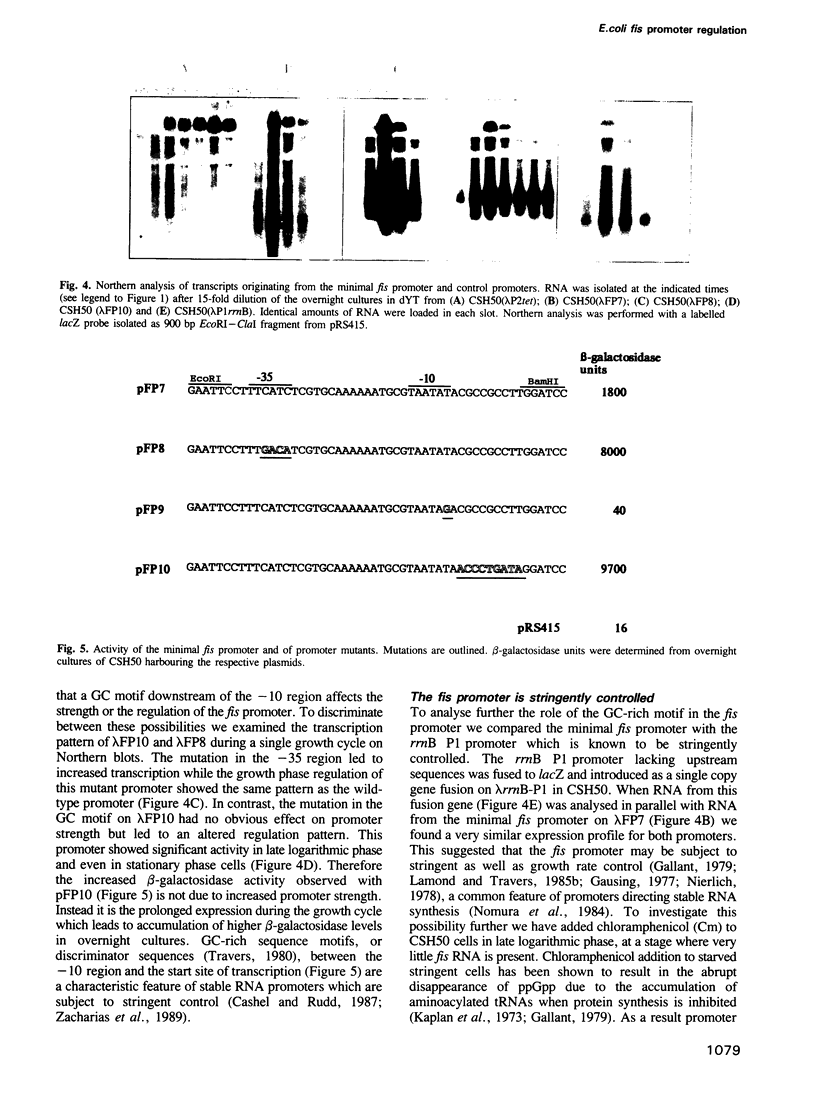
Abstract
The DNA binding protein FIS is involved in processes like site specific DNA inversion, lambda excision and stimulation of stable RNA synthesis in Escherichia coli. The amount of FIS protein is subject to dramatic changes during growth. We demonstrate that fis is part of an operon with one ORF of unknown function preceding the fis gene. Regulation of fis synthesis occurs at the transcriptional level. Within 15 min after nutritional upshift a large burst of fis mRNA is produced which levels off when cells begin to grow. By mutational analysis using promoter-lacZ fusions we demonstrate that the fis promoter is autoregulated by FIS. Growth phase regulation of the fis promoter depends on the presence of a GC motif downstream of the -10 region. We show that the fis promoter is subject to stringent control and discuss this unusual feature with respect to the known and putative functions FIS serves in E. coli.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ball C. A., Johnson R. C. Efficient excision of phage lambda from the Escherichia coli chromosome requires the Fis protein. J Bacteriol. 1991 Jul;173(13):4027–4031. doi: 10.1128/jb.173.13.4027-4031.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baracchini E., Bremer H. Stringent and growth control of rRNA synthesis in Escherichia coli are both mediated by ppGpp. J Biol Chem. 1988 Feb 25;263(6):2597–2602. [PubMed] [Google Scholar]
- Bennett G. N., Schweingruber M. E., Brown K. D., Squires C., Yanofsky C. Nucleotide sequence of region preceding trp mRNA initiation site and its role in promoter and operator function. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2351–2355. doi: 10.1073/pnas.73.7.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L., Smith D. M. Conformational change in the DNA associated with an unusual promoter mutation in a tRNA operon of Salmonella. Cell. 1984 Dec;39(3 Pt 2):643–652. doi: 10.1016/0092-8674(84)90471-9. [DOI] [PubMed] [Google Scholar]
- Bracco L., Kotlarz D., Kolb A., Diekmann S., Buc H. Synthetic curved DNA sequences can act as transcriptional activators in Escherichia coli. EMBO J. 1989 Dec 20;8(13):4289–4296. doi: 10.1002/j.1460-2075.1989.tb08615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruist M. F., Glasgow A. C., Johnson R. C., Simon M. I. Fis binding to the recombinational enhancer of the Hin DNA inversion system. Genes Dev. 1987 Oct;1(8):762–772. doi: 10.1101/gad.1.8.762. [DOI] [PubMed] [Google Scholar]
- Bétermier M., Lefrère V., Koch C., Alazard R., Chandler M. The Escherichia coli protein, Fis: specific binding to the ends of phage Mu DNA and modulation of phage growth. Mol Microbiol. 1989 Apr;3(4):459–468. doi: 10.1111/j.1365-2958.1989.tb00192.x. [DOI] [PubMed] [Google Scholar]
- Chiaramello A. E., Zyskind J. W. Coupling of DNA replication to growth rate in Escherichia coli: a possible role for guanosine tetraphosphate. J Bacteriol. 1990 Apr;172(4):2013–2019. doi: 10.1128/jb.172.4.2013-2019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe H. W., Labahn J., Itoh S., Koch C., Kahmann R., Saenger W. Crystallization of the DNA-binding Escherichia coli protein FIS. J Mol Biol. 1989 Jul 5;208(1):209–210. doi: 10.1016/0022-2836(89)90098-3. [DOI] [PubMed] [Google Scholar]
