F, and I and J). NPA-treated G. gynandra plants showed dramatically larger 1° and 2° veins than NPA-treated T. hassleriana plants (Fig. 4 B and D vs. F and H), likely due to the higher auxin content in developing leaves in C4 G. gynandra than in C3 T. hassleriana (Fig. 1C). Also, the NPA-dependent reduction in vein density was much stronger in C4 G. gynandra than in C3 T. hassleriana (Fig. 4 I–K). In conclusion, inhibition of auxin efflux leads to higher levels of local auxin accumulation and thus also more and thicker 2° veins but fewer higher-order veins because auxin is not efficiently transported to procambial cells that are distant from auxin sources.
Fig. 4.
Effects of NPA, an auxin transport inhibitor, on leaf vascular development and vein densities in G. gynandra and T. hassleriana. (A–D) G. gynandra and (E–H) T. hassleriana plants were grown in half-strength Kimura B nutrient solution without (A, C, E, and G) or with 20 μM NPA (B, D, F, and H). Vein patterns and cross-sections of the fifth newly developed leaves (2.5–3 cm long) are shown. (Scale bar: 100 μm.) (C, D, G, and H) MV: midvein (double arrows). (I and J) Vein density differences between control and NPA-treated leaves of G. gynandra (I) and T. hassleriana (J) (eight biological replicates each; Dataset S7). (K) The total vein density in the fifth mature leaves (eight biological replicates each). The statistical significance was determined by Student’s one-tailed t test (*P < 0.05; **P < 0.0005).
Dosage Effect of Auxin Efflux Transporter PIN1 on Vein Density.
To pursue the above issue further, we considered the dosage effect of PIN1, a major auxin efflux carrier, on vein density. It is known that A. thaliana pin1 homozygous mutant showed thicker 1°, 2°, and 3° veins but fewer higher-order veins (14), similar to the effect of NPA treatment. We found that even heterozygous A. thaliana pin1 (SALK_047613) plants have fewer 4° and higher-order veins than the wild type (Fig. 2A), demonstrating the importance of PIN1 dosage. In addition, in C4 G. gynandra, there are two PIN1 genes and in developing leaves the total expression level of the two PIN1 genes is nearly twice that of the single PIN1 gene in C3 T. hassleriana (Table 1). In maize, three of the four PIN1 genes are expressed at a much higher level in foliar leaf primordia than in husk leaf primordia (Table 1). These observations indicate the need for elevated auxin efflux in developing C4 leaves.
Table 1.
Transcript RPKMs of genes involved in auxin transport in G. gynandra and T. hassleriana, and in maize foliar and husk leaf primordia
| Gene | A. thaliana locus | G. gynandra (Ggy) vs. T. hassleriana (Tha)* | Maize foliar leaf primordia (FP) vs. husk leaf primordial (HP)† | ||||||||
| Ggy S1 | Tha S1 | Ggy S2 | Tha S2 | FP | HP | FP3/4 | HP3/4 | FP5 | HP5 | ||
| Auxin efflux‡ | |||||||||||
| PIN1a | AT1G73590 | 52.35 | 32.65 | 52.33 | 33.92 | 2,853.50 | 1,233.50 | 1,648.50 | 351.50 | 639.50 | 275.00 |
| PIN1b | 32.51 | 33.46 | 1,867.00 | 723.00 | 1,760.00 | 281.00 | 1,034.50 | 526.50 | |||
| PIN1c | 375.50 | 145.50 | 257.00 | 225.50 | 152.50 | 864.50 | |||||
