Abstract
Malfunction of ubiquitin-proteasome system is tightly linked to tumor formation and tumor metastasis. Targeting the ubiquitin-pathway provides a new strategy for anti-cancer therapy. Despite the parts played by ubiquitin modifiers, removal of ubiquitin from the functional proteins by the deubiquitinating enzymes (DUBs) plays an important role in governing the multiple steps of the metastatic cascade, including local invasion, dissemination, and eventual colonization of the tumor to distant organs. Both deregulated ubiquitination and deubiquitination could lead to dysregulation of various critical events and pathways such as apoptosis and epithelial-mesenchymal transition (EMT). Recent TCGA study has further revealed the connection between mutations of DUBs and various types of tumors. In addition, emerging drug design targeting DUBs provides a new strategy for anti-cancer therapy. In this review, we will summarize the role of deubiquitination and highlight the recent discoveries of DUBs with regards to multiple metastatic events including anti-apoptosis pathway and EMT. We will further discuss the regulation of deubiquitination as a novel strategy for anti-cancer therapy.
Keywords: Deubiquitinases, Metastasis, Apoptosis, Anti-cancer treatment
1. Introduction
The ubiquitin modification is required in the regulation of both proteolytic and non-proteolytic events, targeting proteins for proteasomal or lysosomal degradation, protein interactions, protein activity, protein localization to signaling transduction (Swatek & Komander, 2016). Like the balance of phosphorylation events by kinases and phosphatases, ubiquitin modification could be simply divided into two parts: the covalent attachment of ubiquitin mediated by ubiquitination enzymes including E1 activating enzyme, E2 conjugating enzyme and E3 ligase and the reversible removal of ubiquitin catalyzed by deubiquitinating enzymes (DUBs) (Hershko & Ciechanover, 1998; Komander & Rape, 2012).
Recent basic and translational studies have revealed a connection between malfunction of the ubiquitin-proteasome system with both tumor formation and tumor metastasis. While tremendous efforts have been focused on the role of ubiquitin conjugation, especially by the ubiquitin-protein E3 ligase, in orchestrating tumor formation and tumor progression, the impact of deubiquitinase in tumorigenesis, particular in tumor metastasis, remains less well-understood. Results from the recent combinatorial studies comprising functional cancer genomics, tissue array of human cancer specimen and medical chemistry indicate the great potential of targeting a series of deubiquitinating enzymes as novel strategy to combat cancer. Thus, determining the detailed mechanism by which DUBs regulate cancer metastasis and apoptosis pathways will address the knowledge gap about DUBs in regulating tumor metastasis and apoptosis and further provide novel targets for anti-cancer therapy. In this review, we will focus on recent findings of DUBs and their regulatory roles in distinct events of cancer metastasis including EMT, anoikis resistance and microenvironment support, and conclude with a discussion of the potential of DUB inhibitors as therapeutics.
2. A brief introduction to DUBs
The reversal of ubiquitin conjugation of targeted proteins relies on deubiquitinating enzymes (DUBs), which catalytically cleave single Ub or poly-ubiquitin chains from proteins. The human genome encodes approximately 100 potential DUBs which can be classified into six families: ubiquitin-specific proteases (USPs), ubiquitin COOH-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), Josephins, the JAB1/MPN/MOV34 family (JAMMs) and motif interacting with Ub-containing novel DUB family (MINDYs) (Abdul Rehman et al., 2016). USPs, UCHs, OTUs, Josephins and the newly identified MINDYs families are thiol proteases, while the sixth family, JAMMs, comprises zinc metalloproteases (Ronau, Beckmann, & Hochstrasser, 2016).
The mechanism of protein degradation mediated by ubiquitin has been well studied; meanwhile, there are growing evidences that reveal the non-proteolytic roles of ubiquitin modification. Deubiquitination counteracts the ubiquitin cascade, including inhibiting E2 ubiquitin conjugating enzymes and E3 ligases. Proteasome-related DUBs help to prevent degradation of proteins conjugated with an ubiquitin chain. Lysosome-associated DUBs play crucial roles in receptor degradation and recycling. Alternatively, DUBs can remove or edit ubiquitin chains to alter the signals mediated by non-degradation ubiquitin. After severing ubiquitin chains from proteins, DUBs also generate free ubiquitin from ubiquitin precursors and release ubiquitin from unanchored isopeptide-linked ubiquitin chains into the ubiquitin pool (Komander, Clague, & Urbé, 2009).
Growing evidence indicates germline and somatic mutations, as well as alterations in expression frequency of DUBs, correlate with human disease, ranging from immune diseases to many human cancers. Mutations or deletions in DUBs could be involved in tumors. For example, the DUB A20 is a negative regulator of NF-κB pathway. Several studies have reported the deletions or mutations of TNFAIP3 (encoding gene of A20) in lymphomas such as the marginal zone lymphoma and the Non-Hodgkin’s Lymphoma, indicating A20 is a tumor suppressor and/or immune regulator (Honma et al., 2009; Nocturne et al., 2013). Mutated A20 results in truncated proteins which are defective in inhibiting NF-κB pathway, leading to an increased expression of NF-κB-mediated pro-inflammatory cytokines (Zhou et al., 2016).
Many DUBs are associated with tumors by their alterations in protein expression. For instance, increased expression levels of OTUD6B, UCH37, VCPIP1, USP7 and COPS5 are detected in various breast cancers (Luise et al., 2011). USP6 is considered to be an oncogenic protein and is overexpressed in primary aneurysmal bone cyst (ABC) and nodular fasciitis by chromosome translocation, and forms fusion proteins with CDH11, TRAP150, ZNF9, OMD, and COL1A1, which result in promoter swapping and transcriptional up-regulation (Oliveira et al., 2004). However, the roles of the DUBs are poles apart depending upon the different tumor types. In certain types of cancers, such as colon and thyroid cancers, the mRNA level of USP4 is elevated, whereas in lung cancer, the USP4 protein level is decreased (Gupta, Copeland, Gilbert, Jenkins, & Gray, 1993). USP4 has been suggested as an oncogene which could deubiquitinate and stabilize ARF-BP1, counteracting the function of E3 ligase of p53 (Zhang, Berger, Yang, & Lu, 2011). In lung cancer, USP4 was identified as a negative regulator of TNFα-mediated migration in lung cancer cells by deubiquitinating TRAF2 and TRAF6 (Xiao et al., 2012). In prostate carcinoma, USP2 protein is upregulated, whereas in colon cancer, USP2 expression is downregulated. In prostate cancer, USP2 is responsible to the stabilization of Fatty Acid Synthase (FAS), leading to the protection of tumor cell from apoptosis. While lower expression in colon cancer, how it works in colon cancer still remains unclear. (Graner et al., 2004; Priolo et al., 2006).
3. DUBs in regulation of critical steps in tumor metastasis
Tumor metastasis comprises a complicated series of biological processes in terms of the invasion-metastasis cascade that requires cancer cells to (1) migrate and invade from the origin sites through the extracellular matrix (ECM) to the stromal cell layers, (2) ingress into the blood vessels, otherwise known as intravasation, (3) survive during the transportation through the blood vessels, (4) egress out of the vessels termed extravasation into the distant organs, (5) survive and form micrometastasis, and (6) restart the proliferation to generate macrometastasis (Fig. 1 & Table 1) (Valastyan & Weinberg, 2011). In this review, we will discuss the emerging regulatory roles of DUBs in cellular signaling pathways critical for tumor metastasis such as EMT, self-renewal, apoptosis (anoikis) resistance, microenvironmental interactions, which are important processes contributing to the tumor metastasis (Fig. 2 & Table 1). Accordingly, small molecule inhibitors against DUBs have recently received a prominent focus in the field of anti-cancer therapies.
Fig. 1.
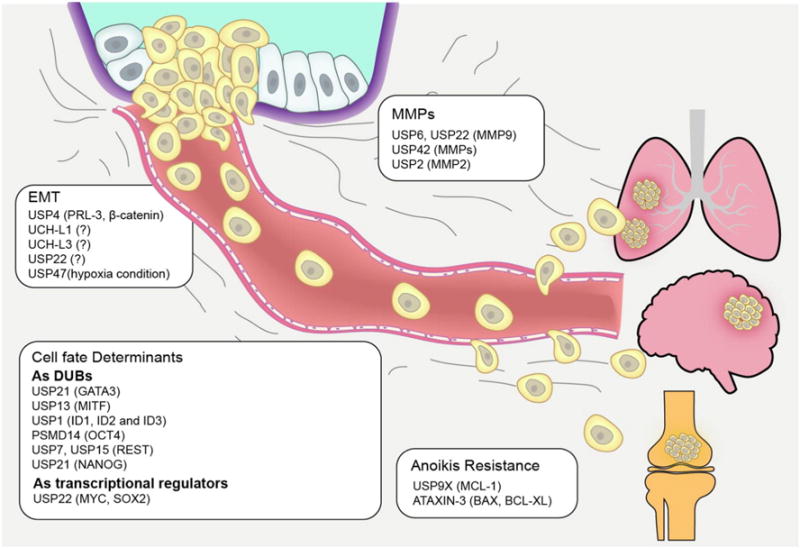
DUBs contribute to key events of metastatic cascade. DUBs play important role in various events during the metastatic progression, such as EMT, ECM degradation, cell fate regulation, as well as resistance of anoikis in the circulation. The critical role of DUBs in regulating EMT is summarized, including USP4, UCHL1, UCHL3, USP22 and USP47. The importance of DUBs participated in MMPs regulation is summarized, including USP6, USP22, USP2 and USP42. The involvement of DUBs in cell fates determinants is summarized, including USP21, USP13, USP1, PSMD14, USP7, USP15 and USP21. The important DUBs that have been reported in regulating anoikis regulation include USP9X and ATAXIN-3. DUBs with known targeting proteins or without clear substrates are all listed in the order of the events in which they are evolved.
Table 1.
List of deubiquitinase and their substrate and their E3 ligase counterparts.
| DUBs | Substrates | E3 ligase | Biological function |
|---|---|---|---|
| ATAXIN-3 | BAX, Bcl-xL | Parkin | Apoptosis |
| PSMD14 | Oct-4 | WWP2 | Cell fate determinants |
| USP1 | ID1, ID2, ID3 | CDH1, SMURF2 | Cell fate determinants |
| USP11 | XIAP | XIAP, Siah-1 | Apoptosis |
| USP13 | MITF | PIAS2 | Invasive growth |
| USP7, USP15 | REST | β-TrCP | Apoptosis |
| USP2 | MMP2 | CPL, SKP2 | Cell fate determinants |
| USP21 | GATA-3, Nanog | Fbw7, FBXW8 | Cell fate determinants |
| USP22 | Myc, Sox2 | Fbw7, SKP2, WWP2 | Cell fate determinants |
| USP30 | TOM20 | Parkin | Apoptosis |
| USP4 | PRL-3, β-catenin, Sox2 | FBXW8 | Cell fate determinants |
| USP42 | MMPs | CPL, SKP2 | Invasive growth |
| USP47 | β-catenin | β-TrCP | Invasive growth |
| USP6, USP22 | MMP9 | SKP2 | Invasive growth |
| USP8 | HIF1α, LRIG1 | MDM2 | Invasive growth |
| USP9X | Mcl-1 | Mule, β-TrCP, Fbw7, CDC20, TRIM17 | Apoptosis |
Fig. 2.
