Abstract
The popularization of genome-wide analyses and RNA sequencing led to the discovery that a large part of the human genome, while effectively transcribed, does not encode proteins. Long non-coding RNAs have emerged as critical regulators of gene expression in both normal and disease states. Studies of long non-coding RNAs expressed in the heart, in combination with gene association studies, revealed that these molecules are regulated during cardiovascular development and disease. Some long non-coding RNAs have been functionally implicated in cardiac pathophysiology and constitute potential therapeutic targets. Here, we review the current knowledge of the function of long non-coding RNAs in the cardiovascular system, with an emphasis on cardiovascular development and biology, focusing on hypertension, coronary artery disease, myocardial infarction, ischemia, and heart failure. We discuss potential therapeutic implications and the challenges of long non-coding RNA research, with directions for future research and translational focus.
Keywords: transcriptomics, RNAs, long non-coding RNAs, cardiovascular system, cardiovascular development, cardiovascular disease, vascular disease, therapy, non-coding RNAs
Main Text
New sequencing technologies, combined with bioinformatics and computational tools, have allowed the scientific community to appreciate the great complexity of the transcriptome.1 In particular, the discovery of various types of non-protein coding RNAs (ncRNAs) and their different functions in regulating developmental and disease processes is expanding our knowledge of molecular biology and could significantly advance therapeutic options for many patients, including those suffering from cardiovascular disease. Thousands of ncRNAs have been described and classified into two large groups: small ncRNAs, which are up to 200 nucleotides long, and long non-coding RNAs (lncRNAs), which are longer than 200 nucleotides.
lncRNAs are a heterogeneous group of transcripts exerting major regulatory roles in gene expression, and their importance in cardiovascular disease has been reinforced.2, 3 The dynamic expression and specific profiles of lncRNAs in different pathophysiological states suggest their functional relevance and potential to be used as non-invasive markers of disease and therapeutic targets.4, 5 However, establishing the biological actions of each lncRNA is proving more complex than investigating microRNAs (miRNAs, the most popular class of small ncRNAs within the biomedical community). This is due to lncRNAs’ multiple modalities of action and their low conservation among vertebrates.6 Therefore, a large gap remains between the number of lncRNAs identified, and then listed in databases, and their functional characterization and implications in pathophysiological situations. A list of databases and their major characteristics, such as number of lncRNAs, species, and association with function and other genes, is given in Table 1.
Table 1.
List of Available Databases on lncRNAs
| Database | What | Species | No. of lncRNAs | Last Update | Association with Function | Association with Protein-Coding RNAs | Association with miRNAs | Reference |
|---|---|---|---|---|---|---|---|---|
| ANGIOGENES | in silico screening of protein-coding genes and lncRNAs in ECs | human, mouse, zebrafish | 24,382 (15,149 in human) | 2016 | X | 164 | ||
| ChIPBase | transcriptional regulation of lncRNA from ChIP-seq data | 10 | 10,200 ChIP-seq datasets | 2016 | X | 165, 166 | ||
| deepBase | identification, annotation, and function prediction of lncRNAs from RNA-seq data | 14 | 191,547 | 2016 | X | 167, 168 | ||
| GENCODE | manually curated human and mouse lncRNA reference based on ENCODE project | human, mouse | 42,302 | 2016 | 169, 170 | |||
| LincSNP | annotated disease-associated SNPs in human lncRNAs | human | 244,545 | 2016 | X | 171 | ||
| LNCipedia | annotated lncRNA sequences, structures, protein coding potential, and miRNA binding sites | human | 118,777 | 2016 | X | X | 172, 173 | |
| lncRNAdb | curated reference database of functionally annotated eukaryotic lncRNAs | 71 | 295 (183 in human) | 2015 | X | 174, 175 | ||
| LncRNADisease | experimentally supported and predicted associations between lncRNAs and diseases | human | 1,564 | 2015 | X | 176 | ||
| lncRNome | annotated human lncRNAs | human | 17,547 | 2013 | X | X | X | 177 |
| NONCODE | integrated annotation of ncRNAs, especially lncRNAs | 16 | 487,164 (167,150 in human) | 2016 | X | X | 178 |
ChIP-seq, chromatin immunoprecipitation sequencing.
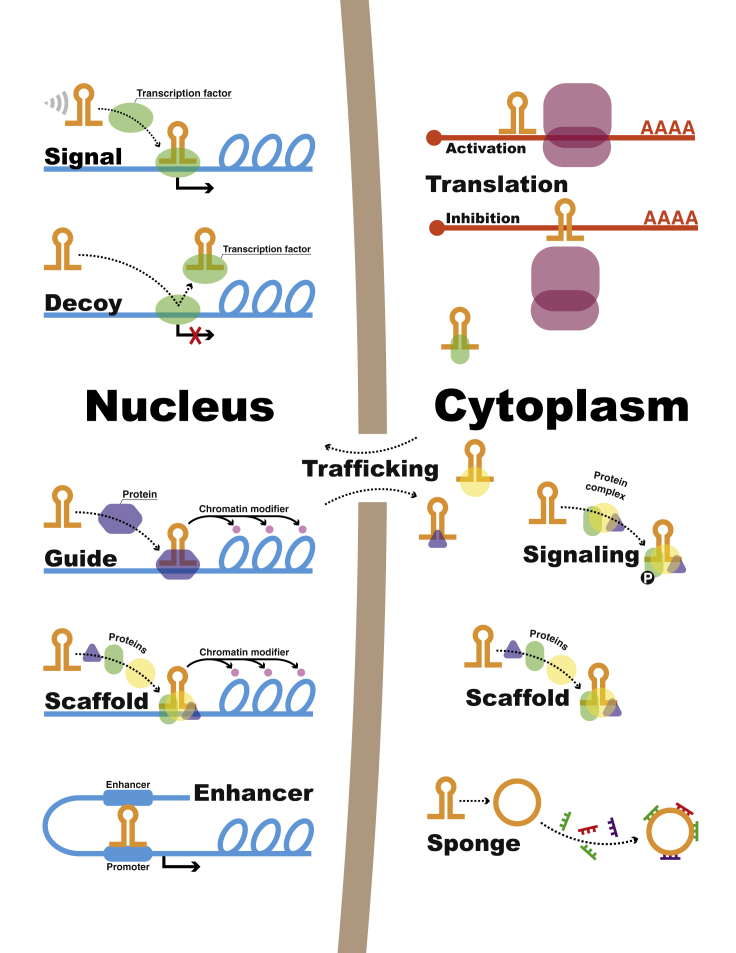
So far, it has been demonstrated that lncRNAs can regulate gene expression through functional mechanisms including epigenetic, transcriptional, and post-transcriptional, either activating or suppressing gene expression. lncRNAs can also mediate signaling, such as phosphorylation, and trafficking of proteins.7, 8 One way to classify lncRNAs is according to their mechanism of action: signal, decoy, guide, scaffold,9 enhancer, or sponge lncRNAs (particularly circular lncRNAs [circRNAs]) (Figure 1).10, 11 In this review, we do not address in detail the lncRNAs mechanisms of action but instead refer to reviews on the subject.2, 12, 13, 14
Figure 1.
Classification of lncRNAs by Mechanism of Action
Signal lncRNAs respond to specific stimuli and thus show expression specific to cell type. Decoys bind transcription factors and other proteins away from their target site, repressing transcription. Guides interact with regulatory proteins, forming ribonucleoprotein complexes, and direct them to their target sites in subcellular locations. Scaffolds serve as platforms to bring different proteins together, both in the cytoplasm and in the nucleus, activating or repressing transcription. Enhancers are regulatory sequences in which transcription factors bind to initiate transcription; these regions of the genome produce several transcripts, enhancer lncRNAs, which act in cis to regulate expression of target genes. In the cytoplasm, lncRNAs can activate or inhibit translation by binding to target mRNAs. They can also regulate protein trafficking and signaling, such as phosphorylation. Sponging miRNAs is another way lncRNAs (including circular RNAs) regulate gene expression post-transcriptionally.
A growing number of lncRNAs are implicated in cardiovascular development and disease, although it is not clearly understood how they participate in pathological processes. Their potential as therapeutic targets has often been raised, and there are a few examples of in vivo modulation of lncRNAs. However, modulating lncRNAs has been a challenging task to date.15, 16, 17, 18, 19 Here, we summarize the current understanding of lncRNA function in cardiovascular pathophysiology and discuss its potential for therapy.
Function of lncRNAs in the Cardiovascular System
Cardiac Development and Biology
Transcriptomics profiling and loss-of-function approaches in progenitor and embryonic stem cells (ESCs) have demonstrated the importance of lncRNAs for cardiac development and cell differentiation. More than 1,000 lncRNAs were reported as being dynamically regulated during differentiation,20, 21 and further transcriptome analyses of embryonic and adult-stage murine hearts identified several lncRNAs specific to tissue and developmental stage.22, 23
Among biologically validated lncRNAs, several have been associated with cardiac development (Table 2). For example, Braveheart (Bvht) has a critical role in cardiac lineage commitment in mouse. It is abundantly expressed in embryonic stem cells and regulates the transition from nascent mesoderm to cardiac progenitor.24 Bvht, by modulating the core cardiovascular gene network and mediating the epigenetic regulation of cardiac fate, is necessary to maintain cardiac commitment.25 Conversely, the lateral mesoderm-specific lncRNA Fendrr (fetal-lethal non-coding developmental regulatory RNA) controls mesodermal differentiation, as well as heart and body wall development, by binding to the histone-remodeling polycomb repressive complex PRC2 and TrxG/MLL to modulate chromatin status.26
Table 2.
lcnRNAs Associated with Cardiovascular Biology
| lncRNA | Expression | Biological Context | Action | Genomic Localization | Organism | Reference |
|---|---|---|---|---|---|---|
| Differentiation and Cardiac Development | ||||||
| Bvht | embryonic stem cells | cardiomyocyte differentiation | signal | intergenic | mouse | 24, 25 |
| Fendrr | lateral plate mesoderm | development of heart and body wall | signal | intergenic | human, mouse, rat | 26 |
| Novlnc6 | embryonic stem cells (particularly left ventricle), cardiomyocytes | cardiac differentiation and maturation | decoy | human, mouse | 27 | |
| CARMEN | cardiac precursor cell | cardiomyocyte differentiation of cardiac precursor cells | enhancer | intergenic | human, mouse, rat | 28 |
| n411949 | embryonic heart | cardiac development | unknown | antisense | mouse | 22 |
| n413445 | embryonic heart | cardiac development | unknown | intronic | mouse | 22 |
| Contractile Function | ||||||
| KCNQ1OT1 | cardiac development | signal | antisense | human, mouse | 30 | |
| β-RNA | adult ventricles | contractile phenotype, pressure overload | unknown | antisense | rat | 33 |
| Vascular Development | ||||||
| SENCR | smooth muscle cells | maintenance of smooth muscle cells’ differentiated state | decoy | antisense | human | 37, 38 |
| SMILR | VSMCs | proliferation of smooth muscle cells | proposed scaffold or enhancer | intergenic | human | 39 |
| MALAT1 | ECs | proliferation of ECs and vascularization | decoy | intergenic | human, mouse | 17 |
| PUNISHER | ECs | identity of ECs | guide | antisense | human, mouse, zebrafish | 42 |
Numerous enhancer-associated lncRNAs have been implicated in cardiogenic differentiation,21, 27 among which the enhancer lncRNA Novlnc6 modulates expression of MKX2.5, a transcription factor critical for cardiac differentiation and maturation.27 CARMEN (cardiac mesoderm enhancer-associated non-coding RNA) is also responsible for cardiogenic specification and differentiation in precursor cells, possibly by regulating PRC2.28 Furthermore, several lncRNAs regulate specific mRNA abundance during heart development, although as yet there is no clear understanding of their role during cardiogenic differentiation. For example, n411949 regulates Mccc1 mRNA, which metabolizes leucine, and n413445 modulates ReIb, which is involved in the nuclear factor κB (NF-κB) pathway.22 Overall, fetal gene program reactivation constitutes a hallmark in multiple cardiovascular diseases. Although a moderate number of dynamically regulated lncRNAs in embryonic cells are equally regulated in the hypertrophic heart,22 some lncRNAs associated with cardiac pathologies may also be implicated in cardiac development.27 As for cell proliferation, a study identified eight lncRNAs putatively implicated in the proliferative capacity of cardiac cells in fetal heart29 that require further investigation.