| PIN1d | 116.00 | 16.00 | 77.00 | 2.00 | 7.50 | 2.00 | |||||
| Total | 84.86 | 32.65 | 85.79 | 33.92 | 5,212.00 | 2,118.00 | 3,742.50 | 860.00 | 1,834.00 | 1,668.00 | |
| PIN2 | AT5G57090 | 11.11 | 0.01 | 3.03 | 0.02 | ||||||
| PIN3a | AT1G70940 | 14.71 | 38.73 | 8.99 | 55.05 | 6.50 | 54.00 | 7.50 | 291.50 | 32.00 | 308.50 |
| PIN3b | 0 | 0 | 0 | 0.50 | 0.50 | 0 | |||||
| Total | 6.50 | 54.00 | 7.50 | 292.00 | 32.50 | 308.50 | |||||
| PIN4a | AT2G01420 | 38.35 | 37.39 | 67.36 | 63.11 | ||||||
| PIN4b | 9.91 | 8.17 | |||||||||
| Total | 38.35 | 47.30 | 67.36 | 71.28 | |||||||
| PIN5a | AT5G16530 | 1.48 | 2.81 | 0.99 | 2.90 | 0 | 0 | 0 | 0 | 0.50 | 0.50 |
| PIN5b | 0 | 0 | 0 | 0 | 2.50 | 4.00 | |||||
| PIN5c | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Total | 0 | 0 | 0 | 0 | 3.00 | 4.50 | |||||
| PIN6 | AT1G77110 | 87.52 | 63.54 | 60.70 | 55.10 | ||||||
| PIN8 | AT5G15100 | 2.02 | 0.37 | 1.02 | 0.24 | 114.50 | 86.00 | 30.00 | 17.00 | 17.00 | 6.50 |
| Auxin influx§ | |||||||||||
| AUX1a | AT2G38120 | 43.92 | 28.87 | 33.97 | 24.00 | 722.50 | 1,749.00 | 172.50 | 3,771.00 | 1,749.00 | 3,851.50 |
| AUX1b | 43.04 | 15.65 | 42.58 | 21.67 | 99.50 | 19.50 | 35.50 | 8.50 | 19.50 | 8.00 | |
| AUX1c | 4.58 | 4.94 | 3.34 | 6.15 | |||||||
| AUX1d | 1.59 | 1.92 | |||||||||
| Total | 91.54 | 51.05 | 79.89 | 53.74 | 822.00 | 1,768.50 | 208.00 | 3,779.50 | 1,768.50 | 3,859.50 | |
| LAX1a | AT5G01240 | 21.95 | 28.94 | 37.03 | 29.74 | ||||||
| LAX1b | 3.68 | 21.03 | 2.13 | 32.60 | |||||||
| Total | 25.63 | 49.97 | 39.16 | 62.34 | |||||||
| LAX2a | AT2G21050 | 170.41 | 63.48 | 186.75 | 114.22 | 5,483.00 | 1,764.00 | 4,751.50 | 632.00 | 1,764.00 | 448.50 |
| LAX2b | 297.00 | 107.50 | 1,085.00 | 257.50 | 107.50 | 503.00 | |||||
| Total | 5,780.00 | 1,871.50 | 5,836.50 | 889.50 | 1,871.50 | 951.50 | |||||
| LAX3 | AT1G77690 | 14.37 | 15.88 | 8.01 | 14.85 | 3.50 | 8.50 | 16.50 | 8.00 | 8.50 | 86.00 |
Two developmental stages, S1 (0.5–0.8 mm) and S2 (∼2 mm), of young fifth leaves were examined.
FP and HP: the midvein initiated; FP3/4 and HP3/4: midvein laterals initiated; and FP5 and HP5: midvein laterals and intermediates initiated. Data are from ref. 20.
PIN1, PIN-FORMED 1.
AUX1, AUXIN RESISTANT 1; and LAX, LIKE AUXIN RESISTANT.
Importance of Auxin Influx in Vein Density Control.
A. thaliana LAX2 encodes an auxin influx transporter. We found that LAX2 was expressed at a higher level in C4 G. gynandra than in C3 T. hassleriana (Table 1). In maize, there are two LAX2 genes and both tend to be expressed at a much higher level in foliar leaf primordia (high vein density) than in husk leaf primordia (low vein density) (Table 1). To further assess the role of LAX2 in vein density control, we examined A. thaliana lax2 mutant (SAIL_178_C02) and found that the mutant had fewer 4° and higher-order veins than the wild type (Fig. 2A). The reduction was not dramatic, likely because there are other auxin influx genes in A. thaliana (Table 1). Nonetheless, the effect is significant, indicating that decreased auxin influx can also reduce the number of higher-order veins.
