Abstract
Mechanical biomarkers associated with cytoskeletal structures have been reported as powerful label-free cell state identifiers. In order to measure cell mechanical properties, traditional biophysical (e.g., atomic force microscopy, micropipette aspiration, optical stretchers) and microfluidic approaches were mainly employed; however, they critically suffer from low-throughput, low-sensitivity, and/or time-consuming and labor-intensive processes, not allowing techniques to be practically used for cell biology research applications. Here, a novel inertial microfluidic cell stretcher (iMCS) capable of characterizing large populations of single-cell deformability near real-time is presented. The platform inertially controls cell positions in microchannels and deforms cells upon collision at a T-junction with large strain. The cell elongation motions are recorded, and thousands of cell deformability information is visualized near real-time similar to traditional flow cytometry. With a full automation, the entire cell mechanotyping process runs without any human intervention, realizing a user friendly and robust operation. Through iMCS, distinct cell stiffness changes in breast cancer progression and epithelial mesenchymal transition are reported, and the use of the platform for rapid cancer drug discovery is shown as well. The platform returns large populations of single-cell quantitative mechanical properties (e.g., shear modulus) on-the-fly with high statistical significances, enabling actual usages in clinical and biophysical studies.
Keywords: mechanophenotype, high-throughput cell screening, microfluidics, inertial cell stretcher, deformability cytometry
Graphical Abstract
A novel inertial microfluidic cell stretcher (iMCS) capable of characterizing and classifying large populations of single-cell deformability near real-time in a fully automated manner is presented. Through this method, statistically robust cell deformability measurements in breast cancer progression and epithelial mesenchymal transition are reported, and the platform usage for cancer drug discovery is also demonstrated.
1. Introduction
The mechanical response of cells is a powerful label-free biophysical marker for identifying cell diseases and cellular states.[1] For example, cancer cells exhibit a more compliant phenotype than their benign counterparts, allowing them to metastasize to distal sites.[2] Plasmodium falciparum infected red blood cells become stiffer, increasing the risk of occlusions in the spleen and peripheral capillaries.[3] Embryonic stem cells were found to be more deformable than their differentiated progeny.[4] All these observations suggest that measuring cellular mechanical properties is an extremely important task, and this principle has been widely applied in biophysical studies,[5] diagnosis,[6] therapies,[7] and drug discovery.[8]
General biophysical approaches for measuring cell stiffness include atomic force microscopy (AFM), micropipette aspiration, and optical stretchers. AFM can precisely measure the stiffness of individual cells, but the process is inherently slow (<1 cell min−1) and difficult to standardize.[9] Micropipette aspiration is another well-known method which effectively evaluates cell viscoelastic properties,[10] but the throughput remains low (<1 cell min−1)[11] due to slow manual operation. Alternatively, optical stretchers are often used which allow contact-free measurements. However, the throughput is limited (~1 cell min−1), and it only allows a small degree of cell deformation (<1 nN).[12] For a meaningful cell state identification, thousands of cells should be analyzed with large strain because cellular characteristics are invariably heterogeneous even within an isogenic cell population.[13]
To address these challenges, recent advancements in microfluidics have allowed a wide variety of microfluidic solutions[14] (a detailed comparison can be found in our recent work).[14a] Briefly, many of these microfluidic devices use either fluid flow or geometric constrictions to achieve cell deformation. For example, Gossett et al.,[15] reported on-chip deformability cytometry (DC), in which cells are hydrodynamically stretched at high strain (>1 μN) via extensional flow analyzing a large number of cell deformability measurements with a cell processing rate (defined as the rate of cells passing through microfluidic device without considering post image processing) of 1,000–2,000 cells sec−1. However, cells experience non-negligible horizontal drifting motion near the stagnation point because cells collide with the compressible (non-rigid) fluidic wall, not allowing high quality imaging. As another drawback, due to flow instability and intrinsic hydraulic resistance asymmetry, cells tend not to be located in the channel center, causing considerable asymmetric cell stretching. Moreover, high-speed imaging returns a large number of image stacks which require a very long image processing time (tens of minutes via cluster computing[15, 17] without considering data transferring time), significantly decreasing the actual throughput (defined as the physical rate from sample injection to full cell analysis) to <10 cells sec−1. Alternatively, a bottlenecked microfluidic channel design is commonly presented, and cells are elongated as they pass through the gap. Recently, a method named real-time deformability cytometry (RT-DC) was reported,[18] which deforms cells in a microchannel with a constriction and analyzes cell deformability in real-time with an actual throughput of 100 cells sec−1. However, only a low degree of cell deformation (<1 nN) is available not suitable for stiff cells and/or capturing subtle cell responses. Furthermore, the method employs burdensome sheath fluids, may suffer from clogging due to a bottleneck design, and an unconventional media component was added for higher cell deformation (a detailed comparison is presented in Table S1).
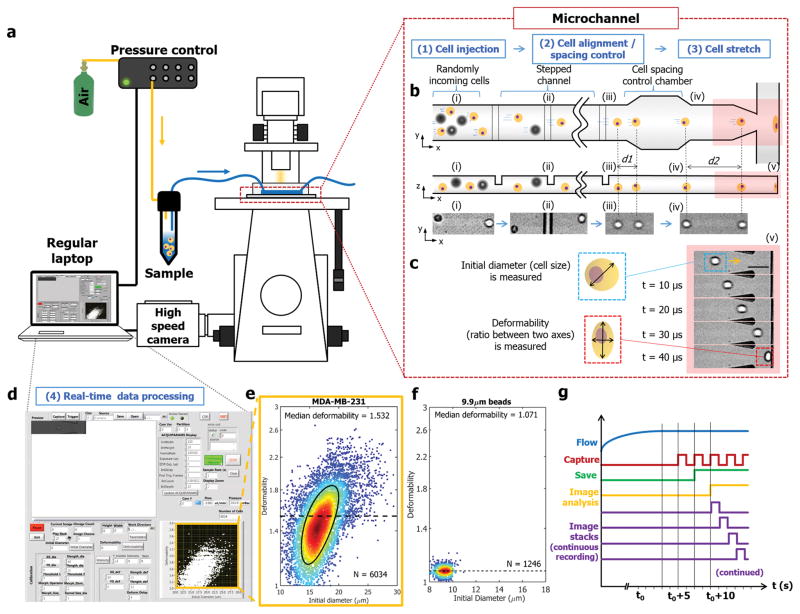
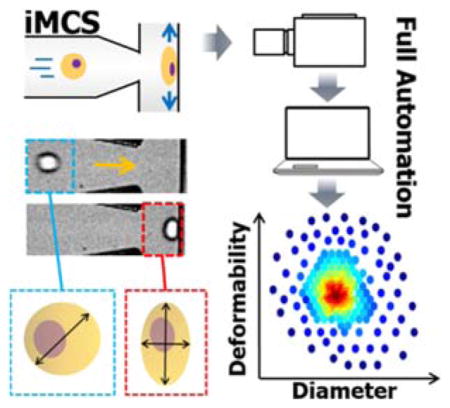
Here, we introduce a novel inertial microfluidic cell stretcher (iMCS): fully automated, high-throughput, and near real-time cell mechanical phenotyping. As shown in Figure 1, the platform is comprised of four sequential processes: (1) Cell injection, where cell suspensions are injected into a microchannel through a programmable pressure control system, (2) Cell alignment and spacing, where inertia passively guides randomly-aligned incoming cells to form a single-stream with controlled spacing (Movie S1), (3) Cell stretch, where a stream of cells deforms upon collision at a T-junction with large strain (Movie S2), and (4) Real-time data processing, where images of dynamic single-cell elongation motions are analyzed, and results are displayed close to real-time (Movie S3) similar to traditional flow cytometry plots (Figure 1e and f). The presented method inherently provides high-degree of cell deformation by utilizing high fluid momentum, and high-throughput cell stiffness characterization through our near real-time image analysis algorithm. Our algorithm effectively calculates cell initial diameter and deformability (the ratio between two axes at the maximum deformation state as shown in Figure 1c) with a rate of 450 cells sec−1. Note that the system operates in a fully automated manner from sample injection to results visualization which makes it user friendly and robust cell deformability measurements. Through iMCS, we successfully measured cell stiffness changes in cancer disease progression and epithelial mesenchymal transition, and demonstrated that the platform can be utilized for rapid drug discovery as well. We also numerically investigated the cell deformation process to extract intrinsic cellular biophysical properties on-the-fly from the experimental measurements, and discussed origins of whole cell deformation responses.