Cardiomyocyte repolarization during the final stage of the action potential needs potassium fluxes mediated by the Kv7.1 channels. In late embryogenesis, lncRNA Kcnq1ot1 (potassium voltage-gated channel, KQT-like subfamily, member 1 opposite strand/antisense transcript 1) regulates the expression of the transcript Kcnq1, which encodes the potassium channel Kv7.1.30 Such regulation fulfills the requirement for increased cardiac contractile activity in this late developmental stage. Dysregulation of KCNQ1OT1 expression has been associated with left ventricular (LV) dysfunction after myocardial infarction (MI),31 thereby potentially linking this lncRNA to cardiac contractility and arrhythmia in a clinical setting.
A switch in myosin heavy chain (MHC) isoforms accompanies the acquisition of the adult cardiac contractile phenotype. The expression of α-MHC in adult left and right ventricles is associated with higher filament sliding velocity, while the slower β-MHC confers higher force at lower energy cost.32 In rodent models (which mainly express the α-MHC isoform in the normal adult stage), the intergenic region between the two genes has been shown to regulate the transition from β- to α-MHC during cardiac development through co-transcription of an antisense RNA, called β-RNA, targeting and inhibiting the myosin heavy chain 7 (MYH7) transcript (encoding the β-MHC isoform).33, 34 This mechanism is responsive to thyroid status and further implicated in the response to pressure overload.35, 36
Vascular Development and Biology
Growing evidence describes lncRNAs as key molecular players of vascular and endothelial cell (EC) biology. SENCR (smooth muscle and EC-enriched migration/differentiation-associated lncRNA) was among the first lncRNAs to be identified in human vascular smooth muscle cells (VSMCs) and ECs, being involved in their differentiation.37, 38 The lncRNA SMILR (smooth muscle-induced lncRNA) appears to promote VSMC proliferation and may achieve this by regulating expression of adjacent transcripts, HAS2 (hyaluronan synthase 2).39 HAS2 plays a role in proliferation in saphenous vein-derived VSMCs, in which small interfering RNA (siRNA) targeting of HAS2 resulted in reduced proliferation ability.40 SMILR could be a target for therapy of atherosclerosis because VSMC aberrant function is one of the defining features of atherosclerotic plaques (see Coronary Artery Disease and Atherosclerosis). MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) is one of the most abundant lncRNAs in mammalian cells, and it was observed in ECs. Due to its increased expression in hypoxia, MALAT1 is proposed to have a role in vascularization, although knockout mice did not present severe developmental abnormalities.17 MALAT1 is involved in alternative splicing41 and possibly regulates gene expression during vascular development and disease.17
In vitro differentiation of embryonic stem cells can recapitulate the development process, with transcriptome studies being employed to identify novel lncRNAs whose role is important for either commitment of cardiovascular progenitors or endothelial commitment.38, 42 A study that identified hundreds of novel ncRNA transcripts in the vascular setting functionally characterized the lncRNA PUNISHER, which is expressed in mature ECs.42 Morpholino targeting of PUNISHER in zebrafish culminated in extensive vascular defects, including abnormal branching and vessel formation. PUNISHER silencing in human umbilical vein ECs (HUVECs) using short hairpin RNA (shRNA) revealed similar defects. Overall, PUNISHER compromises EC function, but its mechanism of action or its involvement in vascular pathologies have not yet been elucidated.42
lncRNAs in Cardiovascular Disease
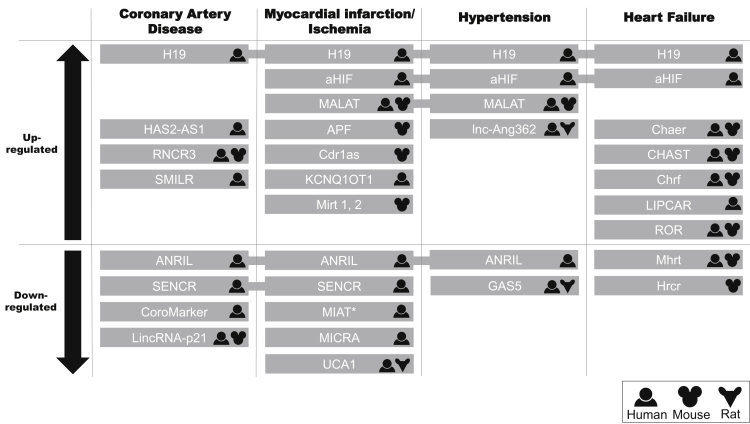
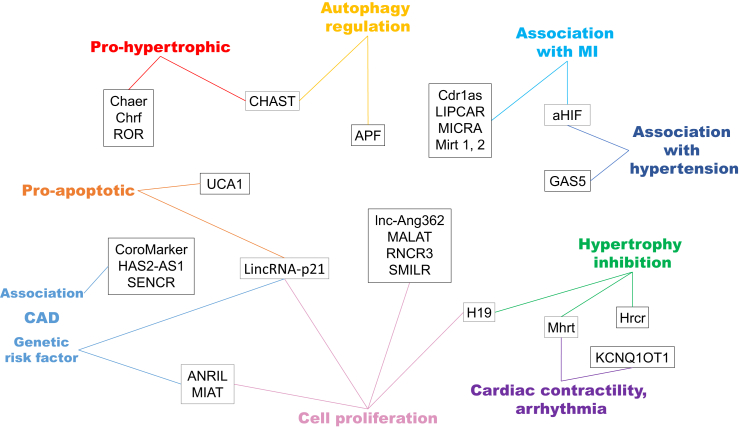
The association between lncRNAs and cardiovascular disease is just coming to light with several reports about their specific expression in different cardiac diseases. Dysregulation of certain lncRNAs has been shown in both human and rodent models (Figure 2), in which some studies present encouraging results for disease prognosis and therapy. The biological context of lncRNAs discussed in this section is illustrated in Figure 3. However, because of their poor conservation across species, translation of animal findings to human applications should be approached with caution. Here we overview the role of lncRNAs in hypertension, coronary artery disease (CAD), MI, ischemia, and heart failure.
Figure 2.
lncRNAs that Are Up- or Downregulated in Cardiovascular Diseases
The human, mouse, or rat symbol indicates in which organism the lncRNA has been described. *MIAT is downregulated in ST-elevation myocardial infarction (STEMI) patients compared to non-ST-elevation myocardial infarction (NSTEMI) patients; it correlates with hypertension, but there is no change in regulation.
Figure 3.
Biological Context of lncRNAs Associated with Cardiovascular Disease and Their Mechanism of Action
CAD, coronary artery disease; MI, myocardial infarction.
Some lncRNAs differentially expressed during cardiovascular development also participate in a pathological setting, while others are involved in more than one cardiovascular disease. SENCR and H19, for example, are widely implicated in cardiovascular disease. SENCR, besides playing a role in VSMC and EC differentiation during development, has also been suggested to affect CAD. This lncRNA was found to be downregulated in VSMCs from a type 2 diabetes mellitus murine model, promoting proliferation and migration.43 In addition, SENCR overexpression protected against the effects of high glucose stress on mouse VSMCs,43 and its reduced expression has been associated with premature CAD in humans.38
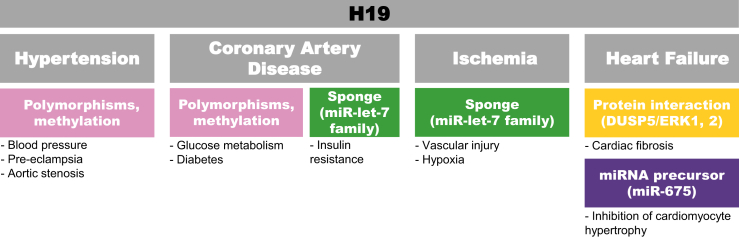
H19 is an important regulator of mammalian development and disease in that it inhibits cell proliferation.44 It is normally highly expressed during in utero development and downregulated at birth; however, studies reveal a re-expression of lncRNA H19 in cardiovascular disease settings, although not all mechanisms and involved players have been described.45, 46, 47, 48, 49, 50, 51, 52, 53 Human genome-wide association studies (GWASs) have demonstrated significant associations between H19 locus and systolic or mean arterial blood pressure.54 High H19 expression has been linked to hyperhomocysteinemia, a known risk factor for CAD,45, 51 and polymorphisms correlate with CAD risk.47 Methylation at the IGF2/H19 locus have been implicated in regulation of glucose metabolism and development of diabetes,46, 53, 55 renal development,56 pre-eclampsia,57 and aortic stenosis,58 indicating possible links. Furthermore, H19 was reported to sponge let-7 family miRNAs,59 which are believed to have atheroprotective60, 61, 62, 63 or proatherosclerotic64 roles and are downregulated in CAD patients.65 In addition, H19 was identified as differentially expressed in normoxic versus hypoxic ECs.66 Finally, it is a precursor of miR-675, which inhibits cardiomyocyte hypertrophy52 and contributes to cardiac fibroblast proliferation and fibrosis, acting through repression of DUSP5/ERK1/2 (Figure 4).67
Figure 4.
H19 Is Associated with Hypertension, CAD, Atherosclerosis, Ischemia, and Heart Failure
Although mechanistic insights into the role of H19 in cardiovascular disease are lacking, methylation regulation and sponging of miRNAs have been suggested and may overlap among diseases. Polymorphisms have been correlated with blood pressure and CAD. H19 action as a sponge for the miRNA let-7 family has been linked to CAD and could be a possible mechanism in hypoxia. In heart failure, it acts by interacting with protein to regulate cardiac fibrosis and is a precursor of miR-675, which targets an inducer of hypertrophy.
Hypertension
Hypertension has a complex etiology, involving an interplay of environmental and genetic components, and is a major risk factor for other cardiovascular diseases. Although many miRNAs have been shown to act in the pathogenesis of hypertension,68 reports on lncRNAs that relate to hypertension are scarce. However, studies based on animal and cell models in this area are emerging. One lncRNA involved in hypertension and vascular remodeling is GAS5 (growth arrest-specific 5).69 GAS5 was found to be downregulated in the plasma of human hypertensive patients and in the arteries and retina of a rat model, the spontaneously hypertensive rat. In these rats, GAS5 knockdown exacerbated the hypertensive phenotype, arterial remodeling, and microvascular dysfunction.69 In HUVECs and VSMCs, proliferation, migration, and resistance to oxidative stress were altered by GAS5 siRNAs. Co-culture experiments suggested that GAS5 participates in extracellular vesicle-mediated cross-talk between ECs and VSMCs.
