Abstract
Advances in computational biology and large-scale transcriptome analyses have revealed that a much larger portion of the genome is transcribed than was previously recognized, resulting in the production of a diverse population of RNA molecules with both protein-coding and non-coding potential. Emerging evidence indicates that a number of RNA molecules have been mis-annotated as non-coding and actually harbor short open reading frames (sORFs) that code for functional peptides, which have evaded detection until now due to their small size. sORF encoded peptides, or micropeptides, have been shown to play important roles in fundamental biological processes and in the maintenance of cellular homeostasis. These small proteins can act independently, for example as ligands or signaling molecules, or they can exert their biological functions by engaging with and modulating larger regulatory proteins. Given their small size, micropeptides may be uniquely suited to fine-tune complex biological systems.
Keywords: micropeptide, short open reading frame, bioactive peptide, ncRNA
Introduction
Innovative work over the past decade using DNA sequencing and proteomic approaches has revolutionized the field of genomics, enabling a comprehensive look into genes, transcripts and their translated protein products. Numerous large-scale genomic studies have revealed that a much larger fraction of the genome is transcribed and translated than was initially appreciated and increasing attention has been placed on identifying the complete set of mammalian genes, both protein-coding and non-protein-coding[1–17].
Proteins are obtained from the translation of an open reading frame (ORF) on an mRNA transcript, which consists of a sequence of in-frame codons beginning at a start codon and ending with a stop codon. A major challenge in the field of gene annotation is the ability to accurately identify ORFs that code for bona fide protein products and distinguish them from the exponentially higher number of spurious non-coding ORFs that occur randomly throughout the genome simply by chance and are not translated. This obstacle becomes especially pronounced when focusing on short ORFs (sORFs) that code for small proteins or peptides. Since the likelihood that an ORF encodes a genuine protein increases with its length, most ORF-finding algorithms have historically set a threshold length of 300 nucleotides, or 100 amino acids, as the minimum size for detection. An unintended reprocussion of this filtering was that some transcripts with legitimate protein coding potential were erroneously classified as members of a much larger class of non-coding RNAs (ncRNAs). A critical and defining feature of all ncRNAs is their inherent lack of protein coding potential. Therefore, recent findings by several groups that some ncRNA transcripts actually harbor sORFs that code for functional small proteins, often referred to as micropeptides, underscores the likelihood that additional transcripts currently annotated as ncRNAs encode proteins with important biological activities[1, 10, 18–30].
In this review, we provide an overview of the methods that have been developed and fine-tuned to identify novel coding sORFs. We will show how these innovative techniques have led to the discovery of biologically active small proteins that play important roles in a number of cellular processes and highlight the exciting implications of these studies. Lastly, we will detail some of the experimental techniques that have been developed and successfully implemented to verify the coding potential of sORFs and decipher the biological function of the micropeptides they generate.
Identification of Protein-Coding sORFs
Recent technological advances have increased our ability to identify sORFs and reliably predict the likelihood that these sequences are translated to produce stable protein products. Both computational and experimental approaches have been successfully developed and implemented to infer protein coding potential, analyze the transcriptional and translational state of a given region, and detect the putative protein product generated from translation. Recently, combinations of these techniques have been used to generate robust data sets[1–3, 8, 10, 12, 13].
Computational Approaches
One complicating factor in distinguishing protein-coding mRNAs from lncRNAs is that the transcripts resemble each other on many levels. Just like protein-coding mRNAs, lncRNAs are transcribed by RNA polymerase II, capped on their 5′ end, spliced via canonical splice motifs, and frequently polyadenylated (Figure 1A). LncRNAs are also typically associated with epigenetic signatures common to protein-coding genes, such as trimethylation of histone H3 lysine 4 (H3K4me3) at the transcriptional start site and trimethylation of histone H3 lysine 36 (H3K36me3) throughout the gene body[31]. One important difference between lncRNAs and protein-coding mRNAs is their low level of nucleotide sequence conservation [4, 5, 32]. Similar to other protein-coding transcripts, a hallmark of functional sORFs is evolutionary conservation of the protein sequence. As a result, computational techniques that are largely based on sequence conservation have been developed to attempt to identify true protein-coding sORFs from non-coding regions.
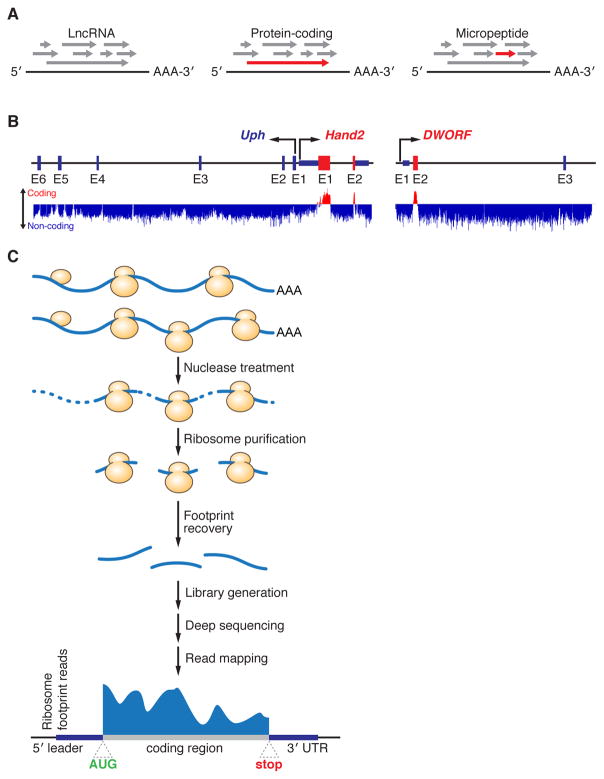
Figure 1. Tools and Methods for the Identification of Micropeptides.
