Abstract
Purpose
To devise a comprehensive multi-platform genetic testing strategy for inherited retinal disease and describe its performance in 1,000 consecutive families seen by a single clinician.
Methods
The clinical records of all patients seen by a single retina specialist between January 2010 and June 2016 were reviewed and all patients who met the clinical criteria for a diagnosis of inherited retinal disease were included in the study. Each patient was assigned to one of 62 diagnostic categories and this clinical diagnosis was used to define the scope and order of the molecular investigations that were performed. The number of nucleotides evaluated in a given subject ranged from two (a multiplex allele-specific assay for the most common mutations in BBS1 and BBS10) to nearly 900,000 (the coding sequences, and splice junctions of 305 genes known to cause inherited retinal disease).
Results
Disease-causing genotypes were identified in 760 families (76%). These genotypes were distributed across 104 different genes. More than 70% of these 104 genes have coding sequences small enough to be efficiently packaged into an adeno-associated virus. Mutations in ABCA4 were the most common cause of disease in this cohort (173 families) while mutations in 80 genes caused disease in five or fewer families (i.e., 0.5% or less). Disease-causing genotypes were identified in 576 of the families without next generation sequencing (NGS). This included 23 families with mutations in the repetitive region of RPGR exon 15 that would have been missed by NGS. Whole exome sequencing of the remaining 424 families revealed mutations in an additional 182, and whole genome sequencing of four of the remaining 242 families revealed two additional genotypes that were invisible by the other methods. Performing the testing in a clinically-focused tiered fashion would be 6.1% more sensitive, 17.7% less expensive and have a significantly lower average false genotype rate than using whole exome sequencing to assess more than 300 genes in all patients (7.1 vs. 128%; p<0.001).
Conclusions
Genetic testing for inherited retinal disease is now more than 75% sensitive. A clinically-directed tiered testing strategy can increase sensitivity and improve statistical significance without increasing cost.
When the first inherited retinal disease genes were discovered in the late 1980s and early 1990s 1-3, ophthalmic genetics was largely a descriptive subspecialty. The primary goals of the ophthalmologist were to give the patient's condition a name and to try to discern the inheritance pattern so that one could give the patient and their family members a reasonably accurate estimate of the risk that other family members would be affected with a similar disease. At that time, the chance that a molecular diagnosis could be accomplished for the average patient with an inherited retinal disease was less than 5%, and such tests were only performed by a few research laboratories.
The main limitations to molecular diagnosis in the early 1990's were the overall lack of knowledge of the human genome and the relatively crude and laborious methods for investigating it. An often underappreciated positive effect of those limitations was that molecular tests in the early 1990s tended to be very focused by the clinical features of the family being studied. For example, one would not have sequenced the rhodopsin gene in a person with the clinical features of Best disease and thus would not have been in a position to observe a rare non-disease-causing polymorphism in the rhodopsin gene and incorrectly conclude that it was disease-causing in that patient.
Many things have changed in ophthalmic genetics in the past 25 years, perhaps most notably the successful use of gene therapy for inherited retinal disease 4-6, the more widespread availability of preimplantation genetic testing to reduce the recurrence of severe genetic diseases, and the introduction of CRISPR-based genome editing 7-9, which, when coupled with induced pluripotent stem cells 10,11 and in vitro retinal differentiation have the potential to generate immunologically-matched genetically-corrected cells for therapeutic transplantation 10,11 (see Table 1 for a glossary of abbreviations and acronyms used in this paper). The advent of these gene-directed interventions have increased both the value and the risks of genetic testing. For these treatments to work, one must know the disease-causing gene and in some cases the exact disease-causing mutations with complete accuracy. The diagnostic goal of the clinician is no longer to just give the clinical findings a name, it is to identify the patient's disease-causing genotype with sufficient accuracy that the probability of a gene-directed intervention helping the patient is significantly greater than the possibility that it will cause harm.
Table 1. List of abbreviations and acronyms.
| Abbreviation | Definition |
|---|---|
| AD | Autosomal Dominant |
| ADNIV | Autosomal Dominant Neovascular Inflammatory Vitreoretinopathy |
| AR | Autosomal Recessive |
| AR -1 | Autosomal Recessive - 1 Allele Identified |
| ARMS | Amplification Refractory Mutation System |
| AZOOR | Acute Zonal Occult Outer Retinopathy |
| BBS | Bardet-Biedl Syndrome |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| CSNB | Congential Stationary Night Blindness |
| CSSD | Congenital Stationary Synaptic Dysfunction |
| DDND | Developmental Delay and/or Neuromuscular Degeneration; |
| ECORD | Early Childhood Onset Retinal Dystrophy |
| ERG | Electroretinogram |
| EV | Erosive Vitreoretinopathy |
| ExAC | Exome Aggregation Consortium |
| FEVR | Familial Exudative Vitreoretinopathy |
| FGR | False Genotype Rate |
| HMA | Homocystinuria with Macular Atrophy |
| HPCD | Helicoid Peripapillary Chorioretinal Degeneration |
| ISCEV | International Society for Clinical Electrophysiology of Vision |
| IVS | Intervening Sequence |
| L/M Opsin | Long/Medium Wave Length Opsin |
| LCA | Leber Congenital Amaurosis |
| LCHAD | Long-chain 3-hydroxyacyl-coenzyme A Dehydrogenase Deficiency |
| LHON | Leber Hereditary Optic Neuropathy |
| MCLMR | Microcephaly Congenital Lymphedema and Chorioretinopathy |
| MDPD | Mutation Detection Probability Distribution |
| MIDD | Maternally Inherited Diabetes and Deafness |
| MIS | Missense |
| Mito | Mitochondrial |
| NGS | Next Generation Sequencing |
| PCR | Polymerase Chain Reaction |
| PV | Plausible Variants |
| RP | Retinitis Pigmentosa |
| RPE | Retinal Pigment Epithelial |
| SECORD | Severe Early Childhood Onset Retinal Dystrophy |
| TERM | Terminating |
| VVD | Vision Variation Database |
| XL | X-Linked |
Fortunately, genetic testing methods have also changed dramatically in recent years. What was once considered to be the largest scientific undertaking of mankind, the sequencing of the entire human genome 12,13, can now be accomplished in an individual patient in just a few weeks time for a few thousand dollars. This has led some to believe that experienced clinicians are now less necessary for the care of patients with inherited diseases and that the tests themselves are so powerful that they can provide the correct answer in almost any clinical situation regardless of the quality or quantity of accompanying clinical information. Actually, the reverse is true. As genetic tests have become larger in scope and sensitivity, the need for exceptionally detailed and accurate clinical information has also increased. This is primarily because there is a lot of normal variation in the human genome – millions of genetic differences between any two healthy individuals – and as a result, very broad investigations will always result in multiple plausibly-disease-causing findings that will need to be winnowed to one on clinical grounds.
We undertook this study for several related purposes: 1) to determine the current overall sensitivity of genetic testing for inherited retinal disease, 2) to determine the relative frequencies of inherited retinal disorders seen in a single North American eye clinic, 3) to determine the proportions of these diseases caused by mutations in specific genes, 4) to develop a teachable algorithm for pretest clinical diagnosis, 5) to evaluate the efficiency of a clinically-driven tiered genetic testing strategy, and 6) to provide practicing ophthalmologists some insight into the complexity of next generation sequencing data and the obligation to apply corrections for multiple measurements to these data.
Methods
Human Subjects
This study was approved by the Institutional Review Board of the University of Iowa and adhered to the tenets set forth in the Declaration of Helsinki. All patients seen by a single clinician (EMS) in the Retina Clinic of the University of Iowa Department of Ophthalmology and Visual Sciences between January 2010 and June 2016, who were judged by that clinician to have a monogenic heritable component to their eye disease, and who were 60 years of age or younger when first symptomatic, were offered inclusion in the study. Those who chose to participate (more than 99% of those invited) provided written informed consent. In many cases, additional family members were also invited to participate in the study either at the time of the original clinic visit, at a later visit, or by sending samples and records by mail. Patients with the following clinical diagnoses were excluded from the cohort: age-related macular degeneration, central serous retinopathy, autoimmune retinal disease, and acute zonal occult outer retinopathy (AZOOR).
Clinical Assessment
All probands and available family members underwent a complete eye examination including visual acuity assessment, intraocular pressure measurement, evaluation of ocular motility and pupils, slit lamp biomicroscopy, and indirect ophthalmoscopy. The majority of patients also underwent Goldmann perimetry, color fundus photography, and spectral domain optical coherence tomography. A subset also had an assessment of color vision, reduced intensity autofluorescence imaging 14, fluorescein angiography and/or electroretinography according to ISCEV standards 15.
Diagnostic Classification
All available historical, clinical, electrophysiological and imaging data from each participant were digitized and re-reviewed by a single clinician (EMS) for the purpose of placing each of the 1,000 probands into an objectively defined clinical category. A patient's genotype was never used to place them into a category. Even when a clinical diagnosis appeared to be “wrong” after genetic testing (such as the de novo rhodopsin mutation that mimicked autosomal recessive ECORD in a young female – patient # 442 in Supplemental Table 1), the original clinical diagnosis was retained. The purpose of this objective assignment was to allow us to determine how many patients out of 1000 would fall into each specific clinical category and, which genes were responsible for disease in each of these objectively defined categories. The names and inclusion criteria for the majority of the 96 resulting diagnostic categories (Figure 1, Table 2) are for the most part well defined in the existing clinical literature. However, in a few cases, some empiric rules were established to more clearly define the borders between categories (see Results). Also, the higher order grouping of the individual categories was somewhat non-standard and was chosen to minimize the number of decisions or clinical tests that were needed to place a patient into their category.
Figure 1. Distribution of patients and molecular findings across all levels of the clinical classification system.