A screening approach using angiotensin II (Ang II)-treated rat VSMCs identified differentially expressed lncRNAs, one of which, lnc-Ang362, is proximal to miR-221 and miR-222.70 These two miRNAs, which regulate the proliferation of rat VSMCs71 and the migration of HUVECs,72 were also upregulated in response to Ang II and appeared to be co-transcribed with the lncRNA. Consistently, lnc-Ang362 siRNA knockdown decreased miR-221/222 expression and reduced VSMCs proliferation.70 Thus, together with miR-221/222, lnc-Ang362 may represent an interesting therapeutic target that deserves further investigation, because it is conserved in human.
Given the poor evolutionary conservation of many lncRNAs, any approach based solely on animal models may limit discovery. In a cohort of patients with acute MI, expression of four lncRNAs—ANRIL (antisense non-coding RNA in the INK4 locus), aHIF (hypoxia-inducible factor 1A antisense RNA 2), MIAT (MI-associated transcript), and MALAT1—was found to be significantly associated with hypertension.31 Although their mechanisms of action in hypertension are not yet described, ANRIL affects cell adhesion, proliferation, and apoptosis,73 and MALAT1 modulates EC migration, sprouting, and proliferation.17 A novel bioinformatics tool, LncDisease, has been used to predict four human lncRNAs associated with hypertension, three of which were validated as dysregulated in Ang II-treated human VSMCs.74 Further investigation is required to firmly establish their validity in human hypertension.
CAD and Atherosclerosis
Atherosclerosis is typified by the formation of a fibro-fatty plaque in the arterial vessel wall and involves the molecular and functional dysregulation of ECs, VSMCs, macrophages and other leukocytes, and platelets.75 When atherosclerosis occurs in the epicardial vessels of the heart, it is referred to as CAD, which is a leading cause of death worldwide. Dysregulated expression of specific lncRNAs has been reported to contribute to CAD.
The lncRNA RNCR3 (retinal non-coding RNA 3) expression is altered during atherosclerosis, being overexpressed in atherosclerotic VSMCs and ECs compared with non-atherosclerotic tissue in mouse and human.76 Compared with control mice, downregulation of RNCR3 with shRNA aggravated atherosclerosis in thoracic aorta tissue and increased inflammatory factors in plasma. In vitro treatment with oxidized low-density lipoprotein (ox-LDL) increased RNCR3 levels in HUVECs and VSMCs, reducing proliferation and viability and increasing apoptosis. These data suggest that RNCR3 is atheroprotective. Moreover, the same study demonstrated in vitro that exosomes derived from ECs are rich in RNCR3, which is transferred to VSMCs and induces their proliferation and migration. The proposed mechanism of action in ECs is that RNCR3 regulates the transcription factor KLF2 by sponging miR-185-5p, which targets KLF2. Thus because of the atheroprotective role of RNCR3 in atherosclerosis, its induced upregulation potentially represents a therapeutic intervention.76
LincRNA-p21 was identified as a transcriptional target of p53.77 In VSMCs, this lncRNA disrupted the binding between p53 and its inhibitor, mouse double minute 2 (MDM2), with consequent effects on cell proliferation and apoptosis.78 LincRNA-p21 protected against neointimal hyperplasia in the carotid artery injury mouse model and was downregulated in aortic atherosclerotic plaques and in coronary artery tissues from CAD patients.78 Furthermore, polymorphisms in lincRNA-p21 have been associated with CAD risk.79 The therapeutic potential of lncRNA-p21 in acute vascular injury is suggested by its regulatory role of cell proliferation and apoptosis in CAD.
Remodeling of the extracellular matrix and neointimal formation are additional key features of CAD. Deposition of hyaluronan, synthesized by HAS2, contributes to this process. Two lncRNAs independently regulate HAS2: HAS2-AS1 (HAS2 antisense RNA 1) and SMILR. The antisense transcript HAS2-AS1 was increased in atherectomy samples collected from severely diseased carotid arteries and appears to promote HAS2 transcription in VSMCs in the presence of O-GlcNAcylation by altering chromatin configuration.80 This may be particularly relevant to neointimal formation in diabetic patients. SMILR, which promotes VSMC proliferation, was significantly upregulated in VSMCs stimulated with interleukin 1α and platelet-derived growth factor compared to unstimulated cells. In addition, its levels in plasma correlated with the inflammatory marker C-reactive protein and were upregulated in human carotid artery atherosclerotic plaques compared with adjacent healthy tissue.39
Several other lncRNAs have been associated with inflammation,81, 82, 83, 84, 85, 86, 87, 88 including in the context of diabetes,89, 90, 91 although direct links to CAD remain to be established. Downregulation of the lncRNA termed CoroMarker, originally identified as a biomarker of CAD,92 has been shown to decrease pro-inflammatory cytokine secretion from THP-1 monocytic cells,93 while ANRIL has been linked to the inflammatory response in ECs.94 CoroMarker, for instance, is a prominent example of lncRNA as biomarker for CAD, because it correctly identified with high sensitivity most CAD patients in a large cohort.92
MI and Ischemia
Although advances in treatment and diagnosis of MI have increased patient survival and quality of life, MI is still a major cause of mortality and morbidity worldwide.95 Several lncRNAs have been uncovered as being dysregulated in MI, and some may play roles in pathological angiogenesis and ischemic cardiac injury. Several GWASs have revealed an association between the INK4 locus and the risk of CAD, including MI. This locus is important for cell-cycle progression and cell growth and encodes tumor-suppressor genes CDKN2A and CDKN2B and the lncRNA ANRIL.96, 97, 98 Several linear and circular isoforms of ANRIL have been identified, and their expression differs according to specific tissues and disease conditions.99, 100 Despite contradictions in the literature regarding its mechanisms of action,73, 101, 102, 103, 104, 105 there is a consensus that ANRIL affects cell proliferation.73, 101, 102, 103, 106, 107 Moreover, increased blood circulating ANRIL expression was associated with LV dysfunction.31 The same study identified the lncRNAs MIAT and MALAT1 as similarly associated.31 A large-scale GWAS identified MIAT and six variants in its locus that confer susceptibility to MI. One variant was associated with upregulation of MIAT expression.108 So far, the molecular mechanisms by which this lncRNA regulates MI remain largely unknown, although it may be involved in alternative splicing in the diseased heart.109
In a mouse model of MI induced by coronary ligation, several lncRNAs were dysregulated in the heart, among which the two most strongly upregulated were named Mirt1 and Mirt2 (myocardial infarction-associated transcript 1 and 2).110 Their levels peaked 24 hr after MI and returned to baseline after 2 days, indicating that lncRNAs may be dynamically regulated in pathological processes. Mirt1 and Mirt2 levels correlated with expression of genes involved in reversing LV remodeling and preserved ejection fraction, suggesting a protective role in LV function. This study evidenced a therapeutic potential of these two lncRNAs. However, no human homologs of Mirt1 and Mirt2 have been described so far.
In addition to their potential as therapeutic targets, lncRNAs could serve as clinical biomarkers. One of the first studies to provide evidence of the feasibility of using lncRNAs as biomarkers for cardiovascular disease identified and validated LIPCAR (long intergenic non-coding RNA predicting cardiac remodeling) in a large number of patients.111 LIPCAR levels were increased in plasma samples from patients with LV remodeling after acute MI111 and in patients with type 2 diabetes mellitus.112 Circulating LIPCAR may also have prognostic value because its levels correlated with higher mortality risk in patients with heart failure. LIPCAR’s therapeutic potential remains to be investigated.
Myocardial infarction-associated circular RNA (MICRA) was the first circRNA to be identified as a potential biomarker of LV dysfunction after MI, with its predictive value confirmed in two independent cohorts.113 Therapeutic applications for this circRNA are yet to be discovered. A few circRNAs with relevance to cardiovascular disease have been unveiled and reviewed.114, 115, 116, 117 For example, Cdr1as (cerebellar degeneration-related protein 1 antisense transcript) was uncovered as a miR-7a sponge in cardiomyocytes, showing upregulation in MI mice and cardiomyocytes under hypoxia.118
The lncRNA UCA1 (urothelial carcinoma-associated 1), proposed as a biomarker as well, presented altered expression in MI patients.119 In rats with ischemia and reperfusion (I/R)-induced heart injury, another study found that UCA1 contributes to cardiac injury by enhancing apoptosis of cardiomyocytes.120 However, its application as a biomarker seems limited, because it performed worse than classical markers of MI (e.g., creatine kinase).119 Nonetheless, because it has a role in I/R injury, future studies could reveal a therapeutic potential to UCA1. Another lncRNA playing a role in the heart response to ischemia is HIF1A-AS2, also known as aHIF. It destabilizes the mRNA producing the hypoxia-inducible factor 1-α (HIF1α), which is considered the master transcriptional regulator of cellular response to hypoxia, including post-ischemic angiogenesis.121 Besides being overexpressed in the failing heart,121 aHIF was found to be dysregulated in the blood of patients after MI.31
Although a certain level of autophagy has been shown to be cardio-protective in ischemia,122 reports suggest that the accumulation of autophagosomes can trigger cardiomyocyte death, particularly during post-ischemic reperfusion.123, 124 The lncRNA APF (autophagy-promoting factor) participates in the regulation of autophagy and MI in mice.125 APF is increased during I/R injury and sequesters miR-188-3p, resulting in an upregulation of the miR-188-3p target gene ATG7, a promoter of autophagy. APF is important in determining myocardial I/R injury. Because inhibition of autophagy can be protective in the setting of MI, both APF and miR-188-3p represent potential targets for therapy.125
Several studies have identified lncRNAs involved in limb ischemia. Inhibition of MALAT1 using GapmeRs led to worse outcomes following experimental hindlimb ischemia. This suggested the possibility that MALAT1 plays a reparative, proangiogenic role.17 Moreover, SENCR was found to be reduced in human critical limb ischemia muscles.38 Several additional lncRNAs, including H19, were identified as differentially expressed in normoxic versus hypoxic ECs, followed by expressional and functional validation in the mouse limb ischemia model.66
Heart Failure
Heart failure is a complex condition of declined cardiac function in response to various pathophysiological stresses that cause cardiac remodeling, characterized by maladaptive hypertrophy. Maladaptive remodeling in the failing heart is considered an important target for therapy, and several lncRNAs are implicated in this process.126, 127
CHRF (cardiac hypertrophy-related factor), which is conserved between humans and mice, stimulates cardiac hypertrophy and was the first lncRNA reported to have implications in heart failure.126 It acts as a sponge to miR-489, hence upregulating its downstream target, MYD88, which is a key gene in activating cardiac hypertrophy.126 This was observed in cardiomyocytes of mice with pressure overload-induced cardiac hypertrophy, but the authors also reported a natural overexpression of CHRF in human heart failure tissue, emphasizing that this transcript may have a similar function in human.126
The heart-enriched lncRNA Chaer (cardiac hypertrophy-associated epigenetic regulator) is also required for cardiac hypertrophy and is functionally conserved between mouse and human.128 By interacting with PRC2, Chaer inhibits histone lysine methylation at the promoter regions of pro-hypertrophic genes, thus allowing their expression.128 Similarly, CHAST (cardiac hypertrophy-associated transcript) plays a role in promoting hypertrophy and is functionally conserved.15 CHAST levels were endogenously increased during cardiac hypertrophy in mice and in hypertrophic heart tissue from patients with aortic stenosis, a cause of cardiac hypertrophy and fibrosis. Induced overexpression of Chast in mice led to cardiomyocyte hypertrophy, while its suppression attenuated remodeling and hypertrophy without signs of toxicity.15 This is one of the most prominent examples of the strong therapeutic potential of lncRNAs for cardiac remodeling, showing that manipulation of a specific lncRNA can improve cardiac function.