(A) Regardless of their coding potential, all mRNA transcripts contain multiple different open reading frames (ORFs) of varying lengths (grey arrows). Typically, the longest ORF within the transcript codes for the functional protein product (red arrow), and this is most readily seen in large protein coding genes (middle). However, in the case of very small proteins like micropeptides (right), finding the correct ORF is extremely challenging because the longest one is frequently not the actual coding region, and the coding ORF gets lost in the noise of other spurious non-coding ORFs. (B) Computational tools, such as PhyloCSF, have been developed to help identify potential coding genes based on the evolutionary conservation of their nucleotide sequence. The mouse Upperhand (Uph)-Hand2 locus (Left) is a perfect example of a region of the genome that contains a conserved protein coding gene (Hand2) and a non-coding transcript (Uph). As depicted, Hand2 scores positively on PhyloCSF (red color, upward deflection) specifically in the region that codes for the functional Hand2 protein (exon 1 and 2, E1 and E2). Conversely, Uph scores negatively throughout its sequence as illustrated by the negative (blue) score. PhyloCSF has been used to identify several novel micropeptides including dwarf open reading frame (DWORF, right), whose strong sequence conservation can be seen prominently in exon 2 (E2). (C) Experimental methods such as ribosome profiling have also been developed that aid in the identification of novel protein coding genes. In this technique, active translation is halted by the addition of translation inhibitors and samples are treated with nucleases to generate ribosome protected fragments (RPFs), or footprints, that are protected from digestion by the presence of the ribosome. These footprints are then recovered, sequenced and mapped to the genome to reveal their origin.
Cross-species comparisons are a powerful technique in gene prediction as most genes are subject to evolutionary pressure to maintain sequence conservation and display a prevalence of synonymous codon substitutions (nucleotide substitutions that do not change the coded amino acid) versus nonsynonymous substitutions[33–35]. Metrics have been developed that calculate the ratio of nonsynonymous (KA) to synonymous (KS) substitutions (KA/KS) and a value of <1 typically satisfies the requirement for protein-coding potential[36]. However, in the case of micropeptides, it can be difficult to score statistically significant values due to the very short length of the sequences and the fact that the number of possible changes is low[11]. As a result, other techniques are required to adequately distinguish coding potential for small peptides hidden in lncRNAs.
PhyloCSF is a particularly vigorous computational method that has been integrated into the UCSC Genome Browser, which makes for free and easy access for all researchers[2, 13, 37]. The program examines evolutionary signatures characteristic of alignments of conserved coding regions, similar to the methods described above, and provides a phylogenic assessment of codon substitution frequencies. PhyloCSF provides a conservation score for all six potential reading frames (three on the positive strand and three on the negative strand) of a given stretch of nucleotides providing a robust way to evaluate evolutionarily conserved protein-coding regions (Figure 1B).
Recent studies have utilized these computational methods to identify multiple sORFs embedded in the genome. Mackowiak et al. developed and implemented an integrated pipeline that computationally identified sORFs with high accuracy by using conservation features specific to known micropeptides[12]. The authors concentrated on the idea that evolutionary conservation is a strong indicator for functionality and focused on features including depletion of non-synonymous mutations, an absence of frame-shifting indels, and characteristic steps in sequence conservation around start and stop codons to identify true sORFs from random non-coding ORFs[12]. Using this approach, the authors identified hundreds of previously unknown conserved sORFs in major model organisms (both vertebrate and invertebrate). While computation approaches have been invaluable in aiding the search for sORFs, there are other techniques that have also helped shed light on the protein coding potential of sORFs embedded in non-coding elements.
Ribosome profiling
Ribosomes are complex molecular machines that link amino acids together in the exact order specified by the nucleotide code within a transcript in order to produce a protein product. Ribosome profiling (Ribo-Seq) is a deep-sequencing-based tool that provides a genome-wide snapshot of active translation with single nucleotide resolution. The method hinges on the ability of translating ribosomes to protect RNA segments of 20–30 nucleotides in length from nuclease digestion[7–9, 38–41]. Cytoplasmic lysates are prepared in the presence of translation inhibitors and mRNA-ribosome complexes are treated with nucleases to generate ribosome protected fragments (RPFs), also frequently referred to as ribosome footprints (Figure 1C). These RPFs can be isolated and purified for subsequent sequencing, which allows for the identification of the precise position of the ribosome at the time at which translation was halted. In addition to revealing the identity of ribosome-bound transcripts, measuring the density of RPFs on a given transcript can provide valuable information on the quantitative dynamics of translation and the rates of protein synthesis within translated regions[9, 10]. Additionally, to eliminate technical noise, other features are analyzed such as length and trinucleotide periodicity, positioning of the ORF within a transcript and responsiveness to translation inhibitors[1, 3, 8, 42–44]. Poly-Ribo-Seq is a modified ribosome profiling method that enriches in polysomes, which are complexes of mRNA molecules and two or more ribosomes, has also been developed and was successfully implemented to identify a number of sORFs in the Drosophila genome[22, 45, 46].