The structure of the classification system is shown at left with the common clinical terms for each phenotypic group shown in the adjacent column. The Total column provides the number of probands assigned to each clinical group, while the Solved column shows the number of probands in each group with a disease-causing genotype identified. The Genes columns provide the number of genes that have been observed to cause the diseases of that clinical group in the published literature and/or at the University of Iowa. The false genotype rate (FGR) columns give the percentage of normal individuals that would be expected to harbor a plausible disease-causing complete genotype by chance in any of the genes assigned to each clinical category in the published literature and/or at the University of Iowa. PV is the average number of plausible disease-causing variants one would expect to observe in a normal individual by chance in any of the genes assigned to each clinical category in the published literature. The bar lengths represent the percent of solved cases for each clinical category while the alternating shades represent the proportional contributions of each gene in descending order. Gene names are given for any genes that cause at least 15% of the disease in a given category. Blue bars indicate categories with an FGR less than 5% while grey bars indicate categories with an FGR greater than or equal to 5%. Abbreviations: AD, Autosomal Dominant; ADNIV, Autosomal Dominant Neovascular Inflammatory Vitreoretinopathy; AR, Autosomal Recessive; CSNB, Congenital Stationary Night Blindness; CSSD, Congenital Stationary Synaptic Dysfunction; DDND, Developmental Delay and/or Neuromuscular Degeneration; ECORD, Early Childhood Onset Retinal Dystrophy; EV, Erosive Vitreoretinopathy; FEVR, Familial Exudative Vitreoretinopathy; HMA, Homocystinuria with Macular Atrophy; HPCD, Helicoid Peripapillary Chorioretinal Degeneration; LHON, Leber Hereditary Optic Neuropathy; MCLMR, Microcephaly Congenital Lymphedema and Chorioretinopathy; MIDD, Maternally Inherited Diabetes and Deafness; SECORD, Severe Early Childhood Onset Retinal Dystrophy; XL, X-linked.
Table 2.
Inherited retinal disease categories.
| I – Photoreceptor Disease |
| A – Isolated |
| 1 – Acquired/Progressive |
| a – Retinitis Pigmentosa |
| i – X-linked |
| ii – Autosomal Dominant |
| iii – Autosomal Recessive |
| iv – Other Multiplex |
| b – Cone and Cone Rod Dystrophy |
| i – X-linked |
| ii – Autosomal Dominant |
| iii – Autosomal Recessive |
| iv – Other Multiplex |
| 2 – Congenital/Stationary |
| a – LCA |
| b – SECORD |
| c – ECORD |
| d – Achromatopsia (Congenital Stationary Cone Dysfunction) |
| e – Blue Cone Monochromacy |
| f – Congenital Stationary Night Blindness |
| i – X-linked |
| ii – Autosomal Dominant |
| iii – Autosomal Recessive with normal fundus |
| iv – Enhanced S-cone Syndrome |
| v – Fundus Albipunctatus |
| vi – Oguchi Disease |
| g – Congenital Stationary Synaptic Dysfunction |
| h – Delayed Retinal Maturation |
| B – Syndromic |
| 1 – Usher Syndrome |
| a – Type I |
| b – Type II |
| c – Type III |
| 2 – Bardet-Biedl Syndrome |
| 3 – Neuronal Ceroid Lipofucinosis |
| 4 – Senior-Loken Syndrome |
| 5 – Joubert Syndrome |
| 6 – Microcephaly Congenital Lymphedema and Chorioretinopathy |
| 7 – Retinitis Pigmentosa with Ataxia |
| 8 – Peroxisomal Biogenesis Disorders |
| 9 – Cohen Syndrome |
| II – Macular Diseases |
| A – Autosomal Recessive Stargardt Disease |
| B – Best Disease |
| C – Pattern Dystrophy |
| D – Autosomal Dominant Stargardt Disease |
| E – Sorsby Fundus Dystrophy |
| F – Malattia Leventinese |
| G – North Carolina Macular Dystrophy |
| H – Syndromic Macular Diseases |
| 1 – Macular Dystrophy, Diabetes and Deafness |
| 2 – Pseudoxanthoma Elasticum |
| 3 – Homocystinuria with Macular Atrophy |
| 4 – Spinocerebellar Atrophy |
| I – Benign Fleck Retina |
| III – Third Branch Disorders |
| A – Choroidopathies |
| 1 – Choroideremia |
| 2 – Gyrate Atrophy |
| 3 – Late Onset Retinal Dystrophy |
| 4 – Nummular Choroidal Atrophy |
| 5 – Helicoid Peripapillary Chorioretinal Degeneration |
| B – Retinoschisis |
| 1 – X-linked |
| 2 – Recessive |
| C – Optic Neuropathies |
| 1 – Nonsyndromic |
| a – Autosomal Dominant |
| b – Autosomal Recessive |
| c – Leber Hereditary Optic Neuropathy |
| 2 – Syndromic |
| a – Wolfram Syndrome |
| b – Hearing Loss |
| D – Tumors |
| 1 – von Hippel Lindau |
| 2 – Retinoblastoma |
| 3 – Tuberous Sclerosis |
| 4 – Gardner Syndrome |
| E – Vitreoretinopathies |
| 1 – Stickler Syndrome |
| 2 – Familial Exudative Vitreoretinopathy |
| a – Norrie Disease |
| b – Autosomal Dominant |
| 3 – AD Neovascular Inflammatory Vitreoretinopathy |
| 4 – Wagner Disease (Erosive Vitreoretinopathy) |
| 5 – Knobloch Syndrome |
| 6 – Heritable Vascular Tortuosity |
| a – Autosomal Dominant Retinal Vascular Tortuosity |
| b – Cerebroretinal Vasculopathy |
| c – Fascioscapulohumeral Dystrophy |
| F – Albinism |
| 1 – X-linked Ocular Albinism |
| 2 – Oculocutaneous Albinism |
| a – Nonsyndromic |
| b – Hermansky Pudlak |
| c – Chediak Higashi |
| G – Isolated Foveal Hypoplasia |
Disease Genes
The published literature was reviewed to identify all genes that had been convincingly shown to cause genetic retinal disease. These 305 genes (Supplemental Table 2) were divided into two groups based on whether a gene was only known to cause severe progressive loss of cognition and/or neuromuscular control and/or significantly shortened life expectancy (43 genes) or not (262 genes). The published literature was also reviewed to identify the retinal phenotypes that had been previously associated with each of these 305 genes and these data were used to associate each gene with one or more of the 96 diagnostic categories shown in Figure 1, Table 2 and Supplemental Table 2. The 43 genes associated with the more severe systemic diseases were only included in the analysis when clinical features suggestive of a debilitating systemic phenotype were already manifest.
DNA Extraction
Blood samples were obtained from all probands (n = 1,000) and available family members (n = 2,348) and DNA was extracted by using the manufacturer's specifications for whole-blood DNA extraction using Gentra System's Autopure LS instrument.
First Tier Genetic Testing
A preliminary mutation detection probability distribution (MDPD 16) was established for each of the 96 clinical categories using a combination of the published literature and the anonymized summary experience of the Carver Nonprofit Genetic Testing Laboratory at the University of Iowa. These MDPDs were used to devise focused screens designed to detect the most common disease-causing alleles of the most common genes associated with each of the diagnostic categories. These screens each employed one or more of the following approaches: automated Sanger sequencing with an ABI 3730xl sequencer, allele-specific genotyping with a Fluidigm EP1, amplification refractory mutation system (ARMS 17), chromosomal microarray analysis, and/or plasmid cloning of PCR products followed by Sanger sequencing. Variants were considered “disease-causing” if they met our previously published criteria 18 for an Estimate of Pathogenic Probability (EPP) of 2 or 3. A genotype was considered “convincing,” and the patient included in the calculation of the solve rate for that diagnostic category, if it consisted of a heterozygous mutation with an EPP of 2 or 3 in a gene known to cause a dominant disease, a hemizygous mutation with an EPP of 2 or 3 in a gene known to cause X-linked disease; or, two mutations (suspected to lie on separate alleles by direct observation or statistical inference) each with an EPP of 2 or 3, in a gene known to cause recessive disease.
Cloning and Sequencing of RPGR Exon 15
To detect mutations in the low complexity region of RPGR exon 15, Sanger sequencing of TA cloned PCR products was performed. Patient DNA was PCR amplified and the products were gel purified and TA cloned into the pCR®2.1 TOPO® Vector using the TOPO® TA Cloning® Kit (Invitrogen, Grand Island, NY). TA cloned PCR products were transformed using One Shot® TOP10 chemically competent cells (Invitrogen). Transformed cells were subsequently streaked and cultured on AIX plates (AIX; Aachen, Germany) for blue-white colony screening. Validated clones were picked, expanded in LB broth, purified and Sanger sequenced on the ABI 3730xl sequencer using optimized sequencing chemistry.
Next Generation Sequencing
Whole exome sequencing was performed using the Agilent v5 exome kit with the addition of custom xGen Lockdown probes (IDT, Coralville IA) to target regions of the genome relevant to eye disease that are not well covered in the standard exome kit. These regions cover known non-coding mutations in CEP290, USH2A, ABCA4, and the L/M opsin cluster, in addition to insufficiently covered coding exonic sequence in genes such as ABCC6 (all bait sequences available upon request). Whole exome sequencing was performed using the Illumina HiSeq 2500 or 4000. Whole Genome Sequencing was performed using the HiSeq X (Hudson Alpha; Huntsville, AL). Sequences were aligned to the genome using BWA 19. Single nucleotide variations and small insertions and deletions were called using GATK 20. Structural variants were called using Conifer 21 and Manta 22.