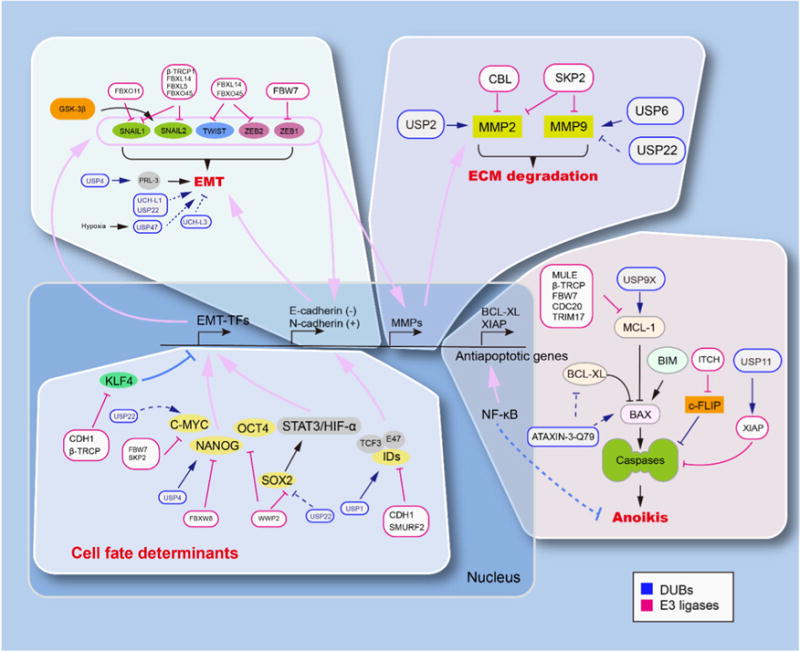
Ubiquitin modifications are involved in tumor metastasis. In the processes of tumor metastasis, the major regulatory proteins including core transcription factors are under tight regulation of ubiquitin modification, orchestrated by both E3 ligases and DUBs. The EMT related upstream regulatory factors (EMT-TFs) including KLF4, c-Myc, Nanog, Oct-4, Sox2, IDs are depicted in cell fate determinants panel. The EMT related regulators that control N-Cadhein/E-Cadherin expression including Snail-1, Snail-2, Twist, Zeb1, Zeb2 are depicted in EMT panel. The MMP2 and MMP9 related regulators are depicted in ECM degradation panel. The regulation of anti-apoptotic genes such as Bcl-xL, Bax, Bim, c-FLIP, XIAP are depicted in anoikis panel. The regulatory effect of E3 ligases and DUBs are denoted as activation symbols (arrow-headed lines) and inhibition symbols (bar-headed lines). The dotted lines indicate the effect is indirect or without clear mechanism.
3.1. DUBs affecting epithelial-mesenchymal transition (EMT)
During embryonic development, cells can transition between epithelial and mesenchymal states in a highly plastic and dynamic manner. A shift toward the mesenchymal state, in a process referred to as epithelial-mesenchymal transition (EMT), changes the adhesion molecules expressed by the cell, allowing it to adopt a migratory and invasive behavior. The reverse of this process, called mesenchymal-epithelial transition (MET), is associated with a loss of this migratory freedom, with cells adopting an apico-basal polarization and expressing the junctional complexes that are hallmarks of epithelial tissues (Nieto, Huang, Jackson, & Thiery, 2016).
The EMT is initiated in response to pleiotropic signaling factors that induce the expression of specific transcription factors (TFs) called EMT-TFs and miRNAs together with epigenetic and post-translational regulators, many of which are involved in embryonic development, wound healing, fibrosis, and cancer metastasis. The activities of the major EMT-TFs, such as SNAI1, SNAI2, ZEB1, ZEB2, and TWIST1, have been described and reviewed extensively. At the protein level, EMT is regulated by post-translational modifications including phosphorylation and the ubiquitin-mediated degradation (Hong et al., 2011; Zhou et al., 2004). For instance, F-box proteins target the major EMT-TFs to be degraded by the proteasome (Diaz & de Herreros, 2016). Other factors that maintain the epithelial phenotype include proteins such as the ubiquitin ligases FBXW7 and Siah, which prevent EMT by targeting Zeb1 for degradation (Chen et al., 2015; Yang et al., 2015).
Surprisingly, there are no examples of DUBs which were identified as regulators of major EMT-TFs. There still is the possibility that the abundance and stability of Snail, Twist and Zeb could be affected by one or several DUBs. Xing et al. reported that USP4 interacted with and catalytically stabilized PRL-3 (Xing et al., 2016). Stabilization of PRL-3 resulted in activation of Akt and reduction of E-cadherin. Besides, USP4 was also found to be highly expressed in brain metastatic lung adenocarcinoma PC14PE6/LvBr4 cell lines as a β-catenin-specific deubiquitinating enzyme, involved in the increased stability of β-catenin protein (Hwang et al., 2016). Several reports revealed that some DUBs were involved in EMT event but the identities of the substrates remain unclear. UCH-L1 promotes EMT but similarly, the target substrates have not yet been identified (Jang, Baek, & Kim, 2011). UCH-L3 was found to be downregulated in the highly metastatic prostate cell lines, and knockdown of UCH-L3 induced EMT process (Song, Lee, & Kim, 2014). In lung adenocarcinoma cells, USP22 was implicated to promote cell invasion by the induction of EMT (Hu et al., 2015). Choi et al. reported that USP47 was induced in human colon cancer cells (DLD-1, HT-29, and SW-480) under the hypoxic condition, and could be regarded as a novel regulator of EMT under hypoxic conditions (Choi & Surh, 2015).
Recently studies demonstrated the connection between DNA damage repair and EMT in cancer development and metastasis (Mani, Guo, Liao, et al., 2008; Zhang, Wei, Wang, et al., 2014). In the presence of genotoxic stress, phosphorylation of ZEB1 (an EMT regulator) by ATM facilitates its interaction with USP7 that results in its stabilization and promotion of EMT (Zhang et al., 2014). ZEB1 also enhances DNA damage response and increases tumor resistance to DNA damage. Thus, it is thought that ZEB1 could be a convergent regulator for both EMT and DDR (Zhang et al., 2014). In addition, recent studies indicated that downregulation of RAP80 leads to elevation of ZEB1 protein expression and activation of EMT signaling (Park, Korm, Chung, et al., 2016). In vivo, Rap80 could form two distinct complexes, including RAP80-BRCA1 containing complex as well as RAP80-Abraxas-RCC36-BRCA1 complex (Shao et al., 2009). Rap80 complex could recognize and disassemble K63-ubiquitin chain upon targeting to DNA double-strand breaks (Yan, Li, Wang, et al., 2015). The BRCC36 in the Rap80 complex acts as a component of multi-protein isopeptidase (BRCC36 isopeptidase complex or BRISC), which contains ABRO1/KIAA0157, BRCC36, MERIT40/NBA1, and BRCC45/BRE. BRISC complex functions as a deubiquitinase specifically targeting K63–linked ubiquitin hydrolysis (Yan et al., 2015). In BRISC complex, BRCC36 and ABRO1 are the two of the most important subunits, which orchestrate BRISC DUB activity and cytoplasmic localization, whereas the other two subunits render the integrity and stability of the complex (Yan et al., 2015).
3.2. DUBs and cell fate determinants
During tumorigenesis, the metastatic potential of tumors with different cellular origins may be shaped by the dominant lineage-specific cell fate regulators expressed in the originating cells. In addition, alteration or loss of differentiation control may result in dedifferentiation, acquisition of stem cell-like activities, and cellular plasticity that facilitate the development of metastatic traits (Ben-Porath et al., 2008; Reya, Morrison, Clarke, & Weissman, 2001). Accumulating evidence supports the notion that loss of differentiation factors leads to dedifferentiation and acquisition of stem cell-like traits that correlate with metastasis initiation properties (Cao, Cheung, & Nguyen, 2011). Mutation, epigenetic silencing, or reduced expression of luminal differentiation factors in the mammary gland (GATA3 and ELF5) has been shown to promote breast cancer metastasis (Chakrabarti et al., 2012; Kouros-Mehr et al., 2008). Similarly, the loss of MITF, a melanocyte differentiation factor, is sufficient to increase metastasis of melanoma (Cheli et al., 2012). On the other hand, opposing the function of lineage-specific differentiation factors, the increased activity of stem cell factors has been shown to promote metastasis. For example, inhibitor of differentiation/DNA binding (ID)-1 increases breast cancer lung metastasis (Gupta et al., 2007), and the mammary stem cell marker PROCR has been reported to be involved in self-renewal and metastasis (Wang et al., 2015). Interestingly, other factors that support tumor initiation activity seem to work only in the malignant context and are not involved in the regulation of normal adult tissue stem cells. For example, MTDH, an essential factor to support tumor initiation and metastasis in breast, prostate, and liver cancers, is dispensable for embryonic and postnatal development (Wan et al., 2014). Such factors are ideal candidates for therapeutic targeting to prevent metastasis initiation. Not only are tissue-specific cell fate determinants critical in metastasis initiation, embryonic cell fate regulators also play important roles. With the discovery of the Yamanaka factors—Sox2, Myc, Klf4, Oct3/4, and others—as potent reprogramming factors, these genes have also garnered much attention in cancer research. Each of these factors has been linked to increased aggressiveness and poor prognosis for tumors (Ben-Porath et al., 2008; Li et al., 2013; R, N., et al., 2014; Tai et al., 2011; Wang et al., 2009). Although Sox2, NANOG, OCT3/4, and KLF4 have been shown to increase metastasis of bladder cancer, breast cancer, lung cancer, and head and neck squamous carcinoma cells (Habu et al., 2015; Lu, Mazur, Lin, Appella, & Xu, 2014; Vaira et al., 2013), none of these factors has been specifically studied vis-à-vis metastasis initiation. Based on current knowledge, these factors may also facilitate metastatic initiation by promoting cell plasticity, adaptability, survival ability, and self-renewal as they do in primary tumors. Therefore, future research should be conducted to study these cell fate regulators during metastasis initiation.
GATA3 is a necessary transcription factor for the cell differentiation. Proteasomal degradation of GATA3 is mediated by the E3 ligase Fbw7 (Kitagawa et al., 2014). Zhang et al. have reported that in Treg cells, the DUB USP21 interacted with and stabilized GATA3 via deubiquitination with the overexpression of USP21 obviously decreasing the ubiquitination status of GATA3. In addition, the presence of FOXP3 together with GATA3 results in the inhibition of Th-2 cytokines expression. Expression of USP21 leads to stabilize GATA3. Thus, USP21, GATA3, and FOXP3 could form a positive loop in promoting FOXP3 expression that in turns modulates Treg cell activity (Zhang et al., 2013).
The microphthalmia-associated transcription factor (MITF) is fundamental for melanocyte development. Post-translational modifications of MITF such as phosphorylation and/or ubiquitination are mediated by mutation-induced MAPK pathway activation (Kim, Hwang, et al., 2003; Xu et al., 2000). The E3 ligase involved in MITF ubiquitination has yet to be identified. However, Zhao et al. reported the identification of a deubiquitinating enzyme USP13 which could stabilize and upregulate MITF protein through deubiquitination. By modulate the expression of MITF downstream genes, USP13 appeared to be essential for melanoma growth both in vivo and in vitro, indicating the possibility of targeting USP13 for melanoma therapy (Zhao, Fiske, Kawakami, Li, & Fisher, 2011).
ID proteins are transcription factors controlling differentiation and cell fate determination. The functions of IDs in tumor metastasis lie in the induction of EMT of primary tumor cells as well as the enhancement of tumor colonization in secondary site (Fong et al., 2003). E3 ligases APC/C-Cdh1 (targeting ID1, ID2, ID4) and SMURF2 (targeting ID1 and ID3) appear to be responsible for the regulation of IDs abundance and stability (Kong, Cui, & Zhang, 2011; Lasorella et al., 2006). USP1 was reported to deubiquitinate and stabilize ID1, ID2 and ID3, leading to the maintenance of the stem cell-like properties in osteosarcoma cells (Williams et al., 2011).
