Abstract
Biglycan (BGN), a proteoglycan of the extracellular matrix, is included in mRNA signatures for prostate cancer aggressiveness. To understand the impact of BGN on prognosis and its relationship to molecularly defined subsets, we analyzed BGN expression by immunohistochemistry on a tissue microarray containing 12,427 prostate cancers. Seventy-eight percent of 11,050 interpretable cancers showed BGN expression, which was considered as low intensity in 47.7% and as high intensity in 31.1% of cancers. BGN protein expression rose with increasing pathological tumor stage, Gleason grade, lymph node metastasis and early PSA recurrence (P < .0001 each). Comparison with our molecular database attached to the TMA revealed that BGN expression was linked to presence of TMPRRS2:ERG fusion and PTEN deletion (P < .0001 each). In addition, BGN was strongly linked to androgen-receptor (AR) levels (P < .0001), suggesting a hormone-depending regulation of BGN. BGN up-regulation is a frequent feature of prostate cancer that parallels tumor progression and may be useful to estimate tumor aggressiveness particularly if combined with other molecular markers.
Abbreviations: BGN, Biglycan; CHD1, Chromodomain-helicase-DNA-binding protein 1; ERCC1, Excision repair cross-complementation; ERG, Erythroblast transformation-specific (ETS) related gene; FISH, Fluorescence in situ hybridization; FOXP1, Fork head box protein P1; Ki67LI, Ki67 labeling index; MAP3K7, Mitogen-activated protein kinase kinase kinase 7; PSA, Prostate specific antigen; PTEN, Phosphatase and tensin homolog; SLRP, Small leucine-rich proteoglycan; TGF-β, Transforming growth factor β; TMA, Tissue microarray; TMPRSS2, Trans membrane protease, serine 2
Introduction
Prostate cancer is the most prevalent cancer in men in Western societies. Although most prostate cancers have a rather indolent clinical course, this disease still represents the third most common cause of cancer related death in men [1]. A reliable distinction between indolent and aggressive forms of the disease is highly desirable to enhance the quality of therapeutic decisions. Despite recent advances, the only established pretreatment prognostic parameters currently remain Gleason grade and tumor extent on biopsies, preoperative prostate-specific antigen (PSA), and clinical stage. Because these data are statistically powerful but not sufficient for optimal individual treatment decisions, it can be hoped that a better understanding of disease biology will eventually lead to the identification of clinically applicable molecular markers that enable a more reliable prediction of prostate cancer aggressiveness.
Biglycan (BGN) is a member of the small leucine-rich repeat proteoglycans (SLRP) family characterized by a core protein with leucine-rich repeats attended by cysteine clusters [2]. BGN is normally secreted from extracellular matrix fibroblasts and facilitates assembly of collagen fibrils and bone matrix [3]. In addition, BGN up-regulation has been implicated in the response of inflammatory processes triggered by transforming growth factor β (TGF-ß) [4], [5], [6]. More importantly, BGN up-regulation has been reported from many malignant epithelial tumors, including a large variety of gastro-intestinal cancers [7], [8], [9], [10] as well as gynecological tumors [11]. In some of these cancers, BGN up-regulation has been linked to advanced [8], [12] and metastatic [11], [13], [14] cancers or adverse patient prognosis. That opposite observations have been made in other tumor types [12], [15] may suggest tumor type specific roles of BGN expression. Earlier work has demonstrated that the prostate is an abundant secretor of glycoproteins, including many types of proteoglycans [16]. In prostate cancer, BGN has gained interest because it is part of a commercial RNA expression signature for estimating prostate cancer aggressiveness [17].
To understand the role of BGN for prostate cancer biology and is association to molecular features of the disease, we analyzed our large tissue microarray (TMA) resource including more than 12,400 prostate cancers for immunohistochemical BGN expression. The database attached to our TMA contains pathological and clinical follow-up data, as well molecular data of key molecular alterations of this disease.
Materials and Methods
Patients
Radical prostatectomy specimens were available from 12,427 patients, undergoing surgery between 1992 and 2012 at the Department of Urology and the Martini Clinics at the University Medical Center Hamburg-Eppendorf. The local ethics committee (Ethics commission Ärztekammer Hamburg, WF-049/09 and PV3652) approved the use of specimens and data for research. According to local laws (HmbKHG, §12,1), informed consent was not required for this study. Patient records/information was anonymized and de-identified prior to analysis. All work has been carried out in compliance with the Helsinki Declaration. Follow-up data were available for a total of 11,665 patients with a median follow-up of 50.0 months (range: 1 to 264 months; Table 1). Prostate specific antigen (PSA) values were measured following surgery and PSA recurrence was defined as a postoperative PSA of 0.2 ng/ml and increasing at first of appearance. All prostate specimens were analyzed according to a standard procedure, including a complete embedding of the entire prostate for histological analysis [18]. The TMA manufacturing process was described earlier in detail [19], [20]. In short, one 0.6 mm core was taken from a representative tissue block from each patient. The tissues were distributed among 27 TMA blocks, each containing 144 to 522 tumor samples. For internal controls, each TMA block also contained various control tissues, including normal prostate tissue. The molecular database attached to this TMA contained results on ERG expression in 10,678 [21], ERG break apart fluorescence in situ hybridization (FISH) analysis in 7099 (expanded from [22]) and deletion status of 5q21 (CHD1) in 7932 (expanded from [23]), 6q15 (MAP3K7) in 6069 (expanded from [24]), PTEN (10q23) in 6704 (expanded from [25]) and 3p13 (FOXP1) in 7081 (expanded from [26]) cancers.
Table 1.