Calculation of the False Genotype Rate (FGR)
Genetic variations that cause rare, high-penetrance, monogenic diseases are also rare in the population and most genotyping pipelines, including ours, remove variants that are too common to cause the rare diseases under study. For this project, the cutoff for recessive variants was set at 0.006 (the frequency of the more common well-established disease-causing mutations in ABCA4), the cutoff for mitochondrial variants was 0.004 (the frequency of the most common LHON variant, 11778), and the cutoff for dominant disease was set at 0.0001 (the frequency of the most common well-established mutations in RHO). The frequency at which one would encounter a variant at or below these thresholds in healthy people is proportional to the amount of exomic sequence analyzed and was directly measured by applying the pipeline cutoff values to the whole exome data from the 60,000 healthy individuals collected by the Exome Aggregation Consortium (ExAC 23) and mitochondrial variants observed in 32,000 healthy individuals in MitoMap 24. We defined the false genotype rate (FGR) as the frequency with which one would encounter a plausibly disease-causing recessive or dominant complete genotype when sequencing the coding regions of a specific set of genes in a healthy person. We used the ExAC data to calculate FGR values for each group of genes mapped to each specific clinical category (Figure 1). The FGR is conceptually very similar to the commonly used false discovery rate (FDR). We chose the term FGR for the current analysis because: 1) we had recessive complete genotypes for some genes and dominant mutations for others; 2) the recessive genotypes were not directly observed, but modeled from data from the ExAC database; and, 3) we wanted to fully convey the associated risk of an incorrect genetic test result.
Calculation of Genetic Test Costs
For each diagnostic category, a specific sequence of tests was devised based upon the mutation detection probability distribution 16 for that category obtained from both the published literature and the anonymized experience of the Carver Nonprofit Genetic Testing Laboratory at the University of Iowa. During development, each step in the testing sequence was optimized by subjecting it to a cost analysis (available upon request). For the analysis in this paper, the research cost of the currently recommended sequence of tests for each patient was calculated based upon their pre-test clinical diagnosis (details of the specific testing order, primer sequences, PCR conditions, etc., for any diagnostic category is available by request to the authors). The current research costs of the test components are: DNA extraction and quality control genetic markers -$40; Amplification Refractory Mutation System reaction - $38; one set of 44 alleles assayed using the Fluidigm system - $35; bidirectional Sanger sequencing of one PCR amplimer - $20; chromosomal microarray analysis - $500; TA-cloning and bidirectional sequencing of RPGR exon 15 codons 762 to 1,100 - $650 for males and $975 for females; whole exome sequencing, analysis and confirmation - $1,200; whole genome sequencing - $2,450.
Results
The 1,000 probands in this study came to our clinic from 40 different states, the District of Columbia and seven foreign countries (Supplemental Figure 1). Four hundred eighty-nine were female and 511 were male. The average age at entry into the study was 37.3 years (36.3 years for males and 38.5 years for females); the range was 8 months to 88 years. Plausible disease-causing genotypes were identified in 760 of these probands, 393 males and 367 females (Supplemental Table 1). The average age at entry into the study was very slightly younger for those in whom a disease-causing genotype was identified (34.9 years for males and 37.7 years for females).
The clinical classification system (Figure 1, Table 2) used in this study was devised as a means for clinicians who see adults and older children with inherited retinal diseases to 1) efficiently communicate their clinical impressions to the molecular diagnostic laboratory charged with identifying the patients' disease-causing mutations; and 2) to narrow the pre-test hypothesis to the smallest number of genes possible at our current level of clinical understanding. For the most part, the names used to refer to the individual clinical entities in the classification system are in common clinical use, and only the higher order grouping of these terms is in any way unusual. This grouping was chosen to keep entities with similar genetic causes as close to one another as possible in the diagnostic tree so that if the initial screening was negative, a laboratory could recursively enlarge the molecular hypothesis in the most statistically efficient manner. For example, RDS-associated pattern dystrophy and ABCA4-associated Stargardt disease can cause almost identical clinical findings in selected patients. As a result, these categories are adjacent to each other in the classification scheme. If a screen of ABCA4 were negative, the lab would screen RDS (even without a dominant family history) before moving on to screen larger genomic spaces. The most clinically homogeneous and genetically heterogeneous groups were those affected with non-syndromic acquired photoreceptor degeneration (retinitis pigmentosa – group IA1a; and, cone or cone rod dystrophy – group IA1b; Figure 1). Multiplex kindreds belonging to these large categories were subdivided according to their pedigree structure as follows: i – X-linked (affected males in multiple sibships connected to each other through unaffected or mildly affected females with no instances of male to male transmission), ii – autosomal dominant (a minimum of three generations with at least one instance of male to male transmission), iii – autosomal recessive (multiple affected individuals in a single sibship with normal parents), and iv – other multiplex (all other multiplex kindreds).
Placement into one of the first three categories of congenital/stationary photoreceptor disease (IA2a-c) required clear historical evidence of parental or physician awareness of significant visual dysfunction – more than just night blindness – before the patient's fourth birthday. These patients were further divided into: a – Leber congenital amaurosis (LCA) if their visual acuity was so poor that they did not use it for education or activities of daily living; b – Severe Early Childhood Onset Retinal Dystrophy (SECORD 25) if they had useful vision but became legally blind before age 10; or, c – Early Childhood Onset Retinal Dystrophy (ECORD) if they were not legally blind before age 10. Patients were diagnosed with Congenital Stationary Synaptic Dysfunction (IA2g) if they had stable reduced acuity from birth, selective loss of the b-wave on the scotopic ERG, diminished b-wave amplitudes on the photopic ERG, but no difficulties with vision in dim light.
As the clinical records for the 1,000 patients and their relatives were reviewed to place them into these categories, it became evident that ten types of easily obtainable historical information were of particular value in reproducibly assigning patients to these categories and a form was created to assist physicians in acquiring these historical data in a prospective manner (Supplemental Table 4).
For this study, patients could be assigned to a higher-order point in the classification system if there were insufficient data to make a more specific assignment. For example, isolated patients with retinitis pigmentosa were assigned to IA1a while a member of an autosomal dominant family with affected individuals in three generations and clear male to male transmission was assigned to IA1aii. Of the 96 possible locations a proband could be placed in this classification system, only 62 were used at least once when subdividing the 1,000 probands in this study (Figure 1 and Table 2).
Figure 2 shows the breakdown of the 1,000 probands among the most common diagnostic categories while Figure 1 shows the frequency with which a convincing disease-causing genotype could be identified in each of these categories. In this cohort, 64.7% of the probands had photoreceptor disease (Category I), 28.2% had a macular dystrophy (Category II) and 7.1% had one of the 42 entities of the Category III. Overall, convincing genotypes were identified in 76% of the probands with the highest positivity among those with a macular dystrophy (88.3%). With four exceptions, patients with autosomal recessive disease were only considered positive if they had both disease alleles identified. The exceptions were patients with a clinical diagnosis of Stargardt disease (23 probands), Usher syndrome (two Type-1 and five Type-2 probands), achromatopsia (one proband) and homocystinuria (one proband), who were each found to have a convincing disease-causing mutation on only one allele (the FGR was less than 1% for each of these pre-test hypotheses).
Figure 2. Graphical depiction of the distribution of 1,000 consecutive probands among the larger diagnostic categories.

The center chart indicates the proportion of probands assigned to each of the three main branches of the classification system. The outer charts show the fraction of probands assigned to the larger diagnostic categories within each branch.
Figure 1 also shows the genetic heterogeneity of each of the 62 clinical categories with at least one patient assigned to it in this study. The most heterogeneous category was simplex RP (IA1a) which had disease-causing mutations identified in 36 different genes and a total solve rate of only 56.7%. The least heterogeneous category with at least ten probands in it was choroideremia (IIIA1), which had a 100% solve rate in 14 patients, all with mutations in a single gene (CHM).
Figure 3, Table 3 and Supplemental Table 3 show the frequencies of disease-causing genotypes in each of the 104 genes that were found to cause disease in at least one family in the cohort. ABCA4 was the single most common disease-causing gene and was responsible for disease in 173 families. Twelve additional genes, USH2A, RPGR, RHO, PRPH2, BEST1, CRB1, BBS1, CEP290, PRPF31, CHM, RS1, and RP1 each caused disease in 1% or more of the cohort and these 13 genes were collectively responsible for disease in almost one half of the families (497). The remaining 91 genes each caused disease in less than 1% of the cohort and collectively caused disease in 26.2% of the total. Thirty of the genes each caused disease in a single family and one family had a de novo chromosomal translocation. This cohort is certainly not a random sample of the US population. However, it was ascertained consecutively, and was drawn from 40 of the 50 US states. Thus, we felt that it would be reasonable to use these data to provide a rough estimate of the total number of individuals affected with each gene-specific disease in the country. Assuming that mutations in ABCA4 cause disease in 1/10,000 people 26, Table 3 gives an estimate for the total number of people of all ages in the US with mutations in this gene. Similarly, by using 2010 US census data that show 20.2 million people in the US under the age of 5 27, one can also estimate the number of new cases of each gene-specific retinal disease in the US per year. Collectively, these data suggest that there are currently about 140,000 people in the US affected with one of the diseases evaluated in this study and about 1,700 new cases per year.
Figure 3. Distribution of the number of probands per gene.

Thirteen genes each caused disease in 1% or more of the probands in this study (left of dashed vertical line) while the other 91 each caused disease in less than 1%. These data are presented in more detail in Table 3.
Table 3.