Discussion
The above observations provide a molecular basis for high vein density in C4 leaves. C4 leaves have higher vein density than in C3 leaves because developing C4 leaves have higher auxin content, owing to elevated expressions of genes promoting auxin biosynthesis and reduced expressions of negative regulators such as MYC2 (Fig. 1). The observations that exogenous application of auxin led to formation of additional leaf veins (9, 15) and that the two myc2 mutants of A. thaliana studied showed both elevated auxin content in developing leaves and higher leaf vein densities suggest that increasing auxin biosynthesis alone is sufficient to increase vein density. However, the increase would be limited if auxin transport is not coordinately enhanced in C4 leaves relative to C3 leaves. This reasoning is based on four observations. First, in developing leaves, the total expression level of the two PIN1 auxin efflux carrier genes in C4 G. gynandra (high vein density) was twice that of the single PIN1 gene in C3 T. hassleriana (low vein density), and the difference in the total PIN1 gene expression was even more conspicuous between maize foliar (high vein density) and husk (low vein density) leaf primordia (Table 1). Second, a similar conclusion applies to the auxin influx carrier LAX2 expression levels (Table 1). Third, both A. thaliana pin1 and lax2 null mutants have reduced leaf vein density, suggesting the importance of not only auxin efflux (PIN1) but also influx (LAX2) in the formation of higher-order vein. Fourth, new leaves in NPA-treated G. gynandra showed a vein density even lower than that in the wild-type T. hassleriana (Fig. 4K), despite the fact that G. gynandra has a higher auxin content in developing leaves than T. hassleriana (Fig. 1C). This observation suggests that, without a sufficiently high level of auxin transport, few higher-order veins can develop in a leaf, even if its auxin content is high. As the brassinosteroid (BR) signaling pathway interacts with the auxin signaling pathway (25), mutations in the BR biosynthesis pathway in A. thaliana and sorghum have been found to reduce the leaf vein density (26, 27). However, as we have little functional data from monocots, whether our hypothesis holds for monocots remains to be tested.
Although the full picture is yet to emerge, this work advances our understanding of the molecular events leading to the high vein density in C4 leaves and paves the way to unravel the genetic control of C4 leaf development in the future. There has been much interest in engineering C3 rice to express C4 traits to increase its photosynthetic efficiency and productivity (16, 17). A critical step in the C4 rice project would be to increase its leaf vein density. This study suggests that an elevated vein density can be achieved by genetically engineering the rice genome to increase auxin biosynthesis and transport in developing leaves.
Materials and Methods
Plant Material and RNA Isolation.
Seeds of G. gynandra were collected from southern Taiwan and seeds of T. hassleriana and maize were purchased from Known-You Seed Company (Taiwan). G. gynandra and T. hassleriana were grown in growth chambers under the light–dark cycle: 12-h light (200–250 μmol⋅m−2⋅s−1) at 27 °C and 12-h darkness at 25 °C. Maize was cultivated in the greenhouse in February to April 2017.
For isolation of total RNA, the fifth leaves of G. gynandra and T. hassleriana that were ∼0.5–0.8 mm long (S1) and ∼2 mm long (S2) were individually harvested 2 h after dawn and immediately frozen in liquid nitrogen. Total RNA was isolated by TRIzol reagent (Invitrogen) according to the manufacturer’s instructions, using a 1:2 ratio of sample:TRIzol reagent. RNA samples were treated with DNase I at 37 °C for 30 min to eliminate contaminating genomic DNA.
The jin1-9 (SALK_017005), jin1-10 (SALK_083483), pin1 (SALK_047613), and lax2 (SAIL_178_C02) mutants were from The Arabidopsis Biological Resources Center (ABRC). Plants of A. thaliana wild-type Columbia-0 (Col-0) and myc2 mutants (i.e., jin1-10 and jin1-9) were grown in growth chambers at 23°C with a light–dark cycle: 12-h light (200–250 μmol⋅m−2⋅s−1) and 12-h darkness.
cDNA Library Preparation and Sequencing.
RNA samples of G. gynandra and T. hassleriana fifth leaves at stage S1 and stage S2 were prepared separately. The cDNA libraries were then constructed and sequenced by the High-Throughput Genomics Core at Biodiversity Research Center, Academia Sinica, using Illumina HiSeq 2500 with the 150-bp paired-end format. The library construction protocol was optimized for milder RNA fragmentation for longer insert library to facilitate transcriptome assembly. The raw reads were processed as in Liu et al. (18). The numbers of raw reads and processed reads for each RNA sample are given in Dataset S1.
Assembly of Reads and Construction of ORF Databases.
For each RNA sample, the processed paired-end reads were merged to generate longer reads by FLASH (28). The merged and unmerged paired-end reads of the S1 and S2 samples in each species were assembled de novo, using the CLC Genomics Workbench with default options, which was found to produce better assemblies than other programs (29).