Pathological and Clinical Data of the Arrayed Prostate Cancers
| No. of Patients (%) |
||
|---|---|---|
| Study Cohort on TMA(n = 12,427) | Biochemical Relapse Among Categories | |
| Follow-up (mo.) | ||
| n | 11,665 (93.9%) | 2769 (23.7%) |
| Mean | 62.9 | - |
| Median | 50.0 | - |
| Age (y) | ||
| ≤50 | 334 (2.7%) | 81 (24.3%) |
| 51–59 | 3061 (24.8%) | 705 (23%) |
| 60–69 | 7188 (58.2%) | 1610 (22.4%) |
| ≥70 | 1761 (14.3%) | 370 (21%) |
| Pretreatment PSA (ng/ml) | ||
| <4 | 1585 (12.9%) | 242 (15.3%) |
| 4–10 | 7480 (60.9%) | 1355 (18.1%) |
| 10–20 | 2412 (19.6%) | 737 (30.6%) |
| >20 | 812 (6.6%) | 397 (48.9%) |
| pT stage (AJCC 2002) | ||
| pT2 | 8187 (66.2%) | 1095 (13.4%) |
| pT3a | 2660 (21.5%) | 817 (30.7%) |
| pT3b | 1465 (11.8%) | 796 (54.3%) |
| pT4 | 63 (0.5%) | 51 (81%) |
| Gleason grade | ||
| ≤3 + 3 | 2848 (22.9%) | 234 (8.2%) |
| 3 + 4 | 6679 (53.8%) | 1240 (18.6%) |
| 3 + 4 Tertiary 5 | 433 (3.5%) | 115 (26.6%) |
| 4 + 3 | 1210 (9.7%) | 576 (47.6%) |
| 4 + 3 Tertiary 5 | 646 (5.2%) | 317 (49.1%) |
| ≥4 + 4 | 596 (4.8%) | 348 (58.4%) |
| pN stage | ||
| pN0 | 6970 (91%) | 1636 (23.5%) |
| pN+ | 693 (9%) | 393 (56.7%) |
| Surgical margin | ||
| Negative | 9990 (81.9%) | 1848 (18.5%) |
| Positive | 2211 (18.1%) | 853 (38.6%) |
Percent in the column “Study cohort on TMA” refers to the fraction of samples across each category. Percent in column “Biochemical relapse among categories” refers to the fraction of samples with biochemical relapse within each parameter in the different categories. NOTE: Numbers do not always add up to 12,427 in different categories because of cases with missing data. Abbreviation: AJCC, American Joint Committee on Cancer.
Immunohistochemistry
Freshly cut TMA sections were stained in 1 day and in one experiment. Slides were deparaffinized and exposed to heat-induced antigen retrieval for 5 minutes in an autoclave at 121 °C in pH 7.8 Tris-EDTA-citrate buffer. Primary antibody specific for BGN (rabbit polyclonal antibody, Sigma-Aldrich, Germany; cat#HPA003157; dilution 1:1350) was applied at 37 °C for 60 minutes. Bound antibody was then visualized using the EnVision Kit (Dako, Glostrup, Denmark) according to the manufacturer's directions. BGN protein expression was typically seen in the cytoplasm of all (100%) tumor cells. Accordingly, the staining intensity in prostate epithelial cells was recorded in three categories for each cancer, including negative (no to weak detectable staining), low (moderate staining) and high (strong staining).
Statistics
For statistical analysis, the JMP 12.0 software (SAS Institute Inc., NC, USA) was used. Contingency tables were calculated to study association between BGN expression categories and clinicopathological variables, and the chi-square (Likelihood) test was used to find significant relationships. Kaplan–Meier curves were generated using biochemical (PSA) recurrence as the clinical endpoint. The log-rank test was applied to test the significance of differences between stratified survival functions. Cox proportional hazards regression analysis was performed to test the statistical independence and significance between pathological, molecular, and clinical variables.
Results
Technical Issues
A total of 11,050 (89.0%) tumor samples were interpretable in our TMA analysis. Reasons for non-informative cases (1377 spots; 11.0%) included lack of tissue samples or absence of unequivocal cancer cells in the TMA spot.
BGN Expression in Normal and Cancerous Prostate Epithelium
Normal prostate epithelium did not show detectable BGN staining under the selected experimental conditions. In cancers, BGN staining was localized in the cytoplasm. Positive BGN staining was seen in 8701 of our 11,050 (78.7%) interpretable prostate cancers and was considered as low intensity in 47.7% and as high intensity in 31.1% of cancers. Representative images of negative and positive BGN staining are given in Figure 1.
Figure 1.
Representative pictures of BGN staining in prostate cancer with negative (A), low (B), and high-intensity (C) staining at 100×, insets 400× magnification and spot size of 600 μm.
Association with TMPRSS2:ERG Fusion Status, ERG Protein Expression and PTEN Deletion
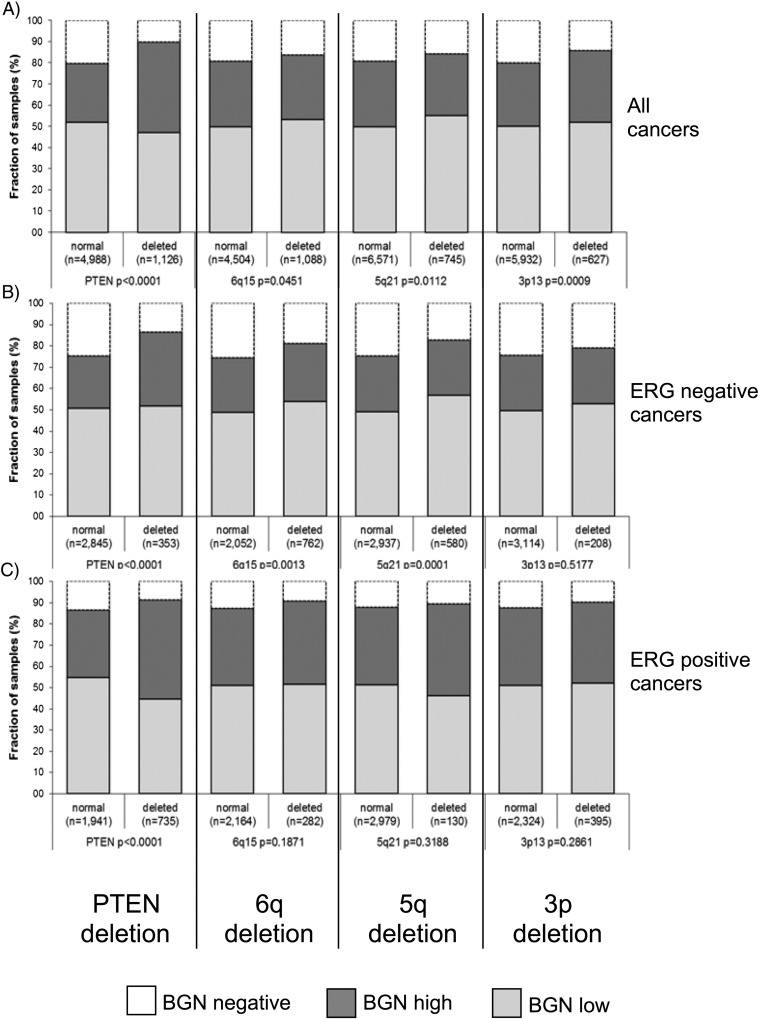
Data on TMPRSS2:ERG fusion status obtained by FISH were available from 6462 and by immunohistochemistry from 9686 tumors with evaluable BGN staining. Data on both ERG FISH and IHC were available from 6201 cancers, and an identical result (ERG IHC-positive and break by FISH or ERG IHC-negative and missing break by FISH) was found in 5919 of 6201 (95.5%) cancers. High-level BGN staining was linked to TMPRSS2:ERG rearrangement and ERG positivity in prostate cancers. BGN staining was seen in 87.3% and 86.8% of cancers with TMPRSS2:ERG fusion detected by IHC and FISH, but found in only 73.8% of cancers without ERG staining and 76.7% of cancers without ERG rearrangements detected by FISH (P < .0001 each). High-level BGN staining was also significantly linked to PTEN deletion (Figure 2). The effect was additive and of similar size as the ERG fusion effect.