Estimate of the total number of people of all ages in the United States with mutations in genes observed in this study.
| Gene | No. in Cohort | Freq. in U.S. | No. in U.S. | New Cases per Year |
|---|---|---|---|---|
| ABCA4 | 173 | 1 / 10,000 | 32,440 | 404 |
| USH2A | 76 | 1 / 22,763 | 14,251 | 177 |
| RPGR | 48 | 1 / 36,042 | 9,001 | 112 |
| RHO | 34 | 1 / 50,882 | 6,376 | 79 |
| PRPH2 | 32 | 1 / 54,062 | 6,000 | 75 |
| BEST1 | 25 | 1 / 69,200 | 4,688 | 58 |
| CRB1 | 20 | 1 / 86,500 | 3,750 | 47 |
| BBS1 | 19 | 1 / 91,053 | 3,563 | 44 |
| CEP290 | 18 | 1 / 96,111 | 3,375 | 42 |
| PRPF31 | 15 | 1 / 115,333 | 2,813 | 35 |
| CHM | 14 | 1 / 123,571 | 2,625 | 33 |
| RS1 | 13 | 1 / 133,077 | 2,438 | 30 |
| RP1 | 10 | 1 / 173,000 | 1,875 | 23 |
| FAM161A | 9 | 1 / 192,222 | 1,688 | 21 |
| MYO7A | 8 | 1 / 216,250 | 1,500 | 19 |
| OPA1 | 8 | 1 / 216,250 | 1,500 | 19 |
| PCDH15 | 8 | 1 / 216,250 | 1,500 | 19 |
| RP2 | 8 | 1 / 216,250 | 1,500 | 19 |
| GUCA1A | 7 | 1 / 247,143 | 1,313 | 16 |
| IMPG2 | 7 | 1 / 247,143 | 1,313 | 16 |
| MAK | 7 | 1 / 247,143 | 1,313 | 16 |
| PDE6B | 7 | 1 / 247,143 | 1,313 | 16 |
| EYS | 6 | 1 / 288,333 | 1,125 | 14 |
| PROM1 | 6 | 1 / 288,333 | 1,125 | 14 |
| RDH12 | 6 | 1 / 288,333 | 1,125 | 14 |
| CLN3 | 5 | 1 / 346,000 | 938 | 12 |
| CNGB3 | 5 | 1 / 346,000 | 938 | 12 |
| IQCB1 | 5 | 1 / 346,000 | 938 | 12 |
| NR2E3 | 5 | 1 / 346,000 | 938 | 12 |
| VHL | 5 | 1 / 346,000 | 938 | 12 |
| BBS2 | 4 | 1 / 432,500 | 750 | 9 |
| CACNA1F | 4 | 1 / 432,500 | 750 | 9 |
| CDH23 | 4 | 1 / 432,500 | 750 | 9 |
| CDHR1 | 4 | 1 / 432,500 | 750 | 9 |
| FLVCR1 | 4 | 1 / 432,500 | 750 | 9 |
| GUCY2D | 4 | 1 / 432,500 | 750 | 9 |
| KIF11 | 4 | 1 / 432,500 | 750 | 9 |
| KLHL7 | 4 | 1 / 432,500 | 750 | 9 |
| NMNAT1 | 4 | 1 / 432,500 | 750 | 9 |
| BBS10 | 3 | 1 / 576,667 | 563 | 7 |
| CERKL | 3 | 1 / 576,667 | 563 | 7 |
| CNGA3 | 3 | 1 / 576,667 | 563 | 7 |
| COL2A1 | 3 | 1 / 576,667 | 563 | 7 |
| CRX | 3 | 1 / 576,667 | 563 | 7 |
| ELOVL4 | 3 | 1 / 576,667 | 563 | 7 |
| IFT140 | 3 | 1 / 576,667 | 563 | 7 |
| INPP5E | 3 | 1 / 576,667 | 563 | 7 |
| L/M Opsin Cluster | 3 | 1 / 576,667 | 563 | 7 |
| MERTK | 3 | 1 / 576,667 | 563 | 7 |
| MT-TL1 | 3 | 1 / 576,667 | 563 | 7 |
| PRPF8 | 3 | 1 / 576,667 | 563 | 7 |
| RPE65 | 3 | 1 / 576,667 | 563 | 7 |
| VPS13B | 3 | 1 / 576,667 | 563 | 7 |
| ABCC6 | 2 | 1 / 865,000 | 375 | 5 |
| ACO2 | 2 | 1 / 865,000 | 375 | 5 |
| ADGRV1 | 2 | 1 / 865,000 | 375 | 5 |
| CNGB1 | 2 | 1 / 865,000 | 375 | 5 |
| DHDDS | 2 | 1 / 865,000 | 375 | 5 |
| IMPDH1 | 2 | 1 / 865,000 | 375 | 5 |
| KCNV2 | 2 | 1 / 865,000 | 375 | 5 |
| MKKS | 2 | 1 / 865,000 | 375 | 5 |
| NYX | 2 | 1 / 865,000 | 375 | 5 |
| PEX1 | 2 | 1 / 865,000 | 375 | 5 |
| PPT1 | 2 | 1 / 865,000 | 375 | 5 |
| PRDM13 | 2 | 1 / 865,000 | 375 | 5 |
| PRPF3 | 2 | 1 / 865,000 | 375 | 5 |
| RPGRIP1 | 2 | 1 / 865,000 | 375 | 5 |
| SNRNP200 | 2 | 1 / 865,000 | 375 | 5 |
| TIMP3 | 2 | 1 / 865,000 | 375 | 5 |
| TRNT1 | 2 | 1 / 865,000 | 375 | 5 |
| TRPM1 | 2 | 1 / 865,000 | 375 | 5 |
| USH1C | 2 | 1 / 865,000 | 375 | 5 |
| WDR19 | 2 | 1 / 865,000 | 375 | 5 |
| ZNF408 | 2 | 1 / 865,000 | 375 | 5 |
| ABHD12 | 1 | 1 / 1,730,000 | 188 | 2 |
| AIPL1 | 1 | 1 / 1,730,000 | 188 | 2 |
| ATXN7 | 1 | 1 / 1,730,000 | 188 | 2 |
| BBS9 | 1 | 1 / 1,730,000 | 188 | 2 |
| CABP4 | 1 | 1 / 1,730,000 | 188 | 2 |
| CEP78 | 1 | 1 / 1,730,000 | 188 | 2 |
| CLRN1 | 1 | 1 / 1,730,000 | 188 | 2 |
| GPR143 | 1 | 1 / 1,730,000 | 188 | 2 |
| HADHA | 1 | 1 / 1,730,000 | 188 | 2 |
| IFT172 | 1 | 1 / 1,730,000 | 188 | 2 |
| Karyotypic | 1 | 1 / 1,730,000 | 188 | 2 |
| LCA5 | 1 | 1 / 1,730,000 | 188 | 2 |
| MAN2B1 | 1 | 1 / 1,730,000 | 188 | 2 |
| MFRP | 1 | 1 / 1,730,000 | 188 | 2 |
| MFSD8 | 1 | 1 / 1,730,000 | 188 | 2 |
| MT-ND4 | 1 | 1 / 1,730,000 | 188 | 2 |
| MT-ND6 | 1 | 1 / 1,730,000 | 188 | 2 |
| MTR | 1 | 1 / 1,730,000 | 188 | 2 |
| NDP | 1 | 1 / 1,730,000 | 188 | 2 |
| NPHP1 | 1 | 1 / 1,730,000 | 188 | 2 |
| OAT | 1 | 1 / 1,730,000 | 188 | 2 |
| PAX6 | 1 | 1 / 1,730,000 | 188 | 2 |
| PEX6 | 1 | 1 / 1,730,000 | 188 | 2 |
| PNPLA6 | 1 | 1 / 1,730,000 | 188 | 2 |
| POMGNT1 | 1 | 1 / 1,730,000 | 188 | 2 |
| RLBP1 | 1 | 1 / 1,730,000 | 188 | 2 |
| RPIA | 1 | 1 / 1,730,000 | 188 | 2 |
| SLC24A1 | 1 | 1 / 1,730,000 | 188 | 2 |
| TULP1 | 1 | 1 / 1,730,000 | 188 | 2 |
| USH1G | 1 | 1 / 1,730,000 | 188 | 2 |
| WFS1 | 1 | 1 / 1,730,000 | 188 | 2 |
Figure 4 depicts a cost and yield comparison of two different strategies one could employ for genotyping the 1,000 probands of this study. In one case, whole exome sequencing would be performed on every proband and the resulting data would be evaluated for mutations in the 301 non-mitochondrial genes selected for inclusion in this study (see Methods). In this case, the cost to genotype each patient would be the same ($1,200, see methods) and the overall yield would be 70%. The mutations that would be missed would be those that lie in noncoding sequences (e.g., non-exomic mutations that cause Stargardt disease 28, Usher syndrome 29, retinitis pigmentosa 30, Leber congenital amaurosis 31,32 and North Carolina Macular Dystrophy 33), mitochondrial DNA (e.g., mutations that cause Leber hereditary optic neuropathy 34 and maternally inherited diabetes and deafness 35), and repetitive regions (e.g., the repetitive region of RPGR exon 15 36-38). In the other case, one would use a tiered testing strategy in which the testing was customized based upon the clinical findings and testing was stopped once a complete genotype was identified. With the latter strategy, the cost would range from $80 per patient (for those in whom a complete genotype was identified in the initial tier of testing) to more than $2,500 for those who did not have a complete genotype found on prescreening and were judged suitable for whole genome testing (see methods). With this tiered strategy, the average cost per patient would be 17.7% less than performing whole exome sequencing in everyone ($990) and the sensitivity would be a 6.1% higher because of the findings one would make in the non-coding regions, mitochondrial genes, and repetitive DNA that were specifically included in the clinically focused tests.
Figure 4. Financial cost and diagnostic yield of tiered testing strategy.
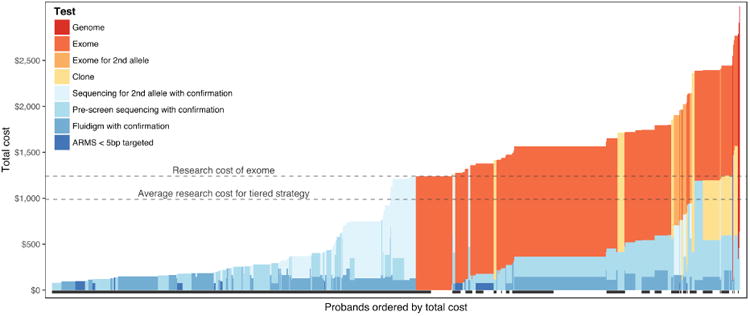
Patients are ordered from lowest cost to highest cost with colors representing the component costs our currently recommended series of genetic tests for each clinical category. A black bar beneath a patient indicates that a causative genotype was discovered in this individual. The horizontal lines highlight the higher cost of uniform whole exome sequencing (upper line) as compared to the average cost of clinically-focused individualized tests (lower line).