Studies of differentiation factors, stem cell factors, and embryonic cell regulators have revealed the impact of ubiquitin modification on those factors. The ubiquitination of OCT3/4, KLF4, and c-Myc has been extensively studied and recent studies of a few putative DUBs, specifically USP22 and Psmd14 have revealed roles for DUBs in regulating stem cell transcription factors and their influence on the efficiency of cellular reprogramming.
Ubiquitination of Oct3/4 is regulated by E3 ligase Wwp2 in mouse cells (WWP2 in human cells). Wwp2 mediates the degradation of Oct3/4 protein by attaching either Lys-48 or Lys-63 linked poly-ubiquitin chains (Liao & Jin, 2010; Xu, Wang, et al., 2009). Buckley et al. mapped the ubiquitinated protein landscape during ESC differentiation and induced pluripotency using a shotgun proteomics approach (Buckley et al., 2012). Additionally, using an ubiquitin-proteasome system-targeted RNAi screening method, they identified several regulators, including Psmd14 (Rpn11), involved in the protein degradation of core stem cell transcription factors. Depletion of Psmd14 leads to a significant decrease in Oct4 protein expression coupled with abnormal ESCs morphology. Psmd14 is a critical protein interacting with lid of the 19S proteasome that regulates the proteasomal lid on/off. Depletion of Psmd14 disrupted the activation of proteasomal lid therefore leading to accumulation of both K48- and K63-linked polyubiquitinated proteins.
Ubiquitination of Klf4 is an important posttranslational modification that is responsible for regulating its protein turnover in the cells. Recently, our lab has shown that Klf4 expression is downregulated in response to TGF-β-signaling, which is mediated by the ubiquitin-proteasome pathway (Hu, Zhou, Davidson, Huang, & Wan, 2012). Furthermore, Cdh1/APC, a putative E3 ubiquitin ligase, was found to interact with Klf4 and to regulate TGF-β-induced Klf4 proteolysis. Mutation of the two destruction boxes within Klf4 resulted in reduced ubiquitination and subsequent protein stabilization. Additionally, the phosphorylation of Klf4 by ERK1 recruits β-TrCP1 or β-TrCP2, both are F-box proteins with E3 ubiquitin ligase activity, to its N-terminal region, and it prepares Klf4 for protein degradation (Kim et al., 2012).
Ubiquitination and proteolysis of c-Myc are also important post-translational modifications regulating the stability and function of c-Myc. The interdependence between S-phase kinase associated protein 2 (Skp2) and c-Myc was been reported (Kim, Herbst, Tworkowski, Salghetti, & Tansey, 2003). Besides facilitating the turnover rate of c-Myc through ubiquitination, Skp2 acts as a cofactor to enhance the Myc-induced transcription of targeted gene promoters. In turn, Skp2-induced activation of the same promoters requires c-Myc (von der Lehr et al., 2003). Fbw7 is another E3 ligase of c-Myc. Unlike Skp2, Fbw7 inhibits c-Myc transcriptional activity in a dose-dependent manner after the phosphorylation of c-Myc catalyzed by GSK-3 (Welcker et al., 2004).
The RE1-silencing transcription factor (REST) is a critical stem cell transcription factor in neural differentiation, as well as identified as a tumor suppressor. USP7 was reported to deubiquitinate REST protein to counterbalance the SCF/β-TrCP-mediated ubiquitination, thereby maintaining quality and status of a stem cell (Huang et al., 2011). Another DUB, USP15 was found to stabilize only the newly synthesized REST, which is accumulated on mitotic exit (Faronato et al., 2013).
Recent studies have indicated that the phosphorylation of the Nanog protein by ERK1 decreases the stability of Nanog, through the binding and ubiquitination of the E3 ligase FBXW8 (Kim et al., 2014). USP21 was reported to deubiquitinate and stabilize Nanog as well as be recruited by Nanog to deubiquitinate H2A, thus facilitating Nanog’s target gene expression (Jin et al., 2016). In addition, the phosphorylation of Sox2 protein by AKT1 enhances the stabilization of Sox2 by antagonizing protein degradation and promotes the self-renewal capacity of mouse ESCs, while Set7 monomethylates Sox2 to induce the recruitment of E3 ligase WWP2 which mediate ubiquitination and degradation of Sox2 (Fang et al., 2014).
Several DUBs have been reported to play roles in transcriptional regulation of human embryonic stem cells based on a genome-scale location analysis. Instead of regulating the stability of targeted proteins, these DUBs are reported as transcriptional repressors of the locus encoding embryonic factors in ESCs and their repression is required for efficient differentiation. USP22, an enzymatic subunit of the hSAGA transcriptional cofactor complex, could be recruited by Myc to specific target gene loci and that USP22 is required for Myc-mediated transcription (Zhang et al., 2008). In addition, USP22 occupies the Sox2 promoter and hydrolyzes mono-ubiquitin from ubiquitinated H2B (uH2B), and blocks transcription of the Sox2 locus (Sussman et al., 2013).
3.3. DUBs and anoikis resistance
Cancer cells often encounter multiple apoptotic death signals in the new environment. Quite likely, cancer cells have already acquired anti-apoptotic mechanisms, such as elevated expression of caspase inhibitors and other anti-apoptotic genes, while they were at the primary tumor sites (Su, Yang, Xu, Chen, & Yu, 2015). Anoikis denotes the cell apoptosis after detaching from the appropriate extracellular matrix (Tan, Goldstein, Crowe, & Yang, 2013). Anoikis resistance enables metastatic cells to survive in the processes of intra-/extravasation and transportation of cells in the vessels. In human cancers, the upregulation of Mcl-1, XIAP, Cav-1, Bcl-xL, cFLIP and 14-3-3ζ and downregulation of Bit1 confer anoikis resistance in cancer cells (Daisy & Saipriya, 2012).
Ubiquitin modification of anoikis regulators has proven to be essential for anoikis resistance during tumor metastasis. Rapid proteasomal degradation of Mcl-1 ensures the triggering of cell death in response to various cellular events (Adams & Cooper, 2007). Mcl-1 is a Bcl-2 family member which is anti-apoptotic and necessary for the survival of human cancer cells. Numbers of studies elucidated different E3 ligases of Mcl-1 such as Mule, SCF/β-TrCP, SCF/Fbw7, APC/C-CDC20 and Trim17 in various cellular contexts, indicating the complexity and importance of Mcl-1 regulation (Mojsa, Lassot, & Desagher, 2014). Deubiquitinase USP9X conversely mediates and opposes Mcl-1 ubiquitination (Schwickart et al., 2010).
Detachment of epithelial cells from the extracellular matrix triggers both pro- and antiapoptotic signals. The non-malignant intestinal epithelial cells enhance the upregulation of inhibitors of apoptosis protein (IAP) family, such as XIAP and cIAP2 (Liu et al., 2006). Increased expression of apoptosis inhibitor protein XIAP and cIAP2 favors anoikis resistance of tumor cells (Berezovskaya et al., 2005; Esposito et al., 2007). Recently, USP11 has been uncovered as a novel deubiquitinase for both XIAP and cIAP2 that promotes cell escape of anoikis (Zhou et al., 2017). Patients with USP11 overexpression shows short distant metastasis-free survival (DMSF) time and disease-free survival (DFS) following neoadjuvant therapy in women with breast cancer (Bayraktar et al., 2013; Zhou et al., 2017).
Mutant of spinocerebellar ataxia type 3 (SCA3) gene encodes a mutant DUB Ataxin-3-Q79 which is associated with induction of apoptotic signals in neuronal cells. Ataxin-3-Q79 was shown to be responsible for the activation of the mitochondrial apoptotic pathway by up-regulating Bax and down-regulating Bcl-xL mRNA expression (Chou et al., 2006).
3.4. DUBs and MMPs
Stromal cells play vital roles in the early steps of metastatic processes vis-à-vis local invasion, intravasation, and extravasation. Matrix metalloproteinases (MMPs) mediate the proteolysis which facilitates the loss of basement membrane (BM) barrier. In tumor cells, the carefully controlled post-translational modifications of MMPs could be derailed in numerous ways. MMP-expressing cells not only facilitate the degradation of the BM and other ECM, but also liberate growth factors to foster cancer cell proliferation (Kessenbrock, Plaks, & Werb, 2010). In human cancer, high expression of MMP-2 and MMP-9 in metastatic cancer cells have been observed.
c-Cbl is a multifunctional adaptor and an E3 ubiquitin ligase related to the cytoskeleton regulation; thus, it is essential for cell adhesion and migration. In glioma SNB19 cells, RNAi-mediated depletion of c-Cbl decreases both cell invasion and expression of MMP2 (Lee & Tsygankov, 2010). In lung cancer cells, it was established that overexpression of E3 ligase Skp2 enhances migration and invasion of A549 lung cancer cells via increasing MMP-2 and MMP-9 expression (Yamada et al., 2016). USP6/TRE17 was shown to induce cell migration in the TRE17-RhoA/ROCK or IKK-NF-κB-MMP-9 signaling pathway. The induction of MMP expression is dependent on the USP activity of USP6 (Ye et al., 2010). However, further study is required to identify the targeting protein of USP6 in this signaling pathway. In gastric cancer cells, USP42 is regarded as an invasion promotor since the evidence showed that silencing of USP42 upregulated the expression of the E-cadherin, and downregulated the expression of MMPs, β-catenin, Twist, and Snail1 (Hou et al., 2016). In breast cancer, overexpression of USP2 in TNBC cell lines MDA-MB-468 and MDA-MB-231 enhances migration and invasion with the elevated MMP2 protein level (Qu et al., 2015). Several studies also indicate that USP2 expression is associated with poor survival of breast cancer patients and may serve as a prognostic biomarker and therapeutic target for TNBC. In colon cancer, USP22 was reported to attenuate the invasion capacity of colon cancer cells by inhibiting the STAT3/MMP9 signaling pathway (Ao, Liu, Bian, Feng, & Liu, 2015).
4. DUBs in regulation of apoptosis pathways in cancers
The intrinsic connection between apoptosis is limited to the tumor initiation but tumor metastasis (Mehlen & Puisieux, 2006). Results from the previous works demonstrated several avenues for apoptosis in regulating cancer metastasis, including (1) blockade of metastatic dissemination by killing misplaced cells (Su et al., 2015); (2) death escape from the immune suppression enhances the survival of metastatic cells (Levy, Roberti, & Mordoh, 2011); (3) survival from the environment of ROS produced by endothelial cells when crossing the vessel or tissue barrier during the extravasation step (Eccles & Welch, 2007); and (4) survival from the hypoxic conditions and insufficiency of cytokines to achieve successful colonization at their destination sites (Azab, De La Puente, Vij, & Azab, 2013).
Apoptosis is a cellular self-destruction program in response to multiple cellular stresses to maintain cellular homeostasis. Two routes of apoptosis, the extrinsic and intrinsic, both involve a complex network of signaling transduction leading to the activation of caspases, which could be simply divided to two subtypes: the ‘initiator’ caspases (caspase-2, -8, -9, or -10) and ‘executioner’ caspases (e.g., caspase-3 or -7). Activation of initiator caspases upon apoptotic stimuli followed by cleavage of executioner caspases are the key events of apoptosis (Fernald & Kurokawa, 2013).