Figure 2.
Association between BGN staining, ERG status determined by immunohistochemistry and PTEN deletion by fluorescence in situ hybridization.
Associations with Tumor Phenotype
High grade BGN expression was significantly linked to advanced pT stage, high Gleason grade, lymph node metastases, high preoperative PSA-levels (P < .0001 each) and surgical margin positivity (P = .0007, Table 2). Subgroup analysis revealed that these associations held also true in subsets of cancers with and without ERG fusion (Supplementary Tables 1 and 2).
Table 2.
Association between BGN Staining Results and Prostate Cancer Phenotype
| BGN IHC Result (%) |
|||||
|---|---|---|---|---|---|
| Parameter | n Evaluable | Negative | Low | High | P Value |
| All cancers | 11,050 | 21.3 | 47.7 | 31.1 | |
| Tumor stage | |||||
| pT2 | 7182 | 23.8 | 49.0 | 27.2 | <0.0001 |
| pT3a | 2430 | 17.6 | 47.1 | 35.3 | |
| pT3b-pT4 | 1395 | 14.6 | 42.2 | 43.3 | |
| Gleason grade | |||||
| ≤3 + 3 | 2460 | 27.0 | 48.6 | 24.4 | <0.0001 |
| 3 + 4 | 5663 | 21.4 | 48.6 | 29.9 | |
| 3 + 4 Tert.5 | 410 | 16.8 | 46.1 | 37.1 | |
| 4 + 3 | 949 | 18.8 | 46.6 | 34.7 | |
| 4 + 3 Tert.5 | 580 | 14.1 | 48.4 | 37.4 | |
| ≥4 + 4 | 474 | 15.8 | 42.0 | 42.2 | |
| Lymph node metastasis | |||||
| N0 | 6271 | 19.5 | 47.7 | 32.8 | <0.0001 |
| N+ | 634 | 13.7 | 41.3 | 45.0 | |
| Preoperative PSA level (ng/ml) | |||||
| <4 | 1366 | 20.8 | 50.2 | 29.0 | <0.0001 |
| 4–10 | 6618 | 22.1 | 49.1 | 28.7 | |
| 10–20 | 2186 | 20.1 | 43.6 | 36.2 | |
| >20 | 759 | 17.8 | 42.6 | 39.7 | |
| Surgical margin | |||||
| Negative | 8801 | 21.7 | 48.1 | 30.2 | 0.0007 |
| Positive | 2047 | 19.5 | 46.1 | 34.4 | |
Association with Other Key Genomic Deletions
To study whether BGN expression might be particularly associated with genomic deletions, BGN data were compared to preexisting findings on PTEN (10q23), 3p13 (FOXP1), 6q15 (MAP3K7) and 5q21 (CHD1) deletion. This analysis revealed that high BGN expression was significantly linked to all of these deletions (Figure 3A) and that the association was most striking for PTEN deletions. Subset analysis of cancers with and without ERG fusion (Figure 3, B and C) revealed that only PTEN deletion was strongly linked to high BGN expression in both subgroups. For the other deletions, the association with high level BGN expression was largely limited to the subset of ERG-negative cancers.
Figure 3.
Association between positive BGN staining and 10q23 (PTEN), 5q21 (CHD1), 6q15 (MAP3K7), 3p13 (FOXP1) deletions in all cancers (A), in ERG-negative (B), and ERG-positive cancers (C).
Association with Tumor Cell Proliferation
High-level BGN staining was significantly linked to high cell proliferation as measured by Ki67 labeling index (LI). The average Ki67LI increased from 2.1 ± 0.07 in cancers lacking BGN expression to 2.9 ± 0.05 in cancers with low and to 3.0 ± 0.06 in cancers with high BGN levels (P < .0001). This association held true in all tumor subsets with identical Gleason score (≤3 + 3: P < .001, 3 + 4: P < .0001, 4 + 3: P = .0085, ≥4 + 4: P = .0137; data not shown).