A study identified in mice a cluster of cardiac-specific lncRNAs termed Mhrt (myosin heavy-chain-associated RNA transcripts) that are transcribed from the Myh7 locus, which is critical for cardiac contraction.129 Besides being transcribed from the same locus as β-RNA (antisense lncRNA involved in cardiac development), the transcripts have different sequences. Although highly expressed in adult hearts, Mhrt transcripts were suppressed during pathological stress. Induced restoration of Mhrt levels prevented cardiac hypertrophy and failure, revealing their cardio-protective role. Mhrt acts as a decoy to inhibit the aberrant expression of pathogenic genes involved in cardiac contractility, thus maintaining cardiac function. Finally, the authors found that the human version of MHRT was repressed in different cardiomyopathies (hypertrophic, ischemic, or idiopathic), indicating a conserved mechanism.129 This study provides further evidence in support of lncRNAs as potential therapeutic targets, for which development of a related therapy is facilitated by the conserved epigenetic regulation in human and mouse.
It has been uncovered that the lncRNA ROR (regulator of reprogramming) plays a role in cardiac hypertrophy. This transcript was naturally overexpressed in murine hypertrophic heart and cardiomyocytes, and its knockdown with siRNA attenuated hypertrophy. ROR enhanced cardiac hypertrophy by interacting with miR-133, a muscle-enriched miRNA that plays a role in hypertrophy.130 Both RNA molecules could be investigated as anti-hypertrophic therapeutic targets.
The circRNA Hrcr (heart-related circular RNA) has a protective role in cardiac hypertrophy and heart failure in mice. It acts as an endogenous sponge for miR-223, thus upregulating the expression of ARC (apoptosis repressor with caspase recruitment domain).117 ARC protein is normally highly expressed in the heart and is involved in cardiomyocyte hypertrophy and apoptosis.131, 132 Overexpression of miR-223 in mice using adenovirus-induced cardiac hypertrophy, and levels of Hrcr were downregulated in failing mouse hearts, indicating that Hrcr might constitute another target to treat heart failure if the mechanism is conserved in humans.117
Therapeutic Applications of lncRNAs
Although lncRNAs offer a multitude of prospective targets due to the diversity of actions and cellular processes implicated, few practical examples of therapeutic applications of lncRNAs have been reported so far. The up- or downregulation of specific lncRNA abundance have been the most thoroughly investigated approaches. Strategies for upregulation of lncRNAs include the use of recombinant adeno- or lentiviruses. Adeno-associated viral (AAV) vectors may represent a more promising approach due to their low pathogenicity.133, 134, 135, 136 This strategy used for targeting miRNAs in preclinical models showed promising results137, 138, 139 and reached successful clinical trials to deliver protein coding genes,133, 140, 141 but its use to deliver lncRNAs remains to be determined.
lncRNA downregulation can be obtained using shRNA or siRNA, more suitable for cytoplasmic lncRNAs,142 antisense oligonucleotides (ASOs)-mediated knockdown using aptamers,143 or GapmeRs forming heteroduplexes with their target lncRNAs that are then recognized and cleaved by the RNase H.144 The latter application, more suitable for nuclear lncRNAs and with fewer off-target effects than shRNA,145 is already undergoing testing in cell and animal models,15, 17, 18 while ASO and siRNA have been used to deplete MALAT1 in human cancer cells and animal models, reducing metastasis.146, 147, 148 Hopefully, it will be possible to transfer some findings in other diseases to help accelerate the development of therapies for cardiovascular disease. Ribozymes or deoxyribozymes, catalyzing the cleavage of the flanked region of the RNA target, represent an additional tool to knock down lncRNAs.149, 150 Finally, small molecules that compete with ligands to bind lncRNAs or induce conformational change in lncRNAs are being identified through large screening efforts.151
The most promising lncRNA targets for therapeutic applications in cardiovascular disease are those for which mechanism of action and effect are well described and preferably cell specific. One example is CHAST, for which the GapmeR-mediated silencing attenuated transverse aortic constriction-induced cardiac remodeling in mice.15 No apparent side effects were observed due to treatment with GapmeRs. Future experiments will determine whether cardiac hypertrophic remodeling may similarly be targeted through the downregulation of CHRF, which regulates Myd88, a factor associated with hypertrophy development,126 or by restoring expression levels of MHRT, thus preventing cardiomyopathy by restricting stress-associated aberrant gene expression mediated by the chromatin-remodeling factor, Brg1.129 Other approaches to decrease cardiac cell death following MI could target apoptotic or autophagic processes by downregulating APF.125 In vascular disease, prevention of MALAT1 upregulation could be used as anti-angiogenic therapy to prevent diabetes-associated microvascular complications.17, 90, 91
Challenges and Next Steps
As interest in the role of lncRNAs increases and technology to detect them becomes more sophisticated, widespread use of RNA sequencing (RNA-seq) screens has identified promising candidates for therapy. lncRNAs potentially represent a powerful tool for personalized medicine due to their specific expression patterns associated with distinct pathologies. The detection of lncRNAs in circulating exosomes152 opened interesting perspectives both scientifically, in terms of signaling regulation and intercellular communication, and for further translational applications to diagnostics.
Several limitations and challenges remain to be resolved before lncRNAs can reach clinical application. Foremost is target specificity, given the pleiotropic implications of a single lncRNA in pathophysiological processes throughout the human body. Although lncRNAs may show dysregulation specific to certain diseases, they exhibit various functions in the organism and some lncRNAs may act through more than one mechanism. As an example, modulation of ANRIL, for which SNPs are associated with CAD,106, 153 is probably hazardous given its implication in cancer development and progression.154, 155
Second, the low conservation of lncRNAs across evolution156 makes both the identification of human lncRNAs and their clinical testing real challenges, because rodents may not be an adequate model. The hurdles to translate animal findings to human are illustrated by Mirt1 and Mirt2, which may have a protective role in LV function, but no homologs in human have been described so far.110 However, it has been suggested that it is the secondary structure of lncRNAs that is conserved and functional, rather than the primary sequence. For example, GAS5 acting as decoy or signal may depend on its secondary structures and their affinities for different ligands.157, 158 This may explain why some lncRNAs with important mechanistic roles have not been observed in other species. If structure is more critical to function than sequence, then lncRNAs previously considered non-conserved may have structural homologs in other species, which would enable the use of existing animal models.
Third, before therapeutic application, the structure-function relationship of each lncRNA must be further elucidated, using newly developed methods to resolve secondary and tertiary structures.24, 159, 160 Finally, treatments must ensure the tissue-specific delivery of the ASOs or pharmacological agents, as well as their penetration into the intracellular compartment of interest. The translation of lncRNA-based therapy into clinical applications should also consider challenges such as route of delivery, low immunogenicity of the delivery system, stability of RNA drug in both circulation and cells, duration of treatment, dosage adjustment, and off-target effects.
Future work is required for a thorough functional characterization of lncRNAs in cardiovascular pathology, both at the molecular and at the cellular level. The role of lncRNAs as epigenetic regulators is critical for gene regulation and disease pathogenesis, yet the fine molecular mechanisms involved remain to be fully elucidated. Although several RNA-seq experiments have been conducted and many potential candidates have been identified,27, 66, 83, 152, 161 few have been sufficiently characterized, either in terms of regulation of the disease or in the ability to be externally regulated. Therefore, much remains to be done to retrieve the most promising candidates for therapeutic development from the huge amount of sequencing data available.
The involvement of circRNAs in cardiovascular pathologies has emerged.113, 117, 162 Although appealing due to their resistance to degradation by exoribonucleases, their use as therapeutic targets requires further investigation. Considering the number of previously characterized circRNAs and the plethora of circRNAs that remain to be characterized, this new branch of the ncRNA family constitutes an invaluable reservoir of therapeutic targets and may be useful to move theranostics a step forward, because they may be used for both diagnostics (biomarkers) and therapeutic purposes. Finally, gene editing with the CRISPR system appeared as an appealing approach for therapy, and a study reported an efficient downregulation of MALAT1 with this system,163 representing a potential tool for therapeutic applications in cardiac disease through modulation of the expression of lncRNAs.
Consortia
The authors Clarissa P.C. Gomes, Helen Spencer, Kerrie L. Ford, Y.M. Michel, Andrew H. Baker, Constanza Emanueli, Jean-Luc Balligrand, and Yvan Devaux are writing on behalf of the Cardiolinc network.
Acknowledgments
C.P.C.G. is funded by the Eurostars E! 9686 MIPROG project. Y.D. is supported by the National Research Fund (grant INTER/EUROSTARS/15/10282117) and the Ministry of Higher Education and Research of Luxembourg. A.H.B. is funded by the British Heart Foundation and European Research Council (VASCMIR). C.E. is a BHF Professor in Cardiovascular Science, and her non-coding RNA studies are supported by the BHF and the Leducq Foundation (MIRVAD transatlantic network grant).