Figure 5 depicts the more important difference between the two screening strategies: the effect on the false genotype rate. Genetic variants that are rare enough in the general population to cause a Mendelian retinal disease are surprisingly common in whole exome sequencing data. In this study, the population frequency cutoff was set according to the most common well-established retinal disease-causing mutations (see methods). If one applies these criteria to the sequence data of the 60,000 healthy individuals in the ExAC database 23, one observes an average of 1.28 plausible disease-causing genotypes per person among the coding sequences of the 301 non-mitochondrial candidate genes considered in this study. Another way to state this is that with a coding sequence hypothesis 301 genes in size, there is an average FGR (see Methods) of 128%. For most medical tests, one would want a positive result to occur by chance no more than 5% of the time and for tests that would be used as the basis of preimplantation genetic testing or subretinal gene therapy, one might argue that it should be even less.
Figure 5. Statistical cost.

The false genotype rate (FGR) is the average number of complete genotypes one would expect to observe by chance in a healthy individual in a specified genomic space, based on data from 60,000 normal individuals 23. The probands in this study are shown ordered according to the FGR associated with their clinical category (see Figure 1). The red line indicates the FGR associated with the genes observed to cause disease in this cohort (see also Supplemental Figure 2). The dashed line indicates an FGR of 5% (i.e., the threshold at which one in 20 people would be expected to harbor a plausibly pathogenic, complete genotype by chance). The black bars at the bottom of the figure indicate that a disease-causing genotype was identified in this proband. Assessing the coding sequences of all 301 non-mitochondrial genes in all probands (green line) would result in an average FGR of 128% (i.e., these probands would be expected to harbor an average of 1.28 plausible, complete genotypes by chance).
Figure 5 shows that one can reduce the false genotype rate to clinically useful levels by narrowing the pre-test hypothesis to a relatively small number of genes. A tiered testing strategy linked to the clinical classification system in this study would identify plausible disease-causing genotypes in 48.7% of the cohort with an FGR less than 5%. Supplemental Figure 2 shows that one can also reduce the FGR per category at a given institution by first considering the genes that have been previously observed to cause disease in patients seen at that institution, and then if negative, considering a larger literature-based group of candidates and adding a statistical penalty for the additional hypothesis. The rationale for this two-step analysis is that the previous 1,000 patients seen in a given institution are likely to be more genetically similar to the next 1,000 patients seen there than they will be to the entire world population represented in the published literature.
For patients whose FGR is greater than 5%, which using the tiered strategy is most commonly due to our current inability to reduce the genetic heterogeneity of categories like simplex retinitis pigmentosa on clinical grounds (Figure 1), it is especially important to confirm the phase and/or segregation of their putative disease-causing variant(s) and to be a bit more skeptical of molecularly weaker genotypes such as those comprised entirely of novel missense variants. Table 4 shows the distribution of the 760 disease-causing genotypes identified in this study among inheritance patterns and mutation types. 2.5% of the genotypes involved molecularly confirmed de novo variants, which is a considerable underestimate of the actual de novo rate given that sufficient family samples to evaluate both parental alleles were available in fewer than 65% of families.
Table 4. Variation Distribution Across Inheritance Types.
| MIS | TERM | MIS + MIS | MIS + TERM | TERM + TERM | TOTALS | |
|---|---|---|---|---|---|---|
| AD | 140 | 32 | - | - | - | 172 |
| XL | 20 | 74 | - | - | - | 94 |
| MITO | 5 | - | - | - | - | 5 |
| AR-1 | 17 | 15 | - | - | - | 32 |
| AR | - | - | 146 | 160 | 151 | 457 |
| TOTALS | 182 | 121 | 146 | 160 | 151 | 760 |
AD – Autosomal Dominant
XL – X-Linked
AR-1 – Autosomal Recessive 1-Allele Identified
AR - Autosomal Recessive
MIS = Missense
TERM = Terminating
Five Illustrative Patients
Patient A is 47-year-old male who first noticed difficulty following the flight of a ball in his 20's (Supplemental Table 1, #375). In his early 40's he was examined and felt to have a cone dystrophy. He has no family history of a similar disorder. Our examination revealed a best corrected visual acuity of 20/50 OD and 20/80 OS. Ophthalmoscopy revealed an iridescent golden sheen to the entire posterior pole with the exception of a reddish atrophic circular area 1.5 mm in diameter centered on the fovea OU (Figure 6A). Optical coherence tomography showed a sharply demarcated loss of photoreceptors and RPE corresponding to the atrophic area seen on fundus examination (Figure 6B). Goldmann perimetry revealed a loss of the I2e isopter as well as a central scotoma to the III4e (Figure 6C). Plasmid cloning and DNA sequencing of the repetitive portion of RPGR exon 15 revealed a two base pair deletion in codon 1059. RPGR codons 800-1070 are poorly covered by whole exome sequencing and this mutation is undetectable with this method. It is also interesting that some frameshifting mutations in this exon are associated with a late onset cone selective disease 37,38, as seen in this patient, while similar mutations elsewhere in the gene cause severe rod predominant retinitis pigmentosa.
Figure 6. A 47-year-old male with RPGR-associated X-linked cone dystrophy. A.

Fundus photograph of the right eye. B: Optical coherence tomogram of the right eye. C: Goldmann visual field of the right eye.
Patient B is an 8-year-old male who first noted difficulties seeing in dim light in early childhood. His maternal grandfather had been diagnosed with choroideremia. On our examination his visual acuity was 20/32-1 OD and 20/32-2 OS. Ophthalmoscopy revealed extensive nummular areas of RPE and choriocapillaris loss each surrounded by a thin rim of hyperpigmented RPE (Figure 7A). The retinal arterioles were near normal in caliber. The Goldmann visual fields were surprisingly well preserved for this degree of retinal loss (Figure 7B). His mother (Supplemental Table 1, #938) and sister both exhibited “mud spattered” pigment mottling of the fundus consistent with the carrier state of an X-linked disease. Conventional DNA sequencing failed to detect a mutation in the CHM gene. However, the phenotype and history were so convincing that whole genome sequencing was performed in the child which revealed a complete duplication of CHM exons 6-8 which had been invisible to the non-quantiptative PCR-based DNA sequencing.
Figure 7. An 8-year-old male with choroideremia. A.

Fundus photograph of the right eye. B: Goldmann visual field of the right eye.
Patient C is a 48-year-old female who first had macular pigment mottling noticed incidentally on fundus examination at age 33 (Supplemental Table 1, #920). Her best corrected visual acuity on our examination was 20/20 OD and 20/60+2 OS. Ophthalmoscopy revealed patchy loss of the RPE and choriocapillaris OS more than OD (Figure 8A and 8B). Fundus autofluorescence revealed more extensive involvement than was visible ophthalmoscopically (Figure 8C and 8D). She developed gestational diabetes at age 27 and was diagnosed with type 2 diabetes at age 32. Her mother and maternal aunt are both diabetic as well. She developed hearing loss in her mid 30's and now wears hearing aids. PCR-based conventional DNA sequencing revealed a heteroplasmic mutation in the mitochondrial DNA at position 3243, which is known to cause an atrophic maculopathy with maternally inherited diabetes and deafness 35. Whole exome sequencing does not routinely assess mitochondrial DNA and as a result this mutation would have been missed unless it was specifically sought because of her phenotype.
Figure 8. A 48-year-old female with maternally inherited diabetes and deafness. A.

Fundus photograph of the right eye. B: Fundus photograph of the left eye. C-D: Fundus autofluorescence images of both the right eye (C) and left eye (D).
Patient D is a 10-year-old female who first had difficulty seeing the blackboard in school at 7 years of age. She has a family history of a similar disease in her father. Her best corrected visual acuity on our examination was 20/200+2 OD and 20/200+1 OS. Ophthalmoscopy revealed a circular area of retinal pigment epithelial (RPE) atrophy 1 mm in diameter centered on fixation OU and yellow pisciform flecks throughout the posterior pole OU (Figure 9A). Optical coherence tomography revealed thinning of the outer nuclear layer and disruption of the ellipsoid zone in an area somewhat larger than the area of RPE atrophy (Figure 9B). Goldmann perimetry was normal except for small central scotomas to the I4e OU (Figure 9C). Sanger sequencing of the coding portions of the ABCA4 gene revealed a single heterozygous missense mutation (Leu2229Pro). Sequencing of non-exomic regions previously shown to harbor disease-causing mutations revealed a previously described 28 cryptic splice activator on the allele opposite the missense variation (IVS36+1216 C>A). The non-exomic mutation would not have been captured by any currently available commercial exome capture reagents.
Figure 9. A 10-year-old female with ABCA4-associated Stargardt disease. A.

Fundus photograph of the right eye. B: Optical coherence tomogram of the right eye. C: Goldmann visual field of the right eye.
Patient E is the 42-year-old father of patient D who first noticed difficulty with his central vision at age 6 (Supplemental Table 1, #804). The following year he was diagnosed with Stargardt disease and by age 13 his acuity had fallen to 20/400 OD and 20/240 OS. On this visit, his acuity was 20/800 OD and 20/250 OS. Ophthalmoscopy revealed an elliptical zone of RPE and choriocapillaris atrophy centered on fixation, very narrowed arterioles, and extensive bone-spicule like pigment in the midperiphery OU (Figure 10A). Optical coherence tomography (Figure 10B) revealed preservation of inner retinal lamination even in the area of macular atrophy. Goldman perimetry revealed complete loss of sensitivity to the I4e stimulus throughout the visual field and an absolute scotoma inferonasally OU (Figure 10C). This patient shared the IVS36+1216 C>A non-exomic mutation with his daughter and harbored an Arg2077Trp variant on his other allele. Schindler et al., (2010) found the Arg2077Trp variant to be the most severe Stargardt allele of the sixteen they evaluated 39. This is consistent with the more severe RP-like phenotype in this individual.
Figure 10. A 42-year-old male with ABCA4-associated Stargardt disease. A.