In extrinsic pathway, in response to the engagement of extracellular ligands by cell surface receptors, a death-inducing signal complex (DISC) is formed to activate the initiator caspases-8 and -10. The DISC is a complex structure containing different components (such as c-FLIP, FADD, and RIP) that together determine whether apoptosis becomes activated in response to different stimuli in different cell types. The efficiency of apoptosis signal transduction is dependent on the DISC formation since the activation of initiator caspases relies heavily on proximity for autoactivation (Yang, 2015). The expression of death signal sensors can be regulated independently from the protein synthesis and degradation in an ubiquitin-dependent manner. The conjugated ubiquitin can serve as an address and transport death signaling sensors to their designated cellular compartments (Menges, Altomare, & Testa, 2009).
The intrinsic pathway, on the other hand, is initiated by the activation of BH3-only proteins which are usually transcriptionally activated by apoptosis stimuli. But there is one exception: BID is activated (termed tBID) by caspase cleavage (by caspases-2 or -8/10) (Czabotar, Lessene, Strasser, & Adams, 2014). The activated multi-BH domain proteins induced by activation of BH3-only proteins then form oligomers to induce the permeabilization of mitochondrial outer membrane (MOMP) which allows the characterized release of cytochrome c and second mitochondria-derived activator of caspases (SMAC) into cytoplasm (Green & Llambi, 2015). Cytochrome c then binds to the adaptor protein apoptotic protease-activating factor 1 (APAF-1) to form a multi-protein complex termed apoptosome, within which the caspase-9 initiates caspase-3 and caspase-7 leading to apoptosis. SMAC, as an inhibitory protein for inhibitor of apoptosis protein (IAPs), especially XIAP, enhances the activity of caspase cascade activated by cytochrome c (Hamacher-Brady & Brady, 2015; Hamacher-Brady, Choe, Krijnse-Locker, & Brady, 2014).
In normal cells, the apoptotic stimuli lower the expression level of antiapoptotic proteins by increasing turnover rate and preventing the constant degradation of proapoptotic proteins. Thus, the dysregulation of anti-/pro-apoptotic proteins will confer the survival of cancer cells via enhancing the degradation of proapoptotic proteins or stabilizing the antiapoptotic proteins. Recent studies on the molecular and cellular functions of different linkage types of ubiquitin chains have revealed that not only proteasome-dependent protein degradation, which is the classical function of ubiquitination, but also signaling roles are played by ubiquitin chains, especially in the regulation of apoptosis signaling cascade (Fig. 3).
Fig. 3.
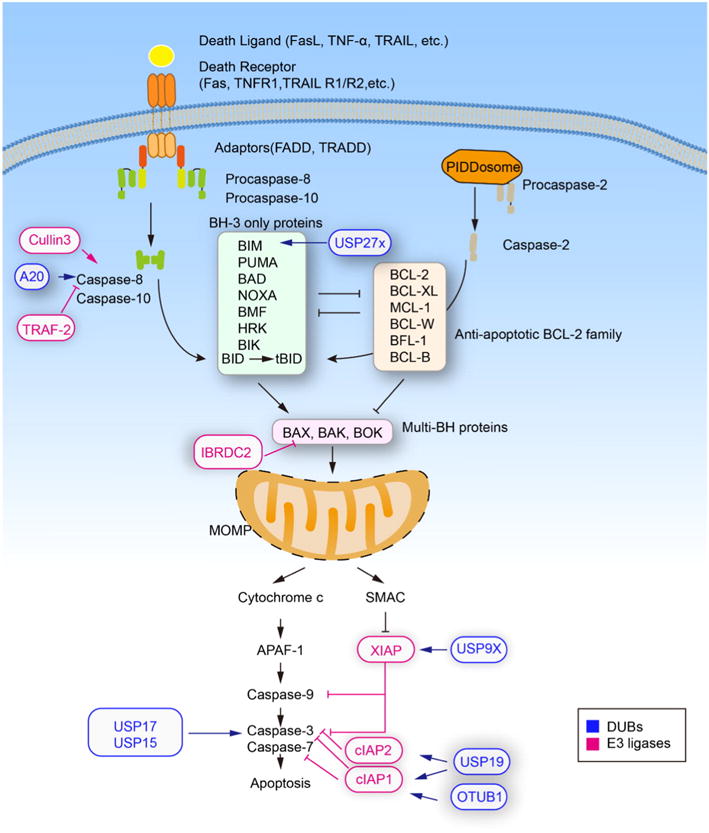
E3 ligases and DUBs that are known to regulate apoptosis pathways. Ubiquitin modifications contribute to both intrinsic and extrinsic apoptosis pathways. In some cases, the polyubiquitination of targeted protein mediated by E3 ligases will result in proteasomal degradation, while some ubiquitination will upregulate the protein activity. The E3 ligase CUL3/RBX1 allows the interaction of caspase-8 with p62, while polyubiquitinates caspase-8 for proteasomal degradation. The DUBs regulated apoptotic components including A20/Caspase8, USP27x/Bim, USP9X/XIAP, USP15/USP17/Caspase-3, USP19/cIAP1/cIAP2, and OTUB1/cIAP1. The regulatory effect of E3 ligases and DUBs are denoted as activation symbols (arrow-headed lines) and inhibition symbols (bar-headed lines). Colored lines denote that the effects are mediated by E3 ligases (blue) and DUBs (pink). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4.1. Caspase-9
E3 ligase XIAP is reported to ubiquitinate the active form of caspase-9, but not the procaspase-9 (Eckelman, Salvesen, & Scott, 2006; Morizane, Honda, Fukami, & Yasuda, 2005). However, further studies are required to elucidate the regulatory mechanism of ubiquitination of caspase-9, both in the normal and cancer context.
4.2. Caspase-8
Polyubiquitination of proteins also alter the activity of targeted proteins. After the recruitment to the DISC, ubiquitination of caspase-8 by a cullin3-based E3 ligase allows caspase-8 to bind to the poly-Ub binding protein p62 (Jin et al., 2009). The TNF receptor-associated factor (TRAF)-2 E3 ligase can also interact with caspase-8 at the DISC and mediates Lys 48-linked polyubiquitination of the large catalytic domain of activated caspase-8. This action prepares the initiator caspase for 26S proteasomal degradation; thereby TRAF2 sets a threshold for apoptosis commitment (Gonzalvez et al., 2012). A20, serving as a DUB, could interact directly with caspase-8 to reverse the Cullin3 mediated ubiquitination and inhibit the caspase activity upon TRAIL-ligand signaling (Jin et al., 2009). Independently, A20 could mediate Lys 63-linked polyubiquitination of receptor-interacting protein kinase 1 (RIPK1/RIP1) as an E3 ligase at the plasma membrane prior to TRAIL ligation (Bellail, Olson, Yang, Chen, & Hao, 2012).
4.3. Caspase-3
DUB3/USP17 was reported to induce apoptosis through caspase-3 activation (Burrows et al., 2004). cIAP2 monoubiquitinates caspase-3 and caspase-7 in vitro but additional studies are required to reveal the underlying mechanism and physiological relevance (Huang et al., 2000). More recent work has pointed out the effect of cIAP1 as an E3 ligase of caspase-3 in the enhancement of proteolysis of caspase-3 and the resistance to TRAIL-induced apoptosis (Choi et al., 2009). XIAP was also reported to polyubiquitinate active form of caspase-3, leading to its proteasomal degradation (Suzuki, Nakabayashi, & Takahashi, 2001). In response paclitaxel treatment, USP15 was demonstrated to regulate the stability of procaspase-3 and the SCF complex to activate caspase-3 and trigger apoptosis (Xu, Takanashi, et al., 2009).
4.4. Caspase-7
XIAP could directly bind and inhibit caspase 7 in a non-degradation manner (Nagano, Hashimoto, Nakashima, Kikkawa, & Kamada, 2012). cIAP1 was reported to bind to and mediate proteasomal degradation of caspase-7, thereby suppressing apoptotic cell death (Choi et al., 2009).
4.5. IAPs
Inhibitor of apoptosis proteins (IAPs) are a class of conserved proteins characterized by the presence of the Baculovirus IAP repeat domain (BIR). Besides BIR, the really interesting new gene (RING) domain, which provides an E3 ligase property, also present in most of the family members (Crook, Clem, & Miller, 1993). IAPs inhibit apoptosis through different mechanisms. In the case of caspase-3 and caspase-7, XIAP binds to the active sites; as for caspase-9, XIAP prevents the dimerization of the apoptosome. As mentioned, Smac/DIABLO and Omi/HtrA2 antagonize the inhibitory function of XIAP by direct binding. Apart from its stoichiometric inhibitory activity, XIAP has been recently shown to be involved in a positive regulatory feedback loop under different death stimuli. Upon the combination of TRAIL and UVB treatment, the release of SMAC distracts the inhibitory function of XIAP, leading to the activation of caspase-3, which in turns cleaves XIAP for proteasomal degradation, further enhancing apoptosis (Hörnle et al., 2011). cIAP1 was also reported to inhibit apoptosis by targeting the downstream of cytochrome c, via the interaction with activated caspase-9 and thus disrupting caspase-9 and the downstream activation of procaspase-3 (Burke, Smith, & Smith, 2010). These mitochondrial IAP antagonists are in turn ubiquitinated in the cytosol of apoptosis-stimulated cells by AREL1 E3 ligase to inhibit apoptosis (Kim et al., 2013).
USP9X was reported as the mitotic deubiquitinase of XIAP, with its stabilization leading to increased resistance toward mitotic spindle poisons. The USP9X–XIAP axis could be regarded as a regulator of the mitotic cell fate decision, and USP9X and XIAP are potential prognostic biomarkers and therapeutic targets for aggressive B-cell lymphoma (Engel et al., 2016).
USP19 was also observed to interact with and stabilize cellular IAP 1 (cIAP1) and cIAP2. Although the catalytic activity of USP19 on c-IAPs was observed in vitro, the stabilization of c-IAPs by USP19 in vivo are mainly seen through deubiquitinase-independent mechanisms in response to TNFα-induced caspase activation and apoptosis, implicating DUBs in the regulation of IAP stability (Mei, Hahn, Hu, & Yang, 2011).
4.6. BCL-2 family
Bim is a pro-apoptotic BH3-only protein of which the expression level determines apoptosis susceptibility. USP27x, acting as a tumor suppressor, could bind to Bim upon its anti-apoptotic ERK-dependent phosphorylation and counteract the following proteasomal degradation via its deubiquitinase activity (Weber et al., 2016). In addition, in NSCLC cells, depletion of USP27x reduces apoptosis when treated with an EGFR inhibitor.
Degradation of BAX is regarded as the “last-minute” safeguard for cancer cells to prevent apoptosis since the activation of BAX is directly linked to MOMP (Wolter et al., 1997). The E3 ligase IBR domain-containing 2 (IBRDC2) was shown to ubiquitinate BAX specifically on the mitochondrial outer membrane following apoptotic stimuli (Benard et al., 2010).
4.7. TNF superfamily
Members of the TNF superfamily are capable of activating both intrinsic (TRAIL-Rs) and extrinsic (TNFR1 and CD95) apoptosis pathways. Ubiquitin modification tightly regulates the TNFR signaling pathways to induce both cell survival and death (Gaur & Aggarwal, 2003). Upon TNF stimulation, TNFR1 recruits and binds to TRADD through the intracellular domain. TRADD in turns recruits a group of proteins including TRAFs, cIAPs, RIPK1, HOIP, HOIL-1L and Sharpin to form complex I which induces the NF-κB signaling. Ubiquitination of RIPK1 is critical for the dissociation of the complex I from TNFR1 providing two types of complex II (Peltzer, Darding, & Walczak, 2016). In the complex IIa FADD and procaspase-8 are assembled to induce apoptosis. Complex IIb/n is formed when complex I is impaired and induces necroptosis by activating RIPK1 and RIPK3 (Dondelinger et al., 2013). RIPK1 could activate NF-κB only when it is in an ubiquitinated form. CYLD and A20 are the major DUBs in the ubiquitin network of TNFR-mediated signaling pathway. A20 regulates the ubiquitination status of the adapter protein in complex I, which opposes the activity of E3 ligases ITCH and RNF11 in the ubiquitination of RIPK1 (Shembade, Parvatiyar, Harhaj, & Harhaj, 2009). CYLD is demonstrated to counteract the E3 activity of TRAF2 (Reiley, Zhang, Wu, Granger, & Sun, 2005). Furthermore, CYLD and another DUB OTULIN have been shown to interact with the LUBAC component to counteract the linear ubiquitination in the TNFR1 signaling pathway (Draber et al., 2015). Other DUBs such as OTUB1 and USP4 are also reported in the regulation network. OTUB1, as described, is a DUB of cIAP1 (Goncharov et al., 2013); USP4 negatively regulates RIPK1-mediated NF-κB signaling by its catalytic activity (Hou, Wang, Zhang, Pan, & Zhao, 2013).