Results of Ribo-Seq have challenged our understanding of the protein-coding potential of the genome and have provided evidence for translation of non-annotated ORFs[9, 10, 47]. Additionally, evidence for alternative start and stop codons for canonical proteins has been found as well as the use of non-AUG start codons, further complicating bioinformatic identification of bona fide small peptides[1, 21, 41, 47, 48]. Pseudogenes have also been shown to associate with ribosomes, suggesting that they could also be a potential source of translated proteins[16]. While ribosome profiling itself is an experimental approach, the evaluation of the coding potential of an identified region of interest is in fact mostly computational. An important concept to note is that ribosome occupancy does not necessarily imply true coding potential and function at the protein level. Studies have shown that not all translation events lead to stable, functional polypeptides and that the act of translation itself can have other important regulatory consequences, such as modulation of a downstream ORF[31, 49–51], or could simply represent technical or biological noise. Current techniques are continuously being modified and enhanced to more reliably identify protein-coding transcripts. Recently, the inclusion of a ribosome release score (RRS), which detects the termination of translation at the stop codon at the end of an ORF and has been shown to robustly distinguish protein-coding transcripts from non-coding RNAs[6], adding another tool to identify sORFs. Altogether, a number of excellent algorithms and metrics have been developed to help analyze and process ribosome profiling data to recognize and identify regions of translation including FLOSS [8], ORF score [1], PROTEOFORMER [52] and ORF-RATER[48].
In addition to the studies mentioned above, several databases have been created that collect ribosome profiling data and genome annotations derived from this data including TISdb[53], GWIPS-viz[54], RPFdb[55] and sORFs.org[56] and information about the large amounts of data contained in these databases as well as the techniques used to generate them have been nicely reviewed in several articles[27, 41, 57, 58]. Continued modification of these techniques will likely be used in combination with other emerging technologies to enhance the reliability and power of the data sets they generate to identify functional sORFs.
Mass Spectrometry
Mass spectrometry (MS) peptidomics and proteomics have recently been implemented in the discovery of micropeptides. MS is a powerful technique for direct detection and quantification of proteins and peptides and is the gold standard in proteomics research. MS experiments differ from ribosome profiling in that MS is able to detect polypeptides that are translated from a sORF and can thereby directly validate the protein-coding potential of the transcript. It is interesting to note that in proteomics studies, there are currently many MS fragmentation spectra that are unidentified, and one potential reason for this is some of these signatures may belong to micropeptides that have not yet been annotated.
Proteomic-based discovery of micropeptides is greatly enhanced by the combination of proteomics and genomics (RNA-Seq), referred to as proteogenomics[59–61]. This technique was utilized by Slavoff et al., where the authors combined peptidomics and massively parallel RNA-sequencing using human K562 leukemia cells to identify sORFs[62]. The authors first created a custom database by integrating all of the possible polypeptides based on the annotated human transcriptome available in the Reference Sequence database (RefSeq) [63] and included an experimental RNA-Seq derived K562 transcriptome. They then performed liquid chromatography followed by tandem mass spectrometry (LC-MS/MS) in an adapted design to enrich for small translation products and subsequently matched their proteomics data against their custom sequencing database. Using these custom polypeptide databases and four previously reported micropeptides as positive controls[64], 86 still uncharacterized micropeptides were discovered[62].
While substantial progress has been made with integrating MS-based proteomics studies with the identification of novel micropeptides, there are still technical problems with this method that are of concern. Notably, many small protein products are often lost in the sample preparation steps leading up to MS and therefore are not available for detection. Even when special precautions are taken to ensure small proteins are preserved for detection, there is an inherent bias against observing short proteins by MS due to the fact that there is often only one chance (or sometimes none) to detect the fragmented product, which often results in the protein being missed entirely. Depending on the specific protease used for digestion in sample preparation, micropeptides also may not be fragmented efficiently to generate large enough signatures that are required for identification. Furthermore, micropeptides may be relatively short lived, of low abundance and can have tissue- and time-specific expression patterns, which further impedes their identification.
Therefore, while great progress has been made in MS-based micropeptide identification, there are still major challenges that need to be addressed. While the presence of a micropeptide of interest in MS data can be relied on heavily as proof of its existence, the absence of a sequence should be considered cautiously and should not be taken as hard evidence against a particular protein being produced. As described in detail above, the best strategy for detecting micropeptides is likely a combination of computational and experimental techniques and, although these methods have room for further optimization, they have been successfully used to identify many putative micropeptides that could have very diverse biological functions.
Identification and Characterization of Biologically Active Micropeptides
Despite their diminutive size, peptides and small proteins play critical roles in many biological processes in living organisms[57, 65–67]. Known classical small peptides include neuropeptides and peptide hormones, which are enzymatically cleaved from larger precursor proteins carrying an N-terminal signal sequence targeting them for the secretory pathway[65, 66, 68, 69]. Unlike these examples of larger proteins that are proteolytically processed to generate their biologically active small peptide products, micropeptides are translated directly from their precursor mRNA (Figure 2). A small number of well-studied micropeptides have indicated that these small proteins may act as important regulators in many fundamental events including development[25, 46, 70–73], DNA repair[74], RNA decapping[29], calcium homeostasis[18, 19, 22, 24, 75]), metabolism[26], stress signaling[23], myoblast fusion[76] and cell death[68, 77]. However, given the putative large number of micropeptides, relatively little is known about their biological activities and regulation.