The above de novo assembled contigs were improved with the following three steps. First, we collected the cDNA sequences of A. thaliana (TAIR10), the previously assembled ORFs in T. hassleriana (19), and also our newly assembled contigs in T. hassleriana. Second, the unmerged and merged reads by FLASH in the S1 and S2 samples of T. hassleriana were used to search against all of the sequences collected in the first step using BLAT (30) with default options. The aligned reads were then assembled again using the CLC Genomics Workbench with the default parameters. Because the previously assembled sequences in T. hassleriana (19) only contained the coding sequences, we transformed each assembled sequence into an ORF. Third, we retained a newly assembled ORF sequence if it was (i) the best (lowest E value) of the BLAST (E value < 10−10) hits to one of the previously assembled T. hassleriana ORFs (19) and it has a longer assembly length than the previously assembled ORF; or (ii) the best hit with the alignment of <70% amino acid sequence identity or no significant hit (threshold E value = 10−5) to the previously assembled ORFs. The ORFs in (i) were considered improved ORFs by length extension and were used to replace the corresponding ORFs of ref. 19 to obtain an improved ORF database. The ORFs in (ii) were considered newly assembled ORFs and were added to the improved ORF database. Finally, we removed the ORFs <200 bp and also the (redundant) ORFs that have 100% sequence identity to other ORFs and have shorter lengths to obtain the final ORF database for T. hassleriana.
To assemble the ORFs of G. gynandra, we collected the reads that could be aligned with ORFs in the new ORF database for T. hassleriana. The remaining steps were the same as in the reassembly of ORFs in T. hassleriana. For the unaligned reads, we used them to assemble additional ORFs and integrated them into the G. gynandra ORF database using the criteria in the third step above. The redundant ORFs and ORFs <200 bp were discarded as above. In the end, we identified 35,934 ORFs and 30,076 ORFs for G. gynandra and T. hassleriana, respectively.
Estimating Gene Expression Levels.
To quantify the expression levels of the assembled ORFs at a given developmental stage (S1 or S2) in a species, the Illumina reads after quality trimming with Q30 for that developmental stage were mapped to the corresponding ORF database for that species. The single-end read data were then mapped to the ORFs using Bowtie2 (31) with default settings. Finally, we used the eXpress software (32) to resolve the multiple-mapped reads and calculate the reads per kilobase per million mapped reads (RPKM) to represent the expression levels of the ORFs (Dataset S2).
A gene is defined as expressed in a species if its RPKM is >1 in at least one of the two stages. For T. hassleriana, 24,650 ORFs were expressed. For G. gynandra, there were 20,455 expressed ORFs with >50% coverage of one ORF in the T. hassleriana ORF database and 24,807 expressed ORFs with >25% coverage, indicating a substantial portion of the assembled ORFs in G. gynandra were partial due to the challenge of de novo transcriptome assembly without a reference genome.
To have meaningful comparisons of gene expression levels between two species, the RPKM values were normalized using the upper quartile normalization procedure. First, we collected G. gynandra ORFs with >50% coverage of one ORF in the T. hassleriana ORF database; this criterion was to avoid mistaking different G. gynandra ORF fragments homologous to the same T. hassleriana locus as multiple ORFs. Next, we selected the orthologous ORFs between the collected G. gynandra ORFs and the T. hassleriana ORF database. If there were multiple paralogous ORFs in a species, we took the sum of their RPKM values. We then compared the RPKM values for the two transcriptomes at the upper quantile and found that the value in T. hasslerianna was 1.0052 and 0.9898 times that in G. gynandra at the S1 stage and at the S2 stage, respectively. As both normalization factors were very close to 1, we actually did not do the normalization.
The published Z. mays foliar (FP) and husk leaf (HP) primordia transcriptome datasets (20) were downloaded for comparing the gene expression levels in foliar and husk leaf primordia.
Identification of Auxin Biosynthesis and Transport Genes in T. hassleriana, G. gynandra, and Maize.
The protein sequences of A. thaliana auxin biosynthesis and transport genes (33–36) were used to search the orthologous genes in T. hassleriana and G. gynandra with a BLAST threshold E value <10−10. For maize (Zea mays) genes, their orthologous relationships to A. thaliana genes were as defined in Ensembl Plants (release 31; plants.ensembl.org/index.html) with an additional requirement that the putative orthologous pairs had >30% amino acid sequence identity.