In summary, the balance of ubiquitin modification orchestrated by E3 ligases and DUBs is essential for the apoptosis activation in response to various death stimuli, through regulating protein stabilities and functions.
5. DUBs targeting therapeutics
Specific mechanisms of deubiquitinating enzymes in various diseases have been described. Research should be concentrated on discovering an inhibitor on DUB’s enzyme activity or antagonist which binds the substrates for therapy of cancer and other diseases (Table 2). While growing numbers of drugs have been developed to inhibit or antagonize DUBs, none of these have yet entered clinical trials.
Table 2.
List of inhibitors of deubiquitinase and their biological function.
| DUBs | Inhibitor | Biological function |
|---|---|---|
| USP7 | P022077, P5091,Cpd 14,P22077, HBX 41,108, HBX-19,818 and HBX-28,258 | Cell fate determinants |
| USP8 | HBX 90,397 and Ethyloxyimino-9H-indeno [1,2-b]pyrazine-2,3-dicarbonitrile | Invasive growth |
| USP9X, USP5, USP14, and UCH37 | WP1130 | Apoptosis |
| USP14, UCH37 | b-AP15 and VLX1570 | Tumor cell invasion |
| USP1 | Pimozide | Cell fate determinants |
| UCHL1 | LDN91946, LDN-57444, Δ12-PGJ2 and 15Δ-PGJ2 | EMT |
| UCHL3 | LS1, Δ12-PGJ2 and 15Δ-PGJ2 | EMT |
| USP30 | 15-oxospiramilactone | Apoptosis |
5.1. DUB inhibition by compounds containing Michael acceptors
Compounds containing Michael acceptors such as α, β-unsaturated ketones have the inhibitory effect on some of cysteine DUBs due to the fact that they can potentially form covalent adducts with free thiols in the active site (Santos & Moreira, 2007). Cyclopentenone prostaglandins (PGs) of the PGJ2 class, chalcone compounds and other compounds containing Michael acceptors will be discussed here.
PGJ2 compounds Δ12-PGJ2 and 15Δ-PGJ2 are identified as inhibitors of UCHL3 and UCHL1, respectively (Lee et al., 2010). Chalcone compounds G5 have broad inhibitory spectrum, whereas another chalcone compounds b-AP15 and its analogue VLX1570 are relatively specific to USP14 and UCH37 (Aleo, Henderson, Fontanini, Solazzo, & Brancolini, 2006; Tian et al., 2014). USP14 and UCH37 are also inhibited by curcumin analogue AC17 (Zhou et al., 2013). UCHL1, UCHL3, USP2 and USP8 were found to be inhibited by AM146, RA-9, and RA-14, which did not inhibit Ataxin-3, A20, BAP1, Otubain 1 or USP7 (Issaenko & Amerik, 2012). WP1130 acts as a partially selective DUB inhibitor for USP9X, USP5, USP14, and UCH37, resulting in downregulation of antiapoptotic and upregulation of proapoptotic proteins, such as Mcl-1 and p53 (Kapuria et al., 2010). Eeyarestatin-1 (Eer1) was identified to inhibit p97/VCP-associated DUB activity such as that of Ataxin-3 (Wang, Li, & Ye, 2008).
5.2. Other small molecule DUB inhibitors
Due to the multifaceted roles of USP7, many inhibitors have been developed targeting USP7, such as P022077, HBX 41,108, HBX-19,818, HBX-28,258, P5091, Cpd 14 and P22077, in which the latter two molecules also inhibit USP47 (Chauhan et al., 2012; Colland et al., 2009; Reverdy et al., 2012; Tian et al., 2011; Weinstock et al., 2012). A small molecule IU1 has been described as specific inhibitor of USP14, only binding the activated USP14 (Lee et al., 2010). LDN-57444 is an isatin O-acyl oxime reported to selectively inhibit UCHL1 in a reversible, competitive, and active site-directed manner (Liu et al., 2003). Compared to LDN-57444, LDN91946, 3-Amino-2-keto-7H-thieno [2, 3-b] pyridin-6-one derivative, was discovered as moderately potent, non-competitive inhibitors of UCHL1 (Mermerian, Case, Stein, & Cuny, 2007). Clinical drugs for treating other diseases previously were found as DUB inhibitors. Pimozide (an anti-psychotic drug) was identified as inhibitors of USP1, and auranofin (a rheumatoid arthritis drug) is a proteasome-associated DUB inhibitor (Chen et al., 2011; Liu et al., 2014). Benefiting from high-throughput screening studies, LS1 has been identified as a UCHL3 inhibitor and PR-619 has been characterized as a general DUB enzyme inhibitor (Edelmann, Nicholson, & Kessler, 2011; Ohayon, Spasser, Aharoni, & Brik, 2012). Interestingly, the mitochondria-localized DUB USP30 was found to be inhibited by a diterpenoid derivative 15-oxospiramilactone (S3), leading to the increased Mfn1/2 proteins which promote mitochondrial fusion (Yue et al., 2014).
6. Conclusion
Blockade of tumor metastatic events and induction of apoptosis for tumor cells have been a strategic concept for anti-cancer therapy for a long time. Given the pivotal role of ubiquitin-proteasome system in regulating various steps of tumor metastasis as well as tumor cell apoptosis, targeting the ubiquitin-pathway could provide new opportunities to clinically manage tumor metastasis and cancer cell for apoptosis. While tremendous efforts have been expended on the steps of conjugating the ubiquitin-chain to the substrate proteins such as identifying ubiquitin E3 ligases, and ubiquitin conjugating enzymes (E2s), the study of the importance of deubiquitinases in regulating the ubiquitination of functional proteins by the removal of ubiquitin-chain from the targeting proteins, which orchestrates a variety of cellular processes and pathophysiology of diseases lags behind. Although recent mechanistic study of deubiquitinase sheds a light on the value of deubiquitinases to be potential targets for cancer therapy, only a small fraction of deubiquitinases of the greater deubiquitinase family (about 100 members) have been well studied in regards to their importance in tumorigenesis and metastasis.
Several issues remain obstacles for the translational application target deubuiquitinase, including (1) how the substrate specificity is established for the limited 100 or so DUBs to target thousands of proteins; (2) despite the simple view of removal of an ubiquitin chain from the substrate, DUBs have been demonstrated to modulate/coordinate the activity of E2 and E3 ligases although a systematic study for the 100 or so DUBs remains to be examined; (3) in-depth view of how deubiquitinases recognize substrate and catalyze the ubiquitin-chain removal in terms of 3D structural study is still missing; (4) as more missense mutations of DUBs are identified by TCGA studies, the physiological relevance of individual DUBs and important functional motifs within the proteins will need to be validated by animal model; and (5) while a few DUB small molecule inhibitors have shown their efficacy in anti-cancer therapy, more efforts are needed in drug development. Understanding of the above questions will not only enhance the molecular basis of ubiquitin-proteasome system in regulating tumor metastasis and tumor apoptosis, but also provide new strategies for next era anti-cancer therapy.
Acknowledgments
We thank all members from Wan laboratory for critical reading and discussion of our manuscript. We apologize to colleagues in the field whose work was not included due to space limitation. This work is supported by NIH grant CA154695, CA202948 and CA202963.
Abbreviations
- PGs
Cyclopentenone prostaglandins
- DISC
Death-inducing signal complex
- DUBs
Deubiquitinating enzymes
- EMT
Epithelial-mesenchymal transition
- IAPs
Inhibitor of apoptosis protein
- IDs
Inhibitor of differentiation/DNA binding
- MMPs
Matrix metalloproteinases
- MITF
Microphthalmia-associated transcription factor
- REST
RE1-silencing transcription factor
- RIPK1/RIP1
Receptor-interacting protein kinase 1
- SMAC
Second mitochondria-derived activator of caspases
- SKP2
S-phase kinase associated protein 2
- TFs
Transcription factors
- USPs
Ubiquitin-specific proteases
Footnotes
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- Abdul Rehman SA, Kristariyanto YA, Choi SY, Nkosi PJ, Weidlich S, Labib K, et al. MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Molecular Cell. 2016;63:146–155. doi: 10.1016/j.molcel.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams KW, Cooper GM. Rapid turnover of mcl-1 couples translation to cell survival and apoptosis. The Journal of Biological Chemistry. 2007;282:6192–6200. doi: 10.1074/jbc.M610643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleo E, Henderson CJ, Fontanini A, Solazzo B, Brancolini C. Identification of new compounds that trigger apoptosome-independent caspase activation and apoptosis. Cancer Research. 2006;66:9235–9244. doi: 10.1158/0008-5472.CAN-06-0702. [DOI] [PubMed] [Google Scholar]
- Ao N, Liu Y, Bian X, Feng H, Liu Y. Ubiquitin-specific peptidase 22 inhibits colon cancer cell invasion by suppressing the signal transducer and activator of transcription 3/matrix metalloproteinase 9 pathway. Molecular Medicine Reports. 2015;12:2107–2113. doi: 10.3892/mmr.2015.3661. [DOI] [PubMed] [Google Scholar]
- Azab F, De La Puente P, Vij R, Azab AK. Tumor hypoxia promotes dissemination and tumor colonization in Waldenström macroglobulinemia. Blood. 2013;122:3011. doi: 10.1158/1541-7786.MCR-14-0150. [DOI] [PubMed] [Google Scholar]
- Bayraktar S, Gutierrez Barrera AM, Liu D, Pusztai L, Litton J, Valero V, et al. USP-11 as a predictive and prognostic factor following neoadjuvant therapy in women with breast cancer. Cancer Journal. 2013;19:10–17. doi: 10.1097/PPO.0b013e3182801b3a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellail AC, Olson JJ, Yang X, Chen ZJ, Hao C. A20 ubiquitin ligase-mediated polyubiquitination of RIP1 inhibits caspase-8 cleavage and TRAIL-induced apoptosis in glioblastoma. Cancer Discovery. 2012;2:140–155. doi: 10.1158/2159-8290.CD-11-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard G, Neutzner A, Peng G, Wang C, Livak F, Youle RJ, et al. IBRDC2, an IBR-type E3 ubiquitin ligase, is a regulatory factor for Bax and apoptosis activation. The EMBO Journal. 2010;29:1458–1471. doi: 10.1038/emboj.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nature Genetics. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezovskaya O, Schimmer AD, Glinskii AB, Pinilla C, Hoffman RM, Reed JC, Glinsky GV. Increased expression of apoptosis inhibitor protein XIAP contributes to anoikis resistance of circulating human prostate cancer metastasis precursor cells. Cancer Research. 2005;65:2378–2386. doi: 10.1158/0008-5472.CAN-04-2649. [DOI] [PubMed] [Google Scholar]
- Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K, et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11:783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke SP, Smith L, Smith JB. cIAP1 cooperatively inhibits procaspase-3 activation by the caspase-9 apoptosome. The Journal of Biological Chemistry. 2010;285:30061–30068. doi: 10.1074/jbc.M110.125955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows JF, McGrattan MJ, Rascle A, Humbert M, Baek KH, Johnston JA. DUB-3, a cytokine-inducible deubiquitinating enzyme that blocks proliferation. The Journal of Biological Chemistry. 2004;279:13993–14000. doi: 10.1074/jbc.M311291200. [DOI] [PubMed] [Google Scholar]