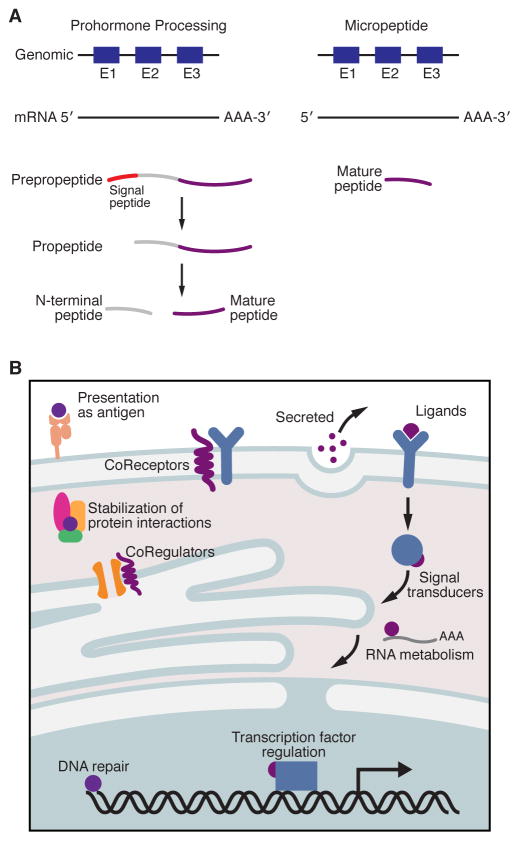
Figure 2. Micropeptide Processing and their Biological Functions.
(A) Unlike classical examples of neuropeptides and peptide hormones that are synthesized as much larger proteins and later proteolytically processed to generate their mature active peptide product (Left), micropeptides are translated directly from their precursor mRNAs as functional molecules (Right). Micropeptides have been shown to work as key regulators of many fundamental biological processes and can act independently or exert their effects by engaging with and modulating much larger regulatory proteins (B).
Evolutionary conservation of a peptide sequence is suggestive of functionality, yet the mere existence of the putatively translated peptide does not necessarily imply that it has a critical biological function. In order to determine the physiological role of an identified micropeptide, experimental demonstration of a biological effect is required. While recent advances in computational biology and experimental techniques have led to the discovery of hundreds or even thousands of potential novel micropeptides, each of these putative proteins needs to be independently authenticated and studied for biological relevance. As a result, it is an exciting time for research in the field of micropeptides because there is a large amount of work that needs to be done to experimentally characterize each of these proteins, which provides many opportunities for researchers from all fields of science to contribute to the growing body of knowledge.
Working with Micropeptides: Validation of Protein Coding Potential
Validation of candidate-translated sORFs can be performed using several approaches[78]. Ideally, an antibody against a peptide of interest is generated and strictly validated to demonstrate its specificity. However, designing effective antibodies against micropeptides is extremely challenging for several reasons, most notably because their small size provides very few peptide choices for optimal antigenicity. Further complicating the matter, several micropeptides have been shown to contain transmembrane domains, which mask relatively large sections of their short sequences, limiting the region available for epitope design. An additional technical concern is that the techniques that rely on the use of antibodies, such as Western blot and immunocytochemistry, are not highly sensitive, and if a peptide is expressed at low levels, even the highest affinity antibody may not be sufficient to produce a strong enough signal for detection.
In cases where an antibody cannot be raised against a micropeptide of interest, there are several alternative methods that can be used to validate its coding potential. CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)–Cas9 (CRISPR-Associated Protein 9)-mediated gene editing strategies can be designed to insert an epitope tag into the endogenous locus of the micropeptide in-frame with the predicted ORF using homology-directed repair in vitro or in vivo (Figure 3A)[79]. This strategy has been used to engineer fusion proteins that can be detected by Western blot and provides convincing evidence that the micropeptide mRNA is actively transcribed from its native chromosomal context and translated into a stable peptide[18, 23, 46]. In addition, in cases where several putative sORFs are identified within a single transcript, this method can be used to systematically distinguish the true micropeptide[23]. Successful implementation of this epitope knock-in technique also allows for useful downstream applications such as immunoprecipitations, immunocytochemistry, and Western blot. As discussed in detail below, when designing epitope tag knock-ins it is critical to consider the position of the tag (N-terminal, C-terminal or internal) as well as the size and biochemical properties of the amino acids it is coded by.
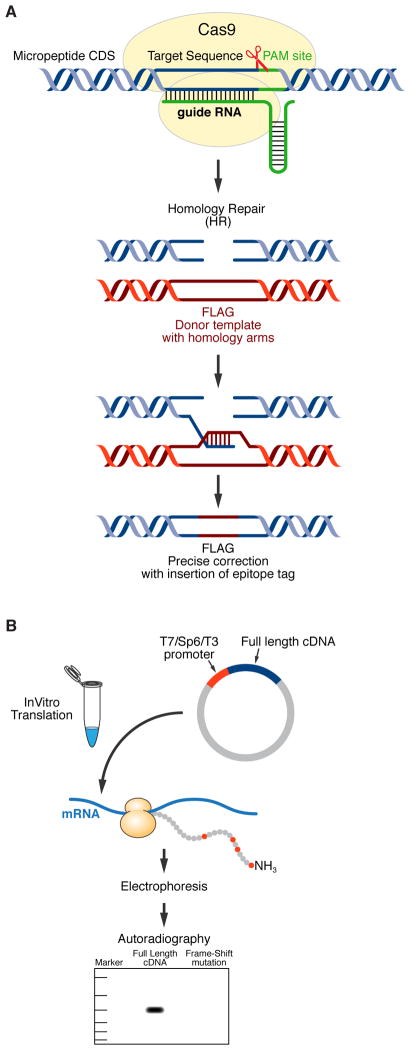
Figure 3. Methods for Verifying Micropeptide Coding Potential.