Figure 1. Inertial microfluidic cell stretcher (iMCS).
a) Schematic illustrating the design and operating principles of the system. b) Cells are inertially controlled for focusing, ordering and deformation in a sequential manner. (i–iii) Randomly incoming cells are focused into a single-stream cell chain through inertial effects in a structured microchannel. (iv) Distance between two closely positioned cells is increased via particle-particle interactions in an expansion-contraction chamber, preventing cells from colliding with the wall simultaneously. c) Time series of high-speed microscope images show a cell deformation process where we quantify cell diameter and deformability; the ratio between two axes. Scale bar represents 50 μm. d) A screen shot of the control panel, and mechanical characterizations of e) MDA-MB-231 cells (black solid line indicates 50%-density contour) and f) 9.9 μm polystyrene microspheres. g) The system control scheme is presented for continuous image recording and analysis. The entire process from cell injection, image acquisition and image analysis to result visualization is controlled in a fully automated manner.
2. Results
2.1 Working principles of inertial microfluidic cell stretcher
Our platform utilizes inertial effects in microfluidic channels to control cell position (focusing) and spacing (ordering) before cells deform during their impact with the channel wall. Traditionally, inertia is neglected in microfluidics due to small channel dimensions and a low flow velocity. However, inertial flow (Re≈O(1–102); Re is the ratio between inertial to viscous forces) is developed in microchannels at high flow rates, and by taking advantage of inertial effects, we can effectively manipulate cell position and spacing. In brief, as shown in Figure 1b, cells flow in a low aspect ratio straight channel interspersed with a series of elevational obstacles arranged orthogonally. Along with inertial lift forces, each obstacle induces a pair of helical secondary flows[19] that direct cells into a single equilibrium position (Movie S1). This inertial cell focusing process is an extremely important step because cells positioned in a single imaging plane are critical for high quality imaging. Having cells located in the center of the fluidic channel is another important characteristic since it allows cells to be compressed under uniform flow and deformed symmetrically. We evaluated our device performance by testing various sizes of polystyrene microparticles and cells at different flow rates (Figure S1), and identified a condition (Re = 125) to form a single-cell stream with near 100% focusing efficiency.
Our second cell position manipulation step is cell-cell spacing control. A cell requires a finite time to reach the maximum deformation state when it collides with the wall at the T-junction. Without appropriate cell-cell spacing control, multiple cells impact the wall simultaneously, preventing single-cell based deformability measurements. To impose proper cell-cell spacing, an expansion and contraction chamber[20] is introduced (Figure 1b). In this chamber, the distance between two closely positioned cells is enlarged due to viscous repulsive interactions (Figure 1b(iii–iv) and S2).[2] It should be noted that the cell-cell spacing is also effected by the cell sample concentration that determines the final throughput of the system (Figure S3), and improperly spaced cells are rejected from image analysis (see Supplementary Note S1).
Using this platform, we first tested the MDA-MB-231 breast cancer cells. The cell suspension was introduced into the platform utilizing the pressure-driven flow control system (Figure 1a). Instead of a syringe pump we use pressure-based flow control to allow rapid flow response, enabling a low volume sample injection (<500 μL). Due to a cell’s high momentum (νcell ≈ 4 m s−1), cells deform with large strain upon cell-wall impact (Figure 1c), allowing highly sensitive measurements. Guiding cells to collide with a rigid PDMS channel is a critical design component for high quality imaging where we eliminate cell drifting motions near stagnation point in DC[15] discussed previously. Note that no cell-wall adhesion was noticed even after a large number of cell-wall impact events (Navg >18,000 cells) and showed comparable cell viability after cell-wall collision (Figure S4).
We capture, store and analyze the dynamic cell elongation process at the T-junction, and display the analyzed results. For full automation, a system control algorithm is developed via LabVIEW and a self-written C++ routine (see Figure 1d–g and Supplementary Note S1). With a single start command, the entire process runs without any human intervention. As shown in Movie S3, upon the onset of sample injection, the high-speed camera is triggered to capture images after the flow rate reaches a set value (450 μL min−1). After 1 second of recording, the image stacks are first saved to the high-speed camera and transferred to a local hard drive. Simultaneously, an image analysis code with a parallel computing capability is initiated during image transfer, quantifying initial diameter and cell deformability and displaying the results close to real-time. As shown in Figure 1e–f, we successfully characterized MDA-MB-231, and rigid 9.9 μm polystyrene microspheres (control). Note typical runs plot as many as 6,300 single-cell data points for 3 seconds of recording, but continuous recording/analysis is available for extreme cell data (until the hard disk space is full). For data visualization, we plot the data in a density scatter plot format that effectively illustrates cell heterogeneity with a high statistical significance and show 50% contour plot (shown in black) to capture bulk cell behaviors. Our routine also finds the median cell deformability value (dotted line in Figure 1e and f) to represent the tested sample’s deformability.
As mentioned, inertial approaches inherently create large image stacks due to high-speed imaging. For a single sample analysis with 3 seconds of recording with a frame rate of 100,000 images sec−1, a total of 300,000 images are recorded. In order to process the resulting large number of image sets with speed and accuracy, we developed new algorithm using C++ codes with the OpenCV library (details in Supplementary Note S1 and Figure S5) for its speed and accuracy[21] over MATLAB based approaches.[15] This aspect is similar to a recently work,[18] but the reported study processing rate (250 μs frame−1) is not fast enough to accommodate the imaging rate required for general inertial systems (<10 μs frame−1). To increase the processing rate, our system transfer images from high-speed camera FPGA to the local hard drive and simultaneously process saved images in parallel (Figure 1g and Supplementary Note S1), avoiding latency.[18] Our developed algorithm effectively calculates cell initial diameter (Dcell) and cell deformability (Scell), and as presented in Movie S3, the whole process only requires approximately 35 seconds from sample injection to display of the fully analyzed results. In order to validate our automated image analysis, we have compared results from our automated routines with manually calculated values using ImageJ. As presented in Figure S6, the two results show very good agreement (<5% error in general). Note that the entire image processing and system control was executed on a commercially available off-the-shelf laptop (HP Zbook 15) which does not require any expensive computational tools, maximizing the potential utility of the system.
2.2 Numerical analysis of cell deformation
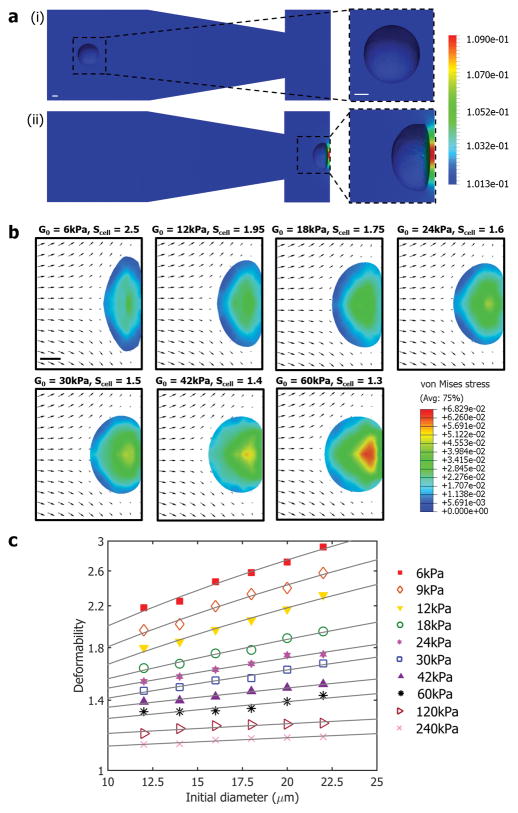
In order to understand the underlying cell deformation phenomena and predict intrinsic cellular biophysical properties from the measured cell diameter and deformability, we performed a series of finite element method (FEM) based numerical analyses. Two separate numerical studies were conducted for far-field (before) and near-field (after cell touches the wall) cases.