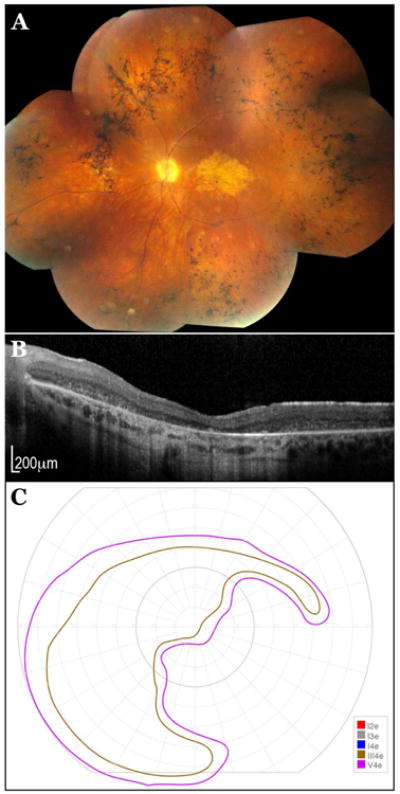
Fundus photograph of the right eye. B: Optical coherence tomogram of the right eye. C: Goldmann visual field of the right eye.
Discussion
Data that are used to arrive at a diagnosis are often incomplete, noisy and somewhat biased. Once a diagnosis is made, treatment outcomes are also dependent upon individual patient variation, the point in the disease course that a treatment is administered, and in some cases, the skill of a surgeon in delivering a treatment to the desired anatomic location. Most physicians effectively combat these challenges with systematic actions, good record keeping and periodic review of their outcomes in the context of new knowledge. The purpose of this study was to review the clinical and molecular findings from 1,000 consecutive families affected with inherited retinal disease – in the context of current technology, public databases and literature – to identify opportunities for improving our accuracy and efficiency in arriving at clinical and molecular diagnoses for patients with inherited retinal diseases. The consecutive nature of the ascertainment allows a rough approximation of the total numbers of individuals in the United States who are affected by various categories of disease (Table 3). These data may be useful as scientists try to devise and implement practical comprehensive strategies for reaching all such patients with some type of useful treatment.
The clinical classification system used in this study (Figure 1, Table 2) is an empiric, internally consistent shorthand that can be used to efficiently communicate clinical observations to the laboratory for the purpose of guiding their molecular investigations, analyses and interpretations, and to align the resulting genotype-phenotype correlations with the constantly changing medical literature. This system was devised by a single clinician over many years and should not be considered a consensus view of how these disorders can be most meaningfully arranged. It is expected and desirable that other physicians will add or subtract categories from this classification scheme as needed to encompass the patients they see in their practice, and to move the clinical entities around to better reflect the order in which they typically pursue a diagnostic workup and the specific diagnostic instruments routinely available to them. The power of this approach lies not in the details of the classification system but in the idea of using clinical information to narrow the pretest hypothesis for the purpose of increasing the sensitivity of the testing and dramatically increasing the statistical significance of the results. To reduce the FGR below 5%, which would be desirable when contemplating a significant intervention such as gene replacement therapy or the preimplantation selection of embryos for disease avoidance, one would need to reduce the pretest hypothesis in most cases to fewer than ten genes (e.g., category IA2b, Figure 1). More than 85% of the terminal categories in the current classification scheme have a FGR of 5% or less (blue bars, Figure 1). The remaining task for clinicians who care for patients with inherited retinal diseases is to carefully scrutinize the ones in the more genetically heterogeneous categories (grey bars, Figure 1) for subtle clinical signs that can be used to further subdivide them into entities associated with a smaller number of genes. Over time, some diagnostic categories and classification arrangements will prove more useful than others for this purpose and an optimal scheme for all inherited eye disease can evolve by combining the best features of many classifications based upon their performance in the pretest prediction of the patients' genotypes.
There are many different strategies that one can use to analyze a patient's DNA for the presence of disease-causing sequence variations and a complete discussion of them is well beyond the scope of this paper. For the present purpose, it is sufficient to think of the many possibilities in terms of four attributes: 1) the degree to which a test can be customized to detect specific variations that would otherwise be missed; 2) the degree to which the test yields a dataset that can be re-analyzed in the future to discover currently unrecognized pathogenic variations; 3) the degree to which multiple platforms are employed to maximize the strengths and minimize the weaknesses of each; and, 4) the degree to which the patients' true disease-causing genotypes will be obscured by normal, non-disease-causing genetic variation.
Next generation sequencing “panels” have now been designed for many diseases and have the advantages that they are relatively focused (compared to whole exome or whole genome tests), they can be customized to include specific non-exomic regions known to cause disease, and they are relatively quick and inexpensive to perform. The disadvantage of such panels is that when negative, they do not allow wider analytical exploration in search of disease-causing mutations outside the genomic space covered by the panel's design. These panels have difficulty in accurately detecting variants within repetitive DNA sequences and can have difficulty detecting deletions larger than 100 nucleotides and smaller than a few exons in size. Moreover, most of these panels evaluate a sufficient number of genes that the false genotype rate associated with them is greater than 5% unless the ordering physician controls this by making a firm and narrow pre-test diagnosis and rigorously evaluates the results in that context.
Whole exome sequencing has the advantage of sampling nearly all of the transcribed sequences in the human genome and can be subjected to very focused analysis to yield statistically meaningful results. If such a focused analysis is negative, the data can be reanalyzed to consider a larger portion of the exome and/or reanalyzed at a later date when new regions of the exome may have been discovered to cause a phenotype similar to the patient under study. The disadvantages of whole exome sequencing are that it is more expensive and time consuming to perform than a next generation sequencing panel and most commercial whole exome reagents are not easily customizable to analyze specific non-exomic regions of interest to specific subspecialties of medicine. Whole exome sequencing also has difficulty with repetitive DNA and can have even greater difficulty detecting single exon deletions than NGS-based panels 40,41. As shown in Figure 5, unless one establishes a narrow pretest hypothesis and evaluates the results accordingly, whole genome sequencing will frequently have a false genotype rate that is so high that the results should be considered hypothesis generating at best.
Whole genome sequencing evaluates nearly all of the non-repetitive sequences in the genome and, although it examines more than fifty times more sequence than whole exome sequencing, is surprisingly only about twice as expensive as the latter method. It is better at detecting deletions, duplications and inversions than whole exome sequencing 42 and can also detect disease-causing variations in non-exomic space 33,43-45. However, the amount of background genetic variation in the nonexomic space is so large, and our current understanding of the function of nonexomic sequences is currently so limited, that pathogenic single nucleotide variations will be completely hidden in the noise unless the pretest hypothesis is limited to only one or two genes and some functional test can be employed to validate the findings functionally 29,33,46. For example, the identification of a number of non-exomic mutations in ABCA4 28 required access to a large cohort of patients with convincing clinical characteristics of Stargardt disease and only a single disease-causing mutation, as well as a rather narrow mechanistic hypothesis, altered splicing, coupled with a convincing assay of this mechanism. Similarly, the discovery of the non-exomic mutations responsible for North Carolina Macular Dystrophy required decades of clinical and molecular genetic work to narrow the genetic interval to less than a million base pairs as well as sufficient families to identify three different mutations tightly clustered in a single regulatory element 33.
It is also important to remember that none of the commonly utilized high-throughput sequencing methods can unambiguously distinguish whether two different mutations observed in a patient were inherited from a single parent, which would not be expected to cause autosomal recessive disease, or whether they were inherited from both parents. The phase of two variants is most reliably established by testing a parent or child of the proband but in many cases can also be determined by testing siblings or more distant relatives. In multiplex families, confirming that all affected individuals actually harbor the genotype found in the proband also increases the likelihood that that genotype is truly disease-causing 18. By reporting such properly segregating genotypes in the literature or through a curated database (e.g., vvd.eng.uiowa.edu), one can strengthen the confidence in those mutations for other physicians caring for other families.
It should go without saying that there is no need to employ the same genotyping strategy for every patient. Some phenotypes are so characteristic that they yield a pre-test hypothesis that can be evaluated with a single conventional DNA sequencing reaction, which costs less than $20 in the research setting (see Methods). Other phenotypes are associated with a small number of genes that can still be analyzed more quickly and with less financial and statistical cost than an entire exome or genome sequence would incur (Figures 4 and 5).
In this study, we divided all of the inherited eye diseases seen by a single clinician over a 5.5 year period into 62 different categories and for all but 7 of these categories were able to devise very focused tests that cost less than an entire exome to perform. We reserved whole exome sequencing for the few clinical categories that were too broad for focused screening and for the cases that were negative after the initial test. We reserved whole genome sequencing for four families that had a phenotype that strongly implicated a single gene (e.g., Patient B, Figure 7) but had no mutations in the coding sequences of that gene. Although this tiered approach resulted in some patients having two or even three molecular evaluations, the focused tests were so inexpensive – less than half the cost of an exome on average – that the tiered strategy was overall less expensive than it would have been if we performed whole exome sequencing on every patient (Figure 4). The very customized nature of the prescreening tests also allowed very challenging portions of the genome to be successfully analyzed, such as the highly repetitive portion of exon 15 in RPGR that is uninterpretable with most next generation sequencing methods. As a result, the sensitivity of our current tiered approach is a 6.1% higher than an all whole exome sequencing strategy would be.
Although the tiered strategy is currently 17.7% less expensive overall in our hands than an all whole exome sequencing approach would be in the same laboratory using the same personnel, this modest overall cost savings is not the main reason that we would employ or recommend this approach. The main reasons are to keep the average FGR as low as possible and to detect important disease-causing mutations that would otherwise be missed (Patients A – C, Figures 6 – 8). The clinical pre-test decision making necessary to achieve the low FGR results in a very low test cost for a large fraction of the patients (Figure 4). This savings in reagent cost and laboratory bandwidth can then be used to pursue much more expensive investigations, such as cloning the repetitive region of RPGR exon 15, in the subset of patients that need it. This results in a higher overall sensitivity of the strategy at a lower cost. It is important to note that as the cost of whole exome sequencing and the associated analysis continues to fall, it will not supplant the value of specific pre-screening tests for many clinical categories until the whole exome sequencing cost falls below that of a single Sanger sequencing reaction. Thirteen of the 25 families with clinical BBS in this cohort had their mutations found in BBS1, and all 13 of these harbored at least one M390R allele. As a result, we would recommend performing a Sanger sequencing reaction in search of this mutation in all BBS patients before proceeding to whole exome sequencing until the total cost of the latter falls below fifty dollars.