(A) CRISPR/Cas9-mediated gene editing can be used to knockin an epitope tag into the endogenous locus of a putative micropeptide in-frame with the predicted sORF to test for coding potential. The Cas9 endonuclease (yellow) is targeted to a specific location on the genome via a single guide RNA (sgRNA, green) which is immediately adjacent to a protospacer adjacent motif (PAM) site. Upon recognition of the appropriate site, Cas9 will then unwind the DNA duplex and create a DNA double strand break. This double strand break can either be repaired by non-homologous end joining (NHEJ) or by homology-directed repair (HDR). To utilize HDR for editing, a donor template with homology to the targeted locus must be provided and this must contain the sequence of the epitope tag you wish to knockin (shown here as FLAG, red). Expression of your epitope tag can then be verified by Western Blot or immunostaining. (B) The coding potential of a sORF can also be assessed by in vitro translation. The full-length cDNA of your peptide of interest must be cloned into a plasmid containing a phage polymerase promoter (shown here as T7, Sp6 or T3) and cell-free protein synthesis is performed in the presence of 35S-methionine, which will radioactively label your micropeptide (35S-methionine is depicted as red circles in the polypeptide chain). These protein products are then subjected to gel electrophoresis and autoradiography and then analyzed to determine if a product of the predicted molecular weight is produced. As a control, a frame-shift mutant of your coding sequence should be cloned and this should not yield an 35S-labeled protein product.
In addition to antibody-based validation of micropeptides, the coding potential of a sORF can also be assessed by in vitro translation assays (Figure 3B)[18, 19]. In these experiments, the full-length cDNA of a putative micropeptide transcript is cloned into a vector containing a phage polymerase promoter (usually T7 or SP6), and then expression of these constructs is evaluated using a cell-free protein synthesizing system in the presence of 35S-methionine. The protein products are then analyzed by gel electrophoresis and autoradiography is performed to visualize the synthesis of an 35S-labeled peptide of the predicted molecular weight. This technique hinges on the concept that ribosomes have the capacity to distinguish between coding and non-coding RNA transcripts, which is supported by several reports that lncRNAs are rarely translated[6, 51, 80] Introducing a frame-shift mutation in the suspected ORF and subsequently abolishing the production of a stable peptide can strengthen results from this method[18, 19]. While this technique can be informative and valuable in the process of screening potential coding sORFs, the results should be interpreted cautiously as it is possible that sequences can by translated in vitro but not in vivo. Conversely, if a construct does not produce a stable peptide in vitro, its coding potential in vivo cannot be ruled out.
Working with Micropeptides: Elucidating Biological Function
Once the coding potential of a micropeptide has been sufficiently demonstrated, the question of its biological relevance still remains. Several of the micropeptides that have been discovered and characterized thus far exert their biological functions by engaging with and modulating larger regulatory proteins. In this way, micropeptides can be thought of as singular protein domains with highly specialized roles and, therefore, the key to elucidating their function often lies in identifying their interacting partner or partners.
Functional proteomics has been successfully used by several groups to help identify interacting proteins of candidate micropeptides[23, 29]. By performing immunoprecipitations and mass spectrometry on the co-precipitated proteins, direct binding partners or components of a specific protein complex can be identified, which is strongly suggestive of the biological function of a micropeptide. As previously discussed, the use of CRISPR/Cas9-mediated gene editing to insert an epitope tag in the endogenous locus of the ORF encoding a micropeptide in frame with its protein product is a powerful technique that simplifies many useful downstream applications including immunoprecipitations to identify binding partners. Epitope tagging of the endogenous allele of a gene also allows for the analysis of the protein in its native context and reduces the chance that artificial binding partners are erroneously identified as a consequence of over-expression. Immunocytochemistry can also be performed in epitope-tagged samples to define the subcellular localization of a micropeptide of interest and contribute additional support for the involvement in a particular biological process.
Alternatively, rather than assessing protein interactions using the endogenous micropeptide, transient overexpression of an epitope-tagged micropeptide in a cultured cell line can be used to identify interacting partners. However, it should be noted that non-physiological transcriptional regulation of proteins of any size often leads to substantial levels of over-expression, which can lead to artificial mis-localization of the protein of interest and perturbations of normal cellular functions[81]. Protein interactions identified from these types of studies needs to be assessed cautiously and strictly validated.
While working with epitope-tagged micropeptides has many advantages, there are several important concerns that should be considered when designing a tagging strategy. Both N- and C-terminal tags should be designed and tested and stable expression and proper localization of your tagged fusion protein should be thoroughly assessed. In general, C-terminal tags are less likely to interfere with N-terminal signal sequences or localization signals, while N-terminal tags often lead to better protein solubility[82]. Internal epitope tags can also be considered, particularly in cases where the N-terminal and C-terminal tagged proteins behave differently. In addition to considering the placement of a tag, the actual epitope used should be carefully deliberated. As micropeptides are extremely small, the tag alone can often be of similar size or even much larger than the micropeptide itself. Therefore, the length and biochemical properties (such as charge and hydrophobicity) of the tag alone should be taken into account. It may even be advantageous to add a small, inert linker sequence to distance the tag from the micropeptide to reduce the possibility that the tag disrupts the structure or function of the peptide[83]. Furthermore, many of the micropeptides that have been discovered thus far contain transmembrane domains that span relatively large portions of their sequences, and these transmembrane domains should be carefully considered when designing epitope tags. Together, these strategies will enable identification of a micropeptide’s subcellular localization, enabling insight into putative biological function.
While many of the functionally characterized micropeptides act as regulators of larger protein complexes, micropeptides have also been shown to act independently in a variety of different manners including as ligands to receptors[25, 73, 84], as cytoplasmic ribosomal proteins[70, 71], as stabilizers of protein-protein interactions[85–89] and as peptides that are presented on the cell surface by Major Histocompatibility Complex class I (MHC I) molecules[90–93](Figure 2B). As is particularly evident in these cases, the characterization of the function of a micropeptide will not only depend on its interaction partners, but also on its specific expression pattern and timing of expression, both of which add additional challenges to deciphering the biological functions of these novel small proteins.