Details of numerical approaches are available in the Numerical Simulation section, but briefly, for the far-field simulation, images from the experimental high-speed image stacks were segmented to describe the shape of the cell at a given instant in time where we can match numerical and simulation results precisely. The flow around the cell was computed by solving the 3D Navier-Stokes (NS) equations using a stabilized FEM.[22] Due to significant cell motion and deformation, an Arbitrary Lagrangian–Eulerian[23] technique was adopted to deform the mesh, in conjunction with a mesh modification scheme.[24] Pressure distribution on the cell surface and the channel were calculated, and as presented in Figure 2a and Movie S4, when the cell is far from the T-junction, the pressure difference is uniform; however, when the cell approaches the channel wall, a large pressure gradient is created between the upstream and downstream faces of the cell due to the stagnating flow near the wall. This imbalance results in a net axial (along the axis of the microchannel) traction acting on the cell, accounting for the cell vertical elongation phenomena seen experimentally.
Figure 2. Numerical analysis of cell deformation process.
a) Far-field simulation was conducted to describe pressure distribution on the surface of the cell and the microchannel. Color bar represents the pressure distribution (MPa). b) Near-field simulation results are presented at the maximum cell deformation states with different shear moduli (G0). Arrows indicate the flow velocity fields, and color bar represents the stress distributions (MPa). c) Numerically generated “look-up figure” for shear modulus as a function of deformability and initial diameter. Solid lines are iso-shear modulus lines. All scale bars represent 5 μm.
When a cell touches the wall, it undergoes further deformation, and a separate near-field simulation was conducted to capture the cell-wall interaction. In our system, cells deform with large strain and therefore an infinitesimal linear strain model[18] cannot be applied. Thus, we numerically model the cell in 3D as a neo-Hookean incompressible hyperelastic solid[25] where a good agreement between numerical and experimental results is reported[26] (see Numerical Simulation section). Numerical analysis results are presented in Figure 2b, and stress distributions are shown at the maximum cell deformation states with different shear moduli and sizes within the range of values reported for breast cancer cells[26a] (see Movie S5 for time-dependent flow velocity fields and stress distributions). As expected, the deformability of a cell decreases with increasing shear modulus, and from the stress distribution information, we calculated that cells are deformed under high compressive forces (>1 μN). We performed a sweep by considering different cell sizes and shear moduli, and generated a plot (Figure 2c) similar to the one generated from experiments. The goal of this effort is to decouple cell size and deformation, and introduced iso-shear modulus lines as additional parameterization in the experimental scatter plots where cell mechanical properties can be extracted on-the-fly as shown in Figure 3–6.
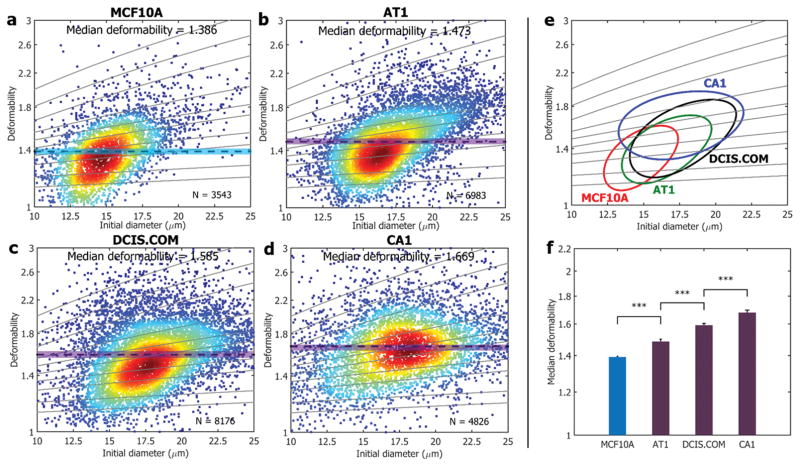
Figure 3. Deformability changes in breast cancer progression in MCF10A series.
Through our iMCS, deformability changes in MCF10A progression are characterized. The model includes four cell types of a) MCF10A, b) MCF10AT1, c) MCF10ADCIS.COM, and d) MCF10ACA1. Solid lines are iso-shear modulus lines from the numerical analysis. e) 50%-density contour plots from all samples a–d). f) Median deformability values from all samples a–d) are compared, showing that median deformability increases with the cell malignancy. *** indicates a P value of less than 0.001, and error bars represent standard deviations (N = 3).
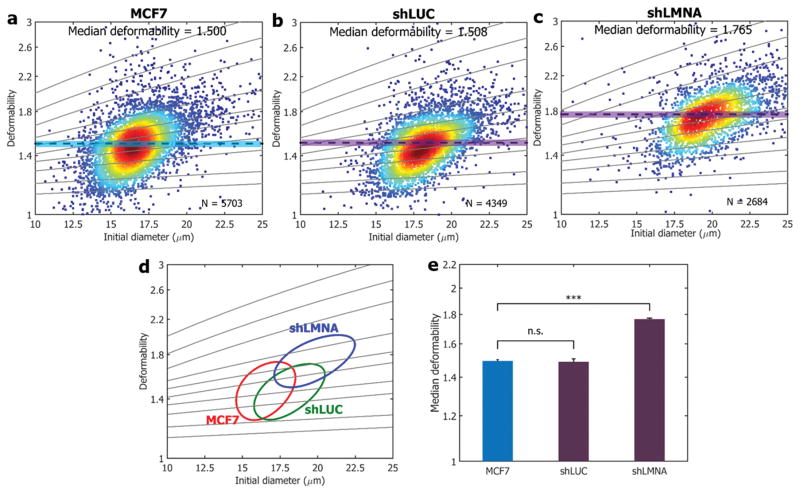
Figure 6. Cell mechanotyping results of Lamin A/C knockdown.
Cell deformability is measured for the following cell lines: a) MCF7 (control) is transfected with shRNA targeting b) luciferase (denoted as shLUC; negative control) and c) Lamin A/C (denoted as shLMNA) knockdown. Solid lines are iso-shear modulus lines from the numerical analysis. d) 50%-density contour plots for all samples. e) A comparison of median deformability for all samples. *** indicates a P value of less than 0.001, and n.s. represents statistical non-significance (P >0.05). All error bars indicate standard deviations (N = 3).
2.3 Mechanical characterization of breast cancer disease progression
To test iMCS sensitivity, we evaluated breast cancer cell lines. It is a well-known fact that cancer cells exhibit discernable stiffness changes as disease progress.[27] We targeted four cell lines specified as MCF10A, MCF10AT1, MCF10ADCIS.COM and MCF10ACA1. These cell lines are prepared based on the invasiveness increases within the MCF10 progression model.[28] Briefly, MCF10A is a normal immortalized mammary epithelial cell line, which is non-tumorigenic. MCF10AT1 is a pre-malignant cell line produced by transfection of MCF10A with constitutively active HRAS.[29] MCF10ADCIS.COM is a cell line cloned from cell culture of a MCF10AT1 xenograft lesion that reproducibly forms DCIS-like comedo lesions.[30] MCF10ACA1 is the most aggressive cell line and readily form tumors that metastasize.[31] We processed these four breast cell types, and each cell line assessment is presented in Figure 3. The deformability median (Scell) values gradually increase (P <0.001), from MCF10A (Scell = 1.386, N = 3,534), MCF10AT1 (Scell = 1.473, N = 6,983), MCF10ADCIS.COM (Scell = 1.585, N = 8,176), to MCF10ACA1 (Scell = 1.669, N = 4,826), which agrees well with the reported breast cancer malignancy tendency.[32] This result clearly demonstrates the presented iMSC is sensitive enough to discriminate breast cancer deformability changes upon the progression.