- Fiil N., Friesen J. D. Isolation of "relaxed" mutants of Escherichia coli. J Bacteriol. 1968 Feb;95(2):729–731. doi: 10.1128/jb.95.2.729-731.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. I. Integration host factor: a protein for all reasons. Cell. 1988 Nov 18;55(4):545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- Gaal T., Barkei J., Dickson R. R., deBoer H. A., deHaseth P. L., Alavi H., Gourse R. L. Saturation mutagenesis of an Escherichia coli rRNA promoter and initial characterization of promoter variants. J Bacteriol. 1989 Sep;171(9):4852–4861. doi: 10.1128/jb.171.9.4852-4861.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant J. A. Stringent control in E. coli. Annu Rev Genet. 1979;13:393–415. doi: 10.1146/annurev.ge.13.120179.002141. [DOI] [PubMed] [Google Scholar]
- Gaston K., Bell A., Kolb A., Buc H., Busby S. Stringent spacing requirements for transcription activation by CRP. Cell. 1990 Aug 24;62(4):733–743. doi: 10.1016/0092-8674(90)90118-x. [DOI] [PubMed] [Google Scholar]
- Gausing K. Regulation of ribosome production in Escherichia coli: synthesis and stability of ribosomal RNA and of ribosomal protein messenger RNA at different growth rates. J Mol Biol. 1977 Sep 25;115(3):335–354. doi: 10.1016/0022-2836(77)90158-9. [DOI] [PubMed] [Google Scholar]
- Gille H., Egan J. B., Roth A., Messer W. The FIS protein binds and bends the origin of chromosomal DNA replication, oriC, of Escherichia coli. Nucleic Acids Res. 1991 Aug 11;19(15):4167–4172. doi: 10.1093/nar/19.15.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse R. L., Stark M. J., Dahlberg A. E. Regions of DNA involved in the stringent control of plasmid-encoded rRNA in vivo. Cell. 1983 Apr;32(4):1347–1354. doi: 10.1016/0092-8674(83)90315-x. [DOI] [PubMed] [Google Scholar]
- Gourse R. L., de Boer H. A., Nomura M. DNA determinants of rRNA synthesis in E. coli: growth rate dependent regulation, feedback inhibition, upstream activation, antitermination. Cell. 1986 Jan 17;44(1):197–205. doi: 10.1016/0092-8674(86)90498-8. [DOI] [PubMed] [Google Scholar]
- Hansen F. G., Atlung T., Braun R. E., Wright A., Hughes P., Kohiyama M. Initiator (DnaA) protein concentration as a function of growth rate in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1991 Aug;173(16):5194–5199. doi: 10.1128/jb.173.16.5194-5199.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heichman K. A., Johnson R. C. The Hin invertasome: protein-mediated joining of distant recombination sites at the enhancer. Science. 1990 Aug 3;249(4968):511–517. doi: 10.1126/science.2166334. [DOI] [PubMed] [Google Scholar]
- Huber H. E., Iida S., Arber W., Bickle T. A. Site-specific DNA inversion is enhanced by a DNA sequence element in cis. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3776–3780. doi: 10.1073/pnas.82.11.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultmark D., Klemenz R., Gehring W. J. Translational and transcriptional control elements in the untranslated leader of the heat-shock gene hsp22. Cell. 1986 Feb 14;44(3):429–438. doi: 10.1016/0092-8674(86)90464-2. [DOI] [PubMed] [Google Scholar]
- Hübner P., Arber W. Mutational analysis of a prokaryotic recombinational enhancer element with two functions. EMBO J. 1989 Feb;8(2):577–585. doi: 10.1002/j.1460-2075.1989.tb03412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Ball C. A., Pfeffer D., Simon M. I. Isolation of the gene encoding the Hin recombinational enhancer binding protein. Proc Natl Acad Sci U S A. 1988 May;85(10):3484–3488. doi: 10.1073/pnas.85.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Bruist M. F., Simon M. I. Host protein requirements for in vitro site-specific DNA inversion. Cell. 1986 Aug 15;46(4):531–539. doi: 10.1016/0092-8674(86)90878-0. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Simon M. I. Hin-mediated site-specific recombination requires two 26 bp recombination sites and a 60 bp recombinational enhancer. Cell. 1985 Jul;41(3):781–791. doi: 10.1016/s0092-8674(85)80059-3. [DOI] [PubMed] [Google Scholar]