References
- 1.Ozsolak F., Milos P.M. RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Devaux Y., Zangrando J., Schroen B., Creemers E.E., Pedrazzini T., Chang C.P., Dorn G.W., 2nd, Thum T., Heymans S., Cardiolinc network Long noncoding RNAs in cardiac development and ageing. Nat. Rev. Cardiol. 2015;12:415–425. doi: 10.1038/nrcardio.2015.55. [DOI] [PubMed] [Google Scholar]
- 3.Bär C., Chatterjee S., Thum T. Long noncoding RNAs in cardiovascular pathology, diagnosis, and therapy. Circulation. 2016;134:1484–1499. doi: 10.1161/CIRCULATIONAHA.116.023686. [DOI] [PubMed] [Google Scholar]
- 4.Devaux Y. Transcriptome of blood cells as a reservoir of cardiovascular biomarkers. Biochim. Biophys. Acta. 2017;1864:209–216. doi: 10.1016/j.bbamcr.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Boon R.A., Jaé N., Holdt L., Dimmeler S. Long noncoding RNAs: From clinical genetics to therapeutic targets? J. Am. Coll. Cardiol. 2016;67:1214–1226. doi: 10.1016/j.jacc.2015.12.051. [DOI] [PubMed] [Google Scholar]
- 6.Johnsson P., Lipovich L., Grandér D., Morris K.V. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta. 2014;1840:1063–1071. doi: 10.1016/j.bbagen.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willingham A.T., Orth A.P., Batalov S., Peters E.C., Wen B.G., Aza-Blanc P., Hogenesch J.B., Schultz P.G. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 8.Wang P., Xue Y., Han Y., Lin L., Wu C., Xu S., Jiang Z., Xu J., Liu Q., Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 9.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ørom U.A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., Lai F., Zytnicki M., Notredame C., Huang Q. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 12.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 13.Zhang K., Shi Z.M., Chang Y.N., Hu Z.M., Qi H.X., Hong W. The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene. 2014;547:1–9. doi: 10.1016/j.gene.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 14.Rashid F., Shah A., Shan G. Long non-coding RNAs in the cytoplasm. Genomics Proteomics Bioinformatics. 2016;14:73–80. doi: 10.1016/j.gpb.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viereck J., Kumarswamy R., Foinquinos A., Xiao K., Avramopoulos P., Kunz M., Dittrich M., Maetzig T., Zimmer K., Remke J. Long noncoding RNA Chast promotes cardiac remodeling. Sci. Transl. Med. 2016;8:326ra22. doi: 10.1126/scitranslmed.aaf1475. [DOI] [PubMed] [Google Scholar]
- 16.Meng L., Ward A.J., Chun S., Bennett C.F., Beaudet A.L., Rigo F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518:409–412. doi: 10.1038/nature13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michalik K.M., You X., Manavski Y., Doddaballapur A., Zörnig M., Braun T., John D., Ponomareva Y., Chen W., Uchida S. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ. Res. 2014;114:1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 18.Krieg A.M. Targeting LDL cholesterol with LNA. Mol. Ther. Nucleic Acids. 2012;1:e6. doi: 10.1038/mtna.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wheeler T.M., Leger A.J., Pandey S.K., MacLeod A.R., Nakamori M., Cheng S.H., Wentworth B.M., Bennett C.F., Thornton C.A. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K., Munson G., Young G., Lucas A.B., Ach R., Bruhn L. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ounzain S., Pezzuto I., Micheletti R., Burdet F., Sheta R., Nemir M., Gonzales C., Sarre A., Alexanian M., Blow M.J. Functional importance of cardiac enhancer-associated noncoding RNAs in heart development and disease. J. Mol. Cell. Cardiol. 2014;76:55–70. doi: 10.1016/j.yjmcc.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matkovich S.J., Edwards J.R., Grossenheider T.C., de Guzman Strong C., Dorn G.W., 2nd Epigenetic coordination of embryonic heart transcription by dynamically regulated long noncoding RNAs. Proc. Natl. Acad. Sci. USA. 2014;111:12264–12269. doi: 10.1073/pnas.1410622111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Werber M., Wittler L., Timmermann B., Grote P., Herrmann B.G. The tissue-specific transcriptomic landscape of the mid-gestational mouse embryo. Development. 2014;141:2325–2330. doi: 10.1242/dev.105858. [DOI] [PubMed] [Google Scholar]
- 24.Xue Z., Hennelly S., Doyle B., Gulati A.A., Novikova I.V., Sanbonmatsu K.Y., Boyer L.A. A G-rich motif in the lncRNA Braveheart interacts with a zinc-finger transcription factor to specify the cardiovascular lineage. Mol. Cell. 2016;64:37–50. doi: 10.1016/j.molcel.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klattenhoff C.A., Scheuermann J.C., Surface L.E., Bradley R.K., Fields P.A., Steinhauser M.L., Ding H., Butty V.L., Torrey L., Haas S. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grote P., Wittler L., Hendrix D., Koch F., Währisch S., Beisaw A., Macura K., Bläss G., Kellis M., Werber M., Herrmann B.G. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ounzain S., Micheletti R., Beckmann T., Schroen B., Alexanian M., Pezzuto I., Crippa S., Nemir M., Sarre A., Johnson R. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur. Heart J. 2015;36 doi: 10.1093/eurheartj/ehu180. 353–68a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ounzain S., Micheletti R., Arnan C., Plaisance I., Cecchi D., Schroen B., Reverter F., Alexanian M., Gonzales C., Ng S.Y. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J. Mol. Cell. Cardiol. 2015;89(Pt A):98–112. doi: 10.1016/j.yjmcc.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 29.Wang J., Geng Z., Weng J., Shen L., Li M., Cai X., Sun C., Chu M. Microarray analysis reveals a potential role of lncRNAs expression in cardiac cell proliferation. BMC Dev. Biol. 2016;16:41. doi: 10.1186/s12861-016-0139-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korostowski L., Sedlak N., Engel N. The Kcnq1ot1 long non-coding RNA affects chromatin conformation and expression of Kcnq1, but does not regulate its imprinting in the developing heart. PLoS Genet. 2012;8:e1002956. doi: 10.1371/journal.pgen.1002956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vausort M., Wagner D.R., Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ. Res. 2014;115:668–677. doi: 10.1161/CIRCRESAHA.115.303836. [DOI] [PubMed] [Google Scholar]
- 32.VanBuren P., Harris D.E., Alpert N.R., Warshaw D.M. Cardiac V1 and V3 myosins differ in their hydrolytic and mechanical activities in vitro. Circ. Res. 1995;77:439–444. doi: 10.1161/01.res.77.2.439. [DOI] [PubMed] [Google Scholar]
- 33.Haddad F., Bodell P.W., Qin A.X., Giger J.M., Baldwin K.M. Role of antisense RNA in coordinating cardiac myosin heavy chain gene switching. J. Biol. Chem. 2003;278:37132–37138. doi: 10.1074/jbc.M305911200. [DOI] [PubMed] [Google Scholar]
- 34.Reiser P.J., Portman M.A., Ning X.H., Schomisch Moravec C. Human cardiac myosin heavy chain isoforms in fetal and failing adult atria and ventricles. Am. J. Physiol. Heart Circ. Physiol. 2001;280:H1814–H1820. doi: 10.1152/ajpheart.2001.280.4.H1814. [DOI] [PubMed] [Google Scholar]
- 35.Haddad F., Qin A.X., Bodell P.W., Jiang W., Giger J.M., Baldwin K.M. Intergenic transcription and developmental regulation of cardiac myosin heavy chain genes. Am. J. Physiol. Heart Circ. Physiol. 2008;294:H29–H40. doi: 10.1152/ajpheart.01125.2007. [DOI] [PubMed] [Google Scholar]
- 36.Haddad F., Qin A.X., Bodell P.W., Zhang L.Y., Guo H., Giger J.M., Baldwin K.M. Regulation of antisense RNA expression during cardiac MHC gene switching in response to pressure overload. Am. J. Physiol. Heart Circ. Physiol. 2006;290:H2351–H2361. doi: 10.1152/ajpheart.01111.2005. [DOI] [PubMed] [Google Scholar]
- 37.Bell R.D., Long X., Lin M., Bergmann J.H., Nanda V., Cowan S.L., Zhou Q., Han Y., Spector D.L., Zheng D., Miano J.M. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler. Thromb. Vasc. Biol. 2014;34:1249–1259. doi: 10.1161/ATVBAHA.114.303240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boulberdaa M., Scott E., Ballantyne M., Garcia R., Descamps B., Angelini G.D., Brittan M., Hunter A., McBride M., McClure J. A role for the long noncoding RNA SENCR in commitment and function of endothelial cells. Mol. Ther. 2016;24:978–990. doi: 10.1038/mt.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ballantyne M.D., Pinel K., Dakin R., Vesey A.T., Diver L., Mackenzie R., Garcia R., Welsh P., Sattar N., Hamilton G. Smooth muscle enriched long noncoding RNA (SMILR) regulates cell proliferation. Circulation. 2016;133:2050–2065. doi: 10.1161/CIRCULATIONAHA.115.021019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van den Boom M., Sarbia M., von Wnuck Lipinski K., Mann P., Meyer-Kirchrath J., Rauch B.H., Grabitz K., Levkau B., Schrör K., Fischer J.W. Differential regulation of hyaluronic acid synthase isoforms in human saphenous vein smooth muscle cells: possible implications for vein graft stenosis. Circ. Res. 2006;98:36–44. doi: 10.1161/01.RES.0000199263.67107.c0. [DOI] [PubMed] [Google Scholar]
- 41.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurian L., Aguirre A., Sancho-Martinez I., Benner C., Hishida T., Nguyen T.B., Reddy P., Nivet E., Krause M.N., Nelles D.A. Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation. 2015;131:1278–1290. doi: 10.1161/CIRCULATIONAHA.114.013303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zou Z.Q., Xu J., Li L., Han Y.S. Down-regulation of SENCR promotes smooth muscle cells proliferation and migration in db/db mice through up-regulation of FoxO1 and TRPC6. Biomed. Pharmacother. 2015;74:35–41. doi: 10.1016/j.biopha.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 44.Ratajczak M.Z. Igf2-H19, an imprinted tandem gene, is an important regulator of embryonic development, a guardian of proliferation of adult pluripotent stem cells, a regulator of longevity, and a ‘passkey’ to cancerogenesis. Folia Histochem. Cytobiol. 2012;50:171–179. doi: 10.5603/fhc.2012.0026. [DOI] [PubMed] [Google Scholar]
- 45.Devlin A.M., Bottiglieri T., Domann F.E., Lentz S.R. Tissue-specific changes in H19 methylation and expression in mice with hyperhomocysteinemia. J. Biol. Chem. 2005;280:25506–25511. doi: 10.1074/jbc.M504815200. [DOI] [PubMed] [Google Scholar]