2.4 Mechanical characterization of EMT cell lines
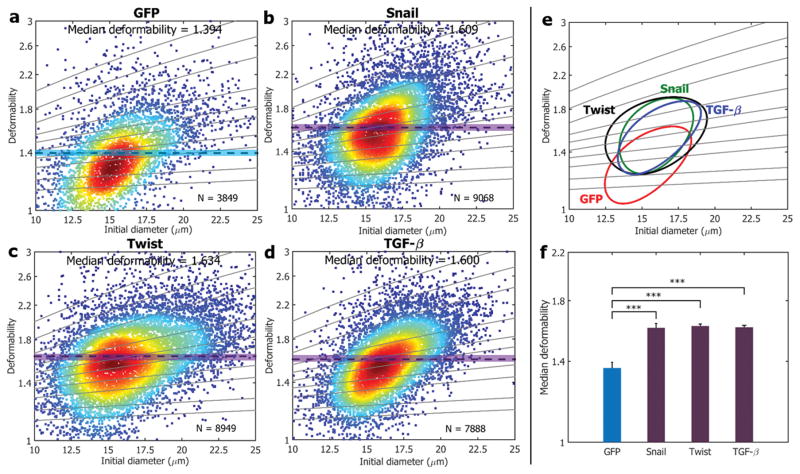
The use of our system can be expanded to identify other cell stiffness change identification, and we targeted the epithelial–mesenchymal transition (EMT) process. The aberrant EMT is a key developmental program endowing cancer cells with capabilities of invasion and metastasis.[33] EMT cells exhibit increased motility and invasiveness by losing cell-to-cell adhesion and cell polarity similar to mesenchymal traits.[34] There is significant evidence suggesting that EMT and mesenchymal-related gene expression are implicated in changes to cell mechanical properties,[35] and we hypothesize that our platform is able to capture the difference between non-EMT and EMT-like cell lines. We employed human mammary epithelial (HMLE) cells and modified HMLE cell lines with the following transcription factors (TFs) known to promote EMT: Snail, Twist and TGF-β.[33] As presented in Figure 4, we tested four cell types: HMLE (GFP tagged control; Scell = 1.394, N = 3,849) and HMLE modified with TFs of Snail (Scell=1.609, N = 9,068), Twist (Scell = 1.634, N = 8,949) and TGF-β (Scell = 1.600, N = 7,888). As indicated, HMLE (control; Figure 4a) are significantly stiffer than cells with EMT traits (Figure 4f; P <0.001).
Figure 4. Mechanical phenotyping of EMT cell lines.
Cell deformability is measured for a) HMLEs that undergo EMT induced by TFs of b) Snail, c) Twist and d) TGF-β. Solid lines are iso-shear modulus lines from the numerical analysis. e) 50%-density contour plots from a–d). f) A comparison of the median deformability values from a–d), confirming EMT cell lines acquire compliant phenotype. *** indicates a P value of less than 0.001, and error bars represent standard deviations (N = 3).
2.5 Rapid cancer drug discovery
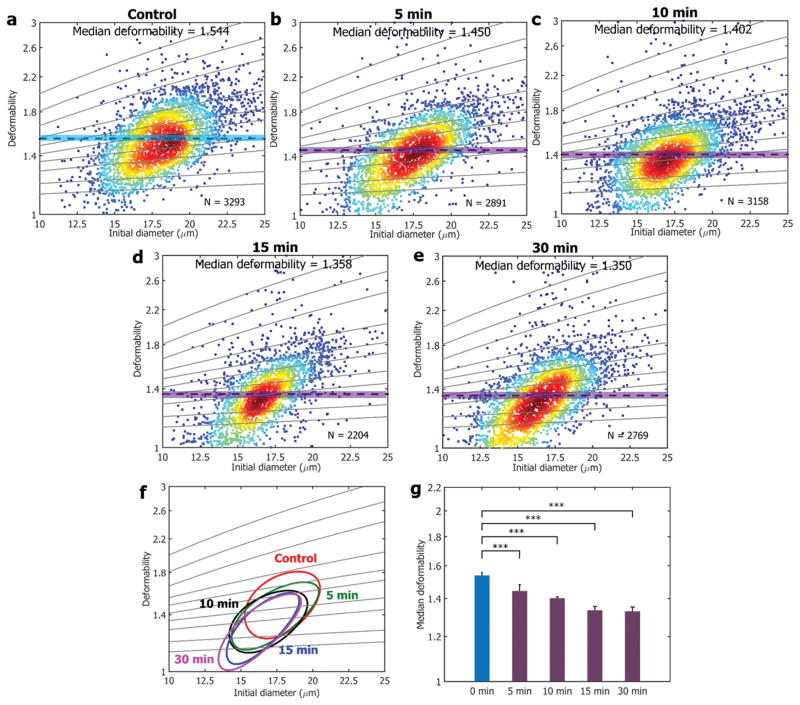
Cells that are sensitive to anticancer drugs are expected to display discernible differences in mechanical properties pre-and post-treatment.[1] Drugs that modify cytoskeletal or nuclear architecture are often applied in cancer treatment,[8, 36] and recently, 4-hydroxyacetophenone (4-HAP) was found to alter pancreatic cancer mechanical properties by stimulating myosin-II and enhancing its cortical localization, which reduces the invasion and migration of pancreatic cancer cells by stiffening them.[8] Micropipette aspiration was used for the study measuring the cell deformability to find the efficacy of 4-HAP; however, due to its low throughput, only tens of cells were characterized to verify the drug responses. Furthermore, it was suggested that 4-HAP can be applied to other cancers, though with a micropipette aspiration approach, it is limited to test other various cancer cell lines at different stages. To address the low throughput issue and test the possibility of 4-HAP for other cancers, we processed MDA-MB-231 cells treated with 4-HAP (see Experimental section) using our system. As presented in Figure 5, the deformability of MDA-MB-231 decreased substantially after 4-HAP treatment. We measured more than 14,000 cells to evaluate the efficacy of 4-HAP, and showed a great possibility of 4-HAP as a potential drug for breast cancer as well. We also measured the cell stiffness changes as a function of treatment time with 4-HAP (Figure 5g) showing how time dependent cell responses can also be measured through our platform.
Figure 5. Time-dependent mechanical responses of MDA-MB-231 cells upon drug treatment of 4-HAP.
a–e) Time-dependent cell deformability changes are presented upon the addition of 4-HAP to MDA-MB-231, showing gradual decrease in the median deformability. Solid lines are iso-shear modulus lines from the numerical analysis. f) 50%-density contour plots from a–e). g) A comparison of median deformability of control a) and all drug treated samples b–e). *** indicates a P value of less than 0.001, and error bars represent standard deviations (N = 3).