Measuring Endogenous Auxin Content by MS.
To quantify the auxin content of developing leaves, plants were grown in growth chambers under the light–dark cycle of 12-h light (200–250 μmol⋅m−2⋅s−1) at 27°C and 12-h darkness at 25 °C. At least 10 fifth leaves of G. gynandra and T. hassleriana and 10 seventh leaves of A. thaliana Col-0 and myc2 mutants (∼3 mm in length) were harvested and immediately frozen in liquid nitrogen. The maize leaf foliar and husk primordia, FP3/4 and HP3/4 (∼2 mm in length), were harvested from 3- to 4-wk-old plants and immediately frozen in liquid nitrogen. All measurements were performed in three biological replicates.
Frozen samples were homogenized using MagNA Lyser Green Beads, MagNA Lyser Instrument (Roche Diagnostics), and extracted in 300 μL of 50 mM sodium phosphate buffer (pH 7.0) containing 10 ng of indole-2,4,5,6,7-d5-3-acetic acid (d5-IAA) (Sigma-Aldrich) as the internal standards. The samples were incubated at 4 °C with continuous shaking for 1 h, and then centrifuged for 5 min at 13,000 × g at 4 °C. The pH was adjusted to 2.7 with 1 M hydrochloric acid. The supernatant was loaded into 10-mg Oasis HLB cartridge (Waters) followed by washing with 1 mL of 20% methanol containing 0.1% acetic acid and eluted with 1 mL of 80% methanol containing 0.1% acetic acid. The eluates were vacuum-dried and dissolved in 60 μL of Milli-Q water. Eluates were analyzed for auxin content using a Waters ACQUITY UPLC/XevoTQ-S tandem quadrupole mass spectrometer (Waters). Characteristic MS transitions were monitored using multiple reaction monitoring for endogenous auxin (m/z, 176 > 130), and d5-IAA (181 > 135) (37). For the LC analysis, we followed the Novak’s procedures (38).
Chemical Treatment and Analyses of Vein Density.
To study the effect of auxin content on G. gynandra leaf vein patterning, 50 or 75 μM yucasin or 20 μM NPA in half-strength Kimura B nutrient solution (39) was used for treating the plants before they developed the third leaf. The control plants were treated with 0.75% DMSO, which was used to dissolve yucasin and NPA. The solution was refreshed every 2 d. After 2 wk of yucasin or 1 wk of NPA treatment, plants were transferred to Kimura B solution for 10 d for the fifth leaf to be fully developed for vein density measurement. NPA treatment in T. hassleriana was conducted as in the NPA treatment of G. gynandra.
All measurements of vein densities of the leaves were made by phenoVein (40) from images of chloral hydrate-cleared leaves (41). Images were taken in a Nikon Eclipse 90i microscope using dark-field by Photometric CoolSNAP HQ2 CCD.
Measurement of Vein Densities in A. thaliana Mutants.
A. thaliana Col-0, myc2 homozygotes, pin1 heterozygotes, and lax2 homozygotes were grown in growth chambers at 23°C with a light–dark cycle of 12-h light (200–250 μmol⋅m−2⋅s−1) and 12-h darkness. At 30 d after germination, the seventh mature leaves (42) were collected and used for measuring their vein densities in the central portion of the leaf.
Cross-Sections of Developing Leaves.
Developing leaves (0.8 or 2 mm long) of G. gynandra and T. hassleriana were fixed by 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.2, and then dehydrated in graded series of ethanol (50%, 70%, 80%, 90%, 95%, and 100%). After dehydration, the leaf samples were embedded in Spurr’s epoxy resin (43), cut into 900-nm sections in a Leica EM UC6 ultramicrotome, mounted on slides, and stained with toluidine blue. Photomicrographs were taken using the Aperio Digital Pathology System.
Supplementary Material
Acknowledgments
We thank John Wang and Jane Langdale for comments. We thank the RNA-sequencing work by the High-Throughput Genomics Core, Biodiversity Research Center, Academia Sinica, and the technical support of Dr. Chih-Yu Lin and Ms. Ting-Hsiang Chang, UPLC-MS/MS and Metabolomics Core Facility, Agricultural Biotechnology Research Center, Academia Sinica. This study was supported by Academia Sinca Thematic project (AS-106-TP-B14) and by the Innovative Translational Agricultural Research Program.