- Cao PD, Cheung WK, Nguyen DX. Cell lineage specification in tumor progression and metastasis. Discovery Medicine. 2011;12:329–340. [PubMed] [Google Scholar]
- Chakrabarti R, Hwang J, Andres Blanco M, Wei Y, Lukacisin M, Romano RA, et al. Elf5 inhibits the epithelial-mesenchymal transition in mammary gland development and breast cancer metastasis by transcriptionally repressing Snail2. Nature Cell Biology. 2012;14:1212–1222. doi: 10.1038/ncb2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan D, Tian Z, Nicholson B, Kumar KG, Zhou B, Carrasco R, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22:345–358. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheli Y, Giuliano S, Fenouille N, Allegra M, Hofman V, Hofman P, et al. Hypoxia and MITF control metastatic behaviour in mouse and human melanoma cells. Oncogene. 2012;31:2461–2470. doi: 10.1038/onc.2011.425. [DOI] [PubMed] [Google Scholar]
- Chen J, Dexheimer TS, Ai Y, Liang Q, Villamil MA, Inglese J, et al. Selective and cell-active inhibitors of the USP1/UAF1 deubiquitinase complex reverse cisplatin resistance in non-small cell lung cancer cells. Chemistry & Biology. 2011;18:1390–1400. doi: 10.1016/j.chembiol.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Wong CSF, Liu MCP, House CM, Sceneay J, Bowtell DD, et al. The ubiquitin ligase Siah is a novel regulator of Zeb1 in breast cancer. Oncotarget. 2015;6:862–873. doi: 10.18632/oncotarget.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YE, Butterworth M, Malladi S, Duckett CS, Cohen GM, Bratton SB. The E3 ubiquitin ligase cIAP1 binds and ubiquitinates caspase-3 and -7 via unique mechanisms at distinct steps in their processing. Journal of Biological Chemistry. 2009;284:12772–12782. doi: 10.1074/jbc.M807550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi BJ, Surh YJ. Abstract 1432: Role of ubiquitin-specific proteases 47 in epithelial-mesenchymal transition. Cancer Research. 2015;75:1432. [Google Scholar]
- Chou AH, Yeh TH, Kuo YL, Kao YC, Jou MJ, Hsu CY, et al. Polyglutamine-expanded ataxin-3 activates mitochondrial apoptotic pathway by upregulating Bax and downregulating Bcl-xL. Neurobiology of Disease. 2006;21:333–345. doi: 10.1016/j.nbd.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Colland F, Formstecher E, Jacq X, Reverdy C, Planquette C, Conrath S, et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Molecular Cancer Therapeutics. 2009;8:2286–2295. doi: 10.1158/1535-7163.MCT-09-0097. [DOI] [PubMed] [Google Scholar]
- Crook NE, Clem RJ, Miller LK. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. Journal of Virology. 1993;67:2168–2174. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: Implications for physiology and therapy. Nature Reviews Molecular Cell Biology. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- Daisy P, Saipriya K. BCL-2 family proteins: The mitochondrial apoptotic key regulators. Current Cancer Therapy Reviews. 2012;8:133–140. [Google Scholar]
- Diaz VM, de Herreros AG. F-box proteins: Keeping the epithelial-to-mesenchymal transition (EMT) in check. Seminars in Cancer Biology. 2016;36:71–79. doi: 10.1016/j.semcancer.2015.10.003. [DOI] [PubMed] [Google Scholar]
- Dondelinger Y, Aguileta MA, Goossens V, Dubuisson C, Grootjans S, Dejardin E, et al. RIPK3 contributes to TNFR1-mediated RIPK1 kinase-dependent apoptosis in conditions of cIAP1/2 depletion or TAK1 kinase inhibition. Cell Death and Differentiation. 2013;20:1381–1392. doi: 10.1038/cdd.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draber P, Kupka S, Reichert M, Draberova H, Lafont E, de Miguel D, et al. LUBAC-recruited CYLD and A20 regulate gene activation and cell death by exerting opposing effects on linear ubiquitin in signaling complexes. Cell Reports. 2015;13:2258–2272. doi: 10.1016/j.celrep.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccles SA, Welch DR. Metastasis: Recent discoveries and novel treatment strategies. Lancet. 2007;369:1742–1757. doi: 10.1016/S0140-6736(07)60781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckelman BP, Salvesen GS, Scott FL. Human inhibitor of apoptosis proteins: Why XIAP is the black sheep of the family. EMBO Reports. 2006;7:988–994. doi: 10.1038/sj.embor.7400795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann MJ, Nicholson B, Kessler BM. Pharmacological targets in the ubiquitin system offer new ways of treating cancer, neurodegenerative disorders and infectious diseases. Expert Reviews in Molecular Medicine. 2011;13:e35. doi: 10.1017/S1462399411002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel K, Rudelius M, Slawska J, Jacobs L, Ahangarian Abhari B, Altmann B, et al. USP9X stabilizes XIAP to regulate mitotic cell death and chemoresistance in aggressive B-cell lymphoma. EMBO Molecular Medicine. 2016;8:851–862. doi: 10.15252/emmm.201506047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito I, Kleeff J, Abiatari I, Shi X, Giese N, Bergmann F, et al. Overexpression of cellular inhibitor of apoptosis protein 2 is an early event in the progression of pancreatic cancer. Journal of Clinical Pathology. 2007;60:885–895. doi: 10.1136/jcp.2006.038257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Zhang L, Wei W, Jin X, Wang P, Tong Y, et al. A methylation-phosphorylation switch determines Sox2 stability and function in ESC maintenance or differentiation. Molecular Cell. 2014;55:537–551. doi: 10.1016/j.molcel.2014.06.018. [DOI] [PubMed] [Google Scholar]
- Faronato M, Patel V, Darling S, Dearden L, Clague MJ, Urbé S, et al. The deubiquitylase USP15 stabilizes newly synthesized REST and rescues its expression at mitotic exit. Cell Cycle (Georgetown, Texas) 2013;12:1964–1977. doi: 10.4161/cc.25035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernald K, Kurokawa M. Evading apoptosis in cancer. Trends in Cell Biology. 2013;23:620–633. doi: 10.1016/j.tcb.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong S, Itahana Y, Sumida T, Singh J, Coppe J-P, Liu Y, et al. Id-1 as a molecular target in therapy for breast cancer cell invasion and metastasis. Proceedings of the National Academy of Sciences. 2003;100:13543–13548. doi: 10.1073/pnas.2230238100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur U, Aggarwal BB. Regulation of proliferation, survival and apoptosis by members of the TNF superfamily. Biochemical Pharmacology. 2003;66:1403–1408. doi: 10.1016/s0006-2952(03)00490-8. [DOI] [PubMed] [Google Scholar]
- Goncharov T, Niessen K, de Almagro MC, Izrael-Tomasevic A, Fedorova AV, Varfolomeev E, et al. OTUB1 modulates c-IAP1 stability to regulate signalling pathways. The EMBO Journal. 2013;32:1103–1114. doi: 10.1038/emboj.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalvez F, Lawrence D, Yang B, Yee S, Pitti R, Marsters S, et al. TRAF2 sets a threshold for extrinsic apoptosis by tagging caspase-8 with a ubiquitin shutoff timer. Molecular Cell. 2012;48:888–899. doi: 10.1016/j.molcel.2012.09.031. [DOI] [PubMed] [Google Scholar]
- Graner E, Tang D, Rossi S, Baron A, Migita T, Weinstein LJ, et al. The isopeptidase USP2a regulates the stability of fatty acid synthase in prostate cancer. Cancer Cell. 2004;5:253–261. doi: 10.1016/s1535-6108(04)00055-8. [DOI] [PubMed] [Google Scholar]
- Green DR, Llambi F. Cell death signaling. Cold Spring Harbor Perspectives in Biology. 2015;7 doi: 10.1101/cshperspect.a006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K, Copeland NG, Gilbert DJ, Jenkins NA, Gray DA. Unp, a mouse gene related to the tre oncogene. Oncogene. 1993;8:2307–2310. [PubMed] [Google Scholar]
- Gupta GP, Perk J, Acharyya S, Candia PD, Mittal V, Todorova-Manova K, et al. ID genes mediate tumor reinitiation during breast cancer lung metastasis. Proceedings of the National Academy of Sciences. 2007;104:19506–19511. doi: 10.1073/pnas.0709185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habu N, Imanishi Y, Kameyama K, Shimoda M, Tokumaru Y, Sakamoto K, et al. Expression of Oct3/4 and Nanog in the head and neck squamous carcinoma cells and its clinical implications for delayed neck metastasis in stage I/II oral tongue squamous cell carcinoma. BMC Cancer. 2015;15:730. doi: 10.1186/s12885-015-1732-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamacher-Brady A, Brady NR. Bax/Bak-dependent, Drp1-independent targeting of X-linked inhibitor of apoptosis protein (XIAP) into inner mitochondrial compartments counteracts Smac/DIABLO-dependent effector caspase activation. The Journal of Biological Chemistry. 2015;290:22005–22018. doi: 10.1074/jbc.M115.643064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamacher-Brady A, Choe SC, Krijnse-Locker J, Brady NR. Intramitochondrial recruitment of endolysosomes mediates Smac degradation and constitutes a novel intrinsic apoptosis antagonizing function of XIAP E3 ligase. Cell Death and Differentiation. 2014;21:1862–1876. doi: 10.1038/cdd.2014.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annual Review of Biochemistry. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hong J, Zhou J, Fu J, He T, Qin J, Wang L, et al. Phosphorylation of serine 68 of Twist1 by MAPKs stabilizes Twist1 protein and promotes breast cancer cell invasiveness. Cancer Research. 2011;71:3980–3990. doi: 10.1158/0008-5472.CAN-10-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma K, Tsuzuki S, Nakagawa M, Tagawa H, Nakamura S, Morishima Y, et al. TNFAIP3/A20 functions as a novel tumor suppressor gene in several subtypes of non-Hodgkin lymphomas. Blood. 2009;114:2467–2475. doi: 10.1182/blood-2008-12-194852. [DOI] [PubMed] [Google Scholar]
- Hörnle M, Peters N, Thayaparasingham B, Vörsmann H, Kashkar H, Kulms D. Caspase-3 cleaves XIAP in a positive feedback loop to sensitize melanoma cells to TRAIL-induced apoptosis. Oncogene. 2011;30:575–587. doi: 10.1038/onc.2010.434. [DOI] [PubMed] [Google Scholar]
- Hou X, Wang L, Zhang L, Pan X, Zhao W. Ubiquitin-specific protease 4 promotes TNF-α-induced apoptosis by deubiquitination of RIP1 in head and neck squamous cell carcinoma. FEBS Letters. 2013;587:311–316. doi: 10.1016/j.febslet.2012.12.016. [DOI] [PubMed] [Google Scholar]
- Hou K, Zhu Z, Wang Y, Zhang C, Yu S, Zhu Q, et al. Overexpression and biological function of ubiquitin-specific protease 42 in gastric cancer. PLoS One. 2016;11 doi: 10.1371/journal.pone.0152997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Yang D, Zhang H, Liu W, Zhao Y, Lu H, et al. USP22 promotes tumor progression and induces epithelial–mesenchymal transition in lung adenocarcinoma. Lung Cancer. 2015;88:239–245. doi: 10.1016/j.lungcan.2015.02.019. [DOI] [PubMed] [Google Scholar]