Working with Micropeptides: Demonstrating Biological Relevance
As with the elucidation of the biological role of any novel protein coding gene, the truly cumbersome work comes in the form of demonstrating a physiological relevance for the protein. One of the most common ways to attempt to understand the function of a gene is to analyze a biological system lacking that gene. This loss-of-function approach can be performed in cell lines or in animal models and rigorous phenotyping must be subsequently implemented. Importantly, it should be shown that rescue of the phenotype observed is accomplished by giving back the mRNA/protein that was lost in order to conclusively prove that the mutant phenotype is indeed due to loss of that particular protein. As discussed above, some micropeptides exert their function by interacting with and modulating much larger proteins and serve as a means to fine-tune complex biological processes. Therefore, loss-of-function studies may not reveal dramatic phenotypes due to potential complementary pathways. In such cases, stressing the physiological system in question in an appropriate manner may help to tease out the requirement for the micropeptide. Having supporting information available, such as potential interacting protein partners, will be extremely useful in helping steer you in the right direction of what to look for, where to look for it, and how to appropriately stress the system of interest.
Concluding Remarks
The recent discovery and characterization of multiple biologically active micropeptides hidden within mRNA transcripts incorrectly classified as ncRNAs indicates an additional level of complexity in the proteome that was previously unappreciated. Intricate computational and experimental techniques have been developed and optimized to identify novel sORF encoded peptides and these methods have uncovered a vast number of putative micropeptides. We have only just begun to decipher the biological roles of these important small proteins and explore the diversity of their functions (see Outstanding Questions). Future efforts will expand upon and refine micropeptide detection techniques, with considerable work needed to validate each candidate individually and elucidate its biological function.
Outstanding Questions.
How can current technology be further optimized to reliably detect translation events from sORFs that generate stable bioactive peptides? Are there novel techniques on the horizon that will surpass what is presently used?
What is the fraction of putative sORF encoded peptides that are actually translated to stable micropeptide products versus those that are unstable byproducts of random translational events?
What is the best strategy for researchers to implement to systematically validate the vast numbers of prospective micropeptides that have been detected?
How will researchers overcome the many unique obstacles that come with working with tiny proteins (i.e. low protein abundance, protein instability, protein loss during sample preparation, lack of currently available antibodies and/or limited epitope options to generate custom antibodies, etc.)?
In cases where in vitro or in vivo loss-of-function studies result in no apparent phenotype, how can researchers gain insights into their system to design ways to induce an appropriate stress to help elucidate the biological function of a micropeptide?
Trends.
Recent advances in computational and experimental techniques have revealed that a much larger portion of the genome is translated than was previously recognized.
Small open reading frames (sORFs) that produce functional, evolutionarily conserved peptides have been found hidden within transcripts annotated as “non-coding”.
It has been demonstrated that these sORF encoded peptides, or SEPs, play essential roles in many important biological processes and have been shown to act independently or as regulators of larger proteins.
To date, biological roles have been assigned to a small fraction of the total putative SEPs that have been identified and a huge amount of work remains to be done to prove their existence and elucidate their functions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bazzini AA, et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014;33(9):981–93. doi: 10.1002/embj.201488411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chew GL, et al. Ribosome profiling reveals resemblance between long non-coding RNAs and 5′ leaders of coding RNAs. Development. 2013;140(13):2828–34. doi: 10.1242/dev.098343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guttman M, et al. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154(1):240–51. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingolia NT, et al. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7(8):1534–50. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ingolia NT, et al. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014;8(5):1365–79. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ingolia NT, et al. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–23. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ingolia NT, et al. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147(4):789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ladoukakis E, et al. Hundreds of putatively functional small open reading frames in Drosophila. Genome Biol. 2011;12(11):R118. doi: 10.1186/gb-2011-12-11-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mackowiak SD, et al. Extensive identification and analysis of conserved small ORFs in animals. Genome Biol. 2015;16:179. doi: 10.1186/s13059-015-0742-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pauli A, et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22(3):577–91. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pauli A, et al. Identifying (non-)coding RNAs and small peptides: challenges and opportunities. Bioessays. 2015;37(1):103–12. doi: 10.1002/bies.201400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frith MC, et al. The abundance of short proteins in the mammalian proteome. PLoS Genet. 2006;2(4):e52. doi: 10.1371/journal.pgen.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim MS, et al. A draft map of the human proteome. Nature. 2014;509(7502):575–81. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 18.Anderson DM, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160(4):595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson DM, et al. Widespread control of calcium signaling by a family of SERCA-inhibiting micropeptides. Sci Signal. 2016;9(457):ra119. doi: 10.1126/scisignal.aaj1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen SM. Everything old is new again: (linc)RNAs make proteins! EMBO J. 2014;33(9):937–8. doi: 10.1002/embj.201488303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji Z, et al. Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife. 2015;4:e08890. doi: 10.7554/eLife.08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magny EG, et al. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science. 2013;341(6150):1116–20. doi: 10.1126/science.1238802. [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto A, et al. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2017;541(7636):228–232. doi: 10.1038/nature21034. [DOI] [PubMed] [Google Scholar]