2.6 Origin of cell deformability
We measure whole-cell responses as a reflection of underlying cellular molecular changes. It is reported that cytosolic cytoskeletal components including actin, intermediate filaments, and microtubules are not mainly responsible for cell deformability changes when cells undergo large deformation;[15] therefore, it still remains to be seen what is the determinant of cellular mechanical responses. Pajerowski et al. [4] found that the extent of human embryonic stem cells deformability can be modulated by the nucleo-skeletal component of Lamin A and C (A/C). As suggested, we investigated the effect of Lamin A/C perturbations on our breast cancer cell lines using our system. We knocked down Lamin A/C (~90%) of MCF7 cell lines via plasmid-based small hairpin RNA (shRNA) mediated interference (more details on the Experimental section and Figure S7), and measured the cell deformability changes upon the presence of the lamina. As shown in Figure 6a and c, with the loss of Lamin A/C, there is a considerable increase in cell deformability, suggesting that manipulation of the nuclear envelop from the lamina accounts for the major cell deformability increase. For our negative control, we used an shRNA targeting luciferase (shLUC), and as presented in Figure 6b, a negligible difference in median deformability values was shown. Moreover, we measured the cell deformability dependence on LaminA/C for a non-tumorigenic mammary epithelial cell line (MCF10A), and similar trends were found (Figure S7) where we postulated that still nuclear contents are attributed as a major determinant for cell deformation read-outs.
3. Discussion
We report a fully automated, high-throughput, near real-time and label-free single-cell mechanotyping platform. Compared to the state-of-the-arts, our system has the following fundamental advantages: (1) Simple operation: A single-command based fully automated process is demonstrated from sample injection, image acquisition and image analysis to results visualization. This unique feature should be highlighted since it allows for an easy user friendly operation, minimized sample handling steps/time for preserving sample integrity, and cost-reduction from not requiring a skilled technician; (2) High-throughput and high-speed analysis: Inertia based approaches inherently offers a rapid cell sample processing capability. Along with our developed high-speed image analysis algorithms, we realize actual high-throughput (~450 cells sec−1) and high-speed (100,000 frames sec−1) cell stiffness analysis, granting a high statistical significance. It should be noted that the reported analysis rate can go up to 1,000 cell sec−1 theoretically, but 100% CPU memory usage causes a system delay (a few seconds) due to interrupt and thread switching latency; (3) High sensitivity: high fluid momentum compresses the cell significantly (>1 μN), enabling high cell strain measurements not easily found in other approaches. High degree of cell deformation is crucial for rigid cell stiffness measurements, and multiple levels of strain rates can be recorded by tuning the flow rate. As our next step, we are currently investigating two cell deformation regimes where deformability is dominated by the cytoskeleton at low strains and the nucleus at higher strains (data not shown); (4) Low sample volume: Inertial approaches normally consume large sample volumes; in contrast, here, a low volume operation (<500 μL) is realized with the programmable pressure-driven flow control, opening high potential for rare sample analysis; (5) Offline and downstream cell manipulation capability: Cells processed with the reported platform can be collected and potentially small subpopulations (e.g., drug resistant cells) can be separated downstream as our previous work[37] but based on their deformability phenotype not by fluorescence which will allow for various deformability based research; (6) Robustness: Due to the full system automation, we eliminate any potential user bias. Consequently, our system returns consistent measurements (Figure S8), and this repeatability is an important feature of the system, increasing analysis accuracy for reliable sample comparison across laboratories and process standardization.
Through our platform, we calculate the cell deformability, a non-dimensional parameter to analyze cell responses. In our plot, the cell distribution especially deformability values are simple enough to be understood similar to general normalized fluorescence intensity measurements with arbitrary units. However, it should be noted that the deformability values are not a random arbitrary numbers. Through our numerical investigations, we show that a cellular mechanical property, the cell shear modulus, can be extracted from the experimentally measured deformability and cell diameter information, opening a possibility of our platform to be used for biophysical studies.
Various samples were processed via our platform, and each examination finds the median cell deformability (other statistical information can be also extracted as illustrated in Figure S9). For breast cancer cell lines, we demonstrated that based on median deformability, cancer states upon malignancy were able be determined. This ability shows great potential that our system could be implemented clinically for rapid cancer diagnosis by processing unknown patient samples. For example, ascites[16] or pleural effusion samples[17] can be directly collected from patients and processed through iMCS where immediate cell deformability results can be obtained, which could potentially guide physicians in a timely manner. Furthermore, we expect to differentiate cancer subtypes as well. Recently, Chang et al.,[38] reported that subtypes of breast cancer display discernable mechanical stiffness which suggests label-free breast cancer subtype identification is possible (data not shown).
Lastly, in this study, we used the system for rapid drug discovery. We postulate that its utility can be expanded for rapid drug screening and personalized medicine. Instead of waiting for days to obtain the efficacy of drug treatment on specific samples at different conditions, our platform offers rapid cell response characterization under various external stimuli (not limited to chemical stimulation), providing a possibility for processing a large number of samples for various drug testing applications.
4. Experimental Section
Microfluidic device fabrication and dimensions
Microfluidic channels were created from Poly(dimethylsiloxane) (PDMS) (Dow Corning Corporation) using standard soft lithography techniques. Replicas were made by pouring PDMS over SU-8 (Microchem Corporation) masters, which were fabricated using standard double-step photolithography.[39] Microchannels were sealed with glass substrates (Fisher Scientific) after being treated by a plasma cleaner (Harrick Plasma) and then incubated overnight in a convection oven (VWR) at 65°C. The microchannel has a total length of 3.5 cm with 80 μm in width and 40 μm in height. It consists of 30 vertical constrictions (20 μm in length, 20 μm in height, and 1 mm in periodicity), followed by an expanded chamber structure (160 μm in width and 800 μm in length with an expanding slope of 2.86 degrees). Before the T-junction, the width of the channel decreased to 40 μm, and the tapering region is 120 μm in length.
Device operation
Cells in suspension were pumped into microchannel at 450 μL min−1 by using a pressure control system (Elveflow). Flowing cells were imaged using a 10× objective on a Zeiss Axio Observer A1 inverted microscope (Carl Zeiss), and cell motions were recorded using a Phantom v2512 high-speed camera (Vision Research) with a frame rate of 100,000 frames sec−1 and an exposure time of 1 μs. All sample comparisons and determinations of statistical significance were performed using the Wilcoxon rank sum test in Matlab.
Microparticle specifications
For particle focusing efficiency tests, 2.9, 6.0, 9.9 and 13 μm polystyrene microspheres with coefficients of variation (CV) of 5%, 12%, 5% and 16%, respectively, were used. All microparticles were purchased from Thermo Fisher Scientific except for 6 μm (Phosphorex).
Cell sample preparation
MCF10A, MCF10AT1, MCF10ADCIS.com and MCF10A-CA1 cells (Karmanos Cancer Institute) were cultured using MCF10A media (DMEM, 5% Horse Serum, 20 ng mL−1 EGF, 0.5 mg mL−1 Hydrocortisone, 100 ng mL−1 Cholera Toxin, 10 μg mL−1 Insulin, antibiotic-antimycotic). For EMT cell lines, HMLE-GFP, HMLE-Twist, HMLE-Snail, HMLE-TGF-β (gift from Sendurai Mani[33]) were cultured as previously described.[33] MDA-MB-231 cells were cultured using standard protocols using DMEM (Life Technologies) with 10% fetal bovine serum (Life Technologies) and 1% penicillin-streptomycin (Life Technologies). In order to establish a baseline level of cell deformability before cells enter the fluidic device, we measured the cell deformability (i.e., roundness) in cell suspension. As shown in Figure S10, more than 90% of the cell population shows deformability less than 1.1 (N >1,000), suggesting cells stay close to spherical. To evaluate the change of deformability during the drug treatment, 0.5 μM 4-HAP was added to cell suspension. Before testing, the 4-HAP added media was replaced with PBS. For Lamin A/C knockdown, oligonucleotides coding for short hairpin RNA (shRNA) against Lamin A/C were cloned into the pLKO.1 TRC plasmid. The sequence targeting Lamin A/C is 5′ GAAGCAACTTCAGGATGAGAT 3′. MCF7 and MCF10A cells were cultured under standard protocols. Lentiviral transduced cells were selected with the antibiotic puromycin (1 μg mL−1 of media for MCF7 and 0.5 μg mL−1 of media for MCF10A). Cell pellets were collected for qRT-PCR, immunofluorescence, and western blot analysis.