- Jones R., Haselkorn R. The DNA sequence of the Rhodobacter capsulatus ntrA, ntrB and ntrC gene analogues required for nitrogen fixation. Mol Gen Genet. 1989 Feb;215(3):507–516. doi: 10.1007/BF00427050. [DOI] [PubMed] [Google Scholar]
- Kahmann R., Rudt F., Koch C., Mertens G. G inversion in bacteriophage Mu DNA is stimulated by a site within the invertase gene and a host factor. Cell. 1985 Jul;41(3):771–780. doi: 10.1016/s0092-8674(85)80058-1. [DOI] [PubMed] [Google Scholar]
- Kanaar R., van de Putte P., Cozzarelli N. R. Gin-mediated DNA inversion: product structure and the mechanism of strand exchange. Proc Natl Acad Sci U S A. 1988 Feb;85(3):752–756. doi: 10.1073/pnas.85.3.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan S., Atherly A. G., Barrett A. Synthesis of stable RNA in stringent Escherichia coli cells in the absence of charged transfer RNA. Proc Natl Acad Sci U S A. 1973 Mar;70(3):689–692. doi: 10.1073/pnas.70.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C., Kahmann R. Purification and properties of the Escherichia coli host factor required for inversion of the G segment in bacteriophage Mu. J Biol Chem. 1986 Nov 25;261(33):15673–15678. [PubMed] [Google Scholar]
- Koch C., Ninnemann O., Fuss H., Kahmann R. The N-terminal part of the E.coli DNA binding protein FIS is essential for stimulating site-specific DNA inversion but is not required for specific DNA binding. Nucleic Acids Res. 1991 Nov 11;19(21):5915–5922. doi: 10.1093/nar/19.21.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C., Vandekerckhove J., Kahmann R. Escherichia coli host factor for site-specific DNA inversion: cloning and characterization of the fis gene. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4237–4241. doi: 10.1073/pnas.85.12.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kostrewa D., Granzin J., Koch C., Choe H. W., Raghunathan S., Wolf W., Labahn J., Kahmann R., Saenger W. Three-dimensional structure of the E. coli DNA-binding protein FIS. Nature. 1991 Jan 10;349(6305):178–180. doi: 10.1038/349178a0. [DOI] [PubMed] [Google Scholar]
- Lamond A. I., Travers A. A. Genetically separable functional elements mediate the optimal expression and stringent regulation of a bacterial tRNA gene. Cell. 1985 Feb;40(2):319–326. doi: 10.1016/0092-8674(85)90146-1. [DOI] [PubMed] [Google Scholar]
- Lamond A. I., Travers A. A. Stringent control of bacterial transcription. Cell. 1985 May;41(1):6–8. doi: 10.1016/0092-8674(85)90050-9. [DOI] [PubMed] [Google Scholar]
- Leirmo S., Gourse R. L. Factor-independent activation of Escherichia coli rRNA transcription. I. Kinetic analysis of the roles of the upstream activator region and supercoiling on transcription of the rrnB P1 promoter in vitro. J Mol Biol. 1991 Aug 5;220(3):555–568. doi: 10.1016/0022-2836(91)90100-k. [DOI] [PubMed] [Google Scholar]
- Liu-Johnson H. N., Gartenberg M. R., Crothers D. M. The DNA binding domain and bending angle of E. coli CAP protein. Cell. 1986 Dec 26;47(6):995–1005. doi: 10.1016/0092-8674(86)90814-7. [DOI] [PubMed] [Google Scholar]