Footnotes
The authors declare no conflict of interest.
Data deposition: Sequences have been deposited in the Sequence Read Archive, www.ncbi.nlm.nih.gov/sra (accession nos. SRR5405059, SRR5407658, SRR5407836, and SRR5407839) and the Transcriptome Shotgun Assemblies, www.ncbi.nlm.nih.gov/Traces/wgs/?view=TSA (accession nos. GFMQ00000000 and GFML00000000).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1709171114/-/DCSupplemental.
References
- 1.Ehleringer JR, Cerling TE, Helliker BR. C4 photosynthesis, atmospheric CO2, and climate. Oecologia. 1997;112:285–299. doi: 10.1007/s004420050311. [DOI] [PubMed] [Google Scholar]
- 2.Sage RF. The evolution of C4 photosynthesis. New Phytol. 2004;161:341–370. doi: 10.1111/j.1469-8137.2004.00974.x. [DOI] [PubMed] [Google Scholar]
- 3.Sage RF. Environmental and evolutionary preconditionsfor the origin and diversification of the C4 photosynthetic syndrome. Plant Biol. 2001;3:202–213. [Google Scholar]
- 4.McKown AD, Dengler NG. Key innovations in the evolution of Kranz anatomy and C4 vein pattern in Flaveria (Asteraceae) Am J Bot. 2007;94:382–399. doi: 10.3732/ajb.94.3.382. [DOI] [PubMed] [Google Scholar]
- 5.Roth-Nebelsick A, Uhl D, Mosbrugger V, Kerp H. Evolution and function of leaf venation architecture: A review. Ann Bot (Lond) 2001;87:553–566. [Google Scholar]
- 6.Fouracre JP, Ando S, Langdale JA. Cracking the Kranz enigma with systems biology. J Exp Bot. 2014;65:3327–3339. doi: 10.1093/jxb/eru015. [DOI] [PubMed] [Google Scholar]
- 7.Amiard V, et al. Anatomical and photosynthetic acclimation to the light environment in species with differing mechanisms of phloem loading. Proc Natl Acad Sci USA. 2005;102:12968–12973. doi: 10.1073/pnas.0503784102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uhl D, Mosbrugger V. Leaf venation density as a climate and environmental proxy: A critical review and new data. Palaeogeogr Palaeoclimatol Palaeoecol. 1999;149:15–26. [Google Scholar]
- 9.Sachs T. The control of the patterned differentiation of vascular tissues. Adv Bot Res. 1981;9:151–262. [Google Scholar]
- 10.Uggla C, Moritz T, Sandberg G, Sundberg B. Auxin as a positional signal in pattern formation in plants. Proc Natl Acad Sci USA. 1996;93:9282–9286. doi: 10.1073/pnas.93.17.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mattsson J, Sung ZR, Berleth T. Responses of plant vascular systems to auxin transport inhibition. Development. 1999;126:2979–2991. doi: 10.1242/dev.126.13.2979. [DOI] [PubMed] [Google Scholar]
- 12.Sieburth LE. Auxin is required for leaf vein pattern in Arabidopsis. Plant Physiol. 1999;121:1179–1190. doi: 10.1104/pp.121.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mattsson J, Ckurshumova W, Berleth T. Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 2003;131:1327–1339. doi: 10.1104/pp.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawchuk MG, Edgar A, Scarpella E. Patterning of leaf vein networks by convergent auxin transport pathways. PLoS Genet. 2013;9:e1003294. doi: 10.1371/journal.pgen.1003294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scarpella E, Marcos D, Friml J, Berleth T. Control of leaf vascular patterning by polar auxin transport. Genes Dev. 2006;20:1015–1027. doi: 10.1101/gad.1402406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hibberd JM, Sheehy JE, Langdale JA. Using C4 photosynthesis to increase the yield of rice-rationale and feasibility. Curr Opin Plant Biol. 2008;11:228–231. doi: 10.1016/j.pbi.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Wang P, Vlad D, Langdale JA. Finding the genes to build C4 rice. Curr Opin Plant Biol. 2016;31:44–50. doi: 10.1016/j.pbi.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 18.Liu WY, et al. Anatomical and transcriptional dynamics of maize embryonic leaves during seed germination. Proc Natl Acad Sci USA. 2013;110:3979–3984. doi: 10.1073/pnas.1301009110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Külahoglu C, et al. Comparative transcriptome atlases reveal altered gene expression modules between two Cleomaceae C3 and C4 plant species. Plant Cell. 2014;26:3243–3260. doi: 10.1105/tpc.114.123752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang P, Kelly S, Fouracre JP, Langdale JA. Genome-wide transcript analysis of early maize leaf development reveals gene cohorts associated with the differentiation of C4 Kranz anatomy. Plant J. 2013;75:656–670. doi: 10.1111/tpj.12229. [DOI] [PubMed] [Google Scholar]