- Hu D, Zhou Z, Davidson NE, Huang Y, Wan Y. Novel insight into KLF4 proteolytic regulation in estrogen receptor signaling and breast carcinogenesis. The Journal of Biological Chemistry. 2012;287:13584–13597. doi: 10.1074/jbc.M112.343566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HK, Joazeiro CA, Bonfoco E, Kamada S, Leverson JD, Hunter T. The inhibitor of apoptosis, cIAP2, functions as a ubiquitin-protein ligase and promotes in vitro monoubiquitination of caspases 3 and 7. The Journal of Biological Chemistry. 2000;275:26661–26664. doi: 10.1074/jbc.C000199200. [DOI] [PubMed] [Google Scholar]
- Huang Z, Wu Q, Guryanova OA, Cheng L, Shou W, Rich JN, et al. Deubiquitylase HAUSP stabilizes REST and promotes maintenance of neural progenitor cells. Nature Cell Biology. 2011;13:142–152. doi: 10.1038/ncb2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SJ, Lee HW, Kim HR, Lee H, Shin CH, Yun S-I, et al. Ubiquitin-specific protease 4 controls metastatic potential through β-catenin stabilization in brain metastatic lung adenocarcinoma. Scientific Reports. 2016;6 doi: 10.1038/srep21596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaenko OA, Amerik AY. Chalcone-based small-molecule inhibitors attenuate malignant phenotype via targeting deubiquitinating enzymes. Cell Cycle (Georgetown, Texas) 2012;11:1804–1817. doi: 10.4161/cc.20174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MJ, Baek SH, Kim JH. UCH-L1 promotes cancer metastasis in prostate cancer cells through EMT induction. Cancer Letters. 2011;302:128–135. doi: 10.1016/j.canlet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- Jin J, Liu J, Chen C, Liu Z, Jiang C, Chu H, et al. The deubiquitinase USP21 maintains the stemness of mouse embryonic stem cells via stabilization of Nanog. Nature Communications. 2016;7:13594. doi: 10.1038/ncomms13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapuria V, Peterson LF, Fang D, Bornmann WG, Talpaz M, Donato NJ. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Research. 2010;70:9265–9276. doi: 10.1158/0008-5472.CAN-10-1530. [DOI] [PubMed] [Google Scholar]
- Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Molecular Cell. 2003b;11:1177–1188. doi: 10.1016/s1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- Kim DS, Hwang ES, Lee JE, Kim SY, Kwon SB, Park KC. Sphingosine-1-phosphate decreases melanin synthesis via sustained ERK activation and subsequent MITF degradation. Journal of Cell Science. 2003a;116:1699–1706. doi: 10.1242/jcs.00366. [DOI] [PubMed] [Google Scholar]
- Kim MO, Kim SH, Cho YY, Nadas J, Jeong CH, Yao K, et al. ERK1 and ERK2 regulate embryonic stem cell self-renewal through phosphorylation of Klf4. Nature Structural & Molecular Biology. 2012;19:283–290. doi: 10.1038/nsmb.2217. [DOI] [PubMed] [Google Scholar]
- Kim SH, Kim MO, Cho YY, Yao K, Kim DJ, Jeong CH, et al. ERK1 phosphorylates Nanog to regulate protein stability and stem cell self-renewal. Stem Cell Research. 2014;13:1–11. doi: 10.1016/j.scr.2014.04.001. [DOI] [PubMed] [Google Scholar]
- Kim JB, Kim SY, Kim BM, Lee H, Kim I, Yun J, et al. Identification of a novel anti-apoptotic E3 ubiquitin ligase that ubiquitinates antagonists of inhibitor of apoptosis proteins SMAC, HtrA2, and ARTS. The Journal of Biological Chemistry. 2013;288:12014–12021. doi: 10.1074/jbc.M112.436113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa K, Shibata K, Matsumoto A, Matsumoto M, Ohhata T, Nakayama KI, et al. Fbw7 targets GATA3 through cyclin-dependent kinase 2-dependent proteolysis and contributes to regulation of T-cell development. Molecular and Cellular Biology. 2014;34:2732–2744. doi: 10.1128/MCB.01549-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbé S. Breaking the chains: Structure and function of the deubiquitinases. Nature Reviews Molecular Cell Biology. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annual Review of Biochemistry. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Kong Y, Cui H, Zhang H. Smurf2-mediated ubiquitination and degradation of Id1 regulates p16 expression during senescence. Aging Cell. 2011;10:1038–1046. doi: 10.1111/j.1474-9726.2011.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouros-Mehr H, Bechis SK, Slorach EM, Littlepage LE, Egeblad M, Ewald AJ, et al. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell. 2008;13:141–152. doi: 10.1016/j.ccr.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasorella A, Stegmüller J, Guardavaccaro D, Liu G, Carro MS, Rothschild G, et al. Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature. 2006;442:471–474. doi: 10.1038/nature04895. [DOI] [PubMed] [Google Scholar]
- Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Tsygankov AY. c-Cbl regulates glioma invasion through matrix metalloproteinase 2. Journal of Cellular Biochemistry. 2010;111:1169–1178. doi: 10.1002/jcb.22839. [DOI] [PubMed] [Google Scholar]
- von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Molecular Cell. 2003;11:1189–1200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- Levy EM, Roberti MP, Mordoh J. Natural killer cells in human cancer: From biological functions to clinical applications. Journal of Biomedicine & Biotechnology. 2011;2011:676198. doi: 10.1155/2011/676198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Xu Y, Chen Y, Chen S, Jia X, Sun T, et al. SOX2 promotes tumor metastasis by stimulating epithelial-to-mesenchymal transition via regulation of WNT/β-catenin signal network. Cancer Letters. 2013;336:379–389. doi: 10.1016/j.canlet.2013.03.027. [DOI] [PubMed] [Google Scholar]
- Liao B, Jin Y. Wwp2 mediates Oct4 ubiquitination and its own auto-ubiquitination in a dosage-dependent manner. Cell Research. 2010;20:332–344. doi: 10.1038/cr.2009.136. [DOI] [PubMed] [Google Scholar]
- Liu Y, Lashuel HA, Choi S, Xing X, Case A, Ni J, et al. Discovery of inhibitors that elucidate the role of UCH-L1 activity in the H1299 lung cancer cell line. Chemistry & Biology. 2003;10:837–846. doi: 10.1016/j.chembiol.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Liu N, Li X, Huang H, Zhao C, Liao S, Yang C, et al. Clinically used antirheumatic agent auranofin is a proteasomal deubiquitinase inhibitor and inhibits tumor growth. Oncotarget. 2014;5:5453–5471. doi: 10.18632/oncotarget.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Li H, Wu X, Yoo BH, Yan SR, Stadnyk AW, et al. Detachment-induced upregulation of XIAP and cIAP2 delays anoikis of intestinal epithelial cells. Oncogene. 2006;25:7680–7690. doi: 10.1038/sj.onc.1209753. [DOI] [PubMed] [Google Scholar]
- Lu X, Mazur SJ, Lin T, Appella E, Xu Y. The pluripotency factor nanog promotes breast cancer tumorigenesis and metastasis. Oncogene. 2014;33:2655–2664. doi: 10.1038/onc.2013.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luise C, Capra M, Donzelli M, Mazzarol G, Jodice MG, Nuciforo P, et al. An atlas of altered expression of deubiquitinating enzymes in human cancer. PLoS One. 2011;6:e15891. doi: 10.1371/journal.pone.0015891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlen P, Puisieux A. Metastasis: A question of life or death. Nature Reviews Cancer. 2006;6:449–458. doi: 10.1038/nrc1886. [DOI] [PubMed] [Google Scholar]
- Mei Y, Hahn AA, Hu S, Yang X. The USP19 deubiquitinase regulates the stability of c-IAP1 and c-IAP2. The Journal of Biological Chemistry. 2011;286:35380–35387. doi: 10.1074/jbc.M111.282020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges CW, Altomare DA, Testa JR. FAS-associated factor 1 (FAF1): Diverse functions and implications for oncogenesis. Cell Cycle (Georgetown, Texas) 2009;8:2528–2534. doi: 10.4161/cc.8.16.9280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermerian AH, Case A, Stein RL, Cuny GD. Structure-activity relationship, kinetic mechanism, and selectivity for a new class of ubiquitin C-terminal hydrolase-L1 (UCH-L1) inhibitors. Bioorganic & Medicinal Chemistry Letters. 2007;17:3729–3732. doi: 10.1016/j.bmcl.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Mojsa B, Lassot I, Desagher S. Mcl-1 ubiquitination: Unique regulation of an essential survival protein. Cell. 2014;3:418–437. doi: 10.3390/cells3020418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizane Y, Honda R, Fukami K, Yasuda H. X-linked inhibitor of apoptosis functions as ubiquitin ligase toward mature caspase-9 and cytosolic Smac/DIABLO. Journal of Biochemistry. 2005;137:125–132. doi: 10.1093/jb/mvi029. [DOI] [PubMed] [Google Scholar]
- Nagano T, Hashimoto T, Nakashima A, Kikkawa U, Kamada S. X-linked inhibitor of apoptosis protein mediates neddylation by itself but does not function as a NEDD8-E3 ligase for caspase-7. FEBS Letters. 2012;586:1612–1616. doi: 10.1016/j.febslet.2012.04.056. [DOI] [PubMed] [Google Scholar]
- Nieto MA, Huang RY-J, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- Nocturne G, Boudaoud S, Miceli-Richard C, Viengchareun S, Lazure T, Nititham J, et al. Germline and somatic genetic variations of TNFAIP3 in lymphoma complicating primary Sjögren’s syndrome. Blood. 2013;122:4068–4076. doi: 10.1182/blood-2013-05-503383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohayon S, Spasser L, Aharoni A, Brik A. Targeting deubiquitinases enabled by chemical synthesis of proteins. Journal of the American Chemical Society. 2012;134:3281–3289. doi: 10.1021/ja2116712. [DOI] [PubMed] [Google Scholar]
- Oliveira AM, Perez-Atayde AR, Inwards CY, Medeiros F, Derr V, Hsi B-L, et al. USP6 and CDH11 oncogenes identify the neoplastic cell in primary aneurysmal bone cysts and are absent in so-called secondary aneurysmal bone cysts. The American Journal of Pathology. 2004;165:1773–1780. doi: 10.1016/S0002-9440(10)63432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Korm S, Chung HJ, et al. RAP80 regulates epithelial-mesenchymal transition related with metastasis and malignancy of cancer. Cancer Science. 2016;107:267–273. doi: 10.1111/cas.12877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltzer N, Darding M, Walczak H. Holding RIPK1 on the ubiquitin leash in TNFR1 signaling. Trends in Cell Biology. 2016;26:445–461. doi: 10.1016/j.tcb.2016.01.006. [DOI] [PubMed] [Google Scholar]
- Priolo C, Tang D, Brahamandan M, Benassi B, Sicinska E, Ogino S, et al. The isopeptidase USP2a protects human prostate cancer from apoptosis. Cancer Research. 2006;66:8625–8632. doi: 10.1158/0008-5472.CAN-06-1374. [DOI] [PubMed] [Google Scholar]
- Qu Q, Mao Y, Xiao G, Fei X, Wang J, Zhang Y, et al. USP2 promotes cell migration and invasion in triple negative breast cancer cell lines. Tumor Biology. 2015;36:5415–5423. doi: 10.1007/s13277-015-3207-7. [DOI] [PubMed] [Google Scholar]
- R N, Dl R, Ws T, A M, S J, S Y, et al. c-Myc and Her2 cooperate to drive a stem-like phenotype with poor prognosis in breast cancer. Oncogene. 2014;33:3992–4002. doi: 10.1038/onc.2013.368. [DOI] [PubMed] [Google Scholar]