- Nierlich D. P. Regulation of bacterial growth, RNA, and protein synthesis. Annu Rev Microbiol. 1978;32:393–432. doi: 10.1146/annurev.mi.32.100178.002141. [DOI] [PubMed] [Google Scholar]
- Nilsson L., Vanet A., Vijgenboom E., Bosch L. The role of FIS in trans activation of stable RNA operons of E. coli. EMBO J. 1990 Mar;9(3):727–734. doi: 10.1002/j.1460-2075.1990.tb08166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M., Gourse R., Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- Osuna R., Finkel S. E., Johnson R. C. Identification of two functional regions in Fis: the N-terminus is required to promote Hin-mediated DNA inversion but not lambda excision. EMBO J. 1991 Jun;10(6):1593–1603. doi: 10.1002/j.1460-2075.1991.tb07680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznikoff W. S., Winter R. B., Hurley C. K. The location of the repressor binding sites in the lac operon. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2314–2318. doi: 10.1073/pnas.71.6.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W., Thompson J. F., Newlands J. T., Gourse R. L. E.coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990 Nov;9(11):3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SADLER J. R., NOVICK A. THE PROPERTIES OF REPRESSOR AND THE KINETICS OF ITS ACTION. J Mol Biol. 1965 Jun;12:305–327. doi: 10.1016/s0022-2836(65)80255-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmientos P., Sylvester J. E., Contente S., Cashel M. Differential stringent control of the tandem E. coli ribosomal RNA promoters from the rrnA operon expressed in vivo in multicopy plasmids. Cell. 1983 Apr;32(4):1337–1346. doi: 10.1016/0092-8674(83)90314-8. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Sokawa J., Sokawa Y. Relaxation effect of chloramphenicol on the stringent control in Escherichia coli. J Biochem. 1978 Jun;83(6):1699–1705. doi: 10.1093/oxfordjournals.jbchem.a132083. [DOI] [PubMed] [Google Scholar]
- Stüber D., Bujard H. Organization of transcriptional signals in plasmids pBR322 and pACYC184. Proc Natl Acad Sci U S A. 1981 Jan;78(1):167–171. doi: 10.1073/pnas.78.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., Moitoso de Vargas L., Koch C., Kahmann R., Landy A. Cellular factors couple recombination with growth phase: characterization of a new component in the lambda site-specific recombination pathway. Cell. 1987 Sep 11;50(6):901–908. doi: 10.1016/0092-8674(87)90516-2. [DOI] [PubMed] [Google Scholar]
- Travers A. A., Lamond A. I., Weeks J. R. Alteration of the growth-rate-dependent regulation of Escherichia coli tyrT expression by promoter mutations. J Mol Biol. 1986 May 5;189(1):251–255. doi: 10.1016/0022-2836(86)90397-9. [DOI] [PubMed] [Google Scholar]
- Travers A. A. Promoter sequence for stringent control of bacterial ribonucleic acid synthesis. J Bacteriol. 1980 Feb;141(2):973–976. doi: 10.1128/jb.141.2.973-976.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgenboom E., Nilsson L., Bosch L. The elongation factor EF-Tu from E. coli binds to the upstream activator region of the tRNA-tufB operon. Nucleic Acids Res. 1988 Nov 11;16(21):10183–10197. doi: 10.1093/nar/16.21.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias M., Göringer H. U., Wagner R. Influence of the GCGC discriminator motif introduced into the ribosomal RNA P2- and tac promoter on growth-rate control and stringent sensitivity. EMBO J. 1989 Nov;8(11):3357–3363. doi: 10.1002/j.1460-2075.1989.tb08498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]