- 21.Dombrecht B, et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell. 2007;19:2225–2245. doi: 10.1105/tpc.106.048017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson JP, et al. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell. 2004;16:3460–3479. doi: 10.1105/tpc.104.025833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishimura T, et al. Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis. Plant J. 2014;77:352–366. doi: 10.1111/tpj.12399. [DOI] [PubMed] [Google Scholar]
- 24.Lomax TL, Muday GK, Rubery PH. Auxin Transport. In: Davies PJ, editor. Plant Hormones: Physiology, Biochemistry and Molecular Biology. Springer; Dordrecht, The Netherlands: 1995. pp. 509–530. [Google Scholar]
- 25.Zhou XY, Song L, Xue HW. Brassinosteroids regulate the differential growth of Arabidopsis hypocotyls through auxin signaling components IAA19 and ARF7. Mol Plant. 2013;6:887–904. doi: 10.1093/mp/sss123. [DOI] [PubMed] [Google Scholar]
- 26.Zhiponova MK, et al. Brassinosteroid production and signaling differentially control cell division and expansion in the leaf. New Phytol. 2013;197:490–502. doi: 10.1111/nph.12036. [DOI] [PubMed] [Google Scholar]
- 27.Rizal G, et al. Two forward genetic screens for vein density mutants in sorghum converge on a cytochrome P450 gene in the brassinosteroid pathway. Plant J. 2015;84:257–266. doi: 10.1111/tpj.13007. [DOI] [PubMed] [Google Scholar]
- 28.Magoč T, Salzberg SL. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bräutigam A, Mullick T, Schliesky S, Weber AP. Critical assessment of assembly strategies for non-model species mRNA-Seq data and application of next-generation sequencing to the comparison of C3 and C4 species. J Exp Bot. 2011;62:3093–3102. doi: 10.1093/jxb/err029. [DOI] [PubMed] [Google Scholar]
- 30.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods. 2013;10:71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodward AW, Bartel B. Auxin: Regulation, action, and interaction. Ann Bot (Lond) 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandler JW. Local auxin production: A small contribution to a big field. BioEssays. 2009;31:60–70. doi: 10.1002/bies.080146. [DOI] [PubMed] [Google Scholar]
- 35.Normanly J. Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb Perspect Biol. 2010;2:a001594. doi: 10.1101/cshperspect.a001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mano Y, Nemoto K. The pathway of auxin biosynthesis in plants. J Exp Bot. 2012;63:2853–2872. doi: 10.1093/jxb/ers091. [DOI] [PubMed] [Google Scholar]
- 37.Chiwocha SD, et al. A method for profiling classes of plant hormones and their metabolites using liquid chromatography-electrospray ionization tandem mass spectrometry: An analysis of hormone regulation of thermodormancy of lettuce (Lactuca sativa L.) seeds. Plant J. 2003;35:405–417. doi: 10.1046/j.1365-313x.2003.01800.x. [DOI] [PubMed] [Google Scholar]
- 38.Novák O, et al. Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J. 2012;72:523–536. doi: 10.1111/j.1365-313X.2012.05085.x. [DOI] [PubMed] [Google Scholar]
- 39.Ma JF, Goto S, Tamai K, Ichii M. Role of root hairs and lateral roots in silicon uptake by rice. Plant Physiol. 2001;127:1773–1780. [PMC free article] [PubMed] [Google Scholar]
- 40.Bühler J, et al. phenoVein-A tool for leaf vein segmentation and analysis. Plant Physiol. 2015;169:2359–2370. doi: 10.1104/pp.15.00974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berleth T, Jurgens G. The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development. 1993;118:575–587. [Google Scholar]
- 42.Kang J, Dengler N. Vein pattern development in adult leaves of Arabidopsis thaliana. Int J Plant Sci. 2004;165:231–242. [Google Scholar]
- 43.Spurr AR. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.