- 46.Ding G.-L., Wang F.-F., Shu J., Tian S., Jiang Y., Zhang D., Wang N., Luo Q., Zhang Y., Jin F. Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes. 2012;61:1133–1142. doi: 10.2337/db11-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao W., Zhu M., Wang H., Zhao S., Zhao D., Yang Y., Wang Z.M., Wang F., Yang Z.J., Lu X., Wang L.S. Association of polymorphisms in long non-coding RNA H19 with coronary artery disease risk in a Chinese population. Mutat. Res. 2015;772:15–22. doi: 10.1016/j.mrfmmm.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 48.Greco S., Zaccagnini G., Perfetti A., Fuschi P., Valaperta R., Voellenkle C., Castelvecchio S., Gaetano C., Finato N., Beltrami A.P. Long noncoding RNA dysregulation in ischemic heart failure. J. Transl. Med. 2016;14:183. doi: 10.1186/s12967-016-0926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han D.K., Khaing Z.Z., Pollock R.A., Haudenschild C.C., Liau G. H19, a marker of developmental transition, is reexpressed in human atherosclerotic plaques and is regulated by the insulin family of growth factors in cultured rabbit smooth muscle cells. J. Clin. Invest. 1996;97:1276–1285. doi: 10.1172/JCI118543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim D.K., Zhang L., Dzau V.J., Pratt R.E. H19, a developmentally regulated gene, is reexpressed in rat vascular smooth muscle cells after injury. J. Clin. Invest. 1994;93:355–360. doi: 10.1172/JCI116967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li L., Xie J., Zhang M., Wang S. Homocysteine harasses the imprinting expression of IGF2 and H19 by demethylation of differentially methylated region between IGF2/H19 genes. Acta Biochim. Biophys. Sin. (Shanghai) 2009;41:464–471. doi: 10.1093/abbs/gmp033. [DOI] [PubMed] [Google Scholar]
- 52.Liu L., An X., Li Z., Song Y., Li L., Zuo S., Liu N., Yang G., Wang H., Cheng X. The H19 long noncoding RNA is a novel negative regulator of cardiomyocyte hypertrophy. Cardiovasc. Res. 2016;111:56–65. doi: 10.1093/cvr/cvw078. [DOI] [PubMed] [Google Scholar]
- 53.Shao W.-J., Tao L.-Y., Gao C., Xie J.-Y., Zhao R.-Q. Alterations in methylation and expression levels of imprinted genes H19 and Igf2 in the fetuses of diabetic mice. Comp. Med. 2008;58:341–346. [PMC free article] [PubMed] [Google Scholar]
- 54.Tragante V., Barnes M.R., Ganesh S.K., Lanktree M.B., Guo W., Franceschini N., Smith E.N., Johnson T., Holmes M.V., Padmanabhan S. Gene-centric meta-analysis in 87,736 individuals of European ancestry identifies multiple blood-pressure-related loci. Am. J. Hum. Genet. 2014;94:349–360. doi: 10.1016/j.ajhg.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao Y., Wu F., Zhou J., Yan L., Jurczak M.J., Lee H.-Y., Yang L., Mueller M., Zhou X.B., Dandolo L. The H19/let-7 double-negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic Acids Res. 2014;42:13799–13811. doi: 10.1093/nar/gku1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanwar Y.S., Pan X., Lin S., Kumar A., Wada J., Haas C.S., Liau G., Lomasney J.W. Imprinted mesodermal specific transcript (MEST) and H19 genes in renal development and diabetes. Kidney Int. 2003;63:1658–1670. doi: 10.1046/j.1523-1755.2003.00905.x. [DOI] [PubMed] [Google Scholar]
- 57.Yu L., Chen M., Zhao D., Yi P., Lu L., Han J., Zheng X., Zhou Y., Li L. The H19 gene imprinting in normal pregnancy and pre-eclampsia. Placenta. 2009;30:443–447. doi: 10.1016/j.placenta.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 58.Hadji F., Boulanger M.-C., Guay S.-P., Gaudreault N., Amellah S., Mkannez G., Bouchareb R., Marchand J.T., Nsaibia M.J., Guauque-Olarte S. Altered DNA methylation of long noncoding RNA H19 in calcific aortic valve disease promotes mineralization by silencing NOTCH1. Circulation. 2016;134:1848–1862. doi: 10.1161/CIRCULATIONAHA.116.023116. [DOI] [PubMed] [Google Scholar]
- 59.Kallen A.N., Zhou X.B., Xu J., Qiao C., Ma J., Yan L., Lu L., Liu C., Yi J.S., Zhang H. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen K.-C., Hsieh I.C., Hsi E., Wang Y.-S., Dai C.-Y., Chou W.-W., Juo S.H. Negative feedback regulation between microRNA let-7g and the oxLDL receptor LOX-1. J. Cell Sci. 2011;124:4115–4124. doi: 10.1242/jcs.092767. [DOI] [PubMed] [Google Scholar]
- 61.Satoh M., Tabuchi T., Minami Y., Takahashi Y., Itoh T., Nakamura M. Expression of let-7i is associated with Toll-like receptor 4 signal in coronary artery disease: effect of statins on let-7i and Toll-like receptor 4 signal. Immunobiology. 2012;217:533–539. doi: 10.1016/j.imbio.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 62.Ding Z., Wang X., Schnackenberg L., Khaidakov M., Liu S., Singla S., Dai Y., Mehta J.L. Regulation of autophagy and apoptosis in response to ox-LDL in vascular smooth muscle cells, and the modulatory effects of the microRNA hsa-let-7 g. Int. J. Cardiol. 2013;168:1378–1385. doi: 10.1016/j.ijcard.2012.12.045. [DOI] [PubMed] [Google Scholar]
- 63.Bao M.H., Zhang Y.W., Lou X.Y., Cheng Y., Zhou H.H. Protective effects of let-7a and let-7b on oxidized low-density lipoprotein induced endothelial cell injuries. PLoS ONE. 2014;9:e106540. doi: 10.1371/journal.pone.0106540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qin B., Xiao B., Liang D., Li Y., Jiang T., Yang H. MicroRNA let-7c inhibits Bcl-xl expression and regulates ox-LDL-induced endothelial apoptosis. BMB Rep. 2012;45:464–469. doi: 10.5483/BMBRep.2012.45.8.033. [DOI] [PubMed] [Google Scholar]
- 65.Fichtlscherer S., De Rosa S., Fox H., Schwietz T., Fischer A., Liebetrau C., Weber M., Hamm C.W., Röxe T., Müller-Ardogan M. Circulating microRNAs in patients with coronary artery disease. Circ. Res. 2010;107:677–684. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- 66.Voellenkle C., Garcia-Manteiga J.M., Pedrotti S., Perfetti A., De Toma I., Da Silva D., Maimone B., Greco S., Fasanaro P., Creo P. Implication of long noncoding RNAs in the endothelial cell response to hypoxia revealed by RNA-sequencing. Sci. Rep. 2016;6:24141. doi: 10.1038/srep24141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tao H., Cao W., Yang J.J., Shi K.H., Zhou X., Liu L.P., Li J. Long noncoding RNA H19 controls DUSP5/ERK1/2 axis in cardiac fibroblast proliferation and fibrosis. Cardiovasc. Pathol. 2016;25:381–389. doi: 10.1016/j.carpath.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 68.Nemecz M., Alexandru N., Tanko G., Georgescu A. Role of microRNA in endothelial dysfunction and hypertension. Curr. Hypertens. Rep. 2016;18:87. doi: 10.1007/s11906-016-0696-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y.N., Shan K., Yao M.D., Yao J., Wang J.J., Li X., Liu B., Zhang Y.Y., Ji Y., Jiang Q., Yan B. Long noncoding RNA-GAS5: a novel regulator of hypertension-induced vascular remodeling. Hypertension. 2016;68:736–748. doi: 10.1161/HYPERTENSIONAHA.116.07259. [DOI] [PubMed] [Google Scholar]
- 70.Leung A., Trac C., Jin W., Lanting L., Akbany A., Sætrom P., Schones D.E., Natarajan R. Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circ. Res. 2013;113:266–278. doi: 10.1161/CIRCRESAHA.112.300849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu X., Cheng Y., Zhang S., Lin Y., Yang J., Zhang C. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ. Res. 2009;104:476–487. doi: 10.1161/CIRCRESAHA.108.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu N., Zhang D., Chen S., Liu X., Lin L., Huang X., Guo Z., Liu J., Wang Y., Yuan W., Qin Y. Endothelial enriched microRNAs regulate angiotensin II-induced endothelial inflammation and migration. Atherosclerosis. 2011;215:286–293. doi: 10.1016/j.atherosclerosis.2010.12.024. [DOI] [PubMed] [Google Scholar]
- 73.Holdt L.M., Hoffmann S., Sass K., Langenberger D., Scholz M., Krohn K., Finstermeier K., Stahringer A., Wilfert W., Beutner F. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 2013;9:e1003588. doi: 10.1371/journal.pgen.1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang J., Ma R., Ma W., Chen J., Yang J., Xi Y., Cui Q. LncDisease: a sequence based bioinformatics tool for predicting lncRNA-disease associations. Nucleic Acids Res. 2016;44:e90. doi: 10.1093/nar/gkw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ross R. Atherosclerosis—an inflammatory disease. N. Engl. J. Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 76.Shan K., Jiang Q., Wang X.Q., Wang Y.N.Z., Yang H., Yao M.D., Liu C., Li X.M., Yao J., Liu B. Role of long non-coding RNA-RNCR3 in atherosclerosis-related vascular dysfunction. Cell Death Dis. 2016;7:e2248. doi: 10.1038/cddis.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huarte M., Guttman M., Feldser D., Garber M., Koziol M.J., Kenzelmann-Broz D., Khalil A.M., Zuk O., Amit I., Rabani M. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu G., Cai J., Han Y., Chen J., Huang Z.-P., Chen C., Cai Y., Huang H., Yang Y., Liu Y. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation. 2014;130:1452–1465. doi: 10.1161/CIRCULATIONAHA.114.011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tang S.S., Cheng J., Cai M.Y., Yang X.L., Liu X.G., Zheng B.Y., Xiong X.D. Association of lincRNA-p21 haplotype with coronary artery disease in a Chinese Han population. Dis. Markers. 2016;2016:9109743. doi: 10.1155/2016/9109743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vigetti D., Deleonibus S., Moretto P., Bowen T., Fischer J.W., Grandoch M., Oberhuber A., Love D.C., Hanover J.A., Cinquetti R. Natural antisense transcript for hyaluronan synthase 2 (HAS2-AS1) induces transcription of HAS2 via protein O-GlcNAcylation. J. Biol. Chem. 2014;289:28816–28826. doi: 10.1074/jbc.M114.597401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Carpenter S., Aiello D., Atianand M.K., Ricci E.P., Gandhi P., Hall L.L., Byron M., Monks B., Henry-Bezy M., Lawrence J.B. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rapicavoli N.A., Qu K., Zhang J., Mikhail M., Laberge R.-M., Chang H.Y. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. eLife. 2013;2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu Y., Ferguson J.F., Xue C., Ballantyne R.L., Silverman I.M., Gosai S.J., Serfecz J., Morley M.P., Gregory B.D., Li M., Reilly M.P. Tissue-specific RNA-seq in human evoked inflammation identifies blood and adipose LincRNA signatures of cardiometabolic diseases. Arterioscler. Thromb. Vasc. Biol. 2014;34:902–912. doi: 10.1161/ATVBAHA.113.303123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li Z., Chao T.-C., Chang K.-Y., Lin N., Patil V.S., Shimizu C., Head S.R., Burns J.C., Rana T.M. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. USA. 2014;111:1002–1007. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cui H., Xie N., Tan Z., Banerjee S., Thannickal V.J., Abraham E., Liu G. The human long noncoding RNA lnc-IL7R regulates the inflammatory response. Eur. J. Immunol. 2014;44:2085–2095. doi: 10.1002/eji.201344126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krawczyk M., Emerson B.M. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-κB complexes. eLife. 2014;3:e01776. doi: 10.7554/eLife.01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chan J., Atianand M., Jiang Z., Carpenter S., Aiello D., Elling R., Fitzgerald K.A., Caffrey D.R. Cutting edge: a natural antisense transcript, AS-IL1α, controls inducible transcription of the proinflammatory cytokine IL-1α. J. Immunol. 2015;195:1359–1363. doi: 10.4049/jimmunol.1500264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Atianand M.K., Hu W., Satpathy A.T., Shen Y., Ricci E.P., Alvarez-Dominguez J.R., Bhatta A., Schattgen S.A., McGowan J.D., Blin J. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell. 2016;165:1672–1685. doi: 10.1016/j.cell.2016.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reddy M.A., Chen Z., Park J.T., Wang M., Lanting L., Zhang Q., Bhatt K., Leung A., Wu X., Putta S. Regulation of inflammatory phenotype in macrophages by a diabetes-induced long noncoding RNA. Diabetes. 2014;63:4249–4261. doi: 10.2337/db14-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Puthanveetil P., Chen S., Feng B., Gautam A., Chakrabarti S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J. Cell. Mol. Med. 2015;19:1418–1425. doi: 10.1111/jcmm.12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu J.Y., Yao J., Li X.M., Song Y.C., Wang X.Q., Li Y.J., Yan B., Jiang Q. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 2014;5:e1506. doi: 10.1038/cddis.2014.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang Y., Cai Y., Wu G., Chen X., Liu Y., Wang X., Yu J., Li C., Chen X., Jose P.A. Plasma long non-coding RNA, CoroMarker, a novel biomarker for diagnosis of coronary artery disease. Clin. Sci. 2015;129:675–685. doi: 10.1042/CS20150121. [DOI] [PubMed] [Google Scholar]