- Reiley W, Zhang M, Wu X, Granger E, Sun SC. Regulation of the deubiquitinating enzyme CYLD by IκB kinase gamma-dependent phosphorylation. Molecular and Cellular Biology. 2005;25:3886–3895. doi: 10.1128/MCB.25.10.3886-3895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverdy C, Conrath S, Lopez R, Planquette C, Atmanene C, Collura V, et al. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chemistry & Biology. 2012;19:467–477. doi: 10.1016/j.chembiol.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Ronau JA, Beckmann JF, Hochstrasser M. Substrate specificity of the ubiquitin and Ubl proteases. Cell Research. 2016;26:441–456. doi: 10.1038/cr.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos MMM, Moreira R. Michael acceptors as cysteine protease inhibitors. Mini Reviews in Medicinal Chemistry. 2007;7:1040–1050. doi: 10.2174/138955707782110105. [DOI] [PubMed] [Google Scholar]
- Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- Shao G, Lilli DR, Patterson-Fortin J, Coleman KA, Morrissey DE, Greenberg RA. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3166–3171. doi: 10.1073/pnas.0807485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shembade N, Parvatiyar K, Harhaj NS, Harhaj EW. The ubiquitin-editing enzyme A20 requires RNF11 to downregulate NF-κB signalling. The EMBO Journal. 2009;28:513–522. doi: 10.1038/emboj.2008.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HM, Lee JE, Kim JH. Ubiquitin C-terminal hydrolase-L3 regulates EMT process and cancer metastasis in prostate cell lines. Biochemical and Biophysical Research Communications. 2014;452:722–727. doi: 10.1016/j.bbrc.2014.08.144. [DOI] [PubMed] [Google Scholar]
- Su Z, Yang Z, Xu Y, Chen Y, Yu Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Molecular Cancer. 2015;14:48. doi: 10.1186/s12943-015-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman RT, Stanek TJ, Esteso P, Gearhart JD, Knudsen KE, McMahon SB. The epigenetic modifier ubiquitin-specific protease 22 (USP22) regulates embryonic stem cell differentiation via transcriptional repression of sex-determining region Y-box 2 (SOX2) The Journal of Biological Chemistry. 2013;288:24234–24246. doi: 10.1074/jbc.M113.469783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Nakabayashi Y, Takahashi R. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proceedings of the National Academy of Sciences. 2001;98:8662–8667. doi: 10.1073/pnas.161506698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swatek KN, Komander D. Ubiquitin modifications. Cell Research. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai SK, Yang MH, Chang SY, Chang YC, Li WY, Tsai TL, et al. Persistent Krüppel-like factor 4 expression predicts progression and poor prognosis of head and neck squamous cell carcinoma. Cancer Science. 2011;102:895–902. doi: 10.1111/j.1349-7006.2011.01859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K, Goldstein D, Crowe P, Yang JL. Uncovering a key to the process of metastasis in human cancers: A review of critical regulators of anoikis. Journal of Cancer Research and Clinical Oncology. 2013;139:1795–1805. doi: 10.1007/s00432-013-1482-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z, D’Arcy P, Wang X, Ray A, Tai Y-T, Hu Y, et al. A novel small molecule inhibitor of deubiquitinating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood. 2014;123:706–716. doi: 10.1182/blood-2013-05-500033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Isamiddinova NS, Peroutka RJ, Goldenberg SJ, Mattern MR, Nicholson B, et al. Characterization of selective ubiquitin and ubiquitin-like protease inhibitors using a fluorescence-based multiplex assay format. Assay and Drug Development Technologies. 2011;9:165–173. doi: 10.1089/adt.2010.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaira V, Faversani A, Martin NM, Garlick DS, Ferrero S, Nosotti M, et al. Regulation of lung cancer metastasis by Klf4-Numb-like signaling. Cancer Research. 2013;73:2695–2705. doi: 10.1158/0008-5472.CAN-12-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastyan S, Weinberg RA. Tumor metastasis: Molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Hu G, Wei Y, Yuan M, Bronson RT, Yang Q, et al. Genetic ablation of metadherin inhibits autochthonous prostate cancer progression and metastasis. Cancer Research. 2014;74:5336–5347. doi: 10.1158/0008-5472.CAN-14-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Cai C, Dong X, Yu QC, Zhang XO, Yang L, et al. Identification of multipotent mammary stem cells by protein C receptor expression. Nature. 2015;517:81–84. doi: 10.1038/nature13851. [DOI] [PubMed] [Google Scholar]
- Wang Q, He W, Lu C, Wang Z, Wang J, Giercksky KE, et al. Oct3/4 and Sox2 are significantly associated with an unfavorable clinical outcome in human esophageal squamous cell carcinoma. Anticancer Research. 2009;29:1233–1241. [PubMed] [Google Scholar]
- Wang Q, Li L, Ye Y. Inhibition of p97-dependent protein degradation by Eeyarestatin I. The Journal of Biological Chemistry. 2008;283:7445–7454. doi: 10.1074/jbc.M708347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Heinlein M, Dengjel J, Alber C, Singh PK, Hacker G. The deubiquitinase Usp27x stabilizes the BH3-only protein Bim and enhances apoptosis. EMBO Reports. 2016;17:724–738. doi: 10.15252/embr.201541392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock J, Wu J, Cao P, Kingsbury WD, McDermott JL, Kodrasov MP, et al. Selective dual inhibitors of the cancer-related deubiquitinating proteases USP7 and USP47. ACS Medicinal Chemistry Letters. 2012;3:789–792. doi: 10.1021/ml200276j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Orian A, Jin J, Grim JE, Grim JA, Harper JW, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SA, Maecker HL, French DM, Liu J, Gregg A, Silverstein LB, et al. USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell. 2011;146:918–930. doi: 10.1016/j.cell.2011.07.040. [DOI] [PubMed] [Google Scholar]
- Wolter KG, Hsu YT, Smith CL, Nechushtan A, Xi XG, Youle RJ. Movement of Bax from the cytosol to mitochondria during apoptosis. The Journal of Cell Biology. 1997;139:1281–1292. doi: 10.1083/jcb.139.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao N, Li H, Luo J, Wang R, Chen H, Chen J, et al. Ubiquitin-specific protease 4 (USP4) targets TRAF2 and TRAF6 for deubiquitination and inhibits TNFalpha-induced cancer cell migration. The Biochemical Journal. 2012;441:979–986. doi: 10.1042/BJ20111358. [DOI] [PubMed] [Google Scholar]
- Xing C, Lu XX, Guo PD, Shen T, Zhang S, He XS, et al. Ubiquitin-specific protease 4-mediated deubiquitination and stabilization of PRL-3 is required for potentiating colorectal oncogenesis. Cancer Research. 2016;76:83–95. doi: 10.1158/0008-5472.CAN-14-3595. [DOI] [PubMed] [Google Scholar]
- Xu W, Gong L, Haddad MM, Bischof O, Campisi J, Yeh ETH, et al. Regulation of microphthalmia-associated transcription factor MITF protein levels by association with the ubiquitin-conjugating enzyme hUBC9. Experimental Cell Research. 2000;255:135–143. doi: 10.1006/excr.2000.4803. [DOI] [PubMed] [Google Scholar]
- Xu M, Takanashi M, Oikawa K, Tanaka M, Nishi H, Isaka K, et al. USP15 plays an essential role for caspase-3 activation during Paclitaxel-induced apoptosis. Biochemical and Biophysical Research Communications. 2009b;388:366–371. doi: 10.1016/j.bbrc.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Xu H, Wang W, Li C, Yu H, Yang A, Wang B, et al. WWP2 promotes degradation of transcription factor OCT4 in human embryonic stem cells. Cell Research. 2009a;19:561–573. doi: 10.1038/cr.2009.31. [DOI] [PubMed] [Google Scholar]
- Yamada S-I, Yanamoto S, Naruse T, Matsushita Y, Takahashi H, Umeda M, et al. Skp2 regulates the expression of MMP-2 and MMP-9, and enhances the invasion potential of oral squamous cell carcinoma. Pathology Oncology Research: POR. 2016;22:625–632. doi: 10.1007/s12253-016-0049-6. [DOI] [PubMed] [Google Scholar]
- Yan K, Li L, Wang X, et al. The deubiquitinating enzyme complex BRISC is required for proper mitotic spindle assembly in mammalian cells. The Journal of Cell Biology. 2015;210:209–224. doi: 10.1083/jcb.201503039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JK. Death effecter domain for the assembly of death-inducing signaling complex. Apoptosis: An International Journal on Programmed Cell Death. 2015;20:235–239. doi: 10.1007/s10495-014-1060-6. [DOI] [PubMed] [Google Scholar]
- Yang H, Lu X, Liu Z, Chen L, Xu Y, Wang Y, et al. FBXW7 suppresses epithelial-mesenchymal transition, stemness and metastatic potential of cholangiocarcinoma cells. Oncotarget. 2015;6:6310–6325. doi: 10.18632/oncotarget.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Pringle LM, Lau AW, Riquelme DN, Wang H, Jiang T, et al. TRE17/USP6 oncogene translocated in aneurysmal bone cyst induces matrix metalloproteinase production via activation of NF-κB. Oncogene. 2010;29:3619–3629. doi: 10.1038/onc.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue W, Chen Z, Liu H, Yan C, Chen M, Feng D, et al. A small natural molecule promotes mitochondrial fusion through inhibition of the deubiquitinase USP30. Cell Research. 2014;24:482–496. doi: 10.1038/cr.2014.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Berger FG, Yang J, Lu X. USP4 inhibits p53 through deubiquitinating and stabilizing ARF-BP1. The EMBO Journal. 2011;30:2177–2189. doi: 10.1038/emboj.2011.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chen C, Hou X, Gao Y, Lin F, Yang J, et al. Identification of the E3 deubiquitinase ubiquitin-specific peptidase 21 (USP21) as a positive regulator of the transcription factor GATA3. The Journal of Biological Chemistry. 2013;288:9373–9382. doi: 10.1074/jbc.M112.374744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, et al. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Molecular Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Wei Y, Wang L, Debeb BG, Yuan Y, Zhang J, et al. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nature Cell Biology. 2014;16:864–875. doi: 10.1038/ncb3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Fiske B, Kawakami A, Li J, Fisher DE. Regulation of MITF stability by the USP13 deubiquitinase. Nature Communications. 2011;2:414. doi: 10.1038/ncomms1421. [DOI] [PubMed] [Google Scholar]
- Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nature Cell Biology. 2004;6:931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Luo A, Shrivastava I, He M, Huang Y, Bahar I, et al. Regulation of XIAP turnover reveals a role for USP11 in promotion of tumorigenesis. eBioMedicine. 2017;15:48–61. doi: 10.1016/j.ebiom.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Wang H, Schwartz DM, Stoffels M, Park YH, Zhang Y, et al. Loss-of-function mutations in TNFAIP3 leading to A20 haploinsufficiency cause an early-onset autoinflammatory disease. Nature Genetics. 2016;48:67–73. doi: 10.1038/ng.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Zuo Y, Li B, Wang H, Liu H, Wang X, et al. Deubiquitinase inhibition of 19S regulatory particles by 4-arylidene curcumin analog AC17 causes NF-κB inhibition and p53 reactivation in human lung cancer cells. Molecular Cancer Therapeutics. 2013;12:1381–1392. doi: 10.1158/1535-7163.MCT-12-1057. [DOI] [PubMed] [Google Scholar]