- 24.Nelson BR, et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351(6270):271–5. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pauli A, et al. Toddler: an embryonic signal that promotes cell movement via Apelin receptors. Science. 2014;343(6172):1248636. doi: 10.1126/science.1248636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee C, et al. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab. 2015;21(3):443–54. doi: 10.1016/j.cmet.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pueyo JI, et al. New Peptides Under the s(ORF)ace of the Genome. Trends Biochem Sci. 2016;41(8):665–78. doi: 10.1016/j.tibs.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Saghatelian A, Couso JP. Discovery and characterization of smORF-encoded bioactive polypeptides. Nat Chem Biol. 2015;11(12):909–16. doi: 10.1038/nchembio.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D’Lima NG, et al. A human microprotein that interacts with the mRNA decapping complex. Nat Chem Biol. 2017;13(2):174–180. doi: 10.1038/nchembio.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith JE, et al. Translation of small open reading frames within unannotated RNA transcripts in Saccharomyces cerevisiae. Cell Rep. 2014;7(6):1858–66. doi: 10.1016/j.celrep.2014.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–46. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ina Y. New methods for estimating the numbers of synonymous and nonsynonymous substitutions. J Mol Evol. 1995;40(2):190–226. doi: 10.1007/BF00167113. [DOI] [PubMed] [Google Scholar]
- 34.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–20. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 35.Makalowski W, Boguski MS. Evolutionary parameters of the transcribed mammalian genome: an analysis of 2,820 orthologous rodent and human sequences. Proc Natl Acad Sci U S A. 1998;95(16):9407–12. doi: 10.1073/pnas.95.16.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 2002;18(9):486. doi: 10.1016/s0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- 37.Lin MF, et al. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27(13):i275–82. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lareau LF, et al. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. Elife. 2014;3:e01257. doi: 10.7554/eLife.01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolin SL, Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988;7(11):3559–69. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ingolia NT. Ribosome profiling: new views of translation, from single codons to genome scale. Nat Rev Genet. 2014;15(3):205–13. doi: 10.1038/nrg3645. [DOI] [PubMed] [Google Scholar]
- 41.Ingolia NT. Ribosome Footprint Profiling of Translation throughout the Genome. Cell. 2016;165(1):22–33. doi: 10.1016/j.cell.2016.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chew GL, et al. Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish. Nat Commun. 2016;7:11663. doi: 10.1038/ncomms11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calviello L, et al. Detecting actively translated open reading frames in ribosome profiling data. Nat Methods. 2016;13(2):165–70. doi: 10.1038/nmeth.3688. [DOI] [PubMed] [Google Scholar]
- 44.Raj A, et al. Thousands of novel translated open reading frames in humans inferred by ribosome footprint profiling. Elife. 2016:5. doi: 10.7554/eLife.13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aspden JL, et al. Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq. Elife. 2014;3:e03528. doi: 10.7554/eLife.03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galindo MI, et al. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 2007;5(5):e106. doi: 10.1371/journal.pbio.0050106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dunn JG, et al. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife. 2013;2:e01179. doi: 10.7554/eLife.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fields AP, et al. A Regression-Based Analysis of Ribosome-Profiling Data Reveals a Conserved Complexity to Mammalian Translation. Mol Cell. 2015;60(5):816–27. doi: 10.1016/j.molcel.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnstone TG, et al. Upstream ORFs are prevalent translational repressors in vertebrates. EMBO J. 2016;35(7):706–23. doi: 10.15252/embj.201592759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol. 2000;20(23):8635–42. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clark MB, et al. The reality of pervasive transcription. PLoS Biol. 2011;9(7):e1000625. doi: 10.1371/journal.pbio.1000625. discussion e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crappe J, et al. PROTEOFORMER: deep proteome coverage through ribosome profiling and MS integration. Nucleic Acids Res. 2015;43(5):e29. doi: 10.1093/nar/gku1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wan J, Qian SB. TISdb: a database for alternative translation initiation in mammalian cells. Nucleic Acids Res. 2014;42(Database issue):D845–50. doi: 10.1093/nar/gkt1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Michel AM, et al. GWIPS-viz: development of a ribo-seq genome browser. Nucleic Acids Res. 2014;42(Database issue):D859–64. doi: 10.1093/nar/gkt1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie SQ, et al. RPFdb: a database for genome wide information of translated mRNA generated from ribosome profiling. Nucleic Acids Res. 2016;44(D1):D254–8. doi: 10.1093/nar/gkv972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olexiouk V, et al. sORFs.org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res. 2016;44(D1):D324–9. doi: 10.1093/nar/gkv1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet. 2014;15(3):193–204. doi: 10.1038/nrg3520. [DOI] [PubMed] [Google Scholar]