It is interesting to consider what would happen to the data shown in Figures 4 and 5 if the research cost of whole exome and whole genome sequencing became one tenth what it is today (i.e., $120 and $245 per person, respectively). At these price points, the cost of the sequencing would be dwarfed by the cost of the sample handling, quality control measures, bioinformatic analysis, report writing and genetic counseling. As a result, our ratio of whole exome sequencing to whole genome sequencing would likely be the inverse of what it is today and we would also perform many fewer “prescreening tests”. Most of the latter would be performed to cover the low complexity parts of the genome that will continue to elude scrutiny by NGS methods. The overall sensitivity of the testing strategy would increase a few percent because whole genome sequencing is better at detecting copy number variations. However, the need for a narrow pre-test hypothesis would be identical to the need today because the average false genotype rate per base pair of investigated genome is an immutable fact of nature that is completely unaffected by the costs of the methods we employ or the speed with which we employ them.
One might expect that our next step in studying the cohort presented in this paper would be to perform whole genome sequencing in the 240 families that have yet to have their disease-causing mutations identified. However, it is important to note that these families harbor an average of 16.5 plausible disease-causing mutations among the 305 candidate genes we considered in this study (Figure 1). It seems most likely to us that the majority of the genotypes remaining to be discovered in this cohort lie at least in part among the coding sequence variations that we have already detected or the coding sequences of other genes and that further clinical investigation of these families is likely to be more fruitful than increasing the number of rare variants to consider by more than two orders of magnitude. The aggressive ascertainment of additional members of these 240 families will allow us to strengthen or rule out many of the plausible disease-causing variants we have already identified on the basis of their segregation within the families. Continued scrutiny of the positive families in this cohort may also reveal some characteristic clinical features that would favor a specific one-allele hypothesis sufficiently that whole genome sequencing would be indicated in that family. This “families first” strategy would not change even if the cost of whole genome sequencing fell ten-fold. As noted above, the reason for this is that the amount of normal genetic variation in the genome is extremely large and independent of sequencing cost. The most powerful resources for overcoming this noise are, and will continue to be, large and well-characterized patient resources 28,33.
The disadvantages of a tiered testing strategy are that it requires very accurate communication between the clinic and the laboratory to gain the benefits described in this study, and tiered tests take much longer to perform than fragment capture panels. Although there are few situations in which a 3 or 4 month difference in testing time is clinically significant for a patient with a slowly progressive retinal degeneration, it is unquestionable that many families are anxious to have the cause of their disease identified as quickly as possible.
The keys to keeping the FGR down and sensitivity high are to 1) make the best clinical diagnosis possible before ordering a genetic test and use this diagnosis to choose the simplest test that is likely to yield a finding for that diagnosis, 2) obtain samples from parents and siblings of simplex families and as many affected individuals as possible from multiplex pedigrees for use in evaluating the results in the proband, 3) know the cost breakpoint between multiple focused prescreens and whole exome sequencing and switch to whole exome sequencing before exceeding that breakpoint, 4) take advantage of the slow progression of most of these diseases by trying to have a result for the patient at their next visit instead of some arbitrarily short turnaround time that will artificially inflate the cost of the test.
Although the 76% sensitivity achieved in this study is a far cry from the zero percent of 1986, it is likely to get even higher as we continue to analyze the 240 probands of this cohort whose molecular pathophysiology has yet to be determined. Some of the probands in this cohort are likely to have had inflammatory insults to their retinas that mimic Mendelian disease and it is possible that a predisposition to such disease may be detectable in their DNA as our knowledge of the genetics of the immune system continues to expand. There will certainly be additional disease-causing genes identified in the future by subjecting cohorts like this one to more sophisticated analysis or by studying multiplex families who lack mutations in currently known genes. There are also likely to be additional examples of non-exomic 29,33,46 and mitochondrial disease discovered as well as convincing cases of multiple genes interacting with one another to cause disease 47,48.
One advantage that we have today over 1986 is the ability to perform many genetic tests recursively, in silico, using inexhaustible data that is stored on servers instead of exhaustible DNA stored in freezers. Another advantage is the ability to derive phenotypically accurate retinal cell cultures from accessible tissues like skin, and to use these cells to test hypotheses that are generated from the DNA analysis 29,40,49. However, the most valuable resources needed to make these new discoveries are unchanged from 1986: relatively large numbers of patients with exceptionally detailed clinical information and large numbers of affected and unaffected family members that can be used to evaluate the many hypotheses that arise when studying the probands. As a result, the astute clinician, who is a good observer and record keeper, and who is willing to do whatever is necessary to find the correct answer and an effective treatment for his or her patient, remains the most valuable component of the entire effort.
Supplementary Material
Supplemental Figure 1. Geographic distribution of 1,000 probands. Colors show the total number of cases who live within each hexagonal area. Individuals living outside the 48 contiguous United States are shown at the bottom left.
Supplemental Figure 2. Statistical costs. The false genotype rate (FGR) is the average number of complete genotypes one would expect to observe by chance in a healthy individual in a specified genomic space, based on data from 60,000 normal individuals 23. The probands in this study are shown ordered according to the FGR associated with their clinical category (see Figure 1). The red line indicates the FGR associated with genes observed to cause disease in this cohort. The blue line indicates the increased FGR associated with also considering genes from the published literature that were not observed to cause disease in this cohort. The dashed line indicates an FGR of 5% (i.e., the threshold at which one in 20 people would be expected to harbor a plausibly pathogenic, complete genotype by chance).
Supplemental Table 1, pdf version. Clinical and genotypic information for all 1,000 probands. The “ExAC” column indicates the number of alleles observed in ExAC. For some variants, the number of homozygotes is enclosed in parentheses. An asterisk (*) in the “No. Affected with Genotype” denotes a pseudodominant case. Abbreviations: N/A, not applicable; RPGR (CL), cloned open reading from in RPGR; AR, autosomal recessive; AR-1, autosomal recessive single allele finding; AD, autosomal dominant; XL, X-linked.
Supplemental Table 1, xlsx version. Clinical and genotypic information for all 1,000 probands. The “ExAC” column indicates the number of alleles observed in ExAC. For some variants, the number of homozygotes is enclosed in parentheses. An asterisk (*) in the “No. Affected with Genotype” denotes a pseudodominant case. Abbreviations: N/A, not applicable; RPGR (CL), cloned open reading from in RPGR; AR, autosomal recessive; AR-1, autosomal recessive single allele finding; AD, autosomal dominant; XL, X-linked.
Supplemental Table 2. Observed and published genes for each phenotype in the classification system. Each gene is shown along with the number of diagnostic categories in the classification tree in which the gene was observed and the inheritance pattern for the gene within the diagnostic category. A single asterisk (*) indicates genes known to be capable of causing severe progressive loss of cognition and/or neuromuscular control and/or significantly shortened life expectancy. For a subset of these genes, a plus (+) indicates that specific variants can cause non-syndromic disease.
Supplemental Table 3: Diagnostic category statistics. The diagnostic category is shown, along with all genes observed for that category, the number of probands who were diagnosed with a given gene at the level of the category, the number of probands solved at the level of the category, and the total number of probands assigned to that category.
Supplemental Table 4. Inherited eye disease history form.
Acknowledgments
National Eye Institute (RO1EY024588, RO1EY026008), Wynn Institute for Vision Research, Carver Non-Profit Genetic Testing Laboratory, Stephen A. Wynn Foundation, Mark J. Daily, M.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cavenee WK, Dryja TP, Phillips RA, et al. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature. 1983;305:779–784. doi: 10.1038/305779a0. [DOI] [PubMed] [Google Scholar]
- 2.Friend SH, Bernards R, Rogelj S, et al. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643–646. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- 3.Dryja TP, McGee TL, Reichel E, et al. A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature. 1990;343:364–366. doi: 10.1038/343364a0. [DOI] [PubMed] [Google Scholar]
- 4.Maguire AM, Simonelli F, Pierce EA, et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacLaren RE, Groppe M, Barnard AR, et al. Retinal gene therapy in patients with choroideremia: initial findings from a phase 1/2 clinical trial. Lancet. 2014;383:1129–1137. doi: 10.1016/S0140-6736(13)62117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Constable IJ, Blumenkranz MS, Schwartz SD, et al. Gene Therapy for Age-Related Macular Degeneration. Asia Pac J Ophthalmol (Phila) 2016;5:300–303. doi: 10.1097/APO.0000000000000222. [DOI] [PubMed] [Google Scholar]
- 7.Ran FA, Hsu PD, Lin CY, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jinek M, East A, Cheng A, et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiley LA, Burnight ER, Songstad AE, et al. Patient-specific induced pluripotent stem cells (iPSCs) for the study and treatment of retinal degenerative diseases. Prog Retin Eye Res. 2015;44:15–35. doi: 10.1016/j.preteyeres.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Wiley LA, Burnight ER, DeLuca AP, et al. cGMP production of patient-specific iPSCs and photoreceptor precursor cells to treat retinal degenerative blindness. Sci Rep. 2016;6:30742. doi: 10.1038/srep30742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 13.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 14.Cideciyan AV, Swider M, Aleman TS, et al. Reduced-illuminance autofluorescence imaging in ABCA4-associated retinal degenerations. J Opt Soc Am A Opt Image Sci Vis. 2007;24:1457–1467. doi: 10.1364/josaa.24.001457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCulloch DL, Marmor MF, Brigell MG, et al. ISCEV Standard for full-field clinical electroretinography (2015 update) Doc Ophthalmol. 2015;130:1–12. doi: 10.1007/s10633-014-9473-7. [DOI] [PubMed] [Google Scholar]
- 16.Stone EM. Leber congenital amaurosis - a model for efficient genetic testing of heterogeneous disorders: LXIV Edward Jackson Memorial Lecture. Am J Ophthalmol. 2007;144:791–811. doi: 10.1016/j.ajo.2007.08.022. [DOI] [PubMed] [Google Scholar]
- 17.Newton CR, Graham A, Heptinstall LE, et al. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS) Nucleic Acids Res. 1989;17:2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stone EM. Finding and interpreting genetic variations that are important to ophthalmologists. Trans Am Ophthalmol Soc. 2003;101:437–484. [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krumm N, Sudmant PH, Ko A, et al. Copy number variation detection and genotyping from exome sequence data. Genome Research. 2012;22:1525–1532. doi: 10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Schulz-Trieglaff O, Shaw R, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 23.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lott MT, Leipzig JN, Derbeneva O, et al. mtDNA Variation and Analysis Using Mitomap and Mitomaster. Curr Protoc Bioinformatics. 2013;44:1, 23, 1–26. doi: 10.1002/0471250953.bi0123s44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weleber RG, Michaelides M, Trzupek KM, et al. The Phenotype of Severe Early Childhood Onset Retinal Dystrophy (SECORD) from Mutation of RPE65and Differentiation from Leber Congenital Amaurosis. Invest Ophthalmol Vis Sci. 2011;52:292–302. doi: 10.1167/iovs.10-6106. [DOI] [PubMed] [Google Scholar]
- 26.Genead MA, Fishman GA, Stone EM, Allikmets R. The natural history of stargardt disease with specific sequence mutation in the ABCA4 gene. Invest Ophthalmol Vis Sci. 2009;50:5867–5871. doi: 10.1167/iovs.09-3611. [DOI] [PubMed] [Google Scholar]
- 27.Population by sex and selected age groups: 2000 and 2010. United States Census Bureau. Available at: https://www.census.gov/prod/cen2010/briefs/c2010br-03.pdf.