- 93.Cai Y., Yang Y., Chen X., Wu G., Zhang X., Liu Y., Yu J., Wang X., Fu J., Li C. Circulating ‘lncRNA OTTHUMT00000387022’ from monocytes as a novel biomarker for coronary artery disease. Cardiovasc. Res. 2016;112:714–724. doi: 10.1093/cvr/cvw022. [DOI] [PubMed] [Google Scholar]
- 94.Zhou X., Han X., Wittfeldt A., Sun J., Liu C., Wang X., Gan L.M., Cao H., Liang Z. Long non-coding RNA ANRIL regulates inflammatory responses as a novel component of NF-κB pathway. RNA Biol. 2016;13:98–108. doi: 10.1080/15476286.2015.1122164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Townsend N., Wilson L., Bhatnagar P., Wickramasinghe K., Rayner M., Nichols M. Cardiovascular disease in Europe: epidemiological update 2016. Eur. Heart J. 2016;37:3232–3245. doi: 10.1093/eurheartj/ehw334. [DOI] [PubMed] [Google Scholar]
- 96.Broadbent H.M., Peden J.F., Lorkowski S., Goel A., Ongen H., Green F., Clarke R., Collins R., Franzosi M.G., Tognoni G., PROCARDIS consortium Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum. Mol. Genet. 2008;17:806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 97.Helgadottir A., Thorleifsson G., Manolescu A., Gretarsdottir S., Blondal T., Jonasdottir A., Jonasdottir A., Sigurdsson A., Baker A., Palsson A. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 98.McPherson R., Pertsemlidis A., Kavaslar N., Stewart A., Roberts R., Cox D.R., Hinds D.A., Pennacchio L.A., Tybjaerg-Hansen A., Folsom A.R. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Folkersen L., Kyriakou T., Goel A., Peden J., Mälarstig A., Paulsson-Berne G., Hamsten A., Hugh Watkins, Franco-Cereceda A., Gabrielsen A., Eriksson P., PROCARDIS consortia Relationship between CAD risk genotype in the chromosome 9p21 locus and gene expression. Identification of eight new ANRIL splice variants. PLoS ONE. 2009;4:e7677. doi: 10.1371/journal.pone.0007677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Burd C.E., Jeck W.R., Liu Y., Sanoff H.K., Wang Z., Sharpless N.E. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Holdt L.M., Stahringer A., Sass K., Pichler G., Kulak N.A., Wilfert W., Kohlmaier A., Herbst A., Northoff B.H., Nicolaou A. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bochenek G., Häsler R., El Mokhtari N.E., König I.R., Loos B.G., Jepsen S., Rosenstiel P., Schreiber S., Schaefer A.S. The large non-coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10. Hum. Mol. Genet. 2013;22:4516–4527. doi: 10.1093/hmg/ddt299. [DOI] [PubMed] [Google Scholar]
- 103.Congrains A., Kamide K., Katsuya T., Yasuda O., Oguro R., Yamamoto K., Ohishi M., Rakugi H. CVD-associated non-coding RNA, ANRIL, modulates expression of atherogenic pathways in VSMC. Biochem. Biophys. Res. Commun. 2012;419:612–616. doi: 10.1016/j.bbrc.2012.02.050. [DOI] [PubMed] [Google Scholar]
- 104.Yap K.L., Li S., Muñoz-Cabello A.M., Raguz S., Zeng L., Mujtaba S., Gil J., Walsh M.J., Zhou M.M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kotake Y., Nakagawa T., Kitagawa K., Suzuki S., Liu N., Kitagawa M., Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Congrains A., Kamide K., Oguro R., Yasuda O., Miyata K., Yamamoto E., Kawai T., Kusunoki H., Yamamoto H., Takeya Y. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis. 2012;220:449–455. doi: 10.1016/j.atherosclerosis.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 107.Aguilo F., Zhou M.M., Walsh M.J. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res. 2011;71:5365–5369. doi: 10.1158/0008-5472.CAN-10-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ishii N., Ozaki K., Sato H., Mizuno H., Saito S., Takahashi A., Miyamoto Y., Ikegawa S., Kamatani N., Hori M. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J. Hum. Genet. 2006;51:1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- 109.Tsuiji H., Yoshimoto R., Hasegawa Y., Furuno M., Yoshida M., Nakagawa S. Competition between a noncoding exon and introns: Gomafu contains tandem UACUAAC repeats and associates with splicing factor-1. Genes Cells. 2011;16:479–490. doi: 10.1111/j.1365-2443.2011.01502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zangrando J., Zhang L., Vausort M., Maskali F., Marie P.Y., Wagner D.R., Devaux Y. Identification of candidate long non-coding RNAs in response to myocardial infarction. BMC Genomics. 2014;15:460. doi: 10.1186/1471-2164-15-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kumarswamy R., Bauters C., Volkmann I., Maury F., Fetisch J., Holzmann A., Lemesle G., de Groote P., Pinet F., Thum T. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ. Res. 2014;114:1569–1575. doi: 10.1161/CIRCRESAHA.114.303915. [DOI] [PubMed] [Google Scholar]
- 112.de Gonzalo-Calvo D., Kenneweg F., Bang C., Toro R., van der Meer R.W., Rijzewijk L.J., Smit J.W., Lamb H.J., Llorente-Cortes V., Thum T. Circulating long-non coding RNAs as biomarkers of left ventricular diastolic function and remodelling in patients with well-controlled type 2 diabetes. Sci. Rep. 2016;6:37354. doi: 10.1038/srep37354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vausort M., Salgado-Somoza A., Zhang L., Leszek P., Scholz M., Teren A., Burkhardt R., Thiery J., Wagner D.R., Devaux Y. Myocardial infarction-associated circular RNA predicting left ventricular dysfunction. J. Am. Coll. Cardiol. 2016;68:1247–1248. doi: 10.1016/j.jacc.2016.06.040. [DOI] [PubMed] [Google Scholar]
- 114.Devaux Y., Creemers E.E., Boon R.A., Werfel S., Thum T., Engelhardt S., Dimmeler S., Squire I., Cardiolinc network Circular RNAs in heart failure. Eur. J. Heart Fail. 2017;19:701–709. doi: 10.1002/ejhf.801. [DOI] [PubMed] [Google Scholar]
- 115.Elia L., Quintavalle M., Condorelli G. Circular RNAs and heart failure: new players for an old disease. Cardiovasc. Res. 2017;113:254–255. doi: 10.1093/cvr/cvx007. [DOI] [PubMed] [Google Scholar]
- 116.Tan W.L., Lim B.T., Anene-Nzelu C.G., Ackers-Johnson M., Dashi A., See K., Tiang Z., Lee D.P., Chua W.W., Luu T.D. A landscape of circular RNA expression in the human heart. Cardiovasc. Res. 2017;113:298–309. doi: 10.1093/cvr/cvw250. [DOI] [PubMed] [Google Scholar]
- 117.Wang K., Long B., Liu F., Wang J.X., Liu C.Y., Zhao B., Zhou L.Y., Sun T., Wang M., Yu T. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 118.Geng H.H., Li R., Su Y.M., Xiao J., Pan M., Cai X.X., Ji X.P. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS ONE. 2016;11:e0151753. doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yan Y., Zhang B., Liu N., Qi C., Xiao Y., Tian X., Li T., Liu B. Circulating long noncoding RNA UCA1 as a novel biomarker of acute myocardial infarction. BioMed Res. Int. 2016;2016:8079372. doi: 10.1155/2016/8079372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu Y., Zhou D., Li G., Ming X., Tu Yf., Tian J., Lu H., Yu B. Long non coding RNA-UCA1 contributes to cardiomyocyte apoptosis by suppression of p27 expression. Cell. Physiol. Biochem. 2015;35:1986–1998. doi: 10.1159/000374006. [DOI] [PubMed] [Google Scholar]
- 121.Zolk O., Solbach T.F., Eschenhagen T., Weidemann A., Fromm M.F. Activation of negative regulators of the hypoxia-inducible factor (HIF) pathway in human end-stage heart failure. Biochem. Biophys. Res. Commun. 2008;376:315–320. doi: 10.1016/j.bbrc.2008.08.152. [DOI] [PubMed] [Google Scholar]
- 122.Gottlieb R.A., Mentzer R.M. Autophagy during cardiac stress: joys and frustrations of autophagy. Annu. Rev. Physiol. 2010;72:45–59. doi: 10.1146/annurev-physiol-021909-135757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ma X., Liu H., Foyil S.R., Godar R.J., Weinheimer C.J., Hill J.A., Diwan A. Impaired autophagosome clearance contributes to cardiomyocyte death in ischemia/reperfusion injury. Circulation. 2012;125:3170–3181. doi: 10.1161/CIRCULATIONAHA.111.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pattingre S., Tassa A., Qu X., Garuti R., Liang X.H., Mizushima N., Packer M., Schneider M.D., Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 125.Wang K., Liu C.Y., Zhou L.Y., Wang J.X., Wang M., Zhao B., Zhao W.K., Xu S.J., Fan L.H., Zhang X.J. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat. Commun. 2015;6:6779. doi: 10.1038/ncomms7779. [DOI] [PubMed] [Google Scholar]
- 126.Wang K., Liu F., Zhou L.Y., Long B., Yuan S.M., Wang Y., Liu C.Y., Sun T., Zhang X.J., Li P.F. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ. Res. 2014;114:1377–1388. doi: 10.1161/CIRCRESAHA.114.302476. [DOI] [PubMed] [Google Scholar]
- 127.Dangwal S., Schimmel K., Foinquinos A., Xiao K., Thum T. Noncoding RNAs in heart failure. Handb. Exp. Pharmacol. 2017;243:423–445. doi: 10.1007/164_2016_99. [DOI] [PubMed] [Google Scholar]
- 128.Wang Z., Zhang X.J., Ji Y.X., Zhang P., Deng K.Q., Gong J., Ren S., Wang X., Chen I., Wang H. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat. Med. 2016;22:1131–1139. doi: 10.1038/nm.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Han P., Li W., Lin C.H., Yang J., Shang C., Nuernberg S.T., Jin K.K., Xu W., Lin C.Y., Lin C.J. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102–106. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jiang F., Zhou X., Huang J. Long non-coding RNA-ROR mediates the reprogramming in cardiac hypertrophy. PLoS ONE. 2016;11:e0152767. doi: 10.1371/journal.pone.0152767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Murtaza I., Wang H.X., Feng X., Alenina N., Bader M., Prabhakar B.S., Li P.F. Down-regulation of catalase and oxidative modification of protein kinase CK2 lead to the failure of apoptosis repressor with caspase recruitment domain to inhibit cardiomyocyte hypertrophy. J. Biol. Chem. 2008;283:5996–6004. doi: 10.1074/jbc.M706466200. [DOI] [PubMed] [Google Scholar]
- 132.Donath S., Li P., Willenbockel C., Al-Saadi N., Gross V., Willnow T., Bader M., Martin U., Bauersachs J., Wollert K.C., German Heart Failure Network Apoptosis repressor with caspase recruitment domain is required for cardioprotection in response to biomechanical and ischemic stress. Circulation. 2006;113:1203–1212. doi: 10.1161/CIRCULATIONAHA.105.576785. [DOI] [PubMed] [Google Scholar]