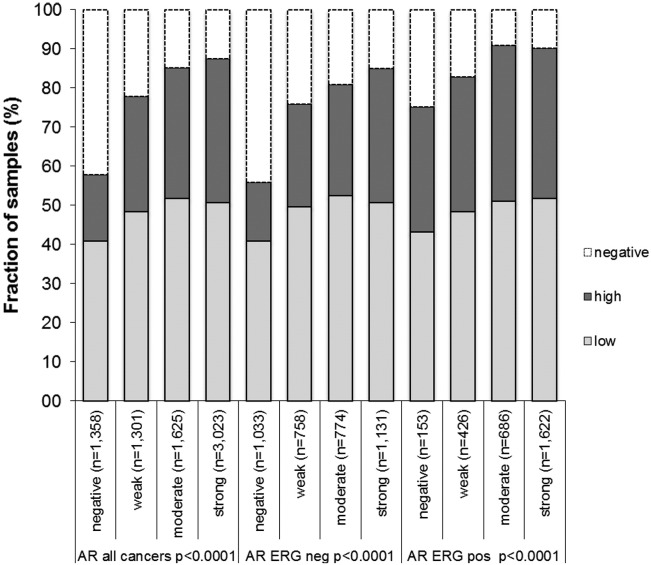
Association with Androgen Receptor
Androgen receptor (AR) expression is a characteristic feature of neoplastic and non-neoplastic prostate epithelial cells. Immunohistochemical data on AR expression were obtained from a previous study [21]. AR expression was strongly correlated to BGN staining. For example, high BGN expression was found in 16.9% of cancers without detectable AR expression, but in 36.8% of tumors with strong AR expression (P < .0001 each). These associations held also true in subsets of cancers with and without ERG fusion (P < .0001 each, Figure 4).
Figure 4.
Association between BGN staining results and androgen receptor (AR) in all cancers, ERG-negative tumors, and ERG-positive tumors.
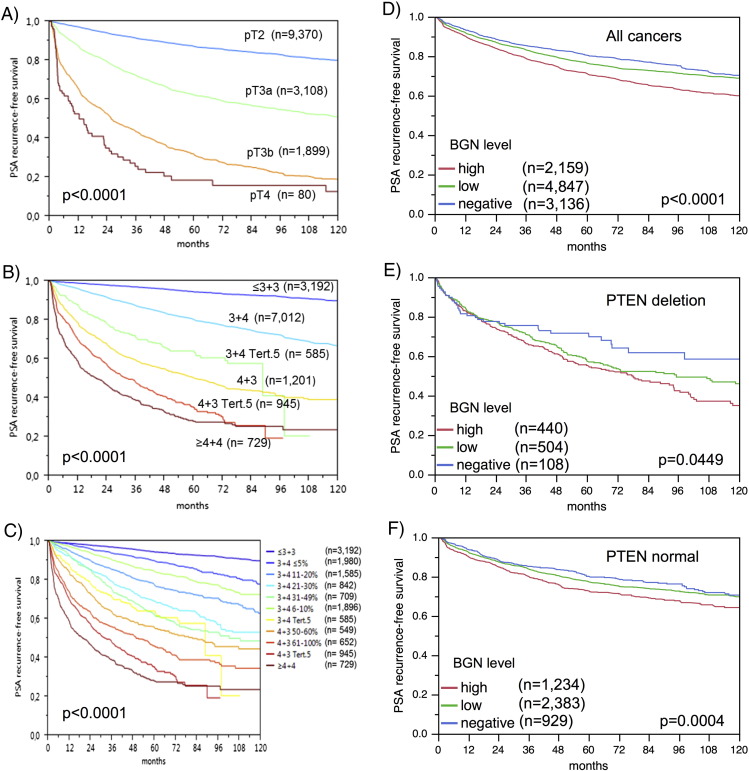
Association with PSA Recurrence
Follow-up data were available for 10,359 patients with interpretable BGN staining on the TMA. The prognostic impact of pT stage, traditional Gleason grade, and quantitative Gleason grade are shown in Figure 5, A–C). High-level BGN expression was significantly associated with early PSA recurrence in all tumors and also in subgroup analyses limited to the subsets of ERG-negative and ERG-positive cancers (P < .0001 each, Figure 5, D–F). To better understand the prognostic power of BGN, we performed further subset analyses in cancers with identical classical and quantitative Gleason scores. Here, BGN staining did not provide prognostic information beyond the Gleason score, neither in any subsets defined by the classical Gleason score (Supplementary Figure 1a) nor by the quantitative Gleason score (Supplementary Figure 1b–h).
Figure 5.
Kaplan–Meier plots of prostate specific antigen (PSA) recurrence after radical prostatectomy and pathological tumor stage (A), classical Gleason score (B), quantitative Gleason score (C), BGN expression in all cancers (D), in PTEN deletion (E), and PTEN normal subset (F).
Multivariate Analysis
Four different types of multivariate analyses were performed evaluating the clinical relevance of BGN expression in different scenarios (Supplementary Table 3). Scenario 1 evaluated all postoperatively available parameters including pathological tumor stage, pathological lymph node status (pN), surgical margin status, preoperative PSA value and pathological Gleason grade obtained after the morphological evaluation of the entire resected prostate. In scenario 2, all postoperatively available parameters with exception of nodal status were included. The rational for this approach was that the indication and extent of lymph node dissection is not standardized in the surgical therapy of prostate cancer and that excluding pN in multivariate analysis can markedly increase case numbers. Two additional scenarios had the purpose to model the preoperative situation as much as possible. Scenario 3 included BGN expression, preoperative PSA, clinical tumor stage (cT stage) and Gleason grade obtained on the prostatectomy specimen. Since postoperative determination of a tumors Gleason grade is “better” than the preoperatively determined Gleason grade (subjected to sampling errors and consequently under-grading in more than one third of cases [27]), another multivariate analysis was added. In scenario 4, the preoperative Gleason grade obtained on the original biopsy was combined with preoperative PSA, cT stage and BGN expression. Overall, these data show, that BGN expression did not provide independent prognostic information, neither in all cancers nor in the subsets of ERG-positive and -negative subgroups.