5. Numerical Simulation
For the three-dimensional far-field simulation, images from the video capture of the cell were segmented to determine the in-plane shape of the cell at a given time. It was assumed that the cell retained its axisymmetric shape corresponding to x-z plane at the center of microchannel (Figure 1b). The velocity of the cell-center at a given instant was determined as the ratio of distance traveled by the cell in between two successive time frames to the time interval. This provided a three-dimensional model of the motion of the cell within the microchannel as it approached the wall. This model was discretized using a stabilized finite element method, and the NS equations were solved in the fluid surrounding the cell. The motion and large-scale deformation of the cell meant that the mesh around the cell had to be updated. This was done by moving the mesh with the cell using the ALE approach at every time-step, and by re-meshing when the elements within the fluid were distorted. As the cell approached the wall, the number of adaptation steps were increased in order to model the small region between the cell and the wall.
The near-field simulations were performed using the three-dimensional finite-element model using Abaqus based on the experimental conditions. The model couples the fluid-structure interaction between the solid domain (cell) and the fluid domain. For the solid domain, the momentum equations are solved in Abaqus/Explicit with an incompressible neo-Hookean constitutive model: U = C10(I1 − 3), where U is the strain energy per unit of reference volume, C10 is a neo-Hookean parameter characterizing the nonlinear material property (C10 = Go/2; Go is the bulk shear modulus), and I1 is a deformation invariant. By estimating a cell shear modulus, we numerically calculated the stress and strain relationship using the following equation with the pressure information from NS equations. , where σij is the stress tensor, Bij is the left Cauchy Green deformation tensor, δij is the Kronecker delta, and p is hydrostatic stress. Since the cell can never impact a perfectly smooth wall due to the divergence of lubrication force, and since the surfaces of the wall are not perfectly smooth in the experimental situation,[40] the cell contacts the wall 1 μm before the stagnation point in the simulation. To save computational cost, the symmetry boundary condition is applied in the x-z plane at the center of microchannel (Figure 1b). A comparison between numerical and experimental results for a cell with an initial diameter of 16 μm is presented in Figure S11.
Supplementary Material
Figure S1. Focusing efficiency of our system. Focusing efficiency characterization using various polystyrene microsphere sizes and MDA-MB-231 breast cancer cells at different flow rates. Larger microspheres reached near 100% focusing efficiency (the ratio of number of particles in the equilibrium position to total particle number) at higher flow rates (Re >100), and we found similar trends were found for cells (higher cell focusing efficiency can be expected with a further channel geometry optimization). Error bars represent standard deviation from the mean (N = 3).
Figure S2. Cell spacing control of our system. Histograms show how the space between closely positioned cells can be enlarged after passing an expansion and contraction chamber through viscous repulsive interactions.[1]
Figure S3. Valid cell count dependence on cell sample concentration. Low concentration exhibits reduced valid cell count since fewer cells were captured in image stacks. In contrary, for overly concentrated samples, multiple cells collide with the wall concurrently, decreasing the total valid cell count. Error bars represent standard deviation from the mean (N = 3).
Figure S4. Cell (MDA-MB-231) viability a) Control cells, and b) cells collected after processing with our platform at Re = 125 were cultured up to day 5 where the two groups show comparable cell proliferation. Scale bar represents 50 μm.
Figure S5. Image analysis. An example of image processing of a cell for initial cell diameter and deformability determination. For cell initial diameter characterization, adaptive thresholding[2] together with morphological operations are applied.[3] The area of the cell’s binary images is first calculated, and the equivalent diameter is obtained using the equation; where Dcell is the equivalent diameter of cells, and Acell is the area of cells in the binary images. For cell deformability, each cell’s boundary is found and the deformability is calculated as , where Scell is the deformability of cells, Lmajor is the major axis length of the deformed cells and Lminor is the minor axis length. Scale bar represents 50 μm.
Figure S6. Image analysis code validation. Each dot represents an error between code analysis and manual measurements in a) cell diameter and b) cell deformability, showing a good agreement.
Figure S7. Cell mechanotyping results of Lamin A/C knockdown. Cell deformability is measured for the following cells: a) MCF10A (control) transfected with an shRNA targeting b) luciferase (denoted as shLUC; negative control) and c) Lamin A/C (denoted as shLMNA). It shows that Lamin A/C knockdown has decreased the stiffness of cell lines. Solid lines are iso-shear modulus lines from the numerical analysis. d) 50%-density contour plots for all samples. e) A comparison of median deformability from all samples. *** indicates a P value (statistical significance) of less than 0.001, and n.s. represents statistical non-significance (P >0.05). All error bars indicate standard deviations (N = 3). Results validating the Lamin A/C knockdown processes are presented via f) qRT-PCR, g) Western blot (GAPDH as a positive control), and h) Immunofluorescence.
Figure S8. Repeatability test of our platform. MCF10A cells in different culture flasks splitted from the same mother cell flask were tested to show the robustness of the system (P >0.05). Solid lines are iso-shear modulus lines from the numerical analysis.
Figure S9. Additional statistical analysis beyond the typical density scatter plot of MCF7 cells. a) Histogram showing the distribution of initial diameter; b) Histogram showing the deformability distribution; c) A typical density scatter plot of MCF7 cells. Solid lines (gray) are iso-shear modulus lines from the numerical analysis. Solid box markers are the median deformability of cells when binned with 1 μm range of initial diameter. Dashed line (black) is the linear fitting of the binned median deformability data; d) Distribution of deformability when binned with 2.5 μm range of initial cell diameter.
Figure S10. Distribution of roundness of un-deformed cells. Baseline level of deformability for a population of cells before entering the device.
Figure S11. Comparison between numerical and experimental analysis for a cell with an initial diameter of 16 μm. Near-field simulation and experimental results are presented at the maximum cell deformation states with different shear moduli (G0) and strain. Color bar represents the stress distributions (MPa). All scale bars represent 20 μm.
Table S1. Comparison of microfluidics based deformability cytometers. Dashed lines in the cell focusing profile column represents the image focal planes.
Movie S1. MDA-MB-231 cell focusing. At Re ≈ 125, inertia passively focuses randomly-aligned incoming cells to a single-stream.
Movie S2. MDA-MB-231 cell deformation. At Re ≈ 125, a stream of single-cell chain formed by inertial effects enters and deforms with large strain upon collision at a T-junction. Our system measures the initial cell diameter and cell deformability (the ratio between two axes at the maximum deformation state) from the recorded image stacks close to real-time.
Movie S3. Fully automated system demonstration. With a single-start command, the entire process runs from sample loading, flow control, image acquisition, analysis to results visualization without any human intervention.
Movie S4. Far field 3D FEM simulation of cell deformation. The simulation movie shows pressure changes around a cell as it approaches to the channel wall.
Movie S5. Near-field 3D FEM simulation of cell deformation. The 3D nearfield simulations are represented for the velocity field and stress distribution during cell deformation process.
Acknowledgments
This work was supported by Rensselaer Polytechnic Institute (RPI), NSF-1444104, DoD-W81XWH-15-1-0495 and NIH-R00CA166815. Device fabrication for this work was performed in part at the Micro and Nano Fabrication Clean Room (MNCR) at RPI.
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Yanxiang Deng, Department of Mechanical, Aerospace, and Nuclear Engineering, Rensselaer Polytechnic Institute (RPI), 110 8th Street, Troy, NY 12180, USA.
Steven P. Davis, Department of Biomedical Sciences, Cancer Research Center, University at Albany, State University of New York, Rensselaer, NY 12144, USA
Fan Yang, Department of Mechanical, Aerospace, and Nuclear Engineering, Rensselaer Polytechnic Institute (RPI), 110 8th Street, Troy, NY 12180, USA.