- 28.Braun TA, Mullins RF, Wagner AH, et al. Non-exomic and synonymous variants in ABCA4 are an important cause of Stargardt disease. Hum Mol Genet. 2013;22:5136–5145. doi: 10.1093/hmg/ddt367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tucker BA, Mullins RF, Streb LM, et al. Patient-specific iPSC-derived photoreceptor precursor cells as a means to investigate retinitis pigmentosa. Elife. 2013;2:e00824. doi: 10.7554/eLife.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan SY, Ali S, Naeem MA, et al. Splice-site mutations identified in PDE6A responsible for retinitis pigmentosa in consanguineous Pakistani families. Mol Vis. 2015;21:871–882. [PMC free article] [PubMed] [Google Scholar]
- 31.Bellingham J, Davidson AE, Aboshiha J, et al. Investigation of Aberrant Splicing Induced by AIPL1 Variations as a Cause of Leber Congenital Amaurosis. Invest Ophthalmol Vis Sci. 2015;56:7784–7793. doi: 10.1167/iovs.15-18092. [DOI] [PubMed] [Google Scholar]
- 32.den Hollander AI, Koenekoop RK, Yzer S, et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet. 2006;79:556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Small KW, DeLuca AP, Whitmore SS, et al. North Carolina Macular Dystrophy Is Caused by Dysregulation of the Retinal Transcription Factor PRDM13. Ophthalmology. 2016;123:9–18. doi: 10.1016/j.ophtha.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howell N, Bindoff LA, McCullough DA, et al. Leber hereditary optic neuropathy: identification of the same mitochondrial ND1 mutation in six pedigrees. Am J Hum Genet. 1991;49:939–950. [PMC free article] [PubMed] [Google Scholar]
- 35.van den Ouweland JM, Lemkes HH, Gerbitz KD, Maassen JA. Maternally inherited diabetes and deafness (MIDD): a distinct subtype of diabetes associated with a mitochondrial tRNA(Leu)(UUR) gene point mutation. Muscle Nerve Suppl. 1995;3:S124–30. doi: 10.1002/mus.880181425. [DOI] [PubMed] [Google Scholar]
- 36.Vervoort R, Lennon A, Bird AC, et al. Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat Genet. 2000;25:462–466. doi: 10.1038/78182. [DOI] [PubMed] [Google Scholar]
- 37.Ayyagari R, Demirci FY, Liu J, et al. X-linked recessive atrophic macular degeneration from RPGR mutation. Genomics. 2002;80:166–171. doi: 10.1006/geno.2002.6815. [DOI] [PubMed] [Google Scholar]
- 38.Yang Z, Peachey NS, Moshfeghi DM, et al. Mutations in the RPGR gene cause X-linked cone dystrophy. Hum Mol Genet. 2002;11:605–611. doi: 10.1093/hmg/11.5.605. [DOI] [PubMed] [Google Scholar]
- 39.Schindler EI, Nylen EL, Ko AC, et al. Deducing the pathogenic contribution of recessive ABCA4 alleles in an outbred population. Hum Mol Genet. 2010;19:3693–3701. doi: 10.1093/hmg/ddq284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tucker BA, Scheetz TE, Mullins RF, et al. Exome sequencing and analysis of induced pluripotent stem cells identify the cilia-related gene male germ cell-associated kinase (MAK) as a cause of retinitis pigmentosa. Proc Natl Acad Sci USA. 2011;108:E569–76. doi: 10.1073/pnas.1108918108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fromer M, Purcell SM. Using XHMM Software to Detect Copy Number Variation in Whole-Exome Sequencing Data. Curr Protoc Hum Genet. 2014;81:7, 23, 1–21. doi: 10.1002/0471142905.hg0723s81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki T, Tsurusaki Y, Nakashima M, et al. Precise detection of chromosomal translocation or inversion breakpoints by whole-genome sequencing. J Hum Genet. 2014;59:649–654. doi: 10.1038/jhg.2014.88. [DOI] [PubMed] [Google Scholar]
- 43.Wildschutte JH, Baron A, Diroff NM, Kidd JM. Discovery and characterization of Alu repeat sequences via precise local read assembly. Nucleic Acids Res. 2015;43:10292–10307. doi: 10.1093/nar/gkv1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merico D, Roifman M, Braunschweig U, et al. Compound heterozygous mutations in the noncoding RNU4ATAC cause Roifman Syndrome by disrupting minor intron splicing. Nat Commun. 2015;6:8718. doi: 10.1038/ncomms9718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Känsäkoski J, Jääskeläinen J, Jääskeläinen T, et al. Complete androgen insensitivity syndrome caused by a deep intronic pseudoexon-activating mutation in the androgen receptor gene. Sci Rep. 2016;6:32819. doi: 10.1038/srep32819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tucker BA, Cranston CM, Anfinson KA, et al. Using patient-specific induced pluripotent stem cells to interrogate the pathogenicity of a novel retinal pigment epithelium-specific 65 kDa cryptic splice site mutation and confirm eligibility for enrollment into a clinical gene augmentation trial. Transl Res. 2015;166:740–749.e1. doi: 10.1016/j.trsl.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dryja TP, Hahn LB, Kajiwara K, Berson EL. Dominant and digenic mutations in the peripherin/RDS and ROM1 genes in retinitis pigmentosa. Invest Ophthalmol Vis Sci. 1997;38:1972–1982. [PubMed] [Google Scholar]
- 48.Kajiwara K, Berson EL, Dryja TP. Digenic retinitis pigmentosa due to mutations at the unlinked peripherin/RDS and ROM1 loci. Science. 1994;264:1604–1608. doi: 10.1126/science.8202715. [DOI] [PubMed] [Google Scholar]
- 49.Tucker BA, Cranston C, Anfinson KR, et al. Using patient specific iPSCs to interrogate the pathogenicity of a novel RPE65 cryptic splice site mutation and confirm eligibility for enrollment into a clinical gene augmentation trial. Transl Res. 2015 Dec;166(6):740–749.e1. doi: 10.1016/j.trsl.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Geographic distribution of 1,000 probands. Colors show the total number of cases who live within each hexagonal area. Individuals living outside the 48 contiguous United States are shown at the bottom left.
Supplemental Figure 2. Statistical costs. The false genotype rate (FGR) is the average number of complete genotypes one would expect to observe by chance in a healthy individual in a specified genomic space, based on data from 60,000 normal individuals 23. The probands in this study are shown ordered according to the FGR associated with their clinical category (see Figure 1). The red line indicates the FGR associated with genes observed to cause disease in this cohort. The blue line indicates the increased FGR associated with also considering genes from the published literature that were not observed to cause disease in this cohort. The dashed line indicates an FGR of 5% (i.e., the threshold at which one in 20 people would be expected to harbor a plausibly pathogenic, complete genotype by chance).
Supplemental Table 1, pdf version. Clinical and genotypic information for all 1,000 probands. The “ExAC” column indicates the number of alleles observed in ExAC. For some variants, the number of homozygotes is enclosed in parentheses. An asterisk (*) in the “No. Affected with Genotype” denotes a pseudodominant case. Abbreviations: N/A, not applicable; RPGR (CL), cloned open reading from in RPGR; AR, autosomal recessive; AR-1, autosomal recessive single allele finding; AD, autosomal dominant; XL, X-linked.
Supplemental Table 1, xlsx version. Clinical and genotypic information for all 1,000 probands. The “ExAC” column indicates the number of alleles observed in ExAC. For some variants, the number of homozygotes is enclosed in parentheses. An asterisk (*) in the “No. Affected with Genotype” denotes a pseudodominant case. Abbreviations: N/A, not applicable; RPGR (CL), cloned open reading from in RPGR; AR, autosomal recessive; AR-1, autosomal recessive single allele finding; AD, autosomal dominant; XL, X-linked.
Supplemental Table 2. Observed and published genes for each phenotype in the classification system. Each gene is shown along with the number of diagnostic categories in the classification tree in which the gene was observed and the inheritance pattern for the gene within the diagnostic category. A single asterisk (*) indicates genes known to be capable of causing severe progressive loss of cognition and/or neuromuscular control and/or significantly shortened life expectancy. For a subset of these genes, a plus (+) indicates that specific variants can cause non-syndromic disease.
Supplemental Table 3: Diagnostic category statistics. The diagnostic category is shown, along with all genes observed for that category, the number of probands who were diagnosed with a given gene at the level of the category, the number of probands solved at the level of the category, and the total number of probands assigned to that category.
Supplemental Table 4. Inherited eye disease history form.