Discussion
The results of our study identify high BGN expression as a weak prognostic feature in prostate cancer, which is also linked to PTEN deleted cancers. Our immunohistochemical analysis revealed cytoplasmic BGN staining in about 80.0% of 11,050 of the interpretable prostate cancers. The lack of BGN staining in normal prostate epithelium indicates that BGN becomes unregulated during prostate cancer development in a large fraction of patients. Comparable data on immunohistochemical BGN expression in prostate cancer are currently lacking in the literature. However, tumor associated up-regulation of BGN has been also reported from esophageal squamous cell carcinoma [9], pancreatic [7], [15], gastric [10], colorectal [8], odontogenic [28], endometrial [11], [29] and ovarian cancer [30]. BGN overexpression was significantly associated with an unfavorable tumor phenotype in our study, including advanced pT stage, high Gleason grade, accelerated cell proliferation, lymph node metastases and early biochemical recurrence (P < .0001 each). These findings are in line with data from studies on various other cancer types. For example, BGN overexpression has been linked to tumor aggressiveness and shortened patient survival in pancreatic, gastric and endometrial cancer [7], [10], [29]. Reasons for the tumor-associated up-regulation may include the role of BGN as a downstream target of various growth and signal transduction pathways like TGFb, Wnt and Akt signaling [31], [32], [33], activation of which is frequently found in malignant tumors [34], [35], [36], [37]. The general importance of these pathways in prostate cancer [38], [39], [40], [41] may not only explain the high fraction of BGN expressing cancers but also its comparatively low prognostic impact. That BGN up-regulation has also been linked to reduce cell proliferation in some cell lines from cancers and fibroblasts [12], [15], [42] is consistent with the cell-type dependent effects of BGN expression. The molecular database attached to our TMA allowed us to draw conclusions on some molecular mechanisms associated with BGN up-regulation. It is well known that about 50% of all prostate cancers carry a gene fusion linking the androgen-regulated serine protease TMPRSS2 with the ETS-transcription factor ERG resulting in an androgen-related overexpression of ERG with subsequent transcriptional deregulation of more than 1600 ERG target genes [21], [43], [44]. Others and we have shown that activation of the TGF-ß signaling pathway is one important consequence of ERG fusion in prostate cancer [44], [45], [46]. That the extracellular matrix modulator TGF-β1 is an important stimulator of BGN transcriptional and post translational modifications [47], [48] provides a mechanistic explanation for the higher fraction of cancers with BGN up-regulation in ERG-positive as compared to ERG-negative cancers. In addition, earlier work involving computational promoter analyses revealed a binding site for ETS transcription factors [49], suggesting that activated ERG might also directly contribute to the up-regulation of BGN in prostate cancer. Another interesting finding was the strong association of BGN overexpression with AR levels, which suggests an androgen-dependent regulation of BGN in prostate cancer. This is supported by studies reporting a role of androgen in regulating collagen and proteoglycan organization in the cervix [50] as well as an androgen depending synthesis of BGN in vascular smooth muscle cells [51]. Given that increased AR signaling is a hallmark of prostate cancer [52], it is tempting to speculate that androgen-dependent up-regulation of BGN may contribute to changes in the extracellular matrix causing the increased tissue stiffness of cancer areas that is often felt during digital rectal examination. The reaction of the prostate to cancer cell invasion is thought to resemble processes involved in wound healing, including increased cellular density, elevated micro vascularity, and increased collagen deposition in the stroma [53], [54]. In fact, also other markers of collagen synthesis such as serine proteases, matrix metalloproteases and propeptides of type I collagen have been found to be significantly increased not only in prostate cancer cells but also in surrounding unaffected tissues [55].
Recurrent deletions including PTEN, 6q15, 5q21, 3p13, are another hallmark of prostate cancer [56], [57]. PTEN inactivation results in hyperactive AKT signaling and is associated with tumor growth, progression and poor clinical outcome [58]. That BGN overexpression was particularly linked to PTEN deletions is of interest with respect to previous reports linking class I proteoglycans – including BGN — to several growth pathways including AKT signaling [2], [31]). For example, earlier work demonstrates that excess extracellular matrix concentrations of soluble BGN can induce cell growth and survival via activation of multiple growth factor receptors including the AKT-upstream receptor EGFR [31], [59]. That activated AKT is also required for BGN core protein synthesis [33] is seen as further support for a role of BGN in signal regulation networks [2], and might also provide an explanation for the marked up-regulation of BGN in PTEN-deleted and AKT-hyperactivated cancers. Recently Li et al. reported a role of PTEN in induction of type I interferon linking the tumor suppressor with the innate immune system [60]. Since BGN is a prototype endogenous ligand to Toll-like receptors stimulating innate immunity [61], it is tempting to speculate that up-regulation of BGN in the progress to aggressive prostate cancer may be counteracted by PTEN loss. Although BGN expression was a prognostic factor in univariate calculations, its prognostic impact was lost in most multivariate analyses including established morphological parameters. The power of morphological methods competing with biomarkers for predicting prostate cancer aggressiveness is best demonstrated by the separate analysis of tumors with comparable morphology. Already within traditional grade groups, the prognostic impact of BGN expression was lost. Based on the large cohort of prostate cancers available at our institution, we had recently shown that prognostic Gleason Grade information can be further refined by using the percentage of Gleason 4 grades as a continuous variable (quantitative Gleason Grade) [62]. Both in biopsies and in prostatectomy samples, prostate cancer prognosis continuously deteriorates with increasing percentage of Gleason 4 pattern. That BGN expression lacks any prognostic impact in all subgroups defined by a comparable quantitative Gleason grade demonstrates how difficult it is for biomarkers to outperform morphological malignancy parameters in prostate cancer. However, it is our anticipation that prognostic gene sets will assist routine clinical decision-making in prostate cancer in the future. It appears likely that combining molecular markers will enable a better and reproducible prognosis prediction than single markers.
In summary, BGN overexpression is a frequent feature of prostate cancer, which parallels tumor progression and is particularly linked to PTEN deleted cancers. Although BGN mRNA measurement is a part of a commercial gene signature estimating prostate cancer aggressiveness the prognostic power of BGN protein measurement is weak and may have clinical use only if combined with other molecular markers.
Acknowledgements
The authors appreciate the excellent technical support of Christina Koop, Sylvia Schnöger and Sasha Eghtessadi.
Footnotes
Disclosure/Conflict of interest: There are no proprietary interests and no financial support was received. No conflicts of interest regarding the article exist.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neo.2017.06.003.