Kevin S. Paulsen, Department of Mechanical, Aerospace, and Nuclear Engineering, Rensselaer Polytechnic Institute (RPI), 110 8th Street, Troy, NY 12180, USA
Maneesh Kumar, Department of Biomedical Sciences, Cancer Research Center, University at Albany, State University of New York, Rensselaer, NY 12144, USA.
Rebecca Sinnott DeVaux, Department of Biomedical Sciences, Cancer Research Center, University at Albany, State University of New York, Rensselaer, NY 12144, USA.
Xianhui Wang, Department of Biomedical Sciences, Cancer Research Center, University at Albany, State University of New York, Rensselaer, NY 12144, USA.
Douglas S. Conklin, Department of Biomedical Sciences, Cancer Research Center, University at Albany, State University of New York, Rensselaer, NY 12144, USA
Prof. Assad Oberai, Department of Mechanical, Aerospace, and Nuclear Engineering, Rensselaer Polytechnic Institute (RPI), 110 8th Street, Troy, NY 12180, USA
Jason I. Herschkowitz, Department of Biomedical Sciences, Cancer Research Center, University at Albany, State University of New York, Rensselaer, NY 12144, USA
Prof. Aram J. Chung, Department of Mechanical, Aerospace, and Nuclear Engineering, Rensselaer Polytechnic Institute (RPI), 110 8th Street, Troy, NY 12180, USA
References
- 1.a) Darling EM, Di Carlo D. Annu Rev Biomed Eng. 2014;17:1. doi: 10.1146/annurev-bioeng-071114-040545. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Di Carlo D. J Lab Autom. 2012;17:32. doi: 10.1177/2211068211431630. [DOI] [PubMed] [Google Scholar]
- 2.Suresh S. Acta Mater. 2007;55:3989. [Google Scholar]
- 3.Cranston HA, Boylan CW, Carroll GL, Sutera SP, Williamson JR, Gluzman IY, Krogstad DJ. Science. 1984;223:400. doi: 10.1126/science.6362007. [DOI] [PubMed] [Google Scholar]
- 4.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Proc Natl Acad Sci USA. 2007;104:15619. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Nguyen AV, Nyberg KD, Scott MB, Welsh AM, Nguyen AH, Wu N, Hohlbauch SV, Geisse NA, Gibb EA, Robertson AG, Rowat AC. Integr Biol. 2016;8:1232. doi: 10.1039/c6ib00135a. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Nyberg KD, Scott MB, Bruce SL, Gopinath AB, Bikos D, Mason TG, Kim JW, Choi HS, Rowat AC. Lab Chip. 2016;16:3330. doi: 10.1039/c6lc00169f. [DOI] [PubMed] [Google Scholar]; c) Qi D, Gill NK, Santiskulvong C, Sifuentes J, Dorigo O, Rao J, Taylor-Harding B, Wiedemeyer WR, Rowat AC. Sci Rep. 2015;5:17595. doi: 10.1038/srep17595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Remmerbach TW, Wottawah F, Dietrich J, Lincoln B, Wittekind C, Guck J. Cancer Res. 2009;69:1728. doi: 10.1158/0008-5472.CAN-08-4073. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez-Cruz RD, Fonseca VC, Darling EM. Proc Natl Acad Sci U S A. 2012;109:E1523. doi: 10.1073/pnas.1120349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Surcel A, Ng WP, West-Foyle H, Zhu Q, Ren Y, Avery LB, Krenc AK, Meyers DJ, Rock RS, Anders RA, Freel Meyers CL, Robinson DN. Proc Natl Acad Sci USA. 2015;112:1428. doi: 10.1073/pnas.1412592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costa KD. Dis Markers. 2003;19:139. doi: 10.1155/2004/482680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hochmuth RM. J Biomech. 2000;33:15. doi: 10.1016/s0021-9290(99)00175-x. [DOI] [PubMed] [Google Scholar]
- 11.Lee LM, Liu AP. J Nanotechnol Eng Med. 2014;5:0408011. doi: 10.1115/1.4029936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guck J, Schinkinger S, Lincoln B, Wottawah F, Ebert S, Romeyke M, Lenz D, Erickson HM, Ananthakrishnan R, Mitchell D, Kas J, Ulvick S, Bilby C. Biophys J. 2005;88:3689. doi: 10.1529/biophysj.104.045476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Nature. 2009;459:428. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Chung AJ, Hur SC. High-Speed Microfluidic Manipulation of Cells in Micro-and Nanomanipulation Tools. Wiley; 2015. [Google Scholar]; b) Mao X, Huang TJ. Lab Chip. 2012;12:4006. doi: 10.1039/c2lc90100e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gossett DR, Tse HT, Lee SA, Ying Y, Lindgren AG, Yang OO, Rao J, Clark AT, Di Carlo D. Proc Natl Acad Sci USA. 2012;109:7630. doi: 10.1073/pnas.1200107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peterson VM, Castro CM, Chung J, Miller NC, Ullal AV, Castano MD, Penson RT, Lee H, Birrer MJ, Weissleder R. Proc Natl Acad Sci USA. 2013;110:E4978. doi: 10.1073/pnas.1315370110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tse HT, Gossett DR, Moon YS, Masaeli M, Sohsman M, Ying Y, Mislick K, Adams RP, Rao J, Di Carlo D. Sci Transl Med. 2013;5:212ra163. doi: 10.1126/scitranslmed.3006559. [DOI] [PubMed] [Google Scholar]
- 18.Otto O, Rosendahl P, Mietke A, Golfier S, Herold C, Klaue D, Girardo S, Pagliara S, Ekpenyong A, Jacobi A, Wobus M, Topfner N, Keyser UF, Mansfeld J, Fischer-Friedrich E, Guck J. Nat Methods. 2015;12:199. doi: 10.1038/nmeth.3281. [DOI] [PubMed] [Google Scholar]
- 19.a) Chung AJ, Gossett DR, Di Carlo D. Small. 2013;9:685. doi: 10.1002/smll.201202413. [DOI] [PubMed] [Google Scholar]; b) Chung AJ, Pulido D, Oka JC, Amini H, Masaeli M, Di Carlo D. Lab Chip. 2013;13:2942. doi: 10.1039/c3lc41227j. [DOI] [PubMed] [Google Scholar]; c) Wu Z, Chen Y, Wang M, Chung AJ. Lab Chip. 2016;16:532. doi: 10.1039/c5lc01435b. [DOI] [PubMed] [Google Scholar]
- 20.Lee W, Amini H, Stone HA, Di Carlo D. Proc Natl Acad Sci USA. 2010;107:22413. doi: 10.1073/pnas.1010297107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradski G, Kaehler A. Learning OpenCV: Computer Vision with the OpenCV Library. O’Reilly Media; Newton, MA, USA: 2008. [Google Scholar]
- 22.Whiting CH, Jansen KE. Int J Numer Methods Fluids. 2001;35:93. [Google Scholar]
- 23.Dwight RP. Robust Mesh Deformation using the Linear Elasticity Equations. Springer; 2009. [Google Scholar]
- 24.Shephard M, Jansen K, Sahni O, Diachin L. J Phys: Conf Ser. 2007;78:012053. [Google Scholar]
- 25.Dao M, Lim CT, Suresh S. J Mech Phys Solids. 2003;51:2259. [Google Scholar]
- 26.a) Nguyen N, Shao Y, Wineman A, Fu J, Waas A. Math Biosci. 2016;277:77. doi: 10.1016/j.mbs.2016.03.015. [DOI] [PubMed] [Google Scholar]; b) Mietke A, Otto O, Girardo S, Rosendahl P, Taubenberger A, Golfier S, Ulbricht E, Aland S, Guck J, Fischer-Friedrich E. Biophys J. 2015;109:2023. doi: 10.1016/j.bpj.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Bartel K, Winzi M, Ulrich M, Koeberle A, Menche D, Werz O, Müller R, Guck J, Vollmar AM, von Schwarzenberg K. Oncotarget. 2016:5. doi: 10.18632/oncotarget.14339. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Xavier M, Rosendahl P, Herbig M, Kräter M, Spencer D, Bornhäuser M, Oreffo RO, Morgan H, Guck J, Otto O. Integr Biol. 2016;8:616. doi: 10.1039/c5ib00304k. [DOI] [PubMed] [Google Scholar]
- 27.a) Wirtz D, Konstantopoulos K, Searson PC. Nat Rev Cancer. 2011;11:512. doi: 10.1038/nrc3080. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Li Q, Lee G, Ong C, Lim C. Biochem Biophys Res Commun. 2008;374:609. doi: 10.1016/j.bbrc.2008.07.078. [DOI] [PubMed] [Google Scholar]
- 28.Marella NV, Malyavantham KS, Wang J, Matsui S-i, Liang P, Berezney R. Cancer Res. 2009;69:5946. doi: 10.1158/0008-5472.CAN-09-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dawson PJ, Wolman SR, Tait L, Heppner GH, Miller FR. Am J Pathol. 1996;148:313. [PMC free article] [PubMed] [Google Scholar]