- 58.Brar GA, Weissman JS. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Biol. 2015;16(11):651–64. doi: 10.1038/nrm4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Castellana N, Bafna V. Proteogenomics to discover the full coding content of genomes: a computational perspective. J Proteomics. 2010;73(11):2124–35. doi: 10.1016/j.jprot.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woo S, et al. Proteogenomic database construction driven from large scale RNA-seq data. J Proteome Res. 2014;13(1):21–8. doi: 10.1021/pr400294c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Branca RM, et al. HiRIEF LC-MS enables deep proteome coverage and unbiased proteogenomics. Nat Methods. 2014;11(1):59–62. doi: 10.1038/nmeth.2732. [DOI] [PubMed] [Google Scholar]
- 62.Slavoff SA, et al. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol. 2013;9(1):59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pruitt KD, et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42(Database issue):D756–63. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oyama M, et al. Diversity of translation start sites may define increased complexity of the human short ORFeome. Mol Cell Proteomics. 2007;6(6):1000–6. doi: 10.1074/mcp.M600297-MCP200. [DOI] [PubMed] [Google Scholar]
- 65.Fricker LD. Neuropeptide-processing enzymes: applications for drug discovery. AAPS J. 2005;7(2):E449–55. doi: 10.1208/aapsj070244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boonen K, et al. Bioactive peptides, networks and systems biology. Bioessays. 2009;31(3):300–14. doi: 10.1002/bies.200800055. [DOI] [PubMed] [Google Scholar]
- 67.Cabrera-Quio LE, et al. Decoding sORF translation - from small proteins to gene regulation. RNA Biol. 2016;13(11):1051–1059. doi: 10.1080/15476286.2016.1218589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hashimoto Y, et al. A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer’s disease genes and Abeta. Proc Natl Acad Sci U S A. 2001;98(11):6336–41. doi: 10.1073/pnas.101133498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cunha FM, et al. Intracellular peptides as natural regulators of cell signaling. J Biol Chem. 2008;283(36):24448–59. doi: 10.1074/jbc.M801252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kondo T, et al. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol. 2007;9(6):660–5. doi: 10.1038/ncb1595. [DOI] [PubMed] [Google Scholar]
- 71.Kondo T, et al. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science. 2010;329(5989):336–9. doi: 10.1126/science.1188158. [DOI] [PubMed] [Google Scholar]
- 72.Chanut-Delalande H, et al. Pri peptides are mediators of ecdysone for the temporal control of development. Nat Cell Biol. 2014;16(11):1035–44. doi: 10.1038/ncb3052. [DOI] [PubMed] [Google Scholar]
- 73.Chng SC, et al. ELABELA: a hormone essential for heart development signals via the apelin receptor. Dev Cell. 2013;27(6):672–80. doi: 10.1016/j.devcel.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 74.Slavoff SA, et al. A human short open reading frame (sORF)-encoded polypeptide that stimulates DNA end joining. J Biol Chem. 2014;289(16):10950–7. doi: 10.1074/jbc.C113.533968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nelson BR, et al. Small open reading frames pack a big punch in cardiac calcium regulation. Circ Res. 2014;114(1):18–20. doi: 10.1161/CIRCRESAHA.113.302716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bi P, et al. Control of muscle formation by the fusogenic micropeptide myomixer. Science. 2017 doi: 10.1126/science.aam9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guo B, et al. Humanin peptide suppresses apoptosis by interfering with Bax activation. Nature. 2003;423(6938):456–61. doi: 10.1038/nature01627. [DOI] [PubMed] [Google Scholar]
- 78.Housman G, Ulitsky I. Methods for distinguishing between protein-coding and long noncoding RNAs and the elusive biological purpose of translation of long noncoding RNAs. Biochim Biophys Acta. 2016;1859(1):31–40. doi: 10.1016/j.bbagrm.2015.07.017. [DOI] [PubMed] [Google Scholar]
- 79.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Banfai B, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22(9):1646–57. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gibson TJ, et al. The transience of transient overexpression. Nat Methods. 2013;10(8):715–21. doi: 10.1038/nmeth.2534. [DOI] [PubMed] [Google Scholar]
- 82.Dyson MR, et al. Production of soluble mammalian proteins in Escherichia coli: identification of protein features that correlate with successful expression. BMC Biotechnol. 2004;4:32. doi: 10.1186/1472-6750-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen X, et al. Fusion protein linkers: property, design and functionality. Adv Drug Deliv Rev. 2013;65(10):1357–69. doi: 10.1016/j.addr.2012.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.O’Carroll AM, et al. The apelin receptor APJ: journey from an orphan to a multifaceted regulator of homeostasis. J Endocrinol. 2013;219(1):R13–35. doi: 10.1530/JOE-13-0227. [DOI] [PubMed] [Google Scholar]
- 85.Hubner NC, et al. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol. 2010;189(4):739–54. doi: 10.1083/jcb.200911091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mansfeld J, et al. APC15 drives the turnover of MCC-CDC20 to make the spindle assembly checkpoint responsive to kinetochore attachment. Nat Cell Biol. 2011;13(10):1234–43. doi: 10.1038/ncb2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Planta RJ, Mager WH. The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast. 1998;14(5):471–7. doi: 10.1002/(SICI)1097-0061(19980330)14:5<471::AID-YEA241>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 88.Stamm LV, et al. Cloning of the recA gene from a free-living leptospire and distribution of RecA-like protein among spirochetes. Appl Environ Microbiol. 1991;57(1):183–9. doi: 10.1128/aem.57.1.183-189.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Uzunova K, et al. APC15 mediates CDC20 autoubiquitylation by APC/C(MCC) and disassembly of the mitotic checkpoint complex. Nat Struct Mol Biol. 2012;19(11):1116–23. doi: 10.1038/nsmb.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shastri N, et al. Producing nature’s gene-chips: the generation of peptides for display by MHC class I molecules. Annu Rev Immunol. 2002;20:463–93. doi: 10.1146/annurev.immunol.20.100301.064819. [DOI] [PubMed] [Google Scholar]
- 91.Starck SR, et al. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science. 2012;336(6089):1719–23. doi: 10.1126/science.1220270. [DOI] [PubMed] [Google Scholar]
- 92.Starck SR, Shastri N. Non-conventional sources of peptides presented by MHC class I. Cell Mol Life Sci. 2011;68(9):1471–9. doi: 10.1007/s00018-011-0655-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Starck SR, et al. Translation from the 5′ untranslated region shapes the integrated stress response. Science. 2016;351(6272):aad3867. doi: 10.1126/science.aad3867. [DOI] [PMC free article] [PubMed] [Google Scholar]