Appendix A. Supplementary data
Supplementary materials
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Schaefer L, Iozzo RV. Biological functions of the small leucine-rich proteoglycans: from genetics to signal transduction. J Biol Chem. 2008;283(31):21305–21309. doi: 10.1074/jbc.R800020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xing X, Gu X, Ma T, Ye H. Biglycan up-regulated vascular endothelial growth factor (VEGF) expression and promoted angiogenesis in colon cancer. Tumour Biol. 2015;36(3):1773–1780. doi: 10.1007/s13277-014-2779-y. [DOI] [PubMed] [Google Scholar]
- 4.Nastase MV, Young MF, Schaefer L. Biglycan: a multivalent proteoglycan providing structure and signals. J Histochem Cytochem. 2012;60(12):963–975. doi: 10.1369/0022155412456380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hildebrand A, Romaris M, Rasmussen LM, Heinegard D, Twardzik DR, Border WA, Ruoslahti E. Interaction of the small interstitial proteoglycans biglycan, decorin and fibromodulin with transforming growth factor beta. Biochem J. 1994;302(Pt 2):527–534. doi: 10.1042/bj3020527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaefer L, Babelova A, Kiss E, Hausser HJ, Baliova M, Krzyzankova M, Marsche G, Young MF, Mihalik D, Gotte M. The matrix component biglycan is proinflammatory and signals through Toll-like receptors 4 and 2 in macrophages. J Clin Invest. 2005;115(8):2223–2233. doi: 10.1172/JCI23755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aprile G, Avellini C, Reni M, Mazzer M, Foltran L, Rossi D, Cereda S, Iaiza E, Fasola G, Piga A. Biglycan expression and clinical outcome in patients with pancreatic adenocarcinoma. Tumour Biol. 2013;34(1):131–137. doi: 10.1007/s13277-012-0520-2. [DOI] [PubMed] [Google Scholar]
- 8.Gu X, Ma Y, Xiao J, Zheng H, Song C, Gong Y, Xing X. Up-regulated biglycan expression correlates with the malignancy in human colorectal cancers. Clin Exp Med. 2012;12(3):195–199. doi: 10.1007/s10238-011-0155-4. [DOI] [PubMed] [Google Scholar]
- 9.Zhu YH, Yang F, Zhang SS, Zeng TT, Xie X, Guan XY. High expression of biglycan is associated with poor prognosis in patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2013;6(11):2497–2505. [PMC free article] [PubMed] [Google Scholar]
- 10.Wang B, Li GX, Zhang SG, Wang Q, Wen YG, Tang HM, Zhou CZ, Xing AY, Fan JW, Yan DW. Biglycan expression correlates with aggressiveness and poor prognosis of gastric cancer. Exp Biol Med (Maywood) 2011;236(11):1247–1253. doi: 10.1258/ebm.2011.011124. [DOI] [PubMed] [Google Scholar]
- 11.Sun H, Wang X, Zhang Y, Che X, Liu Z, Zhang L, Qiu C, Lv Q, Jiang J. Biglycan enhances the ability of migration and invasion in endometrial cancer. Arch Gynecol Obstet. 2016;293(2):429–438. doi: 10.1007/s00404-015-3844-5. [DOI] [PubMed] [Google Scholar]
- 12.Niedworok C, Rock K, Kretschmer I, Freudenberger T, Nagy N, Szarvas T, Vom Dorp F, Reis H, Rubben H, Fischer JW. Inhibitory role of the small leucine-rich proteoglycan biglycan in bladder cancer. PLoS One. 2013;8(11):e80084. doi: 10.1371/journal.pone.0080084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maishi N, Ohba Y, Akiyama K, Ohga N, Hamada J, Nagao-Kitamoto H, Alam MT, Yamamoto K, Kawamoto T, Inoue N. Tumour endothelial cells in high metastatic tumours promote metastasis via epigenetic dysregulation of biglycan. Sci Rep. 2016;6:28039. doi: 10.1038/srep28039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu L, Duan YT, Li JF, Su LP, Yan M, Zhu ZG, Liu BY, Yang QM. Biglycan enhances gastric cancer invasion by activating FAK signaling pathway. Oncotarget. 2014;5(7):1885–1896. doi: 10.18632/oncotarget.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber CK, Sommer G, Michl P, Fensterer H, Weimer M, Gansauge F, Leder G, Adler G, Gress TM. Biglycan is overexpressed in pancreatic cancer and induces G1-arrest in pancreatic cancer cell lines. Gastroenterology. 2001;121(3):657–667. doi: 10.1053/gast.2001.27222. [DOI] [PubMed] [Google Scholar]
- 16.Munkley J, Mills IG, Elliott DJ. The role of glycans in the development and progression of prostate cancer. Nat Rev Urol. 2016;13(6):324–333. doi: 10.1038/nrurol.2016.65. [DOI] [PubMed] [Google Scholar]
- 17.Knezevic D, Goddard AD, Natraj N, Cherbavaz DB, Clark-Langone KM, Snable J, Watson D, Falzarano SM, Magi-Galluzzi C, Klein EA. Analytical validation of the Oncotype DX prostate cancer assay - a clinical RT-PCR assay optimized for prostate needle biopsies. BMC Genomics. 2013;14:690–702. doi: 10.1186/1471-2164-14-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erbersdobler A, Hammerer P, Huland H, Henke RP. Numerical chromosomal aberrations in transition-zone carcinomas of the prostate. J Urol. 1997;158(4):1594–1598. [PubMed] [Google Scholar]
- 19.Mirlacher M, Simon R. Recipient block TMA technique. Methods Mol Biol. 2010;664:37–44. doi: 10.1007/978-1-60761-806-5_4. [DOI] [PubMed] [Google Scholar]
- 20.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4(7):844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 21.Weischenfeldt J, Simon R, Feuerbach L, Schlangen K, Weichenhan D, Minner S, Wuttig D, Warnatz HJ, Stehr H, Rausch T. Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell. 2013;23(2):159–170. doi: 10.1016/j.ccr.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Minner S, Enodien M, Sirma H, Luebke AM, Krohn A, Mayer PS, Simon R, Tennstedt P, Muller J, Scholz L. ERG status is unrelated to PSA recurrence in radically operated prostate cancer in the absence of antihormonal therapy. Clin Cancer Res. 2011;17(18):5878–5888. doi: 10.1158/1078-0432.CCR-11-1251. [DOI] [PubMed] [Google Scholar]