- 30.Miller FR, Santner SJ, Tait L, Dawson PJ. J Natl Cancer Inst. 2000;92:1185a. doi: 10.1093/jnci/92.14.1185a. [DOI] [PubMed] [Google Scholar]
- 31.So JY, Lee HJ, Kramata P, Minden A, Suh N. Mol Cell Pharmacol. 2012;4:31. [PMC free article] [PubMed] [Google Scholar]
- 32.Kadota M, Yang HH, Gomez B, Sato M, Clifford RJ, Meerzaman D, Dunn BK, Wakefield LM, Lee MP. PLoS One. 2010;5:e9201. doi: 10.1371/journal.pone.0009201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. Cell. 2008;133:704. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hollier BG, Evans K, Mani SA. J Mammary Gland Biol Neoplasia. 2009;14:29. doi: 10.1007/s10911-009-9110-3. [DOI] [PubMed] [Google Scholar]
- 35.Gjorevski N, Boghaert E, Nelson CM. Cancer Microenviron. 2012;5:29. doi: 10.1007/s12307-011-0076-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kavallaris M. Nat Rev Cancer. 2010;10:194. doi: 10.1038/nrc2803. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y, Chung AJ, Wu TH, Teitell MA, Di Carlo D, Chiou PY. Small. 2014;10:1746. doi: 10.1002/smll.201302885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang JM, Park IA, Lee SH, Kim WH, Bae MS, Koo HR, Yi A, Kim SJ, Cho N, Moon WK. Eur Radiol. 2013;23:2450. doi: 10.1007/s00330-013-2866-2. [DOI] [PubMed] [Google Scholar]
- 39.McDonald JC, Whitesides GM. Acc Chem Res. 2002;35:491. doi: 10.1021/ar010110q. [DOI] [PubMed] [Google Scholar]
- 40.Vigolo D, Griffiths IM, Radl S, Stone HA. J Fluid Mech. 2013;727:236. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Focusing efficiency of our system. Focusing efficiency characterization using various polystyrene microsphere sizes and MDA-MB-231 breast cancer cells at different flow rates. Larger microspheres reached near 100% focusing efficiency (the ratio of number of particles in the equilibrium position to total particle number) at higher flow rates (Re >100), and we found similar trends were found for cells (higher cell focusing efficiency can be expected with a further channel geometry optimization). Error bars represent standard deviation from the mean (N = 3).
Figure S2. Cell spacing control of our system. Histograms show how the space between closely positioned cells can be enlarged after passing an expansion and contraction chamber through viscous repulsive interactions.[1]
Figure S3. Valid cell count dependence on cell sample concentration. Low concentration exhibits reduced valid cell count since fewer cells were captured in image stacks. In contrary, for overly concentrated samples, multiple cells collide with the wall concurrently, decreasing the total valid cell count. Error bars represent standard deviation from the mean (N = 3).
Figure S4. Cell (MDA-MB-231) viability a) Control cells, and b) cells collected after processing with our platform at Re = 125 were cultured up to day 5 where the two groups show comparable cell proliferation. Scale bar represents 50 μm.
Figure S5. Image analysis. An example of image processing of a cell for initial cell diameter and deformability determination. For cell initial diameter characterization, adaptive thresholding[2] together with morphological operations are applied.[3] The area of the cell’s binary images is first calculated, and the equivalent diameter is obtained using the equation; where Dcell is the equivalent diameter of cells, and Acell is the area of cells in the binary images. For cell deformability, each cell’s boundary is found and the deformability is calculated as , where Scell is the deformability of cells, Lmajor is the major axis length of the deformed cells and Lminor is the minor axis length. Scale bar represents 50 μm.
Figure S6. Image analysis code validation. Each dot represents an error between code analysis and manual measurements in a) cell diameter and b) cell deformability, showing a good agreement.
Figure S7. Cell mechanotyping results of Lamin A/C knockdown. Cell deformability is measured for the following cells: a) MCF10A (control) transfected with an shRNA targeting b) luciferase (denoted as shLUC; negative control) and c) Lamin A/C (denoted as shLMNA). It shows that Lamin A/C knockdown has decreased the stiffness of cell lines. Solid lines are iso-shear modulus lines from the numerical analysis. d) 50%-density contour plots for all samples. e) A comparison of median deformability from all samples. *** indicates a P value (statistical significance) of less than 0.001, and n.s. represents statistical non-significance (P >0.05). All error bars indicate standard deviations (N = 3). Results validating the Lamin A/C knockdown processes are presented via f) qRT-PCR, g) Western blot (GAPDH as a positive control), and h) Immunofluorescence.
Figure S8. Repeatability test of our platform. MCF10A cells in different culture flasks splitted from the same mother cell flask were tested to show the robustness of the system (P >0.05). Solid lines are iso-shear modulus lines from the numerical analysis.
Figure S9. Additional statistical analysis beyond the typical density scatter plot of MCF7 cells. a) Histogram showing the distribution of initial diameter; b) Histogram showing the deformability distribution; c) A typical density scatter plot of MCF7 cells. Solid lines (gray) are iso-shear modulus lines from the numerical analysis. Solid box markers are the median deformability of cells when binned with 1 μm range of initial diameter. Dashed line (black) is the linear fitting of the binned median deformability data; d) Distribution of deformability when binned with 2.5 μm range of initial cell diameter.
Figure S10. Distribution of roundness of un-deformed cells. Baseline level of deformability for a population of cells before entering the device.
Figure S11. Comparison between numerical and experimental analysis for a cell with an initial diameter of 16 μm. Near-field simulation and experimental results are presented at the maximum cell deformation states with different shear moduli (G0) and strain. Color bar represents the stress distributions (MPa). All scale bars represent 20 μm.
Table S1. Comparison of microfluidics based deformability cytometers. Dashed lines in the cell focusing profile column represents the image focal planes.
Movie S1. MDA-MB-231 cell focusing. At Re ≈ 125, inertia passively focuses randomly-aligned incoming cells to a single-stream.
Movie S2. MDA-MB-231 cell deformation. At Re ≈ 125, a stream of single-cell chain formed by inertial effects enters and deforms with large strain upon collision at a T-junction. Our system measures the initial cell diameter and cell deformability (the ratio between two axes at the maximum deformation state) from the recorded image stacks close to real-time.
Movie S3. Fully automated system demonstration. With a single-start command, the entire process runs from sample loading, flow control, image acquisition, analysis to results visualization without any human intervention.
Movie S4. Far field 3D FEM simulation of cell deformation. The simulation movie shows pressure changes around a cell as it approaches to the channel wall.
Movie S5. Near-field 3D FEM simulation of cell deformation. The 3D nearfield simulations are represented for the velocity field and stress distribution during cell deformation process.