- 133.Zsebo K., Yaroshinsky A., Rudy J.J., Wagner K., Greenberg B., Jessup M., Hajjar R.J. Long-term effects of AAV1/SERCA2a gene transfer in patients with severe heart failure: analysis of recurrent cardiovascular events and mortality. Circ. Res. 2014;114:101–108. doi: 10.1161/CIRCRESAHA.113.302421. [DOI] [PubMed] [Google Scholar]
- 134.Gray S.J., Samulski R.J. Optimizing gene delivery vectors for the treatment of heart disease. Expert Opin. Biol. Ther. 2008;8:911–922. doi: 10.1517/14712598.8.7.911. [DOI] [PubMed] [Google Scholar]
- 135.White K., Nicklin S.A., Baker A.H. Novel vectors for in vivo gene delivery to vascular tissue. Expert Opin. Biol. Ther. 2007;7:809–821. doi: 10.1517/14712598.7.6.809. [DOI] [PubMed] [Google Scholar]
- 136.Nathwani A.C., Reiss U.M., Tuddenham E.G., Rosales C., Chowdary P., McIntosh J., Della Peruta M., Lheriteau E., Patel N., Raj D. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med. 2014;371:1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kota J., Chivukula R.R., O’Donnell K.A., Wentzel E.A., Montgomery C.L., Hwang H.W., Chang T.C., Vivekanandan P., Torbenson M., Clark K.R. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Montgomery R.L., Yu G., Latimer P.A., Stack C., Robinson K., Dalby C.M., Kaminski N., van Rooij E. MicroRNA mimicry blocks pulmonary fibrosis. EMBO Mol. Med. 2014;6:1347–1356. doi: 10.15252/emmm.201303604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Quattrocelli M., Crippa S., Montecchiani C., Camps J., Cornaglia A.I., Boldrin L., Morgan J., Calligaro A., Casasco A., Orlacchio A. Long-term miR-669a therapy alleviates chronic dilated cardiomyopathy in dystrophic mice. J. Am. Heart Assoc. 2013;2:e000284. doi: 10.1161/JAHA.113.000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Collins M., Thrasher A. Gene therapy: progress and predictions. Proc. Biol. Sci. 2015;282:20143003. doi: 10.1098/rspb.2014.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jessup M., Greenberg B., Mancini D., Cappola T., Pauly D.F., Jaski B., Yaroshinsky A., Zsebo K.M., Dittrich H., Hajjar R.J., Calcium Upregulation by Percutaneous Administration of Gene Therapy in Cardiac Disease (CUPID) Investigators Calcium Upregulation by Percutaneous Administration of Gene Therapy in Cardiac Disease (CUPID): a phase 2 trial of intracoronary gene therapy of sarcoplasmic reticulum Ca2+-ATPase in patients with advanced heart failure. Circulation. 2011;124:304–313. doi: 10.1161/CIRCULATIONAHA.111.022889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Robb G.B., Brown K.M., Khurana J., Rana T.M. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- 143.Bennett C.F., Swayze E.E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 144.Fluiter K., Mook O.R., Vreijling J., Langkjaer N., Højland T., Wengel J., Baas F. Filling the gap in LNA antisense oligo gapmers: the effects of unlocked nucleic acid (UNA) and 4′-C-hydroxymethyl-DNA modifications on RNase H recruitment and efficacy of an LNA gapmer. Mol. Biosyst. 2009;5:838–843. doi: 10.1039/b903922h. [DOI] [PubMed] [Google Scholar]
- 145.Lennox K.A., Behlke M.A. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016;44:863–877. doi: 10.1093/nar/gkv1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fan Y., Shen B., Tan M., Mu X., Qin Y., Zhang F., Liu Y. TGF-β-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin. Cancer Res. 2014;20:1531–1541. doi: 10.1158/1078-0432.CCR-13-1455. [DOI] [PubMed] [Google Scholar]
- 147.Arun G., Diermeier S., Akerman M., Chang K.C., Wilkinson J.E., Hearn S., Kim Y., MacLeod A.R., Krainer A.R., Norton L. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gutschner T., Hämmerle M., Eissmann M., Hsu J., Kim Y., Hung G., Revenko A., Arun G., Stentrup M., Gross M. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Citti L., Rainaldi G. Synthetic hammerhead ribozymes as therapeutic tools to control disease genes. Curr. Gene Ther. 2005;5:11–24. doi: 10.2174/1566523052997541. [DOI] [PubMed] [Google Scholar]
- 150.Su J.Z., Fukuda N., Hu W.Y., Kanmatsuse K. Ribozyme to human TGF-beta1 mRNA inhibits the proliferation of human vascular smooth muscle cells. Biochem. Biophys. Res. Commun. 2000;278:401–407. doi: 10.1006/bbrc.2000.3814. [DOI] [PubMed] [Google Scholar]
- 151.Pedram Fatemi R., Salah-Uddin S., Modarresi F., Khoury N., Wahlestedt C., Faghihi M.A. Screening for small-molecule modulators of long noncoding RNA-protein interactions using AlphaScreen. J. Biomol. Screen. 2015;20:1132–1141. doi: 10.1177/1087057115594187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Huang X., Yuan T., Tschannen M., Sun Z., Jacob H., Du M., Liang M., Dittmar R.L., Liu Y., Liang M. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Deloukas P., Kanoni S., Willenborg C., Farrall M., Assimes T.L., Thompson J.R., Ingelsson E., Saleheen D., Erdmann J., Goldstein B.A., CARDIoGRAMplusC4D Consortium. DIAGRAM Consortium. CARDIOGENICS Consortium. MuTHER Consortium. Wellcome Trust Case Control Consortium Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pasmant E., Sabbagh A., Vidaud M., Bièche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25:444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 155.Iacobucci I., Sazzini M., Garagnani P., Ferrari A., Boattini A., Lonetti A., Papayannidis C., Mantovani V., Marasco E., Ottaviani E. A polymorphism in the chromosome 9p21 ANRIL locus is associated to Philadelphia positive acute lymphoblastic leukemia. Leuk. Res. 2011;35:1052–1059. doi: 10.1016/j.leukres.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 156.Diederichs S. The four dimensions of noncoding RNA conservation. Trends Genet. 2014;30:121–123. doi: 10.1016/j.tig.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 157.Kino T., Hurt D.E., Ichijo T., Nader N., Chrousos G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Guo X., Gao L., Wang Y., Chiu D.K., Wang T., Deng Y. Advances in long noncoding RNAs: identification, structure prediction and function annotation. Brief. Funct. Genomics. 2016;15:38–46. doi: 10.1093/bfgp/elv022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ding Y., Tang Y., Kwok C.K., Zhang Y., Bevilacqua P.C., Assmann S.M. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 160.Spitale R.C., Crisalli P., Flynn R.A., Torre E.A., Kool E.T., Chang H.Y. RNA SHAPE analysis in living cells. Nat. Chem. Biol. 2013;9:18–20. doi: 10.1038/nchembio.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yang K.C., Yamada K.A., Patel A.Y., Topkara V.K., George I., Cheema F.H., Ewald G.A., Mann D.L., Nerbonne J.M. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation. 2014;129:1009–1021. doi: 10.1161/CIRCULATIONAHA.113.003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Boeckel J.N., Jaé N., Heumüller A.W., Chen W., Boon R.A., Stellos K., Zeiher A.M., John D., Uchida S., Dimmeler S. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ. Res. 2015;117:884–890. doi: 10.1161/CIRCRESAHA.115.306319. [DOI] [PubMed] [Google Scholar]
- 163.Aparicio-Prat E., Arnan C., Sala I., Bosch N., Guigó R., Johnson R. DECKO: single-oligo, dual-CRISPR deletion of genomic elements including long non-coding RNAs. BMC Genomics. 2015;16:846. doi: 10.1186/s12864-015-2086-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Müller R., Weirick T., John D., Militello G., Chen W., Dimmeler S., Uchida S. ANGIOGENES: knowledge database for protein-coding and noncoding RNA genes in endothelial cells. Sci. Rep. 2016;6:32475. doi: 10.1038/srep32475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhou K.R., Liu S., Sun W.J., Zheng L.L., Zhou H., Yang J.H., Qu L.H. ChIPBase v2.0: decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data. Nucleic Acids Res. 2017;45:D43–D50. doi: 10.1093/nar/gkw965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yang J.H., Li J.H., Jiang S., Zhou H., Qu L.H. ChIPBase: a database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-seq data. Nucleic Acids Res. 2013;41:D177–D187. doi: 10.1093/nar/gks1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zheng L.L., Li J.H., Wu J., Sun W.J., Liu S., Wang Z.L., Zhou H., Yang J.H., Qu L.H. deepBase v2.0: identification, expression, evolution and function of small RNAs, lncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res. 2016;44:D196–D202. doi: 10.1093/nar/gkv1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yang J.H., Shao P., Zhou H., Chen Y.Q., Qu L.H. deepBase: a database for deeply annotating and mining deep sequencing data. Nucleic Acids Res. 2010;38:D123–D130. doi: 10.1093/nar/gkp943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mudge J.M., Harrow J. Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mamm. Genome. 2015;26:366–378. doi: 10.1007/s00335-015-9583-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. GENCODE: the reference human genome annotation for the ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ning S., Yue M., Wang P., Liu Y., Zhi H., Zhang Y., Zhang J., Gao Y., Guo M., Zhou D. LincSNP 2.0: an updated database for linking disease-associated SNPs to human long non-coding RNAs and their TFBSs. Nucleic Acids Res. 2017;45:D74–D78. doi: 10.1093/nar/gkw945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Volders P.J., Verheggen K., Menschaert G., Vandepoele K., Martens L., Vandesompele J., Mestdagh P. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43:D174–D180. doi: 10.1093/nar/gku1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Volders P.J., Helsens K., Wang X., Menten B., Martens L., Gevaert K., Vandesompele J., Mestdagh P. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2013;41:D246–D251. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Quek X.C., Thomson D.W., Maag J.L., Bartonicek N., Signal B., Clark M.B., Gloss B.S., Dinger M.E. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015;43:D168–D173. doi: 10.1093/nar/gku988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Amaral P.P., Clark M.B., Gascoigne D.K., Dinger M.E., Mattick J.S. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39:D146–D151. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chen G., Wang Z., Wang D., Qiu C., Liu M., Chen X., Zhang Q., Yan G., Cui Q. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983–D986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bhartiya D., Pal K., Ghosh S., Kapoor S., Jalali S., Panwar B., Jain S., Sati S., Sengupta S., Sachidanandan C. lncRNome: a comprehensive knowledgebase of human long noncoding RNAs. Database (Oxford) 2013;2013:bat034. doi: 10.1093/database/bat034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhao Y., Li H., Fang S., Kang Y., Wu W., Hao Y., Li Z., Bu D., Sun N., Zhang M.Q., Chen R. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016;44:D203–D208. doi: 10.1093/nar/gkv1252. [DOI] [PMC free article] [PubMed] [Google Scholar]