- 23.Burkhardt L, Fuchs S, Krohn A, Masser S, Mader M, Kluth M, Bachmann F, Huland H, Steuber T, Graefen M. CHD1 is a 5q21 tumor suppressor required for ERG rearrangement in prostate cancer. Cancer Res. 2013;73(9):2795–2805. doi: 10.1158/0008-5472.CAN-12-1342. [DOI] [PubMed] [Google Scholar]
- 24.Kluth M, Hesse J, Heinl A, Krohn A, Steurer S, Sirma H, Simon R, Mayer PS, Schumacher U, Grupp K. Genomic deletion of MAP3K7 at 6q12-22 is associated with early PSA recurrence in prostate cancer and absence of TMPRSS2:ERG fusions. Mod Pathol. 2013;26(7):975–983. doi: 10.1038/modpathol.2012.236. [DOI] [PubMed] [Google Scholar]
- 25.Krohn A, Diedler T, Burkhardt L, Mayer PS, De Silva C, Meyer-Kornblum M, Kotschau D, Tennstedt P, Huang J, Gerhauser C. Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancer. Am J Pathol. 2012;181(2):401–412. doi: 10.1016/j.ajpath.2012.04.026. [DOI] [PubMed] [Google Scholar]
- 26.Krohn A, Seidel A, Burkhardt L, Bachmann F, Mader M, Grupp K, Eichenauer T, Becker A, Adam M, Graefen M. Recurrent deletion of 3p13 targets multiple tumour suppressor genes and defines a distinct subgroup of aggressive ERG fusion-positive prostate cancers. J Pathol. 2013;231(1):130–141. doi: 10.1002/path.4223. [DOI] [PubMed] [Google Scholar]
- 27.Epstein JI, Feng Z, Trock BJ, Pierorazio PM. Upgrading and downgrading of prostate cancer from biopsy to radical prostatectomy: incidence and predictive factors using the modified Gleason grading system and factoring in tertiary grades. Eur Urol. 2012;61(5):1019–1024. doi: 10.1016/j.eururo.2012.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Modolo F, Biz MT, Martins MT, Machado de Sousa SO, de Araujo NS. Expression of extracellular matrix proteins in adenomatoid odontogenic tumor. J Oral Pathol Med. 2010;39(3):230–235. doi: 10.1111/j.1600-0714.2009.00846.x. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Li W, Li X, Tai Y, Lu Q, Yang N, Jiang J. Expression and significance of biglycan in endometrial cancer. Arch Gynecol Obstet. 2014;289(3):649–655. doi: 10.1007/s00404-013-3017-3. [DOI] [PubMed] [Google Scholar]
- 30.Pan S, Cheng L, White JT, Lu W, Utleg AG, Yan X, Urban ND, Drescher CW, Hood L, Lin B. Quantitative proteomics analysis integrated with microarray data reveals that extracellular matrix proteins, catenins, and p53 binding protein 1 are important for chemotherapy response in ovarian cancers. OMICS. 2009;13(4):345–354. doi: 10.1089/omi.2009.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iacob S, Cs-Szabo G. Biglycan regulates the expression of EGF receptors through EGF signaling pathways in human articular chondrocytes. Connect Tissue Res. 2010;51(5):347–358. doi: 10.3109/03008200903427695. [DOI] [PubMed] [Google Scholar]
- 32.Wang H, Sun W, Ma J, Pan Y, Wang L, Zhang WB. Biglycan mediates suture expansion osteogenesis via potentiation of Wnt/beta-catenin signaling. J Biomech. 2015;48(3):432–440. doi: 10.1016/j.jbiomech.2014.12.032. [DOI] [PubMed] [Google Scholar]
- 33.Osman N, Getachew R, Burch M, Lancaster G, Wang R, Wang H, Zheng W, Little PJ. TGF-beta stimulates biglycan core protein synthesis but not glycosaminoglycan chain elongation via Akt phosphorylation in vascular smooth muscle. Growth Factors. 2011;29(5):203–210. doi: 10.3109/08977194.2011.615747. [DOI] [PubMed] [Google Scholar]
- 34.Zhao Y, Scott A, Zhang P, Hao Y, Feng X, Somasundaram S, Khalil AM, Willis J, Wang Z. Regulation of paxillin-p130-PI3K-AKT signaling axis by Src and PTPRT impacts colon tumorigenesis. Oncotarget. 2016 doi: 10.18632/oncotarget.10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duchartre Y, Kim YM, Kahn M. The Wnt signaling pathway in cancer. Crit Rev Oncol Hematol. 2016;99:141–149. doi: 10.1016/j.critrevonc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin GS. Cell signaling and cancer. Cancer Cell. 2003;4(3):167–174. doi: 10.1016/s1535-6108(03)00216-2. [DOI] [PubMed] [Google Scholar]
- 37.Padua D, Zhang XH, Wang Q, Nadal C, Gerald WL, Gomis RR, Massague J. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133(1):66–77. doi: 10.1016/j.cell.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma F, Ye H, He HH, Gerrin SJ, Chen S, Tanenbaum BA, Cai C, Sowalsky AG, He L, Wang H. SOX9 drives WNT pathway activation in prostate cancer. J Clin Invest. 2016;126(5):1745–1758. doi: 10.1172/JCI78815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iwasa K, Hayashi S, Fujishiro T, Kanzaki N, Hashimoto S, Sakata S, Chinzei N, Nishiyama T, Kuroda R, Kurosaka M. PTEN regulates matrix synthesis in adult human chondrocytes under oxidative stress. J Orthop Res. 2014;32(2):231–237. doi: 10.1002/jor.22506. [DOI] [PubMed] [Google Scholar]
- 40.Doersch KM, Moses KA, Zimmer WE. Synergistic immunologic targets for the treatment of prostate cancer. Exp Biol Med (Maywood) 2016 doi: 10.1177/1535370216660212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fang F, Qin Y, Hao F, Li Q, Zhang W, Zhao C, Chen S, Zhao L, Wang L, Cai J. CD147 modulates androgen receptor activity through the Akt/Gsk-3beta/beta-catenin/AR pathway in prostate cancer cells. Oncol Lett. 2016;12(2):1124–1128. doi: 10.3892/ol.2016.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Recktenwald CV, Leisz S, Steven A, Mimura K, Muller A, Wulfanger J, Kiessling R, Seliger B. HER-2/neu-mediated down-regulation of biglycan associated with altered growth properties. J Biol Chem. 2012;287(29):24320–24329. doi: 10.1074/jbc.M111.334425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 44.Brase JC, Johannes M, Mannsperger H, Falth M, Metzger J, Kacprzyk LA, Andrasiuk T, Gade S, Meister M, Sirma H. TMPRSS2-ERG -specific transcriptional modulation is associated with prostate cancer biomarkers and TGF-beta signaling. BMC Cancer. 2011;11:507–515. doi: 10.1186/1471-2407-11-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang J, Xu H, Yang C, Kayarthodi S, Matthews R, Rao VN, Reddy ES. Molecular Mechanism of Activation of Transforming Growth Factor Beta/Smads Signaling Pathway in Ets Related Gene-Positive Prostate Cancers. J Pharm Sci Pharmacol. 2014;1(1):82–85. doi: 10.1166/jpsp.2014.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang J, Xu H, Yang C, Morsalin S, Kayarthodi S, Rungsrisuriyachai K, Gunnal U, McKenzie B, Rao VN, Reddy ES. Ets Related Gene and Smad3 Proteins Collaborate to Activate Transforming Growth Factor-Beta Mediated Signaling Pathway in ETS Related Gene-Positive Prostate Cancer Cells. J Pharm Sci Pharmacol. 2014;1(3):175–181. doi: 10.1166/jpsp.2014.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tiede K, Melchior-Becker A, Fischer JW. Transcriptional and posttranscriptional regulators of biglycan in cardiac fibroblasts. Basic Res Cardiol. 2010;105(1):99–108. doi: 10.1007/s00395-009-0049-8. [DOI] [PubMed] [Google Scholar]
- 48.Heegaard AM, Xie Z, Young MF, Nielsen KL. Transforming growth factor beta stimulation of biglycan gene expression is potentially mediated by sp1 binding factors. J Cell Biochem. 2004;93(3):463–475. doi: 10.1002/jcb.20189. [DOI] [PubMed] [Google Scholar]
- 49.Schmitz B, Salomon A, Rotrige A, Ritter M, Ringelstein EB, Fischer JW, Paul M, Brand E, Brand SM. Interindividual transcriptional regulation of the human biglycan gene involves three common molecular haplotypes. Arterioscler Thromb Vasc Biol. 2013;33(4):871–880. doi: 10.1161/ATVBAHA.112.301073. [DOI] [PubMed] [Google Scholar]
- 50.Ji H, Dailey TL, Long V, Chien EK. Androgen-regulated cervical ripening: a structural, biomechanical, and molecular analysis. Am J Obstet Gynecol. 2008;198(5) doi: 10.1016/j.ajog.2007.11.012. 543 e541-549. [DOI] [PubMed] [Google Scholar]
- 51.Hashimura K, Sudhir K, Nigro J, Ling S, Williams MR, Komesaroff PA, Little PJ. Androgens stimulate human vascular smooth muscle cell proteoglycan biosynthesis and increase lipoprotein binding. Endocrinology. 2005;146(4):2085–2090. doi: 10.1210/en.2004-1242. [DOI] [PubMed] [Google Scholar]
- 52.Tan MH, Li J, Xu HE, Melcher K, Yong EL. Androgen receptor: structure, role in prostate cancer and drug discovery. Acta Pharmacol Sin. 2015;36(1):3–23. doi: 10.1038/aps.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bigler SA, Deering RE, Brawer MK. Comparison of microscopic vascularity in benign and malignant prostate tissue. Hum Pathol. 1993;24(2):220–226. doi: 10.1016/0046-8177(93)90304-y. [DOI] [PubMed] [Google Scholar]
- 54.Tuxhorn JA, Ayala GE, Smith MJ, Smith VC, Dang TD, Rowley DR. Reactive stroma in human prostate cancer: induction of myofibroblast phenotype and extracellular matrix remodeling. Clin Cancer Res. 2002;8(9):2912–2923. [PubMed] [Google Scholar]
- 55.Burns-Cox N, Avery NC, Gingell JC, Bailey AJ. Changes in collagen metabolism in prostate cancer: a host response that may alter progression. J Urol. 2001;166(5):1698–1701. doi: 10.1016/s0022-5347(05)65656-x. [DOI] [PubMed] [Google Scholar]
- 56.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun J, Liu W, Adams TS, Sun J, Li X, Turner AR, Chang B, Kim JW, Zheng SL, Isaacs WB. DNA copy number alterations in prostate cancers: a combined analysis of published CGH studies. Prostate. 2007;67(7):692–700. doi: 10.1002/pros.20543. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, He X, Ngeow J, Eng C. GATA2 negatively regulates PTEN by preventing nuclear translocation of androgen receptor and by androgen-independent suppression of PTEN transcription in breast cancer. Hum Mol Genet. 2012;21(3):569–576. doi: 10.1093/hmg/ddr491. [DOI] [PubMed] [Google Scholar]
- 59.Gaspar R, Pipicz M, Hawchar F, Kovacs D, Djirackor L, Gorbe A, Varga ZV, Kiricsi M, Petrovski G, Gacser A. The cytoprotective effect of biglycan core protein involves Toll-like receptor 4 signaling in cardiomyocytes. J Mol Cell Cardiol. 2016;99:138–150. doi: 10.1016/j.yjmcc.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 60.Li S, Zhu M, Pan R, Fang T, Cao YY, Chen S, Zhao X, Lei CQ, Guo L, Chen Y. The tumor suppressor PTEN has a critical role in antiviral innate immunity. Nat Immunol. 2016;17(3):241–249. doi: 10.1038/ni.3311. [DOI] [PubMed] [Google Scholar]
- 61.Frey H, Schroeder N, Manon-Jensen T, Iozzo RV, Schaefer L. Biological interplay between proteoglycans and their innate immune receptors in inflammation. FEBS J. 2013;280(10):2165–2179. doi: 10.1111/febs.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sauter G, Steurer S, Clauditz TS, Krech T, Wittmer C, Lutz F, Lennartz M, Janssen T, Hakimi N, Simon R. Clinical Utility of Quantitative Gleason Grading in Prostate Biopsies and Prostatectomy Specimens. Eur Urol. 2016;69(4):592–598. doi: 10.1016/j.eururo.2015